Abstract
Insights into the expression of pacemaker-specific markers in human induced pluripotent stem cell (hiPSC)-derived cardiomyocyte subtypes can facilitate the enrichment and track differentiation and maturation of hiPSC-derived pacemaker-like cardiomyocytes. To date, no study has directly assessed gene expression in each pacemaker-, atria-, and ventricular-like cardiomyocyte subtype derived from hiPSCs since currently the subtypes of these immature cardiomyocytes can only be identified by action potential profiles. Traditional acquisition of action potentials using patch-clamp recordings renders the cells unviable for subsequent analysis. We circumvented these issues by acquiring the action potential profile of a single cell optically followed by assessment of protein expression through immunostaining in that same cell. Our same-single-cell analysis for the first time revealed expression of proposed pacemaker-specific markers—hyperpolarization-activated cyclic nucleotide-modulated (HCN)4 channel and Islet (Isl)1—at the protein level in all three hiPSC-derived cardiomyocyte subtypes. HCN4 expression was found to be higher in pacemaker-like hiPSC-derived cardiomyocytes than atrial- and ventricular-like subtypes but its downregulation over time in all subtypes diminished the differences. Isl1 expression in pacemaker-like hiPSC-derived cardiomyocytes was initially not statistically different than the contractile subtypes but did become statistically higher than ventricular-like cells with time. Our observations suggest that although HCN4 and Isl1 are differentially expressed in hiPSC-derived pacemaker-like relative to ventricular-like cardiomyocytes, these markers alone are insufficient in identifying hiPSC-derived pacemaker-like cardiomyocytes.
Keywords: human induced pluripotent stem cells, cardiomyocyte subtype, electrophysiology, pacemaker
INTRODUCTION
Treatment of cardiovascular diseases by replacing dysfunctional or lost cardiomyocytes with human pluripotent stem cells (hPSC)-derived cardiomyocytes is a promising strategy that has emerged from the recent advancements in regenerative medicine. The hPSC-derived cardiomyocyte population is heterogeneous and can be classified functionally into pacemaker- and contractile-like subtypes. The appropriate functional subtype should be chosen to restore cardiac function, e.g., pacemaking cardiomyocytes for treatment of sinoatrial node dysfunction and contractile cardiomyocytes for loss of ventricular muscle mass after myocardial infarction. Anatomically, cardiomyocyte subtypes are geographically restricted in the heart, such that pacemaking cardiomyocytes reside in the sinoatrial and atrioventricular nodes, while contractile atrial and ventricular subtypes populate the atria and ventricles, respectively. Subtype-specific genes are invaluable for identifying and tracking cell differentiation and maturation of intermixed, heterogeneous hPSC-derived cardiomyocyte subtypes that have no distinct anatomical separation. Specific genes have been proposed for identification of hPSC-derived cardiomyocyte subtypes; however, many pacemaker- and atrial-specific genes are expressed in immature cardiomyocytes including hPSC-derived cardiomyocytes and only become subtype-specific as they downregulate in maturing ventricular cardiomyocytes [1]. Assessment of these genes in hPSC-derived cardiomyocytes will be important for studies on the development and maturation of cardiomyocyte subtypes and potentially uncovering suitable markers for cardiomyocyte subtype enrichment.
To date, the protein expression of proposed genes in each subtype—pacemaker, atrial, and ventricular—of hPSC-derived cardiomyocytes remains unclear. Bulk analysis methods, such as western blot or qPCR, do not provide detailed information on gene expression for a heterogeneous population of hPSC-derived cardiomyocytes, since the overall expression level can change due to a change in the number of cells expressing the gene and/or a change in the level of expression per cell. Even single cell analysis using flow cytometry is unable to yield information on protein expression in hPSC-derived pacemaking relative to contractile cardiomyocytes as definitive markers that can identify the subtypes have yet to be determined. Electrophysiological classification of hPSC-derived cardiomyocytes into pacemaker-like, atrial-like and ventricular-like subtypes by their action potential profiles that largely resemble those of their adult counterparts is currently the gold standard for subtype determination for these immature cardiomyocytes [2, 3]. Therefore, to analyze the gene expression in each hPSC-derived cardiomyocyte subtype, the gene expression and the action potential profile (for cardiomyocyte subtype classification) need to be assessed in the same cell.
Recently, some proposed subtype-specific genes have been validated electrophysiologically by acquiring the action potential profiles of positive and negative cells for a transgenic fluorescent reporter, such as GFP or mCherry, driven by the promoter of a gene of interest [4–7]. However, there is still a lack of direct correlation of the proposed subtype-specific gene of interest in relation to the cardiomyocyte subtype and electrophysiology in these studies, because the expression level for the gene of interest was determined indirectly based on the fluorescence intensity of the transgenic reporter proteins. Furthermore, acquisition of action potential profiles using traditional patch-clamp recordings rendered the cells unviable for subsequent analysis. We circumvented these issues by first acquiring the action potentials of a single cell optically, then a gene of interest was assessed by immunostaining that same cell. Optical recordings of action potentials allow determination of the subtype for a hPSC-derived cardiomyocyte while leaving it viable and intact for further analysis. In addition, an optical method has a much higher throughput than patch-clamp recordings and eliminates the bias from a patch clamper’s selection of patchable cells.
In this study, we determined the action potential profiles of human induced pluripotent stem cell (hiPSC)-derived cardiomyocytes and the protein expression of proposed pacemaker-specific markers—hyperpolarization-activated cyclic nucleotide-modulated (HCN)4 channel [8] and Islet (Isl)1 [9, 10]—on a per-cell basis. The two proposed pacemaker-specific markers were chosen for their distinct functional roles in pacemaker cardiomyocyte physiology, such that HCN4 is a pacemaking ion channel directly involved in the electrophysiological phenotype that determined the cardiomyocyte subtypes, while Isl1 is a transcription factor recently shown to drive the pacemaker phenotype. Our same-single-cell-based sequential analysis of electrophysiology followed by pacemaker-specific protein expression enables the direct assessment of the level of pacemaker-specific marker expression in each subtype of hiPSC-derived cardiomyocytes or gene analysis by cardiomyocyte subtype. We found that although HCN4 was higher in hiPSC-derived pacemaker-like cardiomyocytes than atrial- and ventricular-like subtypes, the differences dissipated with time in culture as all subtypes downregulated the protein expression of HCN4. In contrast, Isl1 expression was initially not statistically higher in the hiPSC-derived pacemaker-like cardiomyocytes but did become significantly higher relative to the ventricular-like subtype over time in culture. We describe for the first time the pacemaker-specific gene expression among the different subtypes of hiPSC-derived cardiomyocytes and evaluate the specificity of the proposed pacemaker markers.
