ABSTRACT
The pathogenesis of Bombyx mori cytoplasmic polyhedrosis virus (BmCPV) infection is unclear, although accumulating evidence indicates that circular RNAs (circRNAs), which act as competing endogenous RNAs or positive regulators, play important roles in regulating gene expression in eukaryotes and, thus, may play a role in BmCPV infections. To explore the expression and biological functions of circRNAs in the silkworm midgut following BmCPV infection, silkworm circRNA expression profiles of normal midgut tissue (control) and BmCPV-infected midgut tissue (test) were determined using high-through sequencing. A total of 9,753 and 7,475circRNAs were detected from the control and test samples, respectively. The two samples shared 6,085 circRNAs, while 646 and 737 circRNAs were expressed specifically in the control and test samples, respectively. A total of 3,638 circRNAs were shown to be differentially expressed, and 400 circRNAs were substantially differentially expressed with a fold-change ≥ 2.0 (p< 0.05 and a false discover rate < 0.05), of which 294 were up-regulated and 106 were down-regulated following infection. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were conducted to determine the principal functions of the substantially differentially regulated genes. circRNA-miRNA interaction networks were constructed based on a correlation analysis between the differentially expressed circRNAs and the nature of their microRNA (miRNA) binding sites. The network inferred that 13 miRNAs interacting with 193 circRNAs were among the 300 most abundant relationships. bmo-miR-3389-5p, bmo-miR-745-3p, and bmo-miR-3262 were related to 30, 34, and 34 circRNAs, respectively. circRNA_8115, circRNA_9444, circRNA_4553, circRNA_0827, and circRNA_6649 contained six, five, four, four, and four miRNA binding sites, respectively. We further found that alternative circularization of circRNAs is a common feature in silkworms and that the junction sites of many silkworm circRNAs are flanked by canonical GT/AG splicing signals. Our study is the first to show the circRNA response to virus infection. Thus, it provides a novel perspective on circRNA-miRNA interactions during BmCPV pathogenesis, and it lays the foundation for future research of the potential roles of circRNAs in BmCPV pathogenesis.
KEYWORDS: BmCPV, circRNA, expression profile, infection, miRNA, midgut, silkworm
Introduction
Circular RNAs (or circRNAs) are RNAs that form covalently closed, continuous loop structures with neither 5′ to 3′ polarity nor a polyadenylated tail, and they are considered to be a novel class of non-coding RNAs that have reshaped our understanding of RNA biology [1]. Because of their structures, circRNAs are highly stable in vivo compared with their linear counterparts [2]. The covalently closed ring structure of circRNAs is not only advantageous in terms of conferring protection against degradation from exonucleases, but it should also be beneficial to the application of RNAs as antisense RNAs, aptamers, ribozymes, or small interfering RNAs [3–6]. CircRNAs are found in all kingdoms of life, and they have been identified and characterized from viruses [7], plants[8], archaea [9], and animals [2,10–12].
Because of their abundance and stability, circRNAs are believed to be more effective than non-circular RNAs in sponging microRNAs (miRNAs) [13]. Because miRNAs are important for regulating protein expression and cellular physiology, circRNAs may exert roles in modulating cellular physiology such as cell proliferation and differentiation [14]. It was reported that circRNAs function as miRNA sponges that naturally sequester and competitively suppress miRNA activity to regulate target gene expression [13]. The circRNA CiRS-7 was reported to contain many binding sites for miR-7, which enables it to sponge this miRNA [15]. A circRNA called sex-determining region Y contains many miRNA binding sites, and it was reported to function as a miR-138 sponge [16]. CircHIPK3 contains nine miRNAs with 18 potential binding sites, which can directly bind to and inhibit miR-124, a well-known tumor suppressor [2]. Chondrocyte extracellular matrix-related circRNA regulates matrix metalloproteinase 13 expression by functioning as miR-136 sponge, and it participates in chondrocyte extracellular matrix degradation [17].
In addition to binding miRNAs, circRNAs also regulate the alternative splicing and transcription of linear RNAs. For example, the circRNA Mbl is generated from the second exon of the splicing factor MBL, which competes with canonical pre-mRNA splicing, eventhough the expression of parental genes is also regulated by circRNAs [18]. Recent studies have reported that several circRNAs can be translated into proteins in vitro or in vivo because they possess an internal ribosome entry site in the upstream of their start codons [19]. circRNAs have also been reported to modify the cellular destination and/or function of bound factors [20]. Additionally, evidence indicates that stable circRNAs can be reverse transcribed and integrated into the genome, thereby forming RNA-derived pseudogenes [21].
Many studies have reported that circRNA abundance, localization, function, and structure can be altered in response to various stimuli or pathological conditions in cells. Exposure to these conditions could alter circRNA interactions with DNA, RNA, or protein, which could impact the components of the ribonucleoprotein complex [20].
