Abstract
Co-immunoprecipitation can be utilized to study protein-protein interactions from various environments, cell types, or tissues. Herein, we describe a co-immunoprecipitation protocol that can be used to examine protein complexes found in the pathogenic spirochete Borrelia burgdorferi. The method outlined here has successfully identified known and unknown members of borrelial protein complexes and is an efficient method for studying protein interactions in this pathogenic spirochete.
Keywords: Borrelia, Co-immunoprecipitation, Protein-protein interactions, Spirochetes
1 Introduction
Co-immunoprecipitation is a technique by which protein complexes can be isolated from a solution using an antibody specific for one of the proteins in the complex. This technique is similar to immunoprecipitation studies in which a specific protein can be isolated from a solution containing many other proteins such as from eukaryotic or prokaryotic whole-cell lysates. Immunoprecipitation studies typically require that the protein-specific antibody be bound to a solid substrate or resin allowing for the protein to be precipitated from the solution. Co-immunoprecipitation experiments are performed in a similar manner, but co-immunoprecipitation refers to the fact that proteins tightly bound to the antibody-specific protein will also be precipitated or “pulled-down” in the experimental assay. This technique has been utilized to examine interactions of proteins thought to be in complex with one another and/or to identify unknown members of protein complexes. Many co-immunoprecipitation kits are commercially available but may differ in the solid-state resin or beads provided with the kit. These kits typically provide a solid substrate to which antibody binding proteins such as Protein A and/or Protein G are bound, which provides specific attachment of the antibody to the resin through the Fc portion of the immunoglobulin molecule. Subsequently, the resin-antibody complex is incubated with a solution containing the targeted proteins and all unwanted proteins are washed away. Combining co-immunoprecipitation with SDS-PAGE followed by mass spectrometry and/or immunoblotting can lead to identification of all proteins within a protein complex.
In Borrelia burgdorferi, the causative agent of Lyme disease, protein-protein interactions, especially protein complexes located in the borrelial outer membrane (OM), are known to have important implications in virulence and membrane biogenesis [1]. Co-immunoprecipitation has been a vital technique for examining and identifying new members of such complexes. For example, the β-barrel assembly machinery (BAM) complex is an essential protein complex found in the OM of all dual-membraned bacteria and is required for proper assembly and insertion of β-barrel proteins into the bacterial OM [2–5]. The central component of the BAM complex is the OM protein BamA. While the borrelial BamA ortholog was detected through sequence homology [6], co-immunoprecipitation studies were used to identify other accessory proteins that are part of this outer membrane transport complex in B. burgdorferi. These co-immunoprecipitation studies identified two proteins, designated BamB and BamD, which are part of the borrelial BAM transport complex [7, 8]. This led to the important finding that the borrelial BAM complex has only three members as compared to the five-member complex found in Escherichia coli. In other reports, co-immunoprecipitation has been used to demonstrate that two important B. burgdorferi virulence factors, P66 and OspA, actually interact and are members of the same protein complex [9]. All of these co-immunoprecipitation studies were completed using the protocol described herein (outlined in Fig. 1), indicating that this is an effective method for studying protein-protein interactions in Borrelia spirochetes.
Fig. 1.
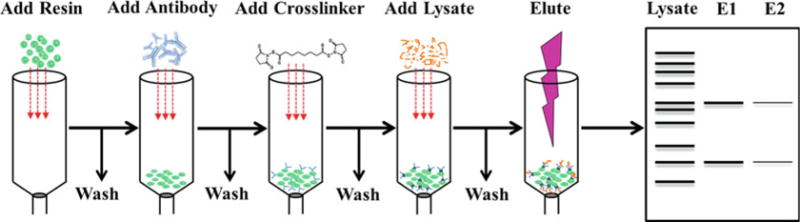
A schematic of the crosslink co-immunoprecipitation (co-IP) protocol. The antibodies are incubated with resin prior to crosslinking with DSS. Borrelial lysate is then added to the crosslinked antibody-resin complex before the specifically bound protein is eluted as described in the protocol. The input lysate and eluates from elution #1 (E1) and elution #2 (E2) can then be analyzed by SDS-PAGE
2 Materials
One liter BSK-II medium containing 6% normal rabbit serum (complete BSK-II medium) [10].
Centrifuge and rotor with 500 ml bottle capacity.
PBS (Phosphate-Buffered Saline), pH 7.4, chilled at 4 °C.
BugBuster Reagent (Novagen, Billerica, MA), supplied at a 10× concentration. Dilute to 1× concentration in PBS, and store at room temperature.
