ABSTRACT
3′ end processing is required for the maturation of all eukaryotic RNAs. Current model suggests that canonical mRNA 3′ processing is carried out exclusively within a protein complex termed mRNA 3′ processing complex. In a recent study, by using RNA-biotin based pull-down assay and high-throughput sequencing, we reported that a subset of small nucleolar RNAs (snoRNAs) were physically associated with this macromolecular machinery. Through detailed characterization of one of these snoRNAs, SNORD50A, we revealed that non-coding RNA, such as snoRNA, may play a regulatory role in mRNA 3′ processing. Our results provided novel insight into both the regulatory mechanism of mRNA 3′ processing and the non-canonical functions of snoRNAs.
KEYWORDS: mRNA 3′ processing, cleavage, polyadenylation, snoRNA
Introduction
Before becoming functional molecules, almost all the eukaryotic pre-mRNAs and many non-coding RNAs are subject to cleavage/polyadenylation at the 3′ end, which takes place in a macromolecular machinery called mRNA 3′ processing complex [1–3]. Recent biochemical purification and proteomic analysis showed that 3′ processing complex is comprised of ∼85 proteins, including several core factors such as poly(A) polymerase and four multi-subunit protein complexes (CPSF, CstF, CF Im, CF IIm) (Fig. 1A, upper panel), as well as non-core factors that may associated with 3′ processing regulation [3]. CPSF recognizes AAUAAA element via Wdr33 and CPSF30 [4,5], CstF interacts with downstream G/U rich element via CstF64 or CstF64tau [6,7], CF Im contributes to mRNA 3′ processing via its interaction with ‘UGUA’ element within polyadenylation site (PAS) [8], The exact functions of CF IIm in mRNA 3′ processing is yet to be confirmed [9]. Notably, almost all protocols that had been previously applied for 3′ processing factors purification do not include the identification of trans-acting RNAs [3,10], if there exist any. To systematically characterize mRNA 3′ processing complex, we have recently purified RNAs that are associated with this complex [11]. Strikingly, snoRNAs were almost all the high-confidence candidates. Importantly, we demonstrated the functionality of one of these snoRNAs, and our results showed SNORD50A can inhibit mRNA 3′ processing via interfering with the interaction of Fip1 and PAS (Fig. 1A, lower panel). The key findings of our study and their significance will be discussed below.
Figure 1.
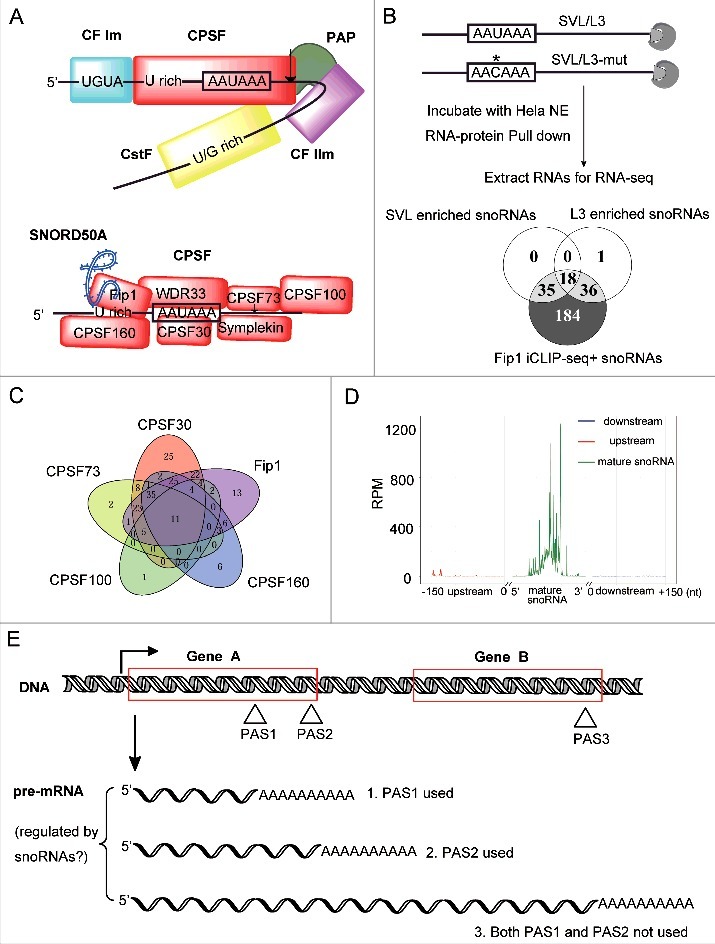
snoRNAs associate with CPSF in vivo and its potential role in gene regulation. (A) Schematic drawing of the assembly of four multi-subunit protein complexes (CPSF, CstF, CF Im, CF IIm) and PAP on PAS RNA sequence (upper panel), the lower panel shows the composition of CPSF complex and the association of SNORD50A with CPSF via Fip1. (B) Schematic representation of the SVL and L3 RNA substrates used in the biotin–streptavidin pull-down assays (upper panel). The AAUAAA hexamer in wild-type RNA substrate and AACAAA in mutant