Highlights
-
•
Digital model of the rat facial nucleus anatomy.
-
•
Precise registration of structural data – obtained across animals – into a digital facial nucleus reference frame.
-
•
3D reconstruction and registration of vibrissal motoneurons with identified target muscle.
-
•
Quantification of morphological cell-to-cell variably within a population of motoneurons that innervate the same muscle.
-
•
Mapping the presynaptic populations of reconstructed vibrissal motoneurons, whose target muscle is known.
Key words: whisker, whisking, brain stem, rabies virus, connectivity
Abstract
The rodent facial nucleus (FN) comprises motoneurons (MNs) that control the facial musculature. In the lateral part of the FN, populations of vibrissal motoneurons (vMNs) innervate two groups of muscles that generate movements of the whiskers. Vibrissal MNs thus represent the terminal point of the neuronal networks that generate rhythmic whisking during exploratory behaviors and that modify whisker movements based on sensory–motor feedback during tactile-based perception. Here, we combined retrograde tracer injections into whisker-specific muscles, with large-scale immunohistochemistry and digital reconstructions to generate an average model of the rat FN. The model incorporates measurements of the FN geometry, its cellular organization and a whisker row-specific map formed by vMNs. Furthermore, the model provides a digital 3D reference frame that allows registering structural data – obtained across scales and animals – into a common coordinate system with a precision of ∼60 µm. We illustrate the registration method by injecting replication competent rabies virus into the muscle of a single whisker. Retrograde transport of the virus to vMNs enabled reconstruction of their dendrites. Subsequent trans-synaptic transport enabled mapping the presynaptic neurons of the reconstructed vMNs. Registration of these data to the FN reference frame provides a first account of the morphological and synaptic input variability within a population of vMNs that innervate the same muscle.
Introduction
Rodents actively move their facial whiskers (vibrissae) to explore their environment. The vibrissal system is thus a common model system to investigate principles of sensory information processing (Feldmeyer et al., 2013), mechanisms that underlie motor control (Deschenes et al., 2016), and for studying interactions between sensory and motor pathways (Schwarz, 2016). Recent studies have provided detailed insight into the central pattern generator pathways that drive rhythmic whisker movements (whisking) at the level of the brain stem (Gao et al., 2001, Hill et al., 2008, Kleinfeld et al., 2014, Deschenes et al., 2016). Furthermore, accumulating evidence suggests that disjoint cortical feedback pathways from both primary motor and somatosensory areas to a variety of brain stem structures may play a crucial role in whisker motor control (Kleinfeld et al., 1999, Berg and Kleinfeld, 2003, Brecht, 2004, Brecht et al., 2004, Sreenivasan et al., 2015, Ebbesen et al., 2017).
Whereas the list of upstream brain structures involved in whisker motor control is constantly expanding (Hattox et al., 2002, Takatoh et al., 2013, Sreenivasan et al., 2015), there is only a single output structure in the rodent nervous system, whose neuronal activity translates directly into whisker movements: the lateral facial nucleus (FN) of the brain stem (Ashwell, 1982). Within the FN, whisker-specific populations of cholinergic motoneurons (vMNs) innervate two classes of muscles that generate the rhythmic whisker movement (Dorfl, 1982, Wineski, 1985). First, extrinsic muscles cause backward movements of multiple whiskers that are located within the same row (i.e. retraction (Wineski, 1985)). In contrast, intrinsic muscles form slings around the follicle (Dorfl, 1982) of individual whiskers (Fig.1A), causing forward movements of single whiskers (i.e. protraction (Carvell et al., 1991)). The intrinsic musculature, which consists almost exclusively of fast contractible, fast fatigable muscle fibers (Jin et al., 2004), thus forms the structural basis for the rapid and coordinated – yet highly flexible – patterns of multi-whisker movements.
Fig. 1.
Geometry and cellular organization of rat FN. (A) Schematic illustration of the musculature underlying whisker motor control (adapted from (Berg and Kleinfeld, 2003)). Injections of retrograde (CTB) and trans-synaptic (rabies virus) tracers into whisker-specific intrinsic muscles were performed to reveal the organization vMNs in rat FN. (B) Maximum projection image across all histological brain sections that comprise the FN. Sections were labeled with NeuN to reveal neuron somata. (C) Zoom-into one optical section from the image stack in panel (B). The outlines of the FN were identified by differences in soma size. Bottom panels show zoom-into illustrate the criteria to delineate the FN by NeuN-labeling. The center locations of all somata within the FN were marked (green spheres). (D) Result of reconstructing the FN outlines and marking all somata from the example experiment shown in panels (B, C). (E) The outlines were transformed into a surface that delineates the volume of the FN. The soma counts were transformed into a 3D density at 50-µm3 resolution. (F) The shape and volume of the FN were approximated by an ellipsoid. The experiment shown in panels (B–E) was repeated for two additional rats and the three FN reconstructions were aligned to yield an average ellipsoid (blue), which represents the FN reference frame. (G) Average (±SDs) volume and soma density of the FN model.
Vibrissal MNs that innervate the extrinsic muscles are located in the dorsal part of the lateral FN. In contrast, vMNs that innervate the intrinsic muscles are located in the ventral part of the lateral FN, where they form a whisker row-specific map (Klein and Rhoades, 1985). In line with these anatomical observations, electrical stimulations of individual vMNs showed that whisker protractions can be evoked primarily for just a single whisker, whereas retractions are usually evoked for multiple whiskers (Herfst and Brecht, 2008). Furthermore, action potentials of vMNs translate into whisker movements in a one-to-one manner (Herfst and Brecht, 2008). The characteristics of the evoked whisker movements depend on the identity of the stimulated neurons. This suggests that the vMNs are heterogeneous with respect to the movement properties they encode. Therefore, the endpoint of all pathways involved in motor control and sensory–motor feedback of a single whisker is represented by a small population of vMNs – approximately 100 per whisker (Klein and Rhoades, 1985) – where each vMN represents a ‘labeled line output’ that codes for specific movements.
Here, we provide a quantitative account of the structural organization that underlies this terminal point of the brain-wide (sensory-) motor pathways of whisker motor control. Similar to our previous work on the rat barrel cortex (Egger et al., 2012), we provide a 3D model of the average FN anatomy, incorporating data from synaptic, cellular and population levels. First, we generated a 3D reference frame for registration of anatomical data into an average model of the FN geometry. Second, we measured the number and 3D distribution of all MNs in the FN. Third, we injected retrograde tracers into intrinsic whisker muscles, determined the locations of the retrogradely labeled vMNs within the FN and used these data – in combination with the reference frame – to generate a digital representation of the vMN map within the FN.
As a first application of the FN model, we injected a replication competent strain (N2c) of rabies virus (Kelly and Strick, 2000) into the intrinsic muscle of a single whisker. This allowed us to retrogradely label a sparse population of whisker-specific vMNs, to reconstruct their respective morphologies and to trans-synaptically label brain stem neurons that provide input to these vMNs. Registration of the rabies data then enabled us to analyze the vMNs’ dendrites in relation to the vMN map of the FN. Our data support previous reports about the number of MNs in the FNs, the number of vMNs per whisker and their row-specific organization, and incorporates this information into a coherent digital model. Furthermore, we provide a first account of the morphological variability within the population of vMNs that innervate the same muscle, reveal that vMNs are not synaptically connected to each other or to other MNs in the FN, and that they receive synaptic inputs from a variety of brain stem nuclei that extend beyond previous reports.
Experimental procedures
Injections of retrograde tracers
All experimental procedures were carried out in accordance with the animal welfare guidelines of the Max Planck Society. Young adult (P28–35) male Wistar rats were anesthetized with a ketamine–xylazine mixture (70/6 mg/kg, i.p.) and placed in a stereotaxic frame (Helmut Saur Laboratories). The rat’s head was rotated 20° upward in the ventral–dorsal direction to give better visual access to the right whisker pad on the snout. Whiskers targeted for injections were identified visually under a surgical stereoscope (Leica MZ6) and marked at the base of the snout with a surgical pen. The fur surrounding the targeted whisker was trimmed and a small incision was made next to the targeted whisker follicle. A steel injection needle attached to a 5-µm glass Hamilton syringe was inserted into the incision approximately 1 mm below the skin of the whisker pad using a manual micromanipulator (Narishige Model BE-8). 500–700 nL of cholera toxin beta subunit (CTB) conjugated to AlexaFluor 488, 594 or 647 (Molecular Probes 1 mg/ml in PBS) was then pressure injected close to the base of the targeted whisker follicle under visual control. Rats underwent a 6-day survival period after tracer injections.
Injections of the N2c strain of rabies virus
All experimental procedures were conducted at the University of Pittsburgh and in accordance with National Institutes of Health guidelines and were approved by the relevant Institutional Animal Care and Use and Biosafety Committees. The procedures for handling rabies virus and animals infected with rabies have been described previously (Kelly and Strick, 2000, Kelly and Strick, 2003) and are in accordance with or exceed the recommendations from the Department of Health and Human Services (Biosafety in Microbiological and Biomedical Procedures). The procedures for injections of rabies virus were identical to those described above for CTB. 500–700 nL of the N2c strain of rabies virus (1 × 109 pfu/mL, 0.25–1.0 mL, provided by M. Schnell, Thomas Jefferson University, Philadelphia) was pressure injected at the base of the follicle of the C3 whisker under visual control. Rats were allowed to survive for 3–5 days. No symptoms were observed during this survival period.
