Abstract
Different physical and chemical stimuli are detected by the peripheral sensory receptors of dorsal root ganglion (DRG) neurons, and the generated inputs are transmitted via afferent fibers into the central nervous system. The gene expression profiles of DRG neurons contribute to the generation, transmission, and regulation of various somatosensory signals. Recently, the single-cell transcriptomes, cell types, and functional annotations of somatosensory neurons have been studied. In this review, we introduce our classification of DRG neurons based on single-cell RNA-sequencing and functional analyses, and discuss the technical approaches. Moreover, studies on the molecular and cellular mechanisms underlying somatic sensations are discussed.
Keywords: Neuron type, Single-cell technology, Somatosensory mechanism, Pain, Gene profiles
Introduction
Peripheral stimuli are detected by the pseudo-unipolar sensory neurons in the dorsal root ganglion (DRG) through molecular sensors at their peripheral nerve terminals. These neurons then transmit peripheral signals to the spinal dorsal horn. Somatosensation comprises different sensory modalities including thermoception, mechanoreception, proprioception, nociception, and pruritoception. There is a notion that different types of DRG neurons are specialized by distinct molecular networks and mediate these sensory modalities. Accumulated experimental evidence has demonstrated that nociceptors mediate other sensory modalities such as mechanoreception [1, 2].
Although the somatosensory molecules have been extensively studied, a global view of the transcriptional profiles of individual DRG neurons has not been obtained. Efforts have been made to understand the gene profiles of these neurons [3–9]. Single-cell RNA-sequencing (scRNA-seq) is an effective technique that has advanced our understanding of single-cell transcriptomes, and the identification of cell clusters of mouse DRG neurons [10, 11]. High-coverage scRNA-seq can provide a better database of single-cell gene profiles [11]. The functions of DRG neurons have been determined by genetic ablation [9, 12]. In vivo whole-cell patch recording plus single-cell real-time PCR has been used to determine the cellular functions corresponding to the neuron types [11]. In this review, the scRNA-seq method is discussed. Then, recent studies on the cell types and functional annotations of somatosensory neurons are summarized.
Methodological Considerations for Neuron-Typing
Methodological Approach of scRNA-seq
For the isolation of single cells, DRG tissue is subjected to enzymatic digestion and mechanical dissociation. Individual cells are isolated manually or using automated equipment. The microfluidic C1 (Fluidigm) is a commercial device, which automatically performs single-cell capture, reverse-transcription, and cDNA amplification in chip chambers. However, the conditions of neuron dissociation cannot be evaluated by the device but by trained experimenters. To fully control the isolation conditions, manual isolation of neurons is preferred. Under a microscope, healthy DRG neurons have a smooth and bright cell membrane. Individual neurons are picked manually and randomly. Generally, 50 cells can be picked by a trained experimenter within 4 h. The efficiency of manual isolation of single cells is acceptable. The single cell is subjected to reverse transcription and cDNA amplification. The method of scRNA-seq was first reported by Tang in 2009 [13]. Sandberg’s lab introduced Smarter-seq, and the subsequently improved Smarter-seq2 [14, 15]. We used the Smarter-seq commercial kit (Clontech) in our studies to identify the gene profiles of individual neurons.
The sequencing depth is important for obtaining the whole transcriptome of a single neuron. Low-coverage scRNA-seq, which detects 4,644 genes in a representative cell, has been suggested to be sufficient for cell-clustering and biomarker identification [16]. However, this number is much smaller than that by high-coverage sequencing, which detects ~10,000 genes per cell [11]. Moreover, the low-coverage sequencing leads to great variation in genes with transcripts of low or medium abundance [16]. As we attempted not only to identify neuron types and their corresponding biomarkers, but also to reveal the whole transcriptomes of individual neurons, high-coverage scRNA-seq was an appropriate choice in our studies.
The number of sequenced neurons is determined by the minimal number of neurons required to cover all neuron types. More neuron samples are better in the process of identifying neuron types, but this also increases the cost. To minimize the number, a specific approach has been designed according to the cellular properties of DRG neurons. One is that a subpopulation of neurons can be labeled by isolectin B4 (IB4). The other is that neuron size is considered to be correlated with the conventional classification of DRG neurons. In our study, IB4-labeling was shown by IB4-fluorescein while neuron size was determined using a scale within the objective lens. Finally, IB4-negative small neurons (cross-sectional area <800 μm2), IB4-positive neurons, and IB4-negative large neurons (cross-sectional area >800 μm2) each accounted for ~1/3 of the selected neurons. Individual neurons were collected manually. When the number of sequenced neurons was close to 100, 9 neuron types were identified. Only one more neuron type was identified when the number approached 200. Thus, a number close to 200 is considered to be acceptable.