substrate (boxes) are shown. The asterisk is used to highlight the single nucleotide change. Venn diagram showing the number of snoRNAs in three datasets (lower panel). Cutoff values of snoRNA reads were set as below. SVL and L3 RNA pull-down assays: fold enrichment >1.5 and average RPM>100; Fip1 iCLIP-seq: RPM>10. (C) Venn diagram showing the number of snoRNAs detected by CLIP-seq experiments of several CPSF factors (reference 13). RPM cutoff value was set as 5. (D) Fip1 binding frequency profile across snoRNA regions based on our Fip1 iCLIP-seq data described in reference 11. Mature snoRNA regions were defined according to reference 14, The regions upstream and downstream of mature snoRNAs were defined accordingly. For reads mapped to mature snoRNA regions, the RPM values were aligned based on the middle position of each snoRNA, the y axis value represents the average RPM at each position for all Fip1 iCLIP-seq+ snoRNAs. For reads mapped to upstream and downstream of mature snoRNAs, RPM values were averaged and plotted at each position for all Fip1 iCLIP-seq+ snoRNAs. (E) Schematic representation of potential regulatory role of snoRNA in gene expression at mRNA 3′ processing level. If a gene has two (gene A) or multiple PASes, snoRNA could affect the level of each transcript and also the APA profile. Moreover, aberrant mRNA 3′ processing for gene A might also lead to transcription of downstream gene B, which might also be regulated by snoRNAs.
snoRNAs do associate with CPSF in vivo
In the recent report [11], we set out to address the question whether there is any trans-acting RNA functioning in mRNA 3′ processing. We reasoned that, if there exist such general non-coding RNAs, they must be directly associated with mRNA 3′ processing complex. Therefore, we purified mRNA 3′ processing complex using the previously well-characterized SVL and L3 pre-mRNA system (Fig. 1B, upper panel) [3,10], and subsequently analyzed the RNA components in the pull-down sample by high-throughput sequencing. Strikingly, almost all the highly enriched RNAs were snoRNAs in both datasets (Fig. 1B, lower panel), indicating snoRNAs may be associated with mRNA 3′ processing complex [11]. We further provided evidence that these snoRNAs associate with mRNA 3′ processing complex at least via Fip1, a CPSF subunit, as iCLIP-seq analysis showed Fip1 could interact with almost all of these enriched snoRNAs (Fig. 1B) [11]. It is important to point out that Fip1 IP was performed under denaturing condition. Therefore, we were detecting bona-fide Fip1-RNA interactions but not RNA interactions with Fip1-associated proteins in vivo. Interestingly, the association of snoRNAs with Fip1 is independent of the canonical snoRNPs, which coincides with a recent report that a large fraction of snoRNAs is present outside the nucleolus [12]. Our main claim is also supported by another independent study of 3′ processing factors in HEK293 cells, showing a subset of snoRNAs interact with CPSF in vivo (Fig. 1C) [13].