Histology
Rats were perfused transcardially with phosphate buffer and brains, as well as the right whisker pads, were removed and fixed with paraformaldehyde. The brains and whisker pads were blocked in 10% gelatin and fixed in 4% PFA overnight. Consecutive 50-µm-thick vibratome sections were cut coronally through the entire brain stem. Consecutive 150-µm-thick vibratome sections were cut sagittally through the entire whisker pad. For rats injected with rabies virus, sections were double-immunolabeled to visualize and count rabies labeled cells with respect to all neurons (NeuN (Mullen et al., 1992)). To do so, slices were permeabilized and blocked in 0.5% Triton X-100 (TX) (Sigma Aldrich) in 100 mM Phosphate Buffer (PB) containing 4% normal goat serum (NGS) (Jackson ImmunoResearch Laboratories) for 2 h at room temperature. Primary antibodies were diluted 1:500 (Rabbit anti-NeuN, EMD Millipore #MAB377) and 1:1000 (Mouse anti-RABV-P 31G10 2 mg/ml, Thomas Jefferson University) in PB containing 1% NGS for 48 h at 4 °C. Secondary antibodies (1:500 goat anti-mouse IgG Alexa-488 and 1:500 goat anti-rabbit Alexa-647) were incubated for 2–3 h at room temperature in PB containing 3% NGS and 0.3% TX. All brain sections were mounted on glass slides, embedded with SlowFade Gold (Invitrogen) and enclosed with a cover slip.
Image acquisition
Images were acquired using either a confocal laser scanning system (Leica Application Suite Advanced Fluorescence SP5; Leica Microsystems) equipped with glycerol/oil immersion objectives (HC PL APO 10× 0.04 N.A., HC PL APO 20× 0.7 N.A., and HCX PL APO 63× 1.3 N.A.), a tandem scanning system (Resonance Scanner), spectral detectors with hybrid technology (GaAsP photocathode) and mosaic scanning software (Matrix Screener, beta version provided by Frank Sieckmann, Leica Microsystems), or with a fluorescence widefield microscope (BX51, Olympus). For quantification of brain stem labeling in rabies injection experiments (Rabies-488, NeuN-647), dual channel confocal mosaic scans of the entire brain stem were acquired using a 10x glycerol objective at a resolution of 0.868 × 0.868 µm per pixel (1.7× digital zoom, 8× line average, 8-kHz scanning speed). For single neuron reconstruction of rabies-positive vMNs in the FN, single channel confocal mosaic image stacks of volumes up to 1 × 1 × 0.05 mm were acquired using a 63× glycerol objective at a resolution of 0.092 × 0.092 × 0.5 µm per voxel (2.5× digital zoom, 8× line average, 8 k Hz scanning speed, 13 × 13 fields of view). For quantification of NeuN-647-labeled cells within the FN, single channel confocal mosaic image stacks of volumes up to 3 × 1.5 × 0.05 mm were acquired using a 20× glycerol objective at a resolution of 0.361 × 0.361 × 1 µm per voxel (2.0× digital zoom, 8x line average, 8-kHz scanning speed, 10 × 6 fields of view). For triple CTB injection experiments, sequential channel (using Olympus filters MNIBA2 for 488, U-MNG2 for 594, U-MWIY2 for 647) widefield images of 2 × 2 fields of view of the FN were acquired using a 10× dry objective at a resolution of 0.926 × 0.926 × 0.5 µm per voxel and sequential channel (same filters as above) widefield images of 7 × 9 fields of view of the whisker pad were acquired using a 4× dry objective at a resolution of 2.303 × 2.303 × 0.5 µm per voxel.
Detection of somata/contours of brainstem structures
Retrogradely and NeuN-labeled somata were detected manually in high resolution images using Amira Software (Stalling et al., 2005). All images were aligned to each other with respect to the midline and ventral edges of the brain stem. Contours of the brain stem and FN were drawn manually using the Filament Editor (Dercksen et al., 2014) in Amira, and contours delineating different nuclei within the brain stem were adopted from the Paxinos Rat Atlas (Paxinos and Watson, 2005).
Reconstruction of dendrites
3D reconstruction of dendrite morphologies was based on a previously described method for semi-automated reconstruction of neuron morphology from bright field microscope images (Oberlaender et al., 2007). Here, the tracing software was adapted to confocal image data, acquired at a lateral/axial resolution of 0.092/0.5 µm as described above. Image stacks were deconvolved using a linear Tikhonov-Miller algorithm and a theoretically computed point spread function of the confocal microscope (Oberlaender et al., 2009) using Huygens software (SVI, Netherlands). Neuronal structures were manually traced in the image stacks using the Filament Editor (Dercksen et al., 2014) in Amira. Alignment and splicing of neuronal branches across consecutive histological slices was also done within the Filament Editor (Dercksen et al., 2014). 3D reconstructions were augmented with FN contours, which were drawn manually in images of all consecutive histological slices.
Generation of the FN reference frame
Brain stem images, including contours of the FN, from three different rats were aligned to the lateral–medial (L–M) and ventral–dorsal (V–D) axes using Amira. The contours of the 3 FN were transformed into isosurfaces using Amira, the respective centers of mass of the resultant FN volumes were calculated, the FN volumes were translated to match their centers of mass, and each FN was then were approximated by an ellipsoid. To do so, we solved the following equation that describes quadric surfaces:
| (1) |
Given the ellipsoid equation:
| (2) |
the coefficients A, B, C represent the inverse of the square of the ellipsoid’s half axes. Let A, B, C be defined as follows:
Substituting this definition of A, B, C into (1) yields:
which can be written in the form of matrices:
| (3) |
where
.
In order to obtain an ellipsoid that best approximates the FN volume (i.e. isosurface), Q must minimize the squared errors between P and R. In closed form this solution is given by:
| (4) |
Eq. (1) can then be written as:
| (5) |
where,
The ellipsoid’s PAs and radii are then computed from the Eigen decomposition of ,where γ is defined as follows:
| (6) |
and where φ is a 3 × 3 matrix whose columns are the PAs of the ellipsoid, and where is a diagonal matrix, whose elements are the inverse of the square roots of the radii (i.e. A, B, C). The FN reference frame was then defined as the average of these three ellipsoids. Specifically, we generated a ‘standardized’ ellipsoid, whose radii and PA orientations were equal to the respective averages across the three individual FN. The origin of the FN reference frame is defined as the center of symmetry of the average FN ellipsoid. The orientation of the standardized FN coordinate system is defined by x′, y′ and z′ axes, which represent the PAs of the average FN ellipsoid that are most parallel to the L–M, C–R and V–D axes, respectively.
Registration to the FN reference frame
For registration of structural data to the FN reference frame, soma distributions and/or morphological reconstructions must be obtained from images of consecutive coronal brain sections, and contours outlining the FN must be obtained from all sections that comprise the FN. The FN contours are (1) converted into an ellipsoid (see above), (2) its center of symmetry is aligned to the one of the FN reference frame, (3) its PAs are rotated to match the respective PAs of the FN reference frame, and (4) its respective radii are linearly scaled to match the respective radii of the FN reference frame. The resultant transformation is then applied to the soma distributions and/or morphological reconstructions. The registration routines were implemented in Matlab (Mathworks, Version R2016a) and can be downloaded (including the FN reference frame and example data) from the Supplementary Material.
Generation of the vMN map within the FN reference frame
3D distributions of retrogradely labeled vMN somata from 36 injections of CTB into intrinsic muscles of different whiskers in 13 rats were registered to the FN reference frame as described above. The soma distributions from injections into whisker muscles of the same row were combined into a single dataset in Amira (i.e. using the ‘Combine Landmarks’ function). Nineteen of the 36 CTB injections were targeted to the C-row, and the distribution of C-row somata was hence used to determine the location and shape of the slabs representing vMNs within the FN. Specifically, we converted the C-row somata into a soma density distribution at a resolution of 100 µm3 (i.e. using the ‘compute point density’ function in Amira with a x/y/z bin size of 100). Then, we approximated the volume around the C-row somata by calculating a surface around 90% of the soma density distribution (i.e. using the ‘IsoSurface’ function in Amira with a threshold of 10% of the maximum density value and enabling the ‘compactify’ option). The resultant surface was smoothed by using the ‘Smooth Surface’ function in Amira (i.e. with iterations set to 4 and lambda to 0.8). This smoothed surface was defined as the average C-row slab. Because the size and orientation of vMN slabs was in general independent of the injected whisker, we duplicated the C-row slab four times and shifted the duplicates along the L–M and V–D axes with respect to each other (i.e. as determined by analyzing the respective center locations of the A-to-E slabs). The vMN map within the FN reference frame thus consists of five slabs of equal shape and volume that are separated by 54 µm and 19 µm along the L–M and V–D axes, respectively (i.e. A-slab is most lateral and most dorsal).
