Abstract
Computed tomography (CT) is commonly used as a diagnostic and treatment planning imaging modality in craniomaxillofacial (CMF) surgery to correct patient’s bony defects. A major disadvantage of CT is that it emits harmful ionizing radiation to patients during the exam. Magnetic resonance imaging (MRI) is considered to be much safer and noninvasive, and often used to study CMF soft tissues (e.g., temporomandibular joint and brain). However, it is extremely difficult to accurately segment CMF bony structures from MRI since both bone and air appear to be black in MRI, along with low signal-to-noise ratio and partial volume effect. To this end, we proposed a 3D deep-learning based cascade framework to solve these issues. Specifically, a 3D fully convolutional network (FCN) architecture is first adopted to coarsely segment the bony structures. As the coarsely segmented bony structures by FCN tend to be thicker, convolutional neural network (CNN) is further utilized for fine-grained segmentation. To enhance the discriminative ability of the CNN, we particularly concatenate the predicted probability maps from FCN and the original MRI, and feed them together into the CNN to provide more context information for segmentation. Experimental results demonstrate a good performance and also the clinical feasibility of our proposed 3D deep-learning based cascade framework.
1 Introduction
A significant number of patients require craniomaxillofacial (CMF) surgery every year for the correction of congenital and acquired condition of head and face [5]. Comparing to computed tomography (CT) that emits harmful ionizing radiation, magnetic resonance imaging (MRI) provides a safer and non-invasive way to assess CMF anatomy. However, it is extremely difficult to accurately segment CMF bony structures from MRI due to unclear boundaries between the bones and air (both appearing to be black) in MRI, low signal-to-noise ratio, and partial volume effect (Fig. 1). Due to these difficulties, even with the time-consuming and labor-intense manual segmentation, the results are often still inaccurate.
Fig. 1.
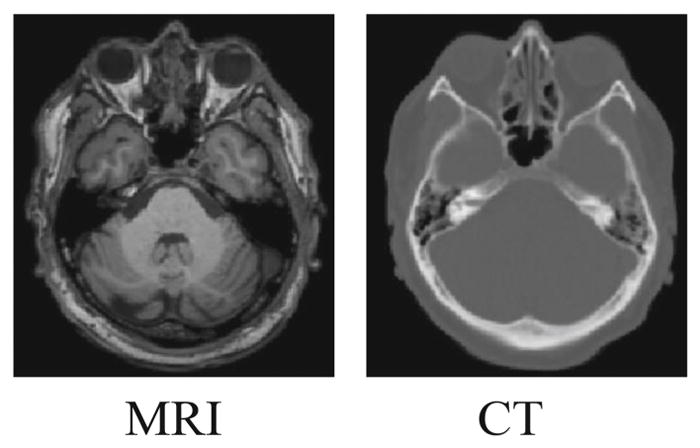
The comparison of MRI and its corresponding CT. Bony structures have unclear bone-to-air boundaries and low signal-to-noise in MRI, comparing to CT.
To date, there are only limited reports on effectively segmenting bony structures from MRI, mainly based on multi-atlas-based approaches [10] and learning-based approaches [9]. In multi-atlas-based approaches, multiple atlas images are rigidly or non-rigidly aligned onto a target image. Based on the derived deformation fields, the corresponding label image from each atlas image is warped into the target image space and then fused (based on a certain image-to-image similarity metric) to obtain the segmentation of the target image. In traditional learning-based approaches, the segmentation task is generally formulated as an optimization problem by selecting the best features from a pool of human-engineered features. However, both type of methods need to pre-define image-to-image similarities or image features, which often require a very careful and time-consuming design.
Recently, there are major breakthroughs in learning-based approaches. Convolutional neural networks (CNN), especially its variant - fully convolutional neural network (FCN), has significantly improved performances in various medical image segmentation tasks [2,7,8]. FCN consists of two types of operations: down-sampling and up-sampling [6]. The down-sampling operation streams (i.e., convolution and pooling) usually result in coarse and global predictions based on the entire input image to the network, while the up-sampling streams (i.e., deconvolution) can generate dense prediction through finer inference. However, in FCN, the input intensity images are heavily compressed to be highly semantic. This will certainly cause serious information loss and subsequently decrease the localization accuracy, even after utilizing the up-sampling streams trying to recover the full image information. Ronneberger et al. [8] partially alleviated this problem by using U-net to aggregate the lower layer feature maps to the higher layer, for compensating the loss of resolution by the pooling operations. Although a more precise localization may be achieved by U-net, it is still extremely challenging to segment those thin and fine bony structures from MRI (Fig. 1).
