Abstract
BACKGROUND:
The Personal Genome Project Canada is a comprehensive public data resource that integrates whole genome sequencing data and health information. We describe genomic variation identified in the initial recruitment cohort of 56 volunteers.
METHODS:
Volunteers were screened for eligibility and provided informed consent for open data sharing. Using blood DNA, we performed whole genome sequencing and identified all possible classes of DNA variants. A genetic counsellor explained the implication of the results to each participant.
RESULTS:
Whole genome sequencing of the first 56 participants identified 207 662 805 sequence variants and 27 494 copy number variations. We analyzed a prioritized disease-associated data set (n = 1606 variants) according to standardized guidelines, and interpreted 19 variants in 14 participants (25%) as having obvious health implications. Six of these variants (e.g., in BRCA1 or mosaic loss of an X chromosome) were pathogenic or likely pathogenic. Seven were risk factors for cancer, cardiovascular or neurobehavioural conditions. Four other variants — associated with cancer, cardiac or neurodegenerative phenotypes — remained of uncertain significance because of discrepancies among databases. We also identified a large structural chromosome aberration and a likely pathogenic mitochondrial variant. There were 172 recessive disease alleles (e.g., 5 individuals carried mutations for cystic fibrosis). Pharmacogenomics analyses revealed another 3.9 potentially relevant genotypes per individual.
INTERPRETATION:
Our analyses identified a spectrum of genetic variants with potential health impact in 25% of participants. When also considering recessive alleles and variants with potential pharmacologic relevance, all 56 participants had medically relevant findings. Although access is mostly limited to research, whole genome sequencing can provide specific and novel information with the potential of major impact for health care.
Rapid technological advances are enabling a view of human genetic variation in ever-increasing detail and at plummeting costs.1 Until recently, analysis has been targeted largely to defined genes, but pan-genomic approaches, such as microarrays, gene-panel testing and exome sequencing, have become mainstream. Now, whole genome sequencing can capture all of the genes (about 1% of the whole genome) and most of the rest of the genome in a single experiment, with the potential to recognize all types of genetic variation and thereby usurp the less comprehensive technologies (Box 1).2 Information from whole genome sequencing can already identify the molecular causes of suspected heritable conditions and cancer;2–7 however, we anticipate that genomic analysis will become a standard component of proactive health care, given its potential to identify predisposition to medically actionable conditions, explain uncharacterized disease and reveal carriers for recessive disorders and predictors of medication safety and response.8 Interpretation of sequence data remains challenging, with unknown clinical utility and predictive value among the general population.9
Box 1: Human genome variation.
The genome is the complete set of genetic material (DNA), contained in the cell’s nucleus and mitochondria. Genes are functional units that instruct the cell to produce specific proteins. They are segmented into exons (coding units) and introns (noncoding spacers), with regulatory sequences at either end and at intron/exon junctions. Noncoding DNA between genes includes various regulatory or structural elements but is largely uncharacterized. Each of 2 versions of a gene (1 maternal and 1 paternal) is called an allele. The Human Genome Project provided the initial draft reference DNA sequence (23 pairs of chromosomes encompassing about 25 000 genes) against which to compare future genome sequences. Despite much similarity, each person’s genome is unique — from variations in the DNA sequence, copy number of genes, its organization and epigenetic changes. Some variations may be inconsequential, contribute to the differences among healthy humans or provide protection against environmental challenges; others have health-related consequences. Genome interpretation involves distinguishing among these. Variant alleles may be null, missense, nonsense, splice variants, deleted, duplicated, disrupted, etc., depending on their effect on the related gene products. Their impact on characteristics of the individual (the phenotype) are described as recessive, semidominant, codominant or dominant. Some traits or diseases result from single-gene variants, with outcomes that are predictable using principles of classical Mendelian genetics. Most involve much more complex interactions among gene variations, with epigenetic and environmental influences. Risk alleles are found more often among people with a particular condition than among those without. Few alleles are deterministic; most have variable expression. Penetrance reflects the proportion of individuals with a particular underlying genetic variant who display a given trait. Mosaicism occurs when a variant arises postfertilization, so that not all cells in the individual have it. Similarly, mitochondrial genomes in each cell may not all be identical, and a variant in only a subset is called heteroplasmy. The size of genetic variants can range from 1 nucleotide pair (bp), into the thousands (kb) or millions (Mb). Canada’s Genetic Non-Discrimination Act S.C. 2017, c.3, which received royal assent on May 4, 2017, prohibits anyone from requiring individuals to undergo a genetic test or disclose the results of a genetic test.
The Personal Genome Project Canada was launched in 2007, and shares the guiding principles and open consent policy of the parent project in the United States.10 It aims to develop a public data set of fully annotated genomic information, connected with human trait information. It can provide control data for other studies, but it also aims to forecast effects of integrating DNA-derived knowledge into routine clinical practice. The project will evaluate the utility of such information, and how best to gather and apply it within Canada’s provincially administered, publicly funded health care system. Participants in this ongoing project are highly motivated to promote genomic research and explicitly forego privacy commitments. We report the data and experiences from whole genome sequencing and medical annotation of genomes of the first 56 participants in the Personal Genome Project Canada.
Methods
Study participants
Information about the Personal Genome Project Canada was posted online (www.personalgenomes.ca) and disseminated through newspaper articles, by word-of-mouth and through Medcan Health Management Inc. Registered volunteers from across Canada underwent an in-person (n = 54) or phone (n = 2) interview and entrance exam (Figure 1), to ensure that they were aware of the potential risks associated with participation and that research results should not substitute for clinical diagnostic testing. To enrol in the project, participants must be over the age of 18 and state their intention to share their genomic data publicly. Self-reported baseline trait data included birth month/year, medications, allergies, vaccines, personal medical history, ethnicity/ancestry, blood pressure, height and weight. We did not exclude individuals based on known health conditions. Blood was drawn at the Medcan clinic (n = 54) or at a community laboratory (n = 2). Participation in the project is an ongoing process, both for the participants described here and for additional volunteers.