Many research groups have conducted studies to better understand the pathogenesis of Bombyx mori cytoplasmic polyhedrosis virus (BmCPV) infections of silkworms from miRNA, mRNA, and protein expression profiles. 58 miRNAs displayed significant differential expression were identified between the BmCPV infected midgut and normal midgut [22]. Another group applied next generation sequencing revealed 167 upregulated and 141 downregulated expression genes in larval instars following BmCPV infection. Several genes that could possibly be involved in B. mori immune response against BmCPV [23]. BmCPV infected midgut of 4008 silkworm strain were investigated with digital gene expression analysis, and a total of 752 differentially expressed genes were detected, of which 649 were upregulated and 103 were downregulated. Most of differentially expressed genes involved in signaling, gene expression, metabolic process, cell death, binding, and catalytic activity changes from the analysis of the expression profiles [24]. The expression profiles of some important differentially expressed genes were involved in signal transduction, enzyme activity and apoptosis changes [25]. The protein expression pattern in the silkworm midgut after BmCPV infection was also investigated with iTRAQ proteomic analysis, the results showed that 11 of the differentially expressed proteins were involved in immunity [26]. However, the alteration of circRNAs expression of silkworms following BmCPV infection remains unclear. In the present study, we performed a comprehensive analysis of the circRNAs in the midgut of silkworms following BmCPV infection to illuminate the potential functions of genome-encoded circRNAs during virus infection.
Results
Properties of circRNAs from silkworm midguts
The circRNAs in the midgut of silkworms were investigated with CircSeq. High- throughput sequencing detected thousands of circRNA transcripts in the midgut of the control silkworms. As shown in Table S1, 100,415,764 clean reads were identified from 101,981,470 raw reads of the control silkworm midgut after discarding low-quality data. A total of 9,753 circRNAs were identified from the silkworm midgut. The average length of the circRNAs in the silkworm was 1,200 nt, while the maximum length was 94,030 nt, and the minimum length was only 54 nt (Table S2). The size of the silkworm circRNA candidates ranged from less than 200 nt to greater than 2,000 nt, most of which ranged from 200 nt to 1,500 nt (Fig. 1A). The GC content of the identified circRNAs ranged from 45% to 65% (Fig. 1B). The biogenesis of the circRNAs via back-splicing differed from the canonical splicing of linear RNAs, but they were all generally formed by alternative splicing of pre-mRNAs. To provide a comprehensive landscape of the origins of the circRNAs, linear transcripts of circRNAs from the corresponding genes were annotated, and the circRNA distribution in the genome was also explored according to the location of the chromosome from which the circRNA sequence overlapped (Table S3). The results showed that according to the localization of the circRNAs in the genome, the circRNAs were classified into five categories, including 93.79% sense overlapping circRNAs, 3.12% exonic circRNAs, 2.29% intergenic circRNAs, 0.29% intronic circRNAs, and 0.34% antisense circRNAs. Most of the circRNAs were derived from the sense-overlapping strategy (Fig. 2). The origins of the circRNAs were also compared between the test and control samples, although no significant disparities were observed. The results also indicated that circRNA formation was not induced by BmCPV infection (Fig. 2). The splicing signals of the circRNAs identified from the midgut of the BmCPV-infected silkworms was investigated, and the results showed that most of the splicing signals had the canonical GT/AG sequence, which is similar to that of mammals (Fig. 3). Eventhough divergent primers were applied to validate the circRNAs from the test samples, alternative splicing events were found in the silkworm midgut, as one pair of the divergent primers identified several circRNAs with different back-spliced junctions. These results indicate that the prevalence of circular alternative splicing events within the circRNAs shared the same splicing acceptors and donors (data not shown).
Figure 1.
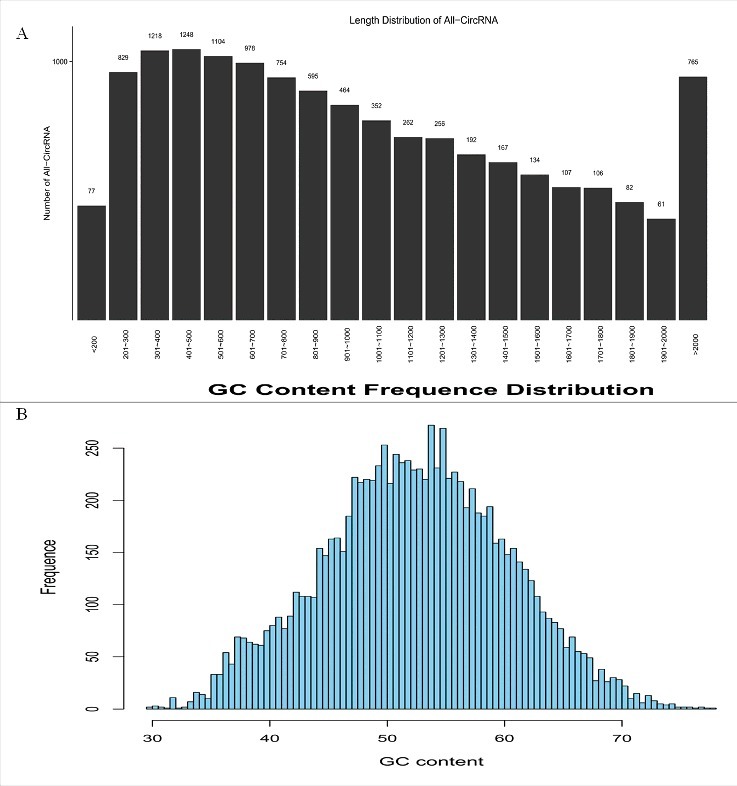
Characterization of the identified circRNAs from the silkworm midgut. (A) The length distribution of the identified circRNAs. (B) The GC content of the identified circRNAs in the midgut.
