SUMMARY
Coenzyme Q (CoQ) is a redox-active lipid required for mitochondrial oxidative phosphorylation (OxPhos). How CoQ biosynthesis is coordinated with the biogenesis of OxPhos protein complexes is unclear. Here, we show that the Saccharomyces cerevisiae RNA-binding protein (RBP) Puf3p regulates CoQ biosynthesis. To establish the mechanism for this regulation, we employed a multi-omic strategy to identify mRNAs that not only bind Puf3p, but also are regulated by Puf3p in vivo. The CoQ biosynthesis enzyme Coq5p is a critical Puf3p target: Puf3p regulates the abundance of Coq5p and prevents its detrimental hyperaccumulation, thereby enabling efficient CoQ production. More broadly, Puf3p represses a specific set of proteins involved in mitochondrial protein import, translation, and OxPhos complex assembly — pathways essential to prime mitochondrial biogenesis. Our data reveal a mechanism for post-transcriptionally coordinating CoQ production with OxPhos biogenesis, and they demonstrate the power of multi-omics for defining genuine targets of RBPs.
eTOC
Mitochondrial biogenesis demands the coordinated integration of metabolites, lipids, and proteins encoded by two genomes into functional organelles. Although transcription factors for this process have been identified, roles of post-transcriptional regulators remain poorly defined. Using a “multi-omic” approach that incorporates measurements of mRNAs, proteins, lipids, and metabolites, we reveal the consequences of regulation by a yeast RNA-binding protein across four ‘omic’ planes. Our data identify a mechanism for the post-transcriptional coordination of mitochondrial coenzyme Q and protein synthesis, and demonstrate the power of multi-omics to identify genuine targets and cellular functions of RNA-binding proteins.
INTRODUCTION
Mitochondria are complex organelles central to cellular metabolism, and their dysfunction is implicated in over 150 human diseases (Vafai and Mootha, 2012). Alterations in metabolic demands induce adaptive changes in mitochondrial mass and composition (Labbe et al., 2014), but precisely how these changes are regulated is unclear. Transcriptional regulators of mitochondria have been identified (Kelly and Scarpulla, 2004), yet the roles of post-transcriptional regulators remain poorly defined.
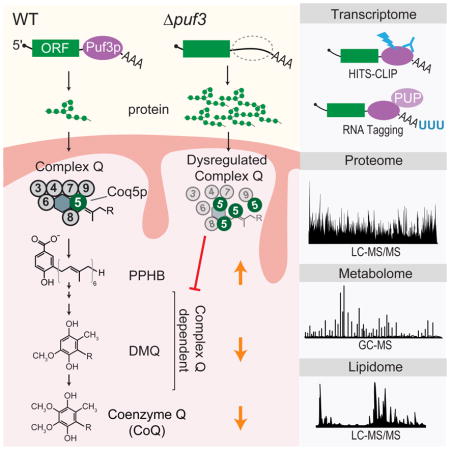
Remodeling and biogenesis of mitochondria is uniquely complicated. It requires synchronized expression of proteins encoded by both nuclear DNA and mitochondrial DNA (mtDNA) (Couvillion et al., 2016), and coordinated assembly of protein complexes, such as those of oxidative phosphorylation (OxPhos), with specific metabolites and lipids, such as coenzyme Q (CoQ). CoQ is a redox active lipid in the electron transport chain whose deficiency causes numerous human diseases (Laredj et al., 2014; Stefely and Pagliarini, 2017). Its biosynthesis likewise requires assembly of a multi-protein complex of enzymes (“complex Q”) and lipids in the mitochondrial matrix (Floyd et al., 2016; He et al., 2014; Stefely et al., 2016b), and production of the water-soluble CoQ head group precursor 4-hydroxybenzoate (4-HB) (Payet et al., 2016; Stefely et al., 2016a). Thus, mitochondrial biogenesis demands integrated remodeling of the proteome, metabolome, and lipidome. The mechanisms for this “multi-omic” regulation are largely uncharacterized.
As described in this report, analysis of our publicly available “Y3K” data set (Stefely et al., 2016a) reveals an unexpected link between CoQ abundance and the Saccharomyces cerevisiae gene PUF3. The encoded protein, Puf3p, belongs to the conserved PUF family of RNA-binding proteins (RBPs), which play key roles throughout Eukarya (Quenault et al., 2011; Spassov and Jurecic, 2003; Wickens et al., 2002). Puf3p binds to mRNAs containing “Puf3p-binding elements” (PBEs), conforming to the consensus UGUANAUA (Gerber et al., 2004; Olivas and Parker, 2000; Zhu et al., 2009), and regulates their translation and stability through a variety of mechanisms (Houshmandi and Olivas, 2005; Jackson et al., 2004; Kershaw et al., 2015; Lee and Tu, 2015; Lee et al., 2010; Miller et al., 2014; Rowe et al., 2014). mRNAs that bind Puf3p have been identified, but which of those mRNAs are genuine “targets” — mRNAs whose activity in vivo is controlled by Puf3p — is an open question. Previously validated targets include mRNAs with connections to OxPhos complexes (e.g., COX17) and the mitochondrial ribosome (Chatenay-Lapointe and Shadel, 2011; Garcia-Rodriguez et al., 2007; Olivas and Parker, 2000). Yeast that lack PUF3 (“Δpuf3 yeast”) exhibit mitochondria-related phenotypes, including reduced respiratory growth (Eliyahu et al., 2010; Gerber et al., 2004; Lee and Tu, 2015) and increased respiratory activity during fermentation (Chatenay-Lapointe and Shadel, 2011). Similarly, Puf3p influences localization of certain mRNAs to the cytoplasmic periphery of mitochondria (Eliyahu et al., 2010; Gadir et al., 2011; Saint-Georges et al., 2008).
Here, we employ a multi-omic strategy that analyzes mRNAs, proteins, lipids, and metabolites to define high-confidence Puf3p targets, and we demonstrate that Puf3p regulates CoQ biosynthesis. In addition to defining a specific Puf3p target responsible for CoQ control — Coq5p — we reveal 90 additional high-confidence direct targets of Puf3p, which provides insight into how CoQ production is coordinated with OxPhos biogenesis pathways. The results demonstrate the power of multi-omic strategies for dissecting the molecular functions of RBPs, which have widespread roles in human health and disease (Gerstberger et al., 2014).
RESULTS
Puf3p Regulates CoQ Biosynthesis
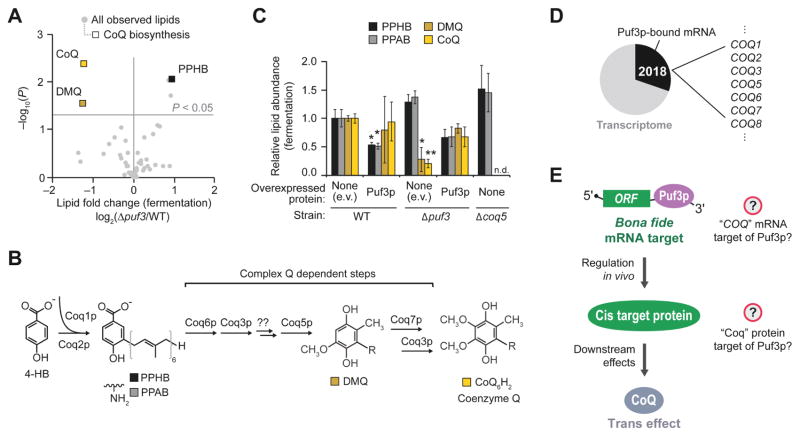
The Y3K data set (Stefely et al., 2016a) reveals Δpuf3 yeast to be significantly (P < 0.05) deficient for CoQ when cultured under fermentation conditions, but not under respiration conditions (Figures 1A, S1A, and S1B). Notably, the early CoQ biosynthesis intermediate polyprenylhydroxybenzoate (PPHB) was elevated, while the later intermediate demethoxy-CoQ (DMQ) was decreased (Figures 1A, 1B, and S1A), suggesting a defect in a step of CoQ production that depends on the CoQ biosynthetic complex (“complex Q” or “CoQ-Synthome”; comprised of Coq3p–Coq9p) (Figure 1B). The accumulation of PPHB in Δpuf3 yeast was the largest such change across all yeast strains in the Y3K study under fermentation growth conditions (Figure S1A), further suggesting a functionally important link between Puf3p and the CoQ pathway.
Figure 1. PUF3 Regulates Coenzyme Q Biosynthesis.
(A) Relative lipid abundances in Δpuf3 yeast compared to WT (mean, n = 3) (fermentation condition) versus statistical significance (P). Raw data from Y3K data set (Stefely et al., 2016a).
(B) Scheme of CoQ biosynthesis. 4-HB, 4-hydroxybenzoate; PPHB, polyprenylhydroxybenzoate; PPAB, polyprenylaminobenzoate; DMQ, demethoxy-CoQ.
(C) Relative lipid abundances in yeast transformed with low-copy plasmids overexpressing Puf3p (or empty vector, e.v.) and cultured in fermentation media (mean ± SD, n = 3). Bonferroni corrected *P < 0.05; **P < 0.01. Two-sided Student’s t-test for all panels.
(D) Pie chart illustrating the fraction of transcribed genes with mRNAs that are putative Puf3p targets, which was derived by aggregating all Puf3p-bound mRNAs reported via HITS-CLIP (Wilinski et al., 2017), RNA Tagging (Lapointe et al., 2015), PAR-CLIP (Freeberg et al., 2013), and RIP-seq (Kershaw et al., 2015). Puf3p-bound mRNAs include 7 mRNAs encoding CoQ biosynthesis enzymes.
(E) Scheme of how Puf3p could impact CoQ production.
In the Y3K project, yeast were cultured in yeast extract-peptone (“YP”) medium (Stefely et al., 2016a). This rich medium possesses an indeterminate amount of CoQ head group precursors, such as 4-HB or para-amino-benzoate (pABA), but was not further supplemented with these molecules. Here, to test the idea that Puf3p impacts CoQ biosynthesis under varying media conditions, we cultured wild-type (WT) or Δpuf3 yeast in synthetic media containing either pABA only, or pABA and 4-HB together. Significant (P < 0.05) alterations in CoQ or CoQ intermediates were observed in each media condition (Figure S1C). Synthetic media with pABA and 4-HB was selected for the further studies because it led to readily quantifiable levels of CoQ, PPHB, and PPAB (the aminated analog of PPHB).
To further test the idea that Puf3p impacts CoQ biosynthesis, we overexpressed Puf3p in WT or Δpuf3 yeast and examined CoQ pathway lipids. Overexpression of Puf3p in WT yeast suppressed production of PPHB and PPAB — striking effects because they are the inverse of those observed in Δpuf3 yeast (Figures 1C and S1D–S1F). Furthermore, plasmid expression of Puf3p recovered CoQ biosynthesis in fermenting Δpuf3 yeast (Figure 1C). Puf3p therefore is involved in CoQ biosynthesis.