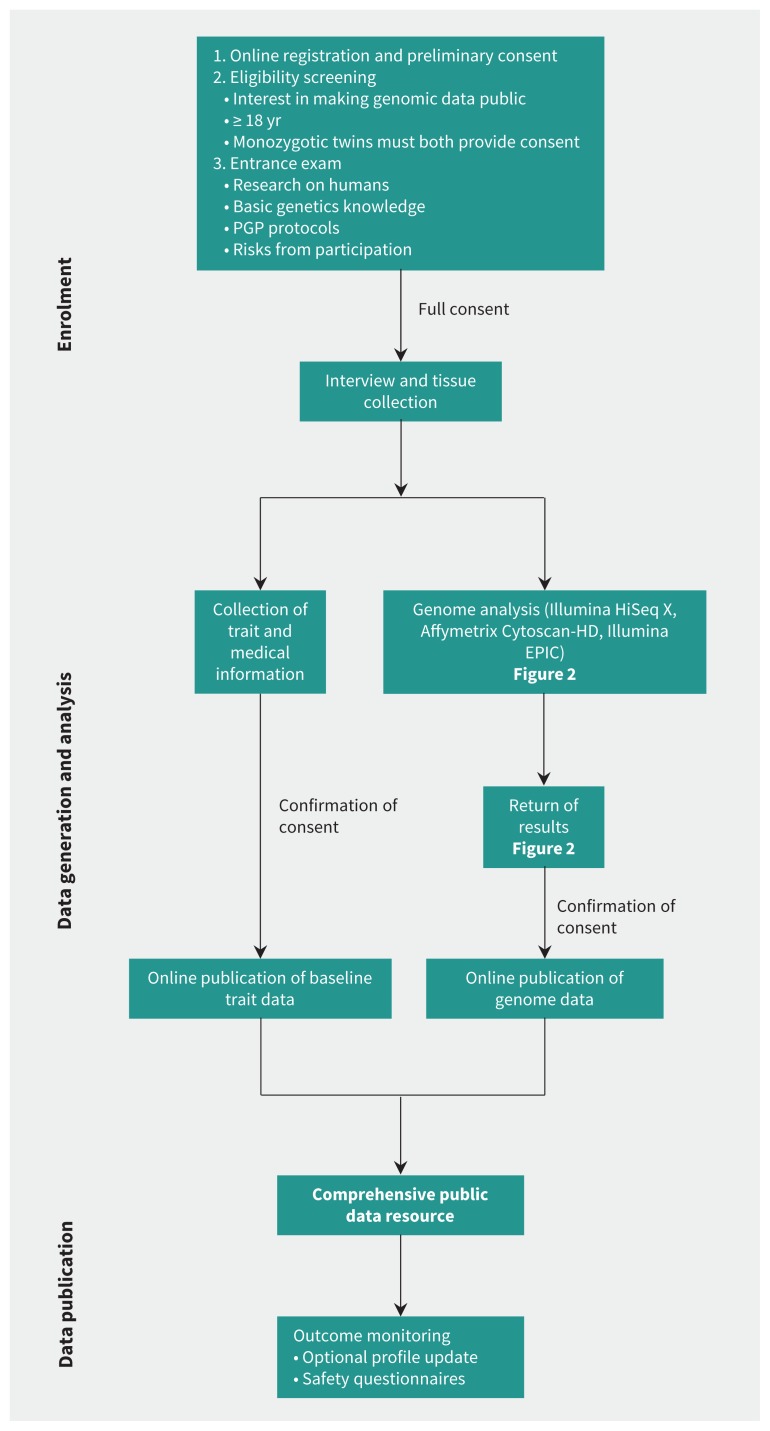
Figure 1:
Personal Genome Project Canada (PGPC) workflow. Potential participants in the project must meet eligibility criteria and undergo an entrance examination. Consent is sought and reaffirmed at stages throughout the process. Research ethics board protocols and consents, and genome data files are available at www.personalgenomes.ca. For adjunct analyses, we also assayed 55 samples using high-resolution microarrays (Affymetrix Cytoscan-HD) (Appendix 1), and we generated induced pluripotent stem cell lines for 3 individuals (Figure S4, Appendix 1).
Data generation and interpretation
We used the Illumina HiSeq X system to sequence DNA extracted from whole blood (median sequence depth of 38× across all 56 samples (Table S4, Appendix 1, available at www.cmaj.ca/lookup/suppl/doi:10.1503/cmaj.171151/-/DC1).
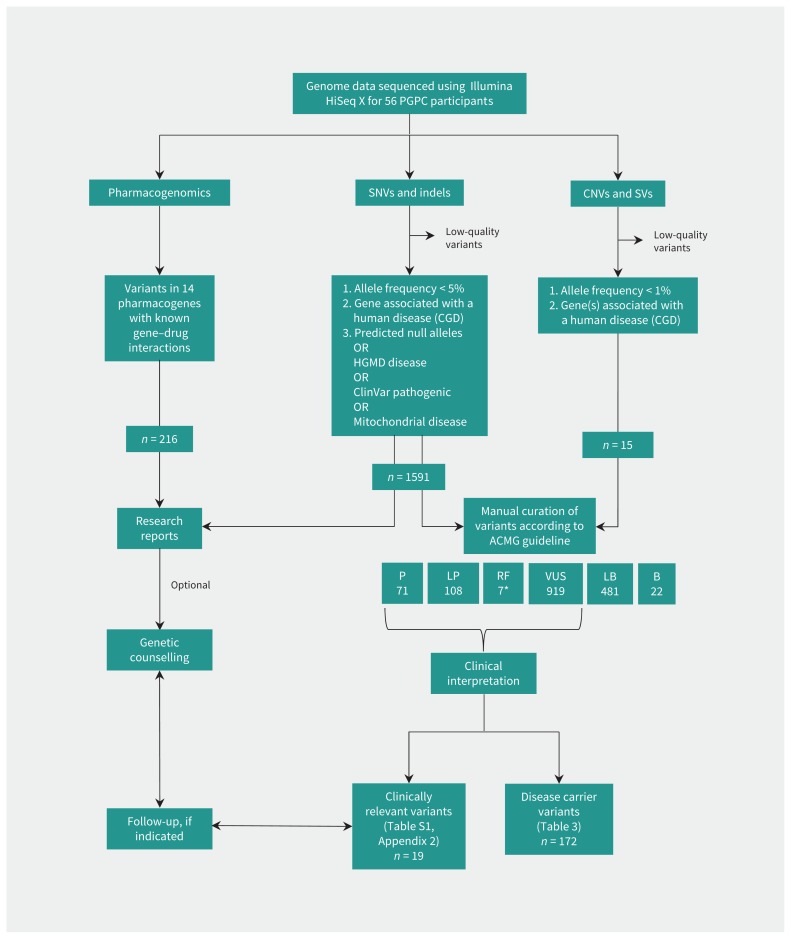
We compared each genome to the Genome Reference Consortium (https://ncbi.nlm.nih.gov/grc) human reference sequence (GRCh37/hg19). Aiming for variants with substantial health impact, we gave first consideration to single nucleotide variants (SNVs) and small insertion/deletion variants (indels) (Figure 2) that are rare (frequencies < 5%) in control cohorts (Supplementary methods, Appendix 1). Preliminary reports described alterations of genes listed in the Clinical Genomic Database (https://research.nhgri.nih.gov/CGD/) where the variant would likely eliminate gene function, and others reported to be disease associated by the Human Gene Mutation Database or ClinVar (Supplementary methods, Appendix 1).2–4,11,12 We returned these reports to participants and offered a genetic counselling session to contextualize the information.
Figure 2:
Analysis and interpretation of whole genome sequencing data. We analyzed data from WGS for variants in the nuclear and mitochondrial genome: single nucleotide variants (SNVs; alternate single bases), insertion/deletions (indels; small segments of DNA that are missing or replicated), structural variants (SVs; variations involving larger segments), including copy number variants (CNVs; deletions/losses or duplications/gains), as well as other rearrangements (inversions or translocations). We also analyzed the data for 391 variants in 14 pharmacogenes (Table S2, Appendix 3). *Two variants (in MUTYH and PCDH15) were both recessive pathogenic and dominant risk factors. Note: B = benign, CGD = Clinical Genomic Database, HGMD = Human Gene Mutation Database, P = pathogenic, LB = likely benign, LP = likely pathogenic, RF = risk factor, VUS = variant of uncertain significance, WGS = whole genome sequencing.
Data were also analyzed for copy number and other structural variations (copy number variants [CNVs]/structural variants [SVs]) by comparison to the Database of Genomic Variants,13 and for variants in mitochondrial DNA. We also extracted information on 14 pharmacogenes from the whole genome sequencing data (Appendix 1), based on guidelines by the Clinical Pharmacogenetics Implementation Consortium, Dutch Pharmacogenetics Working Group and Canadian Pharmacogenomics Network for Drug Safety, and US Food and Drug Administration label recommendations.14,15 To gain further insight into the spectrum of genomic variation, we assessed the disease-causing potential of all disease-associated variants in accordance with guidelines of the American College of Medical Genetics and Genomics. Variants were sorted into categories of standard terminology: “benign,” “likely benign,” “variant of uncertain significance” (VUS), “likely pathogenic” or “pathogenic,”16 by applying specified information from the published literature and various disease- and population-based databases. We expanded the American College of Medical Genetics and Genomics classification to include rare “risk factors,” and predicted how the putative disease-causing variants would influence the health management of the participant. Using a protocol from The Hospital for Sick Children’s Genome Clinic,17,18 a team of researchers, health care professionals and clinicians reviewed the available information and provided input on ambiguous observations, to reach consensus for interpretation. If consensus could not be obtained or the supporting evidence was not sufficient, we designated variants as being of uncertain significance. Those findings that were deemed relevant to health were discussed with participants by a genetic counsellor.