Results
Geometry and cellular organization of rat FN
To define the FN within the brain stem, we sliced brains into 50-µm-thick consecutive coronal sections. Each section was stained with NeuN to reveal neuron somata. We acquired high resolution confocal images of the brain stem from all NeuN-labeled sections that comprised the FN, which typically extended for 1.5 mm along the rostral–caudal (R–C) axis (Fig.1B; i.e. ∼30 consecutive sections). The FN was identifiable by the soma morphology of the MNs. Compared to the somata of neurons in brain stem nuclei adjacent to the FN (Fig.1C), somata of MNs were larger in diameter (30 ± 4 vs. 16 ± 5 µm). Within each image, we marked the center locations of all NeuN-positive somata within the FN of the left hemisphere. In addition, we drew contours around the outmost MNs within each section to delineate the FN (Fig.1D). We repeated the detection of NeuN-positive somata for smaller sub-volumes by acquiring 3D image stacks (i.e. 1 µm optical sectioning). The difference in MN numbers when quantified within a single image plane compared to the 3D stacks was a factor of 2.1 ± 0.4 (mean ± SD). This quantification allowed determining the total number of MNs within the FN by scaling the counting results accordingly. The resultant 3D distribution of MN somata was largely homogeneous throughout the FN, with soma densities being lowest in the intermediate subdivision between the lateral and medial parts of the FN (Fig.1E). The average density of MNs in the FN was 3 ± 1 per 50 µm3.
To assess the variability in FN geometry, volume and soma density, we repeated this quantification for two additional animals and approximated the volumes of the FN with an ellipsoid (see Experimental Procedures), respectively (Fig.1F). The extents of the ellipsoids were 1.3 ± 0.03 mm, 1.6 ± 0.02 mm and 0.9 ± 0.06 mm along the lateral–medial (L–M; x-axis), rostral–caudal (R–C; y-axis), ventral–dorsal (V–D, z-axis) axes, respectively. The difference between the reconstructed FN volume and its approximation by an ellipsoid was 16%. The FN volume determined in this manner was remarkably preserved across animals, deviating on average by less than 5%. We defined the ellipsoid as the 3D geometrical reference frame of the rat FN and generated a standardized coordinate system accordingly. The center of symmetry denotes the origin of the coordinate system and the principal axes (PAs) its 3D orientation. The PAs being most parallel to the L–M, C–R and V–D axes were defined as the x′, y′ and z′ axes, respectively. Using rigid transformations and linear scaling then allowed registering the FN reconstructions and soma density distributions from the three animals into a common reference frame. The differences in orientation (4.6 ± 2.0°) and extent (12.5 ± 7.5 µm) along the three PAs were small across animals. Consequently, when extracting the outlines of the FN, different anatomical datasets can be registered with an accuracy of approximately ±60 µm to the standardized reference frame, allowing for quantitative and precise structural analyses across animals and experimental conditions. Here, we combined the three NeuN datasets and generated an average model of the FN (Fig.1F), which has a volume of 0.91 ± 0.03 mm3 and contains 6136 ± 1020 MNs (Fig.1G).
Labeling of whisker-specific vMNs in rat FN
We injected retrograde tracer agents – Cholera Toxin subunit B conjugated with Alexa488 (CTB-488), 594 and 647 – into the mystacial pad, targeting the intrinsic muscles of three adjacent whiskers within the same arc (B2/C2/D2, N = 3 rats) or row (C1–3, N = 3 rats) (Fig.2A). Following the injections, we removed the pads (Fig.2B) and sliced them into consecutive 150-µm-thick vibratome sections. The sections revealed that the center of the injections was 910 ± 285 µm (median: 950 µm, range: 400–1400 µm) underneath the pads’ surface and 609 ± 415 µm (median: 518 µm, range: 185–1702 µm) posterior to the base of the follicle. At this depth, injection sites overlapped with the intrinsic muscle of the targeted whisker and had diameters of 412 ± 182 µm (median: 374 µm, range: 190–819 µm). The separation between follicles of adjacent whiskers was more than twice the diameter of the injection sites, and significantly smaller within rows than across rows (789 ± 108 µm vs. 1114 ± 173 µm; p < 10−8, n = 22 whiskers in N = 6 rats). The CTB did not infect the follicle (i.e. sensory pathways) or tissue between muscles (Fig.2C). Instead, the CTB spread within the injected muscle, resulting in injection sites whose shapes resembled the sling of the infected intrinsic muscle (Fig.2D). Consequently, CTB injections were precise and small enough to specifically infect the intrinsic muscle of an individual whisker and overlap between the injection site and intrinsic muscles of adjacent whiskers – if at all – was more likely within rows than across rows.
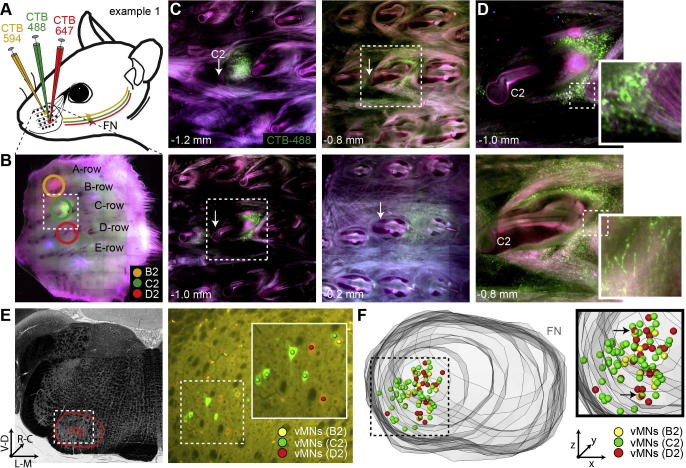
Fig. 2.
Labeling of whisker-specific vMNs in rat FN. (A) Schematic illustration of the injection experiments. Injections of three different versions of a retrograde tracer (i.e. CTB-488, 594 and 647) were targeted to the intrinsic muscles of adjacent whiskers (N = 6). (B–F) Illustrations of the injection sites and quantifications of the retrogradely labeled vMNs for one exemplary experiment, in which injections were targeted to the B2, C2 and D2 whiskers, respectively. (B) Example image of the mystacial pad showing the injections sites. (C) The pad shown in panel (B) was sliced into consecutive 150-µm-thick sections. Top-left panel (1.2 mm underneath the surface of the pad) shows the center location of the injection, which is posterior to the base of the follicle (white arrow) of the targeted C2 whisker. The remaining three panels illustrate that injections did not spread into the follicle and adjacent intrinsic (see panels at 0.8 and 1.0 mm depths) or extrinsic (see panel at 0.2 mm depth) muscles. (D) Zoom-ins to panels in (C) illustrate that the retrograde tracer spreads within the injected muscle, i.e. the shape of the injection site resembles the sling of the injected intrinsic muscle. (E) Left panel: Bright field image of 50-µm-thick coronal section allows delineating the outlines of the FN. Right panel: Widefield fluorescent images (superimposed with the bright field image) illustrate that each CTB injection (i.e. corresponding to panels (B–D)) labels a specific set of vMNs. (F) 3D reconstruction of the FN and distributions of all retrogradely labeled vMNs. The zoom-in shows that vMNs were largely whisker-specific and confined to the ventrolateral part of the FN. Note: The black arrows denote the vMNs that were labeled by two retrograde markers (i.e. C2/D2), likely reflecting the small spread of the C2 injection toward the D2 whisker (see panel (C)).
Following the quantification of the mystacial pads, brains were sliced into 50-µm-thick consecutive coronal sections and high resolution images were acquired for the entire volume of the FN (i.e. N = 3 rats for B2/C2/D2 and C1–3, respectively). Quantification of the retrogradely labeled (i.e. CTB-positive) vMNs confirmed our observations from the mystacial pad analyses. First, vMNs were labeled largely by just a single retrograde marker, indicating that injections were indeed restricted to individual whiskers (Fig.2E). Second, overlap in labeling was observed exclusively for adjacent whiskers (i.e. C2/D2 not B2/D2) and more pronounced for whiskers within the same row (median: 4% dual-labeling, range: 0–33%) than across rows (median: 1%, range: 0–5%), reflecting that the separation of whisker follicles is larger across rows than within rows. Third, retrogradely labeled neurons were restricted to the ventrolateral (VL) part of the FN (Fig.2F), confirming that the CTB did not spread into the extrinsic muscles (Klein and Rhoades, 1985). Finally, as expected from the variability of injection site depth, diameter and overlap with the targeted muscle, the number of retrogradely labeled neurons per injection deviated substantially within and across experiments (mean ± SD: 42 ± 32; n = 18 injections in N = 6 rats).
