Abstract
Methyl CpG Binding Protein 2 (MeCP2) is an important epigenetic factor in the brain. MeCP2 expression is affected by different environmental insults including alcohol exposure. Accumulating evidence supports the role of aberrant MeCP2 expression in ethanol exposure-induced neurological symptoms. However, the underlying molecular mechanisms of ethanol-induced MeCP2 deregulation remain elusive. To study the effect of ethanol on Mecp2/MeCP2 expression during neurodifferentiation, we established an in vitro model of ethanol exposure, using differentiating embryonic brain-derived neural stem cells (NSC). Previously, we demonstrated the impact of DNA methylation at the Mecp2 regulatory elements (REs) on Mecp2/MeCP2 expression in vitro and in vivo. Here, we studied whether altered DNA methylation at these REs is associated with the Mecp2/MeCP2 misexpression induced by ethanol. Binge-like and continuous ethanol exposure upregulated Mecp2/MeCP2, while ethanol withdrawal downregulated its expression. DNA methylation analysis by methylated DNA immunoprecipitation indicated that increased 5-hydroxymethylcytosine (5hmC) and decreased 5-methylcytosine (5mC) enrichment at specific REs were associated with upregulated Mecp2/MeCP2 following continuous ethanol exposure. The reduced Mecp2/MeCP2 expression upon ethanol withdrawal was associated with reduced 5hmC and increased 5mC enrichment at these REs. Moreover, ethanol altered global DNA methylation (5mC and 5hmC). Under the tested conditions, ethanol had minimal effects on NSC cell fate commitment, but caused changes in neuronal morphology and glial cell size. Taken together, our data represent an epigenetic mechanism for ethanol-mediated misexpression of Mecp2/MeCP2 in differentiating embryonic brain cells. We also show the potential role of DNA methylation and MeCP2 in alcohol-related neurological disorders, specifically Fetal Alcohol Spectrum Disorders.
Keywords: MeCP2, DNA methylation, Epigenetics, Neural stem cells, Fetal Alcohol Spectrum Disorders, 5mC, 5hmC
Introduction
Methyl CpG Binding Protein 2 (MeCP2) is a multifunctional epigenetic factor in the brain, which is involved in transcriptional regulation (Zachariah and Rastegar, 2012) and chromatin architecture (Stuss et al., 2013; Thambirajah et al., 2012). MECP2/MeCP2 mutations or altered expression lead to a range of neurodevelopmental disorders including autism spectrum disorders and Rett Syndrome [reviewed in (Ezeonwuka and Rastegar, 2014)]. MeCP2 also plays a role in substance abuse disorders such as drug addiction and alcoholism (Feng and Nestler, 2010; Repunte-Canonigo et al., 2014). On the other hand, MeCP2 expression itself is sensitive to exposure to different environmental insults such as psychostimulants including cocaine and amphetamine, and alcohol (ethanol) [reviewed in (Liyanage and Rastegar, 2014)].
Ethanol is a typical teratogen and prenatal exposure to ethanol leads to a wide range of developmental abnormalities known as Fetal Alcohol Spectrum Disorders (FASD) (Jones and Smith, 1973). Recent studies also implicate the potential of preconception paternal alcohol consumption, which can lead to FASD (Kim et al., 2014; Knezovich and Ramsay, 2012). Previous studies have shown the importance of both alcohol exposure and withdrawal (termination of alcohol consumption) in FASD pathogenesis (Carlson et al., 2012), emphasizing the necessity to study the effect of both ethanol exposure and withdrawal on the central nervous system development (Pierog et al., 1977; Thomas and Riley, 1998). During development, “binge-like” (single time) ethanol exposure as well as continuous ethanol exposure are reported to cause severe effects that contribute to FASD pathobiology (Flegel et al., 2011).
Implicating the potential involvement of MeCP2 in FASD, a MECP2 mutation (R270X) has been found in a FASD patient who showed overlapping phenotypes with both Rett Syndrome and FASD during development (Zoll et al., 2004). Additionally, both increased and decreased MeCP2 expression patterns have been reported in ethanol-fed rodent brain cells in vivo as well as in cultured cells in vitro [reviewed in (Liyanage and Rastegar, 2014)]. The discrepancies in these studies regarding the effect of ethanol on MeCP2 expression could be attributed to multiple factors such as the model of study (in vivo animal models or in vitro cultured cells), stage of embryonic development, specific brain region or cell type within a brain region, concentration and duration of ethanol treatment (Bekdash et al., 2013; Chen et al., 2013; Gangisetty et al., 2014; Guo et al., 2012; Kim et al., 2013, 2014; Perkins et al., 2013; Subbanna et al., 2014; Tunc-Ozcan et al., 2013). Further supporting the role of MeCP2 in alcoholism, a recent study demonstrated the regulatory effect of MeCP2 on sensitivity to ethanol and drinking ethanol (Repunte-Canonigo et al., 2014). Despite this increasingly evident link between changes in MeCP2 expression (increased or decreased) and ethanol exposure, the molecular mechanisms by which ethanol affects MeCP2 expression are understudied. Therefore, a detailed analysis of the effect of ethanol on MeCP2 expression and associated mechanisms is critical, which will be the primary focus of this current study.
DNA methylation is one of the most studied epigenetic mechanisms that are crucial in controlling gene expression during brain development (Barber and Rastegar, 2010; Delcuve et al., 2009; Liyanage and Rastegar, 2014; Olynik and Rastegar, 2012). Two major forms of DNA methylation are 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC). While 5mC methylation of upstream promoter regions is considered to be a gene repressive mark, 5hmC has been detected in active gene regions (Liyanage et al., 2012, 2014). MeCP2 has been shown to be the major protein which binds to both 5mC and 5hmC in the brain (Mellen et al., 2012). MECP2 promoter hypermethylation is associated with MECP2 downregulation in the autistic brain (Nagarajan et al., 2006). Increased Mecp2 promoter methylation is also associated with reduced Mecp2 expression in postnatal mouse brain in response to maternal separation and stress (Franklin et al., 2010). Apart from the regulatory elements (REs) found within the promoter, a silencer element within the Mecp2 intron 1 is known to negatively regulate Mecp2 expression (Liu and Francke, 2006). Recently, we showed that DNA methylation at the REs found within the Mecp2 promoter and intron 1 correlated with the dynamic expression of Mecp2 in differentiating neural stem cells (NSC) and in response to a DNA demethylating drug Decitabine (also called 5-Aza-2′-deoxycytidine) (Liyanage et al., 2013). Recently, our studies in adult murine brain regions demonstrated a correlation between the expression of Mecp2 and DNA methylation at the Mecp2 REs in vivo (Olson et al., 2014). In this current report, we studied whether DNA methylation at these reported Mecp2 REs within the promoter and intron 1 contributes in deregulating Mecp2 expression induced by ethanol.
Previously, we established a differentiating murine embryonic brain-derived NSC system to study the DNA methylation-mediated regulation of Mecp2 (Liyanage et al., 2013). We have used this neural stem cell model to study the regulation of genes involved in neural development including Mecp2 and Meis1 (Barber et al., 2013; Liyanage et al., 2013; Rastegar et al., 2009). We have successfully used the same NSC system to rescue the aberrant neuronal morphologies in Mecp2-deficient NSC by MECP2 gene therapy strategies (Rastegar et al., 2009). We also demonstrated the expression and regulation of Mecp2/MeCP2 by DNA methylation in a similar NSC model (Liyanage et al., 2013). In this current study, we have modified this previously established NSC system to model binge-like and continuous ethanol exposure and ethanol withdrawal. We show that ethanol alters Mecp2/MeCP2 expression in association with interplay between 5mC and 5hmC enrichment at the Mecp2 regulatory elements.
Materials and methods
Ethics statement
All experiments were performed in agreement with the standards of the Canadian Council on Animal Care with the approval of the Office of Research Ethics of University of Manitoba.
Neural stem cell isolation, culture and differentiation
Neural stem cells isolated from the forebrains of embryonic day (E) 14.5 CD-1 mice were cultured as previously described (Barber et al., 2013; Liyanage et al., 2013; Rastegar et al., 2009). In brief, dissected forebrain tissues were homogenized in NSC media Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 1:1 (DMEM/F12; Wisent) containing 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), glutamine, antibiotic/antimycotic, glucose, recombinant human epidermal growth factor (rhEGF; Sigma, 20 ng/ml), basic fibroblast growth factor (bFGF; Upstate, 20 ng/ml), heparin (Sigma, 2 μg/ml) and hormone mix [DMEM:F12, glucose (0.6%), insulin (0.25 mg/ml), transferrin (1 mg/ml), progesterone (0.2 μM), putrescine (0.097 mg/ml), sodium selenite (0.3 μM)] and plated at a density of 105 cells/cm2 in serum-free full NSC media. Cells were cultured for 7 days to generate neurospheres. The dissociated neurosphere cells were plated in growth factor-reduced matrigel coated-plates (BD Biosciences) at a density of 105 cells/cm2 in DMEM media (GIBCO) containing 10% fetal bovine serum (FBS, Invitrogen) in the absence of rhEGF and bFGF. Cells were differentiated for 8 days under these conditions.
Ethanol treatment
Dissociated neurosphere cells were treated at the onset of differentiation at day 0 (D0) with ethanol (Commercial Alcohols) at a final concentration of 70 mM. To study binge ethanol exposure, ethanol treatment was done only once at D0 for 48 hours (h) and cells were collected at day 2 (D2). For continuous ethanol exposure, cells were treated with ethanol starting at D0 for 8 days continuously and ethanol with media was refreshed every two days. To study the effect of ethanol withdrawal, the culture media was exchanged with fresh culture media after 48 h, and cells were kept in culture for an extra 6 days. The media was refreshed every other day. Control cells were cultured under similar experimental conditions in the absence of ethanol. Cells were harvested at D2 and day 8 (D8).
Quantitative real time polymerase chain reaction (qRT-PCR)
Total RNA was extracted using an RNeasy Mini Kit (Qiagen). RNA was converted to cDNA with a reaction set up containing Superscript III Reverse Transcriptase (Invitrogen) using previously described methods (Kobrossy et al., 2006; Liyanage et al., 2013; Nolte et al., 2006; Rastegar et al., 2004). Quantitative RT-PCR was performed using SYBR Green-based RT2 qPCR Master Mix (Applied Biosystems) in a Fast 7500 Real-Time PCR machine (Applied Biosystems) as previously described (Barber et al., 2013; Liyanage et al., 2013; Olson et al., 2014). Transcript levels of Mecp2 and cell type-specific markers were examined using primers listed in Table 1. The Ct values (threshold cycle) for all genes were normalized with reference to the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (Gapdh) to obtain ΔCt values for each sample. Relative quantification analysis was carried out using Microsoft Excel 2010 by comparing the 2−ΔCt of each sample to that of untreated control sample at D2 and D8 as described in our previous studies (Liyanage et al., 2013; Olson et al., 2014).
Table 1.
The list of primers used for qRT-PCR experiments and their sources.
| Gene | Direction | Sequence | Reference |
|---|---|---|---|
| Mecp2 | Forward | GGTAAAACCCGTCCGGAAAATG | Kriaucionis and Bird (2004) |
| Reverse | TTCAGTGGCTTGTCTCTGAG | ||
| β Tubulin III (Tub II) | Forward | TCAGCGATGAGCACGGCATA | Barber et al. (2013) |
| Reverse | CACTCTTTCCGCACGACATC | ||
| Gfap | Forward | GCTCACAATACAAGTTGTCC | |
| Reverse | ACCTAATTACACAGAGCCAGG | ||
| Olig2 | Forward | GTGGCTTCAAGTCATCTTCC | Liyanage et al. (2013) |
| Reverse | GTAGATCTCGCTACACAGTC | ||
| Gapdh | Forward | AACGACCCCTTCATTGAC | Rastegar et al. (2004) |
| Reverse | TCCACGACATACTCAGCAC | ||
| Mbp | Forward | GGCACGCTTTCCAAAATCT | Schneider and d’Adda di Fagagna (2012) |
| Reverse | CCATGGGAGATCCAGAGC | ||
| Ki67 | Forward | GCTGTCCTCAAGACAATCATCA | Schneider and d’Adda di Fagagna (2012) |
| Reverse | GGCGTTATCCCAGGAGACT | ||
| Nestin | Forward | CTGCAGGCCACTGAAAAGT | |
| Reverse | TCTGACTCTGTAGACCCTGCTTC |
Immunocytochemistry (ICC)
ICC for cultured neural stem cells were carried out as described previously (Liyanage et al., 2013; Rastegar et al., 2009; Zachariah et al., 2012), using antibodies in Tables 2 and 3. Briefly, differentiated cells on coverslips were washed with phosphate buffered saline (PBS, GIBCO) and fixed in 4% paraformaldehyde. Cells were permeabilized with 2% NP40 in PBS for 10 min, followed by preblocking with 10% normal goat serum (NGS, Jackson ImmunoResearch) in PBS for 1 h. Appropriate primary antibodies were added and incubated overnight at 4 °C followed by washes with PBS. This was followed by incubation with secondary antibodies that were diluted in 10% NGS (1 h at room temperature). Coverslips were mounted on glass slides with antifade containing 2 μg/ml DAPI. ICC signals were detected by an Axio Observer Z1 inverted microscope. Images were obtained using Zen 2011 software and assembled using Adobe Photoshop and Adobe Illustrator. For cell population determination, approximately 250 DAPI+ cells from D2 and 400–500 DAPI+ cells from D8 collectively from 3 biological replicates were counted using ImageJ program, as described previously (Liyanage et al., 2013).
