Abstract
With the recent advent of immunotherapy, there is a critical need to understand immune cell interactions in the tumor microenvironment in both pan-cancer and tissue-specific contexts. Multi-dimensional datasets have enabled systematic approaches to dissect these interactions in large numbers of patients, furthering our understanding of the patient immune response to solid tumors. Using an integrated approach, we inferred the infiltration levels of distinct immune cell subsets in 23 tumor types from The Cancer Genome Atlas. From these quantities, we constructed a co-infiltration network, revealing interactions between cytolytic cells and myeloid cells in the tumor microenvironment. By integrating patient mutation data, we found that while mutation burden was associated with immune infiltration differences between distinct tumor types, additional factors likely explained differences between tumors originating from the same tissue. We concluded this analysis by examining the prognostic value of individual immune cell subsets as well as how co-infiltration of functionally discordant cell types associated with patient survival. In multiple tumor types, we found that the protective effect of CD8+ T cell infiltration was heavily modulated by co-infiltration of macrophages and other myeloid cell types, suggesting the involvement of myeloid-derived suppressor cells in tumor development. Our findings illustrate complex interactions between different immune cell types in the tumor microenvironment and indicate these interactions play meaningful roles in patient survival. These results demonstrate the importance of personalized immune response profiles when studying the factors underlying tumor immunogenicity and immunotherapy response.
Keywords: Immunology, pan-cancer, genomics, myeloid-derived suppressor cell, T cell
Introduction
Immunotherapy has emerged as a promising modality for the treatment of cancer. Blockade of immune checkpoint proteins, including CTLA4 and PD1/PDL1, has shown clinical efficacy in multiple tumor types, with responders experiencing prolonged remission that can last several years (1). These therapies predominantly function by enhancing and prolonging cytolytic effector functions of infiltrating lymphocytes, resulting in improved anti-tumor immunity. Despite these successes, a large fraction of treated patients is either non-responsive or develops intolerable side effects to immunotherapeutic approaches (2,3). In some cases, this may be due to an absence of cytotoxic cells in the patients’ tumors which negates any potential benefit of checkpoint blockade therapy (4–6). However, several negative checkpoint regulators have now been identified that function non-redundantly to CTLA4 and PD1/PDL1 (7–9). Many of these regulators are expressed by different immune cell subsets that may be present in varying quantities in the tumor microenvironment of individual patients. These cell types interact with effector cells in the tumor microenvironment, altering their otherwise normal functions. Understanding how this interplay differs between patients can provide insights into why some patients have more effective anti-tumor immune responses and greater sensitivity to immunotherapy than others
Recently, high-dimensional datasets, such as The Cancer Genome Atlas (TCGA), have allowed for the large-scale interrogation of immune activity in multiple tumor types. Analyses of these datasets have made important advances in applying genomic data to tumor immunology. DNA sequencing data has been analyzed to connect neoantigen load to T cell response (10) and link somatic mutations to immune infiltration (11). Expression data has been utilized to measure the cytolytic activity in the tumor microenvironment (12) and more recently, to quantify the infiltration levels of individual immune cell subsets (13–17). One common theme across these studies is the integration of several types of genomic and clinical data, allowing for associations to be made between immune activity, gene expression, mutation burden, and patient survival.
Here, we perform a systematic analysis to investigate the immune cell interactions that take place in the tumor microenvironment of a diverse array of tumor types. We apply our previously developed computational framework (16,18) to quantify infiltration from multiple immune cell subsets and apply the resulting scores to address several questions relating to the interplay between immune cells in the context of cancer. First, we examine the rate at which different immune cell subsets co-infiltrate with each other and examine how the quantities of each subset differ across tumor types. Second, we investigate the factors driving tumor immunogenicity by examining the associations between mutation load and immune infiltration in a pan-cancer and tumor type-specific manner. Third, we map the effect infiltration of different immune cell subsets has on patient survival by performing a pan-cancer survival meta-analysis spanning 125 independent datasets. Finally, we explore the clinical effect of immune cell interactions in the tumor microenvironment by performing multi-class survival analyses based on each patient’s personal immune response profile. Through these analyses we demonstrate the complexity of immune cell interactions in the tumor microenvironment, while providing insights into the tumor-intrinsic features that influence immune infiltration and the effect these subsets have on patient survival in both pan-cancer and tissue-specific contexts.
Materials and Methods
Datasets
Datasets comprising RNAseq from 7172 TCGA tumors and mutation annotation files from 6413 TCGA tumors (5061 which overlapped with RNAseq data; Supplementary Table S1) were obtained from the TCGA Data Portal in June 2015 (level 3, RNAseqV2; http://cancergenome.nih.gov/). Absolute expression values obtained from RNAseq data were log10 transformed. Raw whole exome sequencing data from paired tumor-normal samples for four tumor types (colon adenocarcinoma, rectum adenocarcinoma, uterine corpus endometrial carcinoma, uterine carcinosarcoma) were obtained from CGHub (https://cghub.ucsc.edu/). These data were used to infer microsatellite instability by running MSIsensor using default parameters (19). Consensus purity estimates for available TCGA tumors were downloaded from a supplemental file from prior work (20). Flow cytometry data was obtained from the gene expression omnibus (GEO) under accession number GSE65133 (15). Additional gene expression data and the associated survival data from 125 datasets was obtained from PREdiction of Clinical Outcomes from Genomic profiles (PRECOG, https://precog.stanford.edu/precog_data.php; Supplementary Table S2) (21) and a supplementary breast cancer dataset (GSE11121) from GEO (22). All datasets obtained from hematopoietic cancers were removed from PRECOG for further analyses.
Raw gene expression data from the Immunological Genome Project (ImmGen) was obtained from GEO (GSE15907) in October 2015. Robust Microarray Analysis was used for background correction of raw data, followed by quantile normalization. Each probeset (Affymetrix MoGene-1_0-st) was then fitted to a multichip linear model using the ‘expresso’ function from the ‘affy’ library in R (23). Probes were collapsed into gene symbols using the probe with the highest average intensity across all cell types. Murine transcripts were matched to human transcripts on the basis of gene symbol.
Macrophage gene expression profiles used for cell profile correlation analyses were downloaded from a supplemental file from prior work (24) and integrated into the processed ImmGen dataset. This dataset was quantile normalized at the profile level, median normalized at the gene level, reduced to the genes with the top 50% most variable expression across samples, and then z-transformed so that each cell’s resulting profile followed a standard normal distribution. M1-like and M2-like macrophages in this dataset were defined based on clustering analyses performed in the original study.