MATERIALS AND METHODS
HiPSC culture and differentiation to cardiomyocytes
HiPSCs (19-9-7T, WiCell) were cultured in mTeSR1 (Stem Cell Technologies) on hESC-qualified matrigel (Corning) and passaged using ReLeSR (Stem Cell Technologies). HiPSCs were differentiated following a published protocol [11]. Briefly, hiPSCs were treated with 6 µM glycogen synthase kinase (GSK)3 inhibitor, CHIR99021 (Tocris), from day 0–1, followed by 12µM Wnt inhibitor, IWR-1 (Tocris), from day 3–5 in RPMI 1640 with L-glutamine, B-27 supplement without insulin, and penicillin-streptomycin (Invitrogen). After day 7, hiPSC-derived cardiomyocytes were maintained in RPMI 1640 with L-glutamine and B-27 supplement with insulin. HiPSC-derived cardiomyocytes presented in this work were generated from two independent batches of differentiation with 50–60% cardiomyocyte differentiation efficiency. Cells were cultured to day 37 and 57 before dissociation with TrypLE (Invitrogen) for replating as single cells on matrigel-coated gridded glass bottom dishes (ibidi). Replated cells were allowed to recovery for three days before electrophysiological experiments.
Patch-clamp recording of action potentials
Action potentials of single hiPSC-derived cardiomyocytes of 40 and 60 days post-differentiation were recorded at room temperature using perforated patch technique with Axon Axopatch 200B amplifier interfaced with digitizer Digidata 1440A that were controlled by Clampex software (Molecular Devices). The external recording solution was Tyrode’s solution consisted of (in mM): 140 NaCl, 5 KCl, 1 CaCl2, 1 MgCl2, 10 HEPES, and 10 glucose with pH 7.4 adjusted by NaOH. The pipette solution consisted of (in mM): 120 K-glutamate, 25 KCl, 1 CaCl2, 1 MgCl2, 10 HEPES, pH adjusted 7.4 with KOH and back filled with 200 µg/ml amphotericin B (Sigma-Aldrich). The resistance of borosilicate pipettes ranged from 3–5 MΩ.
Optical recording of action potentials
HiPSC-derived cardiomyocytes of day 40 and 60 were plated on gridded glass bottom dishes to allow tracking of the same cells for correlation with immunostaining for genes of interest. The cells were loaded with 5 µM FluoVolt in media with 0.04% Pluronic F-127 and 2 mM probenecid for 15 min at 37°C. Action potentials of single cells were recorded in Tyrode’s solution with excitation-contraction decoupler blebbistatin (Sigma-Aldrich) at room temperature using a Nipkow disk confocal microscope (Olympus IX71 equipped with Yokogawa CSU10, a 10× microscope objective, and FITC filters) with an EM-CCD camera (Andor iXon 897) at ~100 fps. Images were analyzed using Andor iQ2 software. Data for each cell comprised of immunostaining and time-lapsed membrane potential images, quantified action potential tracings, and cell locations were logged into a database for quick and direct comparison of cells after immunostaining.
Classification of cardiomyocyte subtypes
Action potentials recorded optically were used to classify hiPSC-derived cardiomyocytes into pacemaker-like, atrial-like and ventricular-like cardiomyocyte subtypes. The electrophysiological data from independently differentiated batches were pooled for analyses. We defined a duration-slope-curvature (DSC) factor that describes the action potential profile by APD50, the slope between APD50 and APD90, and the curvature between APD10 and APD30. Clustering analyses were performed using the Mclust package in programming language [R] on the DSC factor separating hiPSC-derived cardiomyocytes into ventricular-like (cluster 3 and 4) and non-ventricular-like (cluster 1 and 2) groups. The non-ventricular-like group was further clustered by frequency of automaticity into high and low frequency clusters. The pacemaker-like subtype comprised of cells in the DSC factor 1 or 2 cluster, the frequency high cluster, and with an amplitude of less than 70 mV. The atrial-like hiPSC-derived cardiomyocytes were classified as cells in the DSC factor 1 or 2 cluster and the frequency low cluster or the frequency high cluster with an amplitude greater than 70 mV. Action potentials of all hiPSC-derived cardiomyocytes were manually checked for their assigned subtype. The classification scheme was consistent and accurate in grouping the hiPSC-derived cardiomyocytes into a cardiomyocyte subtype with similar adult action potential profile.
Gene expression assessment of hiPSC-derived cardiomyocytes by immunostaining
HiPSC-derived cardiomyocytes after optical recording of action potentials were immediately fixed with 4% paraformaldehyde. Day 40 and 60 hiPSC-derived cardiomyocytes from the same batch were stained for pacemaker-specific markers HCN4 (NeuroMab) and Isl1 (Developmental Studies Hybridoma Bank) with nuclear counterstain, DAPI (Invitrogen) on the same day with the same lot of antibodies. All dishes were imaged using a confocal microscope (Nikon A1) on the same day with the same laser power and detector setting. The per-cell determination of marker-specific expression were matched to the action potentials profiles recorded in the same cell by referencing the location of each cell within the grid. Intensity of HCN4 and Isl1 was analyzed using ImageJ software.
Cell size and circularity assessment of hiPSC-derived cardiomyocytes
Each optically recorded hiPSC-derived cardiomyocyte was also manually analyzed for cell size and circularity simultaneously with the immunostaining fluorescence analysis using ImageJ software. The cell size was quantified by the cell area. Cell circularity defined as is an indicator for the level of cell elongation, which varies between 1 for a perfect circle to 0 for a line.
Correlation analysis
A database was generated using LibreOffice Base to organize and link the electrophysiological recordings of each hiPSC-derived cardiomyocyte with its corresponding immunostaining, cell size, and circularity data. Pearson’s coefficient (r) was used to quantify the correlation between two variables, such that a positive value means a direct correlation and a negative value means an inverse relationship. The magnitude of the coefficient or absolute value of r (|r|) indicates the strength of correlation. A |r| < 0.3 was considered to have no correlation between the parameters. A |r| ≥ 0.3 that indicates correlation between the parameters is further classified as weak correlation for 0.3 ≤ |r| < 0.5 and strong correlation for |r| ≥ 0.5.
Statistical analysis
All data are reported as mean±SEM with statistical significance determined by two-way ANOVA with Tukey’s post-hoc test (p<0.05), except for action potential parameters for cardiomyocyte subtypes due to changes in the distribution of these parameters after subtype classification, in which case they are reported as median [interquartile range] with statistical significance determined by a nonparametric Kruskal-Wallis test with Dunn’s multiple comparisons test (p<0.05).