Table 2.
List of primary antibodies used in western blot and immunocytochemistry.
| Primary antibody | Application and dilution | Molecular weight (kDa) | Description | Source |
|---|---|---|---|---|
| MeCP2 (C-terminal) | WB 1:1000 | 72 | Mouse monoclonal | Millipore, 07-013 |
| Beta (β) ACTIN | WB: 1:2500 | 42 | Mouse monoclonal | Sigma, A2228 |
| GAPDH | WB: 1:500 | 37 | Rabbit polyclonal | Santa Cruz, Sc-25778 |
| GFAP | IF 1:200 | N/A | Mouse monoclonal | Invitrogen, 421262 |
| Beta (β) TUBULIN III (TUB III) | IF 1:200 | N/A | Mouse monoclonal | Chemicon, MAB1637 |
| MBP | IF 1:100 | N/A | Rabbit polyclonal | Abcam, ab40390 |
| 5mC | Dot blot 1:1000 | N/A | Mouse monoclonal | Abcam, Ab73938 |
| 5hmC | Dot blot 1:1000 | N/A | Rabbit polyclonal | Active Motif, 39769 |
| NESTIN | IF 1:200 | N/A | Rat polyclonal | Developmental Studies Hybridoma Bank, Rat-401c |
| KI67 | IF 1:200 | N/A | Rabbit polyclonal | Sc-15402 |
| OLIG2 | IF 1:200 | N/A | Rabbit polyclonal | Millipore, AB9610 |
| RNA Polymerase (Pol) II | WB 1:1000 | 217 | Mouse monoclonal | Abcam, Ab817 |
| HDAC2 | WB 1:1000 | 55 | Mouse monoclonal | Abcam, Ab12169 |
| Alpha (α) TUBULIN | WB 1:1000 | 50 | Mouse monoclonal | Sigma, T9026 |
Table 3.
List of secondary antibodies used in western blot and immunocytochemistry.
| Secondary antibody | Application and dilution | Source |
|---|---|---|
| Alexa Fluor 488 conjugated goat anti-mouse IgG | IF 1:2000 or 1:3000 | Invitrogen, A11017 |
| Alexa Fluor 594 conjugated goat anti rabbit IgG | IF 1:2000 or 1:3000 | Invitrogen, A11037 |
| Peroxidase-AffiniPure Gt anti-mouse IgG | WB 1:7500, Dot blot 1:7500 | Jackson ImmunoResearch 115-035-174 |
| Peroxidase-AffiniPure donkey anti-rabbit IgG | WB 1:7500, Dot blot 1:7500 | Jackson ImmunoResearch 711-035-152 |
Measurement of neurite branching and glial cell surface area
Immunofluorescence signals for glial fibrillary acidic protein (GFAP) and β Tubulin III (TUB III) were detected by an Axio Observer Z1 inverted microscope and the images were obtained using Zen 2011 (Carl Zeiss). For quantification of the surface area of GFAP+ astrocytes, superimposed Z-stacked images taken at 0.32 μm sections were analyzed. The surface area of GFAP+ astrocytes was measured using the spline contour tool within Zen 2011 software, as shown in a previous study (Schade et al., 2013). The signal intensity was adjusted to observe all the cellular processes of GFAP+ cells and the outer margin of each cell was carefully marked using the spline contour tool, which directly provides the area in μm2. At least 20 GFAP+ astrocytes per biological replicate (three biological replicates) of control and ethanol-treated cells were measured. For the quantification of number of neurites, at least 20 TUB III+ neurons per biological replicate (three biological replicates) were imaged and neurites were marked and counted in ImageJ program as described in our previous study (Rastegar et al., 2009).
Western blot (WB)
Western blotting was performed as previously described (Barber et al., 2013; Rastegar et al., 2000; Wu et al., 2001; Zachariah et al., 2012). NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific) was used to extract nuclear proteins as previously described (Liyanage et al., 2013; Olson et al., 2014). Polyacrylamide gel electrophoresis was performed with 10–30 μg of nuclear extracts and proteins were transferred to nitrocellulose membranes and blocked in 1–2% skim milk in Tris buffered saline with Tween (TBST: TBS+ 0.1% Tween) for 1 h at room temperature. All primary antibody incubations were done overnight at 4 °C. This was followed by three washes with TBST. Subsequently, we incubated the membranes with peroxidase-conjugated secondary antibody at room temperature for 1 h. Next, membranes were washed with TBST (three times). An enhanced chemiluminescence method was used to develop the membranes. The developing X-ray films were blue in color and they were converted into gray color before quantification and assembly of the figures. Note that the brightness or contrast of the bands was not adjusted. RNA Pol II, HDAC2 (nuclear markers) and α-TUBULIN (mostly used as cytoplasmic marker), ACTIN and GAPDH were tested to select the best endogenous loading control. ACTIN and GAPDH were chosen to be the most reliable and suitable loading controls (for further details see the Results section). Quantification of WB bands was done with Adobe Photoshop CS5 software and normalized to ACTIN or GAPDH signals as described previously (Liyanage et al., 2013; Olson et al., 2014). MeCP2 expression in ethanol-treated samples was compared to the corresponding control untreated cells within the same biological replicate. Note that the exposure times were not the same for different experiments.
DNA dot blot assay for 5mC and 5hmC
DNA dot blot was performed using a previously described protocol (Liyanage et al., 2013). Briefly, genomic DNA was isolated using the DNeasy Blood and Tissue kit (Qiagen) as per manufacturer’s instructions. For each sample, 250 ng of DNA in 250 μl final volume was heat denatured in 0.4 M sodium hydroxide (NaOH), 10 mM ethylenedi-aminetetraacetic acid (EDTA) at 100 °C for 10 min and neutralized with equal volume of ice-cold 2 M ammonium acetate (pH 7.0) to obtain 500 μl of final volume. Denatured DNA was loaded onto Zeta-Probe GT blotting membrane (Bio Rad) set up in a dot blot apparatus (Bio Rad). Wells were rinsed with 0.4 M NaOH to keep DNA denatured. The membrane was rinsed with 2× saline–sodium citrate (SSC), air-dried and UV cross-linked. Then the membrane was blocked with 3–5% skim milk in PBST (PBS+ 0.1% Tween) for 3 h at room temperature followed by incubation in primary antibody [5mC or 5hmC (1:1000) in PBST] at 4 °C overnight. After three washes with PBST, the membrane was incubated in secondary antibody for 1 h at room temperature. The signals were visualized by enhanced chemiluminescence. Total DNA levels were detected by staining the same membrane with 0.02% methylene blue (MB) in 0.3 M sodium acetate (pH 5.2). Adobe Photoshop was used to quantify dot blot signals.
Bisulfite pyrosequencing
Bisulfite pyrosequencing was performed as a service at the Hospital for Sick Children, Toronto, Canada using previously described methods (Liyanage et al., 2013; Olson et al., 2014). Genomic DNA was isolated as described using the DNeasy Blood and Tissue kit (Qiagen) as per manufacturer’s instructions. Three regions in Mecp2 promoter and three regions in Mecp2 intron 1 were analyzed, which we have reported to impact Mecp2 expression in differentiating NSC and adult mouse brain (Liyanage et al., 2013; Olson et al., 2014). The primers used for pyrosequencing analysis are listed in Table 4. The control D2 methylation data were extracted from a recent report from our lab (Liyanage et al., 2013).
Table 4.
List of primers used for bisulfite pyrosequencing.
| Mecp2 regions | Sequence |
|---|---|
| R1 | FP: 5′-TGGGTTTTATAATTAATGAAGGGTAA-3′ |
| RP: 5′-CGCCAGGGTTTTCCCAGTCACGACATTTTACCACAACCCTCTCT-3′ | |
| SP: 5′-AGGTGTAGTAGTATATAGG-3′ | |
| R2 | FP: 5′-AGTTTGGGTTTTATAATTAATGAAGGG-3′ |
| RR: 5′-CGCCAGGGTTTTCCCAGTCACGACATTTTACCACAACCCTCTCT-3′ | |
| SR: 5′-AAGGGTAATTTAGATAAAGAGTAAG-3′ | |
| R3 | FP: 5′-GGTGAATTATTTAGTAGGGAGGTTTTAA-3′ |
| RP: 5′-CGCCAGGGTTTTCCCAGTCACGACAAAAAAAAAACCAACCCCATTCAACTAC-3′ | |
| SP: 5′-AGTAGGGAGGTTTTAATAG-3′ | |
| R4 | FP: 5′-GTTTTAAAAAGTTTTGGGAAAAGGTGTAGT-3′ |
| RP: 5′-CGCCAGGGTTTTCCCAGTCACGACCTAAACCCTAACATCCCAACTACCAT-3′ | |
| SP: 5′-AGTTTAATGGGGATTTTTAATT-3′ | |
| R5 | FP: 5′-AGTAGAAGTTATTATTTGTGGTGTGTAT-3′ |
| RP: 5′-CGCCAGGGTTTTCCCAGTCACGACACTATATTACTTCCCAACTCAACTAATT-3′ | |
| SP: 5′-AGAGGTGTAAGGATTTT-3′ | |
| R6 | FP: 5′-GAAGTAGGAAGAATTGAGTTTGAGGATAG-3′ |
| RP: 5′-CGCCAGGGTTTTCCCAGTCACGACATCTATACACTACCCACATATAATACC-3′ | |
| SP: 5′-GTTTGAGGATAGTTTGAAT-3′ |
F: Forward PCR primer, R: Reverse PCR primer (Biotinylated), S: Sequencing primer, R: Region.
Reference: Liyanage et al. (2013).
Hydroxymethylated DNA immunoprecipitation (hMeDIP) and methylated DNA immunoprecipitation (MeDIP)
DNA immunoprecipitation was carried out using hMeDIP and MeDIP kits (Abcam), from 3 to 4 independent experimental replicates. For hMeDIP with D2 samples, 100 ng per reaction was used (due to the limited amount of the D2 samples), while for all D8 samples 500 ng per reaction was used. For MeDIP, 1 μg of DNA per reaction was used. The immunoprecipitated DNA was analyzed by qPCR using the primers specific for each Mecp2 promoter or intron 1 region (Table 5). For each hMeDIP and MeDIP reaction, 2–3 qPCR technical replicates from 4 independent biological replicates were done to confirm the results. In order to confirm the specificity of the signals obtained from 5mC and 5hmC antibodies for both hMeDIP and MeDIP experiments, non-immunogenic IgG antibody was used. Quantitative PCR with Gapdh primers was used as an unmethylated negative control. For hMeDIP, a positive control DNA with 44 5hmC-CpG sites provided within the kit was used as a positive control for immunoprecipitation and was amplified using control primers provided within the kit. For MeDIP, primers specific for Major Satellite Repeats were used as a positive control for methylated DNA. The percentage input for the Ct values obtained from each qPCR was calculated relative to the Ct value of the input DNA used.
Table 5.
List of primers used in qPCR for hMeDIP and MeDIP.
| Mecp2 region | Direction | Sequence (5′ to 3′) | Reference |
|---|---|---|---|
| R1 | Forward | CAGGTGCAGCAGCACACAGGC | Designed in the lab |
| Reverse | CGGACGGGTTTTACCACAGCC | ||
| R2 | Forward | GAACTCCACC AATCCGCAGC | |
| Reverse | CCTGTGTGCTGCTGCACCTG | ||
| R3 | Forward | CAGGCTCCTCAACAGGCAAC | |
| Reverse | GGCTGGTTTTGTGGGCAGCA | ||
| R4 | Forward | CTAGCTGAGCTGGGAAGTAAC | |
| Reverse | GAGCTGGTCTACAGAAGCAAG | ||
| R5 | Forward | CTGTATAGTGTGGTGGAAGAAG | |
| Reverse | GTTACTTCCCAGCTCAGCTAG | ||
| R6 | Forward | GAGTTTGAGGACAGCCTGAAC | |
| Reverse | CTACCCACATGTAGTGCCTGC |
R: Region.
Statistical analysis
All graphs were generated using GraphPad Prism software and represent average of two to four independent experiments. Error bars indicate standard error of mean (SEM). Student’s t-test was used to determine statistical significance in gene expression. Two-way ANOVA was performed to determine statistical significance in percentage methylation (bisulfite pyrosequencing) and percentage input (hMeDIP and MeDIP) between control and ethanol-treated samples. Statistical significance was calculated using p < 0.05*, p < 0.01**, p < 0.001***, and p < 0.0001****.
Results
A differentiating neural stem cell model was used to study the effect of ethanol on Mecp2/MeCP2 expression
To assess the impact of ethanol exposure on Mecp2/MeCP2 transcript and protein expression, we used a previously characterized embryonic NSC differentiation system (Barber et al., 2013; Liyanage et al., 2013; Rastegar et al., 2009). Brain-derived neural stem cells were isolated from dissected forebrains of E14.5 mouse embryos and were cultured for 7 days in the presence of growth factors to generate neurospheres. Primary neurospheres were then dissociated and single cells were differentiated for 8 days, as we have shown to be sufficient for generating neurons, astrocytes and oligodendrocytes (Fig. 1A) (Barber et al., 2013; Liyanage et al., 2013; Rastegar et al., 2009).