Ethics approval
The study was approved by the Research Ethics Board at The Hospital for Sick Children (REB no. 1000053640). Acknowledging that publication of genomic data is associated with both anticipated and unforeseeable risks, such as the potential for re-identification, consent is sought and reaffirmed at stages throughout the process to ensure that participants have the necessary time and information to make informed decisions about their involvement in the project (Figure 1).
Results
Personal genome sequencing and medical annotation
We report on the genome analyses of the first 56 consecutive participants (Table 1). Whole genome sequencing found an average per participant of 3.7 million high-quality SNVs and indels (1198 rare coding) and 491 CNVs (2.3 rare coding) (Table 2). We assessed all rare genomic variations (SNVs, indels, CNVs and SVs) that were either predicted to eliminate the function of genes listed in the Clinical Genomic Database or reported to be disease-associated by the Human Gene Mutation Database or ClinVar (Figure 2). These included 1606 variants: 1591 SNVs or indels, 13 deletions of 2 kb to 1.9 Mb, 1 mosaic X-chromosome loss and 1 large paracentric inversion. Most were interpreted as VUS (919/1606, 57.2%) or benign/likely benign (503/1606, 31.3%) according to the guideline from the American College of Medical Genetics and Genomics. We classified 175 SNVs/indels and 9 CNVs as pathogenic/likely pathogenic or risk factors (Table 2; Table S1, Appendix 2, available at www.cmaj.ca/lookup/suppl/doi:10.1503/cmaj.171151/-/DC1). These represented an average of 3.3 disease-associated alleles per individual (range 0–8), of which most (172; 3.1 per individual) were associated with autosomal recessive or semidominant inheritance. For example, 5 participants carried single-copy pathogenic variants in CFTR (the gene for cystic fibrosis). We found 94.6% (53/56) of participants to be carriers of at least 1 single-copy pathogenic allele.
Table 1:
Demographic characteristics of enrolment and participants in the Personal Genome Project Canada
| Characteristic | No. of participants† |
|---|---|
| Potential registered* | 1123 |
| Enrolled in study* | 63 |
| Included participants | |
| This WGS study | 56 |
| Median age at enrolment (range), yr | 51 (25–81) |
| Female:male | 25:31 |
| Common health conditions | |
| History of cancer | 7 |
| Cardiovascular disease | 7 |
| Neuropsychiatric disorder | 11 |
| Self-reported ancestry | |
| European | 51 |
| Middle Eastern | 3 |
| Canadian Indigenous | 1 |
| East Asian | 1 |
Note: WGS = whole genome sequencing. Potential participants registered online (www.personalgenomes.ca). Enrolment was a multistep process (Figure 1). Ancestry clustering data are available in Appendix 1 (Figure S1, S2).
Reference date: Sept. 1, 2017.
Unless specified otherwise.
Table 2:
Nuclear and mitochondrial results of whole genome sequencing analyses
| Type | No. of variants, mean ± SD | No. of variants, median (range) | No. of participants | Sum (1–56) | Size range (kb) |
|---|---|---|---|---|---|
| Sequence-level variants (SNVs, indels) | |||||
| All | 3 708 264 ± 366 708 | 3 851 444 (2 491 596–4 102 372) | 56 | 207 662 805 | |
| Rare* coding | 1198 ± 115 | 1188 (939–1457) | 56 | 67 101 | |
| Rare potentially disease-associated† | |||||
| All | 28.41 ± 8.45 | 29.5 (11–43) | 56 | 1591 | |
| Autosomal dominant | 6.77 ± 3.24 | 6 (1–15) | 56 | 379 | |
| Semidominant | 6.79 ± 3.35 | 7 (0–14) | 55 | 380 | |
| Autosomal recessive | 13.68 ± 4.71 | 14 (4–25) | 56 | 766 | |
| X-linked | 0.79 ± 1.06 | 1 (0–6) | 30 | 44 | |
| Mitochondrial‡ | 0.39 ± 0.68 | 0 (0–3) | 17 | 22 | |
| Rare (likely) pathogenic§ or risk factor | |||||
| All | 3.04 ± 1.68 | 3 (0–8) | 53 | 175 | |
| Autosomal dominant | 0.05 ± 0.30 | 0 (0–2) | 2 | 3 | |
| Semidominant | 0.20 ± 0.48 | 0 (0–2) | 9 | 11 | |
| Autosomal recessive | 2.77 ± 1.63 | 2.5 (0–8) | 52 | 155§§ | |
| X-linked | 0 ± 0 | 0 (0) | 0 | 0 | |
| Mitochondrial | 0.02 ± 0.13 | 0 (0–1) | 1 | 1 | |
| Risk | 0.11 ± 0.31 | 0 (0–1) | 6 | 6§§ | |
| Copy number variants and other structural variants | |||||
| All CNVs | |||||
| Deletion | 431 ± 45.90 | 438 (275–521) | 56 | 24 140 | 1–1919 (+ mosaic X loss) |
| Duplication | 60 ± 4.80 | 60 (40–73) | 56 | 3354 | 3–863 |
| Rare¶ coding** CNV | |||||
| Deletion | 1.23 ± 0.96 | 1 (0–4) | 41 | 69 | 2–1919 (+ mosaic X loss) |
| Duplication | 1.09 ± 0.93 | 1 (0–4) | 42 | 61 | 7–863 |
| Rare coding CNV, covering CGD genes | |||||
| Deletion | 0.25 ± 0.51 | 0 (0–2) | 13 | 14 | 2–1919 (+ mosaic X loss) |
| Duplication | 0.29 ± 0.45 | 0 (0–1) | 16 | 16 | 15–863 |
| Rare (likely) pathogenic§ or risk factor CNV | |||||
| Deletion | 0.16 ± 0.42 | 0 (0–2) | 8 | 9¶¶ | 4–1919 (+ mosaic X loss) |
| Duplication | 0 ± 0 | 0 (0) | 0 | 0 | 0 |
| Rare inversion | 0.02 ± 0.13 | 0 (0–1) | 1 | 1 | 8630 |
| Clinically relevant pharmacogenomics diplotype†† | |||||
| All | 3.86 ± 1.09 | 4 (1–6) | 56 | 216 | |
| Serious adverse drug reaction‡‡ | 0.23 ± 0.43 | 0 (0–1) | 13 | 13 | |
Note: ACMG = American College of Medical Genetics and Genomics, CGD = Clinical Genomic Database, CNV = copy number variant, HGMD = Human Gene Mutation Database, kb = kilobase pairs, SD = standard deviation, SNV = single nucleotide variant, WGS = whole genome sequencing.
Rare sequence level variants are defined as those with allele frequencies < 5% in control databases (Supplementary methods in Appendix 1).
Variants in CGD genes, either predicted null alleles or disease-associated in HGMD or ClinVar (Supplementary methods in Appendix 1).
All mitochondrial variants detected at > 5% heteroplasmy.
Interpretation according to the ACMG guideline.16
Rare CNVs are defined as those with < 50% overlap with all gold standard variants in the Database of Genomic Variants (http://dgv.tcag.ca/) and occurring at a frequency of < 1% among all WGS-CNVs in the unrelated parents in the Autism Speaks MSSNG cohort (http://www.mss.ng/).
Coding CNVs are defined as those overlapping coding exons.