We hypothesized that Puf3p controls an enzyme in the CoQ biosynthesis pathway by binding to and regulating the mRNA that encodes it. To explore candidate enzymes, we examined existing data sets from recent studies that used high-throughput sequencing-based technologies to identify mRNAs bound by Puf3p (Freeberg et al., 2013; Kershaw et al., 2015; Lapointe et al., 2015; Wilinski et al., 2017), which built on a formative microarray study (Gerber et al., 2004). In aggregate, the four studies reported an astounding 2,018 putative mRNA “targets” of Puf3p — representing nearly a third of the yeast transcriptome (Table S1). Putative Puf3p targets include seven enzymes directly involved in CoQ biosynthesis: COQ1, COQ2, COQ3, COQ5, COQ6, COQ7, and COQ8 (Figure 1D). We suspected that Puf3p regulated only a fraction of those 2,018 putative targets in vivo — both for this specific case of Puf3p-mediated regulation of CoQ (Figure 1E) and for other biological functions as well.
Multi-omic Analyses Define Puf3p Targets
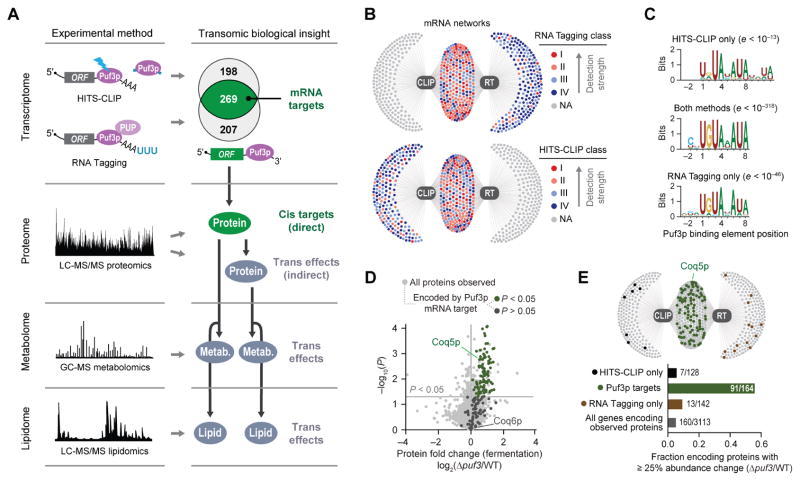
To identify high-confidence Puf3p targets, we integrated data from five experimental methods across four omic planes (Figure 2A). We first identified Puf3p target mRNAs using data from two independent approaches: HITS-CLIP (Licatalosi et al., 2008), which uses UV-crosslinking and immunopurification of RBP-RNA complexes, and RNA Tagging (Lapointe et al., 2015), which employs an RBP-poly(U) polymerase fusion protein to covalently tag mRNAs bound in vivo with 3′ uridines (“U-tags”) (Figure S2A). We focused on the HITS-CLIP and RNA Tagging datasets because they were generated in-house and rely on distinct strategies to identify mRNAs bound by a protein. HITS-CLIP (Wilinski et al., 2017) and RNA Tagging (Lapointe et al., 2015) co-identified 269 Puf3p-bound mRNAs, out of the 467 and 476 mRNAs identified via each technique on its own, respectively (Figure 2A) (hypergeometric test, P < 10−211). Separating Puf3p-bound mRNAs into classes based on detection (I–IV, with class I representing the strongest detection) (Lapointe et al., 2017) showed that co-identified mRNAs were the most robustly detected mRNAs in each method alone (Figures 2B, S2B, and S2C), and 42% of mRNAs identified via both methods were class I or II in each individual approach (Figure S2D). Moreover, the 3′ untranslated regions (UTRs) of mRNAs identified via both methods were strongly enriched for sequences that conform to high-affinity PBEs (UGUAHAUA), and a remarkable 81% also contained an upstream (−1 or −2) cytosine, which is required for particularly high-affinity interactions and regulation in vivo (Lapointe et al., 2015; Zhu et al., 2009) (Figures 2C and S2E). In contrast, RNAs unique to one method had more degenerate PBEs, with much less frequent upstream cytosines.
Figure 2. Multi-omics Maps High-Confidence Puf3p Targets.
(A) Multi-omics approach overview. The Venn diagram indicates Puf3p-bound mRNAs co-identified by RNA Tagging and HITS-CLIP.
(B) Network maps of Puf3p-bound mRNAs (dots) detected by RNA Tagging (RT) and/or HITS-CLIP (CLIP) (edges).
(C) Enriched Puf3p-binding elements identified by MEME for the indicated groups of Puf3p-bound RNAs.
(D) Relative protein abundances in Δpuf3 yeast compared to WT (mean, n = 3) versus statistical significance (P, fermentation condition), highlighting proteins encoded by Puf3p mRNA targets. P value cutoff is for protein abundance changes.
(E) Bar graph and network map of Puf3p-bound mRNAs indicating mRNAs encoding proteins detected in the Y3K proteomics data set with ≥ 25% protein abundance change (P < 0.05, two-sided Student’s t-test).
This figure includes new, integrated analyses of publicly available raw data from the RT (Lapointe et al., 2015), HITS-CLIP (Wilinski et al., 2017), and Y3K multi-omic (Stefely et al., 2016a) data sets generated in our labs.
Integration of HITS-CLIP and RNA Tagging data thus yielded a high-confidence list of co-identified Puf3p-bound mRNAs, referred to here as “Puf3p target mRNAs” (Figure 2A and Table S1). Importantly, our analyses identified the CoQ-related mRNAs COQ2, COQ5, and COQ6 as high-confidence Puf3p targets, but not COQ1, COQ3, COQ7, or COQ8. In searching for the target responsible for the Δpuf3 CoQ deficiency (Figure 1A), the observation of elevated PPHB and decreased CoQ suggested a defect in a complex Q dependent step (Figure 1B) and allowed us to narrow our focus to COQ5 and COQ6. Both mRNAs contain consensus PBEs in their 3′ UTRs, with the COQ5 PBE also possessing a −1 cytosine. However, which of these Puf3p-mRNA interactions ultimately leads to regulation in vivo is unclear from the transcriptomic data alone.
To determine how Puf3p impacts the abundance of proteins encoded by its target mRNAs, we integrated these mRNA data with the proteomics feature of the Y3K data set (Stefely et al., 2016a). We identified protein abundance changes due to loss of PUF3 in two metabolic conditions. In fermentation culture conditions, 160 significant proteome changes were observed in Δpuf3 yeast (P < 0.05 and fold change [FC] > 25%) (Figure S2F). In contrast, only 24 such changes in protein abundance were observed in respiration culture conditions. Thus, we primarily leveraged the fermentation proteomics data set to reveal Puf3p functions. The more drastic protein changes observed in fermentation mirror those of CoQ pathway lipids, which were likewise significantly (P < 0.05) altered only in fermentation (Figures 1A and S1B).
Of the 165 proteins encoded by Puf3p target mRNAs detected in fermenting yeast, 91 (55%) were significantly more abundant by at least 25% in Δpuf3 yeast relative to WT yeast (P < 0.05) (Figure 2D). We collectively refer to these proteins as “Puf3p cis target proteins” (“cis targets”) (Figure 2A), and they include Coq5p but not Coq6p. The 91 cis targets account for a striking 57% (91/160) of significantly (P < 0.05) altered proteins in the Δpuf3 proteome. In contrast, Puf3p-bound RNAs uniquely identified by a single RNA method, especially those detected weakly, were much less likely to be upregulated (Figures 2E and S2G). Thus, the combination of these distinct “omic” approaches identifies RNA-binding events likely to have regulatory effects in the cell. Collectively, these analyses provide high-confidence Puf3p targets across both the transcriptome and proteome (Figure 2A and Table S2). They also demonstrate that Puf3p regulates Coq5p abundance, suggesting a mechanistic link between loss of Puf3p and dysregulation of CoQ pathway lipids.
Coq5p Overexpression Inhibits CoQ Biosynthesis and Yeast Growth
To examine our finding that Puf3p regulates Coq5p, we generated yeast strains with genomic mutations to the 3′ UTR of COQ5 (Figure S3A). Removal of the COQ5 3′ UTR or the site-specific mutation of the Puf3p-binding element (PBE) in COQ5 at its endogenous genomic loci significantly reduced yeast growth in both fermentation and respiration (P < 0.05) (Figures S3A, S3B, and S3C). These growth defects could be due to Coq5p over expression, under expression, or mislocalization. Attempts to compare the abundance of Coq5p in WT and COQ5 PBE mutants were complicated by the large growth rate differences between the strains combined with changes in Coq5p abundance across differing metabolic states (see below). However, disruption of two genes that flank COQ5 (BUL2 and ZDS2) had no discernible effect on yeast growth, therefore ensuring that the growth defects we observed are specific to loss of the PBE in COQ5. Together, these findings demonstrate that the COQ5 3′ UTR and its PBE are important for yeast function.
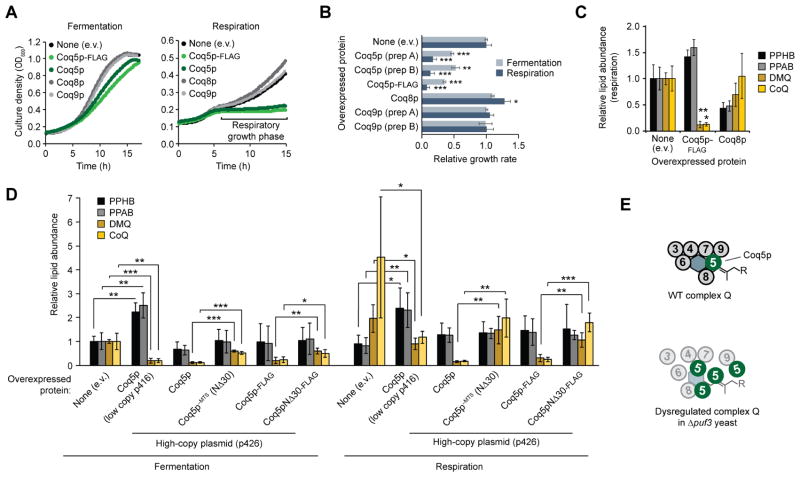
To test our hypothesis that Puf3p-mediated regulation of Coq5p abundance is required for proper CoQ production, we analyzed how Coq5p overexpression (i.e., overriding Puf3p regulation) impacted yeast growth and the CoQ biosynthetic pathway. Markedly reduced CoQ levels can be sufficient for supporting respiratory yeast growth (Stefely et al., 2015), and alterations in CoQ biosynthesis can be difficult to track with growth assays. Thus, to facilitate mechanistic studies, we overexpressed Coq5p from a plasmid with a strong promoter to make associated phenotypes more readily observable. Overexpression of Coq5p in WT yeast slowed fermentative growth and essentially eliminated respiratory growth (Figures 3A and 3B). Although plasmid overexpression likely drives Coq5p to a much higher abundance than that observed in Δpuf3 yeast, plasmid overexpression of Coq8p or Coq9p, which are also complex Q members (Floyd et al., 2016; He et al., 2014; Stefely et al., 2016b), did not inhibit yeast growth, supporting the hypothesis that regulation of Coq5p is particularly important for complex Q function (Figures 3A, 3B, and S3D). To examine whether Coq5p overexpression inhibits respiratory yeast growth by disrupting CoQ production, we examined CoQ pathway intermediates. Indeed, Coq5p overexpression in WT yeast recapitulated the CoQ-related phenotypes of Δpuf3 yeast in fermentation growth conditions — deficiency of DMQ and CoQ, and elevation of PPAB and PPHB (Figures 3C, S3E, and S3F). Similar to the observed growth effects, CoQ intermediates were most affected by overexpression of Coq5p when compared to the overexpression of the other proteins.