Given the fact that almost all the human snoRNAs reside in the intron of the host genes and mRNA processing factors associate with RNA polymerase II during transcription [14,15], the detected association of snoRNAs and Fip1 might be the relics of Fip1 loading on RNA polymerase II across snoRNA locus. However, Fip1 iCLIP-seq detected little signal outside the mature snoRNA region (Fig. 1D) [11], implying Fip1 interacts with snoRNAs post-transcriptionally but not co-transcriptionally. It will be of great interest for future study to investigate how processed snoRNAs are incorporated into CPSF and what is the stoichiometry of snoRNAs and Fip1/CPSF during the entire 3′ processing period. Moreover, given our overlapping and distinct snoRNA enrichment profiles of SVL and L3 PAS RNA substrates (Fig. 1B), it seems snoRNAs associate with CPSF in a PAS-specific manner. Therefore, it will also be of great interest to discern the causes and effects of this context-dependent association of snoRNA and Fip1/CPSF in the future studies.
Function by blocking: A snoRNA regulates mRNA 3′ processing
Unlike the usual mode of recognition of snoRNA substrate through RNA-RNA interactions [14,16], SNORD50A competes with SVL PAS RNA for binding to Fip1 to block mRNA 3′ processing [11]. However, we did not provide sufficient data to prove that the proposed mechanism can be applied for all endogenous PASes. It is possible that U/A rich SNORD50A could bind to specific PASes to block the Fip1/CPSF-PAS interaction. Thus, higher binding frequency of Fip1 across PAS regions was observed upon SNORD50A depletion [11]. To comprehensively characterize more detailed mechanism of SNORD50A function in endogenous mRNA 3′ processing, we believe that recently developed techniques to detect RNA-RNA interactions at transcriptomic level, such as LIGR-seq, SPLASH and PARIS [17–19], will undoubtedly be valuable to address this question. Moreover, little is currently known about the spatial organization of snoRNA in CPSF, a crystal structure or a cryo-electronic microscopy (cryo-EM) analysis of SNORD50A within CPSF will also be helpful to elucidate detailed mechanism of SNORD50A function in mRNA 3′ processing regulation.
In addition to SNORD50A, it is possible that other snoRNAs associated with Fip1/CPSF are also functional in mRNA 3′ processing. Herein, we propose several hypotheses concerning the snoRNA functions within CPSF, though we do not have any experimental evidence to prove these thus far. First, the mammalian PAS sequences are relative degenerate except the A(A/U)UAAA element [20]. Despite progress in deciphering global protein-RNA interactions within mRNA 3′ processing complex, it remains poorly understood how the 3′ processing complex is stabilized on heterogenous PASes, one possibility is that CPSF-associated snoRNAs contribute to the integration of 3′ processing complex on diverse PASes via facilitating protein-RNA or protein-protein interactions. Second, CPSF recognizes A(A/U)UAAA element in vitro, however, this core cis-element is not found in 30% of human endogenous polyadenylation sites [21], raising the possibility that some snoRNAs may facilitate CPSF assembly on these non-canonical polyadenylation sites via RNA-RNA interactions, similar to the role of Y3** ncRNA or U7 snRNA in histone mRNA 3′ processing [22,23]. Third, snoRNAs such as SNORD50A, could also be a negative factor in mRNA 3′ processing [11]. The significance of this negative effect is still unclear. Inspired by the role of U1 snRNA in protecting pre-mRNA from premature cleavage and polyadenylation [24], one of our conjectures is that, some snoRNAs might also have similar function in suppressing the usage of cryptic PASes.
