Abstract
Background
Cartilage degeneration affects millions of people but preventing its degeneration is a big challenge. Although RNA interference (RNAi) has been used in human trials via silencing specific genes, the cartilage RNAi has not been possible to date because the cartilage is an avascular and very dense tissue with very low permeability.
Purpose
The objective of this study was to develop and validate a novel lipid nanoparticle (LNP)-siRNA delivery system that can prevent cartilage degeneration by knocking down specific genes.
Methods
LNP transfection efficiency was evaluated in vitro and ex vivo. Indian Hedgehog (Ihh) has been correlated with cartilage degeneration. The in vivo effects of LNP-Ihh siRNA complexes on cartilage degeneration were evaluated in a rat model of surgery-induced osteoarthritis (OA).
Results
In vitro, 100% of chondrocytes were transfected with siRNA in the LNP-siRNA group. In accordance with the cell culture results, red positive signals could be detected even in the deep layer of cartilage tissue cultures treated by LNP-beacon. In vivo data showed that LNP is specific for cartilage, since positive signals were detected by fluorescence molecular tomography and confocal microscopy in joint cartilage injected with LNP-beacon, but not on the surface of the synovium. In the rat model of OA, intraarticular injection of LNP-Ihh siRNA attenuated OA progression, and PCR results showed LNP-Ihh siRNA exerted a positive impact on anabolic metabolism and negative impact on catabolic metabolism.
Conclusion
This study demonstrates that our LNP-RNAi delivery system has a significantly chondroprotective effect that attenuates cartilage degeneration and holds great promise as a powerful tool for treatment of cartilage diseases by knocking down specific genes.
Keywords: cartilage diseases, lipid nanoparticle, RNA interference, delivery system
Introduction
Cartilage is the tough but flexible tissue that covers the ends of bones at joints, and it also gives shape and support to other parts of the body, such as the ears, nose, and windpipe. Healthy cartilage helps joint articulation by allowing bones to glide smoothly over each other. It protects bones by preventing them from rubbing against each other. Injured, inflamed, or damaged cartilage can cause symptoms such as pain and limited movement, even leading to joint damage and deformity. Causes of cartilage problems include tears and injuries, such as sports injuries, genetic factors, and different types of arthritis. Cartilage diseases affected 52.5 million patients in the United States.1 Here, we target osteoarthritis (OA) to validate our lipid nanoparticle (LNP) as a novel cartilage therapeutic tool.
OA, which affects over 200 million people worldwide,2 is a degenerative disease of the articular joints characterized by slow but progressive loss of cartilage that causes joint pain, stiffness, and loss of mobility.3–6 A previous study showed that an increase of Indian Hedgehog (Ihh) was involved in OA progression in humans.7,8 Further studies demonstrated that Ihh blockade could be a therapeutic approach to prevent or delay articular cartilage degeneration.2,9 However, Ihh gene deletion is currently not a therapeutic option as it is lethal in animals. To date, several small compounds have been reported to inhibit Ihh signaling. However, all these have significant side effects including holoprosencephaly, cleft lip and palate, weight loss, hypotension, and dyspnea, even at relatively low dose in young mice.10–13
RNA interference (RNAi), a natural cellular process that has been identified as a crucial biomolecular means to regulate gene expression through a highly precise mechanism of sequence-directed gene silencing, could theoretically be used to target any disease-associated pathogenic gene of interest.14 Accordingly, RNAi-based therapies targeting those genes that have been identified to negatively regulate cartilage degeneration could facilitate translational therapy for treating cartilage diseases.15 In addition, several Phase I–III clinical trials demonstrated that RNAi-based therapy holds great promise as a powerful means to silence specific genes to treat human diseases.16,17 Thus, an efficient way to knock down a specific gene might be achieved by local delivery of a specific small interference RNA (siRNA) as an effective strategy to slow or halt cartilage diseases without side effects caused by chemical inhibitors. However, cartilage siRNA delivery has not been possible to date because cartilage is an avascular and very dense tissue with very low permeability. Furthermore, the lack of an efficient and safe cartilage RNAi delivery system has largely limited the clinical translation of RNAi approach in cartilage diseases. Thus, the development of a specific delivery system for RNAi-based therapies that addresses this issue is highly desirable.
Recently, we modified an attractive positively charged LNP, which is a lipid-based particle with diameter ~67 nm formed by mixing an ethanol stream containing the lipid mixture with an aqueous stream containing the siRNA.18–20 LNP has been linked with bone formation and used for anabolic therapy,16,21 but it has never been used in the treatment of cartilage diseases. Indeed, the LNP-siRNA system showed an exciting and promising silencing effect in vitro. It also presented many advantages, such as easy preparation as well as positively charged particles, and small particle size with narrow distribution, which could contribute to diffusion through spaces of negatively charged cartilage. The objective of this study is to determine whether the LNP-siRNA delivery system is an efficient and safe tool that can be used to knock down cartilage-specific genes in vivo. We chose Ihh to test whether the delivery system can attenuate surgery-induced OA as a specific therapy for cartilage diseases because of the evidence linking Ihh with OA cartilage degeneration.
Materials and methods
This study was approved by the Institutional Review Board and the Institutional Animal Care and Use Committee of the Rhode Island Hospital (RIH CMTT#: 0111–12). Animals were cared for according to the Public Health Service Policy and the Guide for the Care and Use of Laboratory Animals.
LNPs preparation
The LNPs were prepared by spontaneous vesicle formation with minor modifications.16 Briefly, the lipids, including Dlin-KC2-DMA, dipalmitoyl phosphatidylcholine (DPPC), cholesterol, and C16 ceramide-mPEG2000, were solubilized in ethanol at a concentration of 28.5, 35, 9.5, and 50 mg/mL, respectively. Then, the lipid solution was mixed with 10 μL Dlin-KC2-DMA, 2 μL DPPC, 15 μL cholesterol, 2 μL C16 ceramide-mPEG2000, and 6 μL ethanol. LNP-siRNA (2.5 μM) was made by injecting 35 μL of lipids’ solution into 65 μL of siRNA solution (0.2 nmol siRNA mixed in the 55 μL citrate buffer) drop by drop under ultrasound. LNP-beacon (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) (2.5 μM) was prepared with 0.2 nmol fluorescence molecular GAPDH beacons (a novel fluorescent oligonucleotide probe only showing a positive signal-red when it binds to specific GAPDH messenger RNA [mRNA]); LNP-siRNA (Ihh) (5.0 μM) was prepared with 0.4 nmol Ihh siRNA and LNP-siRNA (Ihh+GAPDH) with 0.2 nmol Ihh siRNA and 0.2 nmol GAPDH siRNA. Then, 400 μL PBS was added to the mixture and ultracentrifugation was performed using a Amicon Ultra-0.5 filter device (Millipore Corporation, UFC501024, Darmstadt, Germany) at 12,000× g for ~5 minutes. The process yielded ~80 μL LNP-siRNA/beacon complexes. The hydrodynamic diameters were measured by laser light scattering following their dilution in distilled water using DELSA 440 SX Zeta Potential Analyzer (Beckman Coulter Corporation, Brea, CA, USA) at 25°C. The morphology of LNPs was visualized by cryo-transmission electron microscopy. The encapsulation efficiency was assessed by the Quanti-RiboGreen RNA assay (Molecular Probes, Thermo Fisher Scientific, Waltham, MA, USA) with or without 0.5% Triton X-100.22
LNP-siRNA/beacon in vitro and ex vivo transfection experiments
Chicken chondrocytes were isolated from tibia cartilage and seeded in 24-well culture plates at a concentration of 1×106 cells/well as previously described.23 At 90% confluence, the cells were cultured overnight under serum-free conditions and treated with 8 μL free siRNA (2.5 μM) or 8 μL LNP-siRNA (2.5 μM); siRNA used in this experiment is control siRNA from Business Company (Qiagen, Waltham, MA, USA). Green fluorescence protein, phase-contrast, three-dimensional (3D) pictures of images obtained by fluorescence microscopy were acquired after 24-hour treatment, and transfection rates were evaluated using a flow cytometer (MACSQuant Analyzer; Miltenyi Biotec, Bisley, UK) by gating fluorescein isothiocyanate-positive events. The same experiments were repeated using astrocytes, chondrocytes cell line C28/I2,24 chondrogenic cell line ATDC5,25 breast cancer cell line MCF-7 (American Type Culture Collection, MD, USA), and human OA chondrocytes. A similar experiment was performed with pig knee cartilage tissues treated with free beacon or LNP–beacon complexes and observed by confocal microscopy 48 hours after treatment. All experiments were repeated three times.