Haplotype pairs on homologous chromosomes that are associated with risk for altered drug efficacy or adverse drug reactions.
HLA-A*3101 and HLA-B*5701 associated hypersensitivity reactions; TPMT- poor/intermediate metabolizers with myelotoxicity risk.
MUTYH variant was interpreted both as likely pathogenic (autosomal recessive) and a risk factor for colorectal cancer.
One likely pathogenic deletion (25 kb) was identified by microarray only; 3 likely pathogenic deletions (4–7 kb) were identified by WGS only.19
Based on expert consensus, we considered 19 variants in 14 of 56 participants (25.0%) to have overt health implications (Table 3). Six variants in 5 participants were pathogenic or likely pathogenic (4 SNVs, 1 CNV). In a 65-year-old man, we identified a pathogenic BRCA1 variant, which is reportable according to the recommendations by the American College of Medical Genetics and Genomics.50 The participant was of Ashkenazi Jewish descent, a population with higher frequencies of pathogenic BRCA1 and BRCA2 variants compared with the general population. His father had died of prostate cancer in his 70s, but the participant had limited knowledge of the medical history of his extended family.
Table 3:
Rare variants with potential health impact identified in the study, by participant ID no.
| Participant ID no. | Gene (accession no.) | Variant, zygosity | Associated disease, inheritance | Interpretation: evidence* | Clinical follow-up | Management implications |
|---|---|---|---|---|---|---|
| PGPC-43 | BRCA1‡ (NM_007300.3) | c.68_69delAG, p.(Glu23Valfs*17), het | Breast and ovarian cancer, AD | Pathogenic: PVS1, PS3, PS4, PP520 | FHx of prostate cancer (70–80 yr) | 1, 2, 3 |
| PGPC-16† | ELN (NM_001278914.1) | c.455–1G > A, p.?, het | Supravalvular aortic stenosis, AD | Likely pathogenic: PVS1, PM221 | No relevant PHx/FHx, normal heart CT | 1, 2, 3 |
| PGPC-25† | LMNA§ (NM_170707.3) | c.1748C > T, p.(Ser583Leu), het | Lipodystrophy, familial partial, AD/AR | Likely pathogenic: PM1, PM2, PP1, PP322 | PHx of hypercholesterolemia, hyperpigmented patch | 1, 2, 3 |
| PGPC-16† | LZTR1 (NM_006767.3) | c.774delT, p.(Phe258Leufs*93), het | Schwannomatosis, AD | Likely pathogenic: PVS1, PM223 | No relevant PHx/FHx | 1, 2, 3 |
| PGPC-40† | SLC7A9 (NM_001243036.1) | c.614dupA, p.(Asn206Glufs*3), het | Cystinuria, AD/AR | Pathogenic: PVS1, PS4-M, PP524,25 | No relevant PHx/FHx | 1, 2, 3 |
| PGPC-27† | Multiple | Seq[GRCh37] Xp22.33q28(1_155270560)x1[0.7] |
Mosaic Turner syndrome | Pathogenic26 | No obvious clinical manifestation | 1, 2 |
| PGPC-02, PGPC-27† | CHEK2 (NM_145862.2) | c.470T > C, p.(Ile157Thr), het | Cancer susceptibility, AD/AR | Risk factor: PS4, PP527,28 | 27: FHx of breast, prostate, throat cancer | 1, 2, 3 |
| PGPC-25,† PGPC-29 | F2 (NM_000506.3) | c.*97G > A (20210G > A), het | Thrombophilia, AD/AR | Risk factor: PS3, PS429 | 29: No relevant PHx/FHx | 1, 2, 3 |
| PGPC-24 | LPL (NM_000237.2) | c.953A > G, p.(Asn318Ser), het | Dyslipidemia, AD/AR | Risk factor: PS3, PS430–32 | PHx/FHx of hypercholesterolemia, FHx of CAD (60 yr) | 1, 2, 3 |
| PGPC-36 | MUTYH¶ (NM_001048171.1) | c.892–2A > G, p.?, het | Familial adenomatous polyposis, AR; colorectal cancer risk, AD | Risk factor (AD), pathogenic (AR): PVS1, PS3, PP5, BS133,34 | FHx of gastric cancer (70–80 yr) | 1, 2, 3 |
| PGPC-48 | PCDH15 (NM_001142769.1) | chr10:g. [55741501_57660800del], het (1.9 Mb) | Neuropsychiatric disease risk, AD; deafness/Usher syndrome, AR | Risk factor (AD), likely pathogenic (AR): PVS1, PM235–38 | FHx of ADHD | 1, 3 |
| Variants of uncertain significance | ||||||
| PGPC-40† | ANK2 (NM_001148.4) | c.4373A > G, p.(Glu1458Gly), het | Long QT syndrome, cardiac arrhythmia, AD | VUS: PS3, PP1, BS139–41** | No relevant PHx/FHx | 1, 2 |
| PGPC-40† | CDH1 (NM_004360.3) | c.2343A > T, p.(Glu781Asp), het | CDH1-related cancer, AD | VUS: PM242,43 | FHx of gastric cancer (75–80 yr) | 1, 2 |
| PGPC-19 | CHMP2B (NM_014043.3) | c.85A > G, p.(Ile29Val), het | Amyotrophic lateral sclerosis, Frontotemporal dementia, AD | VUS: PS3, PP5, BP444–46 | No relevant PHx/FHx | 1 |
| PGPC-14 | KCNE2 (NM_172201.1) | c.29C > T, p.(Thr10Met), het | Long QT syndrome, Atrial fibrillation, AD | VUS: PP5, BS147,48 | ND | 1, 2 |
| Uncertain significance, genetic counselling recommended | ||||||
| PGPC-32 | MT-TV | m.1659T > C, 7% heteroplasmy | Childhood neurologic disorder | Likely pathogenic††49 | ND | 1, 4 |
| PGPC-22 | Multiple | Seq[GRCh37] inv(20) (q11.23q13.12), chr20:g. [35583655_44214109inv], het (8.6 Mb) | Likely minimal risk for unbalanced aberrations in family members | Large, rare structural variant | ND | 1 |
| 53 of 56 PGPC participants | 172 recessive carrier variants (e.g., 5 pathogenic CFTR alleles) | Multiple | Potential disease risk in family members | Risk factor | ND | 1, 4 |
Note: AD = autosomal dominant, ACMG = American College of Medical Genetics and Genomics, ADHD = attention-deficit/hyperactivity disorder, AR = autosomal recessive, CAD = coronary artery disease, CT = computed tomography, FHx = family history, het = heterozygous, LEP = limited evidence for pathogenicity, Mb = megabase pairs, ND = no data, PGPC = Personal Genome Project Canada, PHx = personal history, VUS = variant of uncertain significance. Management implications: genetic counselling = 1, screen/monitor for medical complications and anticipate risk prevention = 2, identification of family members at risk = 3, Family planning = 4.
Interpretation according to the ACMG guidelines.16
Participants with 2 or more variants.
Gene is included in the ACMG list.50
Gene is included in the ACMG list but associated to a different phenotype (dilated cardiomyopathy).
Gene is included in the ACMG list when biallelic.
This variant was recently interpreted as likely pathogenic in another healthy cohort.51
Pathogenic at a higher level of heteroplasmy.
In addition, we identified a likely pathogenic ELN splice acceptor variant in a 63-year-old healthy man, which is predicted to cause in-frame skipping of a well-conserved exon implicated in intermolecular cross-linking of the tropoelastin polymer.21 Elastin dysfunction is associated with incompletely penetrant supravalvular aortic stenosis and other vascular lesions, none of which were found on examination using cardiac computed tomography. In the same participant, we detected a likely pathogenic frameshift variant in LZTR1, associated with increased risk for schwannomas.23 Penetrance for the disease is uncertain, and the participant had no personal or family history of schwannomas.
