Abstract
We report 48 putative DNA binding with one finger (Dof) TF genes from genome and transcriptome data of finger millet (Eleusine coracana L.; FM), involved in plant developmental process. To characterize seed-specific Dof genes, transcript profiles of 32 EcDof identified from transcriptome data of developing spikes of FM genotypes were further analyzed in different tissues (root, stem, and leaf) and developmental stages of spikes (S1, S2, S3, and S4) in two FM genotypes [GE1437 (low protein genotype; LPG) and GE3885 (high protein genotype; HPG)]. More than 50% of identified EcDof genes showed expression during seed development processes. Among these, seven genes (EcDof 3, EcDof 5, EcDof 15, EcDof 18, EcDof 22, EcDof 23, and EcDof 31) expressed maximally at specific stages of seed development. Fourteen EcDof genes showed that differential transcript accumulation in vegetative tissue as well as in developing spikes suggests involvement during seed filling and also throughout the plant development. In addition, three EcDof genes (EcDof 9, EcDof 25, and EcDof 28) expressed preferentially at root and stem tissue. The 3D structural prediction of EcDof proteins showed variability in structural attributes. Molecular docking results showed strong binding affinity for seed-specific EcDof–EcO2 with α-prolamine promoters. The identified and characterized EcDof genes will help to dissect the roles of FM seed-specific Dof genes.
Electronic supplementary material
The online version of this article (10.1007/s13205-017-1068-z) contains supplementary material, which is available to authorized users.
Keywords: Cis-regulatory elements, Dof transcription factors, Eleusine coracana, Finger millet, Seed-storage protein, Transcriptome
Introduction
In finger millet (Eleusine coracana L.) and other cereal crops, the grain filing is the most important process of seed development that ultimately influences the enhanced cereal production with high nutritive values. Cereal provides more than ~ 70% of world’s caloric intake and protein requirement of our daily diet (Varshney et al. 2006). However, according to world health organization (WHO) recent reports, more than ~ 60% population of the India is under-nourishment and facing serious protein energy malnutrition (PEM) problems mainly in the children and women belonging to rural areas. To meet this challenge, bio-fortification of cereals with essential nutrients along with high yield is essential.
Finger millet (FM; Family Poaceae), also known as Ragi, is an important tropical food crop that offers both nutritional and livelihood security for human being and fodder security (Kumar et al. 2016). As compared to other cereals, FM contains more proteins (~5–12%), essential amino acids (Try, Cys, Met etc.), vitamins, fibre, minerals, low fat, high calcium, and low glycemic index. However, being economically nutraceutical crop, it is still neglected and less explored. One of the most attractive features of FM is that it is a nitrogen efficient crop that capitalizes on low nitrogen inputs, but the protein content of FM grain is comparable to other cereals such as wheat and rice, which consume large amounts of nitrogenous fertilizers (Yanagisawa et al. 2004; Gupta et al. 2013). Therefore, it will be an interesting endeavor to understand the holistic unique molecular mechanism of protein accumulation during the grain filling in FM under low nitrogen conditions by characterization of the DNA binding with one finger (Dof) transcription factors (TFs) gene families through transcriptomic studies for better nutraceutical management.
Knowledge accrued so far has shown that a large number of TFs are involved in transcriptional regulation of seed development and grain filing. Among these, plant-specific Dof TFs have master regulatory properties and have been associated with many biological processes unique to plants, which opened new avenues to engineer crops for better nutraceutical management. The presence of Dof-binding sites in the promoter region of seed-specific genes suggests important roles of Dof TFs in regulatory network of seed development (Gaur et al. 2011). The one type of Dof proteins recognizes the Prolamin-box (P-box or TGTAAAG), therefore, named prolamin-binding factor (PBF). These PBF proteins share a high degree of homology in their protein sequences and are specifically expressed during the grain filling stage (Gupta et al. 2011, Mena et al. 1998). In addition, studies of rice genome sequence have shown that there are 30 genes in Dof family, which play an important role during grain filling and regulate the genes involved in pathways of starch and protein synthesis in an ordered fashion (Gaur et al. 2011). It has also been reported that Opaque2 (O2), a bZIP family TF and PBF Dof family members, jointly functions in regulating expression of seed-storage proteins during seed development (Dong et al. 2007, Marzábal et al. 2008, Yamamoto et al. 2006). PBFs bind to the promoter of storage protein gene and interact with O2 that binds to the same promoter in the adjacent region and regulate the synthesis of zein proteins and endosperm starch synthesis during seed development in maize (Zhang et al. 2016).
This paper reports in silico identification of putative seed-specific Dof TF gene families using genomic and transcriptomic data of FM, its annotation and motif analysis, phylogenetic relationship analysis, 3D protein structure prediction, and elucidating their putative functions. Efforts were made to characterize the candidate Dof TF gene(s) that regulates other genes, which are involved in seed-protein accumulation during grain filling in FM. Molecular docking studies were also carried among seed-specific Dof-Opaque 2 TFs with prolamine genes for investigating the probable interaction between them. Our findings enhance knowledge to identify and understand the role of seed-specific Dof TFs gene(s) in FM that will be replicated into other crops for better nutraceutical management and development of functional designer foods.
Materials and methods
Plant Material and selection of stages for sample collection
The seeds of finger millet (FM) genotypes were collected from CRC, Pantnagar. In the present study, two genotypes of FM were used, namely, GE3885 (HPG) and GE1437 (LPG) that have contrasting grain protein (13.8 and 6.2%), respectively. Seedlings were maintained in polyhouse (37–40 ± 2°C; RH = 40 ± 2%) at G.B.P.U.A. &T., Pantnagar (29°N latitude, 79.3°E longitude; 243.8 m asl), Uttarakhand, India.
Four different developmental stages of the spike, booting, or inflorescence emergence, anthesis, grain filling and grain ripening or maturation were identified on the basis of morphology and development stage of ovary and anthers and were designated as S1, S2, S3, and S4, respectively. For qPCR analysis, root, stem, and leaf tissues were collected at 60 days after sowing. Samples were collected from root tips (1–1.5 cm) and 1/3rd portion from the tip of the third leaf. Stem samples were collected from the top (1–1.5 cm) including the shoot apical meristem (SAM) of the main tiller. Spike samples were collected from the 1/3rd portion from the tip for transcript profiling studies. All these samples were collected in the forenoon (between 0800 and 0900 h). At least three independent biological replicates of each tissue sample were harvested and immediately frozen in liquid nitrogen and stored at –80°C until further use.