Lysonase Bioprocessing Reagent (Novagen), purchased as a ready-to-use solution, contains a combination of rLysozyme Solution and Benzonase Nuclease.
Protease Inhibitor Cocktail prepared according to the manufacturer’s instructions.
Microcentrifuge tubes; 1.5 ml and 2 ml volumes.
Microcentrifuge.
Dry bath incubator at 99 °C.
- Pierce crosslink immunoprecipitation (IP) kit (Thermo Scientific, Waltham, MA). The kit is stored at 4 °C at all times. Use ultrapure water to dilute the buffers from the kit. The following list of solutions and material are provided with the kit as supplied and needed for the co-immunoprecipitation procedure described below:
- Protein A/G Plus Agarose Resin, designed by covalent immobilization of a purified recombinant A/G fusion protein onto crosslinked agarose bead resin.
- Control Agarose Resin (unconjugated agarose resin).
- 20× Coupling Buffer (pH 7.2), comprised of 10 mM sodium phosphate and 150 mM sodium chloride. Diluted with ultrapure water to 1× strength.
- DSS (disuccinimidyl suberate) crosslinker compound supplied as aliquots of solid powder. DSS and subsequently dissolved in DMSO (dimethyl sulfoxide) immediately before use.
- IP Lysis/Wash Buffer (pH 7.4) contains 25 mM Tris, 150 mM NaCl, 1 mM EDTA, 15 NP-40 and 5% glycerol.
- 100× conditioning buffer (pH 7.4), diluted to 1× concentration before use.
- Elution buffer comprised of primary amines in a low pH solution.
- Spin columns.
- Screw caps for spin columns.
- Bottom plugs for spin columns.
- Two milliliter volume microcentrifuge collection tubes.
3 Methods
The co-immunoprecipitation procedure described is outlined in Fig. 1.
3.1 Preparation of B. burgdorferi Lysates
Harvest 1 l of mid to late-log phase B. burgdorferi culture (between 5 × 107 and 1 × 108 organisms/ml; see Note 1). To harvest the culture, pipette the culture into two 500 ml centrifuge bottles (see Note 2) and centrifuge at 5000 × g for 20 min at 4 °C (see Note 3).
Resuspend each pellet in 20 ml cold PBS and transfer to a 50 ml conical tube, and centrifuge at 5000 × g for 15 min at 4 °C.
Repeat the PBS wash and centrifugation (step 2) two times.
Following the third wash, weigh the final pellet.
Resuspend the pelleted organisms in 1× BugBuster Reagent at a concentration of 2.5 ml per gram of total pellet weight (see Note 4).
Add Lysonase Bioprocessing Reagent at a concentration of 3 μl/ml BugBuster reagent (see Note 4).
Add 20 μl of Protease Inhibitor Cocktail to the sample.
Incubate the mixture at room temperature with gentle rotation for 2–3 h.
Transfer the mixture to 1.5 ml microcentrifuge tubes. Pellet cell debris at 16,000 × g for 15 min at 4 °C.
Extract the supernatant from the pelleted debris to either use immediately for co-immunoprecipitation or store 500 μl aliquots at −80 °C for later use.
3.2 Binding of Antibody to Agarose Resin
Before pipetting, gently mix the bottle of Protein A/G Plus agarose resin to resuspend any resin that has settled on the bottom of the bottle. Use a cut-off pipette tip to transfer 40 μl of resin (see Note 5) to a spin column that has been placed into a 2 ml microcentrifuge collection tube. Centrifuge at 1000 × g for 1 min (see Note 6) in a microcentrifuge. Discard the flow-through.
Wash the resin with 300 μl of 1× Coupling Buffer (see Note 7). Centrifuge the column and discard the flow-through and repeat once more.
Remove the spin column from the collection tube and tap the bottom of the column on a Kimwipe before inserting a bottom plug.
Add the antibody of interest directly to the spin column (10–50 μl of antibody is recommended; see Note 8). Adjust the total volume in the spin column to 100 μl by adding 20× Coupling Buffer and ultrapure water to make a 1× Coupling Buffer solution.
Secure the spin column with a screw cap. Incubate the spin column at room temperature with gentle rotation for 1 h.
Remove the bottom plug and screw cap, saving them for the next step. Centrifuge the spin column in a 1.5 ml microcentrifuge tube to remove excess antibody solution.
Wash the spin column with 300 μl of 1× Coupling Buffer. Repeat this step three times. Clean the bottom of the spin column using a Kimwipe before replacing the bottom plug.