Recently, the number of publications with scRNA-seq has markedly increased, and various analytical methods have been developed. Based on our experience, we suggest the use of differentially expressed genes, but not all detected genes, for further analysis of cell clusters. Weighted Gene Co-expression Network Analysis is a good method to cluster highly-correlated genes into gene modules [17]. Visualization and cluster analysis of scRNA-seq data can be facilitated by Principal Component Analysis or t-distributed stochastic neighbor embedding (t-SNE) [18]. Technical progress in single-cell capture and the analysis of scRNA-seq data have been reviewed elsewhere [19, 20].
Evaluation of scRNA-seq Data
Data from scRNA-seq should be evaluated carefully. After bioinformatics analysis, representative genes of a cell or cell type can be identified. By quantitative single-cell RT-PCR, the profiles of multiple genes can be examined in individual cells. In situ hybridization (ISH) can be used to analyze the cell distribution of genes in the tissue. However, fluorescent ISH is less sensitive than single-cell RT-PCR. Notably, modified ISH can enhance the sensitivity. A branched DNA ISH has been developed to detect the expression of single-copy genes [21]. However, although branched DNA ISH is very sensitive, the spread of this technique has been limited by competitive interests and the high cost of commercial kits. Recently, a multiplexed fluorescent hybridization chain reaction (HCR) was developed by Pierce’s lab [22]. The cascades of HCR amplify the fluorescent signals. Fluorescent HCR is an alternative, although it is often not sensitive enough to detect gene expression at low levels. Immunostaining of cells is not ideal for the evaluation of scRNA-seq data, because the immunolabeling intensity is not always concordant with the gene expression level [23]. For example, scRNA-seq data show that Nefh, which encodes neurofilament 200 (NF200) and is a well-known marker of myelinated DRG neurons, is also expressed in DRG neurons positive for MAS-related GPR family member D (Mrgprd). However, previous immunostaining results showed that Mrgprd-positive neurons do not contain NF200 [24].
Somatosensory Neuron Types
DRG neurons are divided into different populations with distinct properties, including small-diameter neurons with unmyelinated C-fibers or thinly-myelinated Aδ-fibers, and large-diameter neurons with thickly-myelinated Aα/β-fibers [25]. The small neurons can be subdivided into two subpopulations: one shows peptidergic characteristics by expressing neuropeptides, substance P and calcitonin gene-related peptide (CGRP) [25]; the other does not express neuropeptides but binds IB4 [25]. In contrast, the large neurons are characterized by the expression of NF200 [25]. Tyrosine hydroxylase (TH) is not expressed in neurons of the three subpopulations above, suggesting that TH-positive neurons comprise another subpopulation [26]. DRG neurons can also be classified by the expression of neurotrophic factor receptors. Peptidergic small neurons express glial-derived neurotrophic factor (GDNF) family receptor α 3 (Gfra3) and ropomyosin receptor-kinase A (TrkA), a nerve growth factor (NGF) receptor. Non-peptidergic small neurons contain Ret receptor and Gfra2. Large neurons express TrkB or TrkC.
Efforts have been made to identify the transcriptional profiles of different subpopulations of DRG neurons. Neuron purification by fluorescent activated cell sorting coupled with gene profiling has been used to characterize the molecular properties of TrkC-positive [7] and TRPV1-positive [6] neurons. Gene profiles of ion channel and GPCRs in trigeminal ganglia and DRGs have been analyzed by RNA-seq [5, 8]. In DRG tissue, glial cells, including satellite glial cells and Schwann cells, outnumber sensory neurons. After magnetic purification, the transcriptomes of the neurons were examined [3]. Recently, Chiu et al. [4] used parallel quantitative RT-PCR to analyze the expression levels of candidate genes in single DRG neurons from three subpopulations including IB4+ Nav1.8-Cre/TdTomato+, IB4− Nav1.8-Cre/TdTomato+, and Pvalb-Cre/TdTomato+. The analysis led to the identification of six distinct subgroups, including Mrgprd +, Nppb +, and Th + neurons [4]. However, these results presented the gene profiles of cell groups, but could not identify the neuron types in an unbiased and integrated manner.
