Abstract
The quest to comprehend genetic, biological, and symptomatic heterogeneity underlying Alzheimer’s disease (AD) requires a deep understanding of mechanisms affecting complex brain systems. Neuroimaging genetics is an emerging field that provides a powerful way to analyze and characterize intermediate biological phenotypes of AD. Here, we describe recent studies showing the differential effect of genetic risk factors for AD on brain functional connectivity in cognitively normal, preclinical, prodromal, and AD dementia individuals. Functional neuroimaging genetics holds particular promise for the characterization of preclinical populations; target populations for disease prevention and modification trials. To this end, we emphasize the need for a paradigm shift towards integrative disease modeling and neuroimaging biomarker-guided precision medicine for AD and other neurodegenerative diseases.
Pathophysiology, Genetics, and Functional Brain Processing Underlying AD
AD is the most prevalent neurodegenerative disease and commonest type of dementia in people aged >65 years. Despite enormous efforts in global biomedical research and development, the number of affected individuals with AD is dramatically increasing [1]. Therefore, effective prevention and disease-modifying therapies are needed to reduce the future global burden of neurodegenerative diseases and dementia [2,3]. The genetic, biological, and symptomatic heterogeneity underlying the spectrum of AD clinical phenotypes as well as the complex non-linear progression of the pathophysiological mechanisms are key factors for a decade of failure of AD clinical trials. Once late-stage clinical symptoms appear, the disease shows extensive, advanced, and potentially irreversible neuropathological alterations – such as inflammatory changes, neuritic plaques (also called senile plaques) and neurofibrillary tangles [4] (see Glossary). An emerging exploration of the long and largely uncharted preclinical stages of AD has begun [5].
To date, the amyloid cascade theory is the prevailing hypothesis on the pathogenesis of AD [6]. It postulates that brain β-amyloid (Aβ) accumulation is the primary mechanistic event, or key pathophysiological threshold, impairing synaptic function, later inducing neuronal damage, and finally leading to widespread neurodegeneration and clinical dementia [7]. The detrimental impact of Aβ is assumed to emerge at the system level, as brain functional and structural connections are progressively disrupted (for review see [8]). Moreover, clinical decline has been associated with alterations in both structural and functional brain connectivity, causing abnormal brain integration [9]. Therefore, AD may be considered a complex brain systems disconnection syndrome [10]. However, it is still unclear which factors induce such disconnection. So far, it is largely accepted that axonal and synaptic contacts can spread dysfunction from a local site through mechanisms of diaschisis and transneuronal degeneration [11], generating pathophysiological cascades [12] and, consequently, propagating the disease processes [13]. In addition, it is possible that brain regions affected by pathophysiological events respond with compensatory mechanisms such as increased activity or functional connectivity, owing to excess neuronal stimulation, and leading to cell damage or death in functionally connected brain sites [13]. Finally, according to evidence derived from studies with AD transgenic mouse models [14], abnormal neural connectivity could arise from the slowing or interruption of the fast axonal transport, which occurs before Aβ plaques formation [14] and potentially contributes to transneuronal degeneration [15].
Resting state functional magnetic resonance imaging (rs-fMRI) studies, which assess functional synchrony in brain networks using fMRI, provide numerous findings highlighting the deep reshaping of a number of functional connectivity networks at each stage of the full clinical AD spectrum [16–19], from preclinical to prodromal to AD dementia (Box 1). These changes can occur even in the absence of cognitive impairments or structural neurodegeneration [20]. Although other networks have also been implicated, a recent review [8] reported consistently decreased functional connectivity in the default mode network (DMN) in the full clinical AD spectrum, including the posterior cingulate cortex (PCC), precuneus (Pcu), lateral temporo-parietal cortex, and the medial temporal lobe (MTL) [21]. The MTL is considered the most prominent candidate brain region for initial histopathological changes in AD [4], but the PCC is consistently recognized as one of the earliest sites showing hypometabolism and hypoperfusion [22,23]. Disrupted connectivity between the hippocampus/entorhinal cortex and PCC may perhaps constitute the first neural change in AD pathophysiology [24].
Box 1. Clinical Diagnostic Criteria – Three Meta Categories for the Global Staging of AD.
Preclinical AD: indicates the asymptomatic stage between the earliest neuropathological events and the appearance of AD-related cognitive impairments (clinical stage). Although the preclinical stage of AD represents a continuum, two in vivo preclinical states can be discerned: (i) the asymptomatic at-risk state for AD, which indicates the presence of pathophysiological markers, such as τ pathology [cerebrospinal fluid (CSF) or PET τ] or amyloid pathology (CSF Ab42 or PET amyloid); and (ii) presymptomatic AD, which refers to individuals who will certainly develop AD, because they carry rare autosomal dominant mutations that cause AD, such as APP, PSEN1, or PSEN2.
Prodromal AD (or MCI-due-to-AD): includes the presence of definite impairment in memory function, for example, measured by Free and Cued Selective Reminding Test [81], along with in vivo positive pathophysiological markers (CSF or PET τ, CSF Ab42 or PET amyloid). Instrumental activities of daily living are preserved.
AD dementia: refers to individuals presenting severe cognitive impairments that interfere with social functioning and instrumental activities of daily living. Clinical symptoms must include progressive deficits in memory and in at least one other cognitive domain, that is, executive functions, language, or visuospatial abilities. In vivo pathophysiological or topographic markers (e.g., hippocampal atrophy or cortical thickness) are supportive evidence for the diagnosis of AD dementia.
The genetic makeup has the potential to significantly and differentially modulate functional brain connectivity in normal aging and may directly interact with disease effects [25] (Box 2). Mutations in the amyloid precursor protein (APP), presenilin 1 (PSEN1) or 2 (PSEN2) genes cause early-onset AD dementia, at an unusually early age (around 30–50 years). By contrast, the risk of developing late-onset AD seems to be associated with allelic variations in apolipo-protein E (APOE), phosphatidylinositol binding clathrin assembly protein (PICALM), clusterin (CLU), and bridging integrator 1 (BIN1) genes. Consequently, these have become the most heavily investigated in functional neuroimaging genetics studies of AD [26].
Box 2. Genetic Risk Factors for AD and Their Potential Functional Connectivity Counterpart.
APOE gene: codes for APOE. Regulates amyloid-β (Aβ) oligomerization, aggregation, and receptor-mediated clearance, brain lipid transport, glucose metabolism, neuronal signaling, and neuroinflammation [26,82,83].
Potential influence on functional connectivity: (i) impaired neurite outgrowth; (ii) cytoskeletal disruption and hyperphosphorylation of τ; (iii) mitochondrial dysfunction in neurons; (iv) impaired synaptogenesis; (v) increased apoptosis in neurons; and (vi) Aβ peptide clearance and/or deposition.