Calculation of immune cell relative expression profiles
Our framework uses the relative gene expression profiles of reference immune cells to determine infiltration levels in human tumors. To calculate these profiles, the absolute expression values from each gene in the processed ImmGen dataset are median normalized across all cell types. These values are then z-transformed so that the expression profile for each cell type follows a standard normal distribution. As a result of this step, each cell now has an associated z-score profile where, for a given cell type, values >0 indicate higher relative expression, while values <0 indicate the opposite. At this stage, replicate cell types are collapsed into an average by taking the mean z-score of each replicate and then renormalizing via z-transformation. To input these profiles into the binding association with sorted expression (BASE) algorithm that drives our framework (25), each cell’s z-score profile is then split into an up- and down-regulated profile. Each cell’s up-regulated profile is comprised of all genes where the z-score is >0, with genes with z-scores <0 being set to 0. The down-regulated profile is comprised of the opposite. The up- and down-regulated profiles for each cell type are then each converted to p-values and −log10 transformed to add more weight to each cell’s differentially expressed genes. The resulting transformed values >10 are trimmed to 10 to avoid outliers, and then divided by the maximum value in the dataset to rescale values from 0 to 1. R code for this process as well as the weight profiles for four representative cell types from the ImmGen dataset are contained with this manuscript (Supplementary Software S1, Supplementary Table S3).
Calculation of the infiltration score
Calculation of the infiltration score is performed using the BASE algorithm, which has been previously described (25). R code for the BASE algorithm is available in a prior publication (16) and here as well (Supplementary Software S2). BASE infers immune infiltration by integrating the transformed reference immune cell profiles with patient gene expression data. Each reference immune cell’s up- and down-regulated profile is used as a weight vector, w = [w1, w2, w3 … wj … wn]; where wj = the transformed expression value for gene j and n = the # of genes in the reference profile. Patient gene expression profiles are quantile-normalized and then ranked from high to low. BASE defines these normalized profiles as a gene expression vector g = [g1, g2, g3 … gj … gn]; where gj = a patient’s normalized expression value for their jth ranked gene. Following this step, RNAseq and one-channel array datasets undergo an additional median normalization step. BASE uses these two vectors to calculate a pre-cell lineage score (pCLS) for the up-regulated (pCLSup) and down-regulated (pCLSdn) profiles from a given cell type. These scores are later used to calculate the overall infiltration score.
To obtain the pCLSup and pCLSdn, the BASE algorithm determines the absolute maximal deviation between a foreground function and a background function calculated using the g and w vectors. The foreground function represents the cumulative distribution of a given patient’s gene expression values weighted by the corresponding transformed immune cell expression values:
| 1 |
The background function represents a given patient’s gene expression values weighted by the corresponding genes’ complementary transformed immune cell expression values (1 − w):
| 2 |
To obtain the maximal deviation between these two functions, the maximum difference between the foreground and background function (pCLS+) must be compared to the minimal (most negative) difference between the foreground and background function (pCLS−). The pCLSup/dn with the largest absolute difference becomes the final pCLSup/dn:
| 3 |
| 4 |
| 5 |
The resulting statistics provide a measure of similarity between a patient’s gene expression profile and the relative expression profiles of different immune cells. A high pCLSup is the result of a large absolute deviation between the foreground and background function. This occurs when a given immune cell’s lineage-specific genes, which are given high weights, are also ranked toward the top of a patient’s gene expression profile. This causes there to be a sharp increase early and then a late plateau in the foreground function, and the opposite in the background function. In the case of the pCLSdn, higher weights correspond to negative differential expression in a given cell type, and thus the role of the background and foreground function will be swapped compared to the pCLSup calculations. Thus, a more negative pCLSdn is indicative of high similarity between a patient and cell type’s down-regulated genes.
The resulting pCLSup and pCLSdn are normalized through a permutation-based method where the gene labels in vector g are permuted 1000 times resulting in 1000 permuted gene expression vectors (g1, g2, …, g1000). A pCLSup/dn is then calculated using each permuted expression vector to obtain a null pCLSup/dn distribution. The original pCLSup and pCLSdn are then divided by the mean of the absolute value of their respective null pCLSup/dn distribution, yielding the normalized CLSup and CLSdn. The two scores are then integrated together by subtracting the CLSdn from the CLSup. This final term, referred to as the cell lineage score (CLS) or infiltration score, represents immune infiltration of a given cell type in a patient’s tumor.
Network and correlation analyses
The immune cell co-infiltration network was constructed using a Spearman correlation matrix that assessed the relationship between all possible ImmGen immune cell pairs. All correlations were adjusted for tumor purity through partial correlations between infiltration scores of each cell type and consensus purity estimates (20). Only immune cells anti-correlated with tumor purity at the pan-cancer level (Rho < −0.1) were included in this analysis. An edge between two nodes in the network represented two immune cells whose infiltration scores were correlated at Rho > 0.45. To obtain genetic similarity measurements between the reference cell types, gene expression values from the ImmGen dataset were median normalized across all cell types and then each profile was z-transformed. A genetic similarity matrix was then constructed of pairwise Spearman correlations between the z-score profiles of all possible immune cell pairs. These genetic similarity scores were used to color the edges between each node in the co-infiltration network. The network was visualized using the “Organic yFilesLayout” in Cytoscape v3.4.0.
Survival analyses
Survival meta-analyses were performed using gene expression data obtained from PRECOG. For each PRECOG dataset, infiltration scores for all cell types were fit to a univariate Cox proportional hazards model using the “coxph” function from the R “survival” package. Two-sided p-values calculated using a Wald test were derived from each Cox proportional hazards model and converted to z-scores. The hazard ratio from each Cox proportional hazards model was used to determine the sign of the z-score, with a hazard ratio > 1 corresponding to a z-score > 0 and a hazard ratio < 1 corresponding to a z-score < 0. Meta-z-scores were calculated by applying Stouffer’s method to z-scores from datasets of the same cancer type (26). Survival distributions for different cell types were visualized using Kaplan-Meier curves created by the “survfit” function from the R “survival” package. The thresholds for each Kaplan-Meier plot were determined based on each dataset’s unique infiltration score distribution. Differences between the survival distributions in each Kaplan-Meier plot were calculated using a log-rank test through the “survdiff” function from the R “survival” package.
Code availability
The R code for the BASE algorithm and immune cell weight calculation can be found in Supplementary Software S1 and S2.
Results
To quantify the levels of tumor-infiltrating immune cells in human cancer, we applied our integrated framework (16), which utilizes the whole transcriptome of 220 murine hematopoietic cell types generated by ImmGen (27) as a reference to infer relative levels of immune infiltration in human cancer patients. The ImmGen dataset provides a rich resource of reference hematopoietic gene expression profiles that have been carefully collected and analyzed through rigorous standardized procedures. The high degree of global conservation between human and mouse hematopoietic gene expression profiles make this dataset an excellent tool for probing immune cell interactions in the tumor microenvironment (28). The first step of our framework is to assign weights to the genes of each reference profile according to how differentially they are expressed compared to other reference cell types. Genes that are up-regulated relative to background become part of the up-regulated set, while down-regulated genes make up the down-regulated set. We then input these two sets of weights, along with patient gene expression profiles, into our BASE algorithm (25). For a given reference cell type, BASE functions by calculating two rank-ordered cumulative distributions representing the product between the value of each gene in a patient expression profile and that gene’s respective up- and down- weights in the reference cell type. It then takes the maximal difference between the two distributions, similarly to a Kolmogorov-Smirnov test. This difference is then normalized relative to a null distribution to obtain the final score, which represents the level of infiltration for that cell type.
