Abstract
K- and H-Ras are the most commonly mutated genes in human tumors and are critical for conferring and maintaining the oncogenic phenotype in tumors with poor prognoses. Here, we design genetically encoded antibody-like ligands (intrabodies) that recognize active, GTP-bound K- and H-Ras. These ligands, which use the 10th domain of human fibronectin as their scaffold, are stable inside the cells and when fused with a fluorescent protein label, the constitutively active G12V mutant H-Ras. Primary selection of ligands against Ras with mRNA display resulted in an intrabody (termed RasIn1) that binds with a KD of 2.1 μM to H-Ras(G12V) (GTP), excellent state selectivity, and remarkable specificity for K- and H-Ras. RasIn1 recognizes residues in the Switch I region of Ras, similar to Raf-RBD, and competes with Raf-RBD for binding. An affinity maturation selection based on RasIn1 resulted in RasIn2, which binds with a KD of 120 nM and also retains excellent state selectivity. Both of these intrabodies colocalize with H-Ras, K-Ras, and G12V mutants inside the cells, providing new potential tools to monitor and modulate Ras-mediated signaling. Finally, RasIn1 and Rasin2 both display selectivity for the G12V mutants as compared with wild-type Ras providing a potential route for mutant selective recognition of Ras.
Keywords: mRNA display, E10FnIII, fibronectin, Ras, intrabody
Introduction
K- and H-Ras have been identified as mediators of malignant characteristics [1,2], and oncogenic mutations in these proteins are commonly observed [3,4]. Missense mutations in human Ras were some of the first oncogenes ever discovered, with two groups reporting that the G12V mutation in H-Ras transforms cell lines [5,6]. Ras overactivation is associated with 71% of pancreatic cancers, 35% of colon cancers, and 19% of lung cancers [4]. Thus, molecules that could regulate oncogenic Ras function would be of immense value as targeted therapeutics. However, despite decades of extensive research, there are currently no therapies directly targeting Ras. As a result, Ras continues to be considered as an “undruggable target” [7].
Recent attempts to inhibit Ras signaling have focused on blocking the Switch regions of Ras to prevent Ras from interacting with downstream effectors. Antibodies [8], compounds that covalently modify mutant Ras [9], and cyclic peptides [10,11] that bind to Ras have been generated, and many of these molecules can disrupt Ras signaling. However, while some these molecules are able to modulate Ras activity under certain conditions, each has drawbacks that limit their potential use, including a lack of state specificity, off-target effects, large size, and/or an inability to be effective inside the cells.
We have previously used mRNA display to target G-proteins in a state-specific fashion [12–14]. We have also developed mRNA display methods for generating fibronectins with high affinity to specific targets [15,16]. The fibronectin scaffold (10FnIII) is based on the 10th fibronectin type III domain of human fibronectin [17] and has an immunoglobulin-like fold with loops that are structurally similar to antibody CDRH1 and CDRH3 regions. The 10FnIII scaffold is small (10 kDa), lacks disulfides, can be expressed in Escherichia coli, and is an alternative to antibodies for generating affinity reagents and intrabodies [16–23]. In addition, we evolved a version of 10FnIII, termed e10FnIII, which improves solubility and expression in E. coli and in the reticulocyte lysate translation system [15,24].
Here, we describe the generation of two “RasIns”, e10FnIII-based and disulfide-free intrabodies against activated (GTP-bound) Ras proteins. These fibronectins are selective for the active state of Ras and are specific for Ras over homologous proteins. At the same time, these fibronectins bind both H- and K-Ras-GTP selectively for mutants. Furthermore, Rasin1 and Rasin2 are functional when expressed inside the cells, and RasIn2 and Raf-kinase RBD domain (Raf-RBD) (the canonical binding partner of Ras [25]) bind Ras-GTP with comparable affinity.
Results
An mRNA display selection against K-Ras(G12V)-GTP
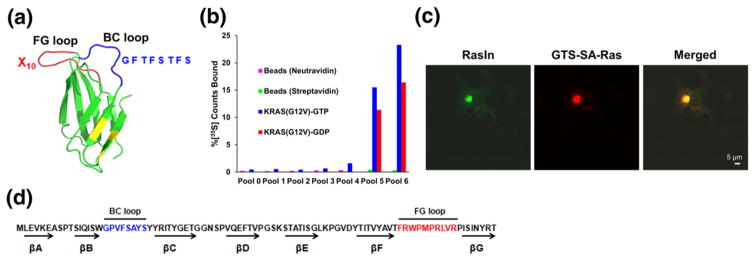
A major goal of this work was to develop state-specific Ras ligands. In order to enhance the targeting of the active state of Ras, our fibronectin library was constructed with a mutagenized sequence (CDRH1) derived from the known K-Ras binder, iDab#6 [26], in the BC loop and a totally random FG loop (Fig. 1a). The initial diversity of the library was ~1012 independent sequences (~5 copies each). mRNA display selection was performed against human K-Ras(G12V) that had been exchanged with GTPγS to create bias for binders to the active state of the target (Fig. 1b). Target specific binding was first detected in round 4 (Fig. 1b). To increase state-selective binding for active Ras, we added a 15-fold molar excess of K-Ras(G12V)-GDP in solution that would compete and remove molecules that were not specific for active Ras during the affinity enrichment step in rounds 5 and 6. The resulting pool 6 has 23% binding to K-Ras(G12V)-GTP, 16% binding to K-Ras(G12V)-GDP, and no detectable binding to the selection matrix (neutravidin-agarose or streptavidin–agarose beads; Fig. 1b). Thus, pool 6 shows a measurable preference for the GTP-bound form of K-Ras(G12V). We then sequenced pool 6 using high-throughput sequencing techniques and ranked the selected sequences by their abundance to determine the sequences that were enriched during selection (data not shown).
Fig. 1.
An mRNA display selection against K-Ras(G12V)-GTPγS using the e10FnIII scaffold. (a) e10FnIII scaffold and library design (PDB ID:1FNF). The BC loop is shown in blue, while the FG loop is shown in red. Backbone mutations relative to wild-type 10FnIII are shown in yellow [15,24]. In order to direct the library to recognize active Ras, we constructed variants of e10FnIII where the BC loop was a doped sequence from a previous Ras ligand (iDab#6) [8]. BC loop doping was ~40% wild-type at each residue. The FG loop was a naïve random sequence. (b) In vitro selection for K-Ras(G12V)-GTPγS. Pool binding was measured for matrix without target (neutravidin or streptavidin agarose) or for immobilized K-Ras exchanged with GTPγS or GDP. No binding is observed in matrix without target, while preferential binding to K-Ras(GTPγS) over K-Ras(GDP) is observed in Round 6. (c) Individual clones were screened for function inside the cells using a cellular colocalization assay [21]. Scale bar represents 5 μm. (d) Screening resulted in the identification of a Ras-specific clone termed RasIn1.