PICALM gene: codes for the phosphatidylinositol binding clathrin assembly protein. Protects neurons from Aβ toxicity by reversing Aβ effects on clathrin-mediated endocytosis and/or by directing amyloid precursor protein transport to the terminal degradation pathway by autophagosomes, which reduces Aβ production [26].
Potential influence on functional connectivity: Aβ peptide clearance and/or deposition.
CLU gene: codes for clusterin. Involved in several biological and pathophysiological mechanisms, including cell death and tumor progression. Moreover, CLU assists clearance of Aβ, interacts with APOE, and promotes neuroinflammation by inhibiting complement activation [26].
Potential influence on functional connectivity: (i) impaired neurite outgrowth; (ii) impaired synaptic integration; and (iii) Aβ peptide clearance.
BIN1 gene: codes for the Bridging integrator 1. Broadly expressed in the brain, where it contributes to retrieve synaptic vesicles, apoptosis, inflammation, and clathrin-mediated Aβ [26,84].
Potential influence on functional connectivity: (i) impaired neurite outgrowth; and (ii) impaired synaptic integration.
APP gene: codes for the amyloid precursor protein. Essential for physiological brain development (neurogenesis and synaptogenesis) and plasticity [26,85].
Potential influence on functional connectivity: (i) Aβ peptide clearance and/or deposition; (ii) impaired neurite outgrowth; and (iii) impaired synaptic integration.
PSEN1 and PSEN2 genes: encode for presenilin 1 and presenilin 2. Presenilins are proteolytic subunits of γ-secretase intramembrane protease complex [26].
Potential influence on functional connectivity: (i) Aβ peptide clearance and/or deposition; (ii) impaired neurite outgrowth; (iii) impaired synaptic integration; and (iv) calcium dyshomeostasis.
Genetic studies of AD have also attempted to integrate multimodal biomarkers to better characterize and stratify populations at risk of developing AD [2]. In this regard, neuroimaging genetics might offer an efficient strategy for characterizing intermediate phenotypes of AD, helping bridge the unexplored biological gap between the cell-level molecular changes and systems-level changes in cognition and behavior. Not surprisingly, several research groups have started to explore the neural underpinnings of genotype-dependent differences in AD.
In the present review, we describe the impact of well-known genetic risk factors of AD on brain functional connectivity alterations in the whole AD spectrum, and critically discuss the key advantages of investigating functional neuroimaging genetics in AD. In particular, we present studies attempting to develop multimodal markers to detect and predict AD [27]. Indeed, to determine when and how brain functional connectivity begins to diverge from expected age-specific norms in individuals with different genetic profiles at risk for AD might be of great value both for the early AD detection and stratification of target populations in clinical trials. These metrics are assumed to be critical for developing and evaluating clinical interventions, to slow or even prevent cognitive decline. This review is focused on addressing new insights in the study of functional brain dysfunction in individuals with genetic susceptibility to AD (Box 2), since extensive literature on AD genetics [26,28] and biomarkers [29] is comprehensively reviewed elsewhere. We provide here a critical overview of recent studies that have addressed the role of AD-related genes in the functional connectivity at rest. In particular, we discuss how autosomal dominant genes APP, PSEN1, and PSEN2, and the major genome-wide associated gene risk variants for AD, that is, APOE, PICALM, CLU, and BIN1, impact resting state functional connectivity in: (i) cognitively normal (CN) individuals, (ii) preclinical AD individuals (including both asymptomatic at risk for AD and presymptomatic diagnostic categories), and (iii) AD dementia patients.
This review is restricted to addressing recent advances in examining the genetic impact on the functionally interacting and integrative networks at rest, which provide new insights on large-scale neuronal communication in the human brain.
CN Individuals at Genetic Risk for AD
Elucidating the neural changes in CN individuals at genetic risk for AD is supposed to provide several advantages: (i) different functional brain patterns in mutation carriers may be identified independently from the disease; (ii) compared to patients, CN individuals can easily perform tasks, making it possible to explore the effective connectivity related to specific cognitive tasks; (iii) the effect of genetic risk variants on brain network functioning can be examined in absence of confounding factors, for example, illness or medication; (iv) all genetic variant profiles are included in the sample; and (v) longitudinal follow-up on CN individuals at increased risk for AD would make it possible to test forms of prevention, trace pathophysiological trajectories from health to dementia, and identify an effective therapeutic window for early preclinical stages of AD.
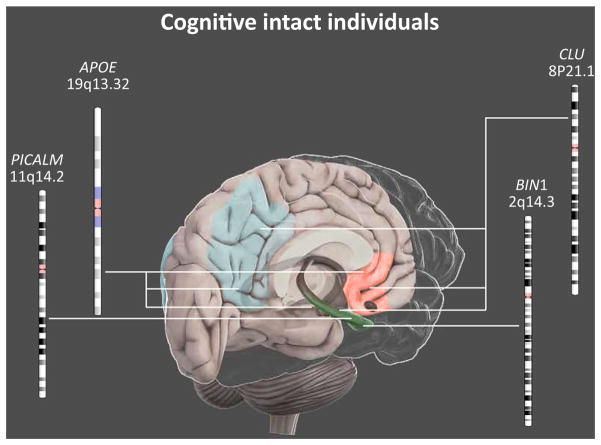
Here, we present data across the lifespan, from childhood to old age, to point out potential temporal trajectories in CN individuals carrying genetic mutations associated with AD (Figure 1) [26].
Figure 1.
Main Effects of Genetic Risk Factors for AD on Brain Functional Connectivity in Cognitively Normal Individuals. Schematic illustration of the main networks influenced by genetic variations in cognitively intact individuals. Mutations in the APOE or CLU genes affect the functional connectivity of (i) the anterior DMN (red), including the anterior cingulate and the middle prefrontal cortices; (ii) the poster DMN (blue), including the posterior cingulate cortex, the precuneus, the inferior parietal lobe, and the retrosplenial cortex; and (iii) the hippocampus (green). By contrast, BIN1 and PICALM genetic variations seem to affect essentially the hippocampal connectivity. AD, Alzheimer’s disease. This figure is a derivative of the work created by Vivid Apps and AXS Biomedical Animation Studio for Cold Spring Harbor Laboratory DNA Learning Center (https://www.dnalc.org/resources/3dbrain.html).