Our analysis also determined that a 70-year-old man (with self-reported hypercholesterolemia) had a heterozygous rare variant in LMNA; the same variant was reported to cause semidominant partial lipodystrophy and metabolic abnormalities, with cardiovascular risk factors particularly pronounced in obese individuals or carriers of 2 pathogenic alleles.22
We also identified a single pathogenic variant in SLC7A9 in a 49-year-old man. The same variant is associated with dominant cystinuria in the stone-forming range;24,25 stone formation is often prevented by adequate hydration and dietary modifications.
Finally, we recognized the mosaic loss of an X chromosome — in about 70% of the blood cells — in a 54-year-old woman with no obvious clinical presentation of Turner syndrome. This could create eligibility for screening for potential late-onset cardiac or endocrine manifestations.
We identified 7 risk factor variants in 5 genes (CHEK2 (×2), F2 (x2), LPL, MUTYH and PCDH15; Table 3). Among those was a CNV deletion (1.9 Mb) of exons 1–23 of PCDH15, which has been reported as a risk factor for neurobehavioural disorders,35–38 in a 44-year-old participant with a family history of attention-deficit/hyperactivity disorder. Despite an extensive literature review, we concluded that 4 other variants — in ANK2, CDH1, CHMP2B and KCNE2 — had uncertain clinical significance (Table 3). We found a missense variant in ANK2 in a 49-year-old man that had been reported previously in a large French pedigree with long QT syndrome, sinus node dysfunction and sudden death,39 and was associated with convincing functional studies in mouse cardiomyocytes. 40 This variant was recently interpreted as likely pathogenic in another healthy cohort.51 However, it is as frequent as 0.1% in some populations (http://gnomad.broadinstitute.org/), which suggests that the variant is either unrelated to disease or functions with incomplete penetrance. Additional variants, as observed in CDH1, CHMP2B and KCNE2, have been published as disease alleles, with functional support, albeit with discordance in the literature and databases.
A 65-year-old woman had a likely pathogenic variant in MT-TV in 7% of her mitochondria,49 and a 65-year-old man had a paracentric inversion (8.6 Mb) on chromosome 20q11.23q13.12. Neither seemed disease-associated in the respective participant but could be relevant to other family members (Table 3).
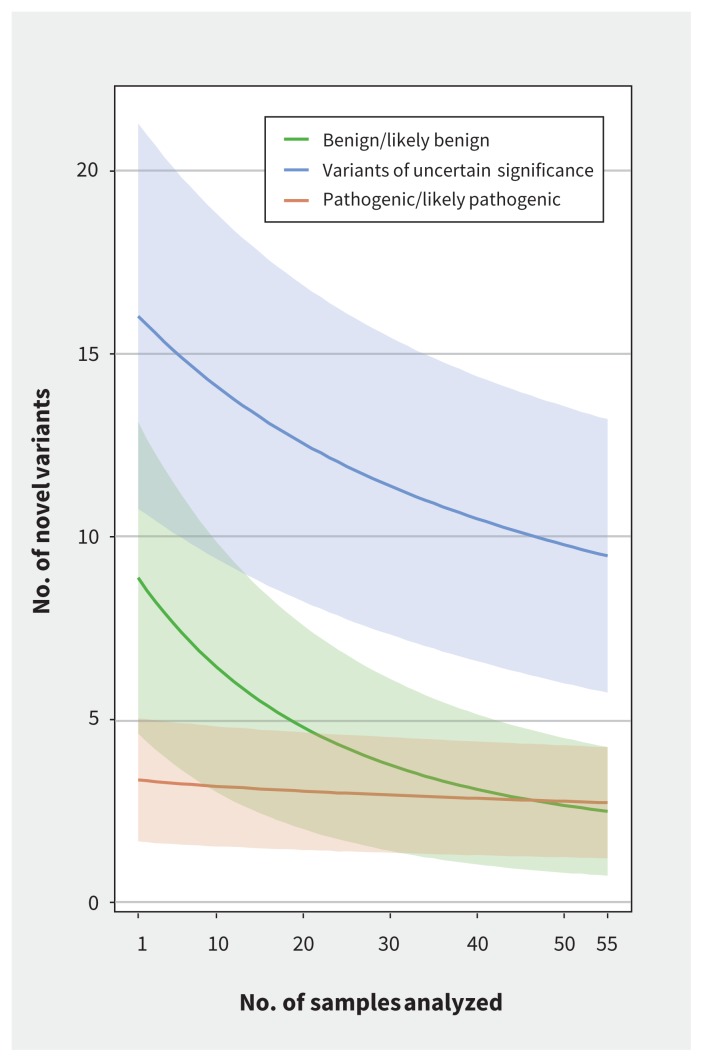
We found multitudes of other data that were potentially relevant to health. For example, there were 172 recessive alleles in 137 disease-associated genes (some have been identified in Canadian studies52,53), and 8 large CNVs(> 100 kb) of uncertain significance but involving genes (e.g., a duplication affecting 16 genes in 1 participant (PGPC-56) (Table S1, Appendix 2). Most variants were interpreted as of uncertain significance or likely benign. Recognition of novel variants declined with each sample analyzed, in particular for the number of variants classified as likely benign or of uncertain significance (Figure 3). Therefore, the burden of variant interpretation becomes lighter with each additional genome interpreted.
Figure 3:
Decline in the number of novel variants as additional samples were analyzed. The burden of new variants drops with each sample analyzed, in particular the number of variants classified as benign/likely benign and uncertain significance. For each of the 55 genetically unrelated participants (excluding the child in a mother–father–child trio) in the Personal Genome Project Canada, single nucleotide variants, insertion/deletions and copy number variants that overlapped genes from the Clinical Genomic Database were classified as benign/likely benign, variant of uncertain significance or pathogenic/likely pathogenic according to American College of Medical Genetics and Genomics interpretation guidelines.16 We then performed 1 million simulations; in each simulation, we randomly assigned the order of the samples, and the number of variants found in sample i that were not found in any of the samples 1, 2, …, i − 1 was calculated for each i, 1 ≤ i ≤ 55. The lines indicate the number of new variants for each value of i (averaged over the million simulations), whereas the shaded areas represent ±1 standard deviation from the mean, for each of the 3 variant categories: benign (green), variant of uncertain significance (blue) and pathogenic (red).
Pharmacogenomics
In addition to risks of genetic disease, we assessed the data for variants in 14 pharmacology-relevant genes (“pharmacogenes”), according to clinical guidelines.15 Participants had an average of 3.9 diplotypes (range 1–6) that were associated with risk for altered drug efficacy and/or adverse reactions (Table S2, Appendix 3, available at www.cmaj.ca/lookup/suppl/doi:10.1503/cmaj.171151/-/DC1). We found 13 participants (23.2%) at risk for severe potentially life-threatening adverse drug reactions (i.e., HLA-A*3101- and HLA-B*5701-associated hypersensitivity reactions, intermediate or low thiopurine methyltransferase activity with myelotoxicity risk). One participant (PGPC-28) had relevant findings in 6 pharmacogenes. These could compromise metabolism of drugs by CYP2D6 or CYP2C19, reduce ability to metabolize thiopurines, increase risk for simvastatin-related muscle toxicity and have implications for initial warfarin dosing. Three participants (PGPC-09, PGPC-16, PGPC-32), who were identified with CYP2C19-metabolizer status indicating favourable response to proton pump inhibitors, had self-reported use of such drugs.