Cell viability measurement
Fluorescein diacetate (FDA) can enter the cells easily and perform intracellular cleavage by esterase that results in free fluorescence; hence, quantification of this fluorescence could reflect metabolic activity of live cells. Propidium iodide (PI) can get into cells only through the membrane of dead or dying cells. Thus, FDA–PI double staining method can provide information about cell viability.26 1) Chicken chondrocytes were seeded in six-well culture plates at 80%–90% confluence and treated with/without 4% (v/v) LNPs (2.5 μM). Three freeze-thaw cycles of the chondrocytes were used as a positive cell death control. The viability of cells was detected by FDA–PI staining method (Thermo Fisher Scientifc). Briefly, stock solutions of FDA and PI were prepared at 5 mg/mL and 40 μg/mL, respectively. Live (green) and dead (red) cells were observed under a fluorescence microscope. 2) Chicken chondrocytes were seeded in six-well culture plates and treated with LNPs (2.5 μM) at different concentrations of 0% (blank), 1% (v/v), 2% (v/v), and 4% (v/v). Twenty-four hours later, cells were harvested and stained by FDA–PI for 15 minutes. The percentages of live/dead cells were quantified by fluorescence-activated cell sorting (FACS) analysis on a MACSQuant instrument (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany).
Cell proliferation assay
The cell proliferation assay was performed using BrdU enzyme linked immunosorbent assay (colorimetric) Kit (Roche Applied Science, Indianapolis, IN, USA) and assays were repeated at least three times. Briefly, chicken chondrocytes were cultured in 96-well plates at a density of 5,000 cells/100 μL/well in complete growth media. After 24 hours, LNPs (2.5 μM) were added to each well at different concentrations of 0% (blank), 1% (v/v), 2% (v/v), and 4% (v/v). The cells were labeled using 10 μL BrdU per well (10 μM BrdU) for 2 hours, before the cell proliferation assay was performed on days 1 and 2. After incubation with BrdU, the cell DNA was denatured in one step by adding FixDenat. Next, the cells were incubated with the anti-BrdU-POD antibody for 90 minutes at room temperature. After removal of the antibody conjugate, the cells were washed and the substrate solution was added. The reaction product was quantified by measuring the absorbance using a SpectraMax 190 Absorbance Microplate Reader (Molecular Devices, Sunnyvale, CA, USA) at 370 nm with a reference wavelength of 492 nm.
Transfection effects in vivo
A pilot study was performed to determine the transfection effects in vivo. We first injected 20 μL alcian blue into the mouse knee joint cavity with an insulin syringe to confirm that mouse intra-articular injection was a reliable method. Then, 8-week-old C57BL6 male mice (n=5) were injected with 20 μL LNP-beacon (2.5 μM) in the right knee joint and 20 μL free beacon (2.5 μM) in the left knee joint. Then, mice were monitored for 72 hours post-intra-articular injection by fluorescence molecular tomography (FMT), a noninvasive and quantitative fluorescence-based technology with high molecular specificity and sensitivity for 3D tissue imaging of live animals,9,27,28 at a wavelength of 680 nm to confirm the LNPs’ transfection reliability. The knee cartilage was harvested 72 hours after injection to confirm the presence of beacon in the cartilage cells by confocal microscopy.
A similar study was performed in rats to verify LNP transfection in vivo. Ten-week-old male Wistar rats (n=10) were randomized into two groups (n=5/group). Rats in one group received 20 μL LNP-beacon (2.5 μM) injection in the right knee joint and 20 μL free beacon (2.5 μM) injection in the left knee joint. Rats in the second group received 20 μL LNP-Cy3-labeled control siRNA (2.5 μM) injection in the right knee joint and 20 μL Cy3-labeled control siRNA (2.5 μM) injection in the left knee joint. After 48 hours, the knee joint cartilages and synovium from rats in the first group were observed under confocal microscope (wavelength =680 nm), whereas knee joint cartilages in the second group were observed under fluorescence microscope (wavelength =570 nm).
LNP-siRNA knockdown experiment in vivo: a pilot study
To investigate in vivo effects of LNP-siRNA, 10-week-old male Wistar rats (n=27) were randomized into three groups (n=9/group): 1) LNP-GAPDH siRNA (2.5 μM) 24-hour-treatment group; 2) LNP-Ihh siRNA (2.5 μM) 24-hour-treatment group; 3) LNP-siRNA (Ihh+GAPDH) 24-hour-treatment group. LNP-siRNA injections were performed on the right knee joint, while the left knee joint was injected with 2.5 μM siRNA without LNP and served as control. The volume of each injection was 40 μL. Animals were euthanized after treatment and GAPDH and Ihh genes expression in the knee cartilage were quantified by real-time quantitative polymerase chain reaction (qPCR).
Rat anterior cruciate ligament transection (ACLT) model of OA and treatment with LNP-Ihh siRNA
To test whether the LNP-siRNA can attenuate surgery-induced OA and thus serve as a specific therapy method for cartilage diseases, we first carried out a pilot study to determine the necessary frequency of intra-articular injection. Five 10-week-old male Wistar rats received 40 μL LNP-beacon (2.5 μM) intra-articular injection in the right knee joint and were monitored dynamically by FMT until the positive signal disappeared. Another nine rats received 40 μL LNP-Ihh siRNA (2.5 μM), and knee cartilage Ihh gene and its downstream genes were quantified by real-time qPCR after 1 week. In our formal study, we chose Ihh siRNA as a target gene because of evidence correlating Ihh with OA.9 One hundred twenty 10-week-old male Wistar rats (180–230 g) were randomized into four treatments (n=30 per group): 1) sham operation and Ihh-siRNA (2.5 μM) injection, 2) ACLT and LNP-Ihh siRNA (2.5 μM) injection, 3) ACLT and LNP-Ihh siRNA (5.0 μM) injection, 4) ACLT and Ihh-siRNA (2.5 μM) injection. ACLTs and sham operations were performed on the right knees, as described previously.29 The volume of each injection was 40 μL. Intra-articular injections were performed 24 hours following surgery and then every other week for a total of five injections per knee. All animals were euthanized 10 weeks after the surgery. OA-related gene expression was quantified by real-time qPCR. The extent of OA progression was graded using the Osteoarthritis Research Society International Osteoarthritis Cartilage Histopathology Assessment System (OOCHAS).30
Histology
After the rats were euthanized with carbon dioxide, the knee joints of the right hindlimbs were harvested and immersed in 10% formalin for 72 hours. The specimens were decalcified in 20% ethylenediaminetetraacetic acid solution (pH 7.2). The tibiae were hemisected in the midsagittal plane and each half was embedded in a single block of Paraplast X-TRA medium (Sigma-Aldrich, St Louis, MO, USA). Blocks were trimmed to expose cartilage using a rotary microtome (Reichert-Jung, Vienna, Austria). Ten adjacent sections were collected at intervals of 0, 100, and 200 μm. Two serial 6-μm-thick sections from each interval of 0, 100, and 200 μm were stained with Safranin-O/fast green and the severity of cartilage damage was assessed using OOCHAS score (OA score = grade × stage; range, 0–24).30 Three independent blinded observers scored each section, and the scores for the medial and lateral tibial plateau were averaged within each joint.