Quantification of immune infiltration in human cancer
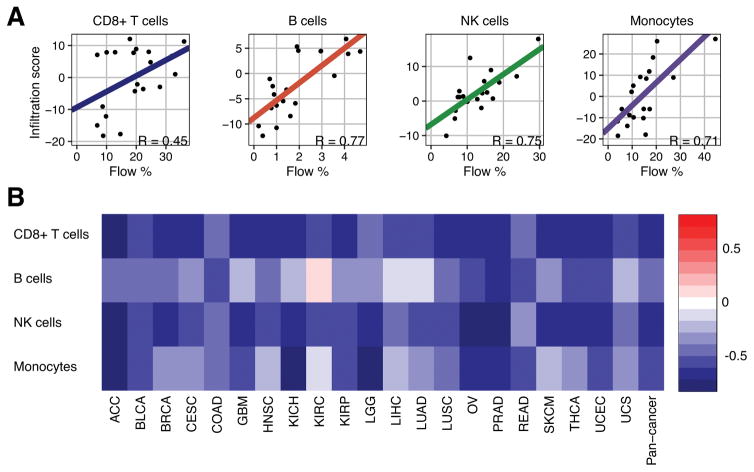
We have previously shown that the scores generated by our algorithm are negatively correlated with expression-based tumor purity (29) and positively correlated with histological lymphocytic infiltration (30) in human breast tumors. We have additionally shown that the murine profiles of ImmGen could serve as proxies for human immune cell profiles (16). To assess the performance of our algorithm on individual cell subsets, we applied it to previously generated gene expression data from human peripheral blood mononuclear cell flow cytometry experiments (15). For most reference profiles, the scores generated by our algorithm were significantly positively correlated with the flow cytometry fraction of their respective cell type and weakly associated with unrelated cell types, indicating good specificity (Supplementary Table S4). To confirm that our method could be extended into tumors, we applied it to RNAseq data from 23 TCGA tumor types (Supplementary Table S3) and correlated the resulting infiltration scores with tumor purity for the 21 tumor types with available consensus purity estimates (20) (Supplementary Table S5). At the pan-cancer level, 60% of the ImmGen cell type infiltration scores were negatively correlated with tumor purity (Rho < −0.1), indicating that the majority of reference profiles could distinguish between tumor-specific and lymphocyte-specific signals. The cell types demonstrating positive tumor purity correlations were primarily from stem cells, pre- and pro- B cells, thymal T cells, and CD8+ effector T cells in the early stages of activation (12 hours – 8 days post-activation), which are known to exhibit high expression of genes involved in tumor-related processes such as dedifferentiation and cell proliferation (31,32). We thus excluded these cell types from the remainder of our analyses, even though many of them exhibited high specificity during flow cytometry analysis.
From the remaining reference profiles, we selected four cell types, CD8+ T cells 45 days post-activation (T.8MEM.SP.OT1.D45.LISOVA), newly-formed B cells (B.FRE.BM), splenic natural killer (NK) cells (NK.DAP12neg.SP), and lung-resident macrophages (MF.LU), that best represented human CD8+ T cells, B cells, NK cells, and macrophages, respectively. Each of these cell types was significantly correlated with the flow cytometry fraction of its corresponding human cell type (R = 0.45, 0.77, 0.75, and 0.71, P all < 0.05; Fig. 1A) and negatively correlated with tumor purity for all tumor types, with one exception (B cells in kidney renal clear cell carcinoma, R = 0.07; Fig. 1B). We additionally compared our representative macrophage cell’s expression profile to the expression profiles of 29 different macrophages that had each been exposed to a different activation condition (24). Expression profile correlation analysis revealed that our representative macrophage was more M2-like than M1-like (P = 0.01, Wilcoxon sum-rank test; Supplementary Figure S1A and S1B), though it exhibited positive associations across all 29 different macrophage conditions. To determine if our representative macrophage could detect immunosuppressive signals from other myeloid cell types, we correlated the resulting infiltration scores from the TCGA data with the expression of genes from a literature-curated myeloid-derived suppressor cell (MDSC)-specific gene signature (14) (Supplementary Figure S1C). We found that 79% of MDSC-specific genes were significantly positively correlated with our macrophage score in a pan-cancer manner, including the traditional MDSC marker gene ITGAM (pan-cancer Rho = 0.54). Based on these results, it is likely that our macrophage infiltration score may better represent immunosuppressive myeloid cell activity than macrophage infiltration, specifically.
Figure 1.
Flow cytometry and tumor purity validation. a Scatterplot of flow cytometry and infiltration score for the four indicated immune cell subsets from 20 subjects. b Spearman correlations between infiltration scores from four indicated immune cell subsets and consensus purity estimates for 21 different cancer types. TCGA abbreviations for each cancer type are listed in Supplementary Table S1.
Immune cell co-infiltration network reveals a high confidence T cell-myeloid cell module
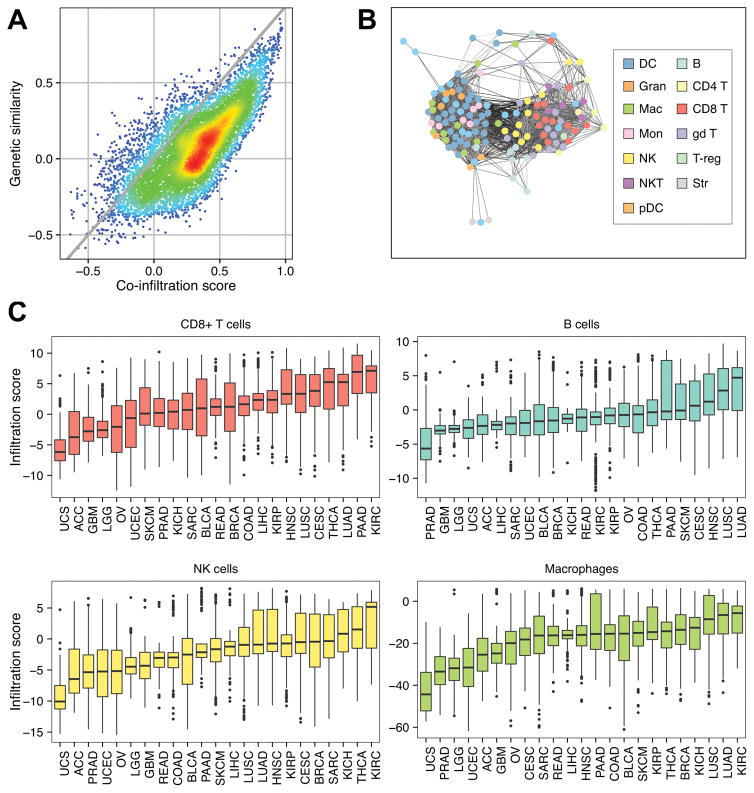
Immune cells of different lineages are often found to co-populate the tumor microenvironment, suggesting potential co-infiltration patterns in solid tumors. By rapidly quantifying cell-type-specific infiltration levels, our framework enables the analysis of potential co-infiltration patterns between immune cell types. However, reference profiles from transcriptionally analogous cell types tend to yield similar scores that may result in artificially high co-infiltration associations between cells. To account for this, we determined the genetic concordance between our reference immune cells by performing pairwise Spearman correlations on their relative expression profiles. As expected, cells from the same lineage tended to have high transcriptomic similarity, whereas cells of distinct subclasses were more discordant. We next calculated pan-cancer co-infiltration scores by performing purity-adjusted pairwise Spearman correlations between the infiltration levels from each pair of cells assessed by our framework. Comparing these two sets of scores revealed several cell-cell pairs that were positively correlated despite having discordant gene expression profiles, suggesting true co-infiltration (Fig. 2A).