To address the aforementioned challenges, we propose a 3D deep-learning cascade framework to automatically segment CMF bony structures from the head MRI. First, we train a 3D U-net model in an end-to-end and voxel-to-voxel fashion to coarsely segment the bony structures. Second, we use a CNN to further refine those coarsely segmented bony structures to acquire more accurate segmentation for the thin bony structures. Also, inspired by the auto-context model [12], we feed the original intensity MR image along with coarsely segmented bony structures as a joint input to the CNN for better guiding the model training during the fine-grained segmentation stage. Our proposed framework has been validated using 16 pairs of MRI and CT images. It is worth indicating the technical contribution of our novel 3D deep-learning based cascade framework, i.e., it can efficiently and accurately segment bony structures from MRI in a coarse-to-fine fashion.
2 Methods
Our proposed deep-learning based cascade framework is mainly designed to solve problem of segmenting CMF bony structures from MR images. The framework includes two stages: (1) coarse segmentation and (2) fine-grained segmentation (Fig. 2).
Fig. 2.
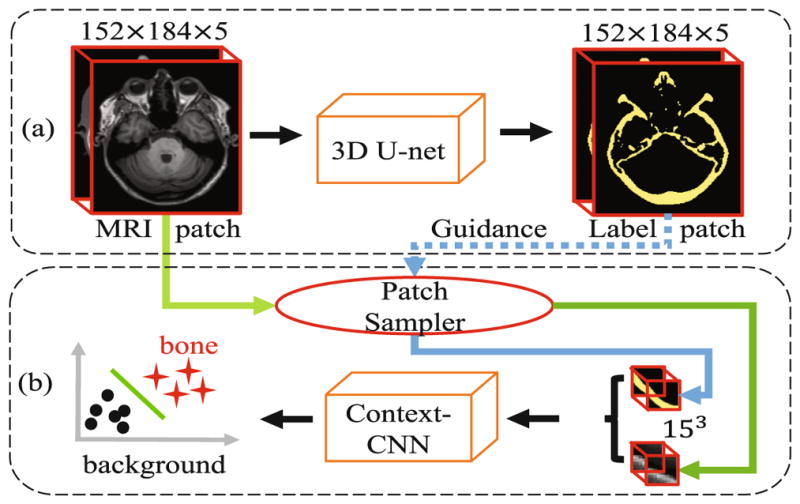
Architecture for our proposed deep-learning based cascade framework. (a) shows the sub-architecture for coarse segmentation by the 3D U-net, which adopts 5 consecutive slices as input; (b) shows another sub-architecture, context-guided CNN, for fine-grained segmentation, based on the patches sampled on the detected bone locations by (a).
2.1 Data Acquisition and Preprocessing
Dataset consists of 16 pairs of MRI and CT scans from Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (see www.adni-info.org for details) were used. The MR scans were acquired using a Siemens Triotim scanner, with the scanning matrix of 126 × 154, voxel size of 1.2 × 1.2 × 1 mm3, TE 2.95 ms, TR 2300 ms, and flip angle 9°, while the CT scans were acquired on a Siemens Somatom scanner, with a scanning matrix of 300×400, and a voxel size of 0.59 × 0.59 × 3 mm3. Due to high contrast for the bony structure, CT scans herein were employed to extract ground truth for MRI. During image preprocessing, both MRI and CT were initially resampled to 1 × 1 × 1 mm3 with final image size of 152 × 184 × 149. Then, each CT image was linearly aligned onto its corresponding MR image. Afterward, the bony structures were segmented from each aligned CT image using thresholding and further refined by an expert. These CT segmentation results provide training labels for the network.