Figure 2.
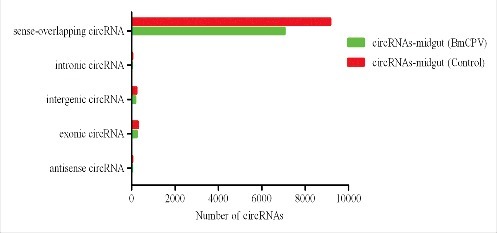
Types and numbers of circRNAs detected from the midguts of silkworms. The circRNAs are classified into five types according to their relationships with associated coding genes, as well as their genomic loci.
Figure 3.

Splice signals of the identified circRNAs in humans and silkworms.
Annotation of the most related linear transcripts and identification of differentially expressed circRNAs
High-throughput sequencing is an efficient approach for investigating the biological function of RNAs. CircSeq detected 9,753 and 7,475 circRNAs in the control and test samples, respectively (Table S2). Expression of circRNAs in BmCPV infected midgut and control silkworm midgut was measured based on RPM (mapped back-splicing junction reads per million mapped reads), which indicated no abnormal expression in the detected samples (Fig. 4A). A total of 6,085 circRNAs were shared between the two samples, while 646 and 737 circRNAs were expressed specifically in the control and test samples, respectively (Fig. 4B).
Figure 4.

Expression profiles of circRNAs detected by CircSeq in normal midguts and BmCPV-infected midguts. (A) Box plots of the RPM value of the circRNAs in two groups. (B) Venn analysis of the circRNAs detected in normal and BmCPV-infected silkworm midguts. (C) MA of differential expression circRNAs. (D) The significant differential expression of circRNAs between normal and BmCPV-infected silkworm midguts (fold change ≥2.0, P<0.05 and FDR<0.05).
CircSeq identified 3,638 circRNAs whose expression was dysregulated in the BmCPV-infected midguts. A filtering analysis at a fold change ≥2.0 (p< 0.05 and a false discovery rate (FDR) < 0.05) showed that 400 circRNAs were substantially differentially expressed between the control and test samples, among which 294 were up-regulated and 106 were down-regulated in the midgut of the BmCPV-infected silkworms (Fig. 4C, D). CircRNA-3112 (fold-change: ∼2,544) was one of the most up-regulated circRNAs. This circRNA was classified as a sense-overlapping circRNA, and the most closely related linear transcript was that of the B. mori spectrin beta chain, non-erythrocytic 5 (XM_012689500.1), while circRNA_4672 was considered to be the most down-regulated circRNA. This circRNA was also classified as a sense-overlapping circRNA, and the most closely related linear transcript was that of the B. mori sodium-dependent nutrient amino acid transporter 1-like (XM_004926983.2). The top 20 circRNAs based on their fold-change are summarized in Table 1, and the circRNA-miRNA network was also predicted. B.mori neuropeptide receptor A35 (NGR-A35) and B. mori protein Wnt-1 (WNT-1) were also among the top 20 up-regulated circRNAs, as revealed by the high-throughput sequencing analysis.
Table 1.
The top 20 dysregulated circRNAs in BmCPV infected midgut.
| Name |
Fold Change |
log2FoldChange |
P value |
Type |
Best related linear transcript (Gene symbol) |
| Upregulation | |||||
| circRNA_3112 | 2544.289157 | 11.31304693 | 0.001624 | sense-overlapping | XM_012689500.1 |
| circRNA_9582 | 1605.325301 | 10.64864996 | 0.002667 | sense-overlapping | XM_012696726.1 |
| circRNA_9081 | 1479.120482 | 10.53052386 | 0.002946 | sense-overlapping | XM_012696281.1 |
| circRNA_7454 | 1206.518072 | 10.23663381 | 0.00267 | sense-overlapping | XM_012694536.1 |
| circRNA_2952 | 999.5421687 | 9.965123622 | 0.00494 | sense-overlapping | XM_012689347.1 |
| circRNA_3493 | 822.8554217 | 9.684495157 | 0.006523 | antisense | XM_012689767.1 |
| circRNA_5655 | 777.4216867 | 9.602553543 | 0.007092 | sense-overlapping | XM_012692464.1 |
| circRNA_4553 | 772.373494 | 9.593154845 | 0.004362 | sense-overlapping | NM_001113265.1 |
| circRNA_3101 | 742.0843373 | 9.535439347 | 0.004579 | sense-overlapping | XM_012689500.1 |
| circRNA_6031 | 731.9879518 | 9.515676092 | 0.007758 | sense-overlapping | XM_012692613.1 |
| Downregulation | |||||