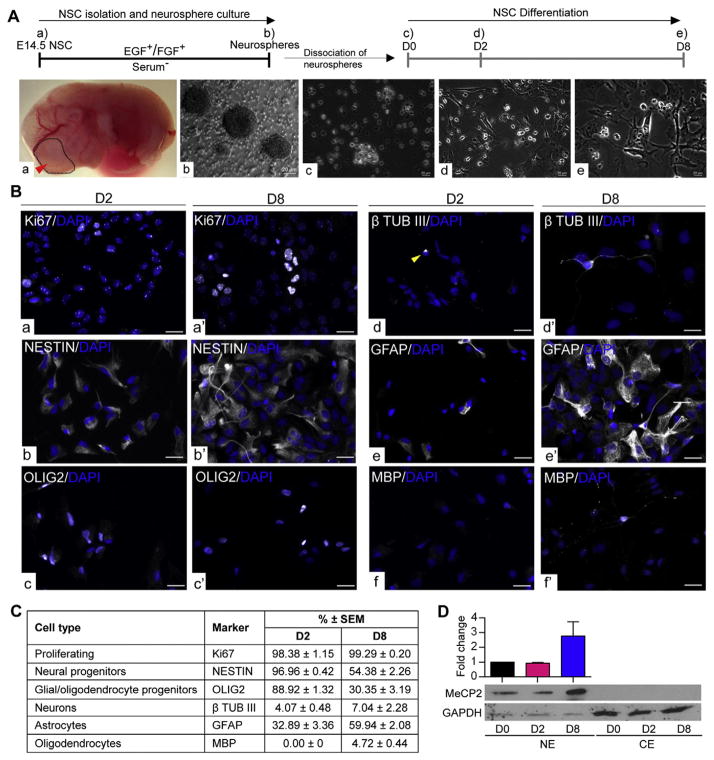
Fig. 1.
Differentiating neural stem cell (NSC) model. A) Schematic representation of NSC isolation, culture and differentiation. NSC are isolated from embryonic day (E) 14.5 forebrain (a) and cultured in the presence of growth factors (EGF and FGF) to generate neurospheres. (b) The neurospheres are dissociated at day zero (D0) (c) and differentiated for 8 days. Samples were collected at early stage of differentiation at day (D) 2 (d) and at a later differentiation stage D8 (e). Scale bars represent 20 μm. B) Representative images of different cell types in D2 and D8 population. a–a′: Ki67, b–b′: NESTIN, c–c′: OLIG2, d–d′: β-TUB III, e–e′: GFAP, f–f′: MBP. Note that no MBP+ cells were found at D2 (f). Scale bars represent 20 μm. C) Quantified percentages of the cellular composition of D2 and D8 differentiating populations. N = 3 ± SEM. D) MeCP2 expression during NSC differentiation detected by western blot. CE: Cytoplasmic extracts and NE: nuclear extracts. GAPDH was used as the loading control. N = 3 ± SEM.
We first determined the cellular composition of D2 and D8 differentiating cell populations by immunocytochemistry. All the images were captured in comparison to the exposure levels set using the primary omission negative control (Fig. S1A). Representative images for each cell type-specific marker at D2 and D8 are shown in Fig. 1B and the quantifications for each cell type marker are shown in Fig. 1C. We first studied Ki67 staining in D2, which marks proliferating cells. We observed two different patterns of Ki67, one that is localized to the nucleoli and another pattern with a diffused localization within the nucleus (extra-nucleolar) (Fig. S1B). Literature shows that both staining patterns are seen in proliferating cells and the localization of Ki67 depends on the stage in cell cycle (Gerdes et al., 1984; MacCallum and Hall, 2000). Therefore, all cells with either diffused, or nucleolar staining patterns were quantified, which resulted in 98.38 ± 1.15 Ki67+ cells (Figs. 1B:a; C). The D2 population was majorly composed of neural progenitor cells positive for NESTIN (96.96% ± 0.41) (Figs. 1B:b; C). D2 was also highly enriched for OLIG2 (88.91% ± 1.31) (Figs. 1B:c; C), which is a marker for glial/oligodendrocyte progenitor cells (Cai et al., 2007; Yokoo et al., 2004). As differentiated cells at D2, we observed 4.07 ± 0.48 TUB III+ neurons and 32.89% ± 3.36 GFAP+ astrocytes (Figs. 1B:d–e; C). The quantification of TUB III+ neurons could presumably be an underestimate due to the very short nature of their neurites. Despite multiple efforts, we did not detect any MBP+ oligodendrocytes at D2. However, these cells were highly enriched for the oligodendrocyte/glial progenitor marker OLIG2 (88.92 ± 1.32). Within the OLIG2+ progenitor population, two types of staining patterns were observed, one which is highly enriched at the nucleus and the second being nucleo-cytoplasmic (Fig. S1C). In the literature, it has been shown that translocation of OLIG2 from cytoplasm to nucleus is required for astrocyte differentiation (Setoguchi and Kondo, 2004). Therefore, it is possible that the OLIG2+ cells in the D2 population are still in the process of committing towards oligodendrocyte/glial lineage.
In D8 population, we found 99.29 ± 0.20 Ki67+ proliferating cells (Figs. 1B:a′; C). At D8 differentiating cells, 54.38% ± 2.26 of cells showed NESTIN expression, indicating that they still carry their neural progenitor lineage signature (Figs. 1B:b′; C). As differentiated cell types at D8, we observed 7.04 ± 2.28 TUB III+ neurons, 59.94 ± 2.08 GFAP+ astrocytes, and 4.72% ± 0.44 MBP+ oligodendrocytes (Figs. 1B: d′–f′; C). This population still had 30.35% ± 3.19 OLIG2+ glial/oligodendrocyte progenitors (Figs. 1B:c′; C). In our previous study, we have shown the detection of MeCP2 in all these cell types (Liyanage et al., 2013).
Collectively, the number of differentiated cell types (TUB III+, GFAP+, and MBP+) increased from D2 to D8, as expected. The number of OLIG2+ glial/oligodendrocyte progenitor cells and neural progenitor cells (NESTIN+) decreased from D2 to D8. These observations confirmed that cells have undergone differentiation from D2 to D8. The number of proliferating cells (Ki67+) more or less remained the same suggesting that D8 cells still are not terminally differentiated (Fig. 1C).
Next, we determined the expression levels of MeCP2 at different stages of NSC differentiation (D0, D2 and D8) by western blot. MeCP2 is a known nuclear protein and yet some studies have reported cytoplasmic expression of MeCP2 (Miyake and Nagai, 2007). Therefore, we first confirmed the expression pattern of MeCP2 in the nuclear and cytoplasmic fractions of D8 differentiated cells, which showed the detection of MeCP2 exclusively in nuclear fractions (Fig. S2A). In order to determine the most suitable endogenous control for our nuclear and cytoplasmic fractions, we screened known nuclear markers RNA Pol II and HDAC2, and α-Tubulin as a known cytoplasmic marker. Additionally, we used GAPDH and ACTIN loading controls, which were detected in both nuclear and cytoplasmic fractions (Fig. S2A). As expected RNA Pol II was detected only in nuclear fraction, while HDAC2 was highly enriched in the nuclear fraction. α-Tubulin was enriched in cytoplasmic fraction. Yet, it is known that α-Tubulin could also be detected in nuclear fraction (Goo et al., 2003; Hayashi et al., 2014). As expected, ACTIN and GAPDH were detected in both fractions (Fig. S2A). This is also in agreement with a previous report from our lab to detect GAPDH in both nuclear and cytoplasmic fractions of murine brain samples (Olson et al., 2014). Re-probe of a membrane with lower levels of MeCP2 in ethanol-withdrawal samples (see next section), indicated a slight induction of RNA Pol II protein levels (Fig. S2B), which suggested that perhaps RNA Pol II is not the best endogenous control to be used for quantification. Therefore, ACTIN and GAPDH were used as loading controls for the rest of the experiments and either ACTIN or GAPDH was used for western blot quantification.
Western blot experiments at D0, D2 and D8 (during differentiation) showed that MeCP2 protein levels are detected at all three stages (Fig. 1D; Fig. S3), which was in agreement with our previous study (Liyanage et al., 2013). MeCP2 expression remained relatively the same from D0 to D2, but increased rapidly by D8 in three biological replicates. A representative blot for MeCP2 expression during NSC differentiation is shown in Fig. 1D. MeCP2 was not detected in the cytoplasmic fractions at any tested differentiation stages (Fig. 1D).
Ethanol exposure and withdrawal show opposing effects on Mecp2/MeCP2 expression in differentiating neural stem cells
At the onset of differentiation (D0), cells were treated with 70 mM ethanol (Fig. 2A). The selected concentration of ethanol was chosen based on previous studies reporting: a) the blood alcohol levels detected in alcoholics (30–100 mM) (Adachi et al., 1991), b) the effects of ethanol on isolated NSC from human fetal brain (20–100 mM) (Vangipuram and Lyman, 2012), and c) conditions which show minimal effects on cell proliferation and survival of cultured NSC (22–69 mM) (Hicks et al., 2010). Control experiments were carried out without ethanol treatment. As established in previous similar studies (Pappalardo-Carter et al., 2013; Vangipuram and Lyman, 2012; Zhou et al., 2011a), cells were treated once with ethanol for 48 h to study the effect of single time or binge-like alcohol exposure. To study the effects of continuous/chronic ethanol exposure, differentiating cells were treated with ethanol for 8 days (until D8). To study the effects of ethanol withdrawal, ethanol was removed from the cultures at D2 and the cells were kept in culture for extra 6 days until D8. Gene expression and DNA methylation analysis were performed after ethanol exposure at D2 and D8 and after ethanol withdrawal at D8 (6 days after ethanol removal) (Fig. 2A). Under each condition, Mecp2/MeCP2 expression levels in ethanol exposure or ethanol-withdrawal conditions were compared to the control untreated cells in each biological replicate.
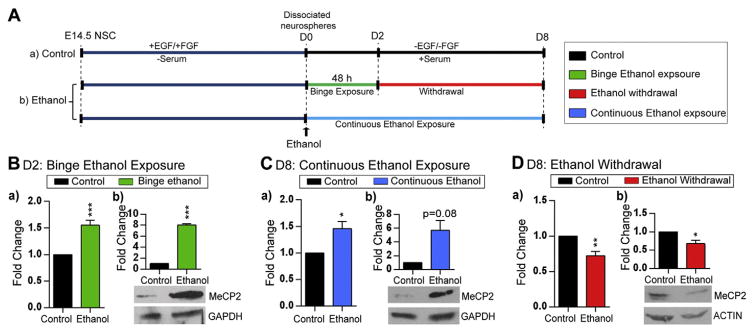
Fig. 2.
Effect of ethanol exposure and withdrawal on Mecp2/MeCP2 expression in differentiating neural stem cells (NSC). A) Schematic representation of NSC differentiation and ethanol treatment. a) Control experiment: NSC are isolated from embryonic day (E)14.5 forebrain and cultured in the presence of growth factors (EGF and FGF) to generate neurospheres. The primary neurospheres were dissociated (D0) and differentiated for 8 days. b) To model binge ethanol exposure and withdrawal, 70 mM of ethanol was added at the onset of NSC differentiation (D0) to the media of cultured dissociated cells for 48 h and ethanol was withdrawn/removed at D2. To model continuous ethanol exposure, dissociated neural stem cells were treated with ethanol for continuously 8 days. Cells were kept in culture till D8 and media was refreshed every other day. B) Effect of binge ethanol exposure on the expression of (a) Mecp2 transcript levels [N = 3 ± SEM], and (b) MeCP2 protein levels [N = 2 ± SEM]. C) Effect of continuous ethanol exposure on the expression of (a) Mecp2 transcript levels [N = 3 ± SEM], and (b) MeCP2 protein levels [N = 2 ± SEM]. D) Effect of ethanol withdrawal on the expression of (a) Mecp2 transcript expression [N = 3 ± SEM], and (b) MeCP2 protein levels [N = 2 ± SEM]. Fold changes are calculated relative to D2 and D8 untreated control cells. Significant differences from controls are indicated with p < 0.001***, p < 0.01** or p < 0.05*. The transcript expression values were normalized to Gapdh expression (endogenous control). ACTIN or GAPDH was used as a loading control for western blots. Note that exposure time for each western blot is different.
Next, we investigated Mecp2 transcript and MeCP2 protein expression in response to ethanol treatments by qRT-PCR and WB. Following ethanol exposure at D2, we observed a slight, but significant induction of Mecp2 transcript levels (1.55-fold, p < 0.001) (Fig. 2B:a), which was translated into a significant induction of MeCP2 at the protein levels by 8-fold (p < 0.001) (Fig. 2B:b). Similarly, continuous ethanol exposure upregulated Mecp2 transcripts (1.46-fold, p < 0.05), and MeCP2 protein levels (5.7-fold, p = 0.08) (Fig. 2C). In contrast, ethanol withdrawal was associated with significantly reduced Mecp2 transcript (1.38-fold, p < 0.01) and MeCP2 protein expression (1.48-fold, p < 0.05) (Fig. 2D). Two biological replicates of western blots for each condition are shown in Fig. S4. Comparison of the Mecp2 transcript and MeCP2 protein levels indicated that even minor changes in Mecp2 transcript levels can be translated into significant changes in MeCP2 protein. Previous reports on MeCP2 expression have shown that both mild over expression (~2-fold) or slightly reduced MeCP2 levels may lead to severe neurological phenotypes (Bodda et al., 2013; Collins et al., 2004). Therefore, the detected alterations in MeCP2 protein levels (5.7-fold and 8-fold induction; and 1.48-fold inhibition) can be biologically important.