Given the central role of the hippocampus in AD neurodegeneration [30], considerable effort has been devoted to study its possible functional connectivity alterations early in life in CN individuals at genetic risk for AD. The influence of the innate genetic patterns on hippocampal connectivity was reported in young individuals [31–33], although results partially disagree. On the one hand, carriers of the G homozygote mutation in BIN1 [33], and the C allele polymorphism in CLU [31] both showed decreased hippocampal–dorsolateral prefrontal cortex (dlPFC) connectivity, while individuals carrying the PICALM risk genotype (G allele) showed reduced strength connectivity between the hippocampus and both the Pcu and the superior frontal gyrus [32]. On the other hand, increased hippocampal connectivity with widespread DMN regions was found in young CLU-C [32] and APOE ε4 carriers [34,35]. Such hippocampal hyperconnectivity was assumed to reflect a compensatory brain response to decreased white matter connections [36,37] and may predict future cognitive decline [38–40]. As hippocampal subfields exhibit specific functional connections [41,42], considering the entire hippocampus may be a major methodological limitation of the above studies. In this regard, Trachtenberg and colleagues [43] reported differences in the anterior hippocampal network (AHN) and posterior hippocampal network (PHN). Hippocampal subfields exhibit specific functional connections, and in line with this, the APOE ε4 genotype more severely affects the connectivity of the AHN rather than the PHN [43]. In particular, the APOE ε4 genotype may more severely affect the connectivity of the AHN rather than PHN [43]. In line with this remark, a variety of parietal and frontal regions, and the basal ganglia, displayed increased connectivity with the AHN and decreased connectivity with the PHN in young CN APOE ε4 carriers. This pattern was recently replicated during memory tasks in a fMRI study with a sample of middle-aged individuals (mean age, 65 years) [44]. Interestingly, only individuals from older adult communities, care centers, and memory clinic groups were included, to increase the chance of recruiting participants with age-related memory concerns and with an increased likelihood of at least one copy of the APOE ε4 allele. There may also be an APOE ε4 ×gender interaction on the DMN [45,46]. Compared to males, female APOE ε4 carriers exhibited reduced functional connectivity of the hippocampus with the posterior regions of DMN (Pcu and PCC) [45]. Further testing revealed a significant interaction between APOE genotype and sex in the precuneus, a major DMN hub [45,46]. The study by Damoiseaux and colleagues revealed lower DMN connectivity in female ε4 carriers compared to either female ε3 homozygotes or male ε4 carriers, whereas males carrying the ε4 phenotype were marginally different from ε3 homozygote males [46].
After extending the analyses of functional brain connectivity at rest in CN middle-aged APOE ε4 carriers to different areas of the DMN, a highly consistent pattern emerged. On the one hand, decreased DMN connectivity was detected in the PCC/Pcu and orbital frontal cortex [47,48]; on the other hand, increased DMN connectivity was found in MTL and PFC structures [47,48]. Almost overlapping results were observed in elderly APOE ε4 carriers [49–53], even before the onset of brain amyloid accumulation processes [20,48].
Nevertheless, the inclusion of both middle-aged adults and elderly in the same sample generated conflicting results: both decreased [51] and increased [35,52] connectivity were found in a number of DMN nodes, including MTL, PCC, and Pcu.
The fact that both decreased and increased functional connectivity were found at rest might be due to differences in methods and analyses, such as the choice of seed region of interest (ROI) derived from an event-related fMRI task [52], independent component analysis (ICA) [35], or graph measures [51]. Further investigations are needed to clarify these discrepancies.
It should be highlighted that, as age increased, ε2 carriers presented a grown DMN functional connectivity, while this was decreased in ε4 carriers [54]. This finding corroborates the hypothesis of antagonistic pleiotropic properties of the APOE ε4 allele, stating that APOE ε4 carriers may enjoy some cognitive benefits during early life, but exhibit impaired brain function in late adulthood [55].
Further analyses revealed that differences in individuals carrying the APOE ε4 allele are not only limited to the DMN. Young adult APOE ε4 carriers showed increased functional connectivity in the sensorimotor network [34] and decreased connectivity between the auditory network and several other brain regions in the frontal, temporal, and parietal cortices, as well as in the basal ganglia [43]. Furthermore, elderly APOE ε4 carriers displayed increased connectivity in the salience network, which is comprised of the dorsal anterior cingulate cortex (dACC), the frontoinsular cortices and subcortical and limbic regions [49,53]. Again, a number of additional brain regions, not typically involved in AD, such as the dorsal occipital cortex and the frontoparietal operculum, showed differences in functional connectivity in CN APOE ε4 carriers compared to non-carriers [50]. The dissimilarities previously described may reflect supplementary effects – either genetically mediated during brain neurodevelopment or caused by an early low degree of amyloid deposition not yet detectable by positron emission tomography (PET) scanning. Indeed, recent studies demonstrated significant associations between τ PET uptake or τ protein concentrations in CSF and alterations in functional connectivity [56,57]. Therefore, investigation of inter-systems dynamics is warranted, such as the interplay of the genetic, molecular, and functional associations is warranted.
In conclusion, existing evidence describes early detectable brain functional connectivity patterns in CN individuals carrying BIN1, CLU-C, PICALM, and APOE genetic polymorphisms that highly correlate with the functional imaging markers found in AD. In particular, neural changes detected in young carriers may trigger late life functional differences.
Preclinical AD
According to the International Working Group (IWG)-2 diagnostic research criteria, individuals carrying an autosomal dominant AD mutation with virtually full penetrance, that is, APP, PSEN1, or PSEN2 mutations, are defined as presymptomatic AD, as they inevitably develop neurodegenerative signs [58].
Functional brain connectivity in individuals with PSEN1 mutations was recently investigated in children (9–17 years old) with altered blood-based and brain imaging biomarkers. Notably, they showed an increased brain activity in parietal regions during a memory tasks and increased rs-fMRI functional connectivity between PCC and MTL regions [59]. Accordingly, young (18–30 years old, [60]) and middle-aged presymptomatic individuals (mean age, 45 years [61–63]) displayed lower intrinsic connectivity in posterior [60–63], and temporal [62] nodes of the DMN compared with controls. Significant correlations were observed between rs-fMRI measures (Z scores) and CSF Aβ42, P-τ181p, and T-τ protein concentrations [63]. Alterations in young and middle-age adults were also observed in frontal regions; however, results are still debated because of decreased [60] as well as increased DMN connectivity [62] results. The heterogeneity of evidence in presymptomatic adults indicates that there is no simple interpretation of autosomal dominant-related changes in resting state functional connectivity. Explanations for such findings may include: (i) compensatory responses related to individual cognitive reserves; (ii) aging-related developmental modifications in the brain networks architecture, independent of the genetic pattern; (iii) interaction with other genes; (iv) neurotransmitter failure; and (v) differential impact on brain function of the different Mendelian AD mutations on brain function. Overall, these data indicate the presence of a relevant genetic impact on functional connectivity due to APP or PSEN1/2 mutations.
Interestingly, reduced DMN functional connectivity, as detected in individuals carrying autosomal dominant mutations, does not differ from the one observed in APOE ε4 carriers [60].