Organization of vMNs in rat FN
We repeated the CTB injections in 13 additional animals. In these experiments, we targeted the intrinsic muscles of 36 whiskers that were located across the entire whisker pad (i.e. from A-to-E row and arc 0–5; note that arc 0 refers to the α–δ whiskers), and that were separated by (at least) one whisker. Specifically, we injected CTB into three whisker muscles within the same row (C1/C3/C5, N = 3 rats), within the same whisker arc (A3/C3/E3, N = 3), along a diagonal axis (A1/C3, N = 3) and into a mix of different rows and arcs (C2/C4/E1, N = 2; B3/D3/β or δ, N = 2). Each injection labeled a specific set of vMNs, with no overlap between the respective populations (Fig.3A). We manually marked the center locations of each retrogradely labeled vMN and traced the outlines of the FN within each brain section. As a consequence of the variability of the injection procedure, the number of vMNs per whisker deviated substantially between experiments (median: 44, range: 5–147). However, the number of vMNs was not significantly correlated with the position of the injected whisker in a particular row (Pearson: R = −0.17) or arc (R = −0.07). Given the efficacy of the CTB (∼80% (Chen et al., 2013)), we thus estimate that the intrinsic muscles of each whisker are represented by approximately 60 vMNs in the ventrolateral FN (mean ± SEM of CTB-labeled vMNs per injection: 46 ± 5; Fig.3B).
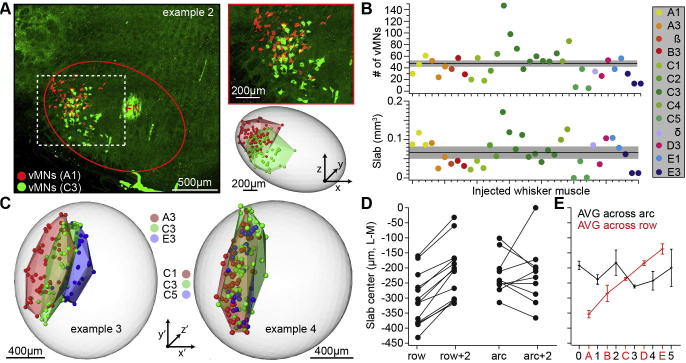
Fig. 3.
Organization of vMNs in rat FN. (A) Example of one of the 13 experiments in which we injected CTB into intrinsic muscles that were separated by (at least) one whisker. Here, injections were targeted to the intrinsic muscle of the A1 and C3 whiskers, respectively. Left panel: Maximum projection confocal image across all histological brain sections that comprised the FN. Vibrissal MNs for both injected whiskers were found throughout the FN along the R–C axis. Vibrissal MNs were whisker-specific and confined to the ventrolateral part. Right panels show zoom-in (top) and the center locations of all retrogradely labeled vMNs (bottom). The volumes occupied by the respective vMN distributions (i.e. shell-shaped slabs) were approximated by calculating a convex hull around the marked somata (light red and green surfaces). (B) The numbers of retrogradely labeled vMNs (top) and the respective slab volumes for all injected whisker muscles (n = 36). The black lines and gray-shaded areas represent mean and SEM, respectively. (C) Reconstruction of retrogradely labeled vMNs and slab volumes for two additional example experiments. Here, injections were targeted to the intrinsic muscles of the A3, C3 and E3 whisker (left panel), and to the C1, C3 and C5 whisker (right panel). Vibrissal MNs representing whiskers within the same row are disjoint, but located within the same slab. (D) Quantification of the slab positions along the L–M axis within each individual experiment (n = 36 injections in N = 13 rats). Slabs representing different whisker rows were always shifted with respect to each other. Slabs representing different whisker arcs (i.e. within the same row) showed no systematic shift with respect to each other (two-sided T-test: p < 0.01). (E) Quantification of the average slab locations along the L–M axis. Error bars denote mean ± SEM.
We calculated a convex hull around the soma distributions of the respective vMNs (Fig.3A). This allowed approximating the volume occupied by the vMNs within the FN (Fig.3B). The resultant volumes resembled elongated shells – in the following referred to as slabs – with a maximal extent approximately parallel to the C–R (i.e. y′) axis (Fig.3C). Similar to the generation of the FN reference frame, we approximated each vMN slab with an ellipsoid. The slab volume (mean ± SEM: 0.065 ± 0.007 mm3) was not significantly correlated with the position of the injected whisker in a particular row (R = −0.12) or arc (R = −0.20). Slabs representing vMNs of different whisker rows were segregated along an axis that was approximately parallel to the L–M (i.e. x′) axis (Fig.3C). In contrast, slabs of vMNs that represented different whiskers within the same row shared largely the same volume. To quantify this row-specific organization of vMNs, we calculated the distances between the center locations of slabs that were separated by one row or arc, respectively (Fig.3D). In experiments where injections were separated by one row (n = 13), the slab representing the whisker closer to the bottom of the snout was always located more medial within the FN. In contrast, slabs representing whiskers of the same row, but of different arcs (n = 10), were not systematically shifted with respect to each other. Slabs of vMNs that represent intrinsic muscles of whiskers within the A-row (i.e. top of the snout) are located in the most lateral part of the FN, approximately 0.35 mm from the center of the FN reference frame. Slabs of the B-to-E row are shifted with respect to each other toward the medial part of the FN (Fig.3E).
Standardized FN reference frame with vMNs slabs
We registered the soma distributions of retrogradely labeled vMNs, and the resultant slab ellipsoids, into the standardized FN reference (Fig.4A–C). The center of symmetry and PAs for each vMN slab ellipsoid were then calculated to determine the relative position and 3D orientation of each slab within the FN (Fig.4D). Except for the center positions along the L–M and V–D axes, none of the slabs’ geometrical properties – 3D extent, R–C position or 3D orientation – were significantly correlated with the position of the injected whisker in a particular row (R2 ≤ 0.05). None of the slabs’ properties were significantly correlated with the position of the injected whisker in a particular arc (R2 ≤ 0.08). Consequently, vMNs that represent intrinsic muscles of individual whiskers form a map of shell-shaped slabs within the ventrolateral part of the FN, with slab size and orientation being independent of whisker identity, and slab centers being shifted in a row-specific manner (i.e. from A-to-E) by 54 and 19 µm along L–M and V–D axes, respectively. Finally, we used the geometrical properties of the slabs and augmented the FN reference frame within an average row-specific map of vMNs. We generated five slabs (i.e. A-to-E) of equal size and orientation (i.e. representing an isosurface around the C-row somata; see Experimental Procedures), and placed each slab at its respective average position within the FN. The center location of the C-row slab varied across animals by 51 µm. Consequently, we have augmented the FN reference frame with an average map of the vMNs that is as precise as the FN reference frame itself (Fig.4E).
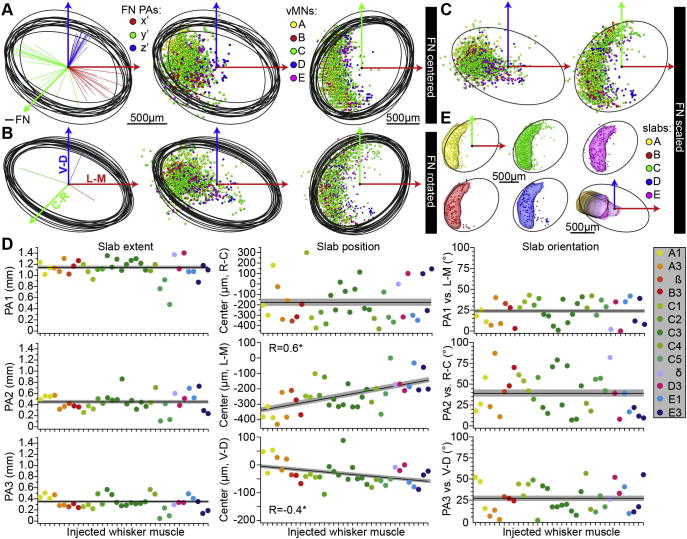
Fig. 4.
Standardized FN reference frame with vMN slabs. (A–C). The FN of all CTB-injected animals (N = 13) were approximated by an ellipsoid, whose centers of symmetry were aligned to the one of the FN reference frame (panel (A)), whose PAs were rotated to match the respective ones of the FN reference frame (panel (B)) and whose dimensions were linearly scaled to match the respective dimensions of the FN reference frame (panel (C)). In all panels, the respective distributions of retrogradely labeled somata were transformed accordingly. (D) After registration, each vMN slab was approximated by an ellipsoid (n = 36) and their respective geometrical parameters are plotted after sorting them by whisker row and then by arc (i.e. from left to right: A1–E3). The black lines and gray-shaded areas represent the mean and SEM of the respective parameter in each panel. Left: The 3D extent of the ellipsoids (i.e. length of the three PAs) did not correlate with the location of a particular whisker along the row or arc. Center: The slab ellipsoids’ center locations along the L–M and V–D axes (but not along the R–C axis) did correlate with the location of a particular whisker along the row (but not along the arc). Right: The 3D orientations of the slab ellipsoids did also not correlate with the location of a particular whisker along the row or arc. (E) Geometrical reference frame of the rat FN including an average map of whisker row-specific slabs. Slab positions represent the mean of the respective parameters shown in panel (D). Slab sizes and orientations are equal for all slabs and based on the C-row slab (see Experimental Procedures).