Figure 2.
Immune cell co-infiltration analyses. a Scatterplot comparing the pairwise infiltration score Spearman correlation coefficients from all possible immune cell combinations (co-infiltration score) to pairwise expression Spearman correlation coefficients from all possible immune cell combinations (genetic similarity score). Gray line represents the trend if the genetic similarity scores were equal to their corresponding co-infiltration scores (y = x). Warmer colors represent higher point density while cooler colors lower point density. b Co-infiltration network representing correlation structure between each reference immune cell. Nodes represent reference immune cells. Edges represent co-infiltration scores > 0.45. Darker edges represent lower transcriptional similarity between reference cell types. c Boxplots comparing the distributions of immune infiltration in four indicated immune cell subsets. Each box spans quartiles with the lines representing the median correlation coefficient for each group. Whiskers represent absolute range excluding outliers. All outliers were included in the plot. TCGA abbreviations for each cancer type are listed in Supplementary Table S1.
To further characterize the landscape of immune cell co-infiltration, we constructed an immune network using the co-infiltration scores (Fig. 2B). Each node in the network represented an ImmGen immune cell, while the edges formed between two nodes were indicative of a co-infiltration score greater than 0.45. Shorter edges represented higher co-infiltration scores. Genetic similarity was not accounted for during edge formation, thus the most immediate modules represented transcriptionally similar cell types including myeloid-derived cells, T cells, and B cells. However, 134 edges existed that linked nodes representing CD8+ T cells and NK cells to those of the myeloid-derived cell module. Of these edges, 22% were between two cell types with a genetic similarity score less than 0, indicating high-confidence co-infiltration between cytolytic cell types and myeloid-derived cells.
While the previous analyses were performed in a pan-cancer manner, we also sought to examine the distribution of immune infiltration across different cancer types. To accomplish this, we stratified the TCGA dataset by cancer type and compared the distribution of immune cell infiltration scores for our four representative cell lineages (Fig. 2C). Tumors with high CD8+ T cell infiltration included kidney renal clear-cell carcinoma, lung adeno- and squamous cell carcinoma and cervical squamous cell carcinoma, which have all been previously noted for their high cytolytic activity (12), as well as pancreatic adenocarcinoma and thyroid carcinoma. NK cell and macrophage infiltration followed similar trends to that of CD8+ T cells while B cells did not, indicating that the trends depicted by the co-infiltration network are a universal feature of cancer. Taken together, these results suggest that some tumor types, such as kidney renal clear-cell carcinoma, are more predisposed to immune infiltration from several different immune cell types, while others may have factors that prevent immune infiltration from occurring, such as in glioblastoma with the blood-brain barrier (33).
Mutation burden is associated with infiltration of multiple immune cell types at the pan-cancer level
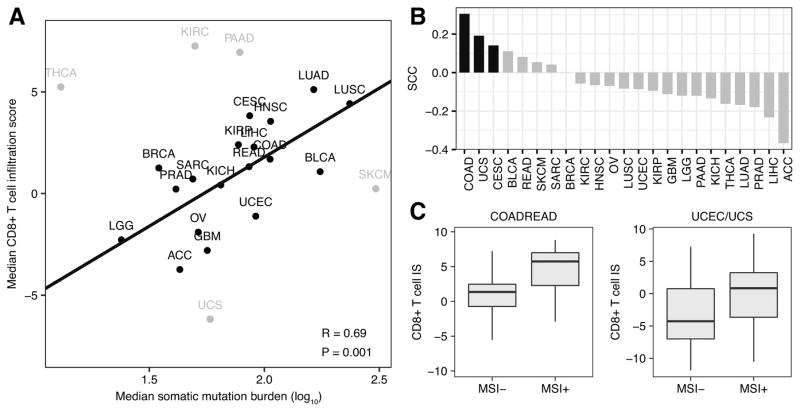
The differences in immune infiltration across tumor types led us to examine the factors that drive tumor immunogenicity. Neoantigens created by nonsynonymous somatic mutations have been linked to higher levels of T cell infiltration and response to immunotherapy in several tumor types (4,6,34–36). To further explore this relationship, we examined the association between the median CD8+ T cell infiltration level and the median mutational burden of each TCGA cancer type and found a weak, positive association between them (R = 0.17, P = 0.44). Notably, five cancer types, kidney renal clear-cell carcinoma, pancreatic adenocarcinoma, thyroid carcinoma, skin cutaneous melanoma, and uterine carcinosarcoma deviated substantially from the remaining tumor types. Dropping these five tumor types considerably improved the relationship between mutation burden and CD8+ T cell infiltration (R = 0.69, P = 1e-3; Fig. 3A). This result supported the link between mutation burden and CD8+ T cell infiltration, though additional factors took precedence in some tumor types.
Figure 3.
Mutation burden and CD8+ T cell infiltration. A, Scatterplot of median somatic mutation number per tumor sample across 23 cancer types (log10 scale) compared with each tumor’s corresponding median CD8+ T cell infiltration score. Pearson correlation coefficient and least-squares regression line presented were calculated excluding the five outlier tumor types in gray. B, Individual Spearman correlation coefficients representing the associations between somatic mutation number and CD8+ T cell infiltration score in 23 different tumor types. Black bars represent statistically significant associations (P < 0.1). C, Box plots comparing the CD8+ T cell infiltration score (CD8+ T cell IS) in MSI− versus MSI+ samples. Left boxplot makes comparison in colorectal adenocarcinoma (COADREAD) samples. Right boxplot makes comparison in combined cohort of uterine corpus endometrial carcinoma (UCEC) and uterine carcinosarcoma (UCS) samples. Each box spans quartiles, with the lines representing the median correlation coefficient for each group. Whiskers represent absolute range excluding outliers. All outliers were included in the plot. TCGA abbreviations for each cancer type are listed in Supplementary Table S1.