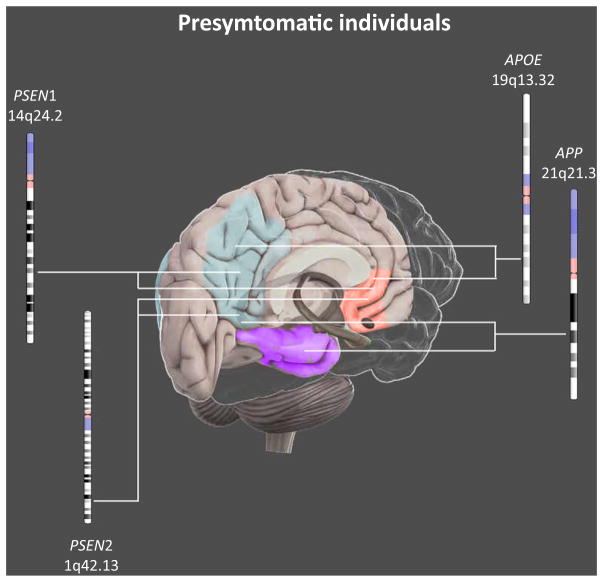
Overall, findings in presymptomatic AD individuals suggest that abnormalities in resting-state networks potentially represent a valuable biomarker to detect early preclinical stages of AD (Figure 2).
Figure 2.
Main Effects of Genetic Risk Factors for AD on Brain Functional Connectivity in Presymptomatic Individuals. In presymptomatic individuals, genetic effects of APP, PSEN1, PSEN2, and APOE were shown in the resting-state functional connectivity of the posterior DMN (blue). In addition, while APP, PSEN2, and APOE influence the anterior DMN (red), PSEN1 mutations affect the temporal lobe (purple). APOE variants affect functional connectivity as well, in sensorimotor, auditory and salience networks (not shown). AD, Alzheimer’s disease. This figure is a derivative of the work created by Vivid Apps and AXS Biomedical Animation Studio for Cold Spring Harbor Laboratory DNA Learning Center (https://www.dnalc.org/resources/3dbrain.html).
To the extent of the existing knowledge, the influence of genetics on the functional architecture in the asymptomatic at-risk state for AD [58], that is, CN individuals showing positivity to AD pathophysiological markers, has yet not been examined.
Patients with AD Dementia
To date, no published studies have identified effects of specific genotypes on functional connectivity patterns in patients with prodromal AD [52] or with mild cognitive impairment (MCI) due to AD [57], that is, MCI individuals with a positive core biomarker signature positive, who have a high likelihood of progressing to AD dementia within a few years.
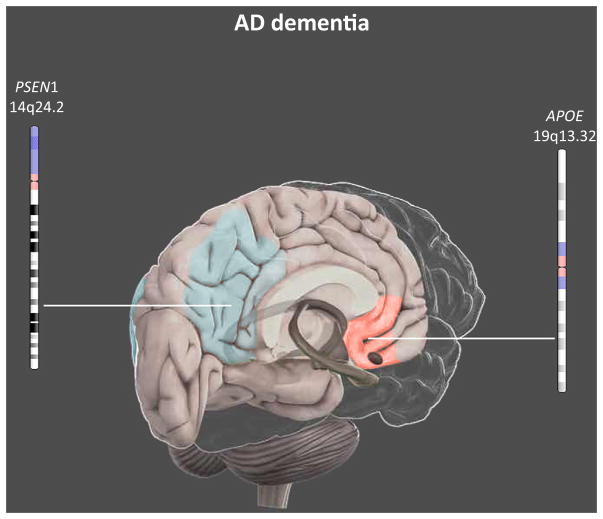
The substantial effects of the APOE ε4 allele on the intrinsic functional architecture have been reported in patients with AD dementia (Figure 3). Specifically, AD demented APOE ε4 carriers exhibited a selective weakness in both intra- and internetwork integration that predominantly resided in the posterior part of the DMN [22,23] and in the executive control network [23]. However, significant results of APOE ε4 effect on the DMN are not consistently reported [22,64,65]. This gap may originate from the high degree of sporadic AD complexity and heterogeneity, which potentially may involve different biological and neurophysiological systems at different levels. For instance, familial autosomal dominant AD individuals with PSEN1 mutations have shown strong decreased frontal connectivity; by contrast, results observed in posterior networks were unclear [61,62].
Figure 3.
Main Effects of Genetic Risk Factors for AD on Brain Functional Connectivity in AD Dementia Individuals. Neuroimaging genetics results in AD dementia patients are still controversial. However, functional alterations at rest resulted in the posterior DMN (blue) in AD diseased individuals with PSEN1 mutations, and in the anterior DMN (red) in APOE ε 4 carriers. Abbreviation: AD, Alzheimer’s disease. This figure is a derivative of the work created by Vivid Apps and AXS Biomedical Animation Studio for Cold Spring Harbor Laboratory DNA Learning Center (https://www.dnalc.org/resources/3dbrain.html).
In conclusion, these findings further support the belief that differences in genetic predispositions could differentially impact on brain function during cellular/molecular pathophysiological stages. Additional research on the interaction among genetics, biology, and environmental factors as well as their influence on brain functional connectivity in AD needs to be conducted.
Genetics of Brain Biomarkers
Some of the issues related to explicate the functional effects of AD risk genotypes in the brain may be addressed by exploiting large consortia linking the areas of neuroimaging and genetics. The use of genome-wide association studies led to identification of >20 genetic susceptibility loci in AD versus CN individuals [66]. In this regard, the Enhancing Neuro Imaging Genetics through Meta-Analysis (ENIGMA) consortium [67–69] (http://enigma.ini.usc.edu/) has recently discovered >20 genetic loci that are consistently associated with brain structural MRI-based measures, in >30 000 individuals worldwide. Loci affecting the risk for neurodegenerative diseases overlap substantially with those affecting brain markers. The authors found that the microtubule associated protein τ gene (MAPT), which is related to Parkinson’s disease, contains polymorphic loci that appear to boost intracranial volume early in life [70]. Similarly, the APOE genotype showed a gradually increasing effect on hippocampal volume ranging from minimal effects in young adults to strong effects in old age [70]. Such evidence supports the antagonistic pleiotropy that some genetic risk factors for neurodegenerative diseases may have a positive influence early in life. Efforts to harmonize functional connectivity phenotypes worldwide should soon reveal whether functional networks implicated in AD show similar or different genetic effects to those seen for structural markers of AD. In this regard, normative data compiled over the lifespan will be useful to stratify into groups with different profiles of genetic risk, as the ENIGMA consortium has done for structural MRI measures. A second benefit of large genetic consortia is their ability to determine the reproducibility of effects in cohorts worldwide. This is crucial as claims of genetic effects in one cohort may not always persist when tested more generally (see, e.g., [71] for an analysis, in >6000 individuals, of genetic markers claimed to affect white matter integrity assessed with diffusion MRI).