Changes to Mecp2/MeCP2 expression in response to ethanol exposure and withdrawal are associated with altered DNA methylation at the Mecp2 regulatory elements
It has been reported that altered gene expression in human alcoholics and alcohol-fed mice is associated with aberrant promoter DNA methylation (Barker et al., 2013; Bleich et al., 2006). In our recent studies, altered Mecp2 expression by a DNA demethylating agent Decitabine was correlated with DNA methylation at the Mecp2 regulatory elements within its promoter and intron 1, in a similar differentiating NSC system. Moreover, Mecp2 expression in adult murine brain regions in vivo was significantly correlated with DNA methylation at the same Mecp2 REs (Liyanage et al., 2013; Olson et al., 2014). This prompted us to investigate the DNA methylation status of the Mecp2 REs upon ethanol exposure and withdrawal. To study whether the same REs are involved in the ethanol-mediated changes in Mecp2 expression, we analyzed DNA methylation patterns of the previously reported three regions within the Mecp2 promoter (R1–R3) and three regions within intron 1 (R4–R6) (Fig. 3A) (Liyanage et al., 2013; Olson et al., 2014).
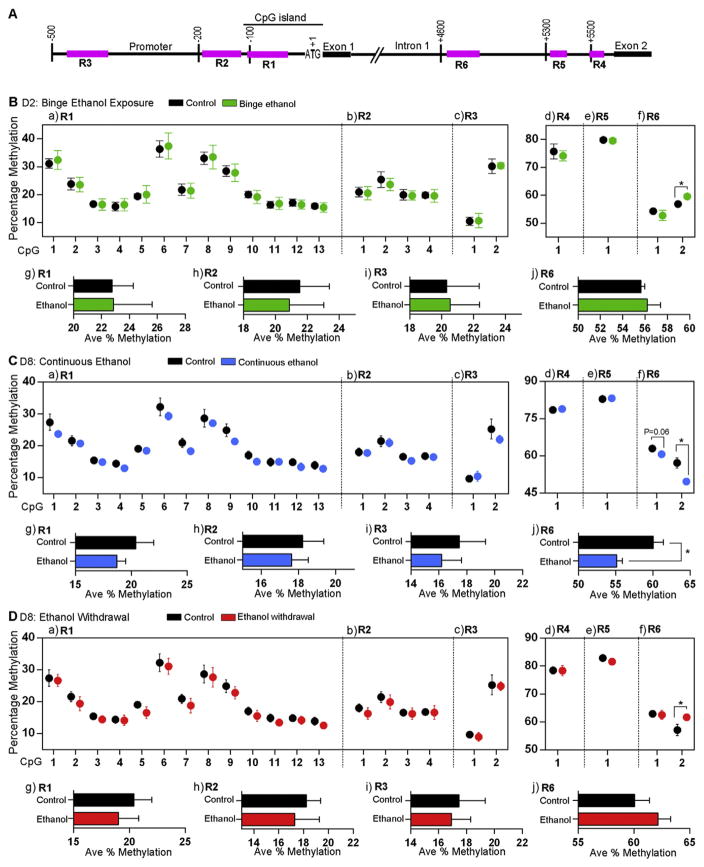
Fig. 3.
Bisulfite pyrosequencing analysis of the effect of ethanol on DNA methylation status at the Mecp2 regulatory elements. A) Schematic representation of the three regions (R1–R3) within the Mecp2 promoter and three regions (R4–R6) within the Mecp2 intron 1 (not drawn to scale). B) Effect of binge ethanol exposure at day (D) 2 on percentage DNA methylation of individual CpG sites within the Mecp2 regulatory regions (a) R1, (b) R2, (c) R3, (d) R4, (e) R5, and (f) R6; and the average percentage DNA methylation (Ave % Methylation) over (g) R1, (h) R2, (i) R3, and (j) R6. C) Effect of continuous ethanol exposure at D8 on percentage DNA methylation of individual CpG sites within the Mecp2 regulatory regions (a) R1, (b) R2, (c) R3, (d) R4, (e) R5, and (f) R6; and the average percentage DNA methylation (Ave % Methylation) over (g) R1, (h) R2, (i) R3, and (j) R6. D) Effect of ethanol withdrawal at D8 on percentage methylation of individual CpG sites within the Mecp2 regulatory regions (a) R1, (b) R2, (c) R3, (d) R4, (e) R5, and (f) R6; and the average percentage DNA methylation (Ave % Methylation) over (g) R1, (h) R2, (i) R3, and (j) R6. Significant differences from controls are indicated with p < 0.05*. N = 3 ± SEM for all D2 samples, ethanol continuous and ethanol withdrawal samples and N = 6 ± SEM for D8 control samples. The control D2 DNA methylation data was extracted from our recent study (Liyanage et al., 2013), which was done at the same time and in parallel to this current study.
First, we performed bisulfite pyrosequencing to analyze DNA methylation at the individual CpG sites within these regions. After binge ethanol exposure at D2, DNA methylation levels at the individual CpG sites within the Mecp2 promoter regions R1–R3 and intronic regions R4 and R5 were not significantly altered from the controls (Fig. 3B:a–e). However, R6: CpG-2 was slightly hypermethylated by 2.7% (p < 0.05) (Fig. 3B:f). Further analysis of average percentage methylation over entire regions R1, R2, R3 and R6 (which contain multiple CpG sites) did not show any significant changes in the average DNA methylation by single ethanol exposure (Fig. 3B:g–j). A similar analysis after continuous ethanol exposure showed significant hypomethylation at both CpG sites within the intron 1 R6 (CpG1: 2.3%, p = 0.06; CpG2: 7.5%, p < 0.05) (Fig. 3C:f). Average methylation over the entire R6 also showed significant demethylation of the R6 (5%, p < 0.05) by continuous ethanol exposure (Fig. 3C:j). However, no changes in DNA methylation at individual CpG sites or average methylation over the entire regions were observed in R1–R5 (Fig. 3C). Comparison of DNA methylation patterns in between D8 control and ethanol withdrawal conditions showed significant hypermethylation of the R6 CpG2 (4.5%, p < 0.05) (Fig. 3D:f) and no change in average DNA methylation of regions 1–6 (Fig. 3D:g–j).
Previous studies have shown that 2–2.5% increase in average MECP2/Mecp2 DNA promoter methylation is correlated with significantly reduced levels of MeCP2 in autistic patients and stressed mouse brain (Franklin et al., 2010; Nagarajan et al., 2006). Our recent studies also showed that minor changes in percentage DNA methylation (2–5%) mediated by Decitabine exposure correlated with significant changes in Mecp2 expression (Liyanage et al., 2013). This indicates that even minor changes in the DNA methylation at the MECP2/Mecp2 REs might be associated with its altered expression. Therefore, it is likely that the detected changes in DNA methylation at the Mecp2 REs are responsible for altered Mecp2/MeCP2 expression in differentiating NSC. However, bisulfite pyrosequencing does not differentiate between 5mC and 5hmC methyl marks (Huang et al., 2010) and might be reflecting cumulative changes in 5mC and 5hmC. Thus, further analysis of 5mC and 5hmC at these regions is required.
Ethanol causes interplay between 5mC and 5hmC enrichment at the Mecp2 regulatory elements
In order to differentiate between changes in 5mC and 5hmC at the Mecp2 REs; we performed hMeDIP for 5hmC and MeDIP for 5mC. Single ethanol exposure at D2 caused significant induction of 5hmC enrichment at the Mecp2 promoter R2 (~3-fold, p < 0.05) (Fig. 4A:b). The increased 5hmC levels at R2 were associated with upregulation of Mecp2 transcript expression, suggesting that the increased 5hmC may contribute to Mecp2 upregulation.
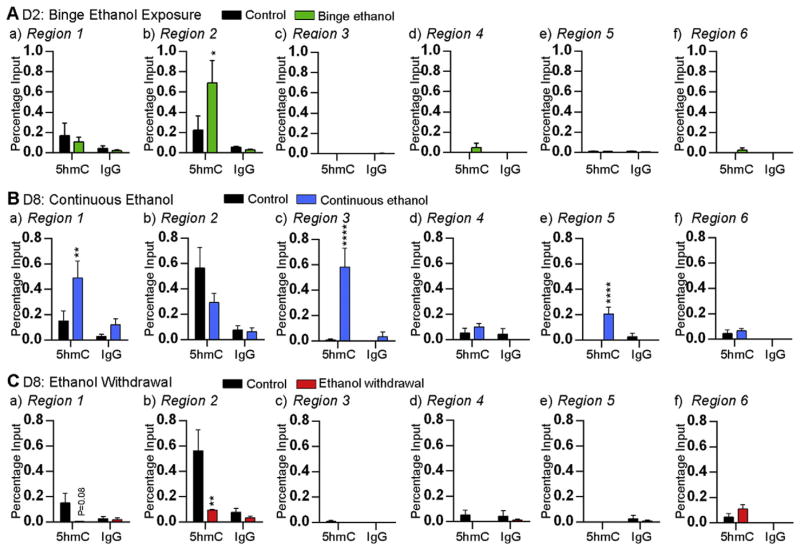
Fig. 4.
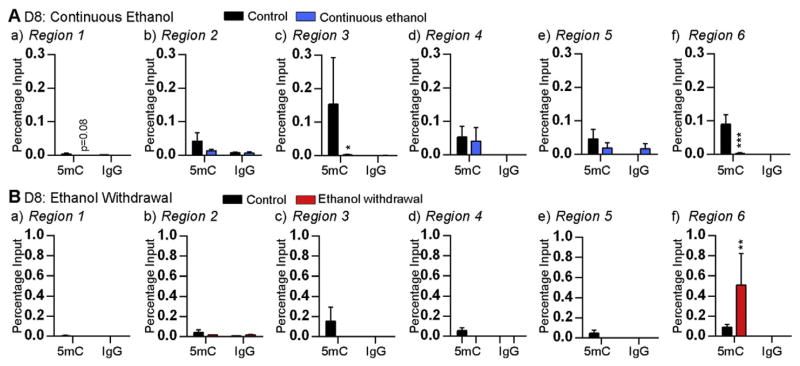
Effect of ethanol on the enrichment of 5hmC at the Mecp2 regulatory elements. Enrichment of 5hmC and non-immunogenic IgG at the Mecp2 promoter (a) region 1, (b) region 2, and (c) region 3, and the Mecp2 intron 1 (d) region 4, (e) region 5, and (f) region 6; after A) binge ethanol exposure at day (D) 2, B) continuous ethanol exposure at D8, and C) ethanol withdrawal at D8. Significant differences from controls are indicated with p < 0.0001****, p < 0.01** or p < 0.05*. N = 4 ± SEM.
Since continuous ethanol exposure and ethanol withdrawal had opposing effects on Mecp2 expression, we analyzed both 5mC and 5hmC enrichment at the Mecp2 REs. In the case of continuous ethanol exposure (associated with increased Mecp2 expression), significant 5hmC enrichment was observed at R1 (~3.2-fold, p < 0.01), R3 (~65-fold, p < 0.0001) and R5 (~69-fold, p < 0.0001) (Fig. 4B). In contrast, reduced 5mC enrichment was observed at R3 (~77-fold, p < 0.05) and R6 (~36.2-fold, p < 0.001) upon continuous ethanol exposure (Fig. 5A). These results suggest that the induced Mecp2 expression by continuous ethanol exposure might be the result of increased 5hmC enrichment and reduced 5mC enrichment at specific Mecp2 REs. Similar analysis after ethanol withdrawal (which caused downregulation of Mecp2 expression) revealed reduced 5hmC enrichment and increased 5mC at specific Mecp2 REs. The 5hmC enrichment at the promoter R1 (~24-fold, p = 0.08) and R2 (~6-fold, p < 0.01) was reduced by ethanol withdrawal (Fig. 4C). In contrast, the 5mC enrichment at the intron 1 R6 was significantly induced by ~5.7-fold (p < 0.01) (Fig. 5B). These results suggested that downregulation of Mecp2 expression by ethanol withdrawal might be the result of reduced 5hmC and increased 5mC enrichment at specific Mecp2 REs.
Fig. 5.
Effect of ethanol on the enrichment of 5mC at the Mecp2 regulatory elements. Enrichment of 5mC and non-immunogenic IgG at the Mecp2 promoter (a) region 1, (b) region 2, and (c) region 3, and the Mecp2 intron 1 (d) region 4, (e) region 5, and (f) region 6; after A) continuous ethanol exposure at day (D) 8 and B) ethanol withdrawal at D8. Significant differences from controls are indicated with p < 0.001***, p < 0.01** or p < 0.05*. N = 4 ± SEM.
In all MeDIP and hMeDIP experiments, the specificity of the detected signals was confirmed by using a non-immunogenic IgG antibody, which showed negligible pull down (Figs. 4 and 5). For hMeDIP, we also used an internal control Gapdh as unmethylated negative control and a control DNA template (44 5hmC-CpGs) as a positive control, which was provided within the kit (Fig. S5A). For MeDIP, the specificity of the detected signals was confirmed by using Gapdh and Major Satellite Repeats as negative and positive controls, respectively (Fig. S5B).
Collectively, the changes we observed in Mecp2/MeCP2 expression after ethanol exposure and withdrawal are associated with dynamic changes in 5mC and 5hmC levels at specific regulatory elements. Overall, these changes in the 5hmc levels were independent of 5mC and vice versa, except for the R3 under continuous ethanol exposure conditions.
Ethanol changes global DNA methylation of differentiating neural stem cells
At the level of Mecp2 gene we observed dynamic changes in 5hmC and 5mC levels upon ethanol exposure and withdrawal. These observations prompted us to further investigate whether ethanol also causes globally changed levels of DNA methylation, as a control experiment for MeDIP and hMeDIP.