Isolation of RasIn1 via a cell-based screen
Ten high abundance sequences were tested for function using a cell-based screen we had previously developed to monitor in vivo protein binding and colocalization [21]. In this assay, the target protein is localized to the Golgi as a fusion protein bearing (1) a Golgi-targeting sequence (GTS), (2) streptavidin (SA), and (3) the target [here, H-Ras(G12V)]. We chose H-Ras for this screen to find those molecules that were Ras specific and would be able to bind to both K- and H-Ras. We also utilized the constitutively active H-Ras(G12V) mutant in order to find state-specific Ras binders that would not be downregulated by endogenous RasGAP that might be present in the COS cells (CV-1 in Origin, and carrying SV40 genetic material). Each candidate was screened as an eGFP fusion that was co-transfected with the target. Candidates were scored for colocalization with the target by immunocytochemistry. The best-performing candidate targeting H-Ras(G12V) was termed RasIn1 (Fig. 1c and d). Images of the COS cells demonstrate that eGFP-labeled RasIn1 (green) and Golgi-targeted SA-H-Ras(G12V) (visualized with rhodamine-biotin; red) accurately colocalize (merged, yellow) with little excess staining elsewhere in the cell. This result indicates that RasIn1 (1) expresses stably and functionally in mammalian cells (COS cells) and (2) has low background and little, nonspecific localization in the rest of the cell. Importantly, this low background binding demonstrates that RasIn1 accurately recognizes H-Ras in the complex environment inside the cytosol.
RasIn1 is selective for the GTP-bound state, competes with Raf-RBD, and is sensitive to mutation of Switch I
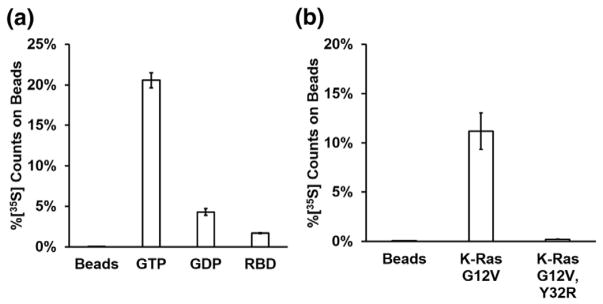
RasIn1 shows efficient pull-down with the active (GTP-bound) form of K-Ras and less pull-down with K-Ras-GDP or beads that lack target (Fig. 2a). To determine where RasIn1 binds K-Ras, we performed competition binding experiments with a natural Ras binding partner, the Raf-RBD. Coincubation of RasIn1 with a molar excess of Raf-RBD decreases the binding 12-fold (Fig. 2a), consistent with the two proteins competing for the same binding interface on Ras.
Fig. 2.
Assessing the binding characteristics of RasIn1 in vitro. (a) RasIn1 preferentially binds active (GTP) over inactive (GDP) forms of H-Ras(G12V) (p = 0.007) and preferentially binds unblocked active H-Ras(G12V) as active H-Ras(G12V) blocked with the c-Raf-kinase RBD domain (Raf-RBD; p = 0.002). (b) Binding of RasIn1 to K-Ras is disrupted by the Y32R mutation in the Switch I region (p = 0.03). Error bars indicate the standard deviation of the mean.
Co-crystal structures of H-Ras with Raf-RBD (PDBID: 3KUD [27]) make it clear that Raf-RBD makes direct contacts with the Switch I region of Ras, proximal to the nucleotide-binding pocket. The Y32R mutation in the Switch I region of Ras has previously been shown to decrease the binding between Ras and Raf-RBD [25]. This mutation also decreases the binding of Ras with Rasin1 (Fig. 2b). Taken together, our data argue that RasIn1 binds Ras in a state-specific fashion via recognition of the Switch I region of Ras in a functionally similar fashion to Raf-RBD.
RasIn1 is specific for K- and H-Ras and discriminates between highly homologous members of the Ras superfamily
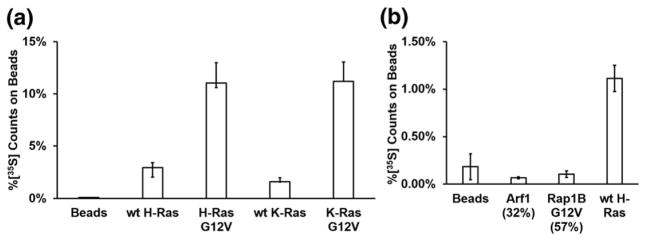
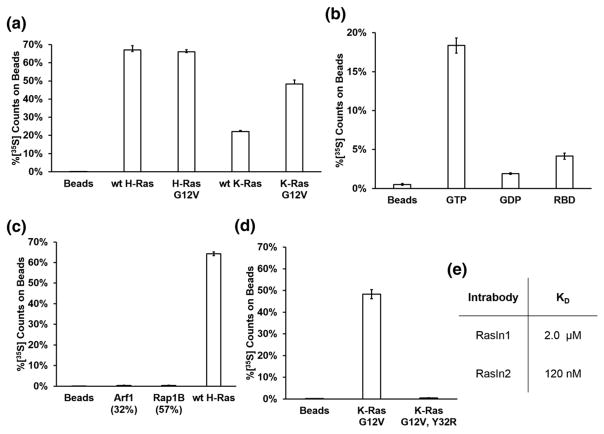
K- and H-Ras are members of the Ras subfamily with high sequence homology and identical Switch I sequences [28,29]. To further characterize the specificity of RasIn1, we tested RasIn1 binding to wild-type K-Ras, K-Ras(G12V), wild-type H-Ras, and H-Ras(G12V) (Fig. 3a). We observed mutant-selective binding for both K- and H-Ras (GTP). We also observe similar binding to both H- and K- Ras(G12V) and to wild-type H- and K-Ras, indicating that RasIn1 is not selective for a particular isoform of Ras.
Fig. 3.
Binding of RasIn1 to different Ras homologs. (a) Radiolabeled RasIn1 shows excellent binding to different versions of Ras (wild-type K- or H-Ras, and G12V mutant K- or H-Ras), showing that the recognition of the active Ras state is robust. (b) Binding specificity of RasIn1 against homologous members of the Ras superfamily. RasIn1 binds specifically to active K-Ras, but less to homologous Ras family members Rap1B(G12V) (p = 0.01) or Arf1 (0.02) (percentage of sequence identity to the K-Ras G domain is shown in parentheses). Error bars indicate the standard deviation of the mean.