Immunohistochemistry (IHC)
To detect the distribution of type II collagen, matrix metalloproteinase 13 (MMP-13), type X collagen, and type II collagen breakdown product in cartilage, we carried out IHC using the 3,3′-diaminobenzidine (DAB) Histostain streptavidin-peroxidase (SP) kit (Novex; Life Technologies, Waltham, MA, USA). Six-micrometer sections were deparaffinized and rehydrated using conventional methods. The sections were digested with 5 mg/mL of hyaluronidase in PBS for 20 minutes. Endogenous peroxidase was blocked by treating the sections with 3% hydrogen peroxide in methanol for 30 minutes. Nonspecific protein binding was blocked by incubation with a serum blocking solution (Li-Cor, Lincoln, NE, USA). The sections were incubated with specific antibodies against rat type II collagen, MMP-13, type X collagen, or type II collagen breakdown product (IBEX Technologies, Mont-Royal, QC, Canada) at 4°C overnight. Thereafter the sections were treated sequentially with biotinylated secondary antibody and SP conjugates, and then developed in DAB chromogen (Invitrogen). The sections were counterstained with hematoxylin (Invitrogen). Photomicrographs were taken with a Nikon E800 microscopy (Nikon, Melville, NY, USA).31
Real-time qPCR
Cartilage samples (tibial plateau and femur condyle) from 15 rats per group were dissected using a scalpel and cartilage samples from each of three rats were combined together. Then, these samples were ground with a mortar and pestle under liquid nitrogen, and total RNA was isolated using an RNeasy isolation kit (Qiagen, Waltham, MA, USA).7 Total RNA (0.5 μg) was reverse transcribed to complementary DNA (cDNA) using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). The resulting cDNA (40 ng/μL) was used as the template to quantify the relative content of mRNA using a QuantiTect SYBR Green PCR kit (Qiagen) with a DNA Engine Opticon 2 Continuous Fluorescence Detection System (MJ Research, Quebec, Canada). The rat primer sequences used were 5′-ACA-GCA-ACA-GGG-TGG-TGG-AC-3′ (forward) and 5′-TTT-GAG-GGT-GCA-GCG-AAC-TT-3′ (reverse) for GAPDH; 5′-CAG-GAA-GGA-CCC-ATT-CCG-TC-3′ (forward) and 5′-AAG-TCA-CAA-ACC-CAG-GTC-CC-3′ (reverse) for Ihh; 5′-AAG-GGA-CAC-CGA-GGT-TTC-ACT-GG-3′ (forward) and 5′-GGG-CCT-GTT-TCT-CCT-GAG-CGT-3′ (reverse) for type II collagen; 5′-CAG-TGC-GAT-GCA-GGC-TGG-CT-3′ (forward) and 5′-CCT-CCG-GCA-CTC-GTT-GGC-TG-3′ (reverse) for Aggrecan; 5′-CCA-GGT-GTC-CCA-GGA-TTC-CC-3′ (forward) and 5′-CAA-GCG-GCA-TCC-CAG-AAA-GC-3′ (reverse) for type X collagen; 5′-GGA-CCT-TCT-GGT-CTT-CTG-GC-3′ (forward) and 5′-GGA-TGC-TTA-GGG-TTG-GGG-TC-3′ (reverse) for MMP-13; 5′-CCG-CAC-GAC-AAC-CGC-ACC-AT-3′ (forward) and 5′-CGC-TCC-GGC-CCA-CAA-ATC-TC-3′ (reverse) for Runx2; and 5′-CGG-CTA-CCA-CAT-CCA-AGG-AA-3′ (forward) and 5′-GCT-GGA-ATT-ACC-GCG-GCT-3′ (reverse) for 18S RNA. The results were normalized to the level of 18s RNA. Relative transcript levels were calculated according to the equation , where ΔΔCt = ΔCtE − ΔCtC (ΔCtE = Ctexp − Ct18S, ΔCtC = CtC − Ct18S).7
Statistical analysis
Data are expressed as means ± standard error of the mean (SEM). Two-tailed paired t-tests were used to compare mRNA levels between the LNP-siRNA and free-siRNA groups. A probability level <5% was considered significant. The OOCHAS scores in different groups were analyzed by one-way analysis of variance with multiple pairwise comparisons made by the Student–Newman–Keuls method (three comparisons or more) at a rejection level of 5% unless otherwise noted. Analyses of variance were also used to compare the cell viability by FACS test and levels of mRNA for type II collagen, aggrecan, MMP-13, Runx2, and type X collagen in the LNP-Ihh siRNA treatment experiment. P-values <0.05 were considered significant. Statistical analyses were performed using SPSS software.
Results
Characterization of LNPs
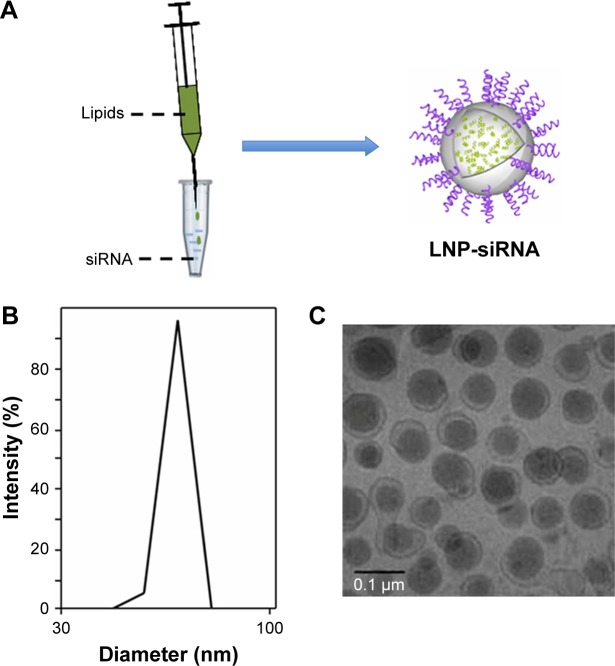
The LNPs delivery system was prepared as showed in Figure 1A. The encapsulation efficiencies of LNPs were above 95% (Figure 1B Y-axis), which suggested that LNPs protected and facilitated the transfection of most siRNAs. The hydrodynamic diameters of LNPs measured by laser light scattering were 67±4.3 nm (Figure 1B X-axis), and the representative morphology of LNPs is shown in Figure 1C. Basic characteristics of LNPs we modified are in agreement with those of Belliveau et al, who reported that encapsulation efficiencies of LNPs are unaffected and remain >95%.19
Figure 1.
LNPs characterization.
Notes: LNPs were produced by mixing lipids’ solution and siRNA solution drop by drop under strong vortex, and ethanol was removed by ultracentrifugation (A). The encapsulation efficiencies of LNPs were above 95% (B). The diameter of LNPs was 67±4.3 nm (C).
Abbreviations: LNP, lipid nanoparticle; siRNA, small interference RNA.