Identification of Dof gene family in FM
The next generation sequencing reads of two FM genotypes (GP-1 and GP-45) were assembled using Trinity assembler (http://www.trinity-software.com/) and made a local transcriptome database for both genotypes of FM (TSA accessions SRR1151079 and SRR1151080). The Dof sequences were retrieved by performing local BLAST against transcriptome local database of FM using rice and sorghum Dof sequences as a query downloaded from available database (http://rice.plantbiology.msu.edu/ and http://www.plantgdb.org/SbGDB/). The sequences of Dof were characterized by the presence and the absence of Dof domain and confirmed by BLASTn.
The deposited genomic sequence data of FM from NCBI were downloaded (accession ID SRP081350) and local tBlastn analysis was performed using Dof domain and complete Dof proteins as query. Dof domain sequence was used as query, since it is intronless. Around 1 kb region, upstream and downstream regions around the resulted local tBlastn hits were extracted and were processed through the FGENESH gene finding algorithm (http://www.softberry.com/berry.phtmltopic=fgenesh). The 76 protein sequence hence generated was analyzed using the expasy tool “decrease redundancy” (https://web.expasy.org/decrease_redundancy/), and finally, 48 non-redundant Dof proteins were identified in FM genome (Supplementary file 1).
Phylogenetic analysis of Dof genes
The putative CDS of Dof genes were translated to protein sequence using the ExPASy Translate tool. Multiple sequence alignment were made using ClustalW2 program (https://www.ebi.ac.uk/Tools/msa/clustalw2/), on the basis of the protein sequences of EcDof genes.
Dof family protein sequences from rice and sorghum were retrieved from NCBI (Supplementary file 1), aligned with putative Dof genes identified from FM genome using ClustalW and phylogenetic tree was constructed in the MEGA 6.0 software (Tamura et al. 2011). Another NJ tree was also constructed using putative EcDOF proteins identified from developing spikes transcriptome data of FM and reported PBF Dof proteins from other crops using MEGA 6.0 software.
Investigation of conserved motifs in EcDof genes
To identify the conserved motifs within putative Dof proteins, the protein sequences of identified EcDof genes were analyzed using multiple expectation maximization for motif elicitation (MEME) tool version 4.11.1(http://meme-suite.org/tools/meme) (Bailey et al. 2006). For the analysis, maximum number of motifs was set to 15 and optimal motif width was set as 6–50 amino acids, while other parameters were set as default and the consensus motif sequences of Dof were generated using motif alignment and search tool (MAST) (http://meme-suite.org/tools/mast).
RNA isolation and cDNA synthesis
Total RNA was isolated from different vegetative tissues (root, stem, and leaf) and all four stages of developing spikes (S1, S2, S3, and S4) of FM genotypes (HPG and LPG), using IRIS total RNA isolation kit (IHBT, Palampur, India). Total RNA (2 µg) was used to synthesize first-strand cDNA using oligo (dT)18 primer with Revert AidTM H-Minus M-MuLV Reverse Transcriptase (RT) (Fermentas, Int. Inc., Canada).
Transcript profiling of the identified Dof TF genes
Real-time PCR (qPCR) was carried out using the 5׳ Real Master Mix SYBR ROX according to manufacturer’s instructions (Eppendorf, India). Gene-specific Dof genes primers were designed using online tool (https://www.genscript.com/tools/real-time-pcr-primer-design-tool). The detailed list of gene-specific primers is given in Supplementary Table 1. Tubulin gene primer was used as internal control. The temperature profiles used for qPCR analysis were 95 °C for 2 min initial denaturation followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30. All samples were amplified in triplicate, and the mean Ct value was considered. The normalized transcript expression was quantified using 2−ΔΔCT method (Livak and Schmittgen 2001) and presented as fold change over control (calibrator).
Table 1.
List of top ten templates used by I-TASSER for 3D structure predictions of DOF and opaque2 proteins
| Gene names | PDB IDs |
|---|---|
| EcDof3 | 2i13A, 3dpaA, 2i13A, 1pdiR, 1pdiR, 3ov0A, 3swrA, 4gatA, 1m11R, 21t7A |
| EcDof8 | 3chnS, 3poyA, 1g9bA, 2zxqA, 2fhbA, 2pffH, 4acqA, 4bedB, 2fgzA, 3ecqB |
| EcDof14 | 3chnS, 4f91B, 1hn0A, 4a5wA, 3cmwC, 2q1fB, 2zxqA, 2fgzA, 4o9xA, 3cmvD |
| EcDof15 | 3chnS, 3poyA, 3cmxD, 3hmjG, 4f91B, 3cmuA, 4bedA, 1g9cA, 3k1gA, 4o9xA |
| EcDof16 | 3chnS, 3j65R, 3j65R, 4bmlA, 3j2kA, 4gatA, 1w0rA, 3gawA, 4bmlA, 4bmlA |
| EcDof19 | 3chnS, 1pclA, 2cseH, 1w0sA, 3j3iA, 4gatA, 1w0rA, 3gawA, 2cseH, 4bmlA |
| EcDof20 | 1pclA, 1pclA, 3iyhA, 1w0sA, 3j3iA, 4gatA, 1w0rA, 1w0sA, 3j65R, 2ocwA |
| EcDof21 | 2ic4A, 3j65 N, 1pdiR, 3k7aM, 4gatA, 1m11R, 2fiyA, 4bmlA, 2cp6A, 1pqvA |
| EcDof22 | 4bmlA, 2xd8B, 2fs3A, 2xyzA, 2xvrA,2e0zA, 3c5bA, 3j4uA, 3bjqJ, 4an5A |
| EcDof24 | 1zlgA, 3j65R, 3j65R, 3j65R, 3j65R, 3ov0A, 4lmhA, 4gatA, 1w0rA, 3askA |
| Opaque 2 | 2nbiA, 2wtyA, 5ijoJ, 1hjbA, 1gd2E, 2bsgA, 3a5tA, 1s58A, 4btgA, 3btaA |
| ZmDof | 5c2vC, 4n16A, 5iybU, 4bmlA, 1s58A, 4n16A, 2nbiA, 5dfzB, 1pqvS, 3j65R |
| ZmOpaque 2 | 2nbiA, 2wtyA, 5dfzC, 1gd2E, 1gd2E, 2ocwA, 3h0gA, 1gd2E, 2nbiA, 2nbiA |
3D structure prediction, validation, and visualization
The three-dimensional structures of EcDof, ZmDof, and O2 proteins sequences were predicted by threading algorithm using I-TASSER (Yang et al. 2015). The predicted models were subjected to structural analysis and verification server (SAVES) (http://services.mbi.ucla.edu/SAVES) for quality checking and validation. The chosen models were then subjected to energy minimization by the Swiss-PDB Viewer software (http://www.expasy.org/spdbv) to validate the protein structure using the steepest descent method. Stability of the protein models was further checked by the structural analysis and verification server (http://nihserver.mbi.ucla.edu/SAVES). PyMol was used for the visualization of modeled protein structures (Seeliger and de Groot 2010).
