Abstract.
Deep learning methods have been shown to improve breast cancer diagnostic and prognostic decisions based on selected slices of dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI). However, incorporation of volumetric and temporal components into DCE-MRIs has not been well studied. We propose maximum intensity projection (MIP) images of subtraction MRI as a way to simultaneously include four-dimensional (4-D) images into lesion classification using convolutional neural networks (CNN). The study was performed on a dataset of 690 cases. Regions of interest were selected around each lesion on three MRI presentations: (i) the MIP image generated on the second postcontrast subtraction MRI, (ii) the central slice of the second postcontrast MRI, and (iii) the central slice of the second postcontrast subtraction MRI. CNN features were extracted from the ROIs using pretrained VGGNet. The features were utilized in the training of three support vector machine classifiers to characterize lesions as malignant or benign. Classifier performances were evaluated with fivefold cross-validation and compared based on area under the ROC curve (AUC). The approach using MIPs outperformed that using central-slices of either second postcontrast MRIs or second postcontrast subtraction MRIs , at statistically significant levels.
Keywords: convolutional neural networks, breast cancer, dynamic contrast-enhanced magnetic resonance imaging, four-dimensional data, maximum intensity projection
1. Introduction
Magnetic resonance imaging (MRI) is one of the imaging modalities for clinical diagnosis and monitoring of breast cancer. Its use has been established for screening of patients with high risk of breast cancer, cancer staging, and monitoring cancer’s response to therapies.1–3 In comparison with more commonly used clinical modalities, such as mammography and ultrasound, MRI offers much higher sensitivity to breast cancer diagnosis.4,5 Despite high sensitivity in breast cancer diagnosis, DCE-MRI has lower specificity, which has room for the future improvements.6 Additionally, dynamic contrast-enhanced MRI (DCE-MRI) provides high-resolution volumetric lesion visualization as well as lesion’s temporal enhancement patterns, the information that carries clinical value for breast cancer management.1 DCE-MRI’s superior diagnostic and prognostic characterization of disease leads to its continuingly increasing use rates.2,7 However, long imaging and image interpretation times in combination with few MRI reading experts make it an expensive clinical procedure.
Automated CAD/radiomics systems are being developed to overcome challenges associated with the image reading time and expertise deficiency. Radiomics systems automatically locate and characterize lesions based on medical images, aiding human readers, reducing image interpretation time, and errors.8,9 Conventional radiomics systems extract predefined features, which characterize lesions in terms of intuitive characteristics, such as size, shape, and morphology.8
Recently, deep learning methods have showed success in various vision tasks, such as image classification, image segmentation, and image generation.9 These techniques have been also adapted to improve diagnostic and prognostic performance based on medical scans.10 However, accurate training of deep models requires large amounts of medical images, which are generally difficult to acquire. To overcome this challenge, pretrained deep convolutional neural networks (CNNs) are often used for medical image classification.9,10–12 Such CNNs can be trained on a large natural image dataset and then, through transfer learning, be applied as feature extractors on medical images. CNN features have achieved state-of-art results across various imaging modalities and clinical questions. However, pretrained CNNs require two-dimensional (2-D) inputs, limiting the amount of 3D–4D image information that can contribute to CNN-based lesion classification.11–14
The goal of this research is to incorporate the dynamic and volumetric components of DCE-MRIs into breast lesion classification with deep learning methods using maximum intensity projection (MIP) images. The classification task in this study is distinguishing benign and malignant lesions. MIP images allow us to collapse the image volumes of postcontrast subtraction MRIs for input to the pretrained CNNs for feature extraction. Our MIPS-CNN methodology demonstrates that incorporating both volumetric and dynamic DCE-MRI components significantly improves CNN-based lesion classification.
2. Methods
2.1. Dynamic Contrast-Enhanced Magnetic Resonance Imaging Dataset
This study was performed on a DCE-MRI dataset, retrospectively collected under an HIPAA-compliant Institutional Review Board protocol. The dataset was collected at the University of Chicago over a 10-year-period, from 2006 till 2016, and includes 690 breast cases, annotated as benign (212 cases) or malignant (478 cases) based on pathology and radiology reports. All of the lesions were clinically biopsy-confirmed, thus, resulting in radiologist’s sensitivity and specificity of 100% based on their BI-RADS. Both primary and secondary lesions were utilized in the study.