Effectiveness of LNPs transfection in vitro and ex vivo
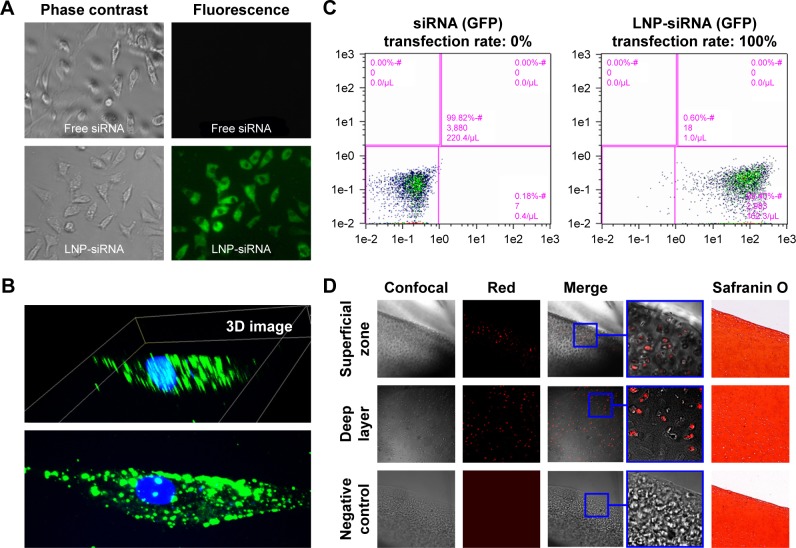
LNP-siRNA and free siRNA were utilized to compare the transfection efficiency. It was shown that none of the chondrocytes were transfected in the free siRNA group, while almost all the chondrocytes were transfected in the LNP-siRNA group (Figure 2A). Meanwhile, 3D images showed that the transfected siRNAs by LNPs were located in the cytoplasm (Figure 2B). Moreover, it was verified by flow cytometry analysis that transfection rate was close to 0 in the free siRNA group and almost 100% in the LNP-siRNA group, indicating that LNP can deliver siRNA into the cytoplasm of chondrocytes with high efficiency (Figure 2C). We also treated astrocytes, chondrocytes cell line C28/I2, chondrogenic cell line ATDC5, and breast cancer cell line MCF-7 with LNP-siRNA, and the results were similar to those obtained using chicken chondrocytes (Figure S1). To test the penetration of LNPs in large animal cartilage, a full thickness of cartilage from pig tibia was incubated with either free beacon or LNP-beacon for 48 hours. A 20-μm frozen section was then analyzed for beacon fluorescence under confocal microscope. The results showed that red fluorescence, a positive signal for beacon, could be detected by confocal microscope in the superficial zone and deep layer of the pig cartilage in the LNP-beacon-treated group. In contrast, no fluorescent signal could be detected in the free-beacon group, which served as control, suggesting that LNPs had an excellent penetration power in the pig cartilage and LNPs could deliver beacon into cartilage cells ex vivo (Figure 2D).
Figure 2.
LNPs transfection efficiency.
Notes: There was no fluorescent signal in the free siRNA-treated chondrocytes, while almost every LNP-siRNA chondrocyte fluoresced green (A); magnification ×10. Three-dimensional images showed that all the fluorescent signals were actually located in the cytoplasm (B), indicating that LNPs can effectively deliver siRNA into the chondrocytes. Flow cytometry analysis further showed that transfection rate was close to 0% in free siRNA-treated group, and almost 100% in the LNP-siRNA-treated group (C). Cartilage tissue cultures showed that fluorescent signal was not only present in the superficial zone but also in the deep layer of the LNP-beacon-treated pig cartilage, whereas no signal was detected in control samples (D); magnification ×4.
Abbreviations: GFP, green fluorescent protein; LNP, lipid nanoparticle; siRNA, small interference RNA.
Effect of LNPs on cell toxicity and proliferation
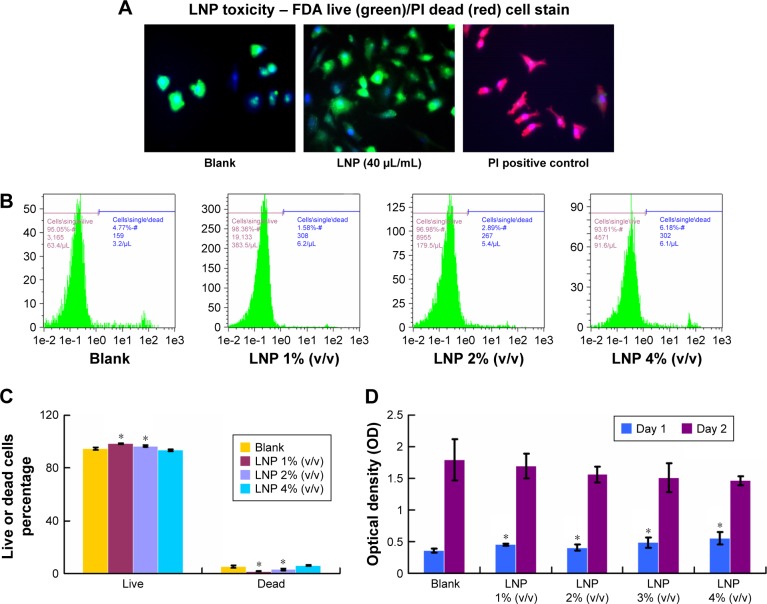
To determine the LNP toxicity, chicken chondrocytes were incubated with LNPs. The live and dead cells were detected by FDA live and PI dead stain (green: live; red: dead; blue: nuclear). The results showed green chondrocytes in the LNPs-treated group and in the blank group, indicating that a high dose of LNPs (40 μL/mL) did not induce cell death (Figure 3A). To further study toxicity, chicken chondrocytes were incubated with different doses of LNPs for 24 hours and then stained by FDA–PI. The relative amounts of live and dead cells were quantified by FACS. We found that more than 90% of the cells were live cells in the blank and LNPs-treated groups (Figure 3B). Furthermore, the percentages of live cells in the 1% (v/v) and 2% (v/v) LNP groups were significantly higher than those in the blank group, and the percentage of live cells in the 4% (v/v) LNP group was not significantly different from that in the blank group. In addition, the percentages of dead cells in the 1% (v/v) and 2% (v/v) LNP groups were significantly lower than those in the blank group, and the percentage of dead cells in the 4% (v/v) LNP group was not significantly different than that in the blank group (Figure 3C). To test the influence of LNP on cell proliferation, chondrocytes were incubated with different concentrations of LNPs, and cell proliferation levels were accessed using a Cell Proliferation BrdU Kit. The data showed that proliferation levels in groups treated with LNPs were significantly higher than in the blank group on day 1, but similar among all five groups on day 2 (Figure 3D).
Figure 3.
Effect of LNPs on cell toxicity and proliferation.
Notes: Green fluorescence indicates live cells and red fluorescence indicates dead cells. We did not detect dead cells in the blank or LNPs-treated groups. All cells in the freeze-thawed group were dead (A); magnification ×10. Fluorescence-activated cell sorting data showed that 1% (v/v) and 2% (v/v) LNP groups increased cell viability (B and C). LNPs were even able to promote cell proliferation after 24 hours of treatment, but there was no statistically significant difference among the blank and LNPs-treated groups after 48 hours of treatment (D). Compared with blank, *P<0.01.
Abbreviations: FDA, Fluorescein diacetate; LNP, lipid nanoparticle; PI, propidium iodide; siRNA, small interference RNA.