Despite great potentials, several key challenges could hinder future studies. First, same snoRNAs could be assembled into different protein/RNA complexes to perform distinct functions [25], which will certainly complicate the data interpretation. Second, snoRNAs might play redundant roles in mRNA 3′ processing. Third, mRNA 3′ processing is coupled with transcription, splicing and RNA turnover in vivo [15], adding another layer of complexity to snoRNA function annotation. Therefore, combinatory approaches are required for the identification and validation of direct associations between specific snoRNAs and mRNA 3′ processing regulation. For example, by using extensive biochemical, molecular and genomic analysis, we provided strong evidence that SNORD50A directly regulates mRNA 3′ processing [11]. It should be noted that, although recently developed CRISPR/Cas9 technology has been becoming popular in gene KO study, this technique should be carefully used in snoRNA KO as gene editing at snoRNA gene locus may interfere with the expression of host gene or other snoRNAs within the same gene cluster. Alternatively, ASO technique may be a method of choice as it proves to be efficient and specific for nuclear non-coding RNAs depletion in mammalian cells [26].
Emerging role of snoRNA in gene regulation
Our report suggests a novel mechanism through which snoRNA regulates gene expression, contributing to the ever-growing list of non-canonical snoRNA cellular roles [11]. Several excellent reviews have discussed the canonical and non-canonical roles of snoRNAs [25,27,28], Below we will only discuss how snoRNA-mediated 3′ processing regulation could be integrated into the eukaryotic transcriptional and post-transcriptional gene regulatory networks. First, mRNA alternative polyadenylation (APA) is increasingly recognized as a critical mechanism for post-transcriptional gene regulation (Fig. 1E, gene A) [15], an important question is how APA is regulated in cells. snoRNA could regulate mRNA 3′ processing and thus may serve as a direct APA regulator in gene expression. Indeed, SNORD50A deletion caused APA changes for 157 genes in HeLa cells [11]. Second, as mRNA 3′ processing is a critical step in gene expression, snoRNA-mediated 3′ processing regulation could directly influence the transcript abundance and/or turnover. For example, 477 genes were up-regulated by 4-fold upon SNORD50A depletion [11]. Finally, as mentioned earlier, mRNA 3′ processing is a co-transcriptional process, its dysregulation may lead to transcription of downstream genes (Fig. 1E), therefore, snoRNA-mediated mRNA 3′ processing regulation could contribute to the delicate gene regulatory network at transcriptional level in eukaryotic cells.
Increasing evidences have indicated that snoRNAs function in gene regulation is under-appreciated. For example, many snoRNAs have been shown to display tissue-specific expression pattern and, more importantly, some snoRNAs have been associated with cell type-specific diseases such as cancer and Prader-Willi syndrome [25]. It is unlikely that this is attributed solely to their housekeeping role of rRNA modification. Analogues to other non-canonical roles such as pre-mRNA splicing regulation and chromatin remodeling [27,28], snoRNA function in mRNA 3′ processing regulation may provide another layer of basic molecular mechanisms underlying these diseases. Indeed, SNORD50A has been implicated in a variety of cancers [29]. Consistently, our results showed that it preferentially targets mRNAs with U rich PASes, and these mRNAs are enriched in function in cell proliferation and apoptosis [11]. Taken together, for snoRNAs function in gene regulation, SNORD50A may be just the tip of the iceberg. Despite many potential challenges, with the fast developing technology and molecular tools, we anticipate more snoRNA functions in mRNA 3′ processing and gene expression will be revealed in the coming years.
Abbreviations
- APA
alternative polyadenylation
- CPSF
cleavage and polyadenylation specificity factor
- CstF
cleavage stimulation factor
- CF Im
cleavage factor Im
- CF IIm
cleavage factor IIm
- iCLIP
individual-nucleotide resolution cross-linking and Immunoprecipitation
- PAS
polyadenylation site
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Chinese national natural science foundation, ID: 31501184; Sun Yat-sen University, ID: 50000-18821101.