Malignant cases included masses and nonmass enhancements; benign cases included masses and foci. Detailed clinical characteristics of the dataset are shown in Table 1. The same population was previously analyzed using CNN features extracted from MRI central slices of each tumor but including precontrast and two postcontrast images.11 The current work builds upon and advances the analysis by incorporating both temporal and volumetric components of DCE-MRIs into the classification by the use of MIP images.
Table 1.
Clinical characteristics of the clinical DCE-MRI dataset.
| Benign/malignant prevalence: no. cases (%) | Benign | 212 (30.7) |
| Malignant | 478 (69.3) | |
| Age: mean (STD) |
54.9 (13.3) Unidentified cases: 103 |
|
| Benign tumor characteristics | ||
| Tumor subtypes: | Fibroadenoma | 87 |
| Fibrocystic change | 77 | |
| Papilloma | 12 | |
| |
Unidentified |
36 |
| Malignant tumor characteristics | ||
| Tumor subtypes: | Invasive ductal carcinoma | 135 |
| Ductal carcinoma in situ | 19 | |
| Invasive ductal carcinoma + ductal carcinoma in situ | 263 | |
| Invasive lobular carcinoma | 18 | |
| Invasive lobular carcinoma mixed | 19 | |
| Unidentified | 24 | |
| Estrogen receptor status: no. of cases | Positive | 328 |
| Negative | 108 | |
| Unidentified | 42 | |
| Progesterone receptor status: no. of cases | Positive | 274 |
| Negative | 159 | |
| Unidentified | 45 | |
| HER2 status: no. of cases | Positive | 72 |
| Negative | 349 | |
| Equivocal | 3 | |
| Unidentified | 54 | |
A part of the contrast-enhanced MR images, 454 cases, was acquired on Philips Achieva 1.5 Tesla (T) scanner. The rest of 236 cases were acquired with Philips Achieva-TX 3T Philips system. A T1-weighted spoiled gradient sequence was utilized during image acquisition. To perform dynamic imaging, patients were injected with the following Gadolinium-based contrast agents: Ominscan was used for patients with GFR over prior to November 25, 2012, and Multihance was used for the patients with GFR and for all patients imaged after November 25, 2012. Each sequence included a precontrast image followed by multiple postcontrast images, with the first postcontrast image obtained 55 to 60 s following the contrast injection. The database consisted of DCE-MRI images of various slice thickness, with two-third cases having slice thickness of 2 mm and one-third cases having slice thickness 1.5 or 1.6 mm.
2.2. Image Preprocessing
Prior to applying pretrained CNN for feature extraction, regions of interest (ROIs) were selected surrounding each lesion on three MRI presentations: (i) the MIP image generated on the second postcontrast subtraction (second postcontrast–precontrast) MRI, (ii) the central slice image of the second postcontrast MRI, and (iii) the central slice image of the second postcontrast subtraction (second postcontrast—precontrast) MRI. To generate an MIP image, a subtracted 3-D MR image is collapsed into a single 2-D image by selecting the voxel having the maximum intensity along the projection through all transverse slices containing the lesion.5,15
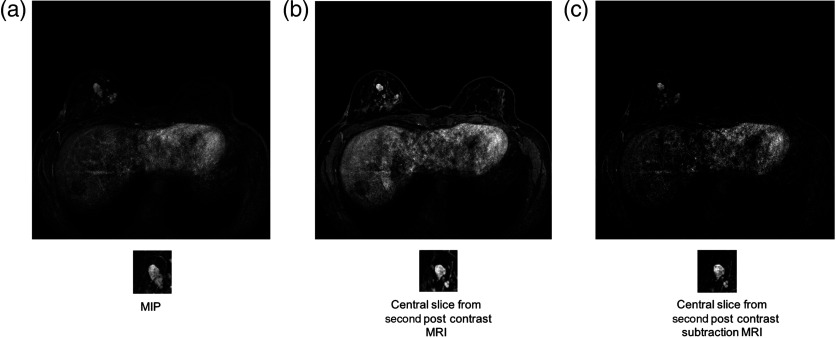
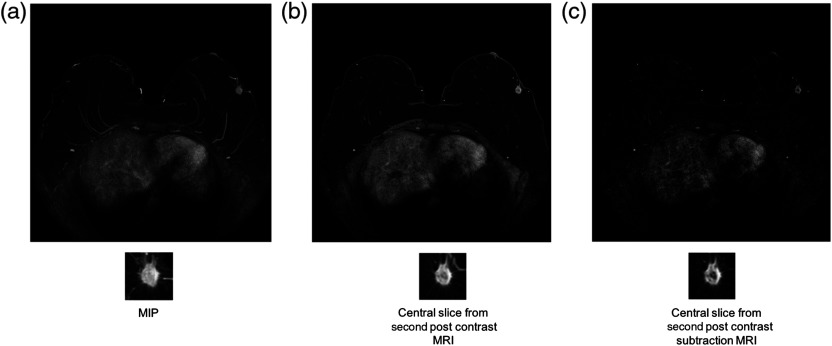
Each lesion was located in the center of its ROI. Each ROI size was selected to be a few pixels greater than the maximum dimension of its lesion, thus insuring that each ROI contained not only a lesion, but also some of the surrounding breast parenchyma. Figures 1 and 2 show examples of the three types of MRI representations with selected ROIs for malignant and benign cases.