Concluding Remarks and Future Perspectives
Overall, evidence is building that several genes associated with AD risk are able to differentially disrupt brain functional connectivity at rest in CN, presymptomatic, and symptomatic AD individuals [72]. Such neural differences are detectable in CN mutation carriers of APOE, PICALM, CLU, and BIN1 genes across the lifespan. Relatively consistent at-rest functional neuroimaging data show decreased connectivity in the middle and posterior DMN regions, including PCC and Pcu, and increased DMN connectivity in the frontal and lateral structures, such as the middle temporal and the prefrontal cortices. Additional functional connectivity alterations associated with the APOE polymorphism were identified in the salience [49] and auditory [43] systems. Accordingly, presymptomatic AD individuals exhibited abnormalities in the DMN [61,62], even at a young age [59,60]. By contrast, significant results were not consistently reported in symptomatic AD dementia patients [61,62], despite two studies reporting a selective alteration of the DMN [22,23] and the executive control network [23].
As a result, existent findings seem to converge in proposing a substantial, although not conclusive, relationship between genetics and functional brain networks in the AD clinical spectrum. However, caution in interpreting the reliability of the outcomes is warranted since large replication studies need to be conducted.
Notably, no direct genetic effect on neural networks was measured in the above-reported studies. Indeed, while they investigated genetic predisposition at the level of polymorphic markers in the genome, complementary data should be produced to identify the gene expression in the known AD functionally-related networks (see Outstanding Questions). In this regard, Richiardi and colleagues [25] indicated a set of 136 genes exhibiting well-orchestrated fluctuations in their expression levels across networks, in healthy adolescents. From a molecular viewpoint, these genes are strictly related to ion channel activity, neurotransmitters, and synaptic function, thus suggesting an intrinsic association of brain functional connectivity with complex synaptic mechanisms. Given the evident convergence of such multimodal dimensions in healthy young individuals, a key future perspective is to define gene expression profiles related to nonpathological variations in structural and functional connectivity networks in CN older adults. Secondly, patterns of altered functional connectivity networks need to be identified in clinical and preclinical cohorts, such as presymptomatic and asymptomatic at-risk for AD individuals (amyloid positive) compared with CN age-matched older controls (amyloid negative). Eventually, the trend in neuroimaging genetics will be to embrace novel approaches, such as the concept of genome-wide association coupled with high-throughput functional neuroimaging [73], or even genome-wide connectome-wide screening [74] to disclose complex genetic traits in CN individuals and across the full AD spectrum.
Outstanding Questions.
Evaluating whether functional connectivity at rest can be developed as a reliable biological marker for establishing and improving early AD stratification, detection, and diagnosis.
Investigating reasons why functional connectivity at rest is decreased in the posterior part of DMN and increased in the anterior part in individuals with increased genetic risk for AD.
Exploring mechanisms whereby the detrimental deposition of proteins in AD, for example, Aβ and hyperphosphorylated τ proteins, spreads to interconnected hub regions.
Elucidating whether the detrimental Aβ deposits cause the functional network alterations or vice versa.
Examining whether (and how) sex, lifestyle, and genetic predisposition interact to affect functional brain networks in preclinical stages of AD.
Compiling large functional neuroimaging genetics data over the lifespan, to stratify the population into subsets of individuals with different genetic risk profiles and functional connectivity patterns, and to assess the reproducibility of the outcomes in cohorts worldwide.
The final goal in AD translational bench-to-bedside-to-bench (reverse translation) research is to develop multimodal neuroimaging-genetic-driven personalized signatures and screenings to enable the development of customized and biomarker-guided targeted therapies, thus improving patient care [3,75]. Recent years have witnessed substantial achievements in biomarker-guided therapeutic strategies in more advanced translational research areas of biomedicine, such as oncology and cardiovascular medicine [76,77]. This path to the paradigm of precision medicine (PM) for detecting, treating, and preventing complex multifactorial neurodegenerative diseases, including AD, will likely transform and revolutionize neurology, psychiatry, and neuroscience via breakthrough advances in sensitive, specific and integrated genomic/epi-genomic, neuroimaging and biofluid biomarker screening, biological staging and patient subset stratification, and earliest biological detection of pathophysiological mechanisms [2,3,78,79]. This will allow both early prevention [79,80] and, ultimately, successful development of combinatorial disease-modifying treatments based on the individuals genetic and pathophysiological profile [76,77].
Significant advances in drug discovery and development programs are still substantially limited by the traditional ‘one-drug-fits-all’ approach, which reductionistically categorizes the continuous genetically and biologically heterogeneous spectrum of different neurodegenerative diseases, including polygenic AD, as hypothesized homogeneous clinicopathological or clinicobiological entities. By contrast, the emerging PM paradigm aims to overcome these historically grown challenges, notably the reductionistic clinically descriptive disease categories [76,77]. Notably, the PM strategy will facilitate a paradigm shift in AD and other neurodegenerative diseases away from the outdated “one-size-fits-all” approach in drug discovery, towards (i) biomarker-guided, molecularly tailored therapies for precise and effective treatment of molecular pathophysiological pathways associated with AD; and (ii) prevention options [76,77,80]. As a result, next-generation neurologists and psychiatrists (as the oncologist today), supported by interdisciplinary colleagues, for example, geneticists, neurochemists, neuro-radiologists, neuropsychologists, together with data science specialists and biostatisticians, will be able to precisely deliver biomarker-guided, targeted and timed interventions adapted to the genetic and biological profiles of individuals at the preclinical stage of AD and other neurodegenerative diseases. Currently, this objective has been conceptualized and operationalized by the international pilot Alzheimer Precision Medicine Initiative Cohort Program (APMI-CP) [76,77].
According to the interdisciplinary and translational systems theory – allowing the implementation of novel and original models to elucidate all brain systems levels – and the PM paradigm, genetically and biologically distinct AD individuals may develop and display converging and/or overlapping clinical phenotypes with distinct combinations of underlying structural and functional neuroimaging genetics patterns that may be subject to dynamic variations across all different stages of the chronically evolving disease spectrum [3,78,79]. As a result, integrating functional brain indices as dynamic biological markers – through integrative disease modeling [76,77] – will complete and further enhance and differentiate the early identification of disease systems endophenotypes [76,77].
Trends.
Neural rs-fMRI differences are detectable in CN mutation carriers of APOE, PICALM, CLU, and BIN1 genes across the lifespan.
CN individuals carrying risk variants of APOE, PICALM, CLU, or BIN1 and presymptomatic AD individuals showed overlapping rs-fMRI alterations: decreased functional connectivity in the middle and posterior DMN regions, and increased the frontal and lateral DMN areas.
No consistent results were reported in AD dementia, despite findings suggest selective alterations in the DMN and in the executive control network.
Multimodal biomarker data – including distinct combinations of underlying functional neuroimaging genetics patterns – are standardized and integrated according to the integrative disease modeling (IDM) concept. The PM paradigm applies IDM to translate biomarker-indicated, individual-specific, molecular pathophysiological mechanisms into tailored clinical applications.