3.3 Crosslinking the Antibody to the Resin
To prevent co-elution of the antibody with the protein complexes of interest, use a 2.5 mM DSS solution dissolved in DMSO to covalently crosslink the antibody to the resin (see Note 9).
In a separate tube, make the following 50 μl solution: 9 μl of 2.5 mM DSS, 2.5 μl of 20× Coupling Buffer, and 38.5 μl of ultrapure water. Gently pipette and transfer the mix to the spin column with antibody bound resin.
Attach the screw cap to the spin column (the bottom plug should already be in place). Allow the mixture to gently rotate at room temperature for 1 h.
Remove and save the bottom plug and screw cap, and transfer the spin column to a microcentrifuge collection tube.
Add 200 μl of elution buffer. Wash and centrifuge the spin column to remove excess antibody and quench the crosslinking reaction. Repeat this step once. The flow-through after the first wash can be saved to verify the antibody-resin crosslinking.
Add 300 μl of the IP Lysis/Wash Buffer. Centrifuge and, discard the flow-through, and repeat once.
Remove excess liquid from the bottom of the spin column using a Kimwipe and place the bottom plug on the spin column.
3.4 Lysate Preclearance
For lysate preclearance, pipette 80 μl of the control agarose resin to a clean spin column (see Note 10). Place the spin column in a microcentrifuge collection tube.
Centrifuge the spin column to remove the excess storage buffer. Add 200 μl of 1× Coupling Buffer to wash the resin. Centrifuge and discard the flow-through.
Add 500 μl of borrelial lysates (see lysate preparation above) to the spin column. Place a clean screw cap and bottom plug onto the spin column and allow the mixture to rotate end-over-end at 4 °C from 2 h to overnight (see Note 11).
Centrifuge the spin column to collect the precleared lysate in a 2 ml microcentrifuge tube. The control resin can be saved to examine the nonspecific binding or loss of target proteins.
3.5 Immunoprecipitation and Elution of Antibody-Bound Antigens
To the spin column containing the antibody-crosslinked resin, add 500 μl of the precleared lysate. Place the screw cap onto the spin column and allow end-over-end mixing of the spin column overnight at 4 °C (see Note 11).
Remove the bottom plug and the screw cap from the spin column. Place the spin column in a 2 ml microcentrifuge collection tube. Centrifuge the column and save the flow-through. The flow-through can be examined for the presence of unbound target antigens.
Add 300 μl of IP Lysis/Wash buffer to the spin column and centrifuge to wash the resin and bound proteins. Discard the flow-through. Repeat a total of four times.
Wash and centrifuge the spin column with 200 μl of 1× Conditioning Buffer, and discard the flow-through. Repeat for a total of two washes.
Place the spin column in a clean 1.5 ml microcentrifuge tube for elution of antibody bound protein complexes.
Add 10 μl of elution buffer to the spin column and centrifuge immediately.
With the spin column placed in the same microcentrifuge tube, add 40 μl of elution buffer (see Note 12) and let the column incubate at room temperature for 10 min before centrifugation. Retain this 50 μl eluate (elution #1) for further analysis (see Note 13).
Place the bottom plug on the spin column. Add 50 μl of Laemmli buffer and loosen the screw cap prior to incubation in a dry bath at 99 °C for 7 min. Remove the bottom plug and place the spin column into a clean microcentrifuge tube. Centrifuge and retain this eluate (elution #2) for further analysis (see Note 13).
3.6 SDS-PAGE Analysis of the Co-immunoprecipitate
Add 5× nonreducing sample buffer and 1 M DTT to elution#1 to make a final concentration of 1× sample buffer with 100 mM DTT.
Boil sample for 5 min. Allow it to cool before loading on a SDS-PAGE gel.
Load between 8–10 μl of elution #1 and elution #2 on 10% or 12% SDS-PAGE gels to separate the eluted fraction for membrane transfer and subsequent immunoblot analysis.
Footnotes
Organisms are enumerated using dark-field microscopy immediately prior to lysate preparation.
Pipetting the culture into centrifuge bottles, as opposed to pouring, will avoid collection of dead and nonviable organisms and cell debris that may settle to the bottom of the culture bottle.
B. burgdorferi has a very fragile outer membrane structure, and it is recommended to harvest these organisms at a low speed. Generally, centrifugation of the organisms should be between 5000 and 6000 × g to keep the organisms from being damaged or the membranes disrupted during the spin. Do not exceed 6000 × g.