Trans-synaptic tracing of whisker muscle-related neuronal networks
As a first application of the standardized FN reference frame, we injected the N2c strain of rabies virus into intrinsic whisker muscles (Fig.5A). Previous studies had shown that ‘fixed’ strains of rabies virus, such as N2c, allow for a very specific tracing of the motor networks involved in the control of single muscles (see (Kelly and Strick, 2000, Ugolini, 2010) for reviews). First, in rats and primates – but not in mice (Coulon et al., 1989, Jackson, 2002) – fixed strains of rabies virus enter the central nervous system exclusively via the motor route following intramuscular injection (Tang et al., 1999, Ugolini, 2008) (but see (Levinthal and Strick, 2012, Dum et al., 2016) for infection of autonomic neurons). Second, rabies virus propagates by trans-synaptic transfer exclusively in the retrograde direction, because intracellular transport of the virus after replication is restricted to the soma and dendrites (Ugolini, 1995, Klingen et al., 2008). Third, trans-synaptic transfer is not gradual, but occurs step-wise at regular intervals of approximately 8–12 h (e.g. (Ugolini, 1995, Tang et al., 1999, Kelly and Strick, 2003)). Rabies tracing thus allows for a precise identification of the trans-synaptic order of labeling, because of the stepwise visualization of successive synaptic relays that is strictly time-dependent, and the synchronous labeling of neuronal populations of the same order (e.g. trans-synaptic spread does not depend on the location of synapses along dendrites or axonal length between pre- and postsynaptic neurons; for a review see (Ugolini, 2010)).
Table 1.
List of abbreviations.
| Sp5C | spinal trigeminal nucleus, caudal part |
| MdD | medullary reticular nucleus, dorsal part |
| MdV | medullary reticular nucleus, ventral part |
| Irt | intermediate reticular nucleus |
| Sp5I | spinal trigeminal nucleus, Interporalis |
| PMn | paramedian reticular nucleus |
| ts | Tectospinal tract |
| Sol | Solitary tract |
| PCRt | parvicellular reticular nucleus |
| Gi | gigantocellular reticular nucleus |
| IOD | inferior olive, dorsal nucleus |
| PreBotC | pre-Botzinger complex |
| LPGi | lateral paragigantocellular nucleus |
| Pr | prepositus nucleus |
| Amb | ambiguus nucleus |
| DPGi | dorsal paragigantocellular nucleus |
| FN | Facial Nucleus |
| SpVe | spinal vestibular nucleus |
| Mve | medial vestibular nucleus |
| Sp5O | spinal trigeminal nucleus, Oralis |
| Pr5 | principal sensory trigeminal nucleus |
| PnC | pontine reticular nucleus |
| vMN | vibrissal motoneuron |
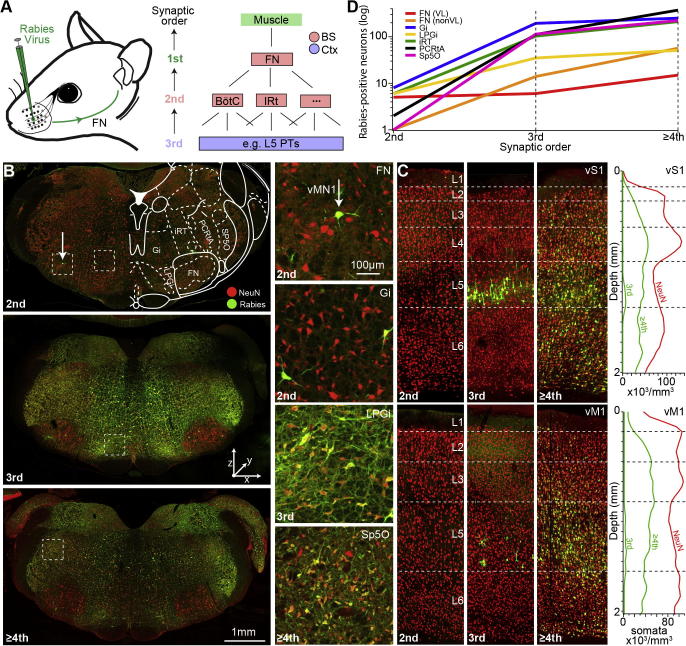
Fig. 5.
Trans-synaptic tracing of whisker muscle-related neuronal networks. (A) Schematic illustration of injection experiments. The N2c strain of rabies virus was injected into the C3 whisker muscle. Right panel: Illustration of defining the trans-synaptic order of rabies labeling. The virus labels vMNs in the FN, then neurons that are presynaptic to those first-order neurons at the level of the brain stem (BS, 2nd order neurons). Third order experiments can be identified by additional labeling that is specific to L5 of the cortex. Forth order experiments show cortical labeling in additional layers. (B) Example images (from top to bottom) represent 2nd (no labeling in cortex), third- (cortical labeling is restricted to L5) and ≥fourth-(cortical labeling in all layers) order spread (see panel (C)). Right panels: within each experiment, a sparse population of vMNs is labeled within the FN, whereas labeling increases with order in different BS nuclei such as the Gi, LPGi and Sp5O (see Table 1 for a list of abbreviations). (C) Example images (left panels) and density distributions of rabies-positive neurons in cortex (right panels) for experiments with second-, third-and ≥fourth-order trans-synaptic spread (images correspond to the brain stem images in panel (B)). Each of the four density profiles (green) represents average from manual quantifications of rabies-positive neurons in three coronal sections through vS1 and vM1, respectively. (D) Quantification of all rabies-positive neurons within the BS depending on the order of rabies spread. In all infected BS nuclei, the number of labeled neurons increases by one order of magnitude from 2nd to 3rd order. The increase from 3rd to ≥4th order is 2–fourfold. The number of rabies-positive neurons within the ventrolateral (VL) FN is independent of the order of spread, but increases outside the ventrolateral FN (nonVL).
Given the properties of fixed strains of rabies virus described above, we were able to determine the order of trans-synaptic spread by the time period between virus injection and perfusion. However, because the initial uptake of the rabies virus is not efficient, and thereby the onset of rabies infection varies across experiments, we additionally determined the order of trans-synaptic spread by quantifying the brain-wide distributions of rabies-positive neurons in each experiment (Fig.5A). Specifically, following the infection of MNs (1st order neurons) that innervate the injected muscle, the virus crosses synapses in the retrograde direction and labels all neurons that provide input to these MNs. In case of the whisker-related musculature, such second-order neurons should be restricted to the level of the brain stem (Hattox et al., 2002, Takatoh et al., 2013). Neurons in the brain stem are typically postsynaptic to pyramidal tract neurons (PTs) that are located in layer 5 (L5) of the cortex (e.g. (Sreenivasan et al., 2015)). Therefore, experiments where the rabies labeling is restricted to L5 of the cortex are likely to reflect third-order trans-synaptic spread. Which specific cortical areas are labeled will depend on the brain stem nuclei that are labeled at 2nd order. Experiments where the rabies labeling has spread to layers above and below L5 represent fourth-order (or higher) trans-synaptic spread.
First-order neurons
Here, we injected rabies virus into the intrinsic muscle of the C3 whisker. The animals were then sacrificed at a regular interval of 12 h, starting 3 days after the injection. The entire rat brains were sliced into consecutive 50-µm-thick coronal sections, which were immunolabeled with NeuN and highly sensitive antibodies against the rabies virus, allowing to reveal the distributions of rabies-positive neurons with respect to all neurons (Fig.5B). In two animals (litter mates with survival times of 72 and 96 h after muscle injection, respectively), rabies labeling was restricted to the level of the brain stem (i.e. no labeling in cortex), and we thus defined them as second-order trans-synaptic spread (Fig.5C). We marked the center locations of the rabies-positive neurons throughout the FN and reconstructed the FN outlines as described above. In both second-order experiments, the number of first-order neurons (i.e. rabies-positive neurons in the FN) was small, with 6 and 3 vMNs being labeled in the two experiments, respectively (Fig.5D). The sparseness of first-order labeling is likely to reflect the fact that fixed strains of rabies virus do not spread within the muscle (Ugolini, 2010), nor do they cause spurious spread or uptake by fibers of passage, even after long-standing infection (Tang et al., 1999, Levinthal and Strick, 2012). Registration to the FN reference frame confirmed that the locations of these first-order vMNs overlapped with those of the CTB labeled C-row somata in the ventrolateral part of the FN.
The sparseness of first-order labeling allowed reconstructing the 3D morphologies of the rabies-positive vMNs (Fig.6A). Here, we reconstructed the soma, dendrite and local axon morphologies of three of the six vMNs from the second-order experiment shown in (Fig.5B). The dendrite reconstructions resembled those reported previously, where individual vMNs that evoked single whisker movements were labeled with biocytin in vivo (Herfst and Brecht, 2008). The dendrites of the three vMNs had similar path lengths (11.4 ± 3.7 mm) and morphological parameters (Fig.6B). Multiple dendrites (6 ± 2) extended radially from the soma within the coronal plane. Most of those dendrites projected beyond the FN borders, where they terminated with sparsely branching arbors. Registration to the FN reference frame revealed substantial differences between the vMNs’ respective 3D dendrite distributions within and outside the FN (Fig.6C). Independent of their soma location, all vMNs projected their dendrites toward and beyond the most caudal part of the FN. Within this caudal region, dendrites innervated a territory more medial than the slabs, with dendritic fields being rotated with respect to each other around the FN axis that is most parallel to the C–R axis. In the sagittal plane, dendritic fields were largely disjoint, with individual vMNs innervating different ‘depth’ within the FN. We calculated the overlap at 50-µm3 resolution between the dendrite morphologies (Fig.6D). The degree of overlap deviated substantially between pairs of vMNs, ranging from 13% of the dendrites from vMN3 that overlap with those of vMN1 to 72% of the dendrites of vMN1 that overlap with those of vMN2 (average overlap: 34%). In addition, the three vMNs differed in their dendritic fields with respect to the whisker row-specific organization of the FN. For example, vMN1 projected almost no dendrites into any of the slabs, whereas 21% of the dendrites of vMN3 overlapped with the E-slab. Dendrites of vMNs are located primarily outside the ventrolateral FN, and those within the ventrolateral FN innervate the slabs in a cell-specific manner (Fig.6D).