DNA structure modeling of prolamine gene promoters
The 1000 bp upstream region of prolamine gene of FM was isolated by 5′RACE using Universal GenomeWalker Kit. The coding DNA sequences of maize were used as query sequences to perform BLASTn searches to extract the 1000 bp upstream region from transcription start site. Due to the limitation of docking server and nucleotide size restriction of the DNA modeling tool, the region of conserved Dof-binding motif was used to model single-stranded DNA structure of these promoters by the make-na server (http://structure.usc.edu/make-na/server.html).
Molecular docking
Dof domain of all protein structures was docked with the Zinc atom by the AutoDockVina (http://vina.scripps.edu). The coordinated structure of Dof protein of FM and maize with ZINC atom was docked with O2 to make hetero-dimer structures and each hetero-dimer structure complex was further docked with modeled Dof-binding motif by Hex (http://www.hex.loria.fr), to identify the probable interacting DNA-binding sites. Pymol (http://www.pymol.org) was used for visualization and analysis of docking results.
Results and discussion
DNA binding with one finger (Dof) constitutes a large family of TFs that are associated with various biological processes unique to plants (Gupta et al. 2013; Kanwal et al. 2014). The synthesis of grain seed proteins (GSPs) during grain filling is controlled by several mechanisms, including transcriptional and post-transcriptional modifications, and is primarily regulated through a network of interacting transcription factors (TFs) (Verdier and Thompson 2008). Several studies support a role for Dof TFs in the regulation of genes encoding seed-storage proteins during seed maturation (Gaur et al. 2011), nutrient partitioning as well as of genes encoding hydrolases involved in the mobilization of reserves upon seed germination (Mena et al. 2002). Finger millet (FM) seeds contain more proteins and essential amino acids even grown under low nitrogen inputs suggesting that this crop is driven by a strong promoter and regulatory elements that enhance accumulation of seed-storage proteins. In the present study, an attempt has been made to gather comprehensive information of Dof gene family in FM using available transcriptome and genome data (Kumar et al. 2015).
Identification of EcDof genes in finger millet
For the first time, sequences of Dof transcription factor (TF) gene family were retrieved in FASTA format from the transcriptome data of pooled developing spikes of FM deposited at NCBI/GenBank (TSA accession SRR1151079 and SRR1151080). The BLAST analysis of FM transcriptome with Dof genes of sorghum and rice identified a total of 32 EcDof genes with 22 partial and 10 full-length genes (Supplementary Table 2). These include earlier isolated full-length EcDof1 (GenBank Acc. No. ACT37358.2) and partial EcDof2 (GenBank Acc. No. AGQ51639.1) in our lab (Gupta et al. 2014).
Recently, the raw sequence reads of whole genome sequencing are deposited in NCBI SRA database with accession number SRP081350 (Hittalmani et al. 2017). A local tblastn analysis was performed using Dof domain and complete Dof proteins as query. The target hit sequence along with 1 kb up and down stream region was extracted and each of the 76 sequence was analyzed using FGENESH gene prediction online tool. To reduce redundancy, the 76 protein sequence was analyzed using the expasy tool decrease redundancy (https://web.expasy.org/decrease_redundancy/), and finally, 48 non-redundant Dof proteins were identified in FM genome. Comparison of 48 EcDof genes (identified using genome sequence) along with 32 EcDof genes identified from transcriptome data (FM developing spikes), suggested that FM genome has 48 non-redundant Dof genes. Among 48 non-redundant EcDof genes, 42 were full length and 6 were partial EcDof genes (Supplementary file 2). However, the number of Dof genes identified in the present study is far less than the number of Dof genes reported by Hittalmani et al. (2017) which was 93 in number. Maybe, the fragmented copies of Dof genes have been misassembled into different contigs resulting into 93 Dof genes. It has recently been reported that although current sequencing methods produce large amounts of data, the genome assemblies based on these data are often error-filled and incomplete assemblies which result in many annotation errors, especially in the number of genes present in a genome (Denton et al. 2014).
There exists great diversity in terms of number of Dof genes as observed in different crops. In Arabidopsis, Sorghum bicolor and Oryza sativa, 38, 28, and 30 different Dof genes, have been annotated, respectively (Kushwaha et al. 2011). Out of them, only 23 AtDof and 17 OsDof genes were found to express during seed development (Gaur et al. 2011). The presence of greater number of Dof genes in FM as compared to other reported crops encourages to further explore the function of Dof gene family member during seed development. The DOF domain (PF002701) was searched against 48 EcDof proteins, and a typical DOF domain was found in the N-terminal region of most of the putative Dof proteins, further verified them as Dof TF genes (Supplementary Fig. S1). Few Dof genes having short sequences at N terminal do not show a DOF domain, but share high similarity to a region of a known Dof protein from other species (data not shown). The evolutionary relationship between different EcDof genes was further analyzed by multiple sequence alignment of the putative EcDof proteins. The aligned figure revealed that DOF domain of most EcDof genes has highly conserved sequences.