Acknowledgments
HH is supported by the AXA Research Fund, the Fondation Université Pierre et Marie Curie and the Fondation pour la Recherche sur Alzheimer, Paris, France. The research leading to these results has received funding from the program ‘Investissements d’avenir’ ANR-10-IAIHU-06 (HH). PT is supported in part by NIH grant U54 EB040203 to the ENIGMA Center for Worldwide Medicine, Imaging and Genomics. The authors wish to thank Dr. Michael Greicius, Associate Professor of Neurology and Neurological Sciences at Stanford University, for critically reading, editing, and discussing our manuscript.
Glossary
- Amyloid β (Aβ)
denotes peptides of different length in terms of amino acids. The 42-amino-acid Aβ peptide (Aβ1–42) is the major component of the neuritic plaques in AD brains and the core biochemical marker for the amyloidogenic process in AD. It derives from the pathological cleavage of APP.
- Diaschisis
a functional interruption of regions remote from the initial insult, caused by the deafferentation of excitatory inputs.
- Functional connectivity
the statistically synchronized or temporal coherent blood oxygen level-dependent (BOLD) activity of remote brain regions that thus share a common functional specialization.
- Functional neuroimaging genetics
identifies genes that contribute to functional alterations in brain networks.
- Integrative disease modeling
is a multidisciplinary approach, which aims to standardize, manage, integrate, and interpret multiple biological quantitative and qualitative data, by applying computational models to support decision-making for translation of patient-specific molecular mechanisms into tailored clinical applications.
- Intermediate phenotype
often referred to as an endophenotype, is a stable phenotype with a clear genetic connection.
- Neuritic plaques
abnormal extracellular deposits primarily composed of Aβ peptides in the grey matter of the brain, also named senile plaques.
- Neurofibrillary tangles
intracellular aggregates of hyperphosphorylated τ proteins. They are generated by the excessive phosphorylation (hyperphosphorylation) of a microtubule-associated protein known as τ, causing it to aggregate in an insoluble form.
- Neuroimaging genetics
methodological approach applied to understand brain structure, function and disease, based on brain imaging modalities and genetic data.
- Precision medicine
biomarker-guided approach based on systems levels that include methodological advancements and findings of wide-ranging pathophysiological profiles of complex, multifactorial neurodegenerative diseases, such as AD. This may allow us to identify and characterize the pathophysiological processes at the preclinical stages, before clinical symptoms appear.
- Resting state functional magnetic resonance imaging (rs-fMRI)
neuroimaging procedure for evaluating synchronous fluctuations of signal intensities across brain regions showing a high degree of temporal correlation, while participants lay with their eyes closed or fix on a visual cue, without performing explicit tasks.
- Structural connectivity
anatomical connections of physical white matter tracts.
- Transneuronal degeneration
process that evolves over time consisting of a progressive structural deterioration of areas remote from the injured site. The damage might first occur in a postsynaptic target, reducing the trophic support to the presynaptic neuron (retrograde), or, alternatively, one neuron may cause the degeneration of its postsynaptic target (anterograde).
References
- 1.Reitz C, et al. Epidemiology of Alzheimer disease. Nat Publ Gr. 2011;7:137–152. doi: 10.1038/nrneurol.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hampel H, et al. Biomarkers for Alzheimer’s disease: academic, industry and regulatory perspectives. Nat Rev Drug Discov. 2010;9:560–574. doi: 10.1038/nrd3115. [DOI] [PubMed] [Google Scholar]
- 3.Hampel H, et al. Development of biomarkers to chart all Alzheimer’s disease stages: the royal road to cutting the therapeutic Gordian Knot. Alzheimer’s Dement. 2012;8:312–336. doi: 10.1016/j.jalz.2012.05.2116. [DOI] [PubMed] [Google Scholar]
- 4.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 5.Dubois B, et al. Preclinical Alzheimer’s disease: definition, natural history, and diagnostic criteria. Alzheimer’s Dement. 2016;12:292–323. doi: 10.1016/j.jalz.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardy JA, Higgins GA. Alzheimer’s Disease: the amyloid cascade hypothesis. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 7.Blennow K, et al. Amyloid biomarkers in Alzheimer’s disease. Trends Pharmacol Sci. 2015;36:297–309. doi: 10.1016/j.tips.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Pievani M, et al. Brain connectivity in neurodegenerative diseases – from phenotype to proteinopathy. Nat Rev Neurol. 2014;10:620–633. doi: 10.1038/nrneurol.2014.178. [DOI] [PubMed] [Google Scholar]
- 9.Matthews PM, et al. Brain structural and functional connectivity and the progression of neuropathology in Alzheimer’s disease. J Alzheimer’s Dis. 2013;33:S163–S172. doi: 10.3233/JAD-2012-129012. [DOI] [PubMed] [Google Scholar]
- 10.Brier MR, et al. Network dysfunction in Alzheimer’s disease: refining the disconnection hypothesis. Brain Connect. 2014;4:299–311. doi: 10.1089/brain.2014.0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fornito A, et al. The connectomics of brain disorders. Nat Rev Neurosci. 2015;16:159–172. doi: 10.1038/nrn3901. [DOI] [PubMed] [Google Scholar]
- 12.Klupp E, et al. In Alzheimer’s disease, hypometabolism in low-amyloid brain regions may be a functional consequence of pathologies in connected brain regions. Brain Connect. 2014;4:371–383. doi: 10.1089/brain.2013.0212. [DOI] [PubMed] [Google Scholar]
- 13.Wu JW, et al. Neuronal activity enhances tau propagation and tau pathology in vivo. Nat Neurosci. 2016;19:1085–1092. doi: 10.1038/nn.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith KDB, et al. In vivo axonal transport rates decrease in a mouse model of Alzheimer’s disease. Neuroimage. 2007;35:1401–1408. doi: 10.1016/j.neuroimage.2007.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartzokis G. Alzheimer’s disease as homeostatic responses to age-related myelin breakdown. Neurobiol Aging. 2011;32:1341–1371. doi: 10.1016/j.neurobiolaging.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Celone KA, et al. Alterations in memory networks in mild cognitive impairment and Alzheimer’s disease: an independent component analysis. J Neurosci. 2006;26:10222–10231. doi: 10.1523/JNEUROSCI.2250-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buckner RL, et al. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer’s disease. J Neurosci. 2009;29:1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Damoiseaux JS, et al. Functional connectivity tracks clinical deterioration in Alzheimer’s disease. Neurobiol Aging. 2012;33:1–20. doi: 10.1016/j.neurobiolaging.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bokde ALW, et al. Assessing neuronal networks: understanding Alzheimer’s disease. Prog Neurobiol. 2009;89:125–133. doi: 10.1016/j.pneurobio.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Thomas JB, et al. Functional connectivity in autosomal dominant and late-onset Alzheimer disease. JAMA Neurol. 2014;71:1111. doi: 10.1001/jamaneurol.2014.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raichle ME. The brain’s default mode network. Annu Rev Neurosci. 2015;38:433–447. doi: 10.1146/annurev-neuro-071013-014030. [DOI] [PubMed] [Google Scholar]