Figure 3. Puf3p Regulates the Potentially Poisonous CoQ Biosynthesis Enzyme Coq5p.
(A) Growth curves for WT yeast transformed with plasmids overexpressing the proteins shown and cultured in either fermentation or respiration media.
(B) Relative growth rates of WT yeast transformed with plasmids overexpressing the proteins shown and cultured in either fermentation or respiration media (mean ± SD, n = 3). **P < 0.01; ***P < 0.001.
(C) Relative lipid abundances in WT yeast transformed with plasmids overexpressing the proteins shown and cultured in respiration media (mean ± SD, n = 3). Bonferroni corrected *P < 0.05; **P < 0.01.
(D) Relative lipid abundances in WT yeast transformed with plasmids overexpressing the indicated Coq5p constructs and cultured under fermentation or respiration growth conditions (mean ± SD, n = 4). *P < 0.05; **P < 0.01; ***P < 0.001.
(E) Model for how Coq5p overexpression dysregulates complex Q.
Two-sided Student’s t-test for all panels.
Coq5p overexpression could be deleterious due to excessive Coq5p methyltransferase activity or non-native Coq5p physical interactions. Overexpression of Coq5p with mutations to putative catalytic residues (D148A or R201A) inhibited yeast growth (Figure S3G), which demonstrates that inhibition occurs independent of Coq5p enzyme activity. Furthermore, overexpression of Coq5p lacking its mitochondrial targeting sequence (MTS) (Coq5p−MTS) reduced its inhibitory effect on growth in respiration (Figure S3H). Similarly, removal of the MTS attenuated the impact of overexpressed Coq5p on the abundance of CoQ and its intermediates in both fermentation and respiration growth conditions (Figure 3D). The lack of full restoration of WT growth and CoQ abundance by Coq5p−MTS suggests that overexpression of Coq5p may have some extra-mitochondrial effects in addition to its mitochondrial effects. Together, our findings imply that overexpressed Coq5p likely disrupts native protein-protein interactions in the mitochondrial matrix, which in turn inhibits yeast proliferation and CoQ biosynthesis.
To further test the idea that Puf3p regulates Coq5p in a metabolic state-dependent fashion, we measured endogenous Coq5p levels in WT and Δpuf3 yeast cultured across the diauxic shift — the transition in cellular metabolism from fermentation to respiration. Prior to the diauxic shift, Coq5p abundance was substantially higher in Δpuf3 yeast in comparison to WT yeast (Figure S3I). In agreement with our proteomic analyses, Coq5p levels returned to near WT levels in post-diauxic shift Δpuf3 yeast (i.e., respiration growth conditions) (Figure S3I).
Our collective findings demonstrate that Puf3p modulates CoQ biosynthesis by regulating Coq5p, a potentially promiscuous protein with detrimental effects when overexpressed (Figures 3E and S3J). By validating Coq5p as a Puf3p target, our findings suggest that the additional 90 cis target proteins identified by our multi-omic analysis are also likely bona fide Puf3p targets in vivo.
Puf3p Coordinates CoQ Production with Mitochondrial Biogenesis Functions
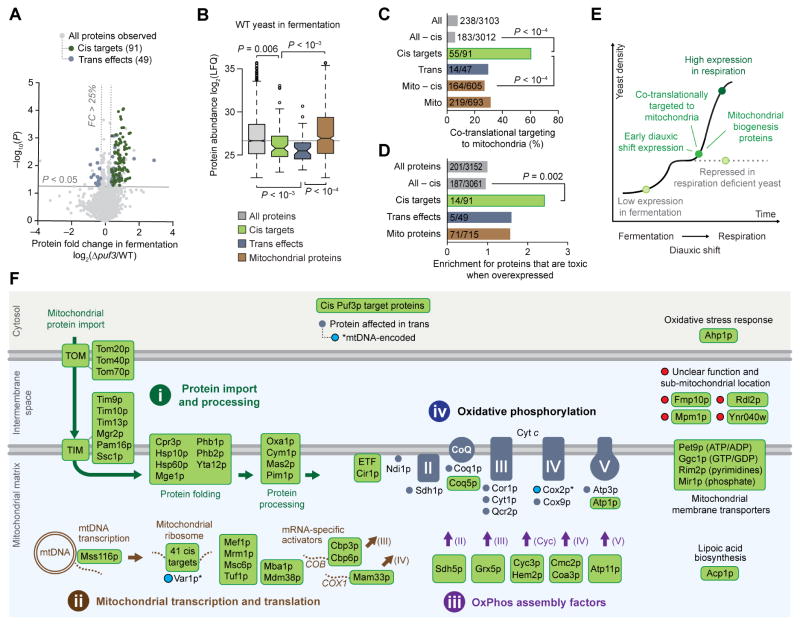
Our multi-omic data set enabled us to map additional Puf3p functions across multiple omic planes (Figure 2A), in a fashion similar to how we mapped Puf3p to its mRNA target COQ5, its cis target protein Coq5p, and its downstream (trans) effect on CoQ lipids. We used the cis Puf3p protein targets to identify downstream “trans effects” in the proteome. By a protein “trans effect,” we refer to proteins whose abundance is dependent on Puf3p, but whose mRNA does not bind Puf3p (see Methods for details). The Δpuf3 proteome changes include the 91 cis targets, which are enriched for mitochondrial organization and translation functions, and 49 trans effects, enriched for mitochondrial OxPhos and electron transport chain (ETC) functions (Figures 4A and S4A and Tables S2 and S3).
Figure 4. Puf3p Targets Prime Biogenesis of Mitochondria and OxPhos.
(A) Relative protein abundances in Δpuf3 yeast compared to WT (mean, n = 3) versus statistical significance (P), highlighting proteins with fold change (FC) > 25% and P < 0.05 (two-sided Student’s t-test) that were either identified as Puf3p targets by both RNA methods (cis targets, green) or neither RNA method (trans effects, blue).
(B) Protein abundances (Stefely et al., 2016a) for each group of proteins shown. LFQ, label free quantitation value. Center lines indicate medians, limits indicate 25th and 75th percentiles, whiskers extend 1.5 times the interquartile range, outliers are represented by dots, and P values were determined with a Student’s t-test (two-tailed, homostatic). Protein set sizes: all proteins n = 3152, mitochondrial proteins n = 715, cis targets n = 91, trans effect proteins n = 49.
(C) Percent of each group of proteins shown that is cotranslationally targeted to mitochondria (Williams et al., 2014). P values determined by a Fisher’s exact test.
(D) Relative fraction of proteins that are toxic when overexpressed across the indicated protein groups. values determined by a Fisher’s exact test.
(E) Cartoon model of a yeast growth curve with key features and dynamics of Puf3p cis targets indicated.
(F) Cartoon map of all identified 91 Puf3p cis targets and select Puf3p trans effect proteins that are elevated in Δpuf3 yeast. Puf3p cis targets include numerous proteins that support (i) mitochondrial protein import and processing, (ii) mitochondrial transcription and translation, and (iii) OxPhos assembly. Trans effects include (iv) an increase in OxPhos proteins.
We quantitatively compared properties of Puf3p cis targets and trans effects to those of either all proteins or mitochondrial proteins. The latter is an informative control set because Puf3p target mRNAs are highly enriched for mitochondrial proteins (Figure S4B). Both cis targets and trans effect proteins have relatively low abundances in fermentation, but not in respiration, reflecting Puf3p-mediated repression in fermentation (Figures 4B and S4C) (Stefely et al., 2016a). In parallel, cis targets are upregulated across early time points in the diauxic shift (Stefely et al., 2016b) (Figure S4D), and yeast with respiration deficient mitochondria, which cannot complete the diauxic shift, exhibit downregulation of Puf3p cis targets as part of their “respiration deficiency response” (Stefely et al., 2016a) (Figure S4E). Cis targets are enriched for proteins that are co-translationally targeted to mitochondria (Williams et al., 2014) (Figures 4C and S4F). Furthermore, they are enriched for proteins that reduce yeast proliferation when overexpressed (Figure 4D), suggesting that regulation of proteins that are toxic when hyperaccumulated — such as that observed with Coq5p — is a general mechanism of Puf3p action. Together, our analyses support a model where Puf3p mediates repression of a set of mitochondrial proteins in fermentation, and this repression is released, or potentially reversed (i.e., cis targets are activated), early during the diauxic shift to help initiate the requisite biological changes, such as mitochondrial biogenesis (Figure 4E).
To reveal the specific biochemical pathways through which Puf3p regulates mitochondrial function, we mapped individual Puf3p cis targets and trans effects (Figures 4F and S5A). Strikingly, 86 of the 87 Puf3p cis targets with known functions fit into pathways that support mitochondrial biogenesis, and in particular converge to generate the OxPhos machinery. The first pathway (i) includes proteins that catalyze the import, folding, and processing of nuclear DNA-encoded mitochondrial proteins, which include many OxPhos subunits. The second pathway (ii) includes proteins that support transcription and translation of mtDNA-encoded genes, which also encode OxPhos subunits. For example, cis Puf3p targets include over half of the mitochondrial ribosomal proteins and critical translational activators (e.g., Cbp3p, Cbp6p, Mam33p, Mba1p, and Mdm38p) (Figures 4F and S5A–S5C). Puf3p trans effects included increased abundance of two proteins encoded by mitochondrial DNA (Var1p and Cox2p, the only two such proteins observed in the proteomics analysis). The third pathway (iii) encompasses assembly factors for each OxPhos subunit, which provide ancillary support for OxPhos complex biogenesis. Collectively, these three pathways of Puf3p cis targets are poised to prime OxPhos biogenesis. Consistently, Puf3p trans effects observed in Δpuf3 yeast include increased abundance of OxPhos complex subunits (iv). Thus, Puf3p regulates production and assembly of both proteins and lipids required for OxPhos.