2.2 Coarse Segmentation with Anatomical Constraint
We first segment the CMF bony structure from MRI by using U-net, as it has been successfully applied in various medical image segmentations tasks. The original U-net in [8] was proposed for 2D image segmentation. In order to extend it for 3D segmentation, it is natural to change its 2D filters (e.g., 3 × 3) to 3D ones (e.g., 3 × 3 × 3), like [1]. The 3D filters are then applied to each training sample, i.e., an entire 3D image. However, in real clinical situation, it is very difficult to obtain unlimited number of MRI dataset, which degrades the final performance. To address this problem, we extract 3D patches as the training samples from each MR image for training, instead of using the entire 3D image as one training sample. This strategy makes the number of patches virtually unlimited. On the other hand, for considering the anatomical shapes of bony structures, we decide to feed a large patch into the network, expecting to form certain anatomical constraint for the network. Specifically, we utilize a patch size of 152 × 184 × 5 to cover the whole bony structure on each slice, by also balancing the number of patches and receptive field. That is, the receptive field is large in the first two dimensions (i.e., 152 × 184), and still reasonable in the 3rd dimension (i.e., covering 5 consecutive slices) which can alleviate the possible inconsistent segmentations across slices. Accordingly, we set up the similar filters in the first two dimensions as the original U-net, while keep the feature map size unchanged through the whole network for the 3rd dimension. We denote this designed network as “3D U-net” (Fig. 2(a)) in the following text.
2.3 Fine-Grained Segmentation
We further utilize the initial probability maps from 3D U-net (in Sect. 2.2) to extract context information for refining the segmentation. Note that the context information can provide important semantic information for segmenting bony structure. In addition, based on the initial probability maps, we densely extract training patches only in the regions with ambiguous probabilities, which allows us to focus on only the potential problematic regions and also save the computational time.
Prior-guided Patch Sampling
Considering the fact that the initial segmentation probability maps already provide sufficient prior information about the locations of bony structures, we propose to use the initial segmentation probability maps to guide the sampling of training patches to the second deep learning architecture, CNN. Specifically, we randomly sample patches around those predicted bone regions. Moreover, since the probability maps also indicate certain information for the locations of difficult-to-segment bony structure (e.g., fine or small bones), we further densely extract the training patches around voxels with ambiguous probabilities.
Context-guided CNN Training
We first train a CNN [11] with a patch size of 15 × 15 × 15 to determine voxels with ambiguous probabilities. Considering large number of training patches, we form our network based on a 3D VGG [11] by removing two pooling layers together with its surrounding convolutional layers (i.e., (we remove the 2nd and 3rd pooling layer groups)), and setting the class number as 2 (bone or not), as shown in Fig. 2(b). Different from the original auto-context model, our context information is from the U-net, instead of the current CNN. This indicates that our proposed method is not iterative, which may significantly shorten the training time. More importantly, since the 3D U-net is performed on full slices, the initial segmentation by the 3D U-net could thus benefit from the long information range dependency, which indicates a voxel in the segmented maps contain information from the full slice of MRI. This can better serve as context information for the CNN (after the 3D U-net). We denote it as “context-CNN” in the text below.
2.4 Implementation Details
We adopt Caffe [4] to implement the proposed deep learning architecture shown in Fig. 2. Cross-entropy loss is utilized as the general loss of the network. Stochastic gradient decrease algorithm is adopted as the optimizer. To train the network, the hyper-parameters need to be appropriately determined. Specifically, the network weights are initialized by xavier algorithm [3], and the weight decay is set as 1e – 4. For the network bias, we initialize it to be 0. The initial learning rate is set as 0.01 for both networks, followed by decreasing the learning rate during the training. To augment the dataset, we flip the images along the 3rd dimension (Note, we only do augmentation for training dataset). We extract approximately 5000 training patches for the first sub-architecture (3D U-net), and 320, 000 patches for the second sub-architecture (context-guided CNN) around the regions that are segmented as bone by the first sub-architecture (3D U-net). The platform we use for training is a Titan X GPU.
3 Experiments and Results
We evaluated the proposed deep-learning based cascade framework on 16 MRI subjects in a leave-one-out fashion. Note that the data used in algorithm development were not used in the evaluation experiment. Dice similarity coefficient (DSC) was used to quantitatively measure the overlap between automated segmentation and the ground truth.
Comparison between coarse and fine-grained segmentation
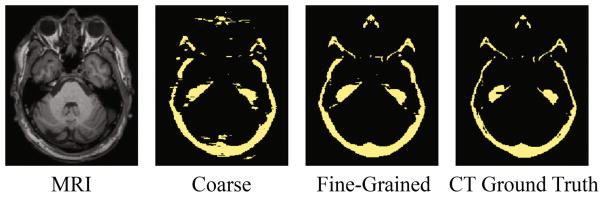
We performed experiments to compare coarse and fine-grained segmentation. The results showed that the initial segmentation method (3D-Unet) achieved an average DSC of 0.8410(0.0315). With the fine-grained segmentation method (context-CNN), the average DSC was improved to 0.9412(0.0316). Figure 3 shows the segmentation results for a randomly selected subject. It clearly demonstrated that the bony structures resulted from fine-grained segmentation became thinner and smoother than the ones resulted from the coarse segmentation. This proves the effectiveness of our proposed cascade model.