ScRNA-seq is an important technology increasingly used to generate cell atlases of the central [27, 28] and peripheral nervous systems [10, 11]. Usoskin et al. reported that peptidergic, non-peptidergic, TH-positive, and NF200-positive mouse DRG neurons can be further classified into 11 subtypes using low-coverage scRNA-seq, which detected ~3,900 ± 1,880 (mean ± SD) genes per neuron [10, 11]. However, the low-coverage scRNA-seq only provided a partial transcriptome, and caused considerable variation in the gene expression profiles, making it difficult to define specific marker genes [10, 11]. In fact, CGRP and NF200 are not good markers for DRG neuron types, because they are expressed in many types [11].
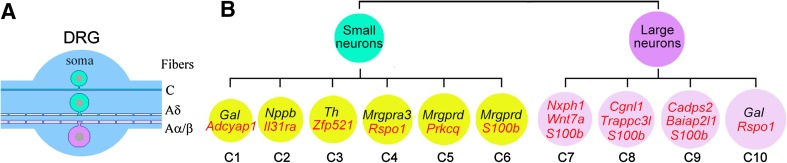
We performed high-coverage scRNA-seq, which detected 10,950 ± 1,218 genes per neuron, and thus identified 10 types and 14 subtypes of DRG neuron [11]. The profiles are available at our database (http://www.ibrainproject.org/en/index.php?c=channel&a=type&tid=84). We not only provided the transcriptome of individual neurons, but also identified distinct marker genes for the neuron types (C1 – C10) and subtypes (Fig. 1) [11]. Neurons in C1 were marked by galanin (Gal), C2 by natriuretic peptide B (Nppb), C3 by Th, C4 by Mrgpra3, C5 by Mrgprd, C7 by neurexophilin 1 (Nxph1) plus S100 Ca2+-binding protein B (S100b), C8 by S100b, and C9 by BAI1-associated protein 2-like 1 (Baiap2l1) plus S100b. C6 neurons expressed a high level of S100b and a low level of Mrgprd. C10 neurons were large neurons expressing Gal. Some subtypes were marked by specific functional genes, such as Mrgprb4 + in the C4-2 subtype of Mrgpra3 + C4, while other subtypes expressed genes representative of myelinated neurons, such as Nefh and S100b in C2-2 of Nppb + C2 [11]. Thus, the number of neuron types and subtypes identified by high-coverage scRNA-seq [11] is greater than that by low-coverage scRNA-seq [10, 11]. Moreover, the specific markers for each neuron type and subtype can be identified by high-coverage scRNA-seq data.
Fig. 1.
Somatosensory neuron types. A morphological characteristics of DRG neurons. B Classification of the types and subtypes of DRG neurons, their markers, and the type-hierarchy. New markers are indicated in red. (Figure adapted and modified from Li et al. [11] with permission).
Growth Factor Receptors and Somatosensory Neuron Types
The timing-specific coordination of multiple transcriptional factors controls the progress of neurogenesis, differentiation, and specification in sensory neuron lineages [29, 30]. Neurogenin 1 (Ngn1) and Ngn2, basic helix-loop-helix transcription factors, play key roles in the neurogenesis and differentiation of primary sensory neurons. Ngn1 determines the formation of TrkA-positive DRG neurons while Ngn2 regulates the formation of TrkB- and TrkC-positive DRG neurons [29, 30]. Runt-related transcription factor 1 (Runx1) and Runx3 regulate the progress of diversification. Runx3 inhibits the expression of TrkB and contributes to the specification of TrkC-positive DRG neurons. Runx1 is restricted to TrkA-positive DRG neurons before the maturation of non-peptidergic neurons [31]. The maturation of non-peptidergic DRG neurons is controlled by NGF signals, because NGF deficiency leads to a dramatic reduction of Runx1 and the loss of Mrgprd [32]. The maintenance of Runx1 expression results in non-peptidergic neurons by inhibiting the expression of neuropeptides, while the loss of Runx1 leads to peptidergic neurons [29, 30]. Neurotrophin receptors represent four types of DRG neurons: TrkA-positive/Runx1-negative, TrkA-negative/Runx1-positive, TrkC-positive/Runx3-positive, and TrkB-positive. GDNF family receptors include Ret, the common receptor, and Gfrα1–4, its co-receptors. Gfrα1–3 and Ret are expressed in different types of DRG neurons. Ret contributes to the diversity of TrkB-positive neurons [29]. TH is specifically expressed in Gfrα2-positive but neither in Gfrα1- nor Gfrα3-positive DRG neurons [26]. Mrgprd is expressed in Gfra2-positive neurons [33]. Gfrα3 is expressed in small peripherin-positive neurons but not in large NF200-positive neurons [34]. Gfrα3 is predominately co-expressed with TrkA or CGRP in DRG neurons [34].