Although the protocol for use of BugBuster reagent recommends using 5 ml reagent per gram of cell pellet, B. burgdorferi has a fragile membrane devoid of lipopolysaccharide and much less peptidoglycan as compared to Gram-negative or Gram-positive organisms, respectively. Therefore, the reagent amount can be reduced and still efficiently rupture the spirochete cells. Lysonase Bioprocessing reagent is complimented with the BugBuster reagent to enhance protein extraction efficiency and ensure maximum retrieval of functional proteins.
Cutting the end of a 20–200 μl pipette tip (2–4 mm) prior to pipetting will allow for the collection of the resin. Large bore pipette tips also can be purchased for this purpose. The volume of resin used for the experiment can vary depending on the specific antigen of interest. However, using between 10 μl and 50 μl of resin is generally suitable for B. burgdorferi co-immunoprecipitations.
Centrifuge at 1000 × g for all the steps that require a benchtop microcentrifuge. Using higher spin speeds can result in clumping of the resin.
The protocol for the crosslink IP kit recommends using smaller volumes of coupling buffer and washing buffers than we have described here. Given that we typically use larger volumes of resin and antibodies than suggested in the manufacturer’s protocol, we adjust our washes accordingly.
We determine the titer of each antibody using either a recombinant form of the antigen or whole cell-lysates as substrate for immunoblot or ELISA-based analyses. For a low-titer antibody (<1:1000 reactivity observed using 100 ng of recombinant protein in an immunoblot or ELISA assay), we will use a volume of up to 50 μl of the antibody or antisera while only 10 μ1 is typically required for high-titer antibodies (>1:1000).
The crosslinking of antibody to the resin prevents elution of antibodies in the final sample. The co-eluted antibody may interfere with the clarity of the immunoblot or for subsequent mass spectrometry analyses; thus, it is recommended to not skip this step.
The preclearance step should be performed immediately before lysate is added to the antibody-crosslinked resin, which could either be the same day as resin preparation or the previous day if preclearing overnight. Preclearance can help reduce nonspecific binding and background; however, this step can be omitted if the target antigen is expressed at low levels and is not detected by specific antibodies upon immunoblotting when preclearance was performed during the experiment.
The lysate can be allowed to mix with the antibody-bound resin between 2 h to overnight depending on the antibody titers as well as the amount of antigen in the lysate.
The amount of elution buffer used can be reduced for a smaller final volume, although this can reduce the final protein yield from the elution step. We recommend using 40–50 μl of elution buffer.
We perform the second elution with reducing buffer for all B. burgdorferi co-immunoprecipitation experiments. Although Elution 1 should contain the target antigens, some proteins may be tightly bound to the antibodies and the low pH elution buffer may not elute all the antibody-bound antigens. Therefore, we heat the antigen-antibody complex in the presence of a reducing buffer for Elution 2. However, this step may also reduce any antibody that was not efficiently crosslinked with the resin. Thus, we recommend examining both eluates via immunoblot to compare the yield of the target antigen(s) and the potential presence of antibody fragments.
References
- 1.Kenedy MR, Lenhart TR, Akins DR. The role of Borrelia burgdorferi outer surface proteins. FEMS Immunol Med Microbiol. 2012;66:1–19. doi: 10.1111/j.1574-695X.2012.00980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 3.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Ricci DP, Silhavy TJ. The bam machine: a molecular cooper. Biochim Biophys Acta. 1818;2012:1067–1084. doi: 10.1016/j.bbamem.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hagan CL, Silhavy TJ, Kahne D. beta-Barrel membrane protein assembly by the bam complex. Annu Rev Biochem. 2011;80:189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- 6.Lenhart TR, Akins DR. Borrelia burgdorferi locus BB0795 encodes a BamA orthologue required for growth and efficient localization of outer membrane proteins. Mol Microbiol. 2010;75:692–795. doi: 10.1111/j.1365-2958.2009.07015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lenhart TR, Kenedy MR, Yang X, Pal U, Akins DR. BB0324 and BB0028 are constituents of the Borrelia burgdorferi beta-barrel assembly machine (BAM) complex. BMC Microbiol. 2012;12:60. doi: 10.1186/1471-2180-12-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn JP, Kenedy MR, Iqbal H, Akins DR. Characterization of the beta-barrel assembly machine accessory lipoproteins from Borrelia burgdorferi. BMC Microbiol. 2015;15:70. doi: 10.1186/s12866-015-0411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kenedy MR, Luthra A, Anand A, Dunn JP, Radolf JD, Akins DR. Structural modeling and physicochemical characterization provide evidence that P66 forms a b-barrel in the Borrelia burgdorferi outer membrane. J Bacteriol. 2014;196:859–872. doi: 10.1128/JB.01236-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbour AG. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]