The Ras superfamily is composed of five subfamilies with related sequence and structure [28,29]. To demonstrate that RasIn1 specifically recognizes Ras, we tested RasIn1 against two other members of the Ras superfamily using a radioactive in vitro pull-down assay (Fig. 3b). RasIn1 shows little to no binding to Rap1B(G12V)-GTP and Arf1-GTP. The ability to discriminate between Ras and Rap1B(G12V) bears comment, as these proteins are 57% sequence identical and the Ras Switch I region (YPDTIED) differs by switching 2 aa from the Rap1B Switch I region (YDPTIED). The Ras Switch I sequence is also similar to the Switch I sequence of Arf1 (TIPTIGF). RasIn1 is thus mutant specific (G12V versus wild-type) and state specific (GTP versus GDP) and differentiates between close Ras homologs.
RasIn1 colocalizes with active Ras in vivo
Our in vitro data indicate that RasIn1 binds to both wild-type and mutant activated Ras proteins (Fig. 3a). Using the COS cell assay (Fig. 1c), we tested if RasIn1 could also recognize wild-type H-Ras and H-Ras(G12V) inside the cells (Fig. 4). Colocalization of RasIn1 is not seen when Golgi-bound target is not expressed (Fig. 4a–c).
Fig. 4.
Colocalization of RasIn1 (green) with various Ras forms (red) in COS-7 cells. RasIn1-EGFP was co-transfected with Golgi-targeting sequence-streptavidin (GTS-SA) (a–c), GTS-SA-wild-type H-Ras (d–f), or GTS-SA-H-Ras(G12V) (g–i). Cells were fixed and stained for EGFP (green) and streptavidin (red). Colocalization (yellow) is indicated in the merged image. Images are representative of at least 15 samples. Scale bar represents 5 μm.
While RasIn1 binds to both H-Ras-GTP and H-Ras(G12V)-GTP in vitro (Fig. 3a), we only observe colocalization in the COS cell assay with H-Ras(G12V). In the cell-based assay, wild-type H-Ras shows poor binding and colocalization, while the H-Ras(G12V) mutant shows good colocalization and binding (Fig. 4). One possible explanation for this observation is that RasIn1 has higher binding for H-Ras(G12V) than wild-type H-Ras. Alternatively, wild-type H-Ras may exist in the cell in a predominantly inactive, GDP-bound state due to downregulation by endogenous Ras-GAP. This hypothesis is consistent with the wild-type protein being a substrate for hydrolysis via endogenous Ras-GAP, whereas the mutant is not [30].
Mutational analysis of RasIn1
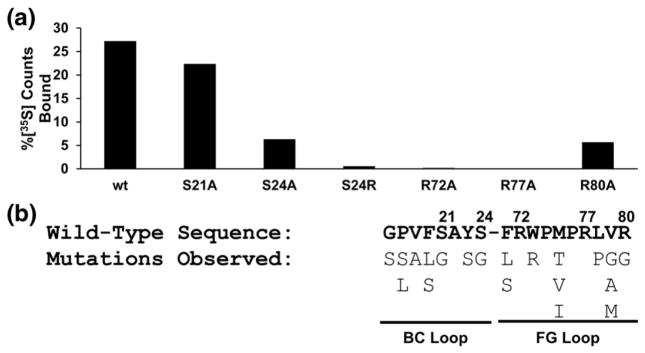
We next constructed several mutants of the RasIn1 loops to determine functionally important residues (Fig. 5a). In the BC loop, mutation of Ser-21 to Ala (S21A) results in little decrease in pull-down efficiency, suggesting that this residue is not critical for the function of the intrabody. On the other hand, the S24A mutation results in a significant decrease in binding, while the S24R mutation that increases steric bulk at this position results in complete loss of binding. These data suggest a specific role for serine at this position. In the FG loop, the R72A and R77A mutations also abolish binding completely, suggesting that these two residues are critical for the function of RasIn1. Likewise, R80A also shows a significant decrease in binding.
Fig. 5.
Mutational analysis of RasIn1 reveals functionally important residues. (a) In vitro pull-down efficiency of RasIn1 point mutants. Radiolabeled point mutants of RasIn1 were tested for binding to immobilized K-Ras-GTP. The percentage of radioactive counts bound to beads is shown. S24, R72, and R77 are critical positions for binding. (b) BC and FG loop mutations observed in the top 20 single point mutants of RasIn1 in high-throughput sequencing of pool 6 (Fig. S2). Residue numbers are shown above the sequence.
High-throughput sequencing also affords new insights into positional scanning. In the high-throughput sequencing data, we observed both the primary clone, RasIn1, and a number of sequences that differ from this clone by a single mutation (Fig. 5b). These mutations likely occurred during the course of the selection, due to inherent error rates in PCR, transcription, and reverse transcription within each mRNA display selection round. Analysis of these point mutants reveals 100% conservation at positions 22, 72, 74, 76, and 77. This point mutational analysis agrees with and complements the mutational analysis above, as both analyses identify R72 and R77 as highly important residues for binding. In addition, point mutational analysis also revealed potentially important contact sites that were not explored in the directed mutational studies, such as A22 and P74. Taken together, the positional scanning data and the high-throughput sequencing mutagenesis data argue that a significant fraction of the residues on the BC and FG loops are involved and important for binding and recognition of active Ras.
Affinity maturation results in a high affinity binder, RasIn2
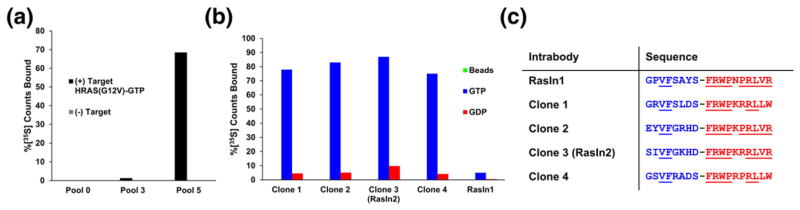
An affinity maturation selection was done to improve the affinity of RasIn1. A doped mRNA display library was constructed based on the loop sequences of RasIn1 such that there was an average of 33% wild-type amino acid at each position in the starting pool. We performed five rounds of affinity enrichment against H-Ras(G12V) in the presence of GppNHp, which, like GTPγS, allowed us to introduce bias for the active state of Ras (Fig. 6a). Additionally, during rounds 4 and 5, the binding temperature during selection was raised to 37 °C to increase selective pressure and to select for sequences with higher affinity and stability. Pool 5 correspondingly showed a pull-down efficiency of 65%, indicating the presence of high affinity molecules in the pool.
Fig. 6.
Selection of RasIn2. (a) Enrichment of Ras-binding sequences through affinity maturation. Pool 5 shows high levels of binding to active H-Ras(G12V). (b) In vitro pull-down efficiency of high abundance sequences from pool 5 in comparison with RasIn1. (c) Sequence comparison of high abundance sequences from pool 5.