Phylogenetic relatedness among Dof genes and motif analysis
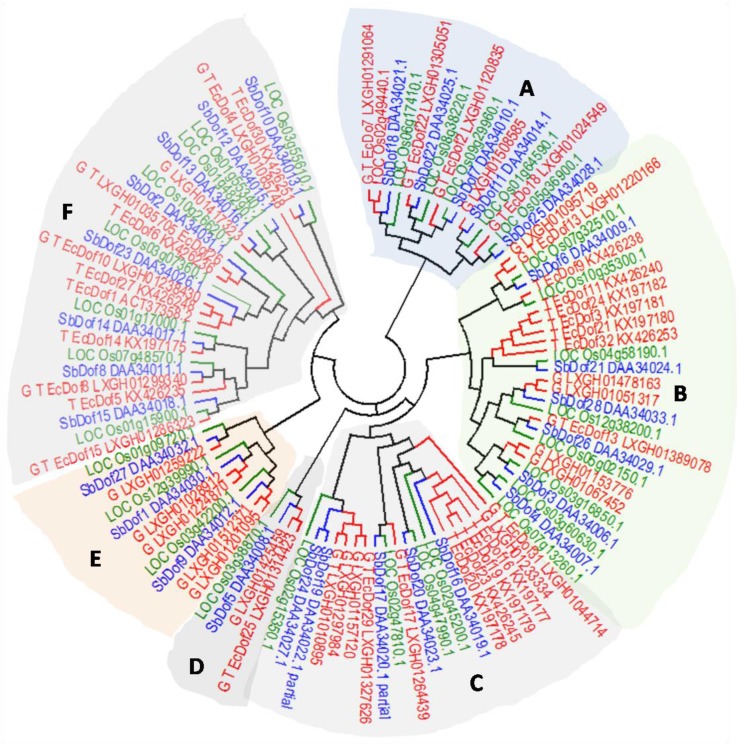
The members of identified FM Dof proteins showed closeness with rice and Sorghum bicolor owing to the monocot nature than to Dof proteins of Arabidopsis representing a dicot. To gain insight into the evolutionary relationship among Dof members, the deduced amino acid sequences of 27 rice, 28 Sorghum, and 48 EcDof genes were aligned for phylogenetic tree construction (Fig. 1). Tree view revealed six groups (A–F) for EcDof proteins. Similarly, total six major groups designated as A–F have been reported for Dof proteins of wheat, rice Arabidopsis, and sorghum (Shaw et al. 2009). Groups B, C, and F represent major groups having 13, 11, and 12 EcDofs, respectively. Groups A, D, and E have 6, 2, and 5 EcDof proteins. The members of group F, i.e., EcDof 14, EcDof15, EcDof8, and EcDof5, show similarity with SbDof14, SbDof15, and SbDof8 that are predicted to be similar to CDF proteins associated with regulation of photoperiodic control of flowering (Kushwaha et al. 2011). Members of group A, EcDof29 and EcDof28, show closeness with SbDof24 and SbDof19 which are associated with the regulation of seed-storage proteins. In addition, EcDof13 showed similarity with OsPBF23 (RPBF) (Washio 2003; Kushwaha et al. 2011).
Fig. 1.
Comparative phylogenetic analysis of 48 Finger millet Dof gene family protein sequences along with Sorghum bicolor and Oryza sativa Dof genes
We further analyzed the conserved motifs in the identified 48 EcDof proteins, using MEME program. Motif analysis revealed the presence of conserved Dof domain (50–52 amino acids) in most of the putative Dof proteins confirming their identity as Dof gene family in FM. Motif analysis supports tree view, as EcDofs in the same group shared similar motifs, suggesting that genes containing the same motifs may be originated from gene expansion of the same group and play crucial roles in group-specific functions (data not shown).
Transcript profiling of EcDof genes in vegetative tissue and developing spikes of finger millet genotypes differing in seed-protein content
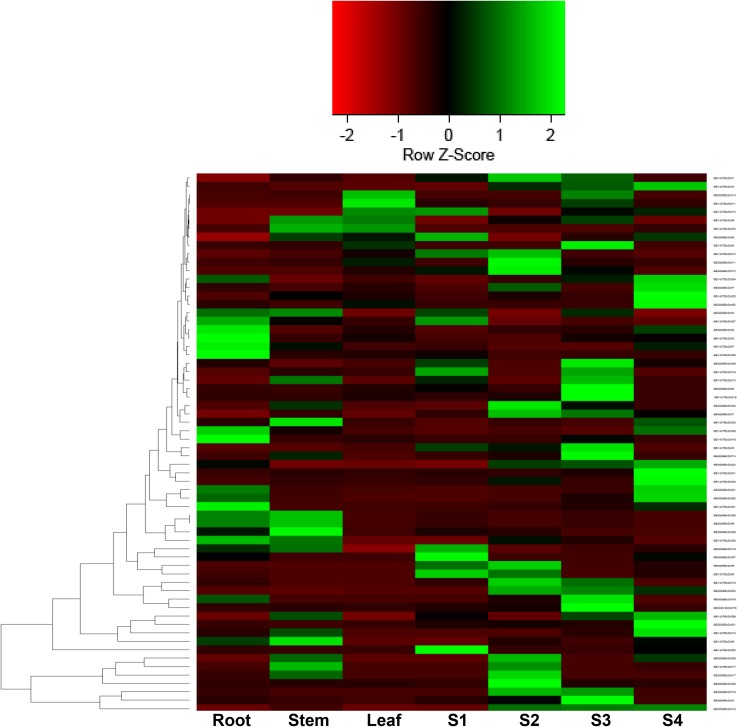
Gene expression patterns can provide important clues for gene function. Since, our aim was to identify seed-specific EcDof genes; therefore, spatial distribution of 32 EcDof genes identified from pooled developing spikes of FM transcriptome was further characterized using qPCR analysis. qPCR analysis of 32 EcDof genes was carried in vegetative tissues (root, stem, and leaf) and developing spikes (S1, S2, S3, and S4 stages) of two FM genotypes differing in seed-protein content [GE-1437 (low protein content; 6.2%) and GE-3885 (high protein content; 13.8%)]. The tubulin gene was used as an internal standard to normalize any variation in the quantity and quality of the starting template cDNA. Transcript analysis revealed spatial variations in the expression of EcDof gens in different tissues in FM genotypes. Heat map revealed that most of the identified EcDof genes showed expression in all tissues whether it is vegetative (root, stem, and leaf) or developing spikes, i.e., S1, S2, S3, and S4 in FM genotypes, although the level of transcript accumulation was differential within genotype as well as tissue wide (Fig. 2). Among genotypes, EcDof transcript accumulation was higher for high seed-protein genotype (GE3885) as compared to low seed-protein genotype (GE1437). On the basis of expression patterns, Dof genes were further categorized having tissue-specific transcript abundance.
Fig. 2.
Heat map of 32 EcDof genes in different tissues (root, stem, and leaf) and developing spikes (S1, S2, S3, and S4 stages) of finger millet (FM) genotypes, GE-1437 (low protein genotype; 6.2%) and GE-3885 (high protein genotype; 13.8%) differing in their grain protein content
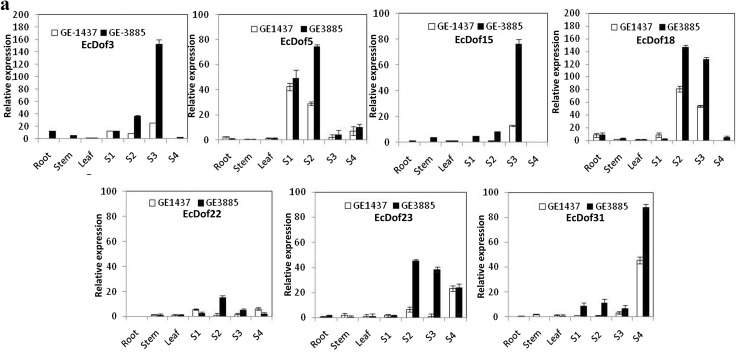
Seven genes EcDof 3, EcDof 5, EcDof 15, EcDof 18, EcDof 22, EcDof 23, and EcDof 31 expressed preferentially in seed developing stages (Fig. 3a). Among them, six genes showed maximal expression at specific stages of seed development viz. EcDof 5 at flowering (S1 stage), five genes (EcDof 3, EcDof 5, EcDof18, EcDof22, and EcDof23) during anthesis (S2 stage), four genes (EcDof 3, EcDof 15, EcDof 18, and EcDof23) at grain filling (S3 stage), and EcDof31 at grain ripening/maturation (S4 stage). The expression pattern depicts that identified EcDof genes were different from earlier identified PBF from maize, which was seed-specific. The involvement of more than one gene at different stages of seed development suggests combinatorial regulation of the downstream genes participating during seed-protein accumulation.