Fig. 1.
Example of a benign lesion. Full MRI images and ROIs for (a) the MIP image of the second postcontrast subtraction MRI, (b) the center slice of the second postcontrast MRI, and (c) the central slice of the second postcontrast subtraction MRI.
Fig. 2.
Example of a malignant lesion. Full MRI images and ROIs for (a) the MIP image of the second postcontrast subtraction MRI, (b) the center slice of the second postcontrast MRI, and (c) the central slice of the second postcontrast subtraction MRI.
2.3. Convolutional Neural Networks Feature Extraction
Following ROI selection, CNN features were extracted from each type of ROI using a pretrained ConvNet VGGNet (Fig. 3).16 The network had been previously trained on ImageNet, a large database on everyday images, and was used as a feature extractor for DCE-MRIs.17,14,15 To extract features from an ROI, we replicated the ROI across the three-color channels of the network. The architecture of the network contains five blocks, each of which consists of two or four convolutional layers and a max-pooling layer, and three fully connected layers. In our methods, CNN features were extracted from the five max-pooling layers, average-pooled, normalized with Euclidean norm,18 and concatenated to form lesion’s CNN feature vector, as described in the previous work.11 Even though, a more common way of applying a pretrained CNN to medical images is to extract features from fully connected layers, the method requires image preprocessing to transform the images to a fixed size. By extracting features from max-pooling layers and average-pooling them, our method allows using images of various sizes, corresponding to enclosed lesion sizes. The architecture details and the feature extraction methodology are demonstrated in Fig. 4.
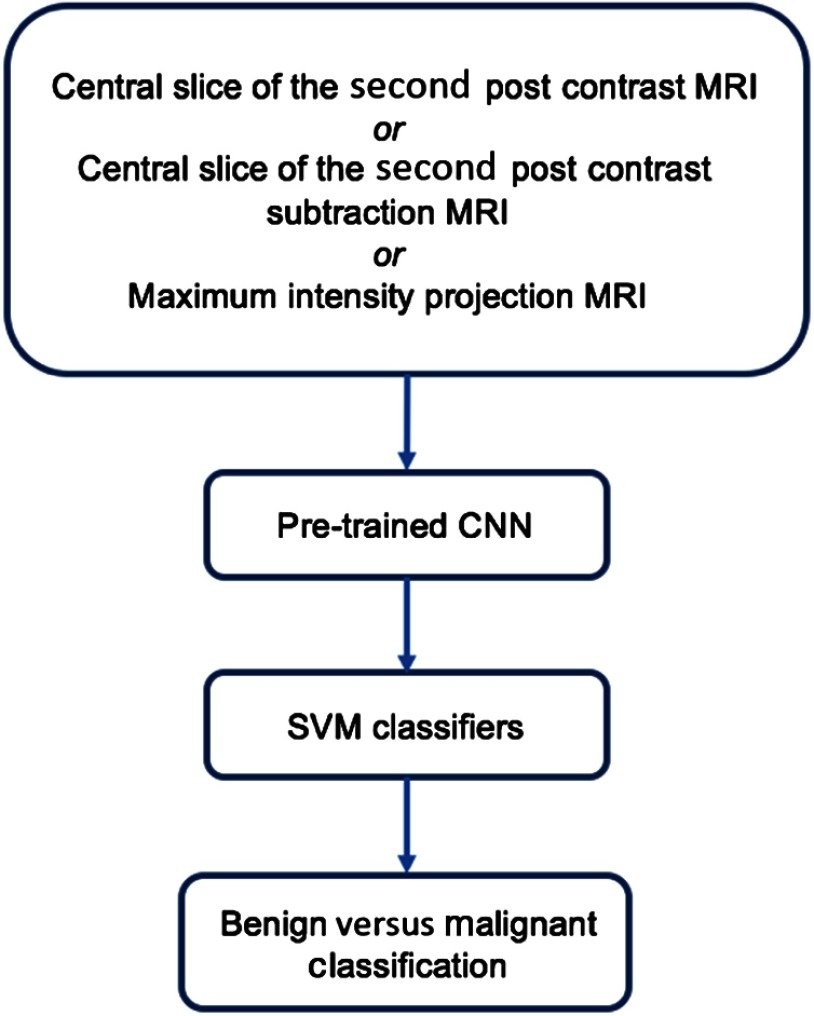
Fig. 3.
Lesion classification pipeline.
Fig. 4.
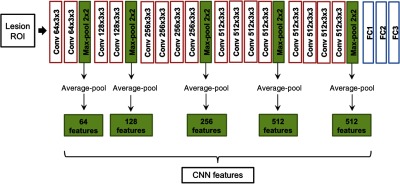
Feature extraction methodology. Lesion ROIs were input into a pretrained VGGNet and features were extracted from five max-pooling layers. The features were then average-pooled, normalized, and concatenated to form the CNN feature vector corresponding to the input ROI.
2.4. Classification
Breast lesions were classified as benign or malignant based on the features extracted using the pretrained CNN. Linear support vector machines were chosen as classifiers for this clinical task (Scikit-learn package, Python Version 2.7.12, Python Software Foundation).19 Linear SVMs require optimization of one hyperparameter, controlling the trade-offs between misclassification errors and model complexity.20 The hyperparameter was optimized on a grid search