Pool 5 of the maturation selection was analyzed by high-throughput sequencing. The four highest abundance clones (Clones 1–4) were selected for further characterization. All clones contain V19 and F20 in the BC loops and highly conserved FG loops, with the only variation in FG occurring at position 79 (V and L). The high sequence conservation suggests that all the clones bind to the same epitope of Ras as RasIn1. These four clones can be further grouped into two pairs (1 and 4 versus 2 and 3) based on the similarity of each clone’s BC loop (Fig. 6c). Five out of seven positions in the BC loop are identical between the two respective clones in both groups.
We then tested the binding of all four clones in a radioactive pull-down assay (Fig. 6b). All clones give high levels of binding compared to the parental RasIn1 clone (>70% versus ~5%, respectively) and also show very good selectivity for Ras-GTP versus Ras-GDP (Fig. 6). We note that RasIn1 shows lower pull-down in this experiment (~5%; Fig. 6b) that uses a relatively small amount of target, whereas in Figs. 2–5, RasIn1 gave a much higher pull-down efficiency due to the much higher target concentration on the beads that were used in those experiments. From these experiments, we chose the highest affinity sequence (Clone 3), termed it RasIn2, and further analyzed the clone.
Characterization of RasIn2
RasIn2 maintained many of the binding characteristics of its parent clone, RasIn1. RasIn2 was matured on H-Ras(G12V) and shows a preference for H-Ras and H-Ras(G12V) over wild-type K-Ras and K-Ras(G12V) (Fig. 7a). RasIn2 is state selective and preferentially binds active Ras (Fig. 7b). RasIn2 competes with Raf-RBD (Fig. 7b) and selectively recognizes K- and H-Ras over Ras family proteins Arf1 and Rap1B (Fig. 7c). RasIn2 also has reduced binding to the Y32R Switch I mutant (Fig. 7d).
Fig. 7.
Characterization of RasIn2 binding. (a) RasIn2 shows binding to different versions of active Ras (wild-type K- or H-Ras, and G12V mutant K- or H-Ras), showing that the recognition of the active Ras state is robust. (b) RasIn2 preferentially binds active (GTP) over inactive (GDP) forms of H-Ras(G12V) (p = 0.003) and preferentially binds unblocked active H-Ras(G12V) over active H-Ras(G12V) blocked with the c-Raf-kinase RBD domain (Raf-RBD; p = 0.002). (c) Binding specificity of RasIn2 against homologous members of the Ras superfamily. RasIn1 binds specifically to active wild-type H-Ras, but less to homologous Ras family members Rap1B(G12V) (p = 0.0001) or Arf1 (p = 0.0002). Percentage of sequence identity to the H-Ras G domain is shown in parentheses. (d) Binding of RasIn2 to K-Ras is disrupted by the Y32R mutation in the Switch I region (p = 0.002). (e) Dissociation constants of RasIn1 and RasIn2 for H-Ras(G12V)-GTP, determined by SPR (Figs. S3 and S4). Error bars indicate the standard deviation of the mean.
Comparison of the affinity-matured clone, RasIn2, to its parent, RasIn1, allows the identification of residues that are important for binding. Affinity maturation resulted in the conservation of 8 of the 10 residues in the FG loop, demonstrating that these residues are highly optimized for binding. These conserved residues agree with the mutational analysis from alanine scanning and from point mutational analysis from high-throughput sequencing (Fig. 5b). However, only two positions are retained in the BC loop after affinity maturation, arguing that the BC loop interactions have been optimized in the context of the new FG loop in the affinity-matured clone and potentially yielded the isoform selectivity of RasIn2 for H-Ras.
We then determined the binding affinity of the two intrabodies by surface plasmon resonance (SPR; Figs. 7e, S3, and S4). These experiments indicate that RasIn1 binds H-Ras(G12V) with a KD of 2.1 μM, while RasIn2 binds with a KD of 120 nM. Thus, affinity maturation provided a nearly 20-fold increase in affinity, an increase of about −1.7 kcal-mol−1, versus the parent clone. This affinity is notable because it is similar to the reported KD value for the Raf-RBD:wild-type H-Ras-GTP complex (KD = 80 nM) [31]. Furthermore, the fact that Raf-RBD binds H-Ras(G12V) with lower affinity than wild-type H-Ras indicates that RasIn2 may have a similar or greater affinity for mutant Ras compared to Raf-RBD [25].
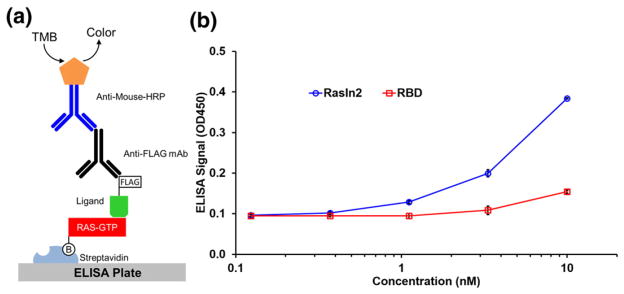
To directly compare the affinities of Raf-RBD and RasIn2 to mutant Ras, we used an ELISA-based activity assay (Fig. 8). In this assay, biotin-labeled H-Ras(G12V) was immobilized on a streptavidin-coated ELISA plate and then incubated with either FLAG-tagged RasIn2 or FLAG-tagged Raf-RBD. After the detection of the FLAG tag with anti-FLAG antibody, followed by a secondary anti-mouse antibody conjugated to horseradish peroxidase (HRP), we observed that RasIn2 gave ~10-fold higher signal as compared to RBD. These data indicate that RasIn2 has a higher affinity to active H-Ras(G12V) than Raf-RBD.
Fig. 8.
ELISA assay indicates that RasIn2 has higher affinity for H-Ras(G12V)-GppNHp than Raf1-RBD. (a) Schematic of the ELISA assay. Biotinylated H-Ras(G12V) was captured on the ELISA plate via streptavidin–biotin link. FLAG-tagged RasIn2 or Raf-RBD was then incubated with the ELISA plate, followed by an anti-FLAG antibody, and then a secondary antibody conjugated to horseradish peroxidase (HRP). The plate was then washed and incubated with tetramethylbenzidine (TMB). (b) The OD450 of the ELISA readout plotted over different concentrations of ligand (RasIn2 or Raf-RBD). RasIn2 shows ~10-fold higher specific signal level than Raf-RBD.