RESULTS
Electrophysiology of hiPSC-derived cardiomyocyte subtypes
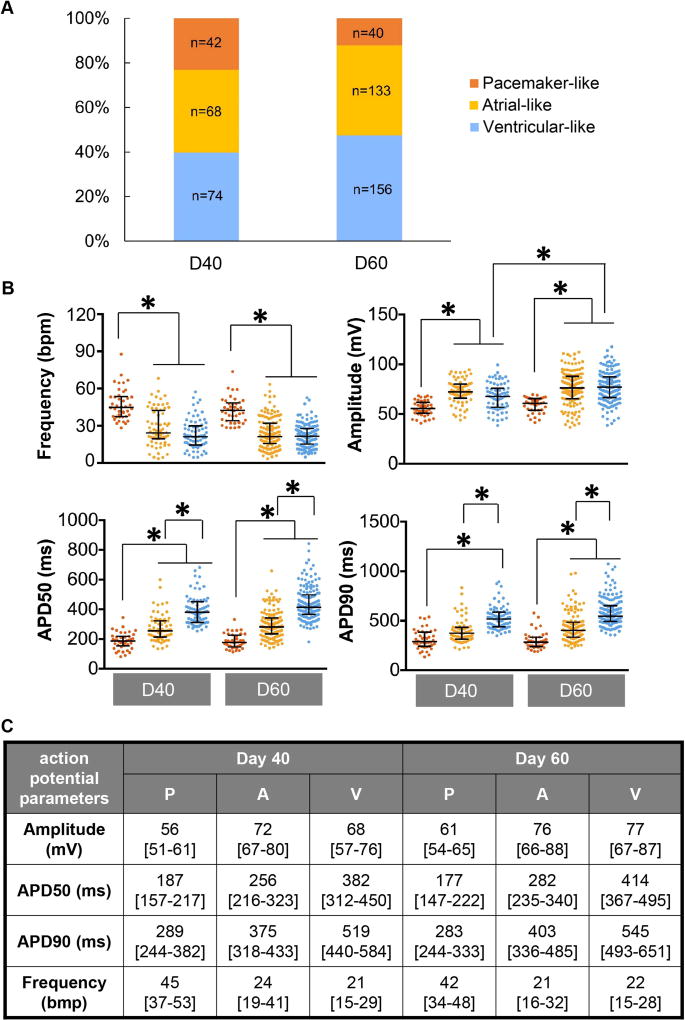
Action potentials of hiPSC-derived cardiomyocytes acquired optically largely recapitulated the dynamics and kinetics of those acquired through patch-clamp recordings (Fig. 1A). These action potential profiles were heterogeneous and resembled those recorded from their adult counterpart in vivo. The distribution of action potential parameters—frequency, amplitude, action potential duration at 50% and 90% repolarization (APD50, APD90)—for hiPSC-derived cardiomyocytes at 40 days (n=184) and 60 days (n=329) post-differentiation exhibited a shift in distribution as cells matured (Fig. 1B). Specifically, frequency of automaticity decreased from day 40 to 60, while increases in APD50, APD90, and amplitude were observed after 20 days of maturation. Mean values of action potential parameters for day 40 and 60 are reported in Figure 1B.
Figure 1. Electrophysiology of hiPSC-derived cardiomyocytes 40 and 60 days post-differentiation.
A) Representative optical and patch-clamp recordings of action potentials. B) Distribution of action potential parameters derived from optical recordings. Action potential parameters are presented as mean±SEM. * indicates p<0.05. (n= 184 for D40 and 329 for D60)
Cluster analysis of action potential profiles classified hiPSC-derived cardiomyocytes at day 40 and 60 post-differentiation into pacemaker-like, atrial-like and ventricular-like subtypes (Supplemental Fig. 1). The pacemaker-like subtype represents the smallest population (23%) followed by the atrial-like (37%) then ventricular-like (40%) subtypes on day 40 (Fig. 2A). Moreover, hiPSC-derived cardiomyocytes at day 60 showed a significant decrease of the pacemaker-like subtype to 12% (p=0.004), while cells of the atrial-like and ventricular-like subtypes increased to 41% and 47%, respectively. The ratio of ventricular- to atrial-like subtype did not change significantly over time.
Figure 2. Electrophysiology of hiPSC-derived cardiomyocyte subtypes.
A) Population distribution of day 40 (D40) and 60 (D60) cardiomyocyte subtypes. B) Action potential parameters derived from optical recordings for D40 and D60 hiPSC-derived cardiomyocytes for pacemaker-like, atrial-like, and ventricular-like subtypes. * indicates p<0.05. C) Table of action potential parameters derived from optical recordings presented as median [interquartile range]. Sampling size for each subtype is shown in bar graph (A).
The frequency of automaticity was significantly faster for pacemaker-like hiPSC-derived cardiomyocytes for day 40 and day 60 (Fig. 2B). The median frequencies of Day 40 atrial-like and ventricular-like hiPSC-derived cardiomyocytes were comparable to each other and did not change appreciably on day 60. The median value for action potential amplitude of the pacemaker-like hiPSC-derived cardiomyocytes was significantly smaller than that for their contractile counterparts (Fig. 2B). The action potential amplitude increased for the ventricular-like subtype over time (p<0.05). The median APD50 for pacemaker-like and atrial-like subtypes on day 40 were significantly shorter than that of ventricular-like group (Fig. 2B). The trend persisted in day 60 hiPSC-derived cardiomyocytes since there was no significant change in magnitude over time. The median APD90 for pacemaker-like subtype was significantly shorter than the ventricular-like subtype on day 40 (Fig. 2B). APD90 did not change appreciably between day 40 and 60 within the different hiPSC-derived cardiomyocyte subtypes. Median and interquartile range for measured action potential parameters of each cardiomyocyte subtype are presented in Figure 2C.
Correlation analysis of action potential parameters (Supplemental Fig. 2) exhibited an inverse relationship between the frequency of automaticity and APD50 for hiPSC-derived cardiomyocytes on day 40 (r=−0.54) and 60 (r=−0.39) post-differentiation. Amplitude also exhibited weak correlation to APD50 for cardiomyocytes of day 40 (r=0.31) and 60 (r=0.39).
Cellular morphology correlation to action potentials
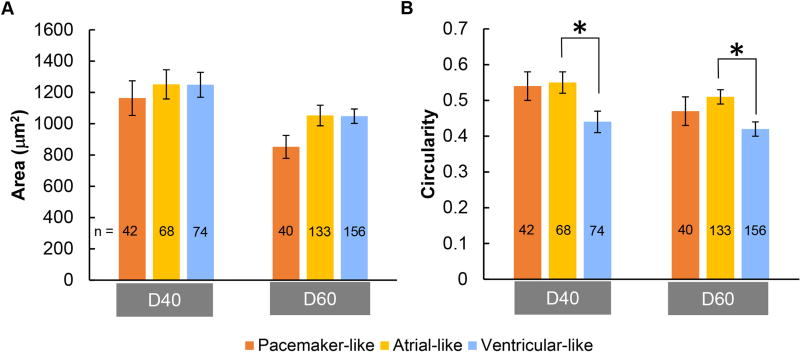
hiPSC-derived pacemaker-like cardiomyocytes were the smallest as measured by cell area compared to the contractile-like cardiomyocytes; however, the difference did not reach statistical significance (Fig. 3A). Circularity, a dimensionless shape index ranging from 0 for a line to 1 for a circle, is the lowest in ventricular-like hiPSC-derived cardiomyocytes, which was statistically different from that of atrial-like hiPSC-derived cardiomyocytes (Fig. 3B).
Figure 3. Cell morphology of hiPSC-derived cardiomyocyte subtypes.
A) Mean cell area for pacemaker-like, atrial-like and ventricular-like subtypes for day 40 and 60 hiPSC-derived cardiomyocytes. B) Mean circularity for each hiPSC-derived cardiomyocyte subtype on day 40 and 60 post-differentiation. * indicates p<0.05. Sampling size for each subtype is shown in bar graphs.