Fig. 3.
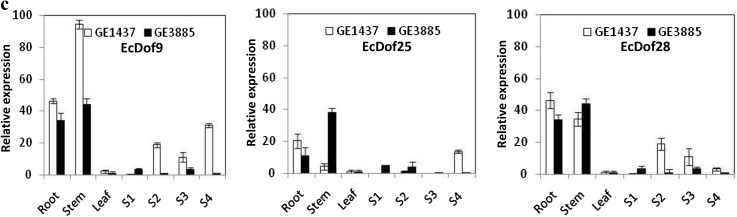
Expression patterns of EcDof genes expressed a preferentially in developing spikes (S1, S2, S3, and S4 stages), b in vegetative tissue and developing spikes, and c in vegetative tissue only (root and stem) of finger millet (FM) genotypes, GE-1437 (low protein genotype; 6.2%) and GE-3885 (high protein genotype; 13.8%) differing in their grain protein content. Error bars represent the standard error of three biological replicates
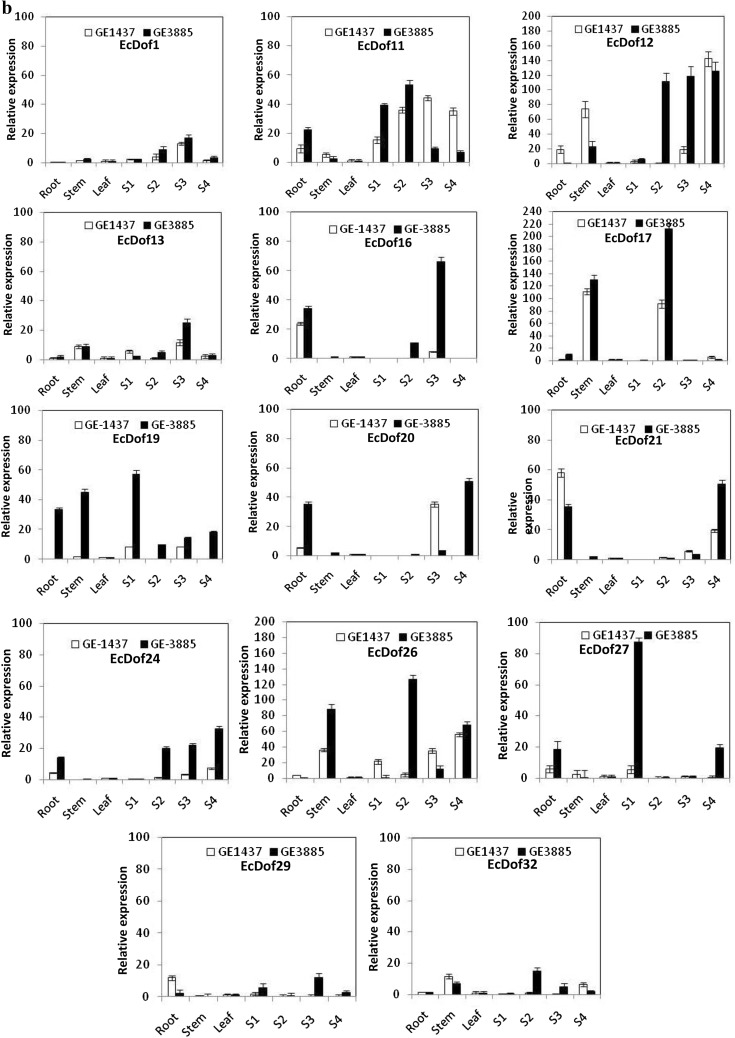
Transcript accumulation of 14 EcDof genes, i.e., EcDof 1, EcDof 11, EcDof 12, EcDof 13, EcDof 16, EcDof 17, EcDof 19 EcDof 20, EcDof 21, EcDof 24, EcDof 26, EcDof 27, EcDof 29, and EcDof 32, was observed in vegetative tissues as well as in developing spikes suggests that these genes are not only involved during grain filling but also associated with other biological processes like in modulating photosynthetic carbon assimilation, flowering time, growth and developmental processes in plants (Fig. 3b). Expression of EcDof 12, EcDof 13, EcDof 17, and EcDof 26 genes was higher in stem and developing spikes, and may be involved preferentially in transcriptional regulation of photosynthetic genes and sucrose transport.
Three EcDof genes, EcDof 9, EcDof 25, and EcDof 28 showed higher expression in root and stem tissue of FM genotypes (Fig. 3c). In root, the transcript abundance was higher for LPG as compared to HPG. Since some of the Dof genes (EcDof2 and ZmDof2) reported to be act as repressor, maybe, these genes act as repressor and inhibit the expression of seed-storage protein genes in FM (Gupta et al. 2014). Transcript abundance of eight EcDof genes, i.e., EcDof 2, EcDof 4, EcDof 6, EcDof 7, EcDof 8, EcDof 10, EcDof 14, and EcDof 30, was low or expression was constant in all stages and further suggests that these genes may be involved in regulation of genes that express at a steady rate throughout the plant development process. Earlier work carried on expression analysis of TaDof family across all major organs revealed that the majority of TaDof members were predominantly expressed in vegetative organs (Shaw et al. 2009).