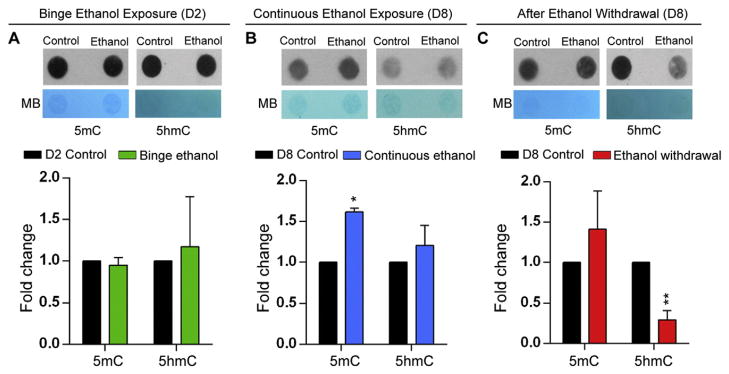
Quantitative DNA methylation analysis by DNA dot blot assay showed that binge ethanol exposure at D2 did not cause any significant alteration in the 5mC and 5hmC levels (Fig. 6A). However, continuous ethanol exposure resulted in significant induction of 5mC levels (1.7-fold, p < 0.05) without significant effect on the 5hmC levels (Fig. 6B). In contrast, ethanol withdrawal significantly reduced 5hmC levels by 4-fold (p < 0.01), with non-significant effects on 5mC levels (Fig. 6C). These findings indicate that the effect of ethanol on global 5mC and 5hmC levels is dependent on the method of ethanol treatment (exposure or withdrawal) and the stage of neural stem cell differentiation. Moreover, the global changes in 5mC and 5hmC levels appear to be independent of each other.
Fig. 6.
Effect of ethanol on global DNA methylation in differentiating neural stem cells (NSC). DNA dot blot experiments to detect global DNA methylation levels for 5mC and 5hmC. MB refers to methylene blue staining for visualizing the total DNA. Quantification of the global 5mC and 5hmc levels normalized to the total DNA levels is also shown. Fold changes were calculated relative to day (D) 2 or D8 control untreated cells. N = 3 ± SEM. Significant differences from controls are indicated with p < 0.05* or p < 0.01**.
Ethanol treatments have minimal effects on neural stem cell fate commitments but cause alterations in neuronal morphology and glial size
Next, we examined whether the observed changes in Mecp2/MeCP2 expression or the DNA methylation in response to ethanol treatments were associated with any changes in the cell population during NSC differentiation.
In order to determine the composition of both control and ethanol-treated populations, we studied the expression of cell type-specific markers that we analyzed in Fig. 1 at the transcript level by qRT-PCR and the number of cells positive for these markers by ICC. Binge ethanol exposure at D2 caused significant induction of Mbp, while changes of the rest of the cell type markers were not statistically significant (Fig. S6A:a). Continuous ethanol exposure at D8 resulted in significant induction of Nestin, Tub III and Mbp. Expression of other cell type-specific marker genes was relatively unaffected by continuous ethanol exposure (Fig. S6B:a). Ethanol withdrawal did not result in significantly altered expression of any of the tested cell type-specific markers (Fig. S6C:a).
Next, in order to determine whether the number of cells expressing these markers was changed by ethanol treatments, immunocytochemical experiments were carried out with the antibodies specific for these markers. Binge ethanol exposure did not significantly change the number of cells positive for TUB III, GFAP, OLIG2, Ki67 and NESTIN. This observation suggested that binge ethanol exposure had minimal effects on cell population at D2 (Fig. S6A:b). Since Mbp expression was upregulated at the transcript level and that was not represented in the number of MBP+ oligodendrocytes determined by ICC, we studied whether the presence of different isoforms of Mbp/MBP has affected our conclusion. Comparison of the mRNA sequence amplified by Mbp-specific primers (Fig. S7A) and the amino acid sequence used to generate the antibody for MBP (Fig. S7B) showed that the qRT-PCR primers and MBP antibody recognize similar sequences within all four isoforms (1, 2, 3 and 5) (Fig. S7). Similar discordance between transcript and protein expression has also been reported for other genes (Velez-Bermudez and Schmidt, 2014).
Similar to binge ethanol exposure, continuous ethanol exposure and ethanol withdrawal did not cause any significant change in the number of cells expressing each tested marker (Figs. S6B:b; C:b). In continuous ethanol-treated population, there was a slight but statistically non-significant increase in the number of MBP+ cells, in accordance with the detected increase in Mbp transcript levels. This detected difference at the transcript levels and the number of cell counts, could once again be due to the discordance between transcript and protein expression or changes in transcript expression within individual cells.
Taken together, these results confirmed that the balance between the numbers of differentiated cell types was not significantly affected under the tested conditions. Thus, the 70 mM ethanol treatments and withdrawal of ethanol had minimal impact on NSC cell fate commitment. Therefore, the observed changes in Mecp2/MeCP2 expression and global DNA methylation are not likely to be due to changes in cell fate commitment.
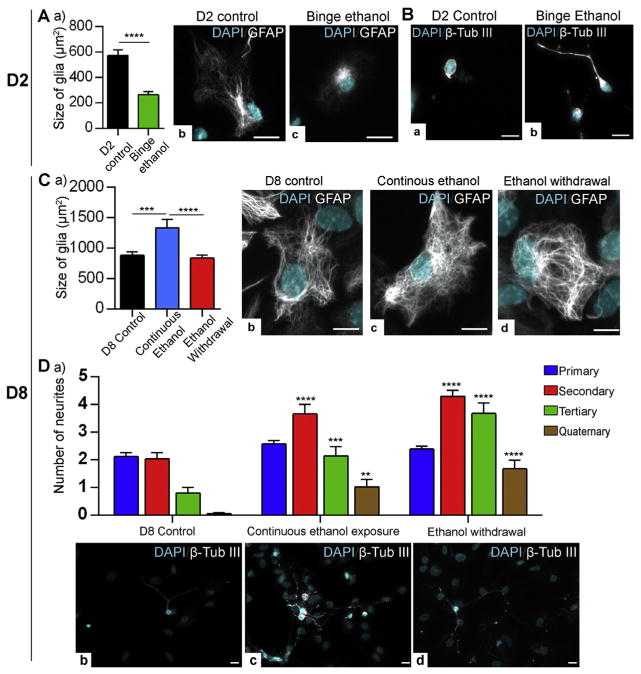
In ethanol-exposed neurons and glia, apart from the altered gene expression, alterations in the neuronal and glial morphology had been reported. Moreover, aberrant neurite outgrowth (both increased and decreased neurite outgrowth) and glial cell growth (mostly reduced glial cell growth) are associated with compromised central nervous system function and abnormal brain size (microcephaly) in FASD (Riley and McGee, 2005; Samson and Grant, 1984). In agreement with these previous reports, we observed changes in neuronal morphology and glial size. As a measurement of glial size, we analyzed the surface area of GFAP+ cells, as other research groups have done to represent the size of glia (Kulijewicz-Nawrot et al., 2012; Rodriguez et al., 2013). Immunofluorescent labeling of binge ethanol-treated astrocytes at D2 showed reduced cellular size (~2.1-fold, p < 0.0001) relative to the control untreated D2 cells (Fig. 7A). Unlike binge ethanol exposure, in the case of continuous ethanol exposure, approximately 1.5-fold (p < 0.001) increase in glial size was observed relative to the untreated D8 control cells. No visible changes in glial size were observed between D8 control and ethanol withdrawal. We detected a 1.59-fold (p < 0.0001) increase in the glial cell size in continuous ethanol exposure as compared to the ethanol withdrawal (Fig. 7C). These results suggest that glial size is affected by ethanol exposure but not by ethanol withdrawal. Also, in the case of ethanol exposure, the binge ethanol and continuous ethanol have opposing effects on the glial cell size.
Fig. 7.
Effect of ethanol on glial size and neuronal morphology. A) Comparison of glial cell size in (μm2) in day (D) 2 control and binge ethanol-treated conditions. a) Graphical representation of the quantification of the glial cell size (μm2). At least 20 GFAP+ cells per biological replicate were quantified under each condition. N = 3 ± SEM. Significant differences from controls are indicated with p < 0.0001****. Representative images of (b) D2 control astrocyte and (c) binge ethanol exposed astrocyte. Scale bars represent 10 μm. B) Immunofluorescent staining of neurons with β Tubulin III in: (a) D2 control and (b) binge ethanol-treated cells. Scale bars represent 10 μm. C) Comparison of glial cell size in (μm2) in D8 control, continuous ethanol-treated and ethanol withdrawal conditions. a) Graphical representation of the quantification of the glial cell size (μm2). At least 20 GFAP+ cells per biological replicate were quantified under each condition. N = 3 ± SEM. Significant differences from controls are indicated with p < 0.001*** or p < 0.0001****. Representative images of (b) D8 control astrocyte, (c) continuous ethanol-exposed astrocyte and (c) ethanol-withdrawn astrocyte are shown. Scale bars represent 10 μm. D) Comparison of neuronal morphology in D8 control, continuous ethanol-treated and ethanol withdrawal conditions. (a) Quantification of number of neurites. At least 20 TUB III+ neuronal cells per biological replicate were quantified under each condition. N = 3 ± SEM. Significant differences from controls are indicated with p < 0.01**, p < 0.001*** or p < 0.0001****. Representative images of (a) D8 control neuron, (b) continuous ethanol-exposed neuron, and (c) ethanol-withdrawn neuron are shown. Scale bars represent 10 μm.
Interestingly, both ethanol exposure and ethanol withdrawal promoted neuronal neurite outgrowth and branching. Compared to the control D2 neurons, the binge ethanol exposed neurons had induced neurite outgrowth. However, quantification of D2 neuronal branching was difficult due to the very short neurites observed in TUB III+ neurons (Fig. 7B). Representative images of D2 neurons are shown in both Fig. 2B and Fig. 7B. Distinct changes in neuronal morphology were detected in continuous ethanol exposed and ethanol withdrawal neurons as compared to D8 control neurons. Quantification of neuronal morphology in D8 control, continuous ethanol exposure and ethanol withdrawal conditions showed that there is increased number of secondary, tertiary and quaternary neurites in both continuous ethanol-exposed neurons and ethanol-withdrawal neurons (Fig. 7D). Comparison of neurite branching between continuous ethanol exposure and ethanol withdrawal indicated significant increase in tertiary neurites in ethanol withdrawal neurons. These observations suggest that exposure to ethanol at the beginning of neural differentiation as well as its subsequent withdrawal may recapitulate key features of ethanol-mediated neuroglial morphological changes and also FASD.
Discussion
Among the epigenetic factors affected by alcohol, MeCP2 has become an emerging target (Chen et al., 2013; Kim et al., 2013; Repunte-Canonigo et al., 2014), mostly due to its solid link with neurodevelopmental disorders (Zachariah and Rastegar, 2012). These previous studies on the detrimental effects of ethanol on MeCP2 also suggest that MeCP2 expression is dependent on the dose and the duration of ethanol exposure. In this study, we demonstrate how Mecp2/MeCP2 expression is affected following a short time single exposure (binge), continuous exposure to ethanol as well as withdrawal of ethanol during neuronal differentiation. We observed that ethanol exposure altered Mecp2/MeCP2 expression in association with altered DNA methylation at the Mecp2 REs. Our results show the contribution of DNA methylation at the promoter and the intron 1 silencer element in deregulating Mecp2/MeCP2 expression upon ethanol exposure. The potential contribution from other mechanisms beyond DNA methylation such as histone acetylation or miRNA activities in deregulation of Mecp2/MeCP2 should not be excluded. In support of this concept, a study showed the association of changes in miRNAs (miR-152, miR-199a-3p, and miR-685) and downregulation of Mecp2 expression followed by both ethanol (75 mM) exposure and withdrawal in E15 primary cortical neurons (Guo et al., 2012).
In our study, in almost all conditions (except for R3 in continuous ethanol exposure), 5hmC levels were changed without affecting the 5mC levels or vice versa. There are several explanations for such changes in DNA methylation patterns in our model. First, the DNA methylation patterns of continuous ethanol exposure and ethanol withdrawal were analyzed 6 days after binge ethanol exposure. During these 6 days, the neural stem cells were differentiating and proliferating with or without ethanol. In order to understand how the two methyl marks are changed gradually, DNA methylation patterns should be analyzed at shorter time points, perhaps every 12–24 h. Secondly, the changes in 5hmC could also occur through either active or passive DNA demethylation (Song et al., 2012). It is unknown which pathways occur in our differentiating NSC model. In congruence with our observations on dynamics of 5mC and 5hmC, other studies suggest the possibility of 5hmC changes occurring independent of 5mC changes (Haffner et al., 2011; James et al., 2014). Even though, our study reports only 5hmC levels, there are other oxidative derivatives of 5mC, namely 5fC and 5CaC which could also contribute to the observed dynamics of 5mC and 5hmC [for review refer to (Liyanage et al., 2014)]. Further analysis is required to study whether altered DNA methylation (5mC and 5hmC) patterns at both Mecp2 gene-specific regions and global levels are taking place gradually from binge ethanol exposure to continuous ethanol exposure or ethanol withdrawal.