HCN4 expression in hiPSC-derived cardiomyocyte subtypes
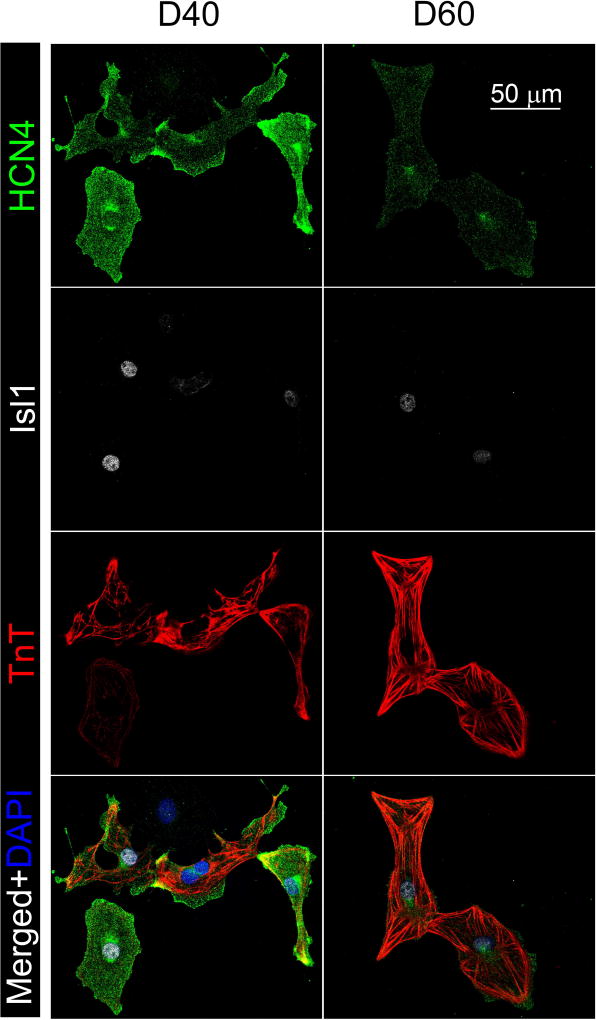
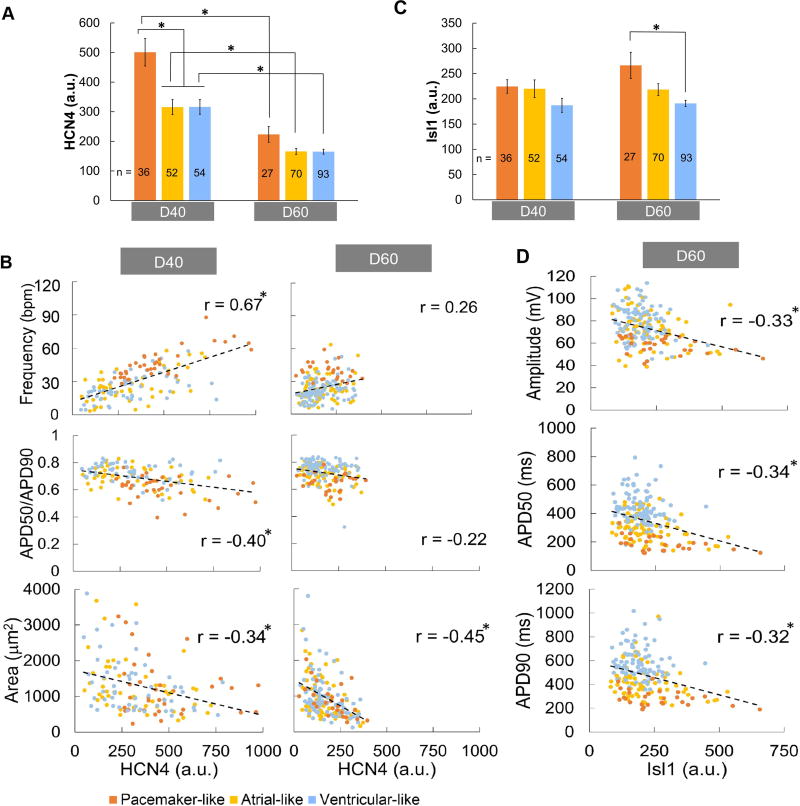
HCN4 was expressed in the majority of the hiPSC-derived cardiomyocytes on day 40 (92%) and 60 (90%) post-differentiation as demonstrated by immunostaining (Fig. 4, Fig. 7B). This is in agreement with our observation that all hiPSC-derived cardiomyocytes we measured optically or using patch-clamp recordings exhibited intrinsic automaticity or phase 4-depolarization. HCN4 expression was analyzed in the same hiPSC-derived cardiomyocyte with known action potential profile for analysis of its expression by cardiomyocyte subtypes (Fig. 5). For day 40 hiPSC-derived cardiomyocytes, the HCN4 mean intensity was significantly higher in pacemaker-like cardiomyocytes (501±46 a.u.) than the atrial- (316±25 a.u.) and ventricular-like (316±25 a.u.) cardiomyocytes (Fig. 6A). There was a two-fold decrease in HCN4 expression with time in all subtypes of hiPSC-derived cardiomyocytes. As of day 60 hiPSC-derived cardiomyocytes, the expression remained the highest in the pacemaker-like cardiomyocytes (223±26 a.u.) compared to the atrial-like (166±10 a.u) and ventricular-like (165±8 a.u.) subtypes.
Figure 4. Immunostaining of HCN4 and Isl1 in hiPSC-derived cardiomyocytes.
Representative HCN4 and Isl1 staining of day 40 and 60 hiPSC-derived cardiomyocytes. Positive troponin T (TnT) staining verifies the identity of hiPSC-derived cardiomyocytes.
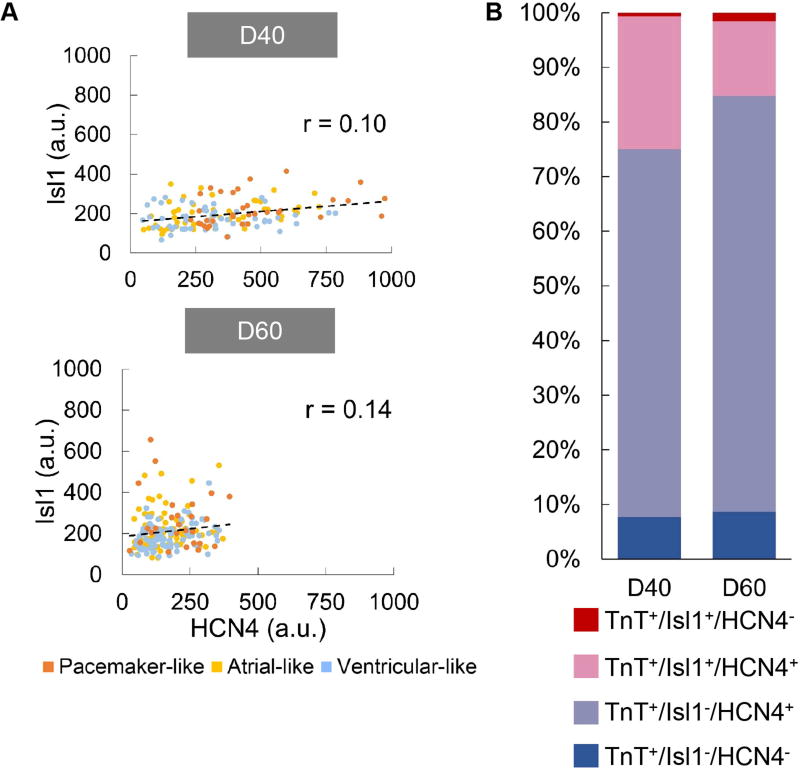
Figure 7. Correlation of Isl1 to HCN4 expression hiPSC-derived cardiomyocytes.