The distributions of Trk receptors and Gfrα1–3 are closely correlated with DRG neuron types, but how they represent different types is not well determined. Morphological studies could help to define the relationships of TrkA–C and Gfrα1–3 with somatosensory neuron types. However, it is not easy to determine the distributions of these six molecules at the same time. ScRNA-seq technology could meet this demand. According to our scRNA-seq data, Gal-positive C1 and C10 neurons highly express Ntrk1 and Gfra3; Nppb-positive C2 neurons prefer to express Ntrk1 or Gfra3; Th-positive C3 neurons express Gfra2 but neither Gfra1 nor Gfra3, consistent with the previous report [26]; Mrgpra3-positive C4 neurons contain high Gfra1 and Ntrk1 but not Gfra2; Mrgprd-positive C5 and C6 neurons contain Gfra2 and Gfra1; C7 neurons contain Ntrk3, while C9 neurons express Ntrk2; and C8 comprises two sub-clusters, one containing Ntrk3 and Gfra1, the other expressing high Ntrk1 (Table 1). Thus, distinct combinations of Ntrk and Gfra contribute to the discrimination of different DRG neuron types.
Table 1.
Expression patterns of growth factor receptors in different types and subtypes of DRG neuron
| Gene | C1 | C2 | C3 | C4-1 | C4-2 | C5 | C6 | C7 | C8-1 | C8-2 | C9 | C10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ntrk1 | +++++ | +++ | +++ | +++++ | + | +++ | +++++ | +++++ | ||||
| Ntrk2 | + | + | + | +++ | +++ | + | +++++ | +++ | ||||
| Ntrk3 | + | +++ | +++ | +++++ | +++++ | +++ | +++ | |||||
| Ngfr | +++ | +++ | + | + | +++ | +++ | +++++ | +++++ | +++++ | |||
| Gfra1 | +++++ | +++++ | + | +++ | + | +++ | ||||||
| Gfra2 | + | +++++ | +++++ | +++ | ||||||||
| Gfra3 | +++++ | +++ | + | +++++ | ||||||||
| ret | + | + | +++++ | +++ | +++ | +++++ | +++ | +++ | +++ |
+++++, high expression; +++, medium expression; +, low expression; blank, lowest expression or not detected
Identification of Rare Neuron Subtypes
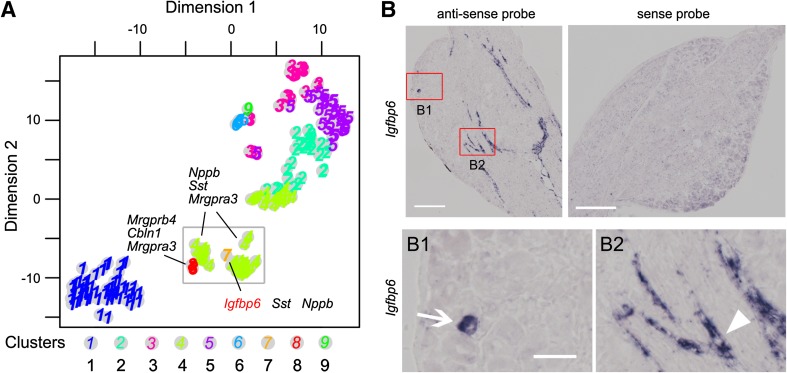
According to our data, 10 types and 14 subtypes of mouse DRG neuron are identifiable. There is always a possibility that a novel type or subtype has not been identified. It has been suggested that the RaceID algorithm can help to identify rare cell types or subtypes within a complex mixture of individual cells [35]. RaceID can identify rare cells expressing outlier genes at levels significantly exceeding the modelled noise [36]. The detailed method was introduced in a report by Grün et al. [35]. Using the R script of the RaceID algorithm kindly supplied by the authors, a total of 1745 differentially-expressed genes from 197 DRG neurons subjected to cell clustering in our report [11] were used for RaceID analysis, which classified the 197 neurons into 9 clusters. Cluster 1 was marked by Mrgprd, as for the C5 cluster; cluster 2 was marked by secretagogin (Scgn) and prokineticin receptor 2 (Prokr2), as in C8-2; cluster 3 was marked by Baiap2l1, as in C9; however, cluster 4 corresponded to a merger of Gal-positive C1, Nppb-positive C2, and Mrgpra3-positive C4; cluster 5 was marked by Ntrk3 and Ptgfr, as in C8-1; cluster 6 was marked by Th, as in C3; and cluster 8 was marked by Mrgprb4 as in C4-2.