Phylogenetic relatedness of EcDof genes with PBF and motif analysis
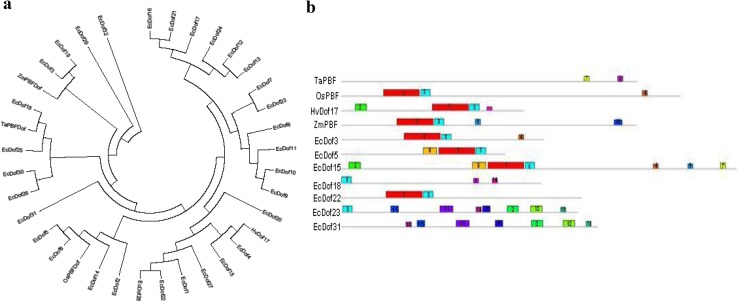
Phylogenetic tree was constructed based on the alignment of amino acid sequences of putative EcDof proteins identified from developing spikes along reported PBF proteins from Zea mays (ZmPBF), Hordeum vulgare (HvPBF), Triticum aestivum (TaPBF), and Oryza sativa (OsPBF). Phylogenetic analysis revealed (Fig. 4a), among 32 Dof genes, that EcDof4 and EcDof 15 show close similarity with HvPBF, EcDof5, and EcDof8 which were close to OsPBF, EcDof18 to TaPBF, EcDof 3, and EcDof19 with ZmPBF, respectively. The identity among EcDof proteins to their respective PBFs was low in the range of 34–58%. Furthermore, motif analysis (Supplementary Table 3) of identified seed-specific EcDofs with reported PBF (Fig. 4b) revealed that position and number of motifs among Dof genes differ, and maybe, these motifs were responsible for difference in binding of EcDof TF to their respective sites and interaction with other proteins (Opaque2) for regulation of downstream genes. Expression of EcDof genes, i.e., EcDof 3, EcDof 5, EcDof 15, and EcDof 18, was predominant in developing spikes of FM genotypes and their phylogenetic relatedness with respective PBFs further supports their role in grain filling and flowering.
Fig. 4.
a Phylogenetic relationship among 32 putative EcDof proteins with reported PBF proteins. An unrooted NJ tree is shown for 32 full-length EcDof proteins along with reference sequences of Zea mays (ZmPBF), Hordeum vulgare (HvPBF; Hv Dof 17), Triticum aestivum (TaPBF), and Rice (RPBF). The scale bar corresponds to 0.05 estimated amino acid substitutions per site. b Distribution of 15 motifs among EcDof proteins using MEME ver.4.9.0. Motif 1 is the conserved Dof domain
Tertiary structure prediction of EcDof and EcO2 proteins
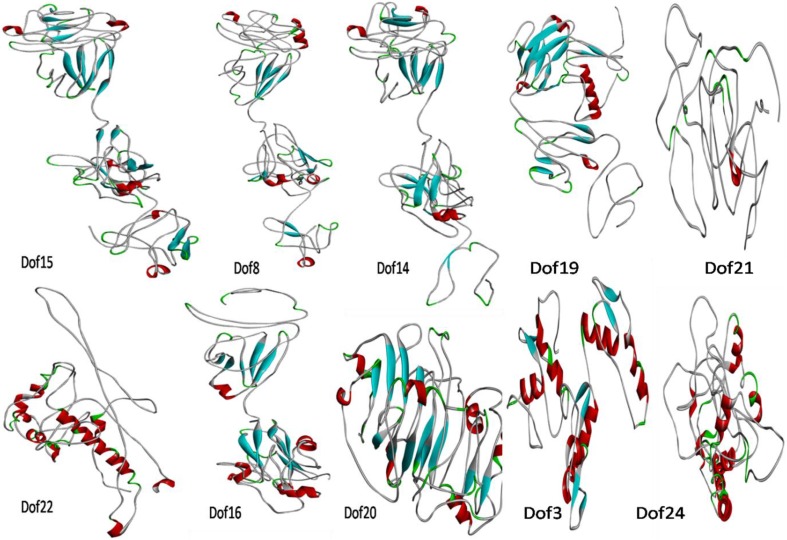
EcDof and EcO2 (finger millet Opaque2, accession no. KX440959), gene sequences, were subjected to BLASTp program against PDB database (http://www.rcsb.org/) for identification of suitable template structures that can be further utilized for comparative 3D structure modeling. Since suitable template was not found using BLAST search, therefore, 3D model of EcDof and EcO2 proteins was constructed using threading algorithm at I-TASSER server (Fig. 5). Three-dimensional structures provide valuable insight into molecular function and putative active site residue identification. 3D models for EcDof proteins along with EcO2 were successfully predicted by different threading templates, as given in Table 1.
Fig. 5.
Predicted 3-D structures of EcDof proteins generated by Discovery studio ver4.1. The helix is represented by red cylinders, sheet by cyan arrows, coils by green, and loops by gray lines
The final results of function predictions are deduced from the consensus of top structural matches with the function scores calculated based on the confidence score (C score) of the I-TASSER structural models. The structural similarity between model and templates was evaluated by template modeling score (TM score), and the sequence identity in the structurally aligned regions was determined.
A total of five top structure models were predicted by I-TASSER server. Only most appropriate predicted structures were chosen for each proteins based on maximum C-score and maximum number of decoys for evaluation and verifications. The selected models have expected TM score ranging from 0.29 ± 0.09 to 0.58 ± 0.14 and RMSD (root mean square deviation) score lies between 9.7 ± 4.6 and 15.1 ± 3.5 Å for the EcDof proteins from I-TASSER server, as shown in Supplementary Table 4. The selected models were found in correct topology based on C-score, TM-score, and RMSD value. Swiss-PDB Viewer was used to stabilizing the stereochemical properties of the chosen models through energy minimization.
Validation of the predicted 3D structure
The stability of Dof and O2 protein models was validated by the Structural Analysis and Verification Server (http://services.mbi.ucla.edu/SAVES/), which have inbuilt tools such as PROCHECK, WHAT_CHEK, ERRAT,VERIFY_3D, and PROVE. The Ramachandran plot statistics of protein models in range of 36.6–78.2% (most favored regions), 14.5–48.1% (additional allowed regions), 3.4–13.7% (generously allowed regions), and 1.5–11.6 (disallowed regions) are shown in Table 2. The results of the PROCHECK analysis revealed that relatively low percentage of residues have phi/psi angles in the disallowed regions, suggesting the acceptability of Ramachandran plots for studied proteins. The stereo chemical quality of the predicted model was found to be satisfactory. Validated models were submitted to Protein Model Database (https://bioinformatics.cineca.it/PMDB/) for further investigation of key structural features through structural bioinformatics techniques such as molecular dynamics simulation that can decode the complexity of Dof proteins and its fundamental roles in seeds. The PMDB IDs of submitted model are PM0080497, PM0080498, PM0080499, PM0080500, PM0080501, PM0080502, PM0080503, PM0080504, PM0080505 and PM0080506 for Dof15, Dof8, Dof14, Dof22, Dof24, Dof16, Dof19, Dof3, Dof21, and Dof20. The 3D structural prediction of EcDof proteins showed variability in structural attributes that further suggest their role in diverse functions.
Table 2.