Two additional pathways of Puf3p cis targets include mitochondrial membrane transporters for nucleotides, which sustain mtDNA transcription, and proteins such as Acp1p that support the TCA cycle (Figures 4F and S5A). Downstream, citrate synthase (Cit1p), the rate limiting gateway enzyme to the TCA cycle and a putative client protein of the Puf3p cis targets Tom70p and Hsp60p (Martin et al., 1992; Yamamoto et al., 2009), and Ptc7p, a phosphatase that reactivates phosphorylated Cit1p (Guo et al., 2017), were also significantly increased trans effect proteins (P < 0.05) (Figure S6A). Accordingly, the abundance of citrate was significantly elevated in Δpuf3 yeast (P < 0.05) (Figure S6B).
The tight functional association of cis Puf3p targets in mitochondrial biogenesis pathways suggests that the four uncharacterized proteins that are cis Puf3p targets — Rdl2p, Ynr040w, Mpm1p, and Fmp10p — might also function in mitochondrial biogenesis (Figure 4F). Previous large-scale screens detected these proteins in mitochondria (Inadome et al., 2001; Reinders et al., 2006; Sickmann et al., 2003; Vogtle et al., 2012). Immunocytochemistry analyses of FLAG-tagged Rdl2p, Ynr040w, Mpm1p, and Fmp10p, validated these observations (Figures S7A and S7B), thereby confirming their classification as mitochondrial uncharacterized (x) proteins (MXPs). While the gene deletion strains for these MXPs are respiration competent (Figure S7C), their overexpression in WT yeast inhibited respiratory growth (Figures S7D), suggesting that they may interact with proteins required for OxPhos and have the potential to be toxic like Coq5p. Given these data, all but one (Ahp1p) of the 91 cis targets localize to mitochondria, further solidifying the role of Puf3p in mitochondrial function. Together, this multi-omic analysis provides a high-resolution map of the molecular targets of Puf3p that will serve as a powerful resource for investigations of mitochondrial biogenesis and function (Tables S1 and S2).
DISCUSSION
Defining Genuine Targets via Multi-omics
Our findings show that multi-omic strategies are powerful in the analysis of RBP function. In our case, despite considerable prior efforts to identify the mRNAs to which Puf3p binds in vivo, it was unclear which of the many binding events were biologically productive — a central, common challenge in the analysis of RBPs (Konig et al., 2012; Licatalosi and Darnell, 2010; Riley and Steitz, 2013). To understand how Puf3p controlled CoQ biosynthesis, including whether there is a direct effect on an mRNA in the CoQ pathway, we generated a high-confidence Puf3p mRNA target list by combining two distinct approaches, and integrated that data with proteomic, metabolomic, and lipidomic studies. This strategy pinpointed Coq5p as the key Puf3p target from a large pool of candidates (> 2,000) that included six other CoQ biosynthesis enzymes (Figure S3J). Our analyses yielded a comprehensive roadmap of specific biochemical processes controlled by Puf3p in vivo. Together, our findings revealed a post-transcriptional mechanism to coordinate CoQ biosynthesis with the broader mitochondrial biogenesis program.
A key first step toward defining biochemical functions of Puf3p was the distillation of large lists of Puf3p-RNA interactions to a smaller set of those mRNAs it controls in vivo — its genuine targets. The HITS-CLIP and RNA Tagging data sets overlapped by ≈50%, which was greatest among the mRNAs detected most robustly in each separate approach. Weakly detected Puf3p-bound mRNAs were unlikely to overlap and may arise through intrinsic biases of each method. Thus, our analyses illustrate that HITS-CLIP and RNA Tagging are complementary, and we demonstrate that integration of their data yields high-confidence RNA targets. Indeed, these analyses rapidly narrowed our focus to COQ5 or COQ6, from the other complex Q candidates COQ3, COQ8, and COQ9. Importantly, our findings also demonstrate that RBP-mRNA interaction data sets are stratified: interactions that lead to biological regulation are concentrated at the top of the rank order. In the absence of a multi-omic data set, a sharp focus on the top tier of candidates has great potential to yield mRNAs that are substantially regulated in vivo.
A second key step was the integration of the Puf3p mRNA target set with Δpuf3 proteome alterations. Our integration revealed 91 Puf3p-mRNA interactions that regulate the abundance of the encoded protein in vivo. The regulated Puf3p targets included Coq5p, but not Coq6p, which yet again narrowed our focus and was critical to reveal how Puf3p controls CoQ biosynthesis. In principle, the remaining 74 Puf3p target mRNAs with unaffected protein abundances could be false positives. However, it is more likely that Puf3p regulates their localization (Gadir et al., 2011; Saint-Georges et al., 2008) independent of their translation, or that the yeast culture systems we used lacked an environmental stress encountered by yeast in nature. Notably, many of these mRNA targets trended toward increased protein abundance (Figure 2D), but did not cross our significance threshold (P < 0.05). They provide an initial foothold for analyzing alternative forms of Puf3p-mediated control (e.g., localization), or Puf3p function under varying environmental or cellular conditions.
Post-Transcriptional Control of CoQ Biosynthesis and Toxic Proteins
Our results reveal a mechanism for post-transcriptional regulation of CoQ biosynthesis. Our biochemical work further demonstrated that Puf3p regulates the abundance of Coq5p, which when overexpressed dramatically reduced yeast growth and CoQ production. These findings suggest that the CoQ biosynthetic complex can be disrupted when particular subunits are too abundant, thereby highlighting the importance of stoichiometry for complex Q activity. Interestingly, however, overexpression of complex Q members Coq8p or Coq9p lacked the same deleterious effects as those observed with Coq5p, demonstrating that some complex Q subunits are particularly poisonous at inappropriately high protein abundance. Together, our findings suggest that Puf3p plays an important role in coordinating expression of complex Q components with the larger program of OxPhos biogenesis, through precise control of proteins that may be toxic when overexpressed. Analysis of the full Puf3p cis target set further suggested that regulation of such poisonous proteins is a broader Puf3p function. This provides an evolutionary rationale for why particular proteins within larger pathways are targeted for regulation by Puf3p.
Puf3p Coordinates Proteins and Lipids in Mitochondrial Biogenesis
By controlling the abundance of proteins that catalyze production of mitochondrial proteins, lipids, and metabolites, Puf3p regulation reaches across three omic planes. Multi-omic coordination may be critical for efficient production of functional mitochondria, for example by linking production of OxPhos protein complexes and the OxPhos lipid CoQ.
A similar mechanism was recently proposed for nuclear- and mitochondrial DNA-encoded proteins via synchronization of cytoplasmic and mitochondrial translation programs (Couvillion et al., 2016). That study suggested a critical role for cytosolic translational control of mitochondrial translational activators in coordinating OxPhos biogenesis and, in particular, identified the COB (complex III subunit) translational activator Cbp6p as a component of the mechanism. Importantly, our work now demonstrates that Puf3p regulates Cbp6p and its binding partner Cbp3p, both of which are cis Puf3p targets. Thus, we propose that Puf3p is a key part of the molecular mechanism that coordinates mitochondrial and cytosolic translation programs. Mammalian RBPs that impact mitochondrial function have been identified (Gao et al., 2014; Schatton et al., 2017), but whether any of these provide Puf3p-like multi-omic regulation of mitochondrial biogenesis is currently unknown.
A Roadmap of Puf3p Functions
Our analyses provide substantial evidence that a primary function of Puf3p is to repress expression of mitochondrial biogenesis factors in fermenting yeast, as opposed to Puf3p having many broad biological functions (Kershaw et al., 2015). We identified 269 target mRNAs and 91 cis target proteins for Puf3p, which can be separated into three pathways that converge to generate the OxPhos machinery. Our strategy furthermore enabled us to map trans Puf3p effects across three omic planes, including protein components of OxPhos complexes, TCA cycle metabolites, and mitochondrial lipids, such as CoQ. Trans Puf3p effect proteins also include splicing factors and epigenetic regulators, and we speculate that Δpuf3 yeast have “sensed” mitochondrial dysfunction through a mitochondria-to-nucleus retrograde signaling pathway. Consistently, the Y3K proteomics data set (Stefely et al., 2016a) shows that Δpuf3 yeast have decreased levels of Ngg1p — a component of the SLIK complex, which regulates gene expression in response to mitochondrial dysfunction (Pray-Grant et al., 2002).
We observed few effects in respiring Δpuf3 yeast. Indeed, Puf3p-mediated destabilization of target mRNAs is inactivated in respiration growth conditions, even though Puf3p remains bound to the mRNAs (Miller et al., 2014). Furthermore, in the earliest timepoints after the diauxic shift, Puf3p activates the translation of target mRNAs (Lee and Tu, 2015). This regulatory switch is very likely mediated by phosphorylation of the N-terminal half of Puf3p (the non-RNA-binding half), which occurs soon after the diauxic shift (Lee and Tu, 2015). At first glance, our findings may seem to contradict the study that reported Puf3p to activate translation of its mRNA targets as yeast enter the diauxic shift (Lee and Tu, 2015). However, our cultures were harvested more than 5 hours after the diauxic shift, by which time the activating function has disappeared (Lee and Tu, 2015). Our analyses of earlier time points also suggest that Puf3p functions early in the diauxic shift (Figure S4D). Yeast that lack PUF3 may only have mild phenotypes because transcriptional control compensates for the lack of Puf3p-mediated regulation, or because other RBPs (e.g., Puf4p or Puf5p) compensate for its absence. Regardless, Puf3p is a critical repressor of mitochondrial biogenesis in fermenting yeast — a function that is conserved across more than 300 million years of evolution (Hogan et al., 2015; Wilinski et al., 2017).
The Puf3p mRNA targets that were not observed in our proteomics studies provide another valuable resource (Tables S1 and S2). For example, they include previously uncharacterized genes (e.g., AIM11, AIM18, YBR292C, YDR286C, YDR381C-A, YGR021W, and YGR161W-C), which we can now potentially link to roles in mitochondrial biogenesis given their identity as Puf3p target mRNAs. Thus, the pathways mapped via multi-omics also provide a framework for retrospective analyses, which is particularly useful since imperfect overlap between omic planes is a common feature of multi-omic studies (e.g., some observed mRNAs are not detected at the protein level). Our high-confidence Puf3p cis target set provides a snapshot of specific mitochondrial biogenesis pathways regulated by Puf3p (Figure 4F) that can guide future analyses of the full set of Puf3p mRNA targets, which include additional OxPhos biogenesis factors.
Collectively, our work reveals post-transcriptional control of CoQ production linked into a larger regulatory network that controls OxPhos biogenesis. Our findings also provide a foundation for further exploring the molecular basis of mitochondrial biogenesis, including potential new roles for Puf3p-regulated MXPs in OxPhos biogenesis. Moreover, our approach presents a generalizable multi-omic strategy to identify biological roles for the many RNA-binding proteins that impact human health and disease.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact: David Pagliarini (dpagliarini@morgridge.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast strains
The parental (wild type, WT) Saccharomyces cerevisiae strain for this study was the haploid MATalpha BY4742. Single gene deletion (Δgene) derivatives of BY4742 were obtained through the gene deletion consortium (via Thermo, Cat#YSC1054). Gene deletions were confirmed by proteomics (significant [P < 0.05] and selective decrease in the encoded protein) or PCR assay.