Effectiveness of LNP transfection in vivo
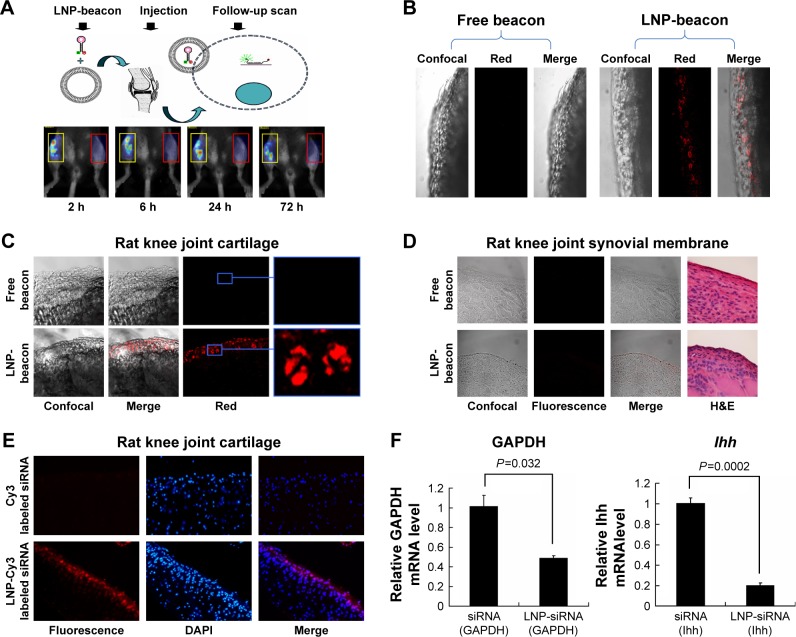
To further test our delivery system in vivo, we first investigated the reliability of our method of mouse intra-articular injection by alcian blue. Gross observation showed that alcian blue adhered to the cartilage and synovial membrane in the joint cavity (Figure S2), suggesting good penetration of the injection. Then, LNP-beacon and free beacon were injected into mice joints for our pilot study. We detected a positive signal by FMT at a wavelength of 680 nm in the LNP-beacon-injected knee (ie, right knee) at different time points (2, 6, 24, and 72 hours) after injection. In contrast, there was no signal in the free-beacon-injected knee (ie, left knee) (Figure 4A). All the mice were euthanized after 72 hours and knee joint cartilage specimens were further observed by confocal microscopy. We were not able to detect fluorescent signal in the free-beacon-injected knee joint cartilage, while we found obvious red fluorescent signals in the LNP-beacon-injected knee joint cartilage, which indicated that only beacon carried by LNP could penetrate through the cartilage and get into cartilage cells in vivo (Figure 4B).
Figure 4.
siRNA could be delivered into cartilage cells by LNPs and functioned in vivo.
Notes: Fluorescent signals could be detected by fluorescence molecular tomography in the LNP-beacon-treated mouse knee joint (red rectangles). In contrast, no fluorescent signal could be detected in the free-beacon-treated mouse knee joint (yellow rectangles) (A). The LNP-beacon-treated cartilage was further observed by confocal microscopy, and red signals were located in all of the cartilage tissue layers. In contrast, no signal was detected in the free-beacon-treated cartilage; magnification ×10 (B). Similar to mice cartilage data, red signals were present in the LNP-beacon-treated rat knee joint cartilage, whereas no signal was found in the free-beacon-treated rat knee joint cartilage; magnification ×10 for panels on the left and magnification ×20 for the insets (C). No positive signal could be detected by confocal microscope from either LNP-beacon-treated or free-beacon-treated rat knee joint synovial membrane; magnification ×10 (D). Fluorescent signals could be detected by fluorescence microscopy in the LNP-Cy3-labeled siRNA-treated rat knee cartilage. In contrast, no fluorescent signal was detected in the free Cy3-labeled siRNA-treated rat knee cartilage; magnification ×10 (E). After 24 hours of in vivo treatment with LNP-GAPDH siRNA or LNP-Ihh siRNA, cartilage GAPDH mRNA decreased by 51.8% and Ihh mRNA decreased by 79.7% (F).
Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Ihh, Indian Hedgehog; LNP, lipid nanoparticle; siRNA, small interference RNA.
In our formal study, there was no signal in the free-beacon knee joint cartilage (left) observed by confocal microscopy, while red positive signals could be observed in the LNP-beacon knee joint cartilage (right) (Figure 4C). Furthermore, there were no positive signals in the synovial membranes from both free-beacon- and LNP-beacon-injected knee joints, indicating that LNPs cannot function in the synovial membrane and that LNP constitutes a specific siRNA delivery tool for cartilage (Figure 4D). In addition, to further validate this result, Cy3-labeled control siRNA was also used in our study. We did not detect a positive fluorescence signal in the Cy3-labeled control siRNA-injected knee joint cartilage by fluorescence microscopy, while we obtained the red positive signals in the LNP-Cy3-labeled control siRNA-injected knee joint cartilage, which further confirmed that LNPs could take siRNA into the cartilage cells in vivo (Figure 4E).
To knock down a specific gene in vivo, we injected LNP-GAPDH siRNA or LNP-Ihh siRNA into the rat right knee joint, and GAPDH siRNA or Ihh siRNA into the left knee joint to serve as control. Real-time qPCR results showed that GAPDH/Ihh mRNA level in the LNP-GAPDH siRNA and Ihh siRNA groups decreased by 51.8% and 79.7%, respectively, when compared with the GAPDH siRNA and Ihh siRNA groups alone 24 hours after intra-articular injection (Figure 4F). This result indicates that LNP-siRNA could not only get into cartilage cells in vivo, but could also exert its impact on a specific gene. We further knocked the Ihh and GAPDH genes down together by LNP-siRNA (Ihh+GAPDH). The data showed that Ihh mRNA level decreased by 41.6% and GAPDH by 19.5%. Nevertheless, type II collagen mRNA level was increased; MMP-13 and type X collagen mRNA levels were decreased (Figure S3).
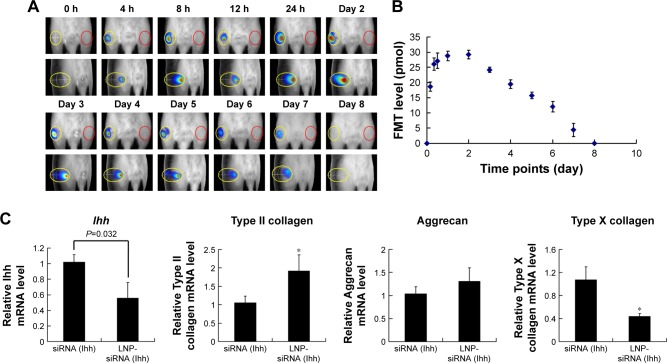
Time duration of LNP-RNAi in the rat knee joint
To determine the frequency of LNP-RNAi treatment, we monitored LNP-beacon signal dynamically in the rat knee joint until the fluorescence disappeared. We found that fluorescence could be detected as soon as 4 hours after injection at a volume of 18.65±1.59 pmol, and the fluorescence intensity reached its climax (29.18±1.39 pmol) on day 2. After that, the FMT value decreased gradually until it disappeared on day 8 (Figure 5A and B). Subsequently, we found that Ihh mRNA level decreased by 44.1% 7 days after treatment with LNP-Ihh siRNA, and type II collagen mRNA level was enhanced, while type X collagen mRNA level was depressed. These findings indicated that LNP-Ihh siRNA could get into cartilage cells in vivo and exert a positive impact on anabolic metabolism and a negative impact on catabolic metabolism and this effect was durable for at least 1 week (Figure 5C).
Figure 5.
LNP-siRNA lasts for 7 days at least in the rat knee joint.
Notes: Fluorescent signals were monitored dynamically by FMT. There was no signal in the left knee (red ovals). In contrast, fluorescent signals lasted for 7 days and reached climax at day 2 in the right knee (yellow ovals) (A). Quantitative data indicated that fluorescence signals could be detected after 4 hours and reached climax on day 2, then decreased gradually (B). Seven days after LNP-Ihh siRNA in vivo treatment, cartilage Ihh mRNA decreased by 44.1% and type II collagen mRNA level significantly increased, while type X collagen mRNA level decreased. Values are mean ± standard error of the mean. *P<0.01 versus siRNA (Ihh) treatment group (C).