We next examined the relationship between mutation load and CD8+ T cell infiltrate within specific tumor types (Fig. 3B). Of the 23 tumor types examined, only colon adenocarcinoma, uterine carcinosarcoma, and cervical squamous cell carcinoma had significant relationships between CD8+ T cell level and mutation load (P < 0.10, Spearman correlation). Two of these tumor types, colon adenocarcinoma and uterine carcinosarcoma, exhibit high incidences of microsatellite instability (MSI), an indicator of genome instability. To investigate the role of MSI in determining immunogenicity, we inferred the MSI status of each colon adenocarcinoma and uterine carcinosarcoma sample using MSIsensor, an algorithm that derives MSI status from paired tumor-normal sequence data (19). For each of these cancer types, MSI+ samples had significantly higher levels of CD8+ T cell infiltration compared to MSI− samples (P = 1.6e-8 and 0.01, Wilcoxon sum-rank test). Combining colorectal adenocarcinoma samples with closely related rectal adenocarcinoma samples and uterine carcinosarcoma samples with their uterine corpus endometrial carcinoma counterparts revealed even stronger differences (P = 1.4e-9 and 7e-5, Wilcoxon sum-rank test; Fig. 3C). Together, these results supported previous findings of a relationship between MSI status and CD8+ T cell infiltrate in colorectal cancer (37), and demonstrated that this relationship holds in endometrial tumor types.
We next extended our mutation analyses to additional cell types by looking at their association with median mutation load across cancer types. (Supplementary Figure S2). Of the three remaining representative cell types, only B cells were significantly associated with median mutation load (R=0.51, P=0.01), with NK cells and macrophages exhibiting positive but insignificant relationships. Removal of the outlier tumor types identified in the T cell analysis improved the correlations for B cells (R = 0.69, P = 2e-3), NK cells (R = 0.37, P = 0.13), and macrophages (R = 0.56, P = 0.02). Stratifying and reanalyzing patients by tumor type revealed that only colon adenocarcinoma exhibited a significant positive relationship between mutation burden and infiltration for each cell type tested (Supplementary Table S6), agreeing with previous findings reporting an association between neoantigen load and overall lymphocytic infiltration of the tumor in colon cancer (38). Interestingly, differences between MSI+ and MSI− colon adenocarcinoma samples were less dramatic for B cells, NK cells, and macrophages compared to CD8+ T cells (P = 0.04, 8e-4, and 0.09 for B cells, NK cells, and macrophages, respectively, Wilcoxon sum-rank test). These associations indicated that mutation burden is a driver of general immune infiltration, but other factors are likely more relevant in some tumor types.
Immune infiltration has varying effects on patient survival across tumor types
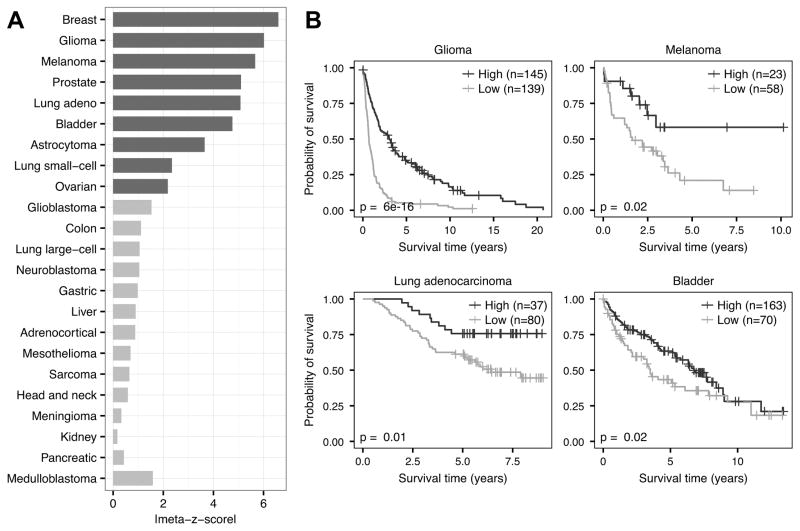
Immune cells have been linked to patient survival in several cancer types, with some cells, such as CD8+ T cells and NK cells, associated with favorable outcomes due to their tumoricidal properties (39). We examined the clinical effect of CD8+ T cell infiltration in a pan-cancer manner by applying our method to the PRECOG meta-dataset, a collection of 166 independent datasets, 125 of which are from solid tumors (21). Importantly, each of these datasets contain overall survival information, enabling large-scale meta-analyses of survival rates across cancer types. Before continuing further, we ensured the pairwise co-infiltration structure between CD8+ T cells and other cell types was consistent between TCGA and PRECOG data by comparing the median pairwise CD8+ T cell co-infiltration scores across all TCGA and solid tumor PRECOG datasets (Spearman correlation R = 0.83, P = 1e-34, Supplementary Figure S3). We then assessed the relationship between CD8+ T cell infiltration and patient survival in each PRECOG dataset using univariate Cox regression. To compare associations across datasets, we derived a z-score from each Cox model and combined the z-scores from datasets representing the same tumor types into a meta-z-score. These meta-z-scores provide a convenient, single metric for performing pan-cancer survival comparisons as has been done previously (21). Using these meta-z-scores, we found that CD8+ T cell infiltration was significantly protective in 9 tumor types and showed protective trends in many others that may have not been powered enough to show a significant association (Fig. 4A). Two-class comparisons of individual datasets from PRECOG (cutoff thresholds in Supplementary Figure S4) showed clear differences in survival time, with high CD8+ T cell infiltration patients experiencing significantly longer overall survival than low CD8+ T cell infiltration patients (Fig. 4B). These results indicate the involvement of CD8+ T cells in the protective anti-tumor immune response and agree with several reports over a wide range of cancer types (40–43).
Figure 4.
Survival meta-analysis of CD8+ T cell infiltration. A, Meta-z-score absolute values indicating prognostic associations of CD8+ T cell infiltration in 23 different tumor types comprising 18,190 samples. Cancers were ranked by weighted meta-z-score. Dark gray bars indicate a weighted absolute meta-z-score >1.96, whereas light gray bars indicate a weighted meta-z-score whose absolute value is <1.96. B, Kaplan–Meier plots depicting the survival probability over time for samples with high (dark gray) and low (light gray) CD8+ T cell infiltration scores. Datasets tested include GSE16011 (glioma), GSE8401 (melanoma), GSE13213 (lung adenocarcinoma), and GSE5479 (bladder). For all Kaplan–Meier plots, samples were stratified into high and low groups based on their infiltration score distributions (thresholds available in Supplementary Figure S4). P values were calculated using the log-rank test. Vertical hash marks indicate censored data.
We extended our analysis to infiltration from other cell types, including our representative B cells, NK cells, and macrophages (Supplementary Table S7, Supplementary Figure S5). Infiltration from our representative B cell lineage was positively associated with survival in 4 tumor types, while negatively associated with breast cancer survival. However, infiltration from other ImmGen B cell subsets was universally protective, suggesting distinct roles between different B cell populations. NK cell infiltration proved to be protective or showed protective trends in every cancer type examined. Most interestingly, macrophage infiltration was associated with poor patient prognosis in 9 cancer types and showed trends toward negative survival associations in several other cancer types. Similar associations were found in other myeloid-derived cells, suggesting immunosuppressive myeloid cell activity.