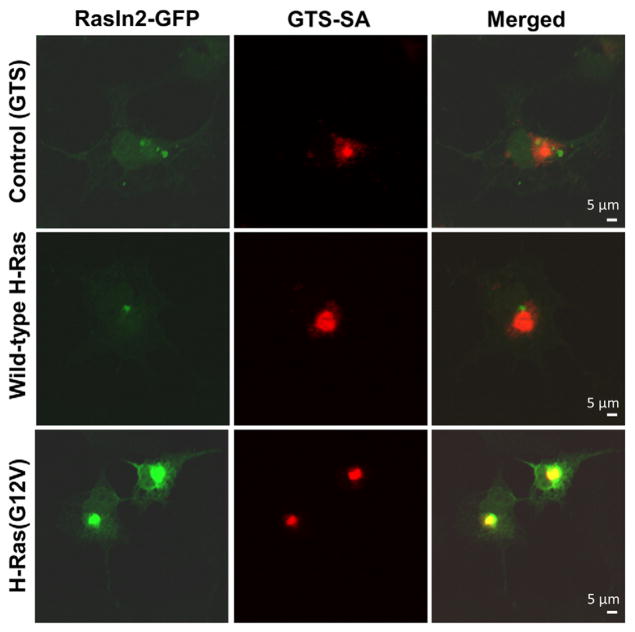
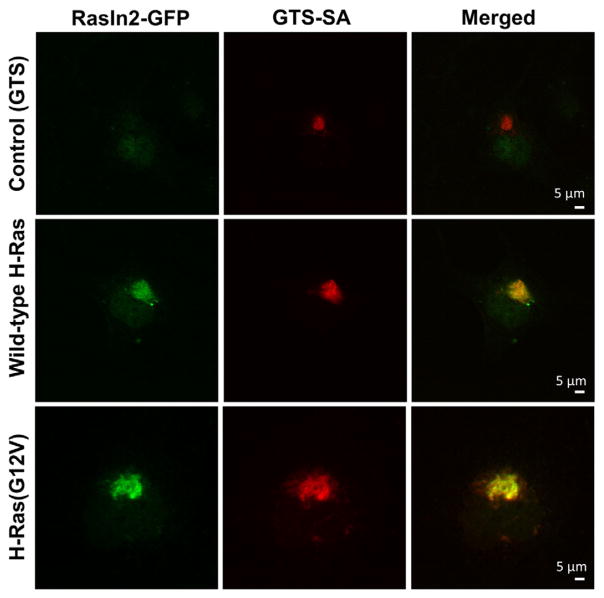
Lastly, we tested RasIn2 in the cell-based colocalization assay against H-Ras(G12V) (Fig. 9) to demonstrate its specific binding inside the cells. Similar to RasIn1, RasIn2 colocalizes well with H-Ras(G12V), arguing that the protein is stable and functional in mammalian cells and recognizes activated Ras in that context. However, unlike RasIn1, RasIn2 also colocalized with wild-type H-Ras inside the cells (Fig. 9d–f). This colocalization could be due to competition between RasIn2 and RasGAP (due to the high affinity of RasIn2), thus preserving or trapping substantial amounts of H-Ras in the GTP-bound state. Alternatively, some colocalization could be from the binding of RasIn2 to the GDP-bound state of H-Ras, possibly due to the high expression levels of transfected RasIn2 and H-Ras present in the cell. We note that colocalization with active Ras can be observed with RasIn1 inside the cells even though the KD is ~2.1 uM (Fig. 4), and in vitro RasIn2 has similar affinity for the GDP form of Ras as RasIn1 has for active H-Ras(G12V)-GTP (Fig. 6b).
Fig. 9.
RasIn2 colocalizes with various Ras forms in COS-7 cells. COS cells were co-transfected with RasIn2-GFP and GTS-SA-H-Ras(G12V) (a–c), GTS-SA-wt H-Ras (d–f), or SA-GTS (no target; g–i). Cells were fixed and stained for EGFP (green) and streptavidin (red). Colocalization (yellow) is visible in the merged image. RasIn2 colocalizes with both active (GTP) and inactive (GDP) H-Ras. Images are representative of at least 15 samples. Scale bar represents 5 μm.
Discussion
The goal of this work was to develop reagents aimed at detecting and modulating activated Ras signaling. The rationale for this effort is that the chronic signaling of K- and H-Ras, by mutation or upstream activation, occurs in more than 50% of human tumors [1]. The result of our work here is the development of two new intrabodies, RasIn1 and RasIn2, which bind to mutant and wild-type K- and H-RAS in a GTP-state-dependent fashion. Furthermore, both RasIn1 and RasIn2 show selectivity for G12V mutants.
In order for these reagents to be considered useful, they should have an array of properties, including (1) selectivity for the GTP-bound state of Ras with little or no binding to related homologs and (2) functionality in the cytosolic environment. Unlike previously generated ligands that bind to Ras, both RasIn1 and RasIn2 satisfy these criteria. RasIn1 has a KD of 2.1 μM, a reasonable outcome from a primary selection with 17 random/randomized residues. It is notable that this affinity is sufficient to provide very good colocalization with the intended target in the COS cells, even though this affinity is less than the downstream partner Raf-RBD for the active state of Ras (KD = 80 nM). RasIn1 also has excellent nucleotide, mutant, and homolog selectivity, showing little or no binding to Ras-GDP (Fig. 2a) or Rap1B-GTP (Fig. 3b). This result is remarkable because Ras and Rap1B are 57% sequence identical, and the Switch I sequences of Ras (YPDTIED) and Rap1B (YDPTIED) differ by only 2 aa (bold). Mutation and competition analysis of the RasIn1 indicates that the binding overlaps the known Ras binding site for Raf-RBD, although the precise sequence interactions must be different—the recognition sequences overall show basic character (R/K rich) and have no sequence homology [32].
Affinity maturation of RasIn1 resulted in the high affinity binder RasIn2, with a KD of 120 nM and a 20-fold improvement gained through sequence optimization of the recognition site. Interestingly, there are 8 conserved positions in the FG loop, even though the loop sequences are mutagenized at >65% per position, such that the 10-residue loop is expected to have between 3 and 4 conserved positions. RasIn2 gives substantially improved pull-down with Ras compared to RasIn1 (Fig. 6b), competes with Raf-RBD (Fig. 7b), shows broad recognition biochemically of K-Ras(G12V)-GTP, wild-type K-Ras-GTP, H-Ras(G12V)-GTP, and wild-type H-Ras-GTP (Fig. 7a), outperforms RBD in an ELISA assay (Fig. 8), and also shows excellent colocalization with H-Ras(G12V) and wild-type H-Ras inside COS cells (Fig. 9). The fact that RasIn1 and RasIn2 show similar intensity in the COS cell assay is likely a result of the high transient expression levels of both the binder (RasIn) and the target (Ras) in the COS cells.
Overall, the two proteins demonstrate that it is possible to use mRNA display to engineer individual, Ras-directed reagents with broad specificity for both the mutant and active state of K- and H-Ras. Furthermore, the selectivity of these binders argues that it should be possible to develop similar reagents for other G-proteins and study other G-protein-mediated pathways.