Ramachandran plot statistics of DOF and opaque2 proteins
| Gene names | Residues in most favored regions (%) | Residues in additional allowed regions (%) | Residues in generously allowed regions (%) | Residues in disallowed regions (%) |
|---|---|---|---|---|
| EcDof3 | 76.1 | 16.0 | 4.8 | 3.2 |
| EcDof8 | 75.1 | 15.9 | 5.8 | 3.2 |
| EcDof14 | 72.1 | 19.7 | 6.0 | 2.2 |
| EcDof15 | 74.6 | 17.6 | 6.0 | 1.8 |
| EcDof16 | 78.2 | 15.4 | 4.9 | 1.5 |
| EcDof19 | 76.2 | 18.6 | 3.4 | 1.7 |
| EcDof20 | 70.3 | 22.5 | 5.1 | 2.0 |
| EcDof21 | 61.6 | 23.8 | 11.0 | 3.7 |
| EcDof22 | 76.3 | 14.5 | 4.8 | 4.3 |
| EcDof24 | 74.2 | 18.4 | 5.7 | 1.6 |
| Opaque 2 | 41.3 | 37.5 | 9.9 | 11.3 |
| ZmDof | 39.5 | 48.1 | 6.8 | 5.6 |
| ZmOpaque 2 | 36.6 | 38.2 | 13.7 | 11.6 |
Molecular docking studies
Molecular docking studies were carried to predict that the interacting pairs of proteins might be regulating the seed-storage protein genes in FM. To understand their interaction, modeled seed-specific EcDof protein structures were docked with Zn++ by AutoDock vina to make Zn++ coordinated functional structures of Dof. These coordinated structures were docked by Opaque2 (EcO2) by Hex to made hetero-dimer complex structures followed by docking of these hetero-dimer structures with prolamin promoter to understanding its binding affinity for decoding the dynamics behavior of EcDof and its role in different biological function involved in accumulation of seed-storage proteins in FM (Supplementary Tables 5 and 6). Docking studies revealed that seed-specific full-length EcDof (15 and 22)-EcO2 proteins have higher binding energy as compared to other EcDof proteins, respectively (Table 3). The EcDof14–EcO2 and Ecdof8–EcO2 with low expression in vegetative and developing spikes have less binding affinity with respective promoters. The energy for EcDof15-EcO2 and EcDof 22-EcO2 proteins was even higher as compared to ZmPBF–ZmO2. Earlier reported, maize endosperm-specific transcription factors opaque2 (O2) and prolamin-box-binding factor (PBF) regulate the expression of α- and β-zein genes by recognizing the O2 box and P box in their promoters (Mena et al. 1998). Multiple prolamine genes viz., α, β, γ, and δ have been isolated in FM might be hetero-dimerization of different seed-specific EcDof and EcO2 TFs are involved in controlling the regulation of multiple prolamine genes during seed-storage protein accumulation in FM. Further in vivo interaction experiment of seed-specific EcDof and EcO2 is required using yeast two-hybrid or BiF system to understand their trans-activation capacity through interaction with its C-terminal domain. Recently, Zhang et al. (2016) have reported that O2 and PBF coordinately not only control regulation of storage protein genes, but also starch synthesis in endosperm.
Table 3.
Molecular docking of Dof-Opaque2 hetero-dimer with prolamin AAGAA motif sequence by Hex
| S. no. | Dof-opaque 2 vs prolamin promoter | Preferential expression | Energy (Kcal/mol) |
|---|---|---|---|
| 1 | EcDof3–EcO2 vs Prolamin promoter | Developing spikes | − 401.60 |
| 2 | EcDof15–EcO2 vs Prolamin promoter | Developing spikes | − 417.76 |
| 3 | EcDof22–EcO2 vs Prolamin promoter | Developing spikes | − 418.07 |
| 4 | EcDof14–EcO2 vs Prolamin promoter | Low expression in vegetative tissues as well as in developing spikes | − 340.75 |
| 5 | EcDof8 –EcO2 vs Prolamin promoter | Low expression in vegetative tissues as well as in developing spikes | − 313.36 |
| 6 | EcPBFDof19–EcO2 vs Prolamin promoter | Expressed in vegetative tissues as well as in developing spikes | − 410.36 |
| 7 | ZmDof–ZmO2 vs zein 1 | Seeds | − 326.52 |
| 8 | ZmDof–ZmO2 vs zein 2 | Seeds | − 354.92 |
Our results revealed that more than 50% of the EcDof genes are expressed during the seed developing process. Sequence similarity/identity between seed-specific EcDofs (EcDof3, EcDof5, and EcDof18) with their respective PBFs is low, suggesting that maybe, some unique regulatory elements are present in EcDof proteins that regulate the seed-storage proteins in FM genotypes. The presence of more seed-specific EcDof genes during seed development revealed the multiplicity of EcDof genes which has to be further dissected. Molecular docking revealed higher binding energy for hetero-dimer seed-specific EcDof–EcO2 onto prolamine promoters in FM. Further identification and characterization of cis-regulatory elements of genes and their validation is required to understand the functional role of such Dof regulatory proteins. In view of their diverse function during seed development and grain filling, it is pertinent to validate and explore the function of these Dof TF genes under investigation using gain-of-function and loss-of-function approaches.
Conclusion
The study provides genome and transcriptome-wide identification of Dof gene family in FM and fundamental information on tissue-specific transcript profiling of EcDof genes. The results indicate that EcDofs are possibly involved in grain filling and other biological functions during plant development. The present study will help to understand the role of Dof TFs and regulatory mechanism involved in seed development and protein accumulation during grain filling in FM. Furthermore, validating the identified Dof TFs genes using knock in and knock out approaches will surely help to explore the precise function of genes involved in seed-storage proteins for better nutraceutical development and designing of functional foods for securing food and nutritional security of rapidly growing world populations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1: Multiple sequence alignment of putative Dof proteins from E. coracana showing the Dof domain. The NLS motif, and conserved cysteine residues (C1–C5) within the Dof domain are indicated at the top of the alignment (PPT 275 kb)
Acknowledgements
This research work was conducted under the research program (Grant No. BT/PR7849/AGR/02/2006) funded by the Department of Biotechnology (DBT), New Delhi at G.B. Pant University of Agriculture and Technology, Pantnagar, India. The financial assistance (Grant No. YSS/2015/00536) provided to SG by Department of Science and Technology (DST), New Delhi is duly acknowledged. The logistic support provided by Director, Experiment Station, G. B. P. U. A. & T., Pantnagar is also acknowledged.