COQ5 PBE mutant strain
The PBE in COQ5 was identified by analysis of the HITS-CLIP data (Wilinski et al., 2017), in which COQ5 had a single peak centered over the sequence: CTGTACATA. Using the haploid MATalpha BY4742 as the parental strain, a yeast strain with a mutant Puf3p-binding element (PBE) in the 3′ untranslated region (UTR) of the COQ5 gene was generated using the Delitto Perfetto method (Storici and Resnick, 2006). Briefly, the 282 nucleotides immediately downstream of the COQ5 coding sequence were replaced with the KanMx4-KlUra3 cassette. Thus, the “coq5Δ3′UTR” strain was created. To enable generation of the endogenous PBE mutant, the COQ5 ORF + 3′ UTR was cloned in p426gpd. The PBE sequence was mutagenized by PIPE cloning. The sequence-confirmed plasmid, containing mutagenized PBE, served as the PCR template in a reaction that generated the cassette that seamlessly replaced KanMx4-KlUra3 with the COQ5 3′ UTR sequence harboring the CTGT --> GACA PBE mutation. Two separate yeast strains with identical COQ5 PBE mutants were independently generated in this manner. One of the two strains maintained a COQ5 3′ UTR identical to that of the WT strain outside of the mutated PBE. In the second strain, the PBE change was scarless, with the exception of the deletion of a single 3′ UTR thymidine nucleotide in a stretch of 10 thymidine nucleotides beginning 184 nucleotides upstream of the desired PBE mutation. As shown in the figures of this report, both of these independently generated strains shared the same biological phenotypes.
METHOD DETAILS
Yeast cultures
General culture procedures and media components
Yeast were stored at −80 °C as glycerol stocks and initially cultured on selective solid media plates at 30 °C. Biological replicates were defined as sep arate yeast colonies after transformation and plating onto solid selective media. Experiments were conducted in biological triplicate (“n = 3”) or quadruplicate (“n = 4”) to afford statistical power. Cell density of liquid media cultures was determined by optical density at 600 nm (OD600) as described (Hebert et al., 2013). Media components included yeast extract (‘Y’) (Research Products International, RPI), peptone (‘P’) (RPI), agar (Fisher), dextrose (‘D’) (RPI), glycerol (‘G’) (RPI), uracil drop out (Ura−) mix (US Biological), histidine drop out (His−) mix (US Biological), Synthetic complete media (SC) mix (US Biological), and G418 (RPI). YP and YPG solutions were sterilized by automated autoclave. G418 and dextrose were sterilized by filtration (0.22 μm pore size, VWR) and added separately to sterile YP or YPG. YPD+G418 plates contained yeast extract (10 g/L), peptone (20 g/L), agar (15 g/L), dextrose (20 g/L), and G418 (200 mg/L). YPD media (rich media fermentation cultures) contained yeast extract (10 g/L), peptone (20 g/L), and dextrose (20 g/L). YPGD media (rich media respiration cultures) contained yeast extract (10 g/L), peptone (20 g/L), glycerol (30 g/L) and dextrose (1 g/L). Synthetic Ura− media and synthetic complete media were sterilized by filtration (0.22 μm pore size). Ura−,D media contained Ura− mix (1.92 g/L), yeast nitrogen base (YNB) (6.7 g/L, with ammonium sulfate and without amino acids), and dextrose (20 g/L). Ura−,GD media contained Ura− mix (1.92 g/L), YNB (6.7 g/L), glycerol (30 g/L), and dextrose (1 g/L). Ura−,D+4HB media contained Ura− mix (1.92 g/L), YNB (6.7 g/L), dextrose (20 g/L), and 4-HB (100 μM). Ura−,GD+4HB media contained Ura− mix (1.92 g/L), YNB (6.7 g/L), glycerol (30 g/L), dextrose (1 g/L), and 4-HB (100 μM). SCD media contained SC mix (2 g/L), YNB (6.7 g/L), and dextrose (20 g/L). SCGD media contained SC mix (2 g/L), YNB (6.7 g/L), glycerol (30 g/L), and dextrose (1 g/L). SCD+4HB media contained SC mix (2 g/L), YNB (6.7 g/L), dextrose (20 g/L), and 4-HB (10 μM). SCGD+4HB media contained SC mix (2 g/L), YNB (6.7 g/L), glycerol (30 g/L), dextrose (1 g/L), and 4-HB (10 μM).
PUF3 rescue cultures
WT or Δpuf3 yeast were transformed with plasmids encoding Puf3p [p423(2μ)-PUF3 or p413(CEN)-PUF3] and cultured on His−,D plates. Starter cultures (3 mL His−,D+4HB) were inoculated with an individual colony of yeast and incubated (30 °C, 230 rpm, 10–15 h). For CoQ quantitation, His−,D+4HB (fermentation) or His−,GD+4HB (respiration) media (100 mL media at ambient temperature in a sterile 250 mL Erlenmeyer flask) were inoculated with 2.5×106 yeast cells and incubated (30 °C, 230 rpm). Samples of the His −,D+4HB cultures were harvested 13 h after inoculation, a time point that corresponds to early fermentation (logarithmic) growth. Samples of His−,GD+4HB cultures were harvested 25 h after inoculation, a time point that corresponds to early respiration growth. For each growth condition, 1×108 yeast cells were pelleted by centrifugation (3,000 g, 3 min, 4 °C), the supernatant was removed, and the cell pellet was flash frozen in N2(l) and stored at −80 °C prior to lipid extractions. For relative growth rate measurements, analogous cultures (initial density of 5×106 cells/mL) were incubated in a sterile 96 well plate with an optical, breathable cover seal (shaking at 1096 rpm). Optical density readings were obtained every 10 min.
Cultures for lipid extract analysis
Cultured WT or Δpuf3 yeast (n = 4) in the following medias: YPD (2% D) (Y3K “F” media), YPGD (3% G, 0.1% D) (Y3K “R” media), SCD (2% D) (fermentation condition), SCGD (3% G, 0.1% D) (respiration condition), SCD+4HB (2% D, 10 μM 4-HB) (fermentation condition), SCGD+4HB (3% G, 0.1% D, 10 μM 4-HB) (respiration condition). Cultured WT yeast harboring empty p426gpd (EV), p426gpd coq5 (high copy), p416gpd coq5 (low copy), p426gpd Coq5p−MTS (NΔ30), p426gpd Coq5p-FLAG, or p426gpd Coq5p−MTS(NΔ30)-FLAG in Ura−D (2% D) (fermentation condition) or Ura−GD (3% G, 0.1% D) (respiration condition). Starter cultures (3 mL YEPD, 3 mL SCD, or 3 mL Ura-, D) were inoculated with an individual colony of yeast and incubated (30 °C, 230 rpm, 10–15 h). Media (100 mL media at ambient temperature in a sterile 250 mL Erlenmeyer flask) was inoculated with 2.5×106 yeast cells and incubated (30 °C, 230 rpm). Sample s of the fermentation cultures were harvested 13 h after inoculation, a time point that corresponds to early fermentation (logarithmic) growth. Samples of respiration cultures were harvested 25 h after inoculation, a time point that corresponds to early respiration growth. For each growth condition, 1×108 yeast cells were pelleted by centrifugation (3,000 g, 3 min, 4 °C), the supernatant was removed, and the cell pellet was flash frozen in N2(l) and stored at −80 °C prior to lipid extractions.
Diauxic shift experiment
Cultured WT or Δpuf3 yeast (n = 3) in 3 mL YEPD. Inoculated 1L YEPD (at ambient temperature in a sterile 4L Erlenmeyer flask) with 2.5×107 yeast cells and incubated (30 °C, 230 rpm). Beginning at 12 hours post-inoculation and proceeding hourly until 23 hours post-inoculation, recorded OD600 for each. At each hourly time point, harvested 2x108 yeast cells (in duplicate) by centrifugation. Pellets were snap frozen and supernatants were saved for glucose analysis. Supernatants were diluted 1:10 and glucose was measured using the Glucose (HK) Assay kit from Sigma (GAHK-20). For Western blot analyses, frozen yeast pellets were lysed in 150 μL of lysis buffer (2 M NaOH, 1 M BME) for 10 min with periodic vortexing. Protein was TCA precipitated with 150 μL of 50% TCA and washed with 1 mL of Acetone. Protein pellet was resuspended in 120 μL of 0.1 M NaOH and 50 μL of 6x LDS sample buffer. 10 μL of this protein extract was run on a gel and subjected to western analysis. Primary antibodies were diluted 1:5000 rabbit anti-coq5 (a generous gift from Catherine Clarke) and mouse anti-actin Sigma #8224. Secondary antibodies were diluted 1:15,000 and included anti-mouse 680 (Licor 925-68020) and anti-rabbit 800 (Licor 925-32211). Western images were obtained using Licor image studio.
Protein overexpression cultures
WT yeast were transformed with plasmids encoding Coq5p, Coq5p-FLAG, Coq8p, Coq9p, Hfd1p, Yjr120w, or Hem25p (p426[2μ]-GPD plasmids) and cultured on Ura−,D plates. Starter cultures (3 mL Ura−,D+4HB) were inoculated with an individual colony of yeast and incubated (30 °C, 230 rpm, 10–15 h). For CoQ quanti tation, Ura−,D+4HB (fermentation) or Ura−,GD+4HB (respiration) media (100 mL media at ambient temperature in a sterile 250 mL Erlenmeyer flask) was inoculated with 2.5×106 yeast cells and incubated (30 °C, 230 rpm). Sample s of the Ura−,D+4HB cultures were harvested 13 h after inoculation. Samples of Ura−,GD+4HB cultures were harvested 25 h after inoculation. For each growth condition, 1×108 yeast cells were pelleted by centrifugation (3,000 g, 3 min, 4 °C), the supernatant was removed, and the cell pe llet was flash frozen in N2(l) and stored at −80 °C prior to lipid extractions. For relative growth rate measurements, analogous cultures (initial density of 5×106 cells/mL) were incubated in a sterile 96 well plate with an optical, breathable cover seal (shaking at 1096 rpm). Optical density readings were obtained every 10 min. Growth rates were determined by fitting a linear equation to the linear growth phase and determining the slope of the line.
Cultures for microscopy
BY4742 S. cerevisiae overexpressing C-terminally FLAG-tagged genes from p416gpd_FLAG plasmids were cultured in Ura−,D media (30 °C, 3 mL starter cultures, ~14 h). Fro m these starter cultures, 1.25×106 cells were used to inoculate Ura−,GD media (50 mL) (respiration culture condition). After incubating 25 hours (30 °C, 230 r .p.m), 1×108 cells were removed from the culture by pipetting and immediately fixed with formaldehyde for microscopy as described below.