Co-infiltrating myeloid cells modulate the protective effect of CD8+ T cells in the tumor microenvironment
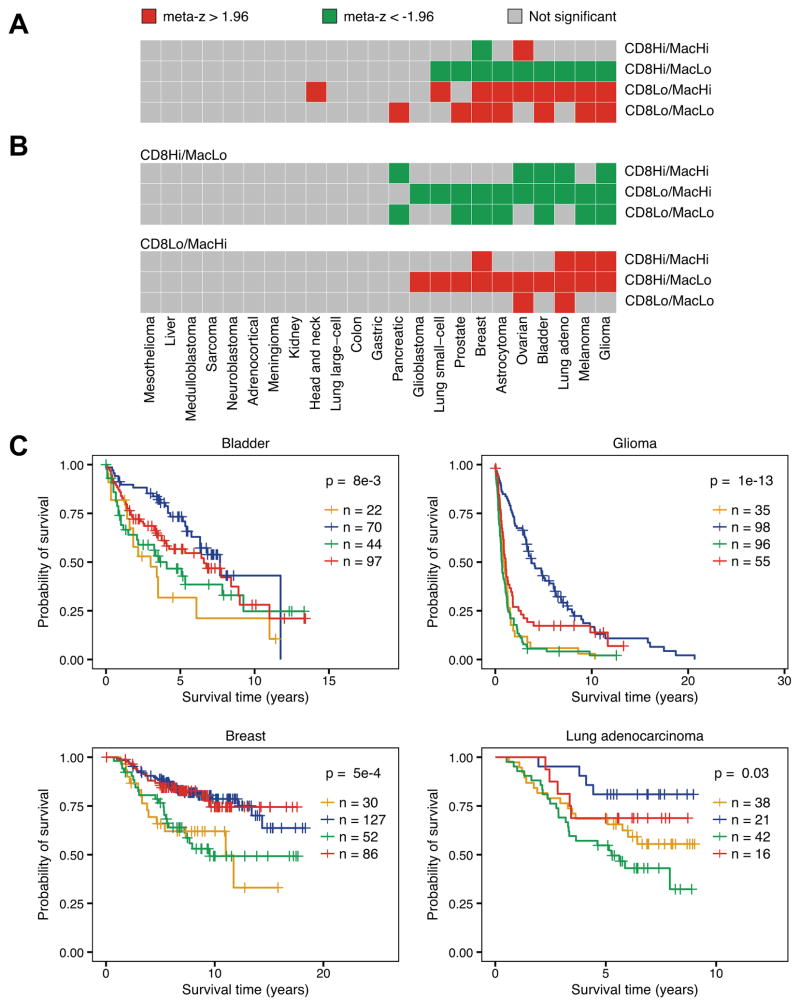
The disparate effects of macrophage and CD8+ T cell infiltration on patient survival suggested that they may act antagonistically. To investigate this, we stratified samples from each PRECOG dataset into four classes based on their levels of CD8+ T cell and macrophage infiltration: high CD8+ T cell/high macrophage, high CD8+ T cell/low macrophage, low CD8+ T cell/high macrophage, and low CD8+ T cell/low macrophage. Cutoffs for each class were made based on the dataset’s median infiltration score for each cell type. We repeated our z-score based survival meta-analysis, this time comparing each class to the three remaining classes using univariate Cox regression (Fig. 5A). In 9 tumor types, patients of the CD8+ T cell high/macrophage low group had significantly longer survival times compared to the other three groups. Interestingly, breast cancer was the only disease where CD8+ T cell high/macrophage high class patients experienced good clinical outcomes relative to the other three classes, contradicting previous findings (44). Patients in the CD8+ T cell low/macrophage high and CD8+ T cell low/macrophage low groups had significantly worse survival time relative to the other three groups in 9 and 8 tumor types, respectively.
Figure 5.
Effect of CD8+ T cell and macrophage infiltration on patient survival. a Weighted meta-z-scores from binary Cox proportional hazard models comparing the indicated class to the three remaining classes. b Weighted meta-z-scores from binary Cox proportional hazards models comparing CD8 T high/macrophage low (top) and CD8 T low/macrophage high (bottom) to the indicated classes. For all heatmaps, high/low status for each cell type was determined using median infiltration score for CD8+ T cells and macrophages. Red boxes indicate cancers where indicated class had significantly worse survival compared to other classes (meta-z > 1.96), green boxes indicate significantly improved survival compared to other classes (meta-z < -1.96), and gray boxes indicate no statistical association. c Kaplan-Meier plots depicting the survival distributions of all four classes: CD8+ T low/macrophage low (orange), CD8+ T high/macrophage low (blue), CD8+ T low/macrophage high (green), and CD8+ T high/macrophage high (red). Colors correspond to the classes as noted in a. Datasets tested include GSE5479 (bladder), GSE16011 (glioma), van de Vijver et al (breast), and GSE13213 (lung adenocarcinoma). For all Kaplan-Meier plots, samples were stratified into high and low groups based on their infiltration score distributions (thresholds available in Supplementary Figures S6 and S7). P-values were calculated using the log-rank test and indicate that at least one curve is significantly different from the rest. Vertical hash marks indicate censored data.
To examine the differences between individual classes, we made two sets of pairwise comparisons using univariate Cox regression (Fig. 5B). In the first set, we examined the difference in survival time between CD8+ T cell high/macrophage low patients and patients from each remaining class. In each comparison, CD8+ T cell high/macrophage low patients exhibited significantly longer survival time in multiple tumor types. Most interestingly, in 4 tumor types, CD8+ T cell high/macrophage low patients had significantly longer survival times relative to CD8+ T cell high/macrophage high class, suggesting antagonistic interactions between the two cell types. In the second set of comparisons, we examined the CD8+ T cell low/macrophage high class and found that in multiple cancer types, this class had a shorter survival time relative to each of the remaining classes, suggesting myeloid cell-mediated tumor growth.
These relationships held at the level of individual datasets, where samples were stratified into the four classes based on their CD8+ T cell and macrophage infiltration levels (Fig 5C; Supplementary Figures S6 and S7). CD8+ T cell high/macrophage low patients demonstrated the longest survival times relative to the other classes in bladder cancer, glioma, and lung adenocarcinoma. Breast cancer was again notable as there was no difference in survival distribution between the two CD8+ T cell high classes. Additionally, while there was minimal separation between the survival distributions of the two CD8+ T cell low classes in bladder cancer, glioma, and breast cancer, patients from the CD8+ T cell low/macrophage high class exhibited shorter survival times than the CD8+ T cell low/macrophage low class in lung adenocarcinoma. To confirm that these survival distributions were not unique to the individual datasets tested, we replicated these results in three additional datasets (Supplementary Figures S8 and S9).