Fig. 6.

Dendrite morphologies of vMNs. (A) Left panel: High-resolution image of the vMN shown in Fig.5B (i.e. vMN1; white arrow). The morphology of vMN1 was reconstructed across consecutive histological sections. Right panel: High-resolution image of histological section adjacent to the one shown in the left panel shows soma, dendrites and axon of a second vMN (i.e. vMN2) and the axon of vMN1 (white arrows). The zoom-in reveals rabies virus particles within the labeled cells. (B) 3D reconstruction of three vMNs from the same animal, including the two vMNs shown in panel (A). Note: Axons (black) of the reconstructed vMNs have no collaterals within the FN. (C) Registration of the rabies-labeled vMNs to the FN reference frame allows quantification of morphological variability with respect to the whisker row-specific map. (D) Quantification of dendrites within the FN reference frame reveal large morphological variability within the population of C3 innervating vMNs. Upper panel: the dendritic fields of the three C3 vMNs overlap only partially with each other (see also colored arrows in panel (C)). Lower panel: dendrites of vMNs extend beyond the FN. Within the FN, dendrites innervate the slabs in a cell-specific manner.
Second-order neurons
The morphological analysis revealed substantial cell-to-cell variability within the population of C3-innervating vMNs. To investigate the degree to which this variability may translate to the populations that provide input to vMNs, we additionally marked the center locations of all rabies-positive neurons throughout the brain stem in both second-order experiments. Rabies-positive neurons were found throughout several brain stem nuclei that were previously reported as being presynaptic to vMNs (Takatoh et al., 2013) or to MNs in the FN in general (Hattox et al., 2002). Labeling in each brain stem nucleus was sparse in second-order experiments, with ∼10–100 neurons per nucleus being rabies-positive. The largest presynaptic (i.e. 2nd order) population to vMNs was found within the gigantocellular reticular nucleus (Gi, ∼10–40 times the number of vMNs) in the ipsilateral hemisphere. The second largest presynaptic populations to vMNs (∼10 times the number of vMNs) were located within the ipsilateral dorsal medullary reticular formation (MdD), the intermediate reticular nucleus (IRt) and the spinal nucleus interpolaris (Sp5I). The most prominent input population within the contralateral hemisphere was located within the Gi (∼5 times the number of vMNs).
Within the brain stem of the second-order experiment that was used to reconstruct the morphologies of the first-order vMNs, 921 putative second-order neurons were labeled throughout 22 different brain stem nuclei (Fig.7). Twenty of those had been described previously as being presynaptic to vMNs or to neurons in the FN in general. The two brain stem regions that were not reported previously are the prepositus nucleus (Pr) and the tectal spinal tract (ts), where sparse labeling on the order of the number of first-order vMNs was identified in both hemispheres (Fig.7A–C). Seven hundred sixteen of the second-order neurons were located within the ipsilateral, 205 within the contralateral hemisphere of the brain stem. The largest populations of second-order neurons within the ipsilateral hemisphere were found within the Gi (258 positive neurons), followed by the IRt (97) and MdD (87). The same nuclei comprised the largest numbers of rabies-positive cells in the contralateral hemisphere (68, 28, 28).
Fig. 7.
Presynaptic populations of vMNs. (A) Maximum confocal projection image across all histological brain sections that comprise the Sp5O of the BS in a second-order experiment after injecting rabies virus into the C3 whisker (same experiment as shown in Fig.5B, C). Images were superimposed with the corresponding pages from the Paxinos rat brain atlas and all rabies-positive neurons were marked and assigned to the respective BS nuclei. Right panel: zoom-into the GI reveals sparse labeling of second- order neurons that are presynaptic to the first-order vMNs in the FN, e.g. the ones in Fig. 6. (B) Maximum confocal projection image across all histological brain sections that comprise the Sp5I of the BS in same experiment as shown in panel (A) Right panel: zoom-into the IRt. (C) Maximum confocal projection image across all histological brain sections that comprise the Sp5C of the BS in the same experiment as shown in panels (A, B). Right panel: zoom-into the MdD. (D) Same BS regions as shown in the zoom-ins in panel (A) for a second second-order experiment after injections into the C3 whisker muscle. Labeling in the second animal was in general sparser than in the first experiment. (E) Quantification of the number of neurons in all rabies-infected BS nuclei in the two second-order C3 injection experiments. The red marked BS nuclei represent those that are consistently labeled in both experiments. The major sources of common input to vMNs are highlighted in green.
We repeated the quantification of rabies-positive cells throughout the brain stem for the second second-order experiment. In this experiment, the total amount of rabies-positive neurons was in general lower throughout the brain stem, with 104 (i.e. 2nd order) neurons being labeled outside the FN (Fig.7D). However the relative proportion of rabies-positive neurons within the two hemispheres was similar across the two second-order experiments, with ∼4 vs. 5 times the number of second-order neurons being labeled in the ipsilateral, compared to the contralateral hemisphere. Also, the nuclei comprising the largest populations of rabies-positive neurons remained unchanged (i.e. the MdD, Gi and IRt). The difference in the overall number of second-order neurons between the two experiments was thus only partly reflected by the lower number of first-order vMNs (i.e. 6 vs 3), but arose primarily from the lack of rabies labeling in some brain stem nuclei in the second second-order experiment. For example, sparse labeling was observed in the ipsilateral pre-Bötzinger complex (PreBotC, 15 cells), ventral medullary reticular formation (MdV, 18 cells) or contralateral parvocellular reticular nucleus (PCRt, 13 cells) in the first second-order experiment, but was lacking in the second one. Among the 22 brain stem nuclei, 11 nuclei showed consistent labeling in both experiments (Fig.7E).
Third-order neurons
In one animal that was sacrificed 84 h after muscle injection, we observed rabies-positive neurons in the brain stem and in cortex. Remarkably in this experiment, cortical labeling was restricted to the vibrissal-related somatosensory (vS1, i.e. barrel cortex and S2) and motor areas (vM1, Fig.5C). In these areas, labeling was restricted to L5. We hence identified this experiment to reflect third-order trans-synaptic spread. In the third-order experiment, rabies-positive neurons were found in virtually every brain stem nucleus and in both hemispheres. In comparison to the second-order experiments, the numbers of rabies-positive neurons in the third-order experiment were by one order of magnitude higher in all brain stem nuclei of that were already labeled in the second-order experiments (Fig.5D). Remarkably, and in contrast to the general increase with trans-synaptic order of the number of rabies-positive neurons within the brain stem (i.e. likely to reflect ‘local’ connectivity), the ipsilateral (and contralateral) FN remained largely void, with a small number of putative first-order neurons (i.e. 6 vMNs) being labeled within, and 15 neurons being labeled outside the ventrolateral part of the FN (i.e. primarily within in the medial part of the FN), respectively.
Fourth-order neurons
In a litter mate of the third-order animal that was sacrificed 24 h later (i.e. 108 h after muscle injection), the rabies virus had spread to all layers of vS1 and vM1 (Fig.5C), to L5 of additional cortical areas, and the number of rabies-positive neurons had further increased in all second-order brain stem nuclei (Fig.5D). This experiment was hence identified as fourth-order (or higher) trans-synaptic spread. Similar to the second-and third-order experiments, even in this ≥fourth-order experiments, the number of rabies-positive neurons within ventrolateral FN was small (Fig.5B). In contrast, the number of rabies-positive cells within the FN, but outside the ventrolateral part, had increased to 57 neurons (i.e. primarily within in the medial part of the FN).
Discussion
We reconstructed the geometry and cellular organization of the FN in the brain stem of the rat, determined the variability of those quantities across animals and used these data to generate an average model of the rat FN. The FN geometry was approximated by an ellipsoid, whose center of symmetry denotes the origin and whose PAs define the 3D orientation of a standardized FN reference frame. The variably of the center of symmetry (∼120 µm), orientation (∼5°) and FN volume (∼5%) was remarkably small across animals. Extracting the outlines of the FN thus allows for precise registration of structural data – obtained across scales and across animals – into a common coordinate system. Furthermore, by injecting retrograde tracers into intrinsic muscles of different whiskers, we determined the organization of vMN somata within the ventrolateral part of the FN. Retrogradely labeled vMNs representing intrinsic muscles of different whisker rows formed concentric shells, referred to as slabs, within the FN. Vibrissal MNs representing the A-row formed the outmost (i.e. far lateral) slab. Vibrissal MNs representing muscles of whiskers from the same row were disjoint, but intermingled within the respective slab without any apparent organization. We augmented the FN model with the average whisker row-specific map of vMNs, allowing to analyze anatomical data with respect to this map at an accuracy of ±60 µm.