In our study, bisulfite pyrosequencing of the studied regions did not reveal any considerable change in DNA methylation between control and ethanol-treated cells (except for intron 1-R6). However, analysis of 5mC and 5hmC enrichment at each region demonstrated significant changes in DNA methylation at these regions, representing a mechanism of DNA methylation-mediated regulation of Mecp2. These results also suggest the importance of investigating the enrichment of 5mC and 5hmC at the gene regulatory elements. This is especially important when no changes of DNA methylation are obtained through bisulfite pyrosequencing, which could be misleading to conclude that DNA methylation might not be involved in regulating certain genes.
Ethanol is known to alter global DNA methylation during neural development and this has been suggested as a potential mechanism for FASD pathogenesis (Chen et al., 2013; Zhou et al., 2011b). One single administration of ethanol at the onset of NSC differentiation did not alter the global 5mC and 5hmC levels. Continuous ethanol exposure induced 5mC levels whereas the withdrawal of ethanol reduced 5hmC levels. Therefore, it is possible that the effects of ethanol on global 5mC and 5hmC methylation is dependent on multiple factors such as the mode of ethanol treatment (binge ethanol, continuous ethanol exposure or ethanol withdrawal), duration of the treatment, stage of neural differentiation and the type of DNA methylation (5mC or 5hmC). Since DNA methylation is a major player in maintaining chromatin architecture and regulating gene expression (Liyanage et al., 2012), altered 5mC and 5hmC levels in turn may alter chromatin structure and gene expression genome-wide. Our findings on ethanol-mediated altered global methylation patterns during NSC differentiation are supported by a recent study showing alteration of the progression of 5hmC marks by ethanol during neural development (Chen et al., 2013).
Neurological conditions caused by prenatal exposure to alcohol can also be considered as consequences of ethanol withdrawal because during pregnancy, the fetus is exposed to alcohol and could be withdrawn from alcohol in utero, or after birth. Therefore, our observations on ethanol withdrawal downregulating Mecp2/MeCP2 expression is in agreement with several other reports where prenatal ethanol exposure as well as preconception ethanol exposure (and subsequent withdrawal) led to decreased level of MeCP2 in a brain region-specific manner (Chen et al., 2013; Kim et al., 2013, 2014; Perkins et al., 2013). Comparing our data with previous reports that were earlier mentioned in the introduction (Bekdash et al., 2013; Chen et al., 2013; Gangisetty et al., 2014; Guo et al., 2012; Kim et al., 2013, 2014; Perkins et al., 2013; Subbanna et al., 2014; Tunc-Ozcan et al., 2013), it is possible that the effect of ethanol on Mecp2/MeCP2 expression is influenced by multiple factors. This could include the type of ethanol treatment (binge exposure, continuous exposure and withdrawal), amount or concentration of ethanol, type of rodent model used in the study (mouse or rats), developmental stage (different embryonic time points or adult), brain regions and cell types within a certain brain region (specific neuronal subtype). For instance, in all previous reports the brain regions studied were either the cortex, striatum or hippocampus whereas our studies were performed using NSC isolated from embryonic forebrain which could also contribute to the differences observed in between the previous studies and our data. Moreover, in primary cortical neurons exposure to ethanol and ethanol-withdrawal with similar concentration (75 mM as compared to 70 mM in our study) resulted in downregulation of Mecp2/MeCP2 expression (Guo et al., 2012). In contrast, our study demonstrates that both binge and continuous ethanol exposure upregulate Mecp2/MeCP2 expression, while withdrawal of ethanol downregulates its expression. Ethanol exposure is considered as an environmental insult that may impact cell types or individuals differently. Additional factors to be considered may include maternal/paternal care, environmental conditions (light/dark, diet) of the rodent in terms of in vivo studies, and in vitro culture conditions.
As a key epigenetic factor for brain development and function, both deficiency and overexpression of MeCP2 lead to severe neurological disorders including autism (Liyanage and Rastegar, 2014). FASD patients also show autistic symptoms (Bishop et al., 2007; Stevens et al., 2013). Hence, our study provides insights on the possible involvement of MeCP2 in associated neurological symptoms in alcoholics and FASD patients. Studying the biological consequences of aberrant MeCP2 expression would be important for further understanding of MeCP2 involvement in ethanol-induced neurological symptoms. For instance, target genes of MeCP2 such as NMDA receptor subunits (Maliszewska-Cyna et al., 2010) are shown to be aberrantly expressed in alcohol-exposed neural cell types (Moykkynen and Korpi, 2012). Therefore, altered MeCP2 expression as well as altered binding of MeCP2 to its target genes may contribute to ethanol teratogenesis. Moreover, it has been postulated that decreased levels of Mecp2/MeCP2 could contribute to the impulsive and hyperactive behaviors of offsprings exposed to ethanol prenatally (Kim et al., 2013). Previous studies show that even minor changes in MeCP2 expression may result in severe neurological conditions [reviewed in (Liyanage and Rastegar, 2014)]. Therefore, it is logical to consider that the ethanol-induced changes in MeCP2 expression in our model are biologically relevant and may contribute to the adverse effects of ethanol on differentiating cells.
Differentiating neural stem cells are frequently used as in vitro models to study the underlying molecular mechanisms of ethanol tera-togenesis and FASD pathogenesis (Pappalardo-Carter et al., 2013; Vangipuram and Lyman, 2012; Zhou et al., 2011a). Alcohol-mediated neurological symptoms and microcephaly are usually associated with disrupted communications between neural cell types due to aberrant neurite outgrowth (Bingham et al., 2004) and glial cell growth or morphology (Guerri et al., 2001). Our observations on the increased neurite outgrowth and changes in glial cellular size/surface area during NSC differentiation are in agreement with these previous reports. Several reports show the suppression of glial growth, glial proliferation as well as decreased expression of GFAP by ethanol (Guerri et al., 2001; Guizzetti and Costa, 1996; Valles et al., 1996). In agreement with these reports, we observed reduced glial cell size after binge ethanol exposure. Similarly, our data on the increased glial size by continuous ethanol treatment is in agreement with the increase in glial size observed after chronic ethanol treatment in vivo in a rat FASD model (Miller and Potempa, 1990). Therefore, our ethanol-treated NSC model system recapitulates two of the cellular phenotypes seen in FASD patients, in vivo FASD models and in vitro cellular models. This indicates that the differentiating neural stem cell model used in our studies represents a suitable system to study the effects of ethanol on gene expression changes, epigenetic mechanisms and molecular mechanisms of ethanol teratogenesis.
Interestingly, there is an overlap between the glial and neuronal cellular phenotypes observed in ethanol-treated conditions and MeCP2 overexpression/deficient-conditions. For example, ethanol exposure (Reiter-Funk and Dohrman, 2005; Roivainen et al., 1995a; Zou et al., 1993) and MeCP2 overexpression (Larimore et al., 2009; Rastegar et al., 2009) have been shown to show similar patterns of increased neurite branching. However, ethanol exposure has also been shown to reduce the neurite outgrowth (Camarillo and Miranda, 2008), similar to few studies on MeCP2-deficiency (Nguyen et al., 2012; Rastegar et al., 2009; Wang et al., 2013). The reduced glial cell growth in ethanol-exposed astrocytes (Guerri et al., 2001) is similar to the glial phenotypes seen in either MeCP2-deficient or MeCP2-overexpressed conditions (Maezawa et al., 2009; Tsujimura et al., 2009). The effects of ethanol on neuronal and glial morphologies/size seem to be influenced by many factors such as cell type, duration and concentration of ethanol exposure. On the other hand, MeCP2 has been shown to either increase or decrease the neurite morphology. Neuronal morphology is also influenced by nearby glial cells (Giordano et al., 2011). The absence of MeCP2 in adjacent glial cells has also been shown to be influencing the neuronal morphology (Ballas et al., 2009). In differentiating neural stem cells, both neurons and glia are present simultaneously, thus any altered MeCP2 expression in glial cells could potentially influence the neuronal morphology. Moreover, MeCP2 expression is cell type-specific and the presence of a mixed cell population might complicate the analysis. Therefore, analysis of pure populations might be beneficial in the future. However, the mixed nature of populations provides much resemblance to the in vivo conditions, where there are cell-to-cell communications and interactions similar to the developing brain. Additionally, ethanol-induced changes in the levels of neural growth factors have also been linked to the impaired neuronal morphology in response to ethanol treatment (Messing et al., 1991; Roivainen et al., 1995b). The MeCP2 target gene brain derived neurotrophic factor (BDNF) is also a key player in neuronal morphology (Kim et al., 2005). Therefore, in addition to the more direct effects of ethanol on neurons and astrocytes, it is possible that MeCP2 and MeCP2-regulatory networks may presumably contribute to the altered neuronal and glial morphology observed in our study reported here.
Further investigations are required to elucidate the impact of 5mC and 5hmC at Mecp2 regulatory elements (R1–R6) which could mediate Mecp2/MeCP2 deregulation by ethanol exposure in vivo.
Conclusion
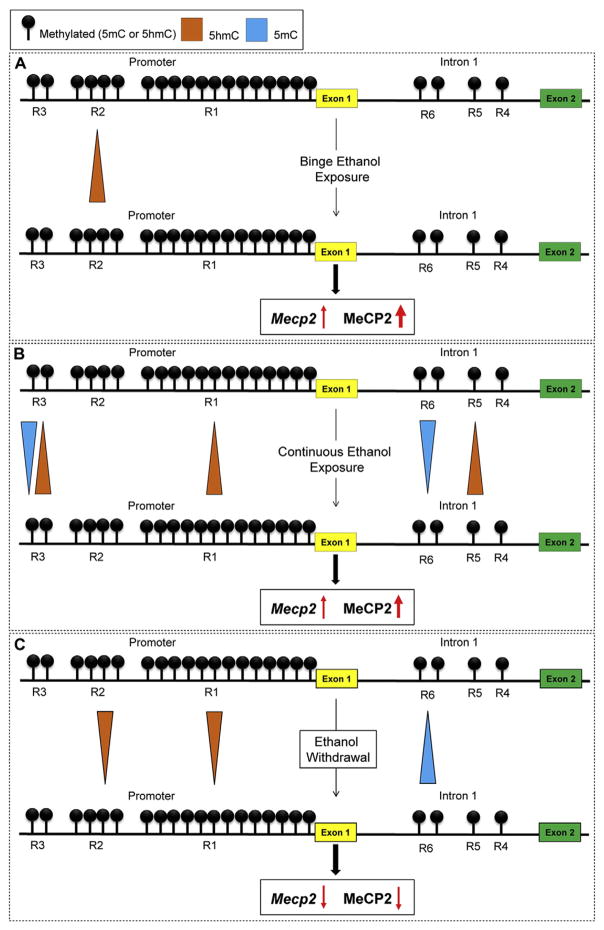
In conclusion, this current study demonstrates deregulation of Mecp2/MeCP2 by binge-like, continuous ethanol exposure and ethanol withdrawal. We report an in vitro ethanol exposure- and withdrawal-model during differentiation of brain-derived neural stem cells, which recapitulates two key features of FASD brain (neuronal morphology and glial size changes) and have minimal effects on NSC cell fate commitments. We show that such altered Mecp2/MeCP2 expression is correlated with dynamic changes of 5hmC and 5mC at the Mecp2 regulatory elements. Fig. 8 illustrates a summary of the correlation between ethanol-induced changes in 5mC/5hmC enrichment at the Mecp2 regulatory elements and Mecp2/MeCP2 expression. Based on our data, the increased 5hmC enrichment at R1, R2, R3 or R5 and decreased 5mC enrichment at R3 or R6 may contribute to the induction of Mecp2/MeCP2 expression. On the other hand, decreased 5hmC at R1 or R2 and increased 5mC enrichment at R6 seem to be contributing to the downregulation of Mecp2/MeCP2 expression. Furthermore, we show how ethanol impacts global DNA methylation at both 5mC and 5hmC levels. Collectively, our studies provide novel insights into the role of epigenetic mechanisms, specifically DNA methylation, and MeCP2 in ethanol-induced neurological symptoms during neural development and ethanol teratogenesis and ultimately on Fetal Alcohol Spectrum Disorders.
Fig. 8.
Summary of the results on DNA methylation and Mecp2 expression in response to ethanol exposure and withdrawal during neural stem cells (NSC) differentiation. The three promoter regions (R1–R3) and three intron 1 regions (R4–R6) within the Mecp2 gene are shown (not to scale). A) Binge ethanol exposure: increased enrichment of 5hmC at the R2 is associated with increased Mecp2/MeCP2. B) Continuous ethanol exposure: increased enrichment of 5hmC at R1, R3 and R5 and decreased 5mC levels at R3 and R6 are associated with increased Mecp2/MeCP2. C) Ethanol withdrawal: increased enrichment of 5mC R6 and decreased 5hmC enrichment at R1 and R2 are associated with downregulated Mecp2/MeCP2.
Supplementary Material
Acknowledgments
This work was supported by funds from the Scottish Rite Charitable Foundation of Canada (SRCFC, Grant 10110), the Natural Sciences and Engineering Research Council of Canada (NSERC Discovery Grant 372405-2009), the Health Sciences Center Foundation (HSCF), the Canadian Institute of Health Research (CIHR) Catalyst Grant CEN-132383, and the Graduate Enhancement of Tri-Council Stipends (GETS). VRBL is a recipient of the Manitoba Health Research Council (MHRC) and University of Manitoba Graduate Fellowship (UMGF) awards. RMZ was a recipient of MHRC/Manitoba Institute of Child Health (MICH)/UMGF awards. JRD is a recipient of a Canada Research Chair. NESTIN monoclonal antibody developed by Susan Hockfield was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by University of Iowa, Biology Department, Iowa City, IA 52242.