Discussion
As immunotherapeutic approaches to cancer treatment become more widespread, it will be increasingly critical to understand the interplay of immune cells in the tumor microenvironment. We have demonstrated, using our computational framework, the utility of using patient gene expression profiles to dissect these interactions in multiple tumor types. Furthermore, we have shown that the presence of infiltrate from a single immune cell type is often not informative with regards to patient survival time. In some cases, the effect of an infiltrating immune cell type may be diminished by other cell types that act antagonistically. While much research has been done to identify the stimulatory and suppressive cell types present in the tumor microenvironment, our approach is the first to our knowledge that has used genomic data to demonstrate clinical differences between patients when adjusting for infiltration of multiple cell types.
Our co-infiltration analyses found that several different immune cell subsets tend to co-populate the tumor microenvironment together. However, using expression data to study cell-cell interactions in the tumor microenvironment can be biased by several confounders. To avoid classifying related cell types as co-infiltrating pairs, we assessed the similarity between our reference gene expression profiles and excluded all correlations that included pairs that were similar at a threshold of Rho > −0.1. Additionally, tumor purity is tied to immune infiltration, meaning that at some level infiltration of all immune cell types will be positively correlated. To address this, we adjusted for tumor purity and only claimed high confidence cell-cell co-infiltration if they were correlated with each other at a level of Rho > 0.45. These rigorous thresholds likely resulted in the exclusion of many true positive cell-cell interactions, but the remaining interactions were likely low in false positives, giving us a high-confidence set of interactions to study further. We recommend that future genomic-based analyses of tumor immune infiltration keep these confounders in mind and correct for them accordingly.
Our expression-based immune infiltration scores can be easily applied to examine the factors underlying tumor immunogenicity. While mutation load was previously linked to CD8+ T cell infiltration (10) and an overall decrease in tumor purity (20), links between mutation load and other immune cell subsets remained unclear. Our initial pan-cancer analyses found that only B cells were significantly associated with somatic mutation count. However, when excluding five outlier tumor types from our analysis, we found that mutation load was correlated with infiltration by CD8+ T cells, B cells, NK cells, and macrophages, implicating a role for mutation load in the overall tumor immune response. Notably, the five tumor types we excluded each deviated from this relationship in a unique way. We could not identify any commonalities between these tumor types that could explain this deviation, suggesting that there could be many additional drivers of tumor immunogenicity that may take precedence over mutation load in certain contexts. In support of this finding, we found that in nearly all cancer types, patients sharing a diagnosis did not exhibit an association between mutation load and immune infiltration. Exceptions to this were in colorectal and endometrial cancers, where MSI-based genomic instability was associated with increased immune infiltration. Going forward, it will be important to identify the immunogenic determinants specific to each tumor type, as current immunotherapeutic approaches are dependent on the presence of immune cells at the tumor microenvironment.
Studying the contribution individual immune cells make on patient survival can provide insights into the roles of each cell type in the context of the tumor microenvironment. However, the co-linear nature of immune cell infiltration can confound the effect a single cell type may have on patient survival. Despite this, our survival meta-analyses found that macrophage infiltration is associated with poor patient survival in many tumor types, while infiltration from CD8+ T cells and NK cells was highly protective. Furthermore, stratifying patients into four groups based on their levels of both T cell and macrophage infiltration revealed that these cell types act antagonistically in regards to patient survival. This result, combined with the negative survival associations we observed with other myeloid cell types and our macrophage phenotype analyses, suggests that the most prominent myeloid signals in the tumor microenvironment come from immunosuppressive myeloid cells and MDSCs. MDSCs have been characterized as potent inhibitors of the host immune response in cancer (45–48). Our results implicate their immunosuppressive role at a clinical level and we thus believe that future immunotherapeutic approaches should focus on inhibiting the myeloid compartment in the tumor microenvironment. Early success has been seen with the blockade of the protein V-domain Ig suppressor of T cell activation (VISTA). VISTA is constitutively and highly expressed in MDSCs and VISTA blockade has been shown to reduce their number, while also resulting in increased maturation of dendritic cells and stimulation of T cell-based anti-tumor immunity (49,50).
In this report, we have demonstrated the utility of genomic data for systematic immunological studies. However, this approach can be improved upon. The high expression of proliferation-associated genes by early stage effector CD8+ T cells led us to choosing a late-stage CD8+ T cell as a proxy for CD8+ T cell infiltration, even though early stage effectors are likely more representative of tumor-infiltrating CD8+ T cells. In addition, expression data provided limited resolution for distinguishing myeloid cells and other closely related cell types. To address these issues, future methods should maximize the differences between closely related cell types when defining weight profiles. Doing so will improve their accuracy, enabling a more comprehensive characterization of the tumor microenvironment. Another issue in this this study is that it does not account for the temporal nature of the tumor immune response. Longitudinal studies will be required to detect the transient immune cell interactions that cannot be captured by a single time point. There are also caveats to consider when performing multi-class survival analyses. The co-linear nature of immune infiltration requires that patients be stratified based on their infiltration levels for different cell types. However, doing so in the data currently available sometimes resulted in small numbers of patients in each class, limiting the power we have to detect associations. This may have explained the lack of survival difference between the CD8+ T cell high/macrophage low and CD8+ T cell high/macrophage high groups in breast cancer, as several studies have indicated that myeloid-based immune suppression of T cell responses takes place in this disease (44). Going forward, larger datasets containing both genomic and clinical information will be required to perform deeper analyses of the molecular interactions between immune cell types.
In conclusion, we have presented a rigorously controlled, comprehensive pan-cancer analysis that details the interplay between immune cells in the tumor microenvironment. Performing these analyses using high-dimensional data has helped delineate the role of individual immune cells across a large number of tumor types. Immune cells co-infiltrate solid tumors, and thus any analyses linking single cell types to patient survival should be carefully adjusted to account for the presence of other cell types. By doing so, we found that CD8+ T cells and myeloid cells exert antagonistic influences toward each other in the tumor microenvironment, emphasizing the importance of targeting the myeloid compartment in future immunotherapies. With the growing availability of genomic data and development of new computational approaches, our understanding of the tumor-immune interface will continue to expand, further improving our knowledge of basic tumor immunology and personalized immunotherapeutic approaches.
Supplementary Material
Acknowledgments
Financial support
This study was supported by the American Cancer Society (IRG-82-003-30) the National Center for Advancing Translational Sciences of the National Institutes of Health (UL1TR001086), and the Geisel School of Medicine at Dartmouth College start-up funding package (C. Cheng). F.S. Varn was additionally supported in part by the National Institute of General Medical Sciences of the National Institutes of Health (T32GM008704).
Footnotes
Disclosure of Potential Conflicts of Interests
The authors declare they have no competing interests.