Interpretation
The Personal Genome Project Canada constitutes a public resource of data from the population at large that supports evaluation of whole genome sequencing and its utility for personalized medical practice in Canada. Although most variants identified by whole genome sequencing were of uncertain significance or likely benign, most participants (53/56) carried at least 1 disease-associated allele (mean 3.3/individual [SD 1.7]). From a clinical research perspective, at this stage, we considered findings to have personal health implications for 14 participants (25%). An additional 172 pathogenic alleles were associated with autosomal recessive or semidominant inheritance patterns (mean 3.1/individual [SD 1.7]), which is close to empirical estimates of the carrier burden for recessive diseases.54 Participants also carried an average of 3.9 pharmacorelevant diplotypes associated with the metabolism of about 50 drugs. This highlights the potential of whole genome sequence data to be used pre-emptively for precision medicine, to reduce risk of adverse drug events or therapeutic failure. In general, we believe our interpretations to be conservative.
Given the variety of potentially relevant findings (Table 3), whole genome sequencing will likely become part of mainstream health care in the foreseeable future. General health care providers will be involved in interpreting and delivering genomic information in the context of personal and family histories. This requires awareness of the spectrum of potential findings, and the technical strengths and limitations of the underlying tests.
Unlike lower-resolution genomic tests, such as karyotyping, microarrays and exome sequencing, whole genome sequencing captures the entire compendium of variation in 1 experiment. Whereas most earlier studies, as well as direct-to-consumer genetic testing, have focused on SNVs and small indels,51,55–57 our study exploited the full potential uncovered by sequencing the entire genome, including copy number, other structural and mitochondrial variants, several of which would not have been detected by other methods (Table S5 in Appendix 1).
For use in the context of clinical diagnostic sequencing, the American College of Medical Genetics and Genomics compiled a list of 56 (revised to 59) genes associated with “actionable” phenotypes50,58 for which functional variants should be reported as “secondary” (incidental or unanticipated) findings. Using these criteria, our analysis identified only 1 pathogenic variant (in BRCA1), a number expected from the rate of incidental findings in larger cohorts.59 However, we found additional variants that we deemed to have health implications (in ANK2, CDH1, ELN, KCNE2, LMNA, LZTR1 and PCDH15; Table 3), and yet others that could be relevant for family planning or newborn screening (in participants PGPC-22 and PGPC-32), or for decisions about appropriate therapies or medications.
Even when using established analysis guidelines,16 variant interpretation is sometimes subjective, and requires considerable manual curation and critical review of the underlying evidence, which may be fraught with discordant interpretation60 and misclassifications.61 Further challenges arise when the a priori probability of disease is low or findings are associated with variable outcomes. All of these issues will become more relevant as the focus for sequencing shifts from diagnostic to predictive/preventive genomics. Improved control data, and even machine-learning approaches (both for variant calling and interpretation), 62 should mitigate some of the subjectivity. However, given the phenotypic spectrum associated with most variation and the influence of environment, concordance is a distant goal.
Once on file, genome sequence data can be reanalyzed as informatics tools improve and novel disease associations emerge.63 Also, new medical concerns, exposures, treatment needs or previously unnoticed familial risks may warrant reinterpretation. Along with the massively increased identification of informative variants by whole genome sequencing, come ever more uncertain findings. Larger collections of genomes, interpreted in the context of thorough and evolving personal and family histories, will help to shift the proportion of VUS into known benign or pathogenic classifications, and enable risk predictions for unbiased cohorts.
Canada’s Genetic Non-Discrimination Act was passed just as we were informing this initial cohort of results and seeking their final consent for publication. Anecdotally, prior perceived limitations to participation seemed to be somewhat relieved once protection afforded by the Act was assured.
Limitations
The 56 inaugural participants of the Personal Genome Project Canada are a small cohort of volunteers, both highly educated and idealistic regarding genomic research. These volunteers do not reflect the diverse Canadian ethnicities, but we explicitly aim to expand diversity as the sample size increases, including participation from Indigenous and recent immigrant peoples. Early personal genome sequencing cohorts were suggested to be enriched for individuals with perceived risk or subtle symptoms of genetic disease;8 although we did not enrol participants who were explicitly seeking genetic information for suspected heritable conditions, neither did we exclude participants with known health conditions (Table 1).
Because of challenges in interpretation, many potentially disease-causing variants were disregarded by our initial analysis (such as novel missense variants, synonymous and noncoding variants, variations in genes not yet associated with a phenotype or variants with allele frequencies > 5%). Certain types of pathogenic alleles are not detected reliably at present through the short-read whole genome sequencing method we used (e.g., those in regions on the Y chromosome and telomeres64–66 or trinucleotide repeat expansions). We analyzed 1 variant at a time and did not consider genetic networks.67 This approach will continue to be appropriate for those genetic variants with substantial discrete impact on phenotypes. However, the full impact of personal genomics in precision medicine will emerge as we recognize those variants with incremental effects and complex interactions, all influenced by recent human adaptation.68,69
Conclusion
In 14 of the 56 participants (25%) — a relatively mature and ostensibly healthy cohort — we identified genomic variants with potential implications for health management of the individuals but also for their families and future generations. We also added recognition, in 100% of participants, of pharmacogene variants, carrier status for recessive alleles and/or copy number variants (some involved in mental health). Coupled with growing knowledge of how such genomic variation relates to health, disease and treatment options, these findings suggest that whole genome sequencing can benefit routine health care in Canada’s future. Despite a considerable burden of uncertainty, and the possibility that false-positive findings may engender follow-up investigations51 and a “worried well” population,70 incorporation of sequence-based family history should serve to enhance personalized patient care.
Acknowledgments
The authors thank the Canadian research volunteers for their enthusiastic participation in this first phase of the project. The authors offer special thanks to George Church and colleagues associated with the Personal Genome Project (United States) for their seminal vision and help to conceptualize and develop this initiative. Jason Bobe provided early helpful discussion. The Centre for Applied Genomics provided expert analytical and technical support. The authors wish to add acknowledgement of some Canadian pioneers in this field who guided them to the personal genome: the late Michael Smith, the late Margaret Thompson, Charles Scriver, Lap-Chee Tsui and Ronald Worton.
See related article at www.cmaj.ca/lookup/doi/10.1503/cmaj.180076
Footnotes
CMAJ Podcasts: author interview at https://soundcloud.com/cmajpodcasts/171151-res
Competing interests: Stephen Scherer serves on the Scientific Advisory Committees of Population Bio and Deep Genomics. Sherilyn Bell, Jo-Anne Herbrick, Jennifer Howe, Ann Joseph-George, Barbara Kellam, Chao Lu, Jeffrey MacDonald, Christian Marshall, Thomas Nalpathamkalam, Rohan Patel, Tara Paton, Giovanna Pellecchia, Sergio Pereira, Miriam Reuter, Stephen Scherer, Lisa Strug, Wilson Sung, Bhooma Thiruvahindrapuram, Susan Walker, Zhuozhi Wang, John Wei, Joe Whitney, Richard Wintle and Ryan Yuen have received grants from Genome Canada/Ontario Genomics; Canada Foundation for Innovation; McLaughlin Centre, University of Toronto; the Government of Ontario, Canadian Institutes of Health Research (CIHR); and the The Hospital for Sick Children Foundation during the conduct of the study. James Ellis, Matthew Hildebrandt, Hin Lee, Peter Pasceri and Wei Wei have received a grant from the McLaughlin Centre, University of Toronto. Daniele Merico is an employee of Deep Genomics. Brett Trost has received a postdoctoral fellowship from CIHR. No other competing interests were declared.
This article has been peer reviewed.