A) Correlation plots for Isl1 vs. HCN4 in hiPSC-derived cardiomyocytes on day 40 (D40, n=142) and 60 (D60, n=190) post-differentiation. Pearson’s coefficient (r) for each correlation is shown. There is no significant correlation between Isl1 and HCN4 as indicated by the r value. B) Distribution of HCN4+ and Isl1+ cells in D40 and D60 hiPSC-derived cardiomyocytes identified as troponin T (TnT)+ cells. (a.u.: arbitrary units)
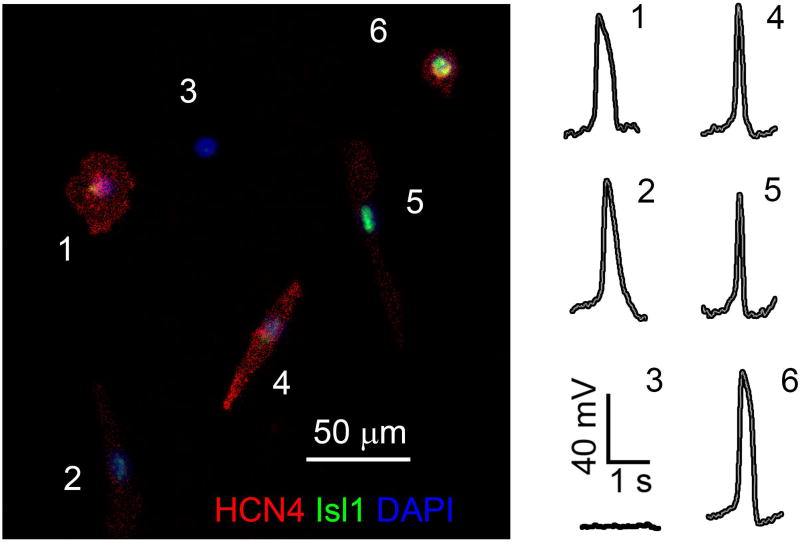
Figure 5. HCN4, Isl1 staining and action potential profile acquired from the same hiPSC-derived cardiomyocytes.
A representative image showing HCN4 and Isl1 staining and action potential profile for each hiPSC-derived cardiomyocyte for direct comparisons enabling analysis of gene expression by cardiomyocyte subtypes.
Figure 6. Analysis of HCN1 and Isl1 expression in hiPSC-derived cardiomyocytes by cardiomyocyte subtypes.
A) D40 and D60 average whole-cell HCN4 fluorescent intensity reflecting level protein expression for each hiPSC-derived cardiomyocyte subtype. B) Correlation of HCN4 in D40 and D60 hiPSC-derived cardiomyocyte to frequency, APD50/APD90 and cell area. C) D40 and D60 average whole-cell Isl1 fluorescent intensity reflecting level protein expression for each hiPSC-derived cardiomyocyte subtype. D) Correlation of Isl1 in D60 hiPSC-derived cardiomyocyte to action potential amplitude, APD50, and APD90. Sampling size for each subtype is shown in bar graphs. * indicates p<0.05 in (A, C) and correlation in (B, D). (a.u.: arbitrary units)
HCN4 expression correlated with the frequency of automaticity in hiPSC-derived cardiomyocytes of 40-day post-differentiation (r=0.67), but the correlation diminished in cardiomyocytes of 60-days post-differentiation (r=0.26) (Fig. 6B). A weak inverse correlation was also observed between HCN4 and APD50/APD90 ratio for day 40 hiPSC-derived cardiomyocytes (r=−0.40) in contrast to those from day 60 exhibiting no correlation (Fig. 6B). In addition, HCN4 expression exhibited a weak inverse relationship to cell size for both day 40 (r=−0.34) and 60 (r=−0.45) hiPSC-derived cardiomyocytes (Fig. 6B).
Isl1 expression in hiPSC-derived cardiomyocyte subtypes
Isl1 was expressed in 25% and 15% of hiPSC-derived cardiomyocytes on day 40 and 60, respectively, as assessed by immunostaining (Fig. 4, Fig. 7B). Isl1 expression was also analyzed in the same hiPSC-derived cardiomyocyte with known action potential profile to enable analysis of its expression by cardiomyocyte subtypes (Fig. 5). Mean nuclear Isl1 staining intensity in pacemaker- and atrial-like day 40 hiPSC-derived cardiomyocytes were comparable (224±13 vs. 220±18 a.u.) and slightly higher than that of ventricular-like subtype (187±14 a.u.), but the difference did not attain statistical significance (Fig. 6C). On day 60, pacemaker-like hiPSC-derived cardiomyocytes did express significantly higher Isl1 (266±26 a.u.) than ventricular-like cells (191±6 a.u.). The Isl1 expression level did increase over time for pacemaker-like subtype but the change was not statistically significant. Clustering analysis separating day 60 hiPSC-derived cardiomyocytes into Isl1-high and Isl1-low clusters revealed a significantly higher percentage of pacemaker-like cells expressing high Isl1 compared to the contractile-like subtypes (30% vs. 10%, p<0.01, Supplemental Fig. 3). Correlation analysis of Isl1 expression to action potential parameters indicated a weak inverse association of Isl1 to amplitude, APD50, and APD90 for hiPSC-derived cardiomyocytes on day 60 (Fig. 6D).
Correlation of Isl1 to HCN4 expression
Correlation of Isl1 to HCN4 protein expression was analyzed by comparing the fluorescence intensity of stained hiPSC-derived cardiomyocytes in the same cell (Fig. 7A). Our analysis showed no correlation between Isl1 and HCN4 on day 40 and 60 as indicated by Pearson’s coefficients of less than 0.3. HiPSC-derived cardiomyocytes were also analyzed for their positive and negative expression for Isl1 and HCN4 (Fig. 7B). The majority of the hiPSC-derived cardiomyocytes identified as troponin T (TnT)+ cells were also HCN4+ (>90%), with only 25% and 15% of these cells being Isl1+ on day 40 and 60, respectively. Of these cells, ~14–24% are double positive for Isl1 and HCN4 and ~70% are HCN4+ only in contrast with ~1–2% being Isl1+ only.