Abbreviations: FMT, fluorescence molecular tomography; Ihh, Indian Hedgehog; LNP, lipid nanoparticle; siRNA, small interference RNA.
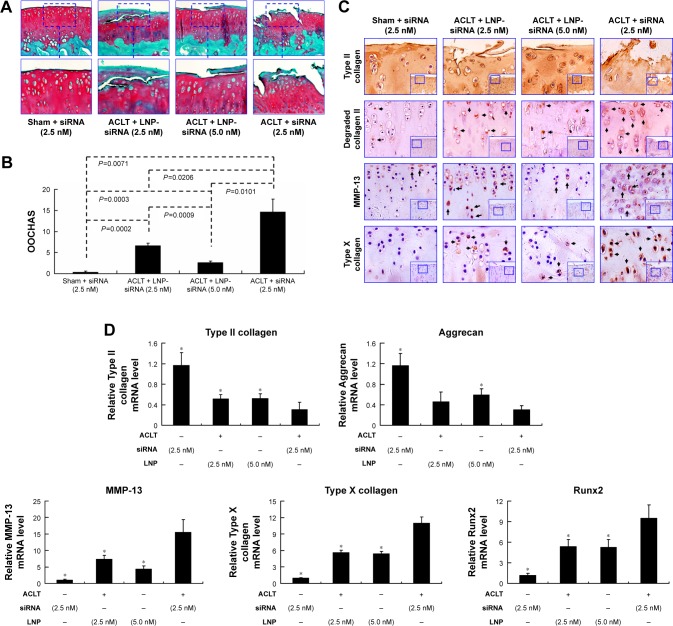
Attenuation of post-traumatic OA pathogenesis using intra-articular LNP-Ihh siRNA in a rat ACLT OA model
Based on the result of LNP-beacon and Ihh mRNA (Figure 5), we took them as references, and treated ACLT OA every other week for a total of five treatments. We found a significant decrease in the OA score in LNP-Ihh siRNA-treated rats as compared with rats that underwent free Ihh siRNA treatment, along with stronger Safranin O staining, more cellularity but less chondrocyte cloning, and less cleft progression (Figure 6A). OOCHAS scores in both LNP-Ihh siRNA-treated groups suggested mild degeneration (mean ± SD, 6.67±0.58 in the 0.1 nmol group and 2.67±0.29 in the 0.2 nmol group; P<0.001), while cartilage damage in rats that underwent ACLT and free-Ihh siRNA treatment was significantly more severe (14.67±3.06; P<0.01). Cartilage from rats that underwent the sham operation had the least amount of damage (0.33±0.29; P<0.01) (Figure 6B). IHC results showed that MMP-13 and type X collagen were elevated in cartilage from the ACLT and free-Ihh siRNA group when compared with LNP-Ihh siRNA-treated cartilage (at 0.1 and 0.2 nmol). This was accompanied by increased OA damage in the ACLT and free-Ihh siRNA group. In contrast, type II collagen content was higher and type II collagen degradation was more inhibited in cartilage from the LNP-Ihh siRNA-treated groups than in the ACLT and free-Ihh siRNA group (Figure 6C). Real-time qPCR results indicated that intra-articular LNP-Ihh siRNA injection enhanced the levels of mRNA for type II collagen and aggrecan, and suppressed the levels of mRNA for MMP-13, Runx2, and type X collagen in our rat model of ACLT (Figure 6D). Type II collagen mRNA levels in rats that underwent ACLT and free-Ihh siRNA treatment were significantly lower than those in rats that underwent ACLT and received LNP-Ihh siRNA (2.5 and 5.0 nM), as well as that in rats that underwent sham operation and free-Ihh siRNA treatment. Aggrecan mRNA levels in rats treated with LNP-Ihh siRNA (5.0 nM) and rats that underwent sham operation with free-Ihh siRNA treatment were significantly higher than in rats that underwent ACLT and free-Ihh siRNA treatment. In contrast, levels of mRNA for MMP-13, Runx2, and type X collagen in rats that underwent ACLT and free-Ihh siRNA treatment were the highest among the four groups (Figure 6D).
Figure 6.
Intra-articular LNP-Ihh siRNA can attenuate the pathogenesis of surgery-induced OA.
Notes: There was less surface damage with stronger Safranin O staining in articular cartilage specimens from LNP-Ihh siRNA-treated animals as compared with those from untreated controls (A). Quantitative data indicated that cartilage damage was reduced in rats that received LNP-Ihh siRNA as compared to ACLT rats with free Ihh siRNA treatment (B). Type II collagen expression in articular cartilage was higher in the LNP-Ihh siRNA-treated and the sham-operated rats, than in rats that underwent ACLT and free Ihh siRNA treatment. In contrast, degraded collagen II, MMP-13, and type X collagen staining were elevated in rats that underwent ACLT and Ihh-siRNA treatment with respect to the LNP-Ihh siRNA-treated and sham-operated rats, which is consistent with reduced OA damage. The bigger inset boxes are original picture (magnification ×10). The smaller inset boxes are the parts shown in the panels (magnification ×20). (C). Real-time polymerase chain reaction indicates that intra-articular LNP-Ihh siRNA inhibits catabolism and enhances anabolic metabolism. Values are the mean ± standard error of the mean. *P<0.01 versus the ACLT and Ihh siRNA treatment group (D).
Abbreviations: ACLT, anterior cruciate ligament transection; Ihh, Indian Hedgehog; LNP, lipid nanoparticle; MMP-13, matrix metalloproteinase 13; OA, osteoarthritis; siRNA, small interference RNA.
Discussion
The treatment of articular cartilage disease is a major problem in clinical practice because of the tissue’s limited self-repair capacity.32 Efforts have been focused on gene therapy to provide a better outcome for cartilage diseases. Currently, RNAi is widely used in the treatment of many diseases including amyloid disease, infectious diseases, myeloid leukemia, bone metabolic disorders, and so on.21,33–35 However, there is no available cartilage-specific targeting delivery system for siRNA delivery in the treatment of cartilage metabolic disorders, because cartilage is a relatively solid and compact tissue that is not easily penetrated by small molecules. In this study, we modified LNP, a positively charged siRNA delivery system, to specifically target articular cartilage, a negatively charged tissue, and to facilitate the delivery of therapeutic cargos to the chondrocytes. This delivery system could establish the foundation for translating RNAi-based therapies from basic science to clinic applications in the field of orthopedic medicine.
In our in vitro and in vivo studies described here, the LNP we modified had a very small diameter of 67±4.3 nm, relative to other RNA nanocarriers,36–38 and a high encapsulation efficiency that remained .95%. These qualities provided a strong base for siRNA delivery and implied the potential to penetrate through the articular cartilage. Our findings demonstrated that LNPs could deliver siRNA into many kinds of cells in culture with a high transfection rate of almost 100%, as well as into porcine cartilage tissue cultures. We further demonstrated that siRNA could be delivered by LNPs into the knee joint to knock down specific genes. It should be noted that our data showed no toxicity of LNPs; in fact, LNPs even promoted cell proliferation and inhibited cell death when used in a moderate concentration. Therefore, LNP may constitute an ideal delivery system with high safety and efficiency for therapeutic use.
In addition to having rather small particles, the LNP is an ionizable cationic delivery system, while proteoglycans, the major molecular components of the extracellular cartilage, are negatively charged (representative proteoglycan molecules include aggrecan, versican, decorin, and perlecan among many).39 Thus, theoretically, LNP in the knee joint will be attracted toward the negatively charged proteoglycans and in this way provide a cartilage-specific delivery system. Furthermore, we have actually demonstrated in our in vivo study that LNP can be only accumulated and function in the extracellular matrix of cartilage, but not in the synovium.