Abbreviations
- 5hmC
5-hydroxymethylcytosine
- 5mC
5-methylcytosine
- bFGF
basic fibroblast growth factor
- Ct
threshold cycle values
- DAPI
4′,6-diamidino-2-phenylindole
- D
day
- DMEM/F12
Dulbecco’s Modified Eagle Medium:Nutrient Mixture F-12
- E
embryonic day
- EDTA
ethylene diaminetetraacetic acid
- FASD
Fetal Alcohol Spectrum Disorders
- FBS
fetal bovine serum
- FP
forward PCR primer
- Gapdh
glyceraldehyde 3-phosphate dehydrogenase
- GFAP
glial fibrillary acidic protein
- H
hours
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- hMeDIP
hydroxymethylated DNA immunoprecipitation
- ICC
immunocytochemistry
- MB
methylene blue
- Mbp
Myelin basic protein
- MeCP2
MethylCpG Binding Protein2
- MeDIP
methylated DNA immunoprecipitation
- NaOH
sodium hydroxide
- NGS
normal goat serum
- NSC
neural stem cells
- OLIG2
oligodendrocyte transcription factor 2
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- qRT-PCR
quantitative real time polymerase chain reaction
- R
region
- REs
regulatory elements
- rhEGF
recombinant human epidermal growth factor
- RP
reverse PCR primer
- SEM
standard error of mean
- SP
sequencing primer
- SSC
saline–sodium citrate
- TBST
Tris buffered saline with Tween
- TUB III
β Tubulin III
- WB
western blotting
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.expneurol.2015.01.006.
Authors’ contributions
Vichithra R.B. Liyanage: Conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, and final approval of manuscript.
Robby M. Zachariah: Conception and design, collection and assembly of data, and final approval of manuscript.
James R. Davie: Scientific input, manuscript revisions, and final approval of manuscript.
Mojgan Rastegar: Conception and design, contributed reagents/materials/analysis tools, data analysis and interpretation, manuscript writing, and final approval of manuscript.
Conflict of interest statement
None declared.
References
- Adachi J, Mizoi Y, Fukunaga T, Ogawa Y, Ueno Y, Imamichi H. Degrees of alcohol intoxication in 117 hospitalized cases. J Stud Alcohol. 1991;52:448–453. doi: 10.15288/jsa.1991.52.448. [DOI] [PubMed] [Google Scholar]
- Ballas N, Lioy DT, Grunseich C, Mandel G. Non-cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nat Neurosci. 2009;12:311–317. doi: 10.1038/nn.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber BA, Rastegar M. Epigenetic control of Hox genes during neurogenesis, development, and disease. Ann Anat. 2010;192:261–274. doi: 10.1016/j.aanat.2010.07.009. [DOI] [PubMed] [Google Scholar]
- Barber BA, Liyanage VR, Zachariah RM, Olson CO, Bailey MA, Rastegar M. Dynamic expression of MEIS1 homeoprotein in E14.5 forebrain and differentiated forebrain-derived neural stem cells. Ann Anat. 2013;195:431–440. doi: 10.1016/j.aanat.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Barker JM, Zhang Y, Wang F, Taylor JR, Zhang H. Ethanol-induced Htr3a promoter methylation changes in mouse blood and brain. Alcohol Clin Exp Res. 2013;37(Suppl 1):E101–E107. doi: 10.1111/j.1530-0277.2012.01906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekdash RA, Zhang C, Sarkar DK. Gestational choline supplementation normalized fetal alcohol-induced alterations in histone modifications, DNA methylation, and proopiomelanocortin (POMC) gene expression in beta-endorphin-producing POMC neurons of the hypothalamus. Alcohol Clin Exp Res. 2013;37:1133–1142. doi: 10.1111/acer.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham SM, Mudd LM, Lopez TF, Montague JR. Effects of ethanol on cultured embryonic neurons from the cerebral cortex of the rat. Alcohol. 2004;32:129–135. doi: 10.1016/j.alcohol.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Bishop S, Gahagan S, Lord C. Re-examining the core features of autism: a comparison of autism spectrum disorder and fetal alcohol spectrum disorder. J Child Psychol Psychiatry. 2007;48:1111–1121. doi: 10.1111/j.1469-7610.2007.01782.x. [DOI] [PubMed] [Google Scholar]
- Bleich S, Lenz B, Ziegenbein M, Beutler S, Frieling H, Kornhuber J, Bonsch D. Epigenetic DNA hypermethylation of the HERP gene promoter induces down-regulation of its mRNA expression in patients with alcohol dependence. Alcohol Clin Exp Res. 2006;30:587–591. doi: 10.1111/j.1530-0277.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- Bodda C, Tantra M, Mollajew R, Arunachalam JP, Laccone FA, Can K, Rosenberger A, Mironov SL, Ehrenreich H, Mannan AU. Mild overexpression of Mecp2 in mice causes a higher susceptibility toward seizures. Am J Pathol. 2013;183:195–210. doi: 10.1016/j.ajpath.2013.03.019. [DOI] [PubMed] [Google Scholar]
- Cai J, Chen Y, Cai WH, Hurlock EC, Wu H, Kernie SG, Parada LF, Lu QR. A crucial role for Olig2 in white matter astrocyte development. Development. 2007;134:1887–1899. doi: 10.1242/dev.02847. [DOI] [PubMed] [Google Scholar]
- Camarillo C, Miranda RC. Ethanol exposure during neurogenesis induces persistent effects on neural maturation: evidence from an ex vivo model of fetal cerebral cortical neuroepithelial progenitor maturation. Gene Expr. 2008;14:159–171. [PMC free article] [PubMed] [Google Scholar]
- Carlson RW, Kumar NN, Wong-Mckinstry E, Ayyagari S, Puri N, Jackson FK, Shashikumar S. Alcohol withdrawal syndrome. Crit Care Clin. 2012;28:549–585. doi: 10.1016/j.ccc.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ozturk NC, Zhou FC. DNA methylation program in developing hippocampus and its alteration by alcohol. PLoS ONE. 2013;8:e60503. doi: 10.1371/journal.pone.0060503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AL, Levenson JM, Vilaythong AP, Richman R, Armstrong DL, Noebels JL, David Sweatt J, Zoghbi HY. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet. 2004;13:2679–2689. doi: 10.1093/hmg/ddh282. [DOI] [PubMed] [Google Scholar]
- Delcuve GP, Rastegar M, Davie JR. Epigenetic control. J Cell Physiol. 2009;219:243–250. doi: 10.1002/jcp.21678. [DOI] [PubMed] [Google Scholar]
- Ezeonwuka C, Rastegar M. MeCP2-related diseases and animal models. Diseases. 2014;2:45–70. doi: 10.3390/diseases2010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Nestler EJ. MeCP2 and drug addiction. Nat Neurosci. 2010;13:1039–1041. doi: 10.1038/nn0910-1039. [DOI] [PubMed] [Google Scholar]
- Flegel K, MacDonald N, Hebert PC. Binge drinking: all too prevalent and hazardous. CMAJ. 2011;183:411. doi: 10.1503/cmaj.110029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin TB, Russig H, Weiss IC, Graff J, Linder N, Michalon A, Vizi S, Mansuy IM. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry. 2010;68:408–415. doi: 10.1016/j.biopsych.2010.05.036. [DOI] [PubMed] [Google Scholar]
- Gangisetty O, Bekdash R, Maglakelidze G, Sarkar DK. Fetal alcohol exposure alters proopiomelanocortin gene expression and hypothalamic–pituitary–adrenal axis function via increasing MeCP2 expression in the hypothalamus. PLoS ONE. 2014;9:e113228. doi: 10.1371/journal.pone.0113228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–1715. [PubMed] [Google Scholar]
- Giordano G, Guizzetti M, Dao K, Mattison HA, Costa LG. Ethanol impairs muscarinic receptor-induced neuritogenesis in rat hippocampal slices: role of astrocytes and extracellular matrix proteins. Biochem Pharmacol. 2011;82:1792–1799. doi: 10.1016/j.bcp.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, Kwak E, Barlev NA, Berger SL, Chow VT, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerri C, Pascual M, Renau-Piqueras J. Glia and fetal alcohol syndrome. Neurotoxicology. 2001;22:593–599. doi: 10.1016/s0161-813x(01)00037-7. [DOI] [PubMed] [Google Scholar]
- Guizzetti M, Costa LG. Inhibition of muscarinic receptor-stimulated glial cell proliferation by ethanol. J Neurochem. 1996;67:2236–2245. doi: 10.1046/j.1471-4159.1996.67062236.x. [DOI] [PubMed] [Google Scholar]
- Guo Y, Chen Y, Carreon S, Qiang M. Chronic intermittent ethanol exposure and its removal induce a different miRNA expression pattern in primary cortical neuronal cultures. Alcohol Clin Exp Res. 2012;36:1058–1066. doi: 10.1111/j.1530-0277.2011.01689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, Toubaji A, Argani P, Iacobuzio-Donahue C, Nelson WG, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Mikami T, Murai Y, Takano Y, Imura J. Alpha-tubulin nuclear overexpression is an indicator of poor prognosis in patients with non-Hodgkin’s lymphoma. Int J Mol Med. 2014;34:483–490. doi: 10.3892/ijmm.2014.1793. [DOI] [PubMed] [Google Scholar]
- Hicks SD, Middleton FA, Miller MW. Ethanol-induced methylation of cell cycle genes in neural stem cells. J Neurochem. 2010;114:1767–1780. doi: 10.1111/j.1471-4159.2010.06886.x. [DOI] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SJ, Shpyleva S, Melnyk S, Pavliv O, Pogribny IP. Elevated 5-hydroxymethylcytosine in the Engrailed-2 (EN-2) promoter is associated with increased gene expression and decreased MeCP2 binding in autism cerebellum. Transl Psychiatry. 2014;4:e460. doi: 10.1038/tp.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KL, Smith DW. Recognition of the fetal alcohol syndrome in early infancy. Lancet. 1973;302:999–1001. doi: 10.1016/s0140-6736(73)91092-1. [DOI] [PubMed] [Google Scholar]
- Kim JH, Sung DK, Park CW, Park HH, Park C, Jeon SH, Kang PD, Kwon OY, Lee BH. Brain-derived neurotrophic factor promotes neurite growth and survival of antennal lobe neurons in brain from the silk moth, Bombyx mori in vitro. Zool Sci. 2005;22:333–342. doi: 10.2108/zsj.22.333. [DOI] [PubMed] [Google Scholar]
- Kim P, Park JH, Choi CS, Choi I, Joo SH, Kim MK, Kim SY, Kim KC, Park SH, Kwon KJ, et al. Effects of ethanol exposure during early pregnancy in hyper-active, inattentive and impulsive behaviors and MeCP2 expression in rodent offspring. Neurochem Res. 2013;38:620–631. doi: 10.1007/s11064-012-0960-5. [DOI] [PubMed] [Google Scholar]
- Kim P, Choi CS, Park JH, Joo SH, Kim SY, Ko HM, Kim KC, Jeon SJ, Park SH, Han SH, et al. Chronic exposure to ethanol of male mice before mating produces attention deficit hyperactivity disorder-like phenotype along with epigenetic dysregulation of dopamine transporter expression in mouse offspring. J Neurosci Res. 2014;92:658–670. doi: 10.1002/jnr.23275. [DOI] [PubMed] [Google Scholar]
- Knezovich JG, Ramsay M. The effect of preconception paternal alcohol exposure on epigenetic remodeling of the h19 and rasgrf1 imprinting control regions in mouse offspring. Front Genet. 2012;3:10. doi: 10.3389/fgene.2012.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobrossy L, Rastegar M, Featherstone M. Interplay between chromatin and trans-acting factors regulating the Hoxd4 promoter during neural differentiation. J Biol Chem. 2006;281:25926–25939. doi: 10.1074/jbc.M602555200. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Bird A. The major form of MeCP2 has a novel N-terminus generated by alternative splicing. Nucleic Acids Res. 2004;32:1818–1823. doi: 10.1093/nar/gkh349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulijewicz-Nawrot M, Verkhratsky A, Chvatal A, Sykova E, Rodriguez JJ. Astrocytic cytoskeletal atrophy in the medial prefrontal cortex of a triple transgenic mouse model of Alzheimer’s disease. J Anat. 2012;221:252–262. doi: 10.1111/j.1469-7580.2012.01536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimore JL, Chapleau CA, Kudo S, Theibert A, Percy AK, Pozzo-Miller L. Bdnf overexpression in hippocampal neurons prevents dendritic atrophy caused by Rett-associated MECP2 mutations. Neurobiol Dis. 2009;34:199–211. doi: 10.1016/j.nbd.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Francke U. Identification of cis-regulatory elements for MECP2 expression. Hum Mol Genet. 2006;15:1769–1782. doi: 10.1093/hmg/ddl099. [DOI] [PubMed] [Google Scholar]
- Liyanage VR, Rastegar M. Rett syndrome and MeCP2. Neruomol Med. 2014;16:231–264. doi: 10.1007/s12017-014-8295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyanage VRB, Zachariah RM, Delcuve GP, Davie JR, Rastegar M. New developments in chromatin research: an epigenetic perspective. In: Simpson NM, Stewart VJ, editors. New Developments in Chromatin Research. Nova Science Publishers; 2012. pp. 29–58. [Google Scholar]