Fig. 3.
Demonstration of coarse and fine-grained segmentation results by 3D-Unet and context-CNN, along with the original MRI (left) and ground-truth bony structures from CT (right).
Impact of context-CNN
We also validated the impact of the context-CNN. Figure 4 shows the convergence curve for the traditional CNN and the context-CNN, which clearly indicates that the context-CNN converges much faster than the traditional CNN. This is because the context-CNN is provided with the context information, which can quickly guide the segmentation. DSC achieved with the traditional CNN is 0.9307(0.0350), which is improved to 0.9412(0.0316) by the context-CNN. This further demonstrates that the context information can not only speed up the convergence process, but also improve the discriminative ability of the network.
Fig. 4.
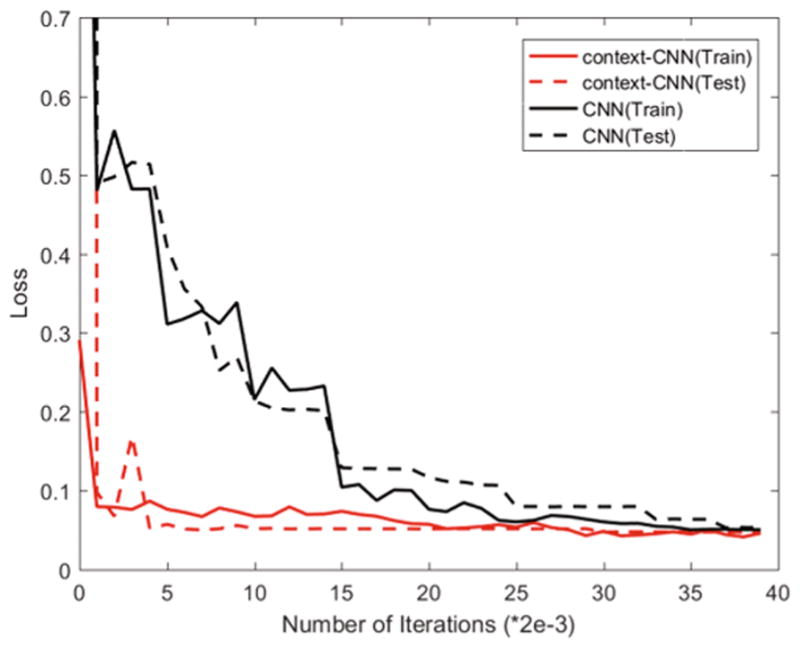
Comparison of convergence curves of the traditional CNN and the context-CNN in both training and testing phases.
Comparison with Other Methods
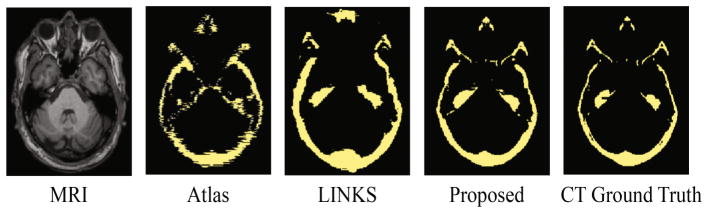
We also compared our method with two widely-used methods, i.e., multi-atlas based [10] (denoted as Atlas) and random forest-based [13] (denoted as LINKS, one of the widely-used learning-based methods), to further demonstrate the advantages of segmentation accuracy by our proposed method. The experimental results are shown in Fig. 5. The result achieved with Atlas method is the worst due to registration errors and difficulty of designing image-to-image similarity. The result achieved with LINKS is better because of the carefully designed features. Compared to LINKS, the result achieved with our proposed method is much more consistent with the CT ground truth. This better result is contributed from the better feature representation and joint optimization for the feature engineering and segmentation. The DSC achieved by the three methods are shown in Fig. 6. The quantitative results are consistent with qualitative results as shown in Fig. 5, which further confirms the superiority of our proposed method over other methods. Furthermore, the time cost of our proposed method is the least among all the three methods. It is mainly due to the use of cascade architecture that can allow fast determination for the categories of most voxels by the coarse segmentation, and thus save a significant computational load for the fine-grained segmentation.