References
- 1.Postow MA, Callahan MK, Wolchok JD. Immune Checkpoint Blockade in Cancer Therapy. J Clin Oncol. 2015;33:1974–82. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maio M, Grob JJ, Aamdal S, Bondarenko I, Robert C, Thomas L, et al. Five-year survival rates for treatment-naive patients with advanced melanoma who received ipilimumab plus dacarbazine in a phase III trial. J Clin Oncol. 2015;33:1191–6. doi: 10.1200/JCO.2014.56.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Champiat S, Lambotte O, Barreau E, Belkhir R, Berdelou A, Carbonnel F, et al. Management of immune checkpoint blockade dysimmune toxicities: a collaborative position paper. Ann Oncol. 2016;27:559–74. doi: 10.1093/annonc/mdv623. [DOI] [PubMed] [Google Scholar]
- 4.van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–42. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–71. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–11. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lines JL, Sempere LF, Broughton T, Wang L, Noelle R. VISTA is a novel broad-spectrum negative checkpoint regulator for cancer immunotherapy. Cancer Immunol Res. 2014;2:510–7. doi: 10.1158/2326-6066.CIR-14-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Yuan Y, Chen W, Putra J, Suriawinata AA, Schenk AD, et al. Immune-checkpoint proteins VISTA and PD-1 nonredundantly regulate murine T-cell responses. Proc Natl Acad Sci U S A. 2015;112:6682–7. doi: 10.1073/pnas.1420370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Mercier I, Lines JL, Noelle RJ. Beyond CTLA-4 and PD-1, the Generation Z of Negative Checkpoint Regulators. Front Immunol. 2015;6:418. doi: 10.3389/fimmu.2015.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24:743–50. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rutledge WC, Kong J, Gao J, Gutman DA, Cooper LA, Appin C, et al. Tumor-infiltrating lymphocytes in glioblastoma are associated with specific genomic alterations and related to transcriptional class. Clin Cancer Res. 2013;19:4951–60. doi: 10.1158/1078-0432.CCR-13-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–95. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Angelova M, Charoentong P, Hackl H, Fischer ML, Snajder R, Krogsdam AM, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015;16:64. doi: 10.1186/s13059-015-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varn FS, Andrews EH, Mullins DW, Cheng C. Integrative analysis of breast cancer reveals prognostic haematopoietic activity and patient-specific immune response profiles. Nat Commun. 2016;7:10248. doi: 10.1038/ncomms10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iglesia MD, Parker JS, Hoadley KA, Serody JS, Perou CM, Vincent BG. Genomic Analysis of Immune Cell Infiltrates Across 11 Tumor Types. J Natl Cancer Inst. 2016:108. doi: 10.1093/jnci/djw144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varn FS, Andrews EH, Cheng C. Systematic analysis of hematopoietic gene expression profiles for prognostic prediction in acute myeloid leukemia. Sci Rep. 2015;5:16987. doi: 10.1038/srep16987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30:1015–6. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971. doi: 10.1038/ncomms9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–45. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt M, Bohm D, von Torne C, Steiner E, Puhl A, Pilch H, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–13. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 23.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 24.Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–88. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng C, Yan X, Sun F, Li LM. Inferring activity changes of transcription factors by binding association with sorted expression profiles. BMC Bioinformatics. 2007;8:452. doi: 10.1186/1471-2105-8-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samuel A, Stouffer EAS, Devinney Leland C, Williams Robin M., Jr . The American Soldier: Adjustment During Army Life. Princeton University Press; 1949. [Google Scholar]
- 27.Jojic V, Shay T, Sylvia K, Zuk O, Sun X, Kang J, et al. Identification of transcriptional regulators in the mouse immune system. Nat Immunol. 2013;14:633–43. doi: 10.1038/ni.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shay T, Jojic V, Zuk O, Rothamel K, Puyraimond-Zemmour D, Feng T, et al. Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc Natl Acad Sci U S A. 2013;110:2946–51. doi: 10.1073/pnas.1222738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan Y, Failmezger H, Rueda OM, Ali HR, Graf S, Chin SF, et al. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med. 2012;4:157ra43. doi: 10.1126/scitranslmed.3004330. [DOI] [PubMed] [Google Scholar]
- 31.Best JA, Blair DA, Knell J, Yang E, Mayya V, Doedens A, et al. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat Immunol. 2013;14:404–12. doi: 10.1038/ni.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gazit R, Garrison BS, Rao TN, Shay T, Costello J, Ericson J, et al. Transcriptome analysis identifies regulators of hematopoietic stem and progenitor cells. Stem Cell Reports. 2013;1:266–80. doi: 10.1016/j.stemcr.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engelhardt B, Ransohoff RM. Capture, crawl, cross: the T cell code to breach the blood-brain barriers. Trends Immunol. 2012;33:579–89. doi: 10.1016/j.it.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–4. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–9. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maby P, Tougeron D, Hamieh M, Mlecnik B, Kora H, Bindea G, et al. Correlation between Density of CD8+ T-cell Infiltrate in Microsatellite Unstable Colorectal Cancers and Frameshift Mutations: A Rationale for Personalized Immunotherapy. Cancer Res. 2015;75:3446–55. doi: 10.1158/0008-5472.CAN-14-3051. [DOI] [PubMed] [Google Scholar]
- 38.Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 40.Nakano O, Sato M, Naito Y, Suzuki K, Orikasa S, Aizawa M, et al. Proliferative activity of intratumoral CD8(+) T-lymphocytes as a prognostic factor in human renal cell carcinoma: clinicopathologic demonstration of antitumor immunity. Cancer Res. 2001;61:5132–6. [PubMed] [Google Scholar]
- 41.Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci U S A. 2005;102:18538–43. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma P, Shen Y, Wen S, Yamada S, Jungbluth AA, Gnjatic S, et al. CD8 tumor-infiltrating lymphocytes are predictive of survival in muscle-invasive urothelial carcinoma. Proc Natl Acad Sci U S A. 2007;104:3967–72. doi: 10.1073/pnas.0611618104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Grainge MJ, Lee AH, et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol. 2011;29:1949–55. doi: 10.1200/JCO.2010.30.5037. [DOI] [PubMed] [Google Scholar]
- 44.Markowitz J, Wesolowski R, Papenfuss T, Brooks TR, Carson WE., 3rd Myeloid-derived suppressor cells in breast cancer. Breast Cancer Res Treat. 2013;140:13–21. doi: 10.1007/s10549-013-2618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–68. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marvel D, Gabrilovich DI. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J Clin Invest. 2015;125:3356–64. doi: 10.1172/JCI80005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khaled YS, Ammori BJ, Elkord E. Myeloid-derived suppressor cells in cancer: recent progress and prospects. Immunol Cell Biol. 2013;91:493–502. doi: 10.1038/icb.2013.29. [DOI] [PubMed] [Google Scholar]
- 49.Lines JL, Pantazi E, Mak J, Sempere LF, Wang L, O’Connell S, et al. VISTA is an immune checkpoint molecule for human T cells. Cancer Res. 2014;74:1924–32. doi: 10.1158/0008-5472.CAN-13-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Mercier I, Chen W, Lines JL, Day M, Li J, Sergent P, et al. VISTA Regulates the Development of Protective Antitumor Immunity. Cancer Res. 2014;74:1933–44. doi: 10.1158/0008-5472.CAN-13-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.