Comparison to previous studies of FN structure
The present data that were used to generate the FN reference frame are in remarkable agreement with several previously reported structural analyses. Specifically, previous studies estimated that the intrinsic muscles of a single whisker are represented by ∼50–100 vMNs (Klein and Rhoades, 1985). We estimate that each whisker is represented by 58 ± 6 vMNs. Moreover, previous functional (Herfst and Brecht, 2008) and structural (Klein and Rhoades, 1985) studies provided evidence that vMNs that represent the intrinsic muscles are confined to the ventrolateral part of the FN, whereas those representing the extrinsic muscles are located within more dorsal areas. Furthermore, within the ventrolateral part of the FN, vMNs were shown to give rise to a whisker row-specific map that spans along the R–C axis of the brain stem (Klein and Rhoades, 1985). Our data support both of these observations, reveals that the map consists of concentric shell-shaped slabs, and that the location, orientation and size of each slab is largely consistent across animals. In conclusion, the average model of the FN generated here, can be regarded as a precise digital reference frame, whose geometry, volume, cellular composition and whisker row-specific organization are in line with the respective data reported previously.
Vibrissal MNs represent terminal points of whisker-related networks
We have previously reported a standardized reference frame for the rat barrel cortex (vS1), and had illustrated how registration of soma density distributions (Meyer et al., 2013) and neuronal morphologies (Narayanan et al., 2015) to the reference frame allows inferring cell type-specific synaptic input patterns (Egger et al., 2014) or relationships between cell type-specific structural properties and whisker-evoked activity patterns (Oberlaender et al., 2012, Egger et al., 2015). Similar to those studies, we envision that the standardized reference frame of the FN will allow incorporating structural data obtained from different experimental approaches, thereby providing quantitative insight into the structural organization of the neuronal networks that underlie whisker motor control. To illustrate how the FN model will help dissecting vibrissal motor networks, we combined injections of replication competent rabies virus into the intrinsic muscles of the C3 whisker with registration to the standardized reference frame. We found that each injection labeled a sparse population of vMNs within the FN slab corresponding to the C-row. The number of rabies-positive neurons within the ventrolateral FN did not increase with the order of trans-synaptic rabies spread. This result is in line with a previous study (Herfst and Brecht, 2008), which found that a vMN that evoked single whisker movements and whose morphology resembles those of the vMNs reconstructed here (i.e. innervating the intrinsic muscles), lacked axon collaterals within the FN (see also Fig.6B). In conclusion, our data reveal that vMNs are not synaptically connected to each other and that they are not presynaptic to any other neuron in the FN. However, the increase in the number of rabies-positive neurons within the FN, but outside the ventrolateral part – in particular from third- to fourth-order trans-synaptic spread – suggests that the lack of local synaptic connectivity may however not generalize from vMNs to all neurons within the FN.
Morphological variability of vMNs may translate into functional variability
Within the population of vMNs, we found substantial variability in dendritic morphologies within the same animal. The overlap between dendritic fields ranged from ∼10 to more than 70%. Moreover, dendrites of the reconstructed vMNs terminated at different depths within the FN and displayed dendritic fields that overlapped with the slabs in a cell-specific manner. These data suggest that vMNs – even when innervating the same muscle – are likely to receive different synaptic input patterns. For example, axonal projection patterns from the different brain stem nuclei that provide input to vMNs may innervate the FN in a slab-specific manner. If this would be the case, synaptic input could depend on the degree of dendrite overlap with these axonal distributions, resulting in cell-specific synaptic input patterns (Takatoh et al., 2013). This interpretation is consistent with our observations from mapping all second-order neurons throughout the brain stem that are putatively presynaptic to the reconstructed vMNs. We found that each vMN receives mono-synaptic input from approximately 50–150 neurons, distributed across a dozen nuclei in both hemispheres of the brain stem. The presynaptic populations identified in the present study are largely in line with those reported previously in mice (Takatoh et al., 2013, Sreenivasan et al., 2015). However, we found that some brain stem nuclei are not consistently labeled across different second-order experiments. The presence and abundance of second-order rabies labeling in the various presynaptic brain stem nuclei may thus reflect the specific dendrite distributions of the vMNs that were labeled at 1st order (which may further reflect the specific muscle fibers that were located at the respective rabies virus injection sites). The variability in dendrite morphology and presumably the resultant variability in synaptic input may – at least in part – represent a structural basis for functional observations, which revealed that each vMN evokes a highly cell-specific whisker movement (Herfst and Brecht, 2008).
Conclusion
By generating a precise model of the FN geometry, we illustrate how to integrate structural data from different experimental conditions – and obtained across animals – into a common reference frame. On the example of rabies virus muscle injections, we show how the reference frame can provide quantitative measurements of the dendritic fields and synaptic input patterns of individual vMNs. The resultant analyses provide a first account of the cell-to-cell variability of those two properties, which is substantial even for vMNs that innervate the same muscle. Future studies need to investigate relationships between the morphology, synaptic input patterns and evoked whisker movements for individual vMNs. The standardized FN reference frame will facilitate such investigations. Furthermore, the FN reference frame sets the stage to generate an anatomically constraint functional model (Lang et al., 2011) of the terminal stage of whisker motor control, which may be a necessary prerequisite for revealing organizational principles of upstream circuits involved in rhythmic whisking and sensory-motor feedback.
Acknowledgments
We thank Rajeev T. Narayanan, Amir Kayvanjoo, Daniel Udvary, Arco Bast, Maksims Ivanovs and Hyoungjun Park for help with reconstructing neuron morphologies; and Dr. M. Schnell (Thomas Jefferson University) for supplying the N2c strain of rabies virus, Dr. A. Wandeler (Animal Disease Research Institute) for supplying the antibody to the rabies virus, M. Page and M. Semcheski for the development of computer programs; and Ms. M. Watach, M. Carrier, and L. Chedwick for technical assistance. Funding was provided by the Center of Advanced European Studies and Research (caesar), the Max Planck Institute for Biological Cybernetics, the Bernstein Center for Computational Neuroscience, funded by German Federal Ministry of Education and Research Grant BMBF/FKZ 01GQ1002, the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No 633428) and the Max Planck Florida Institute for Neuroscience. This work was also supported in part by funds from NIH Grants R01 NS24328 (to P.L.S.) and P40 OD010996 (to P.L.S.) and in part by a grant from the Pennsylvania Department of Health, which specifically disclaims responsibility for any analyses, interpretations, or conclusions.
Acknowledgments
Author contributions
M.O. conceived and designed the study. J.M.G. and E.S.W. carried out experiments with advice from P.L.S. M.M.S. developed analysis and registration routines. All authors performed data analysis. M.O. wrote the paper.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.neuroscience.2017.09.031.
Appendix A. Supplementary data
Supplementary material.