As LNP is a cartilage-specific siRNA delivery system and increased Ihh has been correlated with OA, we knocked down Ihh to demonstrate whether this system is efficient to treat cartilage diseases. We demonstrated that supplementation of LNP-Ihh siRNA is a promising therapeutic strategy to inhibit cartilage degeneration by knocking down Ihh, which depressed cartilage degradative enzymes in an ACLT OA model. Our findings are in agreement with those of Lin et al, who report that inhibition of Ihh signaling has the potential to prevent surgically induced OA by transgenic mice.2,9 These findings support LNP as an effective tool for RNAi therapy in cartilage diseases.
A potential limitation of this study is that surgical transection of the ACL may not be as traumatic as an ACL injury sustained during physical activity. Bone bruises and chondral lesions frequently occur in the latter, and these concomitant injuries may also play a role in the development of post-traumatic OA. Nonetheless, the animal model of ACLT has been frequently used to study OA, and it mimics human OA both macroscopically and biochemically.29,31 In summary, our data indicate that our novel LNP-siRNA delivery system is an effective, promising, and powerful tool to knock down cartilage-specific genes and in this manner provide additional therapeutic approaches for cartilage disease therapy. LNP-Ihh siRNA therapy has a chondroprotective effect and attenuates the pathological progress of OA in a rat model.
Supplementary materials
LNPs transfection efficiency.
Notes: Fluorescence signals were obvious in the LNP-control siRNA-treated cells including astrocytes, chondrocytes cell line C28/I2, chondrogenic cell line ATDC5, and breast cancer cell line MCF-7. In contrast, there were no fluorescent signals when these cells were treated with free control siRNA.
Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; LNP, lipid nanoparticle; siRNA, small interference RNA.
Validated mouse intra-articular injection.
Notes: Twenty microliters of alcian blue solution was injected with an insulin syringe into the mouse knee joint cavity (A). The mouse was sacrificed after 24 hours and the knee joint cavity was opened by a scalpel. Gross observation showed that alcian blue adhered to the cartilage and synovial membrane in the joint cavity, indicating that intra-articular injection was feasible for the study (B).
Targeted multiple genes in vivo.
Notes: Cartilage Ihh and GAPDH genes were knocked down together by LNP-siRNA (Ihh+GAPDH) in vivo. Ihh mRNA level decreased by 41.6% and GAPDH mRNA level decreased by 19.5%. Type II collagen mRNA level increased significantly, but there was no difference in the aggrecan mRNA levels of LNP-siRNA (Ihh+GAPDH)-treated cartilage and free siRNA (Ihh+GAPDH) cartilage. In contrast, MMP-13 and type X collagen mRNA levels significantly decreased in our treatment groups. Values are the mean ± standard error of the mean. *P<0.01 versus the ACLT and Ihh siRNA treatment group.
Abbreviations: ACLT, anterior cruciate ligament transection; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Ihh, Indian Hedgehog; LNP, lipid nanoparticle; mRNA, messenger RNA; MMP-13, matrix metalloproteinase 13; siRNA, small interference RNA.
Acknowledgments
This project was supported by the National Natural Science Foundation of China (grant numbers 81572098 and 81572207), National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR059142), and Shanxi Natural Science Foundation of China (grant number 2015021184). The authors gratefully acknowledge Ericka M Bueno, PhD, for help with the paper preparation and editorial services.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Centers for Disease Control and Prevention (CDC) Prevalence of doctor-diagnosed arthritis and arthritis-attributable activity limitation–United States, 2010–2012. MMWR Morb Mortal Wkly Rep. 2013;62(44):869–873. [PMC free article] [PubMed] [Google Scholar]
- 2.Lin AC, Seeto BL, Bartoszko JM, et al. Modulating hedgehog signaling can attenuate the severity of osteoarthritis. Nat Med. 2009;15(12):1421–1425. doi: 10.1038/nm.2055. [DOI] [PubMed] [Google Scholar]
- 3.Palmer AJ, Brown CP, McNally EG, et al. Non-invasive imaging of cartilage in early osteoarthritis. Bone Joint J. 2013;95-B(6):738–746. doi: 10.1302/0301-620X.95B6.31414. [DOI] [PubMed] [Google Scholar]
- 4.Strombeck B, Petersson IF, Vliet Vlieland TP, EUMUSC.net WP6 group Health care quality indicators on the management of rheumatoid arthritis and osteoarthritis: a literature review. Rheumatology (Oxford) 2013;52(2):382–390. doi: 10.1093/rheumatology/kes266. [DOI] [PubMed] [Google Scholar]
- 5.Iagnocco A, Naredo E. Osteoarthritis: research update and clinical applications. Rheumatology (Oxford) 2012;51(Suppl 7):vii2–vii5. doi: 10.1093/rheumatology/kes328. [DOI] [PubMed] [Google Scholar]
- 6.Brandt KD, Dieppe P, Radin E. Etiopathogenesis of osteoarthritis. Med Clin North Am. 2009;93(1):1–24. xv. doi: 10.1016/j.mcna.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Wei F, Zhou J, Wei X, et al. Activation of Indian hedgehog promotes chondrocyte hypertrophy and upregulation of MMP-13 in human osteoarthritic cartilage. Osteoarthritis Cartilage. 2012;20(7):755–763. doi: 10.1016/j.joca.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C, Wei X, Chen C, et al. Indian hedgehog in synovial fluid is a novel marker for early cartilage lesions in human knee joint. Int J Mol Sci. 2014;15(5):7250–7265. doi: 10.3390/ijms15057250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J, Chen Q, Lanske B, et al. Disrupting the Indian hedgehog signaling pathway in vivo attenuates surgically induced osteoarthritis progression in Col2a1-CreERT2; Ihhfl/fl mice. Arthritis Res Ther. 2014;16(1):R11. doi: 10.1186/ar4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coventry S, Kapur RP, Siebert JR. Cyclopamine-induced holoprosencephaly and associated craniofacial malformations in the golden hamster: anatomic and molecular events. Pediatr Dev Pathol. 1998;1(1):29–41. doi: 10.1007/s100249900004. [DOI] [PubMed] [Google Scholar]
- 11.Lipinski RJ, Dengler E, Kiehn M, Peterson RE, Bushman W. Identification and characterization of several dietary alkaloids as weak inhibitors of hedgehog signaling. Toxicol Sci. 2007;100(2):456–463. doi: 10.1093/toxsci/kfm222. [DOI] [PubMed] [Google Scholar]
- 12.Kimura H, Ng JM, Curran T. Transient inhibition of the Hedgehog pathway in young mice causes permanent defects in bone structure. Cancer Cell. 2008;13(3):249–260. doi: 10.1016/j.ccr.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 13.Lipinski RJ, Hutson PR, Hannam PW, et al. Dose- and route-dependent teratogenicity, toxicity, and pharmacokinetic profiles of the hedgehog signaling antagonist cyclopamine in the mouse. Toxicol Sci. 2008;104(1):189–197. doi: 10.1093/toxsci/kfn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez-Fraga M, Martinez T, Jimenez A. RNA interference technologies and therapeutics: from basic research to products. BioDrugs. 2009;23(5):305–332. doi: 10.2165/11318190-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yonezawa H, Aoki T, Furusawa A. Demonstration of a quantum teleportation network for continuous variables. Nature. 2004;431(7007):430–433. doi: 10.1038/nature02858. [DOI] [PubMed] [Google Scholar]
- 16.Zhang G, Guo B, Wu H, et al. A delivery system targeting bone formation surfaces to facilitate RNAi-based anabolic therapy. Nat Med. 2012;18(2):307–314. doi: 10.1038/nm.2617. [DOI] [PubMed] [Google Scholar]