Only a single neuron was located in clusters 7 and 9 (Fig. 2A). The neuron in cluster 7 expressed insulin-like growth-factor-binding protein 6 (Igfbp6) and Nppb, while that in cluster 9 expressed carcinoembryonic antigen-related cell adhesion molecule 10 (Ceacam10) and Th (Fig. 2A). According to our data, Ceacam10 was expressed by the majority of Th-positive neurons [11], suggesting that Ceacam10 is a potential marker of Th-positive C3 neurons. The neuron of cluster 7 expressed Igfbp6 with Nppb, which is a marker of C2 neurons. Igfbp6 was only contained in a few Nppb-positive C2 neurons. ISH was performed to assess Igfbp6 expression in the DRG. As reported [37], Igfbp6 is expressed in endothelial cells, but was rarely found in neuron(s) of mouse DRG (Fig. 2B). The percentage of Igfbp6-positive DRG neurons was <1%. The results indicated that Igfbp6 is a potential marker of a rare subtype of C2 neuron. The function of Igfbp6-positive DRG neurons needs to be explored. According to previous reports [10, 11] and the present RaceID analysis, it is unlikely that major types or subtypes of DRG neuron have failed to be identified, although the existence of rare types or subtypes cannot be completely ruled out.
Fig. 2.
Rare cell types and subtypes of DRG neuron. A t-SNE map showing cell clusters obtained using the RaceID algorithm. The grey square shows cell cluster 7 and genes corresponding to known markers specifically expressed in the cluster. Cluster numbers are repeated in black. B Gene expression of Igfbp6 in mouse DRG shown by ISH with anti-sense probe (left). No ISH signal is detected with the sense probe (right). Enlargements show the Igfbp6 expression in a neuron (B1, arrow) and a fibroblast in a blood vessel (B2, arrowhead). Scale bars for B, 200 μm; for B1 & B2, 50 μm.
Multiple Functions of Neuron Types Suggested by Their Transcriptional Profiles
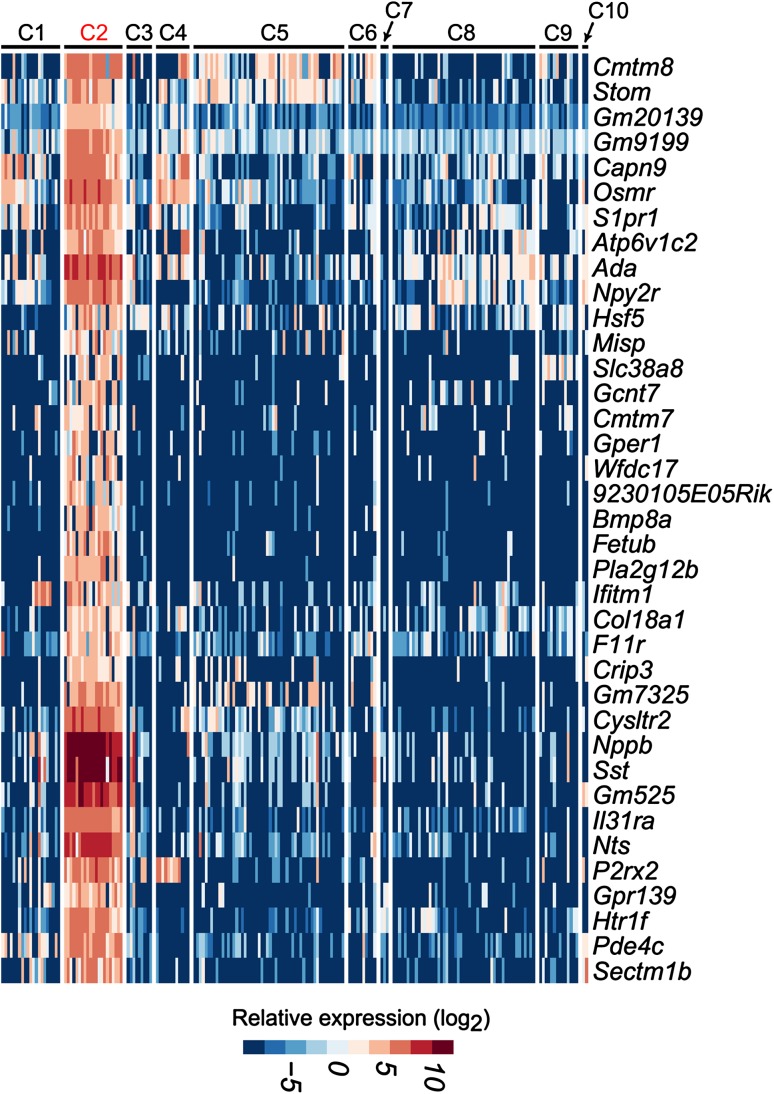
The mechanisms underlying somatosensory functions have been studied for decades, so at least some resources can be used to predict the possible functions of DRG neuron types. For example, C2 neurons selectively express Nppb encoding B-type natriuretic peptide (BNP), Nts encoding neurotensin (NTS), Sst encoding somatostatin (SST), Il31ra encoding interleukin 31 receptor A (Il31ra), and Htr1f encoding 5-hydroxytryptamine (serotonin, 5-HT) receptor 1F (Htr1f) (Fig. 3) [4, 11]. BNP is involved in the transmission of itch signals [38], while Il31ra mediates T helper cell-dependent itch [39]. Moreover, C2 neurons contain other pruritogen receptors, including F2rl1, Hrh1, Htr2a, and Mrgprc11, suggesting that C2 neurons respond to multiple pruritogens [40]. The oncostatin M receptor encoded by Osmr forms a functional receptor complex with Il31ra [41], and is expressed specifically in C2 neurons [11]. Serotonin released from the descending pathway might potentiate itch sensation by activating 5-HT1A receptors on dorsal horn neurons [42]. Serotonin also activates 5-HT1A at the central terminals of C2 neurons [11]. Genetic ablation of C2 neurons might reduce itch induced by IL-31 or 5-HT1F agonist [9].