DISCUSSION
Action potential profiling is currently the only method for determining the cardiomyocyte subtype of a hiPSC-derived cardiomyocyte. There are only a few reports of single-cell analyses for action potential profiles and cardiomyocyte markers in hPSC-derived cardiomyocytes [4, 6, 12]. These studies were performed using patch-clamp recordings from hPSC-derived cardiomyocytes expressing fluorescent proteins driven by the promoter of a gene of interest with an underlying assumption that the reporter expression has an excellent fidelity in reflecting the protein expression for a gene of interest. However, the synthesis and half-life of a fluorescent reporter protein may not faithfully recapitulate the protein expression for a gene of interest as the cardiomyocytes develop. We reason that sequential single-cell analyses of the electrophysiological phenotype and per-cell protein expression pattern originating from the same cell provide a clearer assessment of a highly heterogeneous population of hiPSC-derived cardiomyocytes.
To investigate the expression of markers in different cardiomyocyte subtypes, we first recorded the action potentials optically, such that the cardiomyocyte subtype can be established a priori for each cell before the analyses of the expression of a protein of interest by immunostaining. This one-to-one sequential single-cell analyses enabled a direct determination of the gene expression for each hiPSC-derived cardiomyocyte subtype.
Electrophysiological and morphological development of hiPSC-derived cardiomyocyte subtypes
We classified hiPSC-derived cardiomyocytes into cardiomyocyte subtypes—pacemaker-, atrial-, and ventricular-like—based on the action potential profiles recorded optically. Classification of cardiomyocyte subtypes by action potentials is currently the gold standard for determining the cardiomyocyte subtypes [2, 3, 13]. The subtype distribution likely varies with cell lines and differentiation methods, but our observed distribution of ventricular-like hiPSC-derived cardiomyocytes being the predominant subtype, followed by atrial- and then pacemaker-like subtype, is consistent with previous reports [2, 3, 6, 11]. The characteristics of the action potentials for each subtype are in agreement with those reported in the literature for hiPSC-derived cardiomyocytes and largely resemble those of their adult counterpart in the heart. The pacemaker-like subtype at 40 and 60 days post-differentiation were classified as action potentials with a short APD50, the smallest amplitude, and the fastest frequency of automaticity. The ventricular-like subtype fired action potentials with the longest APD50, a large amplitude, and the slowest frequency of automaticity. The atrial-like subtype exhibited action potentials with characteristics that were intermediate of the two other subtypes. Although hiPSC-derived cardiomyocytes analyzed collectively without distinguishing between the subtypes showed significant changes in all AP parameters between day 40 and 60, statistical differences were not observed between day 40 and 60 when the cells were analyzed by subtypes, except for amplitudes of ventricular-like subtype. These observations suggest that the electrophysiology within each cardiomyocyte subtype did not mature significantly and the collective changes in parameters without distinguishing the subtypes were most likely due to the change in the subtype distribution of the cells, i.e. an increase in the proportion of ventricular- and atrial-like cardiomyocytes as supported by our findings in Figure 2A.
It is important to note that we did exclude ~2% of hiPSC-derived cardiomyocytes with action potentials that did not fall into our defined subtypes. These hiPSC-derived cardiomyocytes fired action potentials characteristically similar to our pacemaker-like subtype with a prominent phase 4-depolarization but a slow frequency of automaticity. It is unclear if these cells are cardiomyocytes destined to become atrioventricular nodal cardiomyocytes or very immature contractile cardiomyocytes.
Action potentials were also recorded from day 20 hiPSC-derived cardiomyocytes (data not shown), but action potentials of these cells were not distinct enough to allow subtype classification based on clustering analysis. Hence, analysis was performed starting with day 40 hiPSC-derived cardiomyocytes with action potentials that more closely resemble those of adult subtypes. Although most action potential parameters within a subtype did not change significantly over time, a significant decrease in the fraction of pacemaker-like cells from day 40 to day 60 suggests that some hiPSC-derived cardiomyocytes went through significant electrophysiological changes resulting in a change in the subtype. Switches in subtype also indicate that cardiomyocyte subtype specification of hiPSC-derived cardiomyocytes may not have been determined by day 40 post-differentiation. Further studies are needed to track the progress of electrophysiology in the same hiPSC-derived cardiomyocytes to determine subtype commitment.
Pacemaking cardiomyocytes in vivo are smaller than atrial and ventricular cardiomyocytes [14]. Similarly, pacemaker-like hiPSC-derived cardiomyocytes did have a smaller mean cell size than contractile-like hiPSC-derived cardiomyocytes, but the difference was not statistically significant even by day 60. Circularity measurement suggests that pacemaker-like and atrial-like hiPSC-derived cardiomyocytes were equally elongated but both less so than the ventricular-like subtype on both day 40 and 60. However, hiPSC-derived ventricular-like cardiomyocytes were only significantly more elongated than atrial-like cardiomyocytes. Our data suggest that cell morphology alone is not sufficient in distinguishing hiPSC-derived pacemaker-like cardiomyocytes from other subtypes.
Expression of pacemaker-specific markers in hiPSC-derived cardiomyocytes
A pacemaker-specific marker can greatly benefit the identification and enrichment of pacemaker-like hiPSC-derived cardiomyocyte subtypes; however, pacemaker-specific markers are especially elusive in immature hiPSC-derived cardiomyocytes because of the nature of these genes, such that they are initially present in all immature cardiomyocytes but become pacemaker-specific after their expression downregulates with maturation in contractile cardiomyocyte subtypes [1]. Currently, there is a significant knowledge gap on the pacemaker-specific genes in hiPSC-derived cardiomyocyte subtypes because the only way to identify hiPSC-derived cardiomyocyte subtypes is through action potential profiling. In this study, we investigated the per-cell expression of two proposed pacemaker-specific genes—HCN4 and Isl1—in the same hiPSC-derived cardiomyocytes with predetermined action potential profiles, enabling us to determine the expression of these pacemaker-specific genes in each subtype. Most proposed pacemaker-specific markers to date are either transcription factors promoting the pacemaking phenotype (e.g. Isl1, Shox2, Tbx3, Tbx18) or channel proteins directly affecting the electrophysiology of pacemaking cardiomyocytes (e.g. CaV1.3, HCN1, HCN4) [9, 10, 15–20]. Therefore, a pacemaker-specific marker from each functional category was evaluated for its specificity in hiPSC-derived cardiomyocytes.