with nested fivefold cross validation. For nested cross validation, each training set was further divided into training and validation sets, which were used for training of the classifier and validation of the hyperparameter, respectively. To evaluate classification performance in the task of distinguishing benign and malignant lesions, receiver operating characteristic (ROC) analysis with fivefold cross validation was utilized.21,22 The dataset was split with 80% for training and 20% for testing subsets within each fold. Area under the ROC curve (AUC) served as a figure of merit in the analysis. AUC is a metric unaffected by class prevalence, which was important given our unbalanced benign/malignant dataset.
Three separate SVM classifiers were trained and evaluated on the corresponding CNN features extracted from ROIs selected on (i) the MIP images generated on the second postcontrast subtraction MRIs, (ii) the central slice images of the second postcontrast MRIs, and (iii) the central slice images of the second postcontrast subtraction MRIs. The significance of differences in classifier malignancy assessment performances was assessed with DeLong tests and corrected for multiple comparisons with Bonferroni–Holm corrections.23,24
3. Results
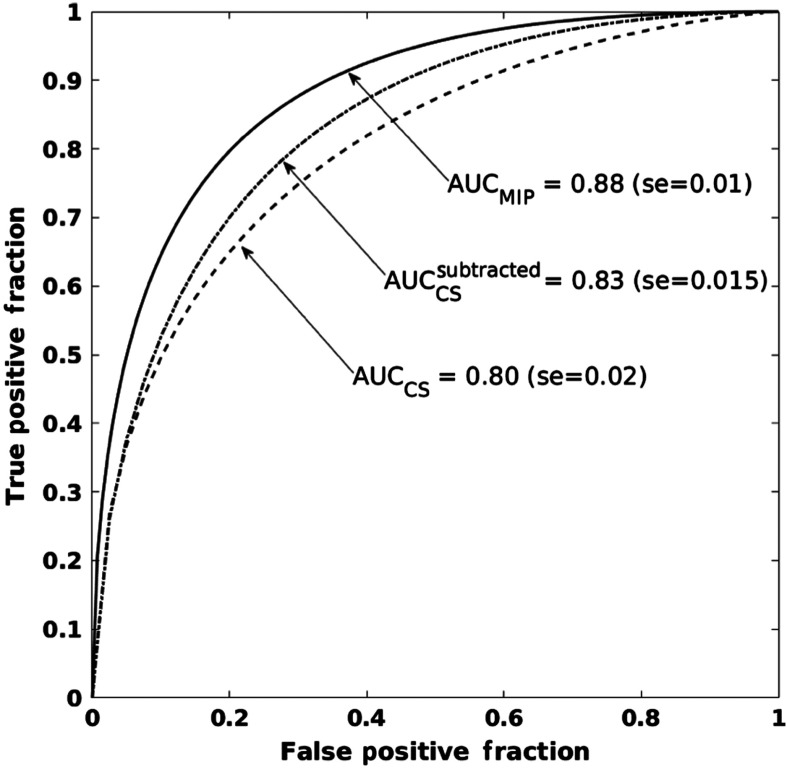
Clinicians often evaluate the extent of the entire tumor using MIPs of DCE-MRIs. Our work utilized this idea and extended it to deep-learning-based tumor evaluations. Our results demonstrate that presenting MIP images, instead of single slices, to a pretrained CNN leads to superior classification of lesion malignancy. Table 2 summarizes AUC values for the performances of the three classifiers in the task of distinguishing benign and malignant lesions. SVMs trained on CNN features extracted from MIP ROIs significantly outperformed classifiers trained on CNN features extracted from ROIs from either the central slice of the second postcontrast MRI or from the central slice of the second postcontrast subtraction MRI. Figure 5 shows the ROC curves corresponding to the classification performances of the three classifiers. Compared with previous work,11 in which we integrated the temporal component of DCE-MRIs by inputting precontrast and two postcontrast MRIs in three-color channels of pretrained CNN, superior performance of breast lesion malignancy assessment was achieved with MIP images. Our results suggest that the diagnostically useful information within MIPs images can be successfully utilized by CNN-based classification methods.
Table 2.