Left open for future work are the cellular effects that can be probed using these binders. The mutant selectivity of RasIn1 and the high affinity of RasIn2 imply that these proteins may be able to modulate or block downstream signaling via Raf kinase activation, a long-sought goal in cancer therapeutics. RasIn1 or RasIn2 could also be developed into active Ras biosensors for in vitro or in vivo studies, histology, or cancer diagnostics. Their small size, high affinity, lack of disulfide bonds, state selectivity, Ras specificity, and ability to be transfected into cells lend RasIn1 and RasIn2 to a wide range of possible future uses that often restrict the application of antibodies or non-genetically encoded molecules.
Materials and Methods
Protein expression and purification
pCDNA 3.1 vector (Invitrogen)-encoding Ras family genes were obtained from Missouri S&T cDNA Resource Center. K-Ras(G12V) and H-Ras (G12V) (1-166) with a 5′ avitag and His tag were cloned into pET16b or pET24a vector. Proteins were expressed in E. coli BL21 (DE3) containing plasmid pBirAcm (Avidity) that overexpresses the birA biotin ligase for in vivo biotinylation. Cells were grown at 30 °C for 4–5 h after induction with 1 mM IPTG at an OD of 0.4, and cells were pelleted and frozen at −80 °C. Cells were lysed with French Press or BPER (Thermo Scientific), and the lysate was cleared by centrifugation and applied to a Ni(II)-NTA column (Qiagen) pre-equilibrated with binding buffer [50 mM Tris–HCl or Hepes (pH 7.5), 100mM NaCl, 0.05% (vol/vol) Tween-20, 1 mM MgCl2, 10 μM GDP, 1 mM DTT, and 20 mM imidazole]. The column was washed with wash buffer (binding buffer with 500 mM NaCl and 40 mM imidazole), and proteins were eluted with a linear gradient of wash buffer and elution buffer (binding buffer with 20 mM NaCl and 400 mM imidazole). The molecular weight and purity of the protein were confirmed by denaturing SDS-PAGE. Purified protein was buffer exchanged into selection buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.05% (vol/vol) Tween-20, 5 mM MgCl2, and 1 mM DTT] using Amicon Ultra spin columns, supplemented with 2 mM GDP and 10% (vol/vol) glycerol, and then frozen in liquid nitrogen. The activity of Ras proteins was confirmed by an in vitro pull-down assay with Raf-RBD
DNA-encoding c-Raf-RBD (c-Raf residues 51–131) was obtained from Integrated DNA Technologies. c-Raf-RBD, RasIn1, and RasIn2 were cloned into the pAO9 vector, which fuses each protein to a C-terminal maltose binding protein [15]. Proteins were expressed in E. coli BL21 (DE3) and purified on a Ni(II)-NTA column as described above. RasIn1 and RasIn2 were subjected to a secondary purification step on an amylose column (New England Biolabs). Proteins were stored at −80 °C in storage buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.05% (vol/vol) Tween-20, and 1 mM DTT].
Rap1B(G12V) (1-166), RhoA(1-181), and Arf1 (18-179) were cloned in pDW363 and expressed in E. coli BL21 (DE3) pBirAcm for 5 h after induction with 0.5 mM IPTG [33]. Cells were pelleted and frozen at −80 °C. Pellets were lysed with BPER, lysate was cleared by centrifugation, and the biotinylated proteins immobilized on neutravidin agarose beads (Thermo Scientific). Rap1B(G12V) functionality was validated with a radioactive pull-down of its canonical binding partner, Ral-GDS (Fig. S1).
K-Ras(G12V, Y32R) mutants were generated by PCR-based site-directed mutagenesis using pDW363 K-Ras(G12V) as template. Proteins were expressed and purified as described for Ras family member proteins.
Nucleotide exchange
For data in Figs. 1, 5, and 6, Ras proteins were immobilized on neutravidin agarose (Thermo Fisher) or Dynabeads® M-280 streptavidin magnetic beads (Life Technologies) and washed three times with exchange buffer [50 mM Tris–HCl (pH 7.5), 5 mM EDTA, 150 mM NaCl, and 1 mM nucleotide]. Beads were incubated at 30 °C for 30 min, transferred on ice, and washed three times with ice-cold selection buffer to stop exchange.
For the binding assays in Figs. 2, 3, and 7, nucleotide exchange was facilitated by incubating the Ras beads in selection buffer (20 mM Hepes, 200 mM NaCl, 200 μM nucleotide, and 0.05% TWEEN) for 3 h.
Library preparation
Libraries were built as described [16], except that the Arg-24 position in e10FnIII was randomized to create a random region totaling eight residues in the BC loop. Oligos encoding the random regions were synthesized by the Yale Keck Oligonucleotide Synthesis facility.
mRNA display selection
RasIn1
The selection protocol was carried out as described by Xiao et al. [22]. The FG loop contained 10 random residues (X10; where X = all 20 natural amino acids, encoded by the NNS codon), and the BC loop of the library was a doped sequence of the CDRH1 loop of iDab#6, an antibody with state-specific affinity for Ras, with approximately 40% frequency at each residue for the initial selection [26]. For the first round, the library was translated in a total volume of 2.5 mL, purified, and reverse transcribed. K-Ras(G12V) was immobilized on neutravidin agarose beads and exchanged with GTPγS as described above. The library was then added to immobilized K-Ras(G12V)GTPγS in selection buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.05% (vol/vol) Tween-20, 5 mM MgCl2, 1 mM DTT, and 10 μM GTPγS] at 4 °C. Following the first round, a FLAG purification step was included after translation to purify mRNA–fibronectin fusions away from unfused mRNA, and the target binding step was performed at 25 °C. At round 3, streptavidin ultralink beads (Thermo Fisher) were used to prevent the enrichment of neurtravidin agarose binding sequences. At round 5, a 15-fold molar excess of K-Ras(G12V)-GDP was added to the selection buffer to enhance selective binding for active Ras.
Affinity maturation
A new library was synthesized containing BC and FG loops that were doped on the RasIn1 sequence (BC = GPVFSAYS and FG = FRWPMPRLVR) for the affinity maturation selection. The frequency of wild-type RasIn1 in the starting pool was estimated to be approximately 1.8 in 108 sequences. For the first round of selection, 200 μL of library was translated. H-Ras(G12V) was immobilized on 40 μL of Dynabeads® M-280 streptavidin magnetic beads and was exchanged with GppNHp. Immobilized H-Ras(G12V) and purified library were incubated as above. Binding for the first round was performed at 4 °C, rounds 2 and 3 were performed at 25 °C, and rounds 4 and 5 were performed at 37 °C to increase binding stringency.