Yeast Transformations
Yeast were transformed with plasmids using a standard lithium acetate protocol (Gietz et al., 1992). Briefly, BY4742 yeast were cultured in YEPD (50 mL) to a density of 2×107 cells/mL. Cells were pelleted and washed twice with water. For each transformation, added PEG 3350 (50% w/v, 240 μL), lithium acetate (1 M, 36 μL), boiled salmon sperm DNA (5 mg/mL, 50 μL), water (30 μL), and plasmid (4 μL) to a pellet containing 1×108 cells, mixed by vortexing, and incubated (42 °C, 4 5 min). The transformed cells were pelleted, resuspended in water (100 μL), and plated on selective media.
DNA Constructs
Yeast gene constructs were generated by amplifying the S. cerevisiae genes FMP10, MPM1, RDL2, and YNR040W from strain BY4742 genomic DNA with primers containing HindIII recognition sequence (forward) and SalI recognition sequence (reverse). Similarly, HEM25 and YJR120W were amplified with BamHI (forward) and EcoRI (reverse) primers. COQ5 (from strain W303) was amplified with SpeI (forward) and SalI or XhoI (reverse) primers. Please see Table S4 for a complete list of primers, including sequences, that were used in this study. PCR reactions contained 1× Accuprime PCR mix, 1 μM forward primer, 1 μM reverse primer, ~250 ng template, and 1× Accuprime Pfx (Invitrogen cat#12344024). After an initial 2 min denaturation at 95 °C, reactions were exposed to 5 cycles of 95 °C for 15 seconds, 55 °C for 30 seconds, and 68 °C for 2 minutes followed by 30 cycles of 95 °C for 15 seconds, 60 °C for 30 seconds, and 68 °C for 2 minutes. Amplicons were purified using a PCR purification kit (Thermo cat#K0702) and digested with the appropriate restriction enzymes and again subjected to PCR purification. Amplified genes were cloned into restriction enzyme digested yeast expression vectors (p426gpd, p416gpd, and/or p416gpd_FLAG). The plasmid p416gpd_FLAG was generated by digesting p416gpd with XhoI and MluI and inserting a double stranded oligonucleotide containing the Flag tag nucleotide sequence and processing XhoI and MluI ends. For PUF3, p423(2μ)-PUF3 and p413(CEN)-PUF3 expression plasmids were constructed in modified p423(2μ) or p413(CEN) backgrounds in which the plasmid promoter and terminators had been removed. The PUF3 gene, including 1,000 upstream nts and 457 downstream nts, was amplified from BY4742 genomic DNA using a standard Phusion DNA polymerase PCR. Amplified products were cleaned via a PCR purification kit (ThermoFisher), digested with the SalI and KpnI restriction enzymes under standard conditions, and cloned into the appropriate plasmid. NΔ30 versions of coq5 constructs were generated by PIPE cloning (Klock and Lesley, 2009). All recombinants were confirmed by DNA sequencing. Cloning was previously reported for p426-GPD-coq8 (Stefely et al., 2015), p426-GPD-COQ9 (Lohman et al., 2014), p426-GPD-HFD1 (Stefely et al., 2016a).
HITS-CLIP Class Definition
Puf3p-bound RNAs identified via HITS-CLIP (Wilinski et al., 2017) were sorted by the number of RNAs detected in their CLIP peak (“peak height”) from most to least. Classes were then defined as follows: the top 10% were designated class I; 11–40% class II; 41–70% class III; and 71–100% class IV. Classes were defined to be of comparable size to the analogous RNA Tagging class to facilitate cross-method comparisons (Lapointe et al., 2017; Lapointe et al., 2015). As a control, the signal detected for these mRNAs [log2(TRPM) for RNA Tagging and log2(peak height) for HITS-CLIP] was correlated across methods (Spearman’s ρ = 0.42, P < 10−12), but not with mRNA abundance (Spearman’s < 0.14, P > 0.01). Thus, we did not normalize RNA Tagging or HITS-CLIP signals to mRNA abundance, though that may be important with other RBPs.
MEME Analyses
For all analyses, the 3′ untranslated regions (UTRs) was defined as the longest observed isoform for a gene (Xu et al., 2009) or 200 nts downstream of the stop codon if not previously defined. MEME was run on a local server with the command: meme [input.txt] -oc [outputdirectory] -dna -mod zoops -evt 0.01 -nmotifs 10 -minw 8 -maxw 12 -maxsize 100000000000.
Definition of Puf3p mRNA Targets
Puf3p HITS-CLIP data were obtained from (Wilinski et al., 2017). Only peaks that were assigned to annotated genes were considered: 467 in total, with 15 genes identified by two peaks. RNA Tagging data were obtained from (Lapointe et al., 2017; Lapointe et al., 2015). All 476 reported RNA Tagging targets were considered. RNA Tagging classes were obtained from (Lapointe et al., 2017) because they were generated using an improved strategy than in the initial report (Lapointe et al., 2015). Genes with mRNAs identified by both approaches, visualized by Venn diagrams, were designated as “Puf3p target mRNAs”. Protein-RNA network maps were constructed using Cytoscape (v. 3.2.1) and the ‘Organic’ ‘yFiles Layouts’ option.
Definition of Puf3p Cis Target Proteins
We defined “Puf3p cis target proteins” (interchangeably referred to as “Puf3p cis targets”) as: proteins encoded by Puf3p mRNA targets with at least a 25% significant (P < 0.05) alteration in protein abundance in yeast that lack PUF3 relative to WT yeast grown in fermentation culture conditions. The proteomic data included 165 proteins encoded by Puf3p mRNA targets (out of 269 total), and 91 proteins were designated as Puf3p cis target proteins (“Puf3p cis targets”) out of the 160 proteins with at least a 25% significant alteration in protein abundance. To ensure rigorous definition, we excluded 20 proteins with significantly altered protein abundances because they were identified as Puf3p-bound mRNAs only via a single method, thus confounding their assignment.
Definition of Puf3p Trans Targets
For the proteome, Puf3p trans effects were defined as proteins that were not encoded by Puf3p mRNA targets with at least a 25% significant (P < 0.05) alteration in protein abundance in yeast that lack PUF3 relative to WT yeast grown in fermentation culture conditions. For the metabolome and lipidome, Puf3p trans effects were defined as metabolites or lipids, respectively, with at least a 25% significant (P < 0.05) alteration in abundance in yeast that lack PUF3 relative to WT yeast grown in fermentation culture conditions.
Gene Ontology Analyses
Analyses were conducted using YeastMine, from the Saccharomyces Genome database (http://yeastmine.yeastgenome.org), with the default settings (Holm-Bonferroni correction).
Gene Property Analyses
To test for characteristic properties of Pufp3 cis targets, Puf3p trans effect proteins, and mitochondrial proteins, we compared our data against numerous publicly available data sets. Briefly, all proteins quantified in Δpuf3 yeast (n = 3152, fermentation growth conditions, Y3K data set (Stefely et al., 2016a)) were assigned to one or more of the following categories where appropriate: Puf3p cis targets (n = 91, as defined in this report), Puf3p trans effect proteins (n = 49, as defined in this report), mitochondrial proteins (Jin et al., 2015) (n = 715), and all profiled proteins (n = 3152) in fermenting Δpuf3 yeast. For each gene property analysis, each protein was assigned a qualitative or quantitative value as reported in a publicly available data set if a corresponding value was reported therein. Protein overexpression toxicity was assigned using data from (Gelperin et al., 2005). Enrichment in protein toxicity amongst cis, trans, and mitochondrial proteins was calculated relative to all proteins. Statistical significance of these enrichments was determined via a Fisher’s exact test. Protein abundance, in fermentation and respiration conditions, was assigned by taking the average log2 label free quantitation (LFQ) value from 36 replicates of WT yeast grown as part of the Y3K study (Stefely et al., 2016a). Data sets have been reported previously for cotranslational import of mitochondrial proteins (Williams et al., 2014), protein abundance changes across the diauxic shift (Stefely et al., 2016b), and average respiration deficiency response (RDR) protein abundance change (change in protein abundance in respiration deficient yeast compared to respiration competent yeast) (Stefely et al., 2016a). For these quantitative variable analyses, statistical significance was calculated using a Student’s t-test (two-tailed, homostatic).
Lipid Extractions
Frozen pellets of yeast (108 cells) were thawed on ice and mixed with glass beads (0.5 mm diameter, 100 μL). CHCl3/MeOH (2:1, v/v, 4 °C) (900 μL) and CoQ10 (10 μL, 10 μM, 0.1 nmol) were added and vortexed (2 × 30 s). HCl (1 M, 200 μL, 4 °C) was added and vortexed (2 × 30 s). The samples were centrifuged (5,000 g, 2 min, 4 °C) to complete phase separation. 555 μL of the organic phase was transferred to a clean tube and dried under Ar(g). The organic residue was reconstituted in ACN/IPA/H2O (65:30:5, v/v/v) (100 μL) for LC-MS analysis.
LC-MS Lipid Analysis
LC-MS analysis was performed on an Acquity CSH C18 column held at 50 °C (100 mm × 2.1 mm × 1.7 μm particle size; Waters) using a Vanquish Binary Pump (400 μL/min flow rate; Thermo Scientific). Mobile phase A consisted of 10 mM ammonium acetate in ACN/H2O (70:30, v/v) containing 250 μL/L acetic acid. Mobile phase B consisted of 10 mM ammonium acetate in IPA/ACN (90:10, v/v) with the same additives. Mobile phase B was held at 40% for 6.0 min and then increased to 60% over 3.0. Mobile phase B was further increased to 85% over 0.25 min and then to 99% for over 1.25 min. The column was then reequilibrated for 3.5 min before the next injection. Ten microliters of sample were injected by a Vanquish Split Sampler HT autosampler (Thermo Scientific). The LC system was coupled to a Q Exactive mass spectrometer by a HESI II heated ESI source kept at 325 °C (Thermo Scientific). The inlet capillary was kept at 350 °C, sheath gas was set to 25 units, and auxiliary gas to 10 units, and the spray voltage was set to 3,000 V. The MS was operated in positive and negative parallel reaction monitoring (PRM) mode acquiring scheduled, targeted PRM scans to quantify key CoQ intermediates. Phospholipids were quantified and identified using a negative dd-Top2 scanning mode.
LC-MS Lipid Data Analysis
CoQ intermediate data were processed using TraceFinder 4.0 (Thermo Fisher Scientific). Discovery lipidomic data were processed using an in-house software pipeline and Compound Discoverer 2.0 (Thermo Fisher Scientific).