Classification performance of classifiers trained on CNN features extracted from three types of ROIs in the task of distinguishing malignant and benign lesions. -values are computed with respect to MIP classifiers and are corrected for multiple comparisons with Bonferroni–Holm corrections.
| ROI type | AUC | -value |
|---|---|---|
| Central slice of second postcontrast | 0.80 () | 0.00058 |
| Central slice of second postcontrast subtracted | 0.83 () | 0.048 |
| MIP | 0.88 () | — |
Fig. 5.
ROC curves showing the performance of three classifiers. The classifiers were trained on CNN features extracted from ROIs selected on: (a) the MIP images of second postcontrast subtraction MRIs, , (b) the central slices of the second postcontrast MRIs, , and (c) the central slices of second postcontrast subtraction MRIs, .
4. Discussion and Conclusions
Recently, deep learning methods have been successfully adapted in clinical decision making based on medical images. One common deep learning-based method involves applying publically available deep CNNs, pretrained on large natural image datasets, to medical images. This can be conducted due to a CNN’s ability to capture image characteristics/features generalizable to various types of images, medical and nonmedical. These features extracted from medical images can be further used to perform automated diagnostic and prognostic tasks. However, most pretrained CNN models demand a 2-D color image as an input, making it challenging to incorporate clinically rich information contained in 4-D medical images. Thus, in this work, we proposed a method to incorporate both volumetric and temporal components of DCE-MRI for classifying lesions as benign or malignant using MIP images with pretrained CNNs.
One of the straightforward ways of classifying 3-D and 4-D MRI images is individually classifying each slice of the full image and averaging the output of the classifiers to attain the final clinical conclusion. However, some DCE-MRI sequences may have over 500 slices. With such a high number of slices, feature extraction and classifier training for each slice might be computationally very expensive, which would greatly increase clinical evaluation time. Our work reports on using MIP images to form a 2-D image from a 4-D DCE-MRI for input into pretrained CNN for the task of classifying lesions as benign or malignant. Specifically, the method involves subtracting a precontrast MRI from the second postcontrast MRI, and then calculating the MIP of the subtraction image. As a result, the MIP image contains information about enhancement changes throughout the lesion volume. This method can be easily adopted in clinical practice since MIP images are already commonly used in the evaluation of breast tumors.