| circRNA_0327 | 0.004432127 | −7.817785029 | 0.018598 | sense-overlapping | XM_012690535.1 |
| circRNA_2873 | 0.003871313 | −8.012961152 | 0.015695 | sense-overlapping | XM_004924781.2 |
| circRNA_8162 | 0.002981583 | −8.389705771 | 0.021277 | sense-overlapping | XM_012695334.1 |
| circRNA_6649 | 0.002857468 | −8.451047141 | 0.008593 | sense-overlapping | XM_004929799.2 |
| circRNA_3450 | 0.00251404 | −8.635776552 | 0.008501 | sense-overlapping | XM_012689737.1 |
| circRNA_1875 | 0.002031466 | −8.943263134 | 0.007605 | sense-overlapping | XM_012697844.1 |
| circRNA_8700 | 0.00194236 | −9.007973916 | 0.007277 | sense-overlapping | XM_004932630.2 |
| circRNA_3508 | 0.001897856 | −9.041413396 | 0.025578 | sense-overlapping | XM_004925402.1 |
| circRNA_7474 | 0.001783184 | −9.131329032 | 0.006708 | sense-overlapping | XM_004930980.2 |
| circRNA_4672 | 0.000545456 | −10.84025027 | 0.003145 | sense-overlapping | XM_004926983.2 |
GO and KEGG pathway analyses
Under the assumption that circRNA function is related to the known function of the host linear transcripts, further analysis of the response of the silkworm circRNAs to the silkworm-specific pathogen BmCPV was conducted, and the top 20 dysregulated GO processes of each subgroup (BP, CC, and MF) were analyzed according to the enriched, dysregulated circRNAs derived from the gene annotation. Prediction terms with a p-value less than 0.05 were selected and ranked by their p-value. According to the routine GO classification algorithms, an enrichment score was used to enrich the significant GO terms of the differentially expressed genes. For the up-regulated circRNAs, the top three GO processes included homophilic cell adhesion via plasma membrane adhesion molecules, phospholipid transport, and cell communication in the BP subgroup. The kinesin complex, microtubule, and myosin complex were the top three processes in the CC subgroup, andmagnesiumion binding, phosphatidylinositol binding, and phospholipid-translocating ATPase activity were the top three processes in the MF subgroup (Fig. 5A, Table S4). The top three GO processes predicted by the down-regulated circRNAs were the same as those predicted for the up-regulated circRNAs. Enrichment scores were also used to enrich the significant GO terms of the differentially expressed circRNAs, where the top three processes were transmembrane transport, oxidation-reduction process, and transport in the BP subgroup, integral component of membrane, membrane, and nucleus in the CC subgroup, and catalytic activity, oxidoreductase activity, and hydrolase activity in the MF subgroup (Fig. 5B, Table S5). Furthermore, 69 and 38 KEGG pathways (p< 0.05) were identified among the up-regulated and down-regulated circRNAs, respectively. The top 20 KEGG pathways in the dysregulated circRNAs are shown in Fig. 6, and they included the Notch signaling pathway, ABC transporters, and the FoxO signaling pathway (Table S6).
Figure 5.
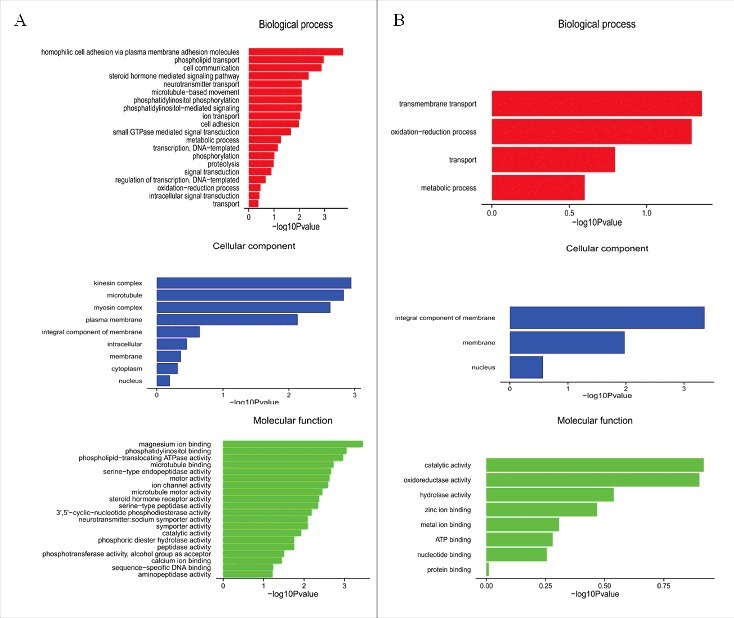
GO analysis of the up-regulated (left panel) and down-regulated (right panel) circRNAs of the control and test groups under the themes of BP, CC, and MF.
Figure 6.
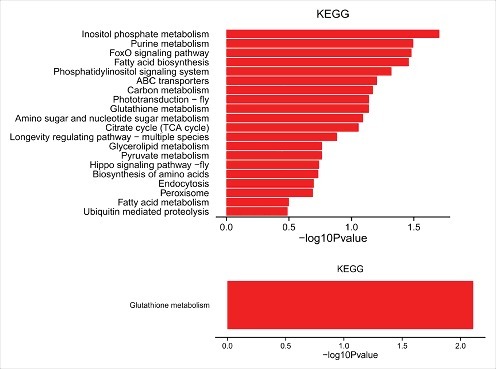
20 KEGG pathways of significantly up-regulated (upper panel) and down-regulated (lower panel) mRNAs.
Construction of a circRNA-miRNAinteraction network
The high-throughput RNA sequencing results were used to reveal the circRNA-miRNA interaction network. The constructed network map contained the top 300 relationships among the circRNAs and the miRNAs, as ranked by the p-value of the hypergeometric distribution. A comparison of the circRNA-miRNA networks of the test and control samples revealed that 13 miRNAs interacting with 193 circRNAs were among the top 300 relationships that were predicted to have closer connections after BmCPV infection (Fig. 7), and Bmo-miR-278-3p bound circRNA_9444, circRNA_8115, circRNA_4553, and circRNA_6649.