Fig. 3.
Genes specifically contained in Nppb-positive mouse C2 DRG neurons. Heatmap showing the expression patterns of selected genes. (Figure adapted and modified from Li et al. [11] with permission).
In addition to the role in itch sensation, C2 neurons are involved in nociception. BNP is shown to inhibit spinal nociceptive transmission [43]. The activation of 5-HT1A inhibits nociceptive neurotransmission presynaptically in spinal dorsal horn [44]. Npy2r encoding neuropeptide Y receptor 2 is predominantly expressed in small CGRP-positive DRG neurons [45]. In fact, this receptor is expressed in all C2 neurons and a few C1 and C8 neurons [11]. The activation of Npy2r-expressing DRG neurons induces abnormally exacerbated pain [46]. Thus, C2 neurons also function in nociceptive neurotransmission.
C2 neurons also specifically express SST, which is co-localized with Il31ra or NTS [10]. Thus, C2 neurons express neuropeptides including BNP, NTS, and SST. Spinal NTS and SST are considered to play inhibitory roles in nociceptive neurotransmission by activating their receptors [47–49]. Sstr2 encoding a subtype of SST receptor (SSTR2) is present in a subtype of C1 neurons, while Ntsr2 encoding a subtype of NTS receptor (NTSR2) is found in C1 and C8 neurons [11]. Thus, SST and NTS secreted from C2 neurons might regulate the activity of C1 neurons by acting at SSTR2 and NTSR2, respectively.
Functional Annotation of DRG Neuron Types and Subtypes
We propose that neuron types should be defined by integrating the transcriptomic, morphological or anatomical, and functional characteristics of neurons. The morphological or anatomical characteristics of neuron types include the sizes of their cell bodies, axons and dendrites, and inputs and outputs of circuits. To find the functions of neuron types, we performed in vivo whole-cell patch recordings to determine the neuronal responses to peripheral stimuli, and then carried out single-cell real-time PCR to identify the neuron type [11]. We found that most types of nociceptors respond to multiple stimulus modalities and others have more specialized response properties. Sensing of noxious heat and mechanical stimuli may be the principal function of many types (C1, C2, C4, C5, and C6) and subtypes of small DRG neurons (mechanoheat nociceptors) that could also be specialized for other somatosensory properties such as itch [11]. Two types of nociceptors more specialized for noxious mechanical stimuli have been found in large DRG neurons (C7 and C9) [11]. Most mechanoheat nociceptors are also polymodal nociceptors because they are also sensitive to various chemical stimuli. The TH-positive C3 neurons function as C-LTMRs (low-threshold mechanoreceptors), and C8 neurons differentially express Trpc1, Kcnk4 encoding K+ channel, subfamily K, member 4, Asic3 encoding acid-sensing (proton-gated) ion channel 3, and Piezo2 encoding piezo-type mechanosensitive ion channel component 2, suggesting roles in mechanoreception. C3 and C8 are two unique neuron types that may serve as mechanoceptors.