Fig. 5.
Comparison of segmentation results by three different methods.
Fig. 6.
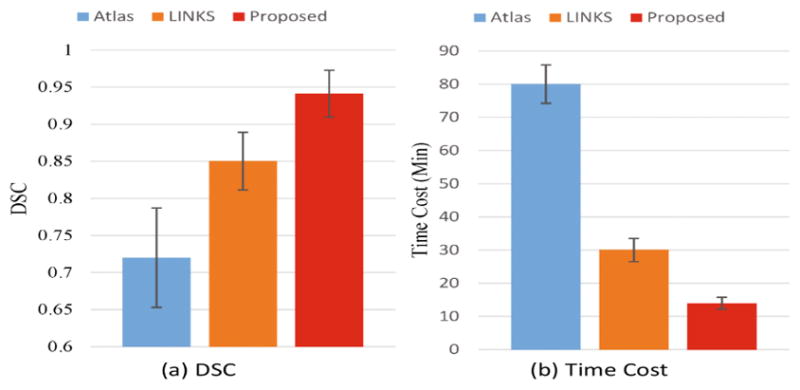
Quantitative comparison of three segmentation methods in terms of (a) DSC and (b) time cost.
4 Discussion and Conclusions
In this paper, we proposed a deep-learning based cascade framework to segment CMF bones from brain MR images. The 3D-Unet is used to perform an initial bone segmentation from MRI. With those initial segmented maps, we then focus on the coarsely predicted bone regions. This can not only provide prior information to support the training patches sampling, but also supply the context information for the context-CNN. With these strategies, our proposed method can work better and fast in segmentation. The experimental results also prove that our proposed method outperforms the other widely used state-of-the-art methods in both segmentation accuracy and computational efficiency.
References
- 1.Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-net: learning dense volumetric segmentation from sparse annotation. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W, editors. MICCAI 2016. LNCS. Vol. 9901. Springer; Cham: 2016. pp. 424–432. [DOI] [Google Scholar]
- 2.Dou Q, Chen H, Jin Y, Yu L, Qin J, Heng P-A. 3D deeply supervised network for automatic liver segmentation from CT volumes. In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W, editors. MICCAI 2016. LNCS. Vol. 9901. Springer; Cham: 2016. pp. 149–157. [DOI] [Google Scholar]
- 3.Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks. AISTATS. 2010:249–256. [Google Scholar]
- 4.Jia Y, et al. ACM Multimedia. ACM; 2014. Caffe: convolutional architecture for fast feature embedding; pp. 675–678. [Google Scholar]
- 5.Kraft A, et al. Craniomaxillofacial trauma: synopsis of 14,654 cases with 35,129 injuries in 15 years. Craniomaxillofacial Trauma Reconstr. 2012;5(01):041–050. doi: 10.1055/s-0031-1293520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long J, et al. Fully convolutional networks for semantic segmentation. CVPR. 2015:3431–3440. doi: 10.1109/TPAMI.2016.2572683. [DOI] [PubMed] [Google Scholar]
- 7.Nie D, et al. ISBI. IEEE; 2016. Fully convolutional networks for multi-modality isointense infant brain image segmentation; pp. 1342–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors. MICCAI 2015. LNCS. Vol. 9351. Springer; Cham: 2015. pp. 234–241. [DOI] [Google Scholar]
- 9.Seim H, et al. Model-based auto-segmentation of knee bones and cartilage in MRI data [Google Scholar]
- 10.Shan L, et al. ISBI. IEEE; 2012. Automatic multi-atlas-based cartilage segmentation from knee MR images; pp. 1028–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 2014 arXiv preprint arXiv:1409.1556. [Google Scholar]
- 12.Zhuowen T, Bai X. Auto-context and its application to high-level vision tasks and 3d brain image segmentation. IEEE TPAMI. 2010;32(10):1744–1757. doi: 10.1109/TPAMI.2009.186. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, et al. Links: learning-based multi-source integration framework for segmentation of infant brain images. NeuroImage. 2015;108:160–172. doi: 10.1016/j.neuroimage.2014.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]