Fluorescence Microscopy
Yeast (1×108 cells) transformed with various FLAG tagged constructs were removed from cultures by pipetting and immediately fixed with formaldehyde (4% final concentration, gentle agitation on a nutator, 1 h, ~23 °C). The fixed cells were harvested by centrifugation (1000 g, 2 min, ~23 °C), washed three times with 0.1 M potassium phosphate pH 6.5 and once with K-Sorb buffer (5 mL, 1.2 M sorbitol, 0.1 M KPi, pH 6.5), and re-suspended in K-Sorb buffer (1 mL). An aliquot of the cells (0.5 mL) was mixed with K-Sorb-BME (0.5 mL, K-Sorb with 140 mM BME) and incubated (~5 min, ~23 °C). Zymolase 100T was added to 1 mg/mL final concentration and incubated (20 min, 30 °C). The resultant spheroplasts were harvested by centrifugation (1000 g, 2 min, ~23 °C), washed once with K-Sorb buffer (1.4 mL), and re-suspended in K-Sorb buffer (0.5 mL). A portion of the cells (0.25 mL) was pipetted onto a poly-D-lysine coated microscope coverslip and allowed to settle onto the slides (20 min, ~23 °C). To permeabilize the cells, the supernatant was aspirated from the coverslips, and MeOH (2 mL, −20 °C) was added immediately and incubated (6 min, on ice). The MeOH was aspirated and immediately replaced with acetone (2 mL, −20 °C) and incubated (30 s, on ice). The acetone was aspirated, and the slides were allowed to air-dry (~2 min). DNA was stained with Hoechst 33342 dye (1 μg/mL in PBS, 2 mL, 5 min incubation, protected from light), and the cells were immediately wash with PBS. The samples were blocked with BSA (“BSA-PBS” [1% BSA in PBS], 2 mL, 30 min incubation at ~23 °C), and incubated with primary antibodies (1 mg/mL stock anti-FLAG primary Ab [Sigma F1804] at a 1:2000 dilution in PBS-BSA; anti-Cit1p antibody [Biomatik Anti-SA160118(Ser)] at a 1:500 dilution in PBS-BSA; 1 mL, 12 h, 4 °C). The anti-Cit1p antibody used in this experiment was generated against the peptide CRPKSFSTEKYKELVKKIESKN (Biomatik, AB001455, Anti-SA160118(Ser), lot # A160414-SF001455, peptide 506543, rabbit RB7668, 0.52 mg/mL). The samples were washed 5 times with PBS-BSA (2 mL, ~23 °C) and incubated with secondary antibodies diluted in PBS-BSA [1 μg/mL working concentration for each: Goat anti-Mouse IgG (H+L) Secondary Antibody, Alexa Fluor 594 conjugate (Thermo A-11005) and Goat anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor 488 (Thermo Cat# A-11008)] (1 mL, 2 h, ~23 °C, in the dark). The samples were washed 5 times with PBS-BSA (2 mL, ~23 °C) and twice with PBS (2 mL, ~23 °C). The last wash was aspirated and the slides were allowed to air dry briefly in the dark. The coverslips were mounted onto slides with 50% glycerol in PBS (8 μL). Fluorescence microscopy was performed on a Keyence BZ-9000 microscope using 100X oil immersion optics at room temperature. Line scan analysis was performed with ImageJ.
QUANTIFICATION AND STATISTICAL ANALYSIS
Overview of Statistical Analyses
For each reported P value (P), the statistical test used is reported in the legend for the corresponding figure panel. The majority of P values in this report were calculated using an unpaired, two-tailed, Student’s t-test. In a few select instances, as noted, P values for hypergeometric tests and Spearman correlation coefficients were calculated using the R software suite. Also, as noted above, a Fischer’s exact test was used for a few select qualitative gene set analyses. For yeast experiments, all instances where n replicates are reported had n biological replicates. As detailed above, for the gene and protein set analyses, n indicates the number of genes or proteins in each set: Puf3p cis targets (n = 91), Puf3p trans effect proteins (n = 49), mitochondrial proteins (n = 715), and all profiled proteins (n = 3152) in fermenting Δpuf3 yeast.
Supplementary Material
(A) Key describing each column in sheets (tables) (B) – (D) of this data set: Gene systematic name, Gene standard name, RNA Tagging Class, Tagging U1 TRPM log2, Tagging PBE, CLIP peak height, CLIP P value, CLIP Class, Number of peaks, Peak location, CLIP PBE, Method. (B) Table of Puf3p mRNA targets, including a summary of RNA Tagging and HITS-CLIP data for each target. (C) Table of all mRNAs identified as a Puf3p-bound mRNA in either approach, including a summary of RNA Tagging and HITS-CLIP data for each mRNA. (D) Table of all genes, highlighting mRNAs identified as a Puf3p-bound mRNA in either approach, including a summary of RNA Tagging and HITS-CLIP data for each mRNA. (E) Table of all genes, highlighting which method identified them as putative Puf3p targets (HITS-CLIP, RNA Tagging, PAR-CLIP, or RIP-seq). It includes a list of the aggregate 2,018 mRNAs identified as putative Puf3p targets.
(A) Key describing each column in sheets (tables) (B) – (D) of this data set: Gene systematic name, Gene standard name, Pathway, Protein effect (cis, trans, or unclear), RNA Tagging Class, Tagging U1 TRPM log2, CLIP peak height, CLIP P value, CLIP Class, Number of peaks, Peak location, Method, Proteomics log2(KO/WT) in fermentation, Proteomics P value in fermentation, and 25% sig protein change. (B) Table of Puf3p cis targets, including a summary of RNA Tagging, HITS-CLIP, and proteomics data for each target. (C) Table of Puf3p trans effect proteins, including a summary of the proteomics data for each protein. (D) Table of all genes and all proteins in the multi-omic analysis, including a summary of RNA Tagging, HITS-CLIP, and proteomics data for each molecule.
(A) Table of Gene Ontology (GO) terms enriched in the Puf3p cis target set. Includes GO terms, P values, −log10(P values), genes in the Puf3p cis target set that overlap with those in the GO term set, and GO term ID number. (B) Table of Gene Ontology (GO) terms enriched in the set of trans effect proteins with increased protein abundance in yeast that lack Puf3p. Includes GO terms, P values, −log10(P values), genes in the Puf3p trans effect set that overlap with those in the GO term set, and GO term ID number.
List of primer names, sequence, and purpose that were used in this study.
HIGHLIGHTS.
The RNA-binding protein (RBP) Puf3p regulates coenzyme Q (CoQ) biosynthesis
Multi-omic analysis of RNAs, proteins, lipids and metabolites defines Puf3p targets
Puf3p regulates the potentially toxic CoQ biosynthesis enzyme Coq5p
Puf3p couples regulation of CoQ with a broader program for controlling mitochondria
Acknowledgments
We thank members of the Pagliarini, Wickens, and Coon laboratories for helpful discussions. We also thank Catherine Clarke (UCLA) for the generous gift of anti-coq5p antibody. This work was supported by a UW2020 award and National Institutes of Health (NIH) grants R01GM112057 and R01GM115591 (to D.J.P.); NIH P41GM108538 (to J.J.C. and D.J.P.); NIH R01GM50942 (to M.P.W.); NIH R35GM118110 (to J.J.C.); Department of Energy Great Lakes Bioenergy Research Center (Office of Science BER DE-FC02-07ER64494 to N.W.K.); NIH fellowship T32GM008349 (to G.M.W.); Wharton and Biochemistry Scholar fellowships (to C.P.L.); and NIH Ruth L. Kirschstein NRSA F30AG043282 (to J.A.S.).
Footnotes
AUTHOR CONTRIBUTIONS
C.P.L., J.A.S., M.P.W., and D.J.P. conceived of the project and its design and wrote the manuscript. J.A.S. and A.J. prepared samples and performed biochemical experiments. P.D.H. and G.W. acquired MS data. C.P.L., J.A.S., A.J., P.D.H., G.M.W., N.W.K., J.J.C., M.P.W., and D.J.P. analyzed data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chatenay-Lapointe M, Shadel GS. Repression of Mitochondrial Translation, Respiration and a Metabolic Cycle-Regulated Gene, SLF1, by the Yeast Pumilio-Family Protein Puf3p. Plos One. 2011:6. doi: 10.1371/journal.pone.0020441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Soto IC, Shipkovenska G, Churchman LS. Synchronized mitochondrial and cytosolic translation programs. Nature. 2016;533:499–503. doi: 10.1038/nature18015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliyahu E, Pnueli L, Melamed D, Scherrer T, Gerber AP, Pines O, Rapaport D, Arava Y. Tom20 Mediates Localization of mRNAs to Mitochondria in a Translation-Dependent Manner. Mol Cell Biol. 2010;30:284–294. doi: 10.1128/MCB.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd BJ, Wilkerson EM, Veling MT, Minogue CE, Xia C, Beebe ET, Wrobel RL, Cho H, Kremer LS, Alston CL, et al. Mitochondrial Protein Interaction Mapping Identifies Regulators of Respiratory Chain Function. Mol Cell. 2016;63:621–632. doi: 10.1016/j.molcel.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeberg MA, Han T, Moresco JJ, Kong A, Yang YC, Lu ZJ, Yates JR, Kim JK. Pervasive and dynamic protein binding sites of the mRNA transcriptome in Saccharomyces cerevisiae. Genome Biol. 2013;14:R13. doi: 10.1186/gb-2013-14-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadir N, Haim-Vilmovsky L, Kraut-Cohen J, Gerst JE. Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. Rna. 2011;17:1551–1565. doi: 10.1261/rna.2621111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Schatton D, Martinelli P, Hansen H, Pla-Martin D, Barth E, Becker C, Altmueller J, Frommolt P, Sardiello M, et al. CLUH regulates mitochondrial biogenesis by binding mRNAs of nuclear-encoded mitochondrial proteins. J Cell Biol. 2014;207:213–223. doi: 10.1083/jcb.201403129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rodriguez LJ, Gay AC, Pon LA. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J Cell Biol. 2007;176:197–207. doi: 10.1083/jcb.200606054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. Plos Biol. 2004;2:342–354. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Niemi NM, Hutchins PD, Condon SG, Jochem A, Ulbrich A, Higbee AJ, Russell JD, Senes A, Coon JJ, et al. Ptc7p Dephosphorylates Select Mitochondrial Proteins to Enhance Metabolic Function. Cell Rep. 2017;18:307–313. doi: 10.1016/j.celrep.2016.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CH, Xie LX, Allan CM, Tran UC, Clarke CF. Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochim Biophys Acta. 2014;1841:630–644. doi: 10.1016/j.bbalip.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Merrill AE, Stefely JA, Bailey DJ, Wenger CD, Westphall MS, Pagliarini DJ, Coon JJ. Amine-reactive neutron-encoded labels for highly plexed proteomic quantitation. Mol Cell Proteomics. 2013;12:3360–3369. doi: 10.1074/mcp.M113.032011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan GJ, Brown PO, Herschlag D. Evolutionary Conservation and Diversification of Puf RNA Binding Proteins and Their mRNA Targets. Plos Biol. 2015:13. doi: 10.1371/journal.pbio.1002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houshmandi SS, Olivas WM. Yeast Puf3 mutants reveal the complexity of Puf-RNA binding and identify a loop required for regulation of mRNA decay. Rna. 2005;11:1655–1666. doi: 10.1261/rna.2168505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inadome H, Noda Y, Adachi H, Yoda K. A novel protein, Mpm1, of the mitochondria of the yeast Saccharomyces cerevisiae. Biosci Biotechnol Biochem. 2001;65:2577–2580. doi: 10.1271/bbb.65.2577. [DOI] [PubMed] [Google Scholar]