Figure 7.

The circRNA-miRNA co-expression network in the silkworm midgut infected with BmCPV compared with that of the normal midgut. Triangle nodes represent circRNAs and circle nodes represent miRNAs. (A) The top 300 miRNA-circRNA interaction networks. (B) circRNAs that sponged more than four miRNAs.
Verification of silkworm circRNAs
According to the sequencing results, 6 circRNAs were selected randomly for validation with divergent primers. Distinct products of the expected size were amplified using the divergent primers and confirmed by Sanger sequencing (Fig. 8). The results showed that the selected 6 circRNAs in the silkworm midgut had covalently closed, continuous loop structures with neither 5′ to 3′ polarity nor a polyadenylated tail, which is consistent with predictions.
Figure 8.

Validation of circRNA expression by RT-PCR and Sanger sequencing. Arrows represent divergent primers binding to the genomic region of circRNAs.
The response of circRNAs to BmCPV infection
To understand the expression level change after BmCPV infection in the midgut, the expression level of five circRNAs (circRNA-1193, 2439, 5655, 6031 and 0962) was investigated and the results showed that the expression level of selected circRNAs were induced by BmCPV infection (Fig. 9). The expression tendency was consistent with the CircSeq results.
Figure 9.
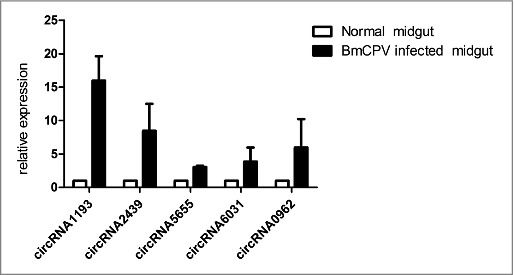
Validation of the expression of circRNAs.
Discussion
BmCPV is a silkworm-specific pathogen, and BmCPV infections can be established orally. Many studies have reported the differential expression levels of mRNAs and miRNAs following BmCPV infection, and they have comprehensively illuminated the functions of these RNAs in the virus life cycle. However, circRNAs, which were identified in the 1990s, are novel non-coding RNAs that are formed by exon back-splicing (circularization) [16,27]. Many circRNAs have been identified in various cell lines and across different species. Whether this type of non-coding RNA is transcribed in invertebrate silkworms, and whether the expressions of these novel circRNAs are affected by BmCPV infection were previously unknown. To better understand these issues, in the present study, BmCPV-infected and normal silkworm midguts were isolated and analyzed by high-throughput sequencing. Thousands of circRNAs were found in the non-infected and BmCPV-infected silkworm midguts, and the characterization of these circRNAs indicated that they are widespread taxonomically, as they were identified in humans and other species. For example, most of the silkworm circRNAs were flanked by canonical GT/AG splicing signals, and the splicing signals of the silkworm circRNAs were similar to those of the circRNAs of other species. Direct back-splicing and exon skipping are two different mechanisms that have been suggested to produce mammalian exonic circRNAs [28]. A canonical spliceosome is used for back-splicing via these two mechanisms [29]. From the comparative analysis, we concluded that the silkworm circRNA formation mechanism uses conserved canonical GT/AG splicing signals and that the mechanism did not change in response to BmCPV infection. This suggests that the alternative circularization of circRNA is a common feature in silkworms. An interesting phenomenon discovered in the present study is that alternative spliced isoforms are present in silkworm circRNAs. CircRNAs derived from back-spliced exons have been shown to be co-expressed with their linear counterparts. A single gene locus can produce multiple circRNAs through alternative back-splice site selection and/or alternative splice site selection [30]. By combining an algorithm with long-read sequencing data and experimental validation, the prevalence of alternative splicing events within circRNAs were revealed in a previous study by comprehensively investigating the internal components of circRNAs in 10 human cell lines and 62 fruit fly samples [31]. Therefore, the discovery of alternative splicing events of circRNAs in silkworms indicates that such events are conserved among different species. Various circRNA isoforms from the same host linear transcripts were expressed in the silkworm midgut during BmCPV infection, which implies that they play important roles in the temporal regulation of midgut circRNAs. RNA misregulation of alternative splicing is considered to be an important factor in several types of cancer. Whether this is involved in BmCPV infection requires further investigation [32].
The expression patterns of many mRNAs and miRNAschange after BmCPV infection. However, the circRNA expression patterns of silkworms following BmCPV infection are still unclear. Therefore, in the present study, circRNA expression levels were investigated with CircSeq, and the results showed that 646 and 737 circRNAs were expressed specifically in the normal and BmCPV-infected midgutsofsilkworms, respectively, and 6,085 circRNAs were shared between the two samples. Among these, 294 up-regulated and 106 down-regulated circRNAs (fold-change ≥ 2.0, p < 0.05, and FDR < 0.05) were identified in BmCPV-infected midguts.