Radioactive in vitro pull-down assays
Ras proteins were immobilized and exchanged with nucleotide as described above. Individual ligands or selection pools were radiolabeled by translation in vitro in rabbit reticulocyte lysate in the presence of [35S] Met. After oligo dT or FLAG purification, radiolabeled ligands were added to Ras beads, incubated for the indicated duration, and washed three times with selection buffer. The percentage of total radioactive counts remaining on the beads was determined by scintillation counting.
Cell-based colocalization assay and immunocytochemistry
The cell-based colocalization assay and immunocytochemistry were done as previously described [21]. Briefly, COS-7 cells (ATCC) were grown to a confluency of ~40–60% on poly-D-lysine-coated 22 × 22 mm glass coverslips in Dulbecco’s modified eagle’s medium (ATCC) supplemented with 10% fetal bovine serum in 5% CO2. pGW mammalian expression plasmids assembled with Golgi-targeted Ras-Streptavidin or 10FnIII-EGFP fusion genes inserted into the MCS were then co-transfected into cells using Effectene transfection reagent with transfection rates of around 10–30% (Qiagen). Following 24 h of plasmid expression, COS cells were washed once with Phosphate Buffered Saline (PBS) [137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH = 7.4], fixed with 4% paraformaldehyde for 5 min, washed three times with PBS, blocked for 30 min in blocking buffer [1% bovine serum albumin, 5% normal goat serum, 0.1% (vol/vol) Triton X-100 in PBS], and stained for 1 h with chicken anti-GFP antibody (Aves), diluted 1:1000 in blocking buffer. After incubation with primary antibody, cells were washed three times with PBS and stained for 30 min with anti-chicken secondary antibody conjugated to Alexa Fluor 488 (Invitrogen), 1:1000, and Biotin-Rhodamine conjugate (VWR), 1:000, in blocking buffer. Cells were then washed three times with PBS and mounted on 75 × 25 mm glass microscope slides in Fluoromount-G (Electron Microscopy Sciences) for imaging. All steps were carried out at room temperature. Images of cells were taken with a 60× water objective at 1.0 zoom on an Olympus IX81 inverted microscope equipped with a GFP/mCherry filter cube (Chroma Technology), an X-cite exacte mercury lamp (Excelitas Technologies), an EM-CCD digital camera (Hamamatsu), and Metamorph software (Molecular Devices). Cells were scored for the colocalization of Rhodamine and Alexa Fluor 488 fluorescence. In each sample, at least 15 cells were imaged, and colocalization was observed either every time or never.
High-throughput sequencing
Selection pools were sequenced at the USC Epigenome Center and the USC Genome & Cytometry Core using MiSeq™ and HiSeq™ and Systems (Illumina). DNA was prepped and sequenced as previously described [34]. Data analysis was done using computer scripts developed in-house.
SPR measurements
Measurements were done in USC NanoBiophysics Core Facility on a Biacore T100 instrument (Biacore). H-Ras(G12V) was immobilized on streptavidin sensor chips at the indicated surface density. Ras protein was exchanged with GppNHp on a chip where indicated by injecting SPR exchange buffer [50 mM Tris–HCl (pH 7.5), 1 mM EDTA, and 0.5 mM GppNHp] at 35 μL/min for 10 min. A concentration series of maltose binding protein–intrabody fusion was injected at a flow rate of 100 μL/min at 25 °C in SPR run buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.01% (vol/vol) Tween-20, 5 mM MgCl2, and 10 μM GppNHp]. Data were analyzed by Biacore T100 Evaluation Software.
ELISA assay
The polystyrene plate was incubated with 50 μL of 30 nM streptavidin in 1X PBS per well overnight at 4 °C. The plate was washed with 1X Tris Buffered Saline (TBS) [50 mM Tris, 150 mM NaCl, pH = 7.5] with 0.1% (vol/vol) Tween-20, filled completely with 5% BSA (wt/ vol) in 1X PBS, incubated for 3 h, and washed. Biotin-labeled H-Ras(G12V)-GppNHp was diluted in sample buffer [1X TBS, 5 mM MgCl2, 1mM DTT, 20 μM GppNHp, 0.1% (vol/vol) Tween-20, and 1 mg/mL BSA]. We added 100 uL of 30 nM biotin-labeled H-Ras(G12V)-GppNHp to each well, and it was incubated for 90 min. After the incubation, 100 μL of exchange buffer (50 mM Tris, 5mM EDTA, 1mM DTT, 600 μM GppNHp, and 1 μM biotin) was added to each well and incubated for 30 min, and the plate was then washed in wash buffer [1X TBS, 5 mM MgCl2, 50 μM GTP, and 0.1% (vol/vol) Tween-20]. RasIn2 protein and RBD were diluted serially in sample buffer and added to the plate. The plate was incubated for 1 h and washed. We added 100 μL of 20 nM Anti-FLAG antibody (Sigma) to the plate. The plate was washed and incubated for 1 h with 100 μL/well of 1:1000 dilution of HRP-conjugated anti-mouse antibody. The plate was washed and incubated with tetramethylbenzidine substrate for approximately 5 min. The reaction was stopped with equal volume of 2 M sulfuric acid, and the OD450 was measured using a plate reader.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (R01CA170820 and R01GM083898). This work acknowledges the instrumentation provided by the USC Nanobiophysics Core Facility.
Abbreviations used
- Raf-RBD
Raf-kinase Ras Binding Domain
- SPR
surface plasmon resonance
- HRP
horseradish peroxidase
- PBS
Phosphate Buffered Saline
- TBS
Tris Buffered Saline
- COS cells
a cell line that is CV-1 in Origin, and carrys SV40 genetic material
Appendix A. Supplementary Data
Supplementary data to this article can be found online at doi:10.1016/j.jmb.2016.11.008.