- 22.Jones DT, et al. Cascading network failure across the Alzheimer’s disease spectrum. Brain. 2015;139:547–562. doi: 10.1093/brain/awv338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang GZ, et al. Correspondence between resting-state activity and brain gene expression. Neuron. 2015;88:659–666. doi: 10.1016/j.neuron.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greicius MD, et al. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci U S A. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richiardi J, et al. Brain networks:Correlated gene expression supports synchronous activity in brain networks. Science. 2015;348:1241–1244. doi: 10.1126/science.1255905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karch CM, Goate AM. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77:43–51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertram L. The role of genetics for biomarker development in neurodegeneration. Progr Neurobiol. 2011;95:501–504. doi: 10.1016/j.pneurobio.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 28.Bettens K, et al. Genetic insights in Alzheimer’s disease. Lancet Neurol. 2013;12:92–104. doi: 10.1016/S1474-4422(12)70259-4. [DOI] [PubMed] [Google Scholar]
- 29.Lista S, et al. Biomarkers in sporadic and familial Alzheimer’s disease. J Alzheimer’s Dis. 2015;47:291–317. doi: 10.3233/JAD-143006. [DOI] [PubMed] [Google Scholar]
- 30.Beason-Held LL. Dementia and the default mode. Curr Alzheimer Res. 2011;8:361–365. doi: 10.2174/156720511795745294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erk S, et al. Hippocampal function in healthy carriers of the CLU Alzheimer’s disease risk variant. J Neurosci. 2011;31:18180–18184. doi: 10.1523/JNEUROSCI.4960-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang P, et al. Impacts of PICALM and CLU variants associated with Alzheimer’s disease on the functional connectivity of the hippocampus in healthy young adults. Brain Struct Funct. 2015;220:1463–1475. doi: 10.1007/s00429-014-0738-4. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, et al. Bridging integrator 1 (BIN1) genotype effects on working memory, hippocampal volume, and functional connectivity in young healthy individuals. Neuropsychopharmacology. 2015;40:1794–1803. doi: 10.1038/npp.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filippini N, et al. Distinct patterns of brain activity in young carriers of the APOE-ε4 allele. Proc Nat Acad Sci U S A. 2009;106:7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westlye ET, et al. Increased hippocampal default mode synchronization during rest in middle-aged and elderly APOE ε4 carriers: relationships with memory performance. J Neurosci. 2011;31:7775–7783. doi: 10.1523/JNEUROSCI.1230-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braskie MN, et al. Common Alzheimer’s disease risk variant within the CLU gene affects white matter microstructure in young adults. J Neurosci. 2011;31:6764–6770. doi: 10.1523/JNEUROSCI.5794-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohannim O, et al. Predicting White Matter Integrity from Multiple Common Genetic Variants. Neuropsychopharmacology. 2012;37:2012–2019. doi: 10.1038/npp.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Brien JL, et al. Longitudinal fMRI in elderly reveals loss of hippocampal activation with clinical decline. Neurology. 2010;74:1969–1976. doi: 10.1212/WNL.0b013e3181e3966e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sweet RA, et al. Effect of Alzheimer’s disease risk genes on trajectories of cognitive function in the cardiovascular health study. Am J Psychiatry. 2012;169:954–962. doi: 10.1176/appi.ajp.2012.11121815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thambisetty M, et al. Alzheimer risk variant CLU and brain function during aging. Biol Psychiatry. 2013;73:399–405. doi: 10.1016/j.biopsych.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strange BA, et al. Functional organization of the hippocampal longitudinal axis. Nat Rev Neurosci. 2014;15:655–669. doi: 10.1038/nrn3785. [DOI] [PubMed] [Google Scholar]
- 42.Fanselow MS, Dong HW. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron. 2010;65:7–19. doi: 10.1016/j.neuron.2009.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trachtenberg AJ, et al. The effects of APOE on the functional architecture of the resting brain. Neuroimage. 2012;59:565–572. doi: 10.1016/j.neuroimage.2011.07.059. [DOI] [PubMed] [Google Scholar]
- 44.Harrison TM, et al. Altered memory-related functional connectivity of the anterior and posterior hippocampus in older adults at increased genetic risk for Alzheimer’s disease. Hum Brain Mapp. 2016;37:366–380. doi: 10.1002/hbm.23036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heise V, et al. Apolipoprotein E genotype, gender and age modulate connectivity of the hippocampus in healthy adults. Neuroimage. 2014;98:23–30. doi: 10.1016/j.neuroimage.2014.04.081. [DOI] [PubMed] [Google Scholar]
- 46.Damoiseaux JS, et al. Gender modulates the APOE4 effect in healthy older adults: convergent evidence from functional brain connectivity and spinal fluid tau levels. J Neurosci. 2012;32:8254–8262. doi: 10.1523/JNEUROSCI.0305-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fleisher AS, et al. Resting-state BOLD networks versus task-associated functional MRI for distinguishing Alzheimer’s disease risk groups. Neuroimage. 2009;47:1678–1690. doi: 10.1016/j.neuroimage.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patel KT, et al. Default mode network activity and white matter integrity in healthy middle-aged ApoE4 carriers. Brain Imaging Behav. 2013;7:60–67. doi: 10.1007/s11682-012-9187-y. [DOI] [PubMed] [Google Scholar]
- 49.Machulda MM. Effect of APOE ε4 status on intrinsic network connectivity in cognitively normal elderly subjects. Arch Neurol. 2011;68:1131–1136. doi: 10.1001/archneurol.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheline YI, et al. APOE4 allele disrupts resting state fMRI connectivity in the absence of amyloid plaques or decreased CSF Aβ42. J Neurosci. 2010;30:17035–17040. doi: 10.1523/JNEUROSCI.3987-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y, et al. Disrupted functional and structural networks in cognitively normal elderly subjects with the APOE ε4 allele. Neuropsychopharmacology. 2014;40:1–31. doi: 10.1038/npp.2014.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matura S, et al. Recognition memory is associated with altered resting-state functional connectivity in people at genetic risk for Alzheimer’s disease. Eur J Neurosci. 2014;40:1–8. doi: 10.1111/ejn.12659. [DOI] [PubMed] [Google Scholar]