Moreover, GO and KEGG pathway analyses were analyzed to explore the biological functions and potential mechanisms of the silkworm circRNAs in response to BmCPV infection. The GO enrichment analysis revealed that some genes are involved in the regulation of BPs, CCs, and MFs. Among the GO terms found in this study, the top three GO processes of the up-regulated circRNAs included magnesium ion binding, phosphatidylinositol binding, and phospholipid-translocating ATPase activity in the MF subgroup. The top three GO processes predicted for the down-regualted circRNAs were activity, oxidoreductase activity, and hydrolase activity in the MF subgroup. Remarkably, the KEGG pathways in the dysregulated circRNAs included the Notch signaling pathway, ABC transporters, and the FoxO signaling pathway. These findings indicate that several circRNAs might be involved in the development and progression of BmCPV infection.
BmCPV specifically infects the epithelial cells of the silkworm midgut, and as the disease progresses, white wrinkles typically occur in the posterior part of the midgut. Consequently, the digestive and absorptive functions of the midgut are severely affected. An analysis of target genes of the downstream miRNAs of the top 20 down-regualted circRNAs showed that most of them are involved in transport and absorption. This suggests that the reduced expression of these genes might lead to the sluggishness of the silkworms. Moreover, because midgut peristalsis is involved in digestion and defecation, BmCPV-infected silkworms lose their appetite and have loosebowels, which might be caused by the downregulation of the expression of the dynein and kinesin genes.
When silkworms are challenged with BmCPV, the silkworm immune system is activated to generate immunoglobulin-like proteins and antiviral factorsthat resist the infection. In the present study, many immune genes were up-regulated, and they were the target genes of the downstream miRNAs of the top 20 up-regulated circRNAs. CircRNAs have been reported to function as miRNA sponges that naturally sequester and competitively suppress miRNA activity to regulate target gene expression. Hundreds of dysregulated circRNAs in the silkworm midgut were identified after BmCPV infection, which might be helpful for investigations of circRNA and host miRNA interactions. Many circular RNAs, such as CiRS-7, sex-determining region Y, and CircHIPK3, contain many miRNA binding sites that can function as sponges [2,13,16]. Given that miRNAs play important roles in the progression of BmCPV infection, some circRNAs are likely be involved in BmCPV infection by interacting with miRNAs. In the present study, circRNA-miRNA interaction networks were established to predict the relationships between circRNAs and miRNAs. One or more miRNA binding sites were found in the majority of the identified circRNAs based on sequences analysis. An examination showed that Bmo-miR-278-3p binds circRNA_9444, circRNA_8115, circRNA_4553 and circRNA_6649, and a previous study showed that miR-278-3p negatively regulates the expression of the insulin-related peptide binding protein 2 gene in silkworm larvae, while it positively regulates the mRNA transcript levels of BmCPV [33]. These results suggest that miR-278-3p may play an important role in BmCPV replication by interacting with host-genome-encoded circRNAs. From the interaction network, the obvious conclusion is that bmo-miR-3389-5p, bmo-miR-745-3p, and bmo-miR-3262 are related to 30, 34, and 34 circRNAs, respectively. circRNA_8115, circRNA_9444, circRNA_4553, circRNA_0827, and circRNA_6649 contain six, five, four, four, and four miRNA binding sites, respectively. These results indicate that circRNAs might regulate the progression of BmCPV infection by binding miRNAs that regulate the expression of associated genes, although the exact regulatory mechanism requires further investigation.
Together, these findings provide potential targets for the future treatment of BmCPV infection,as well as novel insights into BmCPV infection biology. Additional investigations of the circRNAs identified in this study will focus on their association with BmCPV infection and biological functions.
Materials and methods
Silkworm strain
Silkworm larvae of the Dazao strain were fed mulberry (Morus sp.) leaves and kept at 26 ℃ with 70%–85% relative humidity and a 12 h light:12 h dark photoperiod in our laboratory at Soochow University.
Virus inoculation
Silkworm larvae in the test group were inoculated orally with a suspension of 106 polyhedra/ml in distilled water on the 1st day of the 3rd instar. The oral rearing processwas conducted as described previously [34]. The midguts were respectively collected at 6, 12, 24, 48, and 72 h post-inoculation from 30 silkworms. Meanwhile, silkworms fed fresh mulberry leaves coated with sterile water served as a control group.
RNA extraction
All the midguts were mixed, and the total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Total RNA samples were treated with DNase I (using the Ambion Turbo DNA-free kit) and NucAway spin columns (Thermo Fisher Scientific) according to the manufacturer's protocol to remove DNA contamination and salts. Total RNA quality was determined using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
RNase R digestion
Twenty micrograms of total RNA was incubated at 70℃ for 5 min, and then 3 μL of Rnase R was added and incubated at 37℃ for 10 min. After the reaction, treated RNA was purified with the Agencourt RNAClean XP (Beckman Coulter, Brea, CA, USA) and RNeasy MinElute Cleanup kits (Qiagen, Valencia, CA, USA). The quality of the RNA was analyzed with a 2100 Bioanalyzer (Agilent).
Ribo-Zero depletion of rRNA
One microgram of RNA digested with RNase R was treated with the TruSeq Stranded Total RNA with Ribo-Zero Gold Kit (Illumina, San Diego, CA, USA) to remove rRNAs according to the manufacturer's protocol. The treated RNA was fragmented with an Ambion RNA fragmentation kit (Thermo Fisher Scientific).