Footnotes
Author contributions: M.C. performed selections and in vitro pull-down assays and wrote the draft of the manuscript. W.E.E. performed in vitro pull-down assays and assembled the manuscript. G.G.G. performed the COS cell assays. F.J.-Y. performed the ELISA assay. D.K. performed in vitro pull-down assays. T.T.T. advised M.C. and assisted with the Biacore. D.A. advised G.G.G. R.W.R. advised M.C., W.E.E., F.J.-Y., and D.K.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 2.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 5.Reddy EP, Reynolds RK, Santos E, Barbacid M. A point mutation is responsible for the acquisition of transforming properties by the T24 human bladder carcinoma oncogene. Nature. 1982;300:149–152. doi: 10.1038/300149a0. [DOI] [PubMed] [Google Scholar]
- 6.Tabin CJ, Bradley SM, Bargmann CI, Weinberg RA, Papageorge AG, Scolnick EM, Dhar R, Lowy DR, Chang EH. Mechanism of activation of a human oncogene. Nature. 1982;300:143–149. doi: 10.1038/300143a0. [DOI] [PubMed] [Google Scholar]
- 7.McCormick F. KRAS as a therapeutic target. Clin Cancer Res. 2015;21:1797–1801. doi: 10.1158/1078-0432.CCR-14-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanaka T, Lobato MN, Rabbitts TH. Single domain intracellular antibodies: a minimal fragment for direct in vivo selection of antigen-specific intrabodies. J Mol Biol. 2003;331:1109–1120. doi: 10.1016/s0022-2836(03)00836-2. [DOI] [PubMed] [Google Scholar]
- 9.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gareiss PC, Schneekloth AR, Salcius MJ, Seo SY, Crews CM. Identification and characterization of a peptidic ligand for Ras. Chembiochem. 2010;11:517–522. doi: 10.1002/cbic.200900547. [DOI] [PubMed] [Google Scholar]
- 11.Upadhyaya P, Qian Z, Selner NG, Clippinger SR, Wu Z, Briesewitz R, Pei D. Inhibition of Ras signaling by blocking Ras–effector interactions with cyclic peptides. Angew Chem. 2015;54:7602–7606. doi: 10.1002/anie.201502763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ja WW, Wiser O, Austin RJ, Jan LY, Roberts RW. Turning G proteins on and off using peptide ligands. ACS Chem Biol. 2006;1:570–574. doi: 10.1021/cb600345k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ja WW, Roberts RW. In vitro selection of state-specific peptide modulators of G protein signaling using mRNA display. Biochemistry. 2004;43:9265–9275. doi: 10.1021/bi0498398. [DOI] [PubMed] [Google Scholar]
- 14.Austin RJ, Ja WW, Roberts RW. Evolution of class-specific peptides targeting a hot spot of the G alpha s subunit. J Mol Biol. 2008;377:1406–1418. doi: 10.1016/j.jmb.2008.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olson CA, Liao HI, Sun R, Roberts RW. mRNA display selection of a high-affinity, modification-specific phospho-I kappa B alpha-binding fibronectin. ACS Chem Biol. 2008;3:480–485. doi: 10.1021/cb800069c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olson CA, Roberts RW. Design, expression, and stability of a diverse protein library based on the human fibronectin type III domain. Protein Sci. 2007;16:476–484. doi: 10.1110/ps.062498407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J Mol Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 18.Koide A, Abbatiello S, Rothgery L, Koide S. Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor. Proc Natl Acad Sci. 2002;99:1253–1258. doi: 10.1073/pnas.032665299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao HI, Olson CA, Hwang S, Deng H, Wong E, Baric RS, Roberts RW, Sun R. mRNA display design of fibronectin-based intrabodies that detect and inhibit severe acute respiratory syndrome coronavirus nucleocapsid protein. J Biol Chem. 2009;284:17,512–17,520. doi: 10.1074/jbc.M901547200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipovšek D. Adnectins: engineered target-binding protein therapeutics. Protein Eng Des Sel. 2011;24:3–9. doi: 10.1093/protein/gzq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross GG, Junge JA, Mora RJ, Kwon HB, Olson CA, Takahashi TT, Liman ER, Ellis-Davies GCR, McGee AW, Sabatini BL, Roberts RW, Arnold DB. Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron. 2013;78:971–985. doi: 10.1016/j.neuron.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao L, Hung KC, Takahashi TT, Joo KI, Lim M, Roberts RW, Wang P. Antibody-mimetic ligand selected by mRNA display targets DC-SIGN for dendritic cell-directed antigen delivery. ACS Chem Biol. 2013;8:967–977. doi: 10.1021/cb300680c. [DOI] [PubMed] [Google Scholar]
- 23.Binz HK, Amstutz P, Plückthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat Biotechnol. 2005;23:1257–1268. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- 24.Olson CA, Adams JD, Takahashi TT, Qi H, Howell SM, Wu TT, Roberts RW, Sun R, Soh HT. Rapid mRNA-display selection of an IL-6 inhibitor using continuous-flow magnetic separation. Angew Chem Int Ed. 2011;50:8295–8298. doi: 10.1002/anie.201101149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrmann C, Martin GA, Wittinghofer A. Quantitative analysis of the complex between p21 and the ras-binding domain of the human raf-1 protein kinase. J Biol Chem. 1995;270:2901–2905. doi: 10.1074/jbc.270.7.2901. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka T, Williams RL, Rabbitts TH. Tumour prevention by a single antibody domain targeting the interaction of signal transduction proteins with RAS. EMBO J. 2007;26:3250–3259. doi: 10.1038/sj.emboj.7601744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filchtinski D, Sharabi O, Rüppel A, Vetter IR, Herrmann C, Shifman JM. What makes Ras an efficient molecular switch: a computational, biophysical, and structural study of Ras–GDP interactions with mutants of Raf. J Mol Biol. 2010;399:422–435. doi: 10.1016/j.jmb.2010.03.046. [DOI] [PubMed] [Google Scholar]
- 28.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 2004;2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sprang SR. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 30.Gideon P, John J, Frech M, Lautwein A, Clark R, Scheffler JE, Wittinghofer A. Mutational and kinetic analyses of the GTPase-activating protein (GAP)-p21 interaction: the C-terminal domain of GAP is not sufficient for full activity. Mol Cell Biol. 1992;12:2050–2056. doi: 10.1128/mcb.12.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wohlgemuth S, Kiel C, Krämer A, Serrano L, Wittinghofer F, Herrmann C. Recognizing and defining true Ras binding domains I: biochemical analysis. J Mol Biol. 2005;348:741–758. doi: 10.1016/j.jmb.2005.02.048. [DOI] [PubMed] [Google Scholar]
- 32.Clark GJ, Drugan JK, Terrell RS, Bradham C, Der CJ, Bell RM, Campbell S. Peptides containing a consensus Ras binding sequence from Raf-1 and theGTPase activating protein NF1 inhibit Ras function. Proc Natl Acad Sci. 1996;93:1577–1581. doi: 10.1073/pnas.93.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duffy S, Tsao KL, Waugh DS. Site-specific, enzymatic biotinylation of recombinant proteins in Spodoptera frugiperda cells using biotin acceptor peptides. Anal Biochem. 1998;262:122–128. doi: 10.1006/abio.1998.2770. [DOI] [PubMed] [Google Scholar]
- 34.Olson CA, Nie J, Diep J, Al-Shyoukh I, Takahashi TT, Al-Mawsawi LQ, Bolin JM, Elwell AL, Swanson S, Stewart R, Thomson JA, Soh HT, Roberts RW, Sun R. Single-round, multiplexed antibody mimetic design through mRNA display. Angew Chem Int Ed. 2012;51:12,449–12,453. doi: 10.1002/anie.201207005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.