Contributors: Stephen Scherer, Janet Buchanan, Miriam Reuter, Michael Szego, Hin Lee, Christian Marshall, Ronald Cohn, Jo-Anne Herbrick, Richard Wintle, Lisa Strug, Yvonne Bombard, James Ellis and Peter Ray conceived and designed the study. Michael Szego, Miriam Reuter, Hin Lee, Jill Davies, Allison Hazell, Suan Walker, Tara Paton, Sergio Pereira, Jennifer Howe, Chao Lu, Barbara Kellam, Matthew Hildebrandt, Wei Wei, Asli Romm, Peter Pasceri, Ryan Cook and Marc Fiume acquired the data. Stephen Scherer, Miriam Reuter, Janet Buchanan, Susan Walker, Christian Marshall, Bhooma Thiruvahindrapuram, Joe Whitney, Iris Cohn, Neal Sondheimer, Ryan Yuen, Brett Trost, Daniele Merico, Jeffrey MacDonald, Thomas Nalpathamkalam, Wilson Sung, Zhuozhi Wang, Rohan Patel, Giovanna Pellecchia, John Wei, Sherilyn Bell, Anne Bassett, Dimitri Stavropoulos, Sarah Bowdin, Stephen Meyn, Nasim Monfared, Rosanna Weksberg, Cheryl Shuman, S. Mohsen Hosseini, Melanie Mahtani, Ann Joseph-George, Fred Keeley, Jill Davies and Allison Hazell analyzed and interpreted the data. Miriam Reuter, Stephen Scherer, Christian Marshall, Stephen Meyn, Nasim Monfared, Allison Hazell, Ann Joseph-George (chromosomal), Neal Sondheimer (mitochondrial) and Iris Cohn (pharmacogenomics) interpreted the genomic variants. Miriam Reuter, Stephen Scherer and Janet Buchanan drafted the manuscript. All of the authors critically revised the manuscript for important intellectual content, gave final approval of the version to be published and agreed to be accountable for all aspects of the work.
Funding: This work was funded by the University of Toronto McLaughlin Centre, the Canada Foundation for Innovation, Genome Canada/Ontario Genomics, the Government of Ontario, the Canadian Institutes of Health Research, The Hospital for Sick Children Foundation and Medcan Health Management Inc. Anne Bassett is the Dalglish Chair in 22q11.2 Deletion Syndrome at the University Health Network. Stephen Scherer is funded by the GlaxoSmith-Kline–Canadian Institutes of Health Research Chair in Genome Sciences at the University of Toronto and The Hospital for Sick Children.
Data sharing: Research ethics board protocols and consents, and genome data files, are available at www.personalgenomes.ca.
References
- 1.Shendure J, Balasubramanian S, Church GM, et al. DNA sequencing at 40: past, present and future. Nature 2017;550:345–53. [DOI] [PubMed] [Google Scholar]
- 2.Lionel AC, Costain G, Monfared N, et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet Med 2017. August 3 [Epub ahead of print]. 10.1038/gim.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stavropoulos DJ, Merico D, Jobling R, et al. Whole genome sequencing expands diagnostic utility and improves clinical management in pediatric medicine. NPJ Genom Med 2016;1:pii:15012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang YH, Yuen RK, Jin X, et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet 2013; 93:249–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature 2014;511:344–7. [DOI] [PubMed] [Google Scholar]
- 6.Northcott PA, Buchhalter I, Morrissy AS, et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017;547:311–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shlien A, Campbell BB, de Borja R, et al. ; Biallelic Mismatch Repair Deficiency Consortium. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet 2015;47:257–62. [DOI] [PubMed] [Google Scholar]
- 8.Linderman MD, Nielsen DE, Green RC. Personal genome sequencing in ostensibly healthy individuals and the PeopleSeq Consortium. J Pers Med 2016;6:E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ali-Khan SE, Daar AS, Shuman C, et al. Whole genome scanning: resolving clinical diagnosis and management amidst complex data. Pediatr Res 2009;66: 357–63. [DOI] [PubMed] [Google Scholar]
- 10.Lunshof JE, Chadwick R, Vorhaus DB, et al. From genetic privacy to open consent. Nat Rev Genet 2008;9:406–11. [DOI] [PubMed] [Google Scholar]
- 11.Yuen RK, Thiruvahindrapuram B, Merico D, et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med 2015;21:185–91. [DOI] [PubMed] [Google Scholar]
- 12.Yuen RKC, Merico D, Bookman M, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci 2017;20:602–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacDonald JR, Ziman R, Yuen RK, et al. The database of genomic variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 2014;42:D986–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohn I, Paton TA, Marshall CR, et al. Genome sequencing as a platform for pharmacogenetic genotyping: a pediatric cohort study. NPJ Genom Med 2017;2:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 2012;92:414–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowdin S, Ray PN, Cohn RD, et al. The genome clinic: a multidisciplinary approach to assessing the opportunities and challenges of integrating genomic analysis into clinical care. Hum Mutat 2014;35:513–9. [DOI] [PubMed] [Google Scholar]
- 18.Bowdin S, Gilbert A, Bedoukian E, et al. Recommendations for the integration of genomics into clinical practice. Genet Med 2016;18:1075–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trost B, Walker S, Wang Z, et al. A comprehensive workflow for read depth-based identification of copy-number variation from whole-genome sequence data. Am J Hum Genet 2018;102:142–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Struewing JP, Brody LC, Erdos MR, et al. Detection of eight BRCA1 mutations in 10 breast/ovarian cancer families, including 1 family with male breast cancer. Am J Hum Genet 1995;57:1–7. [PMC free article] [PubMed] [Google Scholar]
- 21.Miao M, Reichheld SE, Muiznieks LD, et al. Single nucleotide polymorphisms and domain/splice variants modulate assembly and elastomeric properties of human elastin. Implications for tissue specificity and durability of elastic tissue. Biopolymers 2017;107:e23007. [DOI] [PubMed] [Google Scholar]
- 22.Savage DB, Soos MA, Powlson A, et al. Familial partial lipodystrophy associated with compound heterozygosity for novel mutations in the LMNA gene. Diabetologia 2004;47:753–6. [DOI] [PubMed] [Google Scholar]
- 23.Piotrowski A, Xie J, Liu YF, et al. Germline loss-of-function mutations in LZTR1 predispose to an inherited disorder of multiple schwannomas. Nat Genet 2014;46:182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leclerc D, Boutros M, Suh D, et al. SLC7A9 mutations in all three cystinuria subtypes. Kidney Int 2002;62:1550–9. [DOI] [PubMed] [Google Scholar]
- 25.Frances F, Portoles O, Corella D, et al. SLC7A9 gene variation: impact of 13 frequent mutations in the etiology of cystinuria in a Spanish Mediterranean population [article in Spanish]. Med Clin (Barc) 2006;127:161–6. [DOI] [PubMed] [Google Scholar]
- 26.Russell LM, Strike P, Browne CE, et al. X chromosome loss and ageing. Cytogenet Genome Res 2007;116:181–5. [DOI] [PubMed] [Google Scholar]
- 27.Liu C, Wang Y, Wang QS, et al. The CHEK2 I157T variant and breast cancer susceptibility: a systematic review and meta-analysis. Asian Pac J Cancer Prev 2012;13:1355–60. [DOI] [PubMed] [Google Scholar]
- 28.Han FF, Guo CL, Liu LH. The effect of CHEK2 variant I157T on cancer susceptibility: evidence from a meta-analysis. DNA Cell Biol 2013;32:329–35. [DOI] [PubMed] [Google Scholar]
- 29.Poort SR, Rosendaal FR, Reitsma PH, et al. A common genetic variation in the 3′-untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increase in venous thrombosis. Blood 1996;88:3698–703. [PubMed] [Google Scholar]
- 30.Reymer PW, Gagne E, Groenemeyer BE, et al. A lipoprotein lipase mutation (Asn291Ser) is associated with reduced HDL cholesterol levels in premature atherosclerosis. Nat Genet 1995;10:28–34. [DOI] [PubMed] [Google Scholar]