First and second strand cDNA synthesis
Eight microliters of the First Strand Synthesis Act D Mix and SuperScript II Reverse Transcriptase was added to the Ribo-Zero depleted RNA, and incubated at 25 °C for 10 min, 42°C for 15 min, and 70°C for 15 min. Then, 5 μL of the End Repair control (2 μL of the End Repair control + 98 μL of resuspension buffer) and 20 μL of Second Strand Marking Master Mix were mixed and incubated 16°C for 60 min. RNA purification was conducted using the Bioanalyzer 2100 RNA-6000 Nano Kit (Agilent) according to the manufacturer's instructions.
Adenylation of 3′ends and ligation ofadapters
A-Tailing Control (12.5 μL)(1 μL of A-Tailing Control + 99 μL of resuspension buffer) was mixed with 12.5 μL of A-Tailing Mix and then incubated at 37°C for 30 min and 70°C for 5 min. A ligation control (2.5 μL)(1 μL of the ligation control + 99 μL of resuspension buffer) was mixed with 2.5 μL of the ligation mix and 2.5 μL of the RNA Adapter Index, and then it was incubated at 30°C for 10 min; 5 μL of ligation stop buffer was added to stop the reaction.
RNA sequencing library preparation and Illumina sequencing
Sequencing libraries were constructed according to the manufacturer's instructions (Illumina) and quality controlled on the 2100 Bioanalyzer (Agilent). Illumina sequencing was performed at Shanghai OE Biotech. Co., Ltd. (Shanghai, China). Briefly, after removal ribosomal RNA and then constructing the library, the circRNA sequencing was performed. The clean reads were aligned to the reference genome by Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/manual.shtml). For unmapped reads, the junctions were picked out using back-splice algorithm. Finally, circRNAs were verified with a software developed by OE which were considered as the reference sequence for further analysis. The expression level of circRNAs was measured by “Mapped back-splicing junction reads per million mapped reads” (RPM).
Validation of silkworm circRNAs
To validate the authenticity of the identified circRNAs, the expression levels of 6 circRNAs were selected randomly for validation with divergent primers, which were designed to amplify each circRNA splicing junction, and the results were confirmed by Sanger sequencing. The primers are listed in Table S7.
Bioinformatics analysis
CircRNA Gene Ontology (GO) functional significance and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed with the original genes of the identified, differential expressed circRNAs. The GO analysis (http://www.geneontology.org) was used to construct a meaningful annotation of genes and gene products in a wide variety of organisms. The ontology covered the domains of biological processes (BPs), cellular components (CCs), and molecular functions (MFs). The −log10 value (p-value) denotes the enrichment score representing the significance of the GO enrichment term among the differentially expressed genes. We also performed a KEGG pathway analysis to harvest pathway clusters covering our knowledge of the molecular interaction and reaction networks during differentially regulated gene profiling. Additionally, the −log10 value (p-value) denotes the enrichment score showing the significance of the pathway correlations. miRNA targets were predicted with miRanda software, and the miRNA sequences were obtained from miRBase. For miRNA target gene predictions, we extracted all 3′-untranslated regions of silkworms (Dazao strain), which were downloaded from the National Center for Biotechnology Information UniGene database (http://www.ncbi.nlm.nih.gov/sites/entrez?db = unigene), and the PITA program, which accounts for target accessibility during the interaction of miRNAs and their targets, was employed to predict miRNAs targets using default parameters. We selected and analyzed the target genes with a Gibbs’ standard free energy value of 25 kcal/mol from the original predictions. An miRNA-circRNA network was constructed according to the common target miRNAs of the circRNAs. The interactions of circRNAs and mRNAs with miRNAs were predicted with the Arraystar miRNA target prediction software based on TargetScan and miRanda[35,36].
Validation of the expression of circRNAs
To validate the data from the Circseq, 5 circRNAs (circRNA1193, circRNA2439, circRNA5655, circRNA6031 and circRNA0962) were selected and validated with real-time PCR. Specific divergent primers for each circRNA were designed according to the sequence of the linear transcripts, and all divergent primers were shown in the Table S7. Then total RNA was extracted from the BmCPV infected midgut, digested using RNase R and purified, cDNA was synthesized using the EasyScript One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgen biotech, China). Real-time PCR was performed on a CFX96 Touch Deep Well Real-Time PCR Detection System (Bio-Rad, Berkeley, USA) according to the manufacturer instructions of the iTaq™ Universal SYBR Green Supermix (Bio-Rad, Berkeley, USA). The expression levels of circRNAs were normalized to β-actin (internal standard control) and were calculated using the 2−ΔΔCt method. All experiments were done in triplicate.
Supplementary Material
Funding Statement
National Natural Science Foundation of China (NSFC) (31602007); Suzhou Agricultural Science and Technology Innovation Project (SNG2017046); National Natural Science Foundation of China (NSFC) (31272500).
Disclosure statement
The authors declare that they have no conflict of interest.