- Jackson JS, Houshmandi SS, Leban FL, Olivas WM. Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast. Rna. 2004;10:1625–1636. doi: 10.1261/rna.7270204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin K, Musso G, Vlasblom J, Jessulat M, Deineko V, Negroni J, Mosca R, Malty R, Nguyen-Tran DH, Aoki H, et al. Yeast mitochondrial protein-protein interactions reveal diverse complexes and disease-relevant functional relationships. J Proteome Res. 2015;14:1220–1237. doi: 10.1021/pr501148q. [DOI] [PubMed] [Google Scholar]
- Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- Kershaw CJ, Costello JL, Talavera D, Rowe W, Castelli LM, Sims PFG, Grant CM, Ashe MP, Hubbard SJ, Pavitt GD. Integrated multi-omics analyses reveal the pleiotropic nature of the control of gene expression by Puf3p. Sci Rep-Uk. 2015:5. doi: 10.1038/srep15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klock HE, Lesley SA. The Polymerase Incomplete Primer Extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol Biol. 2009;498:91–103. doi: 10.1007/978-1-59745-196-3_6. [DOI] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives (vol 13, pg 77, 2012) Nat Rev Genet. 2012;13:221–221. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- Labbe K, Murley A, Nunnari J. Determinants and functions of mitochondrial behavior. Annu Rev Cell Dev Biol. 2014;30:357–391. doi: 10.1146/annurev-cellbio-101011-155756. [DOI] [PubMed] [Google Scholar]
- Lapointe CP, Preston MA, Wilinski D, Saunders HAJ, Campbell ZT, Wickens M. Architecture and dynamics of overlapped RNA regulatory networks. Rna. 2017 doi: 10.1261/rna.062687.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe CP, Wilinski D, Saunders HA, Wickens M. Protein-RNA networks revealed through covalent RNA marks. Nat Methods. 2015;12:1163–1170. doi: 10.1038/nmeth.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laredj LN, Licitra F, Puccio HM. The molecular genetics of coenzyme Q biosynthesis in health and disease. Biochimie. 2014;100:78–87. doi: 10.1016/j.biochi.2013.12.006. [DOI] [PubMed] [Google Scholar]
- Lee CD, Tu BP. Glucose-Regulated Phosphorylation of the PUF Protein Puf3 Regulates the Translational Fate of Its Bound mRNAs and Association with RNA Granules. Cell Rep. 2015;11:1638–1650. doi: 10.1016/j.celrep.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Ohn T, Chiang YC, Quigley G, Yao G, Liu YT, Denis CL. PUF3 Acceleration of Deadenylation in Vivo Can Operate Independently of CCR4 Activity, Possibly Involving Effects on the PAB1-mRNP Structure. J Mol Biol. 2010;399:562–575. doi: 10.1016/j.jmb.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang XN, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–U422. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman DC, Forouhar F, Beebe ET, Stefely MS, Minogue CE, Ulbrich A, Stefely JA, Sukumar S, Luna-Sanchez M, Jochem A, et al. Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc Natl Acad Sci U S A. 2014;111:E4697–4705. doi: 10.1073/pnas.1413128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J, Horwich AL, Hartl FU. Prevention of protein denaturation under heat stress by the chaperonin Hsp60. Science. 1992;258:995–998. doi: 10.1126/science.1359644. [DOI] [PubMed] [Google Scholar]
- Miller MA, Russo J, Fischer AD, Leban FAL, Olivas WM. Carbon source-dependent alteration of Puf3p activity mediates rapid changes in the stabilities of mRNAs involved in mitochondrial function. Nucleic Acids Research. 2014;42:3954–3970. doi: 10.1093/nar/gkt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivas W, Parker R. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. Embo J. 2000;19:6602–6611. doi: 10.1093/emboj/19.23.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payet LA, Leroux M, Willison JC, Kihara A, Pelosi L, Pierrel F. Mechanistic Details of Early Steps in Coenzyme Q Biosynthesis Pathway in Yeast. Cell Chem Biol. 2016;23:1241–1250. doi: 10.1016/j.chembiol.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, Workman JL, Yates JR, Grant PA. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol. 2002;22:8774–8786. doi: 10.1128/MCB.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenault T, Lithgow T, Traven A. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011;21:104–112. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res. 2006;5:1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- Riley KJ, Steitz JA. The “Observer Effect” in genome-wide surveys of protein-RNA interactions. Mol Cell. 2013;49:601–604. doi: 10.1016/j.molcel.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe W, Kershaw CJ, Castelli LM, Costello JL, Ashe MP, Grant CM, Sims PFG, Pavitt GD, Hubbard SJ. Puf3p induces translational repression of genes linked to oxidative stress. Nucleic Acids Research. 2014;42:1026–1041. doi: 10.1093/nar/gkt948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, Lemoine S, Tanty V, Devaux F, Jacq C. Yeast Mitochondrial Biogenesis: A Role for the PUF RNA-Binding Protein Puf3p in mRNA Localization. Plos One. 2008:3. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatton D, Pla-Martin D, Marx MC, Hansen H, Mourier A, Nemazanyy I, Pessia A, Zentis P, Corona T, Kondylis V, et al. CLUH regulates mitochondrial metabolism by controlling translation and decay of target mRNAs. J Cell Biol. 2017;216:675–693. doi: 10.1083/jcb.201607019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassov DS, Jurecic R. The PUF family of RNA-binding proteins: Does evolutionarily conserved structure equal conserved function? Iubmb Life. 2003;55:359–366. doi: 10.1080/15216540310001603093. [DOI] [PubMed] [Google Scholar]
- Stefely JA, Kwiecien NW, Freiberger EC, Richards AL, Jochem A, Rush MJ, Ulbrich A, Robinson KP, Hutchins PD, Veling MT, et al. Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling. Nat Biotechnol. 2016a;34:1191–1197. doi: 10.1038/nbt.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefely JA, Licitra F, Laredj L, Reidenbach AG, Kemmerer ZA, Grangeray A, Jaeg-Ehret T, Minogue CE, Ulbrich A, Hutchins PD, et al. Cerebellar Ataxia and Coenzyme Q Deficiency through Loss of Unorthodox Kinase Activity. Mol Cell. 2016b;63:608–620. doi: 10.1016/j.molcel.2016.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefely JA, Pagliarini DJ. Biochemistry of Mitochondrial Coenzyme Q Biosynthesis. Trends Biochem Sci. 2017;42:824–843. doi: 10.1016/j.tibs.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefely JA, Reidenbach AG, Ulbrich A, Oruganty K, Floyd BJ, Jochem A, Saunders JM, Johnson IE, Minogue CE, Wrobel RL, et al. Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthesis. Mol Cell. 2015;57:83–94. doi: 10.1016/j.molcel.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 2006;409:329–345. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- Vogtle FN, Burkhart JM, Rao S, Gerbeth C, Hinrichs J, Martinou JC, Chacinska A, Sickmann A, Zahedi RP, Meisinger C. Intermembrane space proteome of yeast mitochondria. Mol Cell Proteomics. 2012;11:1840–1852. doi: 10.1074/mcp.M112.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′ UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Wilinski D, Buter N, Klocko AD, Lapointe CP, Selker EU, Gasch AP, Wickens M. Recurrent rewiring and emergence of RNA regulatory networks. Proceedings of the National Academy of Sciences. 2017 doi: 10.1073/pnas.1617777114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZY, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–U1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Fukui K, Takahashi H, Kitamura S, Shiota T, Terao K, Uchida M, Esaki M, Nishikawa S, Yoshihisa T, et al. Roles of Tom70 in import of presequence-containing mitochondrial proteins. J Biol Chem. 2009;284:31635–31646. doi: 10.1074/jbc.M109.041756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Stumpf CR, Krahn JM, Wickens M, Hall TM. A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc Natl Acad Sci U S A. 2009;106:20192–20197. doi: 10.1073/pnas.0812079106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Key describing each column in sheets (tables) (B) – (D) of this data set: Gene systematic name, Gene standard name, RNA Tagging Class, Tagging U1 TRPM log2, Tagging PBE, CLIP peak height, CLIP P value, CLIP Class, Number of peaks, Peak location, CLIP PBE, Method. (B) Table of Puf3p mRNA targets, including a summary of RNA Tagging and HITS-CLIP data for each target. (C) Table of all mRNAs identified as a Puf3p-bound mRNA in either approach, including a summary of RNA Tagging and HITS-CLIP data for each mRNA. (D) Table of all genes, highlighting mRNAs identified as a Puf3p-bound mRNA in either approach, including a summary of RNA Tagging and HITS-CLIP data for each mRNA. (E) Table of all genes, highlighting which method identified them as putative Puf3p targets (HITS-CLIP, RNA Tagging, PAR-CLIP, or RIP-seq). It includes a list of the aggregate 2,018 mRNAs identified as putative Puf3p targets.
(A) Key describing each column in sheets (tables) (B) – (D) of this data set: Gene systematic name, Gene standard name, Pathway, Protein effect (cis, trans, or unclear), RNA Tagging Class, Tagging U1 TRPM log2, CLIP peak height, CLIP P value, CLIP Class, Number of peaks, Peak location, Method, Proteomics log2(KO/WT) in fermentation, Proteomics P value in fermentation, and 25% sig protein change. (B) Table of Puf3p cis targets, including a summary of RNA Tagging, HITS-CLIP, and proteomics data for each target. (C) Table of Puf3p trans effect proteins, including a summary of the proteomics data for each protein. (D) Table of all genes and all proteins in the multi-omic analysis, including a summary of RNA Tagging, HITS-CLIP, and proteomics data for each molecule.
(A) Table of Gene Ontology (GO) terms enriched in the Puf3p cis target set. Includes GO terms, P values, −log10(P values), genes in the Puf3p cis target set that overlap with those in the GO term set, and GO term ID number. (B) Table of Gene Ontology (GO) terms enriched in the set of trans effect proteins with increased protein abundance in yeast that lack Puf3p. Includes GO terms, P values, −log10(P values), genes in the Puf3p trans effect set that overlap with those in the GO term set, and GO term ID number.
List of primer names, sequence, and purpose that were used in this study.