Other works have trained neural networks from scratch on the medical image datasets with prior image augmentation. We leave training a 3-D CNN on DCE-MRIs for the future work, as we acquire more images.
For the task of distinguishing malignant and benign lesions, we demonstrated that lesion classification based on MIP images significantly outperforms lesion classification based on either a single slice second postcontrast MRI or a single slice second postcontrast subtraction MRI. Our results also suggest that integration of enhancement changes between precontrast and second postcontrast time-points, without a volumetric component, results in better classification performance compared with single slice MRI at a single time-point. Further explorations are necessary to understand the utility of MIP images in the benign versus malignant lesion classification with pretrained neural networks. One of the directions is controlling for the slice thickness of MRIs. Our DCE-MRI dataset contains two-thirds of the images with a slice thickness of 2 mm and one-third with a slice thickness of 1.5 or 1.6 mm. Therefore, the MIPs are taken over variable depth resolutions. In future work, larger databases need to be collected to perform studies on the effect of the slice thickness.
In summary, DCE-MRI MIPs incorporate clinically useful information about the entire lesion volume as well as the contrast enhancement and can be utilized for malignancy classification with deep CNNs, pretrained on a nonmedical image dataset.
Acknowledgments
This work was supported by the National Institutes of Health Quantitative Imaging Network Grant No. U01CA195564 and the University of Chicago Hodges Award. The authors would like to acknowledge NVIDIA Corporation for donating Titan X Pascal GPU.
Biographies
Natalia Antropova is a PhD student in the medical physics program at the University of Chicago, working in Dr. Maryellen Giger’s computer-aided diagnosis group. Her research focuses on developing deep learning and classical methods for clinical decision-making based on medical images. Her projects involve breast cancer detection, diagnosis, and prognosis using dynamic contrast-enhanced magnetic resonance imaging.
Hiroyuki Abe is a professor of the Department of Radiology at the University of Chicago Medicine. He is a highly experienced breast imager with a strong research track record. His clinical work includes diagnostic interpretation of mammograms, ultrasounds, and MRIs while performing various types of image-guided procedures. He is actively working with medical physicists and clinical colleagues in the translation of methods of acquisition and analysis of breast MRI, ultrasound, and mammographic images.
Maryellen L. Giger is the A. N. Pritzker Professor of radiology/medical physics at the University of Chicago and has been working for multiple decades on computer-aided diagnosis and quantitative image analysis methods in cancer diagnosis and management. Her research interests include understanding the role of quantitative radiomics, computer vision, and deep learning in personalized medicine.
Disclosures
MLG is a stockholder in R2 technology/Hologic and receives royalties from Hologic, GE Medical Systems, MEDIAN Technologies, Riverain Medical, Mitsubishi and Toshiba. She is a cofounder of and stockholder in Quantitative Insights. It is the University of Chicago Conflict of Interest Policy that investigators disclose publicly actual or potential significant financial interest that would reasonably appear to be directly and significantly affected by the research activities.