Acknowledgments
This work has got the financial support of the National Natural Science Foundation of China (grant no. 31602007, 31272500), the Suzhou Agricultural Science and Technology Innovation Project (grant no. SNG2017046), and a project funded by the Priority Academic Program of Development of Jiangsu Higher Education Institutions. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- [1].Kosik KS. Molecular biology: Circles reshape the RNA world. Nature. 2013;495:322-4. [DOI] [PubMed] [Google Scholar]
- [2].Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Abe N, Abe H, Ito Y. Dumbbell-shaped nanocircular RNAs for RNA interference. J Am Chem Soc. 2007;129:15108–9. [DOI] [PubMed] [Google Scholar]
- [4].Bohjanen PR, Colvin RA, Puttaraju M, et al. A small circular TAR RNA decoy specifically inhibits Tat-activated HIV-1 transcription. Nucleic Acids Res. 1996;24:3733–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Umekage S, Kikuchi Y. In vitro and in vivo production and purification of circular RNA aptamer. J Biotechnol. 2009;139:265–72. [DOI] [PubMed] [Google Scholar]
- [6].Abe N, Abe H, Nagai C, et al. Synthesis, structure, and biological activity of dumbbell-shaped nanocircular RNAs for RNA interference. Bioconjugate Chem. 2011;22:2082–92. doi: 10.1021/bc2003154. [DOI] [PubMed] [Google Scholar]
- [7].Kos A, Dijkema R, Arnberg AC, et al. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–60. [DOI] [PubMed] [Google Scholar]
- [8].Flores R, Grubb D, Elleuch A, et al. Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: variations on a theme. RNA Biol. 2011;8:200–6. [DOI] [PubMed] [Google Scholar]
- [9].Lykke-Andersen J, Aagaard C, Semionenkov M, et al. Archaeal introns: splicing, intercellular mobility and evolution. Trends in biochemical sciences. 1997;22:326–31. [DOI] [PubMed] [Google Scholar]
- [10].Zhang X, Yan Y, Lei X, et al. Circular RNA alterations are involved in resistance to avian leukosis virus subgroup-J-induced tumor formation in chickens. Oncotarget. 2017;8:34961–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shen Y, Guo X, Wang W. Identification and characterization of circular RNAs in zebrafish. FEBS Letters. 2017;591:213–20. [DOI] [PubMed] [Google Scholar]
- [12].Zhang C, Wu H, Wang Y, et al. Circular RNA of cattle casein genes are highly expressed in bovine mammary gland. J Dairy Sci. 2016;99:4750–60. [DOI] [PubMed] [Google Scholar]
- [13].Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. [DOI] [PubMed] [Google Scholar]
- [14].Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer (Review). Oncol Reports. 2015;33:2669–74. [DOI] [PubMed] [Google Scholar]
- [16].Capel B, Swain A, Nicolis S, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–30. [DOI] [PubMed] [Google Scholar]
- [17].Liu Q, Zhang X, Hu X, et al. Circular RNA Related to the Chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘Sponge’ in human cartilage degradation. Scientific Reports. 2016;6:22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. [DOI] [PubMed] [Google Scholar]
- [19].Shen T, Han M, Wei G, et al. An intriguing RNA species–perspectives of circularized RNA. Protein Cell. 2015;6:871–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Panda AC, Grammatikakis I, Munk R, et al. Emerging roles and context of circular RNAs. Wiley Interdisciplinary Rev RNA. 2017;8. doi: 10.1002/wrna.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. Rna. 2015;21:172–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wu P, Han S, Chen T, et al. Involvement of microRNAs in infection of silkworm with bombyx mori cytoplasmic polyhedrosis virus (BmCPV). PloS One. 2013;8:e68209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kolliopoulou A, Van Nieuwerburgh F Stravopodis DJ, Deforce D, et al. Transcriptome analysis of Bombyx mori larval midgut during persistent and pathogenic cytoplasmic polyhedrosis virus infection. PloS One. 2015;10:e0121447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gao K, Deng XY, Qian HY, et al. Digital gene expression analysis in the midgut of 4008 silkworm strain infected with cytoplasmic polyhedrosis virus. J Invertebrate Pathol. 2014;115:8–13. [DOI] [PubMed] [Google Scholar]
- [25].Gao K, Deng XY, Qian HY, et al. Cytoplasmic polyhedrosis virus-induced differential gene expression in two silkworm strains of different susceptibility. Gene. 2014;539:230–7. [DOI] [PubMed] [Google Scholar]
- [26].Gao K, Deng XY, Shang MK, et al. iTRAQ-based quantitative proteomic analysis of midgut in silkworm infected with Bombyx mori cytoplasmic polyhedrosis virus. J Proteomics. 2017;152:300–11. [DOI] [PubMed] [Google Scholar]
- [27].Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell. 1991;64:607–13. [DOI] [PubMed] [Google Scholar]
- [28].Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Reports. 2015;10:103–11. [DOI] [PubMed] [Google Scholar]
- [30].Zhang XO, Dong R, Zhang Y, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gao Y, Wang J, Zheng Y, et al. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat Commun. 2016;7:12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genetics. 2016;17:19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wu P, Qin G, Qian H, et al. Roles of miR-278-3p in IBP2 regulation and Bombyx mori cytoplasmic polyhedrosis virus replication. Gene. 2016;575:264–9. [DOI] [PubMed] [Google Scholar]
- [34].Hu X, Zhu M, Wang S, et al. Proteomics analysis of digestive juice from silkworm during Bombyx mori nucleopolyhedrovirus infection. Proteomics. 2015;15:2691–700. [DOI] [PubMed] [Google Scholar]
- [35].Enright AJ, John B, Gaul U, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genetics. 2012;13:271–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