- 17.Pignatello R, Sarpietro MG, Castelli F. Synthesis and biological evaluation of a new polymeric conjugate and nanocarrier with osteotropic properties. J Funct Biomater. 2012;3(1):79–99. doi: 10.3390/jfb3010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crawford R, Dogdas B, Keough E, et al. Analysis of lipid nanoparticles by Cryo-EM for characterizing siRNA delivery vehicles. Int J Pharm. 2011;403(1–2):237–244. doi: 10.1016/j.ijpharm.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 19.Belliveau NM, Huft J, Lin PJ, et al. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol Ther Nucleic Acids. 2012;1:e37. doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Semple SC, Akinc A, Chen J, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28(2):172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 21.Liang C, Guo B, Wu H, et al. Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference-based bone anabolic strategy. Nat Med. 2015;21(3):288–294. doi: 10.1038/nm.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aranda R, 4th, Dineen SM, Craig RL, Guerrieri RA, Robertson JM. Comparison and evaluation of RNA quantification methods using viral, prokaryotic, and eukaryotic RNA over a 10(4) concentration range. Anal Biochem. 2009;387(1):122–127. doi: 10.1016/j.ab.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Wei L, Sun X, Kanbe K, et al. Chondrocyte death induced by pathological concentration of chemokine stromal cell-derived factor-1. J Rheumatol. 2006;33(9):1818–1826. [PubMed] [Google Scholar]
- 24.Wang S, Wei X, Zhou J, et al. Identification of alpha2-macroglobulin as a master inhibitor of cartilage-degrading factors that attenuates the progression of posttraumatic osteoarthritis. Arthritis Rheumatol. 2014;66(7):1843–1853. doi: 10.1002/art.38576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jayasuriya CT, Zhou FH, Pei M, et al. Matrilin-3 chondrodysplasia mutations cause attenuated chondrogenesis, premature hypertrophy and aberrant response to TGF-beta in chondroprogenitor cells. Int J Mol Sci. 2014;15(8):14555–14573. doi: 10.3390/ijms150814555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter DJ, Zhang YQ, Tu X, et al. Change in joint space width: hyaline articular cartilage loss or alteration in meniscus? Arthritis Rheum. 2006;54(8):2488–2495. doi: 10.1002/art.22016. [DOI] [PubMed] [Google Scholar]
- 27.Peterson JD, Labranche TP, Vasquez KO, et al. Optical tomographic imaging discriminates between disease-modifying anti-rheumatic drug (DMARD) and non-DMARD efficacy in collagen antibody-induced arthritis. Arthritis Res Ther. 2010;12(3):R105. doi: 10.1186/ar3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas NP, Li P, Fleming BC, et al. Attenuation of cartilage pathogenesis in post-traumatic osteoarthritis (PTOA) in mice by blocking the stromal derived factor 1 receptor (CXCR4) with the specific inhibitor, AMD3100. J Orthop Res. 2015;33(7):1071–1078. doi: 10.1002/jor.22862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jay GD, Fleming BC, Watkins BA, et al. Prevention of cartilage degeneration and restoration of chondroprotection by lubricin tribosupple-mentation in the rat following anterior cruciate ligament transection. Arthritis Rheum. 2010;62(8):2382–2391. doi: 10.1002/art.27550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pritzker KP, Gay S, Jimenez SA, et al. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthritis Cartilage. 2006;14(1):13–29. doi: 10.1016/j.joca.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 31.Wei L, Fleming BC, Sun X, et al. Comparison of differential biomarkers of osteoarthritis with and without posttraumatic injury in the Hartley guinea pig model. J Orthop Res. 2010;28(7):900–906. doi: 10.1002/jor.21093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tallheden T, Bengtsson C, Brantsing C, et al. Proliferation and differentiation potential of chondrocytes from osteoarthritic patients. Arthritis Res Ther. 2005;7(3):R560–R568. doi: 10.1186/ar1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garber K. Alnylam’s RNAi therapy targets amyloid disease. Nat Biotechnol. 2015;33(6):577. doi: 10.1038/nbt0615-577a. [DOI] [PubMed] [Google Scholar]
- 34.Dyawanapelly S, Ghodke SB, Vishwanathan R, Dandekar P, Jain R. RNA interference-based therapeutics: molecular platforms for infectious diseases. J Biomed Nanotechnol. 2014;10(9):1998–2037. doi: 10.1166/jbn.2014.1929. [DOI] [PubMed] [Google Scholar]
- 35.Wermke M, Camgoz A, Paszkowski-Rogacz M, et al. RNAi profiling of primary human AML cells identifies ROCK1 as a therapeutic target and nominates fasudil as an antileukemic drug. Blood. 2015;125(24):3760–3768. doi: 10.1182/blood-2014-07-590646. [DOI] [PubMed] [Google Scholar]
- 36.Kaneda MM, Sasaki Y, Lanza GM, Milbrandt J, Wickline SA. Mechanisms of nucleotide trafficking during siRNA delivery to endothelial cells using perfluorocarbon nanoemulsions. Biomaterials. 2010;31(11):3079–3086. doi: 10.1016/j.biomaterials.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xue HY, Narvikar M, Zhao JB, Wong HL. Lipid encapsulation of cationic polymers in hybrid nanocarriers reduces their non-specific toxicity to breast epithelial cells. Pharm Res. 2013;30(2):572–583. doi: 10.1007/s11095-012-0902-6. [DOI] [PubMed] [Google Scholar]
- 38.Souto EB, Nayak AP, Murthy RS. Lipid nanoemulsions for anti-cancer drug therapy. Pharmazie. 2011;66(7):473–478. [PubMed] [Google Scholar]
- 39.Yanagishita M, Podyma-Inoue KA, Yokoyama M. Extraction and separation of proteoglycans. Glycoconj J. 2009;26(8):953–959. doi: 10.1007/s10719-008-9138-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LNPs transfection efficiency.
Notes: Fluorescence signals were obvious in the LNP-control siRNA-treated cells including astrocytes, chondrocytes cell line C28/I2, chondrogenic cell line ATDC5, and breast cancer cell line MCF-7. In contrast, there were no fluorescent signals when these cells were treated with free control siRNA.
Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; LNP, lipid nanoparticle; siRNA, small interference RNA.
Validated mouse intra-articular injection.
Notes: Twenty microliters of alcian blue solution was injected with an insulin syringe into the mouse knee joint cavity (A). The mouse was sacrificed after 24 hours and the knee joint cavity was opened by a scalpel. Gross observation showed that alcian blue adhered to the cartilage and synovial membrane in the joint cavity, indicating that intra-articular injection was feasible for the study (B).
Targeted multiple genes in vivo.
Notes: Cartilage Ihh and GAPDH genes were knocked down together by LNP-siRNA (Ihh+GAPDH) in vivo. Ihh mRNA level decreased by 41.6% and GAPDH mRNA level decreased by 19.5%. Type II collagen mRNA level increased significantly, but there was no difference in the aggrecan mRNA levels of LNP-siRNA (Ihh+GAPDH)-treated cartilage and free siRNA (Ihh+GAPDH) cartilage. In contrast, MMP-13 and type X collagen mRNA levels significantly decreased in our treatment groups. Values are the mean ± standard error of the mean. *P<0.01 versus the ACLT and Ihh siRNA treatment group.
Abbreviations: ACLT, anterior cruciate ligament transection; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Ihh, Indian Hedgehog; LNP, lipid nanoparticle; mRNA, messenger RNA; MMP-13, matrix metalloproteinase 13; siRNA, small interference RNA.