References
- 1.Turkbey B., et al. , “The role of dynamic contrast enhanced MR imaging in cancer diagnosis and treatment,” Diagn. Interventional Radiol. 13, e45–e53 (2009).https://doi.org/10.4261/1305-3825.DIR.2537-08.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Cancer Society, Breast Cancer Facts and Figures 2015–2016, American Cancer Society, Inc. (2015). [Google Scholar]
- 3.Pickles M. D., et al. , “Prognostic value of DCE-MRI in breast cancer patients undergoing neoadjuvant chemotherapy: a comparison with traditional survival indicators,” Eur. Radiol. 25, 1097–1106 (2015).https://doi.org/10.1007/s00330-014-3502-5 [DOI] [PubMed] [Google Scholar]
- 4.Morrow M., Waters J., Morris E., “MRI for breast cancer screening, diagnosis, and treatment,” Lancet 378, 1804–1811 (2011).https://doi.org/10.1016/S0140-6736(11)61350-0 [DOI] [PubMed] [Google Scholar]
- 5.Kuhl C. K., et al. , “Abbreviated breast magnetic resonance imaging (MRI): first postcontrast subtracted images and maximum-intensity projection—a novel approach to breast cancer screening with MRI,” J. Clin. Oncol. 32, 2304–2310 (2014).https://doi.org/10.1200/JCO.2013.52.5386 [DOI] [PubMed] [Google Scholar]
- 6.Kneeshaw P. J., Turnbull L. W., Drew P. J., “Current applications and future direction of MR mammography,” Br. J. Cancer 88, 4–10 (2003).https://doi.org/10.1038/sj.bjc.6600713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wernli K. J., et al. , “Patterns of breast magnetic resonance imaging use in community practice,” JAMA Intern. Med. 174, 125 (2014).https://doi.org/10.1001/jamainternmed.2013.11963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giger M. L., Chan H.-P., Boone J., “Anniversary paper: history and status of CAD and quantitative image analysis: the role of medical physics and AAPM,” Med. Phys. 35, 5799–5820 (2008).https://doi.org/10.1118/1.3013555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esteva A., et al. , “Dermatologist-level classification of skin cancer with deep neural networks,” Nature 542, 115–118 (2017).https://doi.org/10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenspan H., van Ginneken B., Summers R. M., “Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique,” IEEE Trans. Med. Imaging 35, 1153–1159 (2016).https://doi.org/10.1109/TMI.2016.2553401 [Google Scholar]
- 11.Antropova N., Huynh B. Q., Giger M. L., “A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets,” Med. Phys. 44(10), 5162–5171 (2017).https://doi.org/10.1002/mp.12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huynh B. Q., Li H., Giger M. L., “Digital mammographic tumor classification using transfer learning from deep convolutional neural networks,” J. Med. Imaging 3, 034501 (2016).https://doi.org/10.1117/1.JMI.3.3.034501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Ginneken B., et al. , “Off-the-shelf convolutional neural network features for pulmonary nodule detection in computed tomography scans,” in IEEE 12th Int. Symp. on Biomedical Imaging (ISBI 2015), pp. 286–289 (2015).https://doi.org/10.1109/ISBI.2015.7163869 [Google Scholar]
- 14.Bar Y., et al. , “Deep learning with non-medical training used for chest pathology identification,” Proc. SPIE 9414, 94140V (2015).https://doi.org/10.1117/12.2083124 [Google Scholar]
- 15.“Maximum intensity projection–MIPAV,” https://mipav.cit.nih.gov/pubwiki/index.php/Maximum_Intensity_Projection (18 September 2017).
- 16.Simonyan K., Zisserman A., “Very deep convolutional networks for large-scale image recognition,” ArXiv Prepr. ArXiv14091556 (2014).
- 17.LeCun Y., Bengio Y., Hinton G., “Deep learning,” Nature 521, 436–444 (2015).https://doi.org/10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 18.Deza M., Deza E., Encyclopedia of Distances, Springer Verlag; (2009). [Google Scholar]
- 19.Russakovsky O., et al. , “ImageNet large scale visual recognition challenge,” Int. J. Comput. Vision 115, 211–252 (2015).https://doi.org/10.1007/s11263-015-0816-y [Google Scholar]
- 20.Shawe-Taylor J., Sun S., “A review of optimization methodologies in support vector machines,” Neurocomputing 74, 3609–3618 (2011).https://doi.org/10.1016/j.neucom.2011.06.026 [Google Scholar]
- 21.Metz C., Pan X., “‘Proper’ binormal ROC curves: theory and maximum-likelihood estimation,” J. Math. Psychol. 43, 1–33 (1999).https://doi.org/10.1006/jmps.1998.1218 [DOI] [PubMed] [Google Scholar]
- 22.Pan X., Metz C. E., “The ‘proper’ binormal model: parametric receiver operating characteristic curve estimation with degenerate data,” Acad. Radiol. 4, 380–389 (1997).https://doi.org/10.1016/S1076-6332(97)80121-3 [DOI] [PubMed] [Google Scholar]
- 23.Holm S., “A simple sequentially rejective multiple test procedure,” Scand. J. Stat. 6(2), 65–70 (1979). [Google Scholar]
- 24.DeLong E. R., DeLong D. M., Clarke-Pearson D. L., “Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach,” Biometrics 44, 837 (1988).https://doi.org/10.2307/2531595 [PubMed] [Google Scholar]