HCN4 expressed in most hiPSC-derived cardiomyocytes is more abundant in pacemaker-like subtype at day 40 but down-regulates at day 60
HCN4 is an ion channel that gives rise to the funny current, If, and contributes to automaticity of pacemaking cells [16]. Although the majority of hiPSC-derived cardiomyocytes are positive for HCN4, day 40 pacemaker-like subtypes did express more HCN4 than the contractile subtypes, with a significant correlation between HCN4 and the frequency of automaticity, but the correlation dissipated with downregulation of HCN4 unexpectedly in all subtypes by day 60 such that the expression level is no longer sufficient to distinguish pacemaker-like from contractile-like subtypes. It is unclear if the observed HCN4 expression changes in hiPSC-derived cardiomyocytes are reflective of development in vivo, but pacemaker-specific genes are thought to be expressed in immature cardiomyocytes and become restricted to pacemaking cardiomyocytes as they downregulate in contractile cardiomyocytes [21]. Therefore, HCN4 downregulation in hiPSC-derived contractile-like cardiomyocytes is in agreement with the findings in vivo. The surprising HCN4 expression downregulation in the hiPSC-derived pacemaker-like cardiomyocytes is in contrast to the retention of high HCN4 expression in pacemaking cardiomyocytes through development and maturation in vivo [8]. The observed HCN4 downregulation is also consistent with previous reports on human pluripotent stem cell-derived cardiomyocytes in vitro analyzed without distinguishing between the pacemaker- and contractile-like cardiomyocyte subtypes [22, 23]. It is possible that the in vitro culture condition is not conducive for preservation of HCN4 expression in these cells. The electrophysiology of stem cell-derived cardiomyocytes has been shown to be affected by the presence of non-cardiomyocytes present in the heterogeneous culture that resulted from differentiation [24]. Although the transcript level of HCN4 was not affected by the non-cardiomyocytes in a previous study, it is still possible that the observed in vitro downregulation of HCN4 protein in pacemaking-like cardiomyocytes is influenced by the 40–50% of non-cardiomyocytes present in our hiPSC-derived cardiomyocyte culture.
Even though age-dependent downregulation of HCN4 expression coincided with a decrease in mean frequency of automaticity for all hiPSC-derived cardiomyocytes, interestingly, the median frequency for hiPSC-derived pacemaker-like cardiomyocytes remained relatively constant with time while the distribution of HCN4 intensity shifted toward a lower mean (Supplemental Fig. 4). Given that the pacemaker-like fraction decreased over time, a loss of HCN4 may have contributed to the loss of pacemaking function in some pacemaker-like hiPSC-derived cardiomyocytes, while others were able to maintain the pacemaking frequency despite a HCN4 downregulation. Automaticity in pacemaking cardiomyocytes can originate from either a “voltage-clock” driven by If encoded by HCN channel expression or a “Ca2+-clock” driven by the cyclic Ca2+-release from the sarcoplasmic reticulum [25]. The sarcoplasmic reticulum in hPSC-derived cardiomyocytes is quite immature but develops with time [26]. An in silico model of pacemaking cardiomyocytes demonstrated that a robust Ca2+-clock with a mature sarcoplasmic reticulum can maintain a comparable frequency of automaticity even with a 50% reduction of If [27]. We propose that day 40 hiPSC-derived pacemaker-like cardiomyocytes may consist of two subgroups—one with automaticity driven by the voltage-clock while the other by the Ca2+-clock. Further investigation is needed to clarify the pacemaking mechanisms in developing hiPSC-derived cardiomyocytes.
Expression of Isl1 is significantly higher in hiPSC-derived pacemaker-like than ventricular-like cardiomyocytes
Isl1 is a transcription factor expressed in cardiac progenitor cells of the second heart field [28, 29]. Recently, Isl1 has been shown to be critical for the development of pacemaking cardiomyocytes possibly through direct upregulation of pacemaker-specific genes and downregulation of atrial-specific genes such as T-box 3 [9, 10]. Interestingly, Isl1 expression initially was not different among the hiPSC-derived cardiomyocyte subtypes; however, by day 60 Isl1 expression was significantly higher in hiPSC-derived pacemaker-like than ventricular-like cardiomyocytes while exhibiting weak correlation to some action potential parameters. Since only 30% of hiPSC-derived pacemaker-like cardiomyocytes expressed high Isl1, if Isl1 is indeed necessary for sustaining the pacemaking phenotype, over time the hiPSC-derived pacemaker-like fraction may further decrease as the Isl1-low pacemaker-like cardiomyocytes transition to a contractile-like subtype.
Both adult pacemaker-specific markers that we tested are not specific in separating hiPSC-derived pacemaker-like from contractile-like cardiomyocytes at the ages examined in this study. Although only one hiPSC line was used in this study, the specificity of a cardiomyocyte subtype marker should be independent of cell lines, differentiation protocols, and differentiation efficiency just as a marker would exhibit the same differential expression among the cardiomyocyte subtypes from hearts of different individuals if it is indeed a specific marker. Based on our data, HCN4, a gene that directly contributes to the electrophysiological phenotype, rather than Isl1, a promoter of pacemaking phenotype, is slightly better at distinguishing the pacemaker-like subtype from the other subtypes as indicated by the positive correlation with frequency of automaticity, a hallmark of pacemaking function. Interestingly, we did not observe any correlation between HCN4 and Isl1 expression in hiPSC-derived cardiomyocytes. The majority of the hiPSC-derived cardiomyocytes were also HCN4+ with fewer than a quarter of these cells being Isl1+ on day 40 and 60, respectively. The lack of correlation may be due to a decrease of HCN4 along with an increase in Isl1 in the pacemaker-like subtype as they develop, as well as possibly the various developmental states that the hiPSC-derived cardiomyocytes were in. Pacemaking cardiomyocytes may terminally mature into a unique population of HCN4+ and Isl1+ cells, but hiPSC-derived cardiomyocyte subtypes have yet to be terminally specified to yield such a population of pacemaking cells. It is important to note that we have observed all TnT+ cells in the porcine sinoatrial node to be HCN4+ but not all of these cells were found to be Isl1+ (Supplemental Fig. 5), suggesting possible subgroups within the pacemaking cardiomyocyte population in vivo similar to the hiPSC-derived pacemaker-like cardiomyocytes. Whether the HCN4+/Isl1− pacemaking cardiomyocytes differ from those that are HCN4+/Isl1+ remain to be elucidated. A better understanding of the pacemaker-specific genes in the adult heart may also facilitate the search for a pacemaker-specific marker for hiPSC-derived cardiomyocytes.
CONCLUSION
This study presents a strategy for directly analyzing the gene expression in each hiPSC-derived cardiomyocyte subtype and revealed for the first time the expression of pacemaker-specific markers—HCN4 and Isl1—in hiPSC-derived pacemaker-, atrial- and ventricular-like cardiomyocyte subtypes. HCN4 expression was observed to be higher in pacemaker-like subtype but the expression downregulated in all subtypes with time in culture. In contrast, Isl1 expression was initially not statistically higher in pacemaker-like subtype but did become significantly higher in the pacemaker-like subtype relative to the ventricular-like subtype with time in culture. The differential level of HCN4 and Isl1 observed among the cardiomyocyte subtypes in hiPSC-derived cardiomyocytes up to 60 days was still evolving and had not yet attained sufficient specificity in identifying hiPSC-derived pacemaker-like cardiomyocytes.