Abbreviations
- Dof
DNA binding with one finger
- FM
Finger millet
- GSPs
Grain seed proteins
- HPG
High protein genotype
- LPG
Low protein genotype
- MAST
Motif alignment and search tool
- MEME
Multiple expectation maximization for motif elicitation
- NJ
Neighbor-joining
- PBF
Prolamine-binding factor
- P box
Prolamin-box
- PEM
Protein energy malnutrition
- qPCR
Real-time PCR
- RIN
RNA integrity number
- SAM
Shoot apical meristem
- TF
Transcription factor
- WHO
World health organization
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13205-017-1068-z) contains supplementary material, which is available to authorized users.
References
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton JF, Lugo-Martinez J, Tucker AE, Schrider DR, Warren WC. Extensive error in the number of genes inferred from draft genome assemblies. PLoS Comput Biol. 2014;10:e1003998. doi: 10.1371/journal.pcbi.1003998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G, Ni Z, Nie X, Sun Q. Wheat DOF transcription factor WPBF interacts with TaQM and activates transcription of an alpha-gliadin gene during wheat sed development. Plant Mol Biol. 2007;63:73–84. doi: 10.1007/s11103-006-9073-3. [DOI] [PubMed] [Google Scholar]
- Gaur VS, Singh US, Kumar A. Transcriptional profiling and in silico analysis of Dof transcription factor gene family for understanding their regulation during seed development of rice Oryza sativa L. Mol Bio Rep. 2011;38:2827–2848. doi: 10.1007/s11033-010-0429-z. [DOI] [PubMed] [Google Scholar]
- Gupta N, Gupta AK, Singh NK, Kumar A. Differential expression of PBF Dof transcription factor in different tissues of three finger millet genotypes differing in seed protein content and color. Plant Mol Biol Rep. 2011;29:69–76. doi: 10.1007/s11105-010-0208-y. [DOI] [Google Scholar]
- Gupta AK, Gaur VS, Gupta S, Kumar A. Nitrate signals determine the sensing of nitrogen through differential expression of genes involved in nitrogen uptake and assimilation in finger millet. Funct Integr Genom. 2013;13:179–190. doi: 10.1007/s10142-013-0311-x. [DOI] [PubMed] [Google Scholar]
- Gupta S, Gupta SM, Gupta AK, Gaur VS, Kumar A. Fluctuation of Dof1/Dof2 expression ratio under the influence of varying nitrogen and light conditions: involvement in differential regulation of nitrogen metabolism in two genotypes of finger millet (Eleusine coracana L.) Gene. 2014;546:327–335. doi: 10.1016/j.gene.2014.05.057. [DOI] [PubMed] [Google Scholar]
- Hittalmani S, Mahesh HB, Shirke MD, Biradar H, Uday G, Aruna YR, Lohithaswa HC, Mohanrao A. Genome and transcriptome sequence of finger millet (Eleusine coracana (L.) Gaertn.) provides insights into drought tolerance and nutraceutical properties. BMC Genom. 2017;18:1–16. doi: 10.1186/s12864-017-3850-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwal P, Gupta S, Arora S, Kumar A. Identification of genes involved in carbon metabolism from Eleusine coracana (L.) for understanding their light mediated entrainment and regulation. Plant Cell Rep. 2014;33:1403–1411. doi: 10.1007/s00299-014-1625-4. [DOI] [PubMed] [Google Scholar]
- Kumar A, Gaur VS, Goel A, Gupta AK. De novo assembly and characterization of developing spikes transcriptome of finger millet (Eleusine coracana): a minor crop having nutraceutical properties. Plant Mol Biol Rep. 2015;33:905–922. doi: 10.1007/s11105-014-0802-5. [DOI] [Google Scholar]
- Kumar A, Metwal M, Kaur S, Gupta AK, Puranik S, Singh S, Singh M, Gupta S, Babu BK, Sood S, Yadav R. Nutraceutical value of finger millet [Eleusine coracana (L.) Gaertn.], and their improvement using omics approaches. Front Plant Sci. 2016;7:934. doi: 10.3389/fpls.2016.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushwaha H, Gupta S, Singh VK, Rastogi S, Yadav D. Genome wide identification of Dof transcription factor gene family in Sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol Biol Rep. 2011;38:5037–5053. doi: 10.1007/s11033-010-0650-9. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marzábal P, Gas E, Fontanet P, Vicente-Carbajosa J, Torrent M, Ludevid MD. The maize Dof protein PBF activates transcription of gamma-zein during maize seed development. Plant Mol Biol. 2008;67:441–454. doi: 10.1007/s11103-008-9325-5. [DOI] [PubMed] [Google Scholar]
- Mena M, Vicente CJ, Schmidt RJ, Carbonero P. An endosperm-specific DOF protein from barley highly conserved in wheat binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. Plant J. 1998;16:53–62. doi: 10.1046/j.1365-313x.1998.00275.x. [DOI] [PubMed] [Google Scholar]
- Mena M, Cejudo FJ, Isabel-Lamoneda I, Carbonero P. A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiol. 2002;130:111–119. doi: 10.1104/pp.005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LM, McIntyre CL, Gresshoff PM, Xue GP. Members of the Dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation. Funct Integr Genom. 2009;9:485–498. doi: 10.1007/s10142-009-0130-2. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Hoisington DA, Tyagi AK. Advances in cereal genomics and applications in crop breeding. Trends Biotechnol. 2006;24:490–499. doi: 10.1016/j.tibtech.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Verdier J, Thompson RD. Transcriptional regulation of storage protein synthesis during dicotyledon seed filling. Plant Cell Physiol. 2008;49:1263–1271. doi: 10.1093/pcp/pcn116. [DOI] [PubMed] [Google Scholar]
- Washio K. Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol. 2003;133:850–863. doi: 10.1104/pp.103.027334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto MP, Onodera Y, Touno SM, Takaiwa F. Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiol. 2006;141:1694–1707. doi: 10.1104/pp.106.082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Akiyama A, Kisaka H, Uchimiya H, Tetuya M. Metabolic engineering with Dof1 transcription factor in plants: improved nitrogen assimilation and growth under low-nitrogen conditions. Proc Natl Acad Sci USA. 2004;101:7833–7838. doi: 10.1073/pnas.0402267101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zheng X, Messing J, Wu Y. Maize endosperm-specific transcription factors O2 and PBF network the regulation of protein and starch synthesis. Proc Natl Acad Sci USA. 2016;113:10842–10847. doi: 10.1073/pnas.1613721113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: Multiple sequence alignment of putative Dof proteins from E. coracana showing the Dof domain. The NLS motif, and conserved cysteine residues (C1–C5) within the Dof domain are indicated at the top of the alignment (PPT 275 kb)