References
- Ashwell K.W. The adult mouse facial nerve nucleus: morphology and musculotopic organization. J Anat. 1982;135:531–538. [PMC free article] [PubMed] [Google Scholar]
- Berg R.W., Kleinfeld D. Vibrissa movement elicited by rhythmic electrical microstimulation to motor cortex in the aroused rat mimics exploratory whisking. J Neurophysiol. 2003;90:2950–2963. doi: 10.1152/jn.00511.2003. [DOI] [PubMed] [Google Scholar]
- Brecht M. What makes whiskers shake? Focus on Current flow in vibrissa motor cortex can phase-lock with exploratory rhythmic whisking in rat. J Neurophysiol. 2004;92:1265–1266. doi: 10.1152/jn.00404.2004. [DOI] [PubMed] [Google Scholar]
- Brecht M., Schneider M., Sakmann B., Margrie T.W. Whisker movements evoked by stimulation of single pyramidal cells in rat motor cortex. Nature. 2004;427:704–710. doi: 10.1038/nature02266. [DOI] [PubMed] [Google Scholar]
- Carvell G.E., Simons D.J., Lichtenstein S.H., Bryant P. Electromyographic activity of mystacial pad musculature during whisking behavior in the rat. Somatosens Mot Res. 1991;8:159–164. doi: 10.3109/08990229109144740. [DOI] [PubMed] [Google Scholar]
- Chen J.L., Carta S., Soldado-Magraner J., Schneider B.L., Helmchen F. Behaviour-dependent recruitment of long-range projection neurons in somatosensory cortex. Nature. 2013;499:336–340. doi: 10.1038/nature12236. [DOI] [PubMed] [Google Scholar]
- Coulon P., Derbin C., Kucera P., Lafay F., Prehaud C., Flamand A. Invasion of the peripheral nervous systems of adult mice by the CVS strain of rabies virus and its avirulent derivative AvO1. J Virol. 1989;63:3550–3554. doi: 10.1128/jvi.63.8.3550-3554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dercksen V.J., Hege H.C., Oberlaender M. The Filament Editor: an interactive software environment for visualization, proof-editing and analysis of 3D neuron morphology. Neuroinformatics. 2014;12:325–339. doi: 10.1007/s12021-013-9213-2. [DOI] [PubMed] [Google Scholar]
- Deschenes M., Kurnikova A., Elbaz M., Kleinfeld D. Circuits in the ventral medulla that phase-lock motoneurons for coordinated sniffing and whisking. Neural Plast. 2016:7493048. doi: 10.1155/2016/7493048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfl J. The musculature of the mystacial vibrissae of the white mouse. J Anat. 1982;135:147–154. [PMC free article] [PubMed] [Google Scholar]
- Dum R.P., Levinthal D.J., Strick P.L. Motor, cognitive, and affective areas of the cerebral cortex influence the adrenal medulla. Proc Natl Acad Sci USA. 2016;113:9922–9927. doi: 10.1073/pnas.1605044113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbesen C.L., Doron G., Lenschow C., Brecht M. Vibrissa motor cortex activity suppresses contralateral whisking behavior. Nat Neurosci. 2017;20:82–89. doi: 10.1038/nn.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger R., Dercksen V.J., Udvary D., Hege H.C., Oberlaender M. Generation of dense statistical connectomes from sparse morphological data. Front Neuroanat. 2014;8:129. doi: 10.3389/fnana.2014.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger R., Narayanan R.T., Helmstaedter M., de Kock C.P., Oberlaender M. 3D reconstruction and standardization of the rat vibrissal cortex for precise registration of single neuron morphology. PLoS Comput Biol. 2012;8:e1002837. doi: 10.1371/journal.pcbi.1002837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger R., Schmitt A.C., Wallace D.J., Sakmann B., Oberlaender M., Kerr J.N. Robustness of sensory-evoked excitation is increased by inhibitory inputs to distal apical tuft dendrites. Proc Natl Acad Sci U S A. 2015;112:14072–14077. doi: 10.1073/pnas.1518773112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmeyer D., Brecht M., Helmchen F., Petersen C.C., Poulet J.F., Staiger J.F., Luhmann H.J., Schwarz C. Barrel cortex function. Prog Neurobiol. 2013;103:3–27. doi: 10.1016/j.pneurobio.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Gao P., Bermejo R., Zeigler H.P. Whisker deafferentation and rodent whisking patterns: behavioral evidence for a central pattern generator. J Neurosci. 2001;21:5374–5380. doi: 10.1523/JNEUROSCI.21-14-05374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattox A.M., Priest C.A., Keller A. Functional circuitry involved in the regulation of whisker movements. J. Comp. Neurol. 2002;442:266–276. doi: 10.1002/cne.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herfst L.J., Brecht M. Whisker movements evoked by stimulation of single motor neurons in the facial nucleus of the rat. J Neurophysiol. 2008;99:2821–2832. doi: 10.1152/jn.01014.2007. [DOI] [PubMed] [Google Scholar]
- Hill D.N., Bermejo R., Zeigler H.P., Kleinfeld D. Biomechanics of the vibrissa motor plant in rat: rhythmic whisking consists of triphasic neuromuscular activity. J Neurosci. 2008;28:3438–3455. doi: 10.1523/JNEUROSCI.5008-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.C. Rabies pathogenesis. J Neurovirol. 2002;8:267–269. doi: 10.1080/13550280290100725. [DOI] [PubMed] [Google Scholar]
- Jin T.E., Witzemann V., Brecht M. Fiber types of the intrinsic whisker muscle and whisking behavior. J Neurosci. 2004;24:3386–3393. doi: 10.1523/JNEUROSCI.5151-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R.M., Strick P.L. Rabies as a transneuronal tracer of circuits in the central nervous system. J Neurosci Methods. 2000;103:63–71. doi: 10.1016/s0165-0270(00)00296-x. [DOI] [PubMed] [Google Scholar]
- Kelly R.M., Strick P.L. Cerebellar loops with motor cortex and prefrontal cortex of a nonhuman primate. J Neurosci. 2003;23:8432–8444. doi: 10.1523/JNEUROSCI.23-23-08432.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein B.G., Rhoades R.W. Representation of whisker follicle intrinsic musculature in the facial motor nucleus of the rat. J. Comp. Neurol. 1985;232:55–69. doi: 10.1002/cne.902320106. [DOI] [PubMed] [Google Scholar]
- Kleinfeld D., Berg R.W., O'Connor S.M. Anatomical loops and their electrical dynamics in relation to whisking by rat. Somatosens Mot Res. 1999;16:69–88. doi: 10.1080/08990229970528. [DOI] [PubMed] [Google Scholar]
- Kleinfeld D., Moore J.D., Wang F., Deschenes M. The brainstem oscillator for whisking and the case for breathing as the master clock for orofacial motor actions. Cold Spring Harb Symp Quant Biol. 2014;79:29–39. doi: 10.1101/sqb.2014.79.024794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingen Y., Conzelmann K.K., Finke S. Double-labeled rabies virus: live tracking of enveloped virus transport. J Virol. 2008;82:237–245. doi: 10.1128/JVI.01342-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang S., Dercksen V.J., Sakmann B., Oberlaender M. Simulation of signal flow in 3D reconstructions of an anatomically realistic neural network in rat vibrissal cortex. Neural Netw. 2011;24:998–1011. doi: 10.1016/j.neunet.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Levinthal D.J., Strick P.L. The motor cortex communicates with the kidney. J Neurosci. 2012;32:6726–6731. doi: 10.1523/JNEUROSCI.0406-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H.S., Egger R., Guest J.M., Foerster R., Reissl S., Oberlaender M. Cellular organization of cortical barrel columns is whisker-specific. Proc Natl Acad Sci U S A. 2013;110:19113–19118. doi: 10.1073/pnas.1312691110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen R.J., Buck C.R., Smith A.M. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- Narayanan R.T., Egger R., Johnson A.S., Mansvelder H.D., Sakmann B., de Kock C.P., Oberlaender M. Beyond Columnar Organization: Cell Type- and Target Layer-Specific Principles of Horizontal Axon Projection Patterns in Rat Vibrissal Cortex. Cereb Cortex. 2015;25:4450–4468. doi: 10.1093/cercor/bhv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlaender M., Broser P.J., Sakmann B., Hippler S. Shack-Hartmann wave front measurements in cortical tissue for deconvolution of large three-dimensional mosaic transmitted light brightfield micrographs. J Microsc. 2009;233:275–289. doi: 10.1111/j.1365-2818.2009.03118.x. [DOI] [PubMed] [Google Scholar]
- Oberlaender M., Bruno R.M., Sakmann B., Broser P.J. Transmitted light brightfield mosaic microscopy for three-dimensional tracing of single neuron morphology. J Biomed Opt. 2007;12:064029. doi: 10.1117/1.2815693. [DOI] [PubMed] [Google Scholar]
- Oberlaender M., de Kock C.P., Bruno R.M., Ramirez A., Meyer H.S., Dercksen V.J., Helmstaedter M., Sakmann B. Cell type-specific three-dimensional structure of thalamocortical circuits in a column of rat vibrissal cortex. Cereb Cortex. 2012;22:2375–2391. doi: 10.1093/cercor/bhr317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G., Watson C. Elsevier Academic Press; Amsterdam: 2005. The Rat Brain and Stereotaxic Coordinates. [Google Scholar]
- Schwarz C. The Slip Hypothesis: Tactile Perception and its Neuronal Bases. Trends Neurosci. 2016;39:449–462. doi: 10.1016/j.tins.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Sreenivasan V., Karmakar K., Rijli F.M., Petersen C.C. Parallel pathways from motor and somatosensory cortex for controlling whisker movements in mice. Eur J Neurosci. 2015;41:354–367. doi: 10.1111/ejn.12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalling D., Westerhoff M., Hege H. Academic Press, London; Visualization Handbook: 2005. Amira-an object oriented system for visual data analysis. [Google Scholar]
- Takatoh J., Nelson A., Zhou X., Bolton M.M., Ehlers M.D., Arenkiel B.R., Mooney R., Wang F. New modules are added to vibrissal premotor circuitry with the emergence of exploratory whisking. Neuron. 2013;77:346–360. doi: 10.1016/j.neuron.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Rampin O., Giuliano F., Ugolini G. Spinal and brain circuits to motoneurons of the bulbospongiosus muscle: retrograde transneuronal tracing with rabies virus. J. Comp. Neurol. 1999;414:167–192. [PubMed] [Google Scholar]
- Ugolini G. Specificity of rabies virus as a transneuronal tracer of motor networks: transfer from hypoglossal motoneurons to connected second-order and higher order central nervous system cell groups. J. Comp. Neurol. 1995;356:457–480. doi: 10.1002/cne.903560312. [DOI] [PubMed] [Google Scholar]
- Ugolini G. Use of rabies virus as a transneuronal tracer of neuronal connections: implications for the understanding of rabies pathogenesis. Dev Biol (Basel) 2008;131:493–506. [PubMed] [Google Scholar]
- Ugolini G. Advances in viral transneuronal tracing. J Neurosci Methods. 2010;194:2–20. doi: 10.1016/j.jneumeth.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Wineski L.E. Facial morphology and vibrissal movement in the golden hamster. J Morphol. 1985;183:199–217. doi: 10.1002/jmor.1051830208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.