Supplementary Material
Acknowledgments
This work is supported by California Institute of Regenerative Medicine (CIRM) Basic Biology Grant (RB4-05764) to DKL. JMG has been supported by a CIRM Postdoctoral Training Fellowship (TG2-01163) and a National Institute of Health T32 Training Program in Basic and Translational Cardiovascular Science (T32HL086350). NC is supported by NIH (R01 HL085727, R01 HL085844, and S10 RR033106) and VA Merit Review Grant I01 BX000576. NC is the holder of the Roger Tatarian Endowed Professor in Cardiovascular Medicine. JWC is supported by National Science Foundation (NSF) grant 1264776.
Nonstandard Abbreviations and Acronyms
- APD
action potential duration
- HCN
hyperpolarization-activated cyclic nucleotide-modulated channel
- hiPSCs
human induced pluripotent stem cells
- hPSCs
human pluripotent stem cells
- If
funny current
- Isl
Islet
- TnT
Troponin T
Footnotes
Author’s Contributions
Yechikov: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Copaciu: collection of data
Gluck: collection of data
Deng: provision of instrumentation
Chiamvimonvat: final approval of manuscript
Chan: provision of instrumentation
Lieu: conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors have no potential conflicts of interest to disclose.
References
- 1.Small EM, Krieg PA. Molecular regulation of cardiac chamber-specific gene expression. Trends Cardiovasc Med. 2004;14:13–18. doi: 10.1016/j.tcm.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Lieu DK, Fu JD, Chiamvimonvat N, et al. Mechanism-based facilitated maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Arrhythm Electrophysiol. 2013;6:191–201. doi: 10.1161/CIRCEP.111.973420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Wilson GF, Soerens AG, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circulation research. 2009;104:e30–41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bizy A, Guerrero-Serna G, Hu B, et al. Myosin light chain 2-based selection of human iPSC-derived early ventricular cardiac myocytes. Stem Cell Res. 2013;11:1335–1347. doi: 10.1016/j.scr.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poon E, Yan B, Zhang S, et al. Transcriptome-guided functional analyses reveal novel biological properties and regulatory hierarchy of human embryonic stem cell-derived ventricular cardiomyocytes crucial for maturation. PLoS One. 2013;8:e77784. doi: 10.1371/journal.pone.0077784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu WZ, Xie Y, Moyes KW, et al. Neuregulin/ErbB signaling regulates cardiac subtype specification in differentiating human embryonic stem cells. Circulation research. 2010;107:776–786. doi: 10.1161/CIRCRESAHA.110.223917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Josowitz R, Lu J, Falce C, et al. Identification and purification of human induced pluripotent stem cell-derived atrial-like cardiomyocytes based on sarcolipin expression. PLoS One. 2014;9:e101316. doi: 10.1371/journal.pone.0101316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vicente-Steijn R, Passier R, Wisse LJ, et al. Funny current channel HCN4 delineates the developing cardiac conduction system in chicken heart. Heart Rhythm. 2011;8:1254–1263. doi: 10.1016/j.hrthm.2011.03.043. [DOI] [PubMed] [Google Scholar]
- 9.Liang X, Zhang Q, Cattaneo P, et al. Transcription factor ISL1 is essential for pacemaker development and function. The Journal of clinical investigation. 2015;125:3256–3268. doi: 10.1172/JCI68257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vedantham V, Galang G, Evangelista M, et al. RNA sequencing of mouse sinoatrial node reveals an upstream regulatory role for Islet-1 in cardiac pacemaker cells. Circulation research. 2015;116:797–803. doi: 10.1161/CIRCRESAHA.116.305913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lian X, Hsiao C, Wilson G, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1848–1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu JD, Jiang P, Rushing S, et al. Na+/Ca2+ exchanger is a determinant of excitation-contraction coupling in human embryonic stem cell-derived ventricular cardiomyocytes. Stem Cells Dev. 2010;19:773–782. doi: 10.1089/scd.2009.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karakikes I, Senyei GD, Hansen J, et al. Small molecule-mediated directed differentiation of human embryonic stem cells toward ventricular cardiomyocytes. Stem cells translational medicine. 2014;3:18–31. doi: 10.5966/sctm.2013-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyett MR, Honjo H, Kodama I. The sinoatrial node, a heterogeneous pacemaker structure. Cardiovascular research. 2000;47:658–687. doi: 10.1016/s0008-6363(00)00135-8. [DOI] [PubMed] [Google Scholar]
- 15.Mangoni ME, Couette B, Bourinet E, et al. Functional role of L-type Cav1.3 Ca2+ channels in cardiac pacemaker activity. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5543–5548. doi: 10.1073/pnas.0935295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siu CW, Lieu DK, Li RA. HCN-encoded pacemaker channels: from physiology and biophysics to bioengineering. J Membr Biol. 2006;214:115–122. doi: 10.1007/s00232-006-0881-9. [DOI] [PubMed] [Google Scholar]
- 17.Kapoor N, Liang W, Marban E, et al. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol. 2013;31:54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiese C, Grieskamp T, Airik R, et al. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circulation research. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- 19.Espinoza-Lewis RA, Yu L, He F, et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2–5. Dev Biol. 2009;327:376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ionta V, Liang W, Kim EH, et al. SHOX2 overexpression favors differentiation of embryonic stem cells into cardiac pacemaker cells, improving biological pacing ability. Stem Cell Reports. 2015;4:129–142. doi: 10.1016/j.stemcr.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spater D, Abramczuk MK, Buac K, et al. A HCN4+ cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nature cell biology. 2013;15:1098–1106. doi: 10.1038/ncb2824. [DOI] [PubMed] [Google Scholar]
- 22.Bosman A, Sartiani L, Spinelli V, et al. Molecular and functional evidence of HCN4 and caveolin-3 interaction during cardiomyocyte differentiation from human embryonic stem cells. Stem Cells Dev. 2013;22:1717–1727. doi: 10.1089/scd.2012.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sartiani L, Bettiol E, Stillitano F, et al. Developmental changes in cardiomyocytes differentiated from human embryonic stem cells: a molecular and electrophysiological approach. Stem Cells. 2007;25:1136–1144. doi: 10.1634/stemcells.2006-0466. [DOI] [PubMed] [Google Scholar]
- 24.Kim C, Majdi M, Xia P, et al. Non-cardiomyocytes influence the electrophysiological maturation of human embryonic stem cell-derived cardiomyocytes during differentiation. Stem Cells Dev. 2010;19:783–795. doi: 10.1089/scd.2009.0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakatta EG, DiFrancesco D. What keeps us ticking: a funny current, a calcium clock, or both? Journal of molecular and cellular cardiology. 2009;47:157–170. doi: 10.1016/j.yjmcc.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Fu JD, Siu CW, et al. Functional sarcoplasmic reticulum for calcium handling of human embryonic stem cell-derived cardiomyocytes: insights for driven maturation. Stem Cells. 2007;25:3038–3044. doi: 10.1634/stemcells.2007-0549. [DOI] [PubMed] [Google Scholar]
- 27.Maltsev VA, Lakatta EG. Yin and yang of the cardiac pacemaker clock system in health and disease. Heart Rhythm. 2010;7:96–98. doi: 10.1016/j.hrthm.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moretti A, Caron L, Nakano A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 29.Bu L, Jiang X, Martin-Puig S, et al. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.