- 31.Reymer PW, Groenemeyer BE, Gagne E, et al. A frequently occurring mutation in the lipoprotein lipase gene (Asn291Ser) contributes to the expression of familial combined hyperlipidemia. Hum Mol Genet 1995;4:1543–9. [DOI] [PubMed] [Google Scholar]
- 32.Hu Y, Liu W, Huang R, et al. A systematic review and meta-analysis of the relationship between lipoprotein lipase Asn291Ser variant and diseases. J Lipid Res 2006;47:1908–14. [DOI] [PubMed] [Google Scholar]
- 33.Tao H, Shinmura K, Hanaoka T, et al. A novel splice-site variant of the base excision repair gene MYH is associated with production of an aberrant mRNA transcript encoding a truncated MYH protein not localized in the nucleus. Carcinogenesis 2004;25:1859–66. [DOI] [PubMed] [Google Scholar]
- 34.Win AK, Dowty JG, Cleary SP, et al. Risk of colorectal cancer for carriers of mutations in MUTYH, with and without a family history of cancer. Gastroenterology 2014;146:1208–11.e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prasad A, Merico D, Thiruvahindrapuram B, et al. A discovery resource of rare copy number variations in individuals with autism spectrum disorder. G3 (Bethesda) 2012;2:1665–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krumm N, O’Roak BJ, Karakoc E, et al. Transmission disequilibrium of small CNVs in simplex autism. Am J Hum Genet 2013;93:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noor A, Lionel AC, Cohen-Woods S, et al. Copy number variant study of bipolar disorder in Canadian and UK populations implicates synaptic genes. Am J Med Genet B Neuropsychiatr Genet 2014;165B:303–13. [DOI] [PubMed] [Google Scholar]
- 38.Georgieva L, Rees E, Moran JL, et al. De novo CNVs in bipolar affective disorder and schizophrenia. Hum Mol Genet 2014;23:6677–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohler PJ, Schott JJ, Gramolini AO, et al. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature 2003;421:634–9. [DOI] [PubMed] [Google Scholar]
- 40.Mohler PJ, Splawski I, Napolitano C, et al. A cardiac arrhythmia syndrome caused by loss of ankyrin-B function. Proc Natl Acad Sci U S A 2004;101:9137–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Scouarnec S, Bhasin N, Vieyres C, et al. Dysfunction in ankyrin-B-dependent ion channel and transporter targeting causes human sinus node disease. Proc Natl Acad Sci U S A 2008;105:15617–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaurah P, MacMillan A, Boyd N, et al. Founder and recurrent CDH1 mutations in families with hereditary diffuse gastric cancer. JAMA 2007;297:2360–72. [DOI] [PubMed] [Google Scholar]
- 43.Figueiredo J, Soderberg O, Simoes-Correia J, et al. The importance of E-cadherin binding partners to evaluate the pathogenicity of E-cadherin missense mutations associated to HDGC. Eur J Hum Genet 2013;21:301–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cox LE, Ferraiuolo L, Goodall EF, et al. Mutations in CHMP2B in lower motor neuron predominant amyotrophic lateral sclerosis (ALS). PLoS One 2010;5:e9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkinson N, Ince PG, Smith MO, et al. ; FReJA Consortium. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B). Neurology 2006;67:1074–7. [DOI] [PubMed] [Google Scholar]
- 46.Keogh MJ, Kurzawa-Akanbi M, Griffin H, et al. Exome sequencing in dementia with Lewy bodies. Transl Psychiatry 2016;6:e728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordon E, Panaghie G, Deng L, et al. A KCNE2 mutation in a patient with cardiac arrhythmia induced by auditory stimuli and serum electrolyte imbalance. Cardiovasc Res 2008;77:98–106. [DOI] [PubMed] [Google Scholar]
- 48.Tester DJ, Will ML, Haglund CM, et al. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm 2005;2:507–17. [DOI] [PubMed] [Google Scholar]
- 49.Blakely EL, Poulton J, Pike M, et al. Childhood neurological presentation of a novel mitochondrial tRNA(Val) gene mutation. J Neurol Sci 2004;225:99–103. [DOI] [PubMed] [Google Scholar]
- 50.Green RC, Berg JS, Grody WW, et al. ; American College of Medical Genetics and Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013;15:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vassy JL, Christensen KD, Schonman EF, et al. ; MedSeq Project. The impact of whole-genome sequencing on the primary care and outcomes of healthy adult patients: a pilot randomized trial. Ann Intern Med 2017. June 27 [Epub ahead of print]. 10.7326/M17-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan EM, Young EJ, Ianzano L, et al. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat Genet 2003;35:125–7. [DOI] [PubMed] [Google Scholar]
- 53.Chardon JW, Smith AC, Woulfe J, et al. LIMS2 mutations are associated with a novel muscular dystrophy, severe cardiomyopathy and triangular tongues. Clin Genet 2015;88:558–64. [DOI] [PubMed] [Google Scholar]
- 54.Morton NE, Crow JF, Muller HJ. An estimate of the mutational damage in man from data on consanguineous marriages. Proc Natl Acad Sci U S A 1956;42: 855–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rego S, Dagan-Rosenfeld O, Zhou W, et al. High frequency actionable pathogenic exome mutations in an average-risk cohort. bioRxiv 2017. June 18 [Epub ahead of print]. 10.1101/151225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ball MP, Thakuria JV, Zaranek AW, et al. A public resource facilitating clinical use of genomes. Proc Natl Acad Sci U S A 2012;109:11920–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzalez-Garay ML, McGuire AL, Pereira S, et al. Personalized genomic disease risk of volunteers. Proc Natl Acad Sci U S A 2013;110:16957–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med 2017;19:249–55. [DOI] [PubMed] [Google Scholar]
- 59.Amendola LM, Dorschner MO, Robertson PD, et al. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res 2015;25:305–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amendola LM, Jarvik GP, Leo MC, et al. Performance of ACMG–AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research consortium. Am J Hum Genet 2016;98:1067–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lek M, Karczewski KJ, Minikel EV, et al. ; Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leung MKK, Delong A, Alipanahi B, et al. Machine learning in genomic medicine: a review of computational problems and data sets. Proc IEEE Inst Electr Electron Eng 2016;104:176–97. [Google Scholar]
- 63.Wenger AM, Guturu H, Bernstein JA, et al. Systematic reanalysis of clinical exome data yields additional diagnoses: implications for providers. Genet Med 2017;19:209–14. [DOI] [PubMed] [Google Scholar]
- 64.Jobling MA, Tyler-Smith C. Human Y-chromosome variation in the genome-sequencing era. Nat Rev Genet 2017;18:485–97. [DOI] [PubMed] [Google Scholar]
- 65.Ledford H. AstraZeneca launches project to sequence 2 million genomes. Nature 2016;532:427. [DOI] [PubMed] [Google Scholar]
- 66.Marx V. The DNA of a nation. Nature 2015;524:503–5. [DOI] [PubMed] [Google Scholar]
- 67.Friend SH, Ideker T. Biomedical technology and the clinic of the future. Nat Biotechnol 2011;29:215. [DOI] [PubMed] [Google Scholar]
- 68.Scriver CR. The human genome project will not replace the physician. CMAJ 2004;171:1461–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Field Y, Boyle EA, Telis N, et al. Detection of human adaptation during the past 2000 years. Science 2016;354:760–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McAlister FA, Laupacis A, Armstrong PW. Finding the right balance between precision medicine and personalized care. CMAJ 2017;189:E1065–8. [DOI] [PMC free article] [PubMed] [Google Scholar]