- 53.Liang Y, et al. Frequency specific effects of ApoE ε4 allele on resting-state networks in nondemented elders. 2017 doi: 10.1155/2017/9823501. https://doi.org/10.1155/2017/9823501. [DOI] [PMC free article] [PubMed]
- 54.Shu H, et al. Opposite neural trajectories of apolipoprotein E ε4 and ε2 alleles with aging associated with different risks of Alzheimer’s disease. Cereb Cortex. 2016;26:1421–1429. doi: 10.1093/cercor/bhu237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tuminello ER, Duke Han S. The apolipoprotein E antagonistic pleiotropy hypothesis: review and recommendations. Res Int J Alzheimer’s Dis. 2011 doi: 10.4061/2011/726197. http://dx.doi.org/10.4061/2011/726197. [DOI] [PMC free article] [PubMed]
- 56.Schultz AP, et al. Phases of hyperconnectivity and hypoconnectivity in the default mode and salience networks track with amyloid and tau in clinically normal individuals. J Neurosci. 2017;37:4323–4331. doi: 10.1523/JNEUROSCI.3263-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sepulcre J, et al. Tau and amyloid-β proteins distinctively associate to functional network changes in the aging brain. Alzheimer’s Dement. 2017 doi: 10.1016/j.jalz.2017.02.011. http://dx.doi.org/10.1016/j.jalz.2017.02.011. [DOI] [PMC free article] [PubMed]
- 58.Dubois B, et al. Advancing research diagnostic criteria for Alzheimer’s disease: the IWG-2 criteria. Lancet Neurol. 2014;13:614–629. doi: 10.1016/S1474-4422(14)70090-0. [DOI] [PubMed] [Google Scholar]
- 59.Quiroz YT, et al. Brain imaging and blood biomarker abnormalities in children with autosomal dominant Alzheimer disease. JAMA Neurol. 2015;72:912. doi: 10.1001/jamaneurol.2015.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Su YY, et al. Lower functional connectivity of default mode network in cognitively normal young adults with mutation of APP, presenilins and APOE ε4. Brain Imaging Behav. 2017;11:818–828. doi: 10.1007/s11682-016-9556-z. [DOI] [PubMed] [Google Scholar]
- 61.Chhatwal JP, et al. Impaired default network functional connectivity in autosomal dominant Alzheimer’s disease: findings from the DIAN study. Ann Neurol. 2012;72:S40–S40. doi: 10.1212/WNL.0b013e3182a1aafe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sala-Llonch R, et al. Evolving brain functional abnormalities in psen1 mutation carriers: a resting and visual encoding FMRI study. J Alzheimer’s Dis. 2013;36:165–175. doi: 10.3233/JAD-130062. [DOI] [PubMed] [Google Scholar]
- 63.Li X, et al. The effects of gene mutations on default mode network in familial Alzheimer’s disease. J Alzheimer’s Dis. 2017;56:327–334. doi: 10.3233/JAD-160730. [DOI] [PubMed] [Google Scholar]
- 64.Koch W, et al. Diagnostic power of default mode network resting state fMRI in the detection of Alzheimer’s disease. Neurobiol Aging. 2012;33:466–478. doi: 10.1016/j.neurobiolaging.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 65.Zhao X, et al. Disrupted small-world brain networks in moderate Alzheimer’s disease: a resting-state fMRI study. PLoS One. 2012;7:e33540. doi: 10.1371/journal.pone.0033540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hibar DP, et al. Novel genetic loci associated with hippocampal volume. Nat Commun. 2017;8:13624. doi: 10.1038/ncomms13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hibar DP, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–229. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson PM, et al. ENIGMA and the individual: predicting factors that affect the brain in 35 countries worldwide. Neuroimage. 2016;145:389–408. doi: 10.1016/j.neuroimage.2015.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adams HHH, et al. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat Neurosci. 2016;19:1569–1582. doi: 10.1038/nn.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jahanshad N, et al. Do candidate genes affect the brain’s white matter microstructure? Large-scale evaluation of 6,165 diffusion MRI scans. bioRxiv. 2017 http://dx.doi.org/10.1101/107987.
- 72.Sanchez-Mut JV, et al. Human DNA methylomes of neurodegenerative diseases show common epigenomic patterns. Transl Psychiatry. 2016;6:e718. doi: 10.1038/tp.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thompson PM, et al. Genetics of the connectome. Neuroimage. 2013;80:475–488. doi: 10.1016/j.neuroimage.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jahanshad N, et al. Genome-wide scan of healthy human connectome discovers SPON1 gene variant influencing dementia severity. Proc Natl Acad Sci U S A. 2013;110:4768–4773. doi: 10.1073/pnas.1216206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hampel H, et al. Recent developments of functional magnetic resonance imaging research for drug development in Alzheimer’s disease. Prog Neurobiol. 2011;95:570–578. doi: 10.1016/j.pneurobio.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 76.Hampel H, et al. Precision medicine – the golden gate for detection, treatment and prevention of Alzheimer’s disease. J Prev Alzheimer’s Dis. 2016;3:243–259. doi: 10.14283/jpad.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hampel H, et al. A precision medicine initiative for Alzheimer’s disease: the road ahead to biomarker-guided integrative disease modeling. Climacteric. 2017;20:107–118. doi: 10.1080/13697137.2017.1287866. [DOI] [PubMed] [Google Scholar]
- 78.Ewers M, et al. Staging Alzheimer’s disease progression with multimodality neuroimaging. Prog Neurobiol. 2011;95:535–546. doi: 10.1016/j.pneurobio.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Teipel SJ, et al. Relevance of magnetic resonance imaging for early detection and diagnosis of Alzheimer disease. Med Clin NA. 2013;97:399–424. doi: 10.1016/j.mcna.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 80.Ewers M, et al. Neuroimaging markers for the prediction and early diagnosis of Alzheimer’s disease dementia. Trends Neurosci. 2011;34:430–442. doi: 10.1016/j.tins.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grober E, et al. Free and cued selective reminding and selective reminding in the elderly. J Clin Exp Neuropsychol. 1997;19:643–654. doi: 10.1080/01688639708403750. [DOI] [PubMed] [Google Scholar]
- 82.Reinvang I, et al. APOE-related biomarker profiles in non-pathological aging and early phases of Alzheimer’s disease. Neurosci Biobehav Rev. 2013;37:1322–1335. doi: 10.1016/j.neubiorev.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 83.Lane-Donovan C, Herz J. ApoE, ApoE receptors, and the synapse in Alzheimer’s disease. Trends Endocrinol Metab. 2017;28:273–284. doi: 10.1016/j.tem.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tan MS, et al. Bridging integrator 1 (BIN1): form, function, and Alzheimer’s disease. Trends Mol Med. 2013;19:594–603. doi: 10.1016/j.molmed.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 85.Bignante EA, et al. Amyloid β precursor protein as a molecular target for amyloid β-induced neuronal degeneration in Alzheimer’s disease. Neurobiol Aging. 2013;34:2525–2537. doi: 10.1016/j.neurobiolaging.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]