References
- [1].Bienroth S, Wahle E, Suter-Crazzolara C, et al.. Purification of the cleavage and polyadenylation factor involved in the 3′-processing of messenger RNA precursors. J Biol Chem. 1991;266:19768–76. [PubMed] [Google Scholar]
- [2].Murthy KG, Manley JL. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem. 1992;267:14804–11. [PubMed] [Google Scholar]
- [3].Shi Y, Di Giammartino DC, Taylor D, et al.. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–76. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chan SL, Huppertz I, Yao C, et al.. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev. 2014;28:2370–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schonemann L, Kuhn U, Martin G, et al.. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev. 2014;28:2381–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yao C, Biesinger J, Wan J, et al.. Transcriptome-wide analyses of CstF64-RNA interactions in global regulation of mRNA alternative polyadenylation. Proc Natl Acad Sci U S A. 2012;109:18773–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yao C, Choi EA, Weng L, et al.. Overlapping and distinct functions of CstF64 and CstF64tau in mammalian mRNA 3′ processing. Rna. 2013;19:1781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yang Q, Gilmartin GM, Doublie S. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing. Proc Natl Acad Sci U S A. 2010;107:10062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chan S, Choi EA, Shi Y. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip Rev RNA. 2011;2:321–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Takagaki Y, Ryner LC, Manley JL. Four factors are required for 3′-end cleavage of pre-mRNAs. Genes Dev. 1989;3:1711–24. [DOI] [PubMed] [Google Scholar]
- [11].Huang C, Shi J, Guo Y, et al.. A snoRNA modulates mRNA 3′ end processing and regulates the expression of a subset of mRNAs. Nucleic Acids Res. 2017;45:8647–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Falaleeva M, Pages A, Matuszek Z, et al.. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc Natl Acad Sci U S A. 2016;113:E1625–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Martin G, Gruber AR, Keller W, et al.. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep. 2012;1:753–63. [DOI] [PubMed] [Google Scholar]
- [14].Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol. 2017;18:18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–2. [DOI] [PubMed] [Google Scholar]
- [17].Sharma E, Sterne-Weiler T, O'Hanlon D, et al.. Global Mapping of Human RNA-RNA Interactions. Mol Cell. 2016;62:618–26. [DOI] [PubMed] [Google Scholar]
- [18].Aw JG, Shen Y, Wilm A, et al.. In vivo mapping of eukaryotic RNA Interactomes reveals principles of higher-order organization and regulation. Mol Cell. 2016;62:603–17. [DOI] [PubMed] [Google Scholar]
- [19].Lu Z, Zhang QC, Lee B, et al.. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell. 2016;165:1267–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shi Y, Manley JL. The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev. 2015;29:889–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tian B, Hu J, Zhang H, et al.. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kohn M, Ihling C, Sinz A, et al.. The Y3** ncRNA promotes the 3′ end processing of histone mRNAs. Genes Dev. 2015;29:1998–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Marzluff WF, Koreski KP. Birth and Death of Histone mRNAs. Trends Genet. 2017;33:745–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kaida D, Berg MG, Younis I, et al.. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Falaleeva M, Welden JR, Duncan MJ, et al.. C/D-box snoRNAs form methylating and non-methylating ribonucleoprotein complexes: Old dogs show new tricks. Bioessays. 2017;39:1600264. doi: 10.1002/bies.201600264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liang XH, Vickers TA, Guo S, et al.. Efficient and specific knockdown of small non-coding RNAs in mammalian cells and in mice. Nucleic Acids Res. 2011;39:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dupuis-Sandoval F, Poirier M, Scott MS. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip Rev RNA. 2015;6:381–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bratkovic T, Rogelj B. The many faces of small nucleolar RNAs. Biochim Biophys Acta. 2014;1839:438–43. doi: 10.1016/j.bbagrm.2014.04.009. [DOI] [PubMed] [Google Scholar]
- [29].Siprashvili Z, Webster DE, Johnston D, et al.. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat Genet. 2016;48:53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]