- Liyanage VR, Zachariah RM, Rastegar M. Decitabine alters the expression of Mecp2 isoforms via dynamic DNA methylation at the Mecp2 regulatory elements in neural stem cells. Mol Autism. 2013;4:46. doi: 10.1186/2040-2392-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyanage VR, Jarmasz JS, Murugeshan N, Del Bigio MR, Rastegar M, Davie JR. DNA modifications: function and applications in normal and disease States. Biol (Basel) 2014;3:670–723. doi: 10.3390/biology3040670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum DE, Hall PA. The location of pKi67 in the outer dense fibrillary compartment of the nucleolus points to a role in ribosome biogenesis during the cell division cycle. J Pathol. 2000;190:537–544. doi: 10.1002/(SICI)1096-9896(200004)190:5<537::AID-PATH577>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Maezawa I, Swanberg S, Harvey D, LaSalle JM, Jin LW. Rett syndrome astrocytes are abnormal and spread MeCP2 deficiency through gap junctions. J Neurosci. 2009;29:5051–5061. doi: 10.1523/JNEUROSCI.0324-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliszewska-Cyna E, Bawa D, Eubanks JH. Diminished prevalence but preserved synaptic distribution of N-methyl-D-aspartate receptor subunits in the methyl CpG binding protein 2(MeCP2)-null mouse brain. Neuroscience. 2010;168:624–632. doi: 10.1016/j.neuroscience.2010.03.065. [DOI] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing RO, Henteleff M, Park JJ. Ethanol enhances growth factor-induced neurite formation in PC12 cells. Brain Res. 1991;565:301–311. doi: 10.1016/0006-8993(91)91662-k. [DOI] [PubMed] [Google Scholar]
- Miller MW, Potempa G. Numbers of neurons and glia in mature rat somatosensory cortex: effects of prenatal exposure to ethanol. J Comp Neurol. 1990;293:92–102. doi: 10.1002/cne.902930108. [DOI] [PubMed] [Google Scholar]
- Miyake K, Nagai K. Phosphorylation of methyl-CpG binding protein 2 (MeCP2) regulates the intracellular localization during neuronal cell differentiation. Neurochem Int. 2007;50:264–270. doi: 10.1016/j.neuint.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Moykkynen T, Korpi ER. Acute effects of ethanol on glutamate receptors. Basic Clin Pharmacol Toxicol. 2012;111:4–13. doi: 10.1111/j.1742-7843.2012.00879.x. [DOI] [PubMed] [Google Scholar]
- Nagarajan RP, Hogart AR, Gwye Y, Martin MR, LaSalle JM. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics. 2006;1:e1–e11. doi: 10.4161/epi.1.4.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MV, Du F, Felice CA, Shan X, Nigam A, Mandel G, Robinson JK, Ballas N. MeCP2 is critical for maintaining mature neuronal networks and global brain anatomy during late stages of postnatal brain development and in the mature adult brain. J Neurosci. 2012;32:10021–10034. doi: 10.1523/JNEUROSCI.1316-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte C, Rastegar M, Amores A, Bouchard M, Grote D, Maas R, Kovacs EN, Postlethwait J, Rambaldi I, Rowan S, et al. Stereospecificity and PAX6 function direct Hoxd4 neural enhancer activity along the antero-posterior axis. Dev Biol. 2006;299:582–593. doi: 10.1016/j.ydbio.2006.08.061. [DOI] [PubMed] [Google Scholar]
- Olson CO, Zachariah RM, Ezeonwuka CD, Liyanage VR, Rastegar M. Brain region-specific expression of MeCP2 isoforms correlates with DNA methylation within Mecp2 regulatory elements. PLoS ONE. 2014;9:e90645. doi: 10.1371/journal.pone.0090645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olynik BM, Rastegar M. The genetic and epigenetic journey of embryonic stem cells into mature neural cells. Front Genet. 2012;3:81. doi: 10.3389/fgene.2012.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappalardo-Carter DL, Balaraman S, Sathyan P, Carter ES, Chen WJ, Miranda RC. Suppression and epigenetic regulation of MiR-9 contributes to ethanol teratology: evidence from zebrafish and murine fetal neural stem cell models. Alcohol Clin Exp Res. 2013;37:1657–1667. doi: 10.1111/acer.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins A, Lehmann C, Lawrence RC, Kelly SJ. Alcohol exposure during development: impact on the epigenome. Int J Dev Neurosci. 2013;31:391–397. doi: 10.1016/j.ijdevneu.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierog S, Chandavasu O, Wexler I. Withdrawal symptoms in infants with the fetal alcohol syndrome. J Pediatr. 1977;90:630–633. doi: 10.1016/s0022-3476(77)80387-9. [DOI] [PubMed] [Google Scholar]
- Rastegar M, Rousseau GG, Lemaigre FP. CCAAT/enhancer-binding protein-alpha is a component of the growth hormone-regulated network of liver transcription factors. Endocrinology. 2000;141:1686–1692. doi: 10.1210/endo.141.5.7478. [DOI] [PubMed] [Google Scholar]
- Rastegar M, Kobrossy L, Kovacs EN, Rambaldi I, Featherstone M. Sequential histone modifications at Hoxd4 regulatory regions distinguish anterior from posterior embryonic compartments. Mol Cell Biol. 2004;24:8090–8103. doi: 10.1128/MCB.24.18.8090-8103.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastegar M, Hotta A, Pasceri P, Makarem M, Cheung AY, Elliott S, Park KJ, Adachi M, Jones FS, Clarke ID, et al. MECP2 isoform-specific vectors with regulated expression for Rett syndrome gene therapy. PLoS ONE. 2009;4:e6810. doi: 10.1371/journal.pone.0006810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter-Funk CK, Dohrman DP. Chronic ethanol exposure increases microtubule content in PC12 cells. BMC Neurosci. 2005;6:16. doi: 10.1186/1471-2202-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repunte-Canonigo V, Chen J, Lefebvre C, Kawamura T, Kreifeldt M, Basson O, Roberts AJ, Sanna PP. MeCP2 regulates ethanol sensitivity and intake. Addict Biol. 2014;19:791–799. doi: 10.1111/adb.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley EP, McGee CL. Fetal alcohol spectrum disorders: an overview with emphasis on changes in brain and behavior. Exp Biol Med (Maywood) 2005;230:357–365. doi: 10.1177/15353702-0323006-03. [DOI] [PubMed] [Google Scholar]
- Rodriguez JJ, Terzieva S, Olabarria M, Lanza RG, Verkhratsky A. Enriched environment and physical activity reverse astrogliodegeneration in the hippocampus of AD transgenic mice. Cell Death Dis. 2013;4:e678. doi: 10.1038/cddis.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roivainen R, Hundle B, Messing RO. Ethanol enhances growth factor activation of mitogen-activated protein kinases by a protein kinase C-dependent mechanism. Proc Natl Acad Sci U S A. 1995a;92:1891–1895. doi: 10.1073/pnas.92.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roivainen R, Hundle B, Messing RO. Ethanol enhances growth factor activation of mitogen-activated protein kinases by a protein kinase C-dependent mechanism. Proc Natl Acad Sci. 1995b;92:1891–1895. doi: 10.1073/pnas.92.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson HH, Grant KA. Ethanol-induced microcephaly in neonatal rats: relation to dose. Alcohol Clin Exp Res. 1984;8:201–203. doi: 10.1111/j.1530-0277.1984.tb05839.x. [DOI] [PubMed] [Google Scholar]
- Schade A, Delyagina E, Scharfenberg D, Skorska A, Lux C, David R, Steinhoff G. Innovative strategy for microRNA delivery in human mesenchymal stem cells via magnetic nanoparticles. Int J Mol Sci. 2013;14:10710–10726. doi: 10.3390/ijms140610710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider L, d’Adda di Fagagna F. Neural stem cells exposed to BrdU lose their global DNA methylation and undergo astrocytic differentiation. Nucleic Acids Res. 2012;40:5332–5342. doi: 10.1093/nar/gks207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi T, Kondo T. Nuclear export of OLIG2 in neural stem cells is essential for ciliary neurotrophic factor-induced astrocyte differentiation. J Cell Biol. 2004;166:963–968. doi: 10.1083/jcb.200404104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Yi C, He C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat Biotechnol. 2012;30:1107–1116. doi: 10.1038/nbt.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SA, Nash K, Koren G, Rovet J. Autism characteristics in children with fetal alcohol spectrum disorders. Child Neuropsychol. 2013;19:579–587. doi: 10.1080/09297049.2012.727791. [DOI] [PubMed] [Google Scholar]
- Stuss DP, Cheema M, Ng MK, Martinez de Paz A, Williamson B, Missiaen K, Cosman JD, McPhee D, Esteller M, Hendzel M, et al. Impaired in vivo binding of MeCP2 to chromatin in the absence of its DNA methyl-binding domain. Nucleic Acids Res. 2013;41:4888–4900. doi: 10.1093/nar/gkt213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbanna S, Nagre NN, Shivakumar M, Umapathy NS, Psychoyos D, Basavarajappa BS. Ethanol induced acetylation of histone at G9a exon1 and G9a-mediated histone H3 dimethylation leads to neurodegeneration in neonatal mice. Neuroscience. 2014;258:422–432. doi: 10.1016/j.neuroscience.2013.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thambirajah AA, Ng MK, Frehlick LJ, Li A, Serpa JJ, Petrotchenko EV, Silva-Moreno B, Missiaen KK, Borchers CH, Adam Hall J, et al. MeCP2 binds to nucleosome free (linker DNA) regions and to H3K9/H3K27 methylated nucleosomes in the brain. Nucleic Acids Res. 2012;40:2884–2897. doi: 10.1093/nar/gkr1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD, Riley EP. Fetal alcohol syndrome: does alcohol withdrawal play a role? Alcohol Health Res World. 1998;22:47–53. [PMC free article] [PubMed] [Google Scholar]
- Tsujimura K, Abematsu M, Kohyama J, Namihira M, Nakashima K. Neuronal differentiation of neural precursor cells is promoted by the methyl-CpG-binding protein MeCP2. Exp Neurol. 2009;219:104–111. doi: 10.1016/j.expneurol.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Tunc-Ozcan E, Ullmann TM, Shukla PK, Redei EE. Low-dose thyroxine attenuates autism-associated adverse effects of fetal alcohol in male offspring’s social behavior and hippocampal gene expression. Alcohol Clin Exp Res. 2013;37:1986–1995. doi: 10.1111/acer.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valles S, Sancho-Tello M, Minana R, Climent E, Renau-Piqueras J, Guerri C. Glial fibrillary acidic protein expression in rat brain and in radial glia culture is delayed by prenatal ethanol exposure. J Neurochem. 1996;67:2425–2433. doi: 10.1046/j.1471-4159.1996.67062425.x. [DOI] [PubMed] [Google Scholar]
- Vangipuram SD, Lyman WD. Ethanol affects differentiation-related pathways and suppresses Wnt signaling protein expression in human neural stem cells. Alcohol Clin Exp Res. 2012;36:788–797. doi: 10.1111/j.1530-0277.2011.01682.x. [DOI] [PubMed] [Google Scholar]
- Velez-Bermudez IC, Schmidt W. The conundrum of discordant protein and mRNA expression. Are plants special? Front Plant Sci. 2014;5:619. doi: 10.3389/fpls.2014.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IT, Reyes AR, Zhou Z. Neuronal morphology in MeCP2 mouse models is intrinsically variable and depends on age, cell type, and Mecp2 mutation. Neurobiol Dis. 2013;58:3–12. doi: 10.1016/j.nbd.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Rastegar M, Gordon J, Safa AR. Beta(2)-microglobulin induces apoptosis in HL-60 human leukemia cell line and its multidrug resistant variants overex-pressing MRP1 but lacking Bax or overexpressing P-glycoprotein. Oncogene. 2001;20:7006–7020. doi: 10.1038/sj.onc.1204893. [DOI] [PubMed] [Google Scholar]
- Yokoo H, Nobusawa S, Takebayashi H, Ikenaka K, Isoda K, Kamiya M, Sasaki A, Hirato J, Nakazato Y. Anti-human Olig2 antibody as a useful immunohistochemical marker of normal oligodendrocytes and gliomas. Am J Pathol. 2004;164:1717–1725. doi: 10.1016/S0002-9440(10)63730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariah RM, Rastegar M. Linking epigenetics to human disease and Rett syndrome: the emerging novel and challenging concepts in MeCP2 research. Neural Plast. 2012;2012:415825. doi: 10.1155/2012/415825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariah RM, Olson CO, Ezeonwuka C, Rastegar M. Novel MeCP2 isoform-specific antibody reveals the endogenous MeCP2E1 expression in murine brain, primary neurons and astrocytes. PLoS ONE. 2012;7:e49763. doi: 10.1371/journal.pone.0049763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Balaraman Y, Teng M, Liu Y, Singh RP, Nephew KP. Alcohol alters DNA methylation patterns and inhibits neural stem cell differentiation. Alcohol Clin Exp Res. 2011a;35:735–746. doi: 10.1111/j.1530-0277.2010.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Chen Y, Love A. Cellular DNA methylation program during neurulation and its alteration by alcohol exposure. Birth Defects Res A Clin Mol Teratol. 2011b;91:703–715. doi: 10.1002/bdra.20820. [DOI] [PubMed] [Google Scholar]
- Zoll B, Huppke P, Wessel A, Bartels I, Laccone F. Fetal alcohol syndrome in association with Rett syndrome. Genet Couns. 2004;15:207–212. [PubMed] [Google Scholar]
- Zou J, Rabin RA, Pentney RJ. Ethanol enhances neurite outgrowth in primary cultures of rat cerebellar macroneurons. Brain Res Dev Brain Res. 1993;72:75–84. doi: 10.1016/0165-3806(93)90161-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.