It is interesting to further analyze how the particular somatosensory functions are mediated by the gene networks in the neuron types and subtypes. For example, we asked whether all types of mechanoheat nociceptors have the same mechanism of mediating noxious heat sensation. In the past decades, intense research led to the discovery of the mechanisms underlying thermal nociception. Although TRPV1 has been proposed to play an important role in heat nociception, Trpv1-knockout mice only show a partial deficit in noxious heat assays [50]. Our study showed that the majority of identified types of small DRG neurons respond to noxious heat stimuli, while Trpv1 is present in C1 and C2 neurons [11]. Importantly, we have found a critical role of fibroblast growth factor 13 (FGF13) in heat nociception. FGF13 is highly expressed in C1, C2, C4, C5, and C6 DRG neurons, and these were identified as mechanoheat nociceptors. Heat nociception, but not mechanical nociception, is completely abolished in Fgf13-deficient mice [51]. One related mechanism is that FGF13 interacts with Nav1.7 and increases its currents. Fgf13-deficiency causes a reduction of Nav1.7 in the plasma membrane and therefore markedly reduces the action potential firing during noxious heat stimulation. Patients with loss-of-function mutations of Nav1.7 show the congenital absence of pain perception [52]. Thus, the FGF13/Nav1.7 interaction may act as a common and final regulatory pathway for most types of mechanoheat nociceptors to transmit noxious heat signaling.
Conclusions and Perspectives
This review underscores the importance of high-coverage scRNA-seq in determining the single-cell transcriptome by discussing our studies using scRNA-seq, analysis of DRG neuron types, and their corresponding functional annotation. High-coverage scRNA-seq can reveal most of the gene expression profile of a single neuron. Functional analysis provides a framework for the somatosensory functions of neuron types and subtypes. However, the gene network mechanisms underlying various somatic sensations remain to be further investigated. The neuron type-specific circuits that transmit sensory information into the central nervous system need to be demonstrated. Moreover, the functions corresponding to different types of DRG neurons should be studied in detail using multiple experimental approaches, including in vivo patch recording, optogenetic techniques, and chemical genetic methods.
Acknowledgements
This review was supported by grants from the National Natural Science Foundation of China (31630033, 31130066, 31671094, and 81300961), the Chinese Academy of Sciences (XDB02010000 and QYZDY-SSW-SMC007), and the Shanghai Science and Technology Committee (16JC1420500).
References
- 1.Meyer RA, Ringkamp M, Campbell JN, Raja SN. Peripheral mechanisms of cutaneous nociception. In: McMahon SB, Koltzenburg M (Eds.). Wall and Melzack’s Textbook of Pain. Elsevier Churchill Livingston, 2006: 3–34.
- 2.Willis WD, Coggeshall RE. Sensory receptors and peripheral nerves. In: Willis WD Jr, Coggeshall RE (Eds.). Sensory Mechanisms of the Spinal Cord. Volume 1. Primary Afferent Neurons and the Spinal Dorsal Horn. New York, Springer, 2004: 19–87.
- 3.Thakur M, Crow M, Richards N, Davey GI, Levine E, Kelleher JH, et al. Defining the nociceptor transcriptome. Front Mol Neurosci. 2014;7:87. doi: 10.3389/fnmol.2014.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiu IM, Barrett LB, Williams EK, Strochlic DE, Lee S, Weyer AD, et al. Transcriptional profiling at whole population and single cell levels reveals somatosensory neuron molecular diversity. Elife. 2014;3:e04660. doi: 10.7554/eLife.04660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flegel C, Schobel N, Altmuller J, Becker C, Tannapfel A, Hatt H, et al. RNA-Seq Analysis of Human Trigeminal and Dorsal Root Ganglia with a Focus on Chemoreceptors. PLoS One. 2015;10:e0128951. doi: 10.1371/journal.pone.0128951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goswami SC, Mishra SK, Maric D, Kaszas K, Gonnella GL, Clokie SJ, et al. Molecular signatures of mouse TRPV1-lineage neurons revealed by RNA-Seq transcriptome analysis. J Pain. 2014;15:1338–1359. doi: 10.1016/j.jpain.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J, Friese A, Mielich M, Sigrist M, Arber S. Scaling proprioceptor gene transcription by retrograde NT3 signaling. PLoS One. 2012;7:e45551. doi: 10.1371/journal.pone.0045551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manteniotis S, Lehmann R, Flegel C, Vogel F, Hofreuter A, Schreiner BS, et al. Comprehensive RNA-Seq expression analysis of sensory ganglia with a focus on ion channels and GPCRs in Trigeminal ganglia. PLoS One. 2013;8:e79523. doi: 10.1371/journal.pone.0079523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stantcheva KK, Iovino L, Dhandapani R, Martinez C, Castaldi L, Nocchi L, et al. A subpopulation of itch-sensing neurons marked by Ret and somatostatin expression. EMBO Rep. 2016;17:585–600. doi: 10.15252/embr.201540983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 11.Li CL, Li KC, Wu D, Chen Y, Luo H, Zhao JR, et al. Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res. 2016;26:83–102. doi: 10.1038/cr.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavanaugh DJ, Lee H, Lo L, Shields SD, Zylka MJ, Basbaum AI, et al. Distinct subsets of unmyelinated primary sensory fibers mediate behavioral responses to noxious thermal and mechanical stimuli. Proc Natl Acad Sci U S A. 2009;106:9075–9080. doi: 10.1073/pnas.0901507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al. mRNA-seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 14.Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 15.Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014;32:1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Maaten L, Hinton G. Visualizing Data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- 19.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58:610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol Cell. 2015;58:598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Player AN, Shen LP, Kenny D, Antao VP, Kolberg JA. Single-copy gene detection using branched DNA (bDNA) in situ hybridization. J Histochem Cytochem. 2001;49:603–612. doi: 10.1177/002215540104900507. [DOI] [PubMed] [Google Scholar]
- 22.Choi HM, Chang JY, le Trinh A, Padilla JE, Fraser SE, Pierce NA. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat Biotechnol. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edfors F, Danielsson F, Hallstrom BM, Kall L, Lundberg E, Ponten F, et al. Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol Syst Biol. 2016;12:883. doi: 10.15252/msb.20167144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zylka MJ, Rice FL, Anderson DJ. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron. 2005;45:17–25. doi: 10.1016/j.neuron.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Rutlin M, Abraira VE, Cassidy C, Kus L, Gong S, et al. The functional organization of cutaneous low-threshold mechanosensory neurons. Cell. 2011;147:1615–1627. doi: 10.1016/j.cell.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 29.Marmigere F, Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat Rev Neurosci. 2007;8:114–127. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- 30.Bhatt S, Diaz R, Trainor PA. Signals and switches in Mammalian neural crest cell differentiation. Cold Spring Harb Perspect Biol. 2013;5:a008326. doi: 10.1101/cshperspect.a008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen CL, Broom DC, Liu Y, de Nooij JC, Li Z, Cen C, et al. Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron. 2006;49:365–377. doi: 10.1016/j.neuron.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 32.Luo W, Wickramasinghe SR, Savitt JM, Griffin JW, Dawson TM, Ginty DD. A hierarchical NGF signaling cascade controls Ret-dependent and Ret-independent events during development of nonpeptidergic DRG neurons. Neuron. 2007;54:739–754. doi: 10.1016/j.neuron.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 33.Kupari J, Airaksinen MS. Different requirements for GFRα2-signaling in three populations of cutaneous sensory neurons. PLoS One. 2014;9:e104764. doi: 10.1371/journal.pone.0104764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orozco OE, Walus L, Sah DW, Pepinsky RB, Sanicola M. GFRα3 is expressed predominantly in nociceptive sensory neurons. Eur J Neurosci. 2001;13:2177–2182. doi: 10.1046/j.0953-816x.2001.01596.x. [DOI] [PubMed] [Google Scholar]
- 35.Grun D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 36.Wen L, Tang F. Computational biology: How to catch rare cell types. Nature. 2015;525:197–198. doi: 10.1038/nature15204. [DOI] [PubMed] [Google Scholar]
- 37.Bach LA. Endothelial cells and the IGF system. J Mol Endocrinol. 2015;54:R1–13. doi: 10.1530/JME-14-0215. [DOI] [PubMed] [Google Scholar]
- 38.Mishra SK, Hoon MA. The cells and circuitry for itch responses in mice. Science. 2013;340:968–971. doi: 10.1126/science.1233765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cevikbas F, Wang X, Akiyama T, Kempkes C, Savinko T, Antal A, et al. A sensory neuron-expressed IL-31 receptor mediates T helper cell-dependent itch: Involvement of TRPV1 and TRPA1. J Allergy Clin Immunol. 2014;133:448–460. doi: 10.1016/j.jaci.2013.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akiyama T, Carstens E. Neural processing of itch. Neuroscience. 2013;250:697–714. doi: 10.1016/j.neuroscience.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hermanns HM. Oncostatin M and interleukin-31: Cytokines, receptors, signal transduction and physiology. Cytokine Growth Factor Rev. 2015;26:545–558. doi: 10.1016/j.cytogfr.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 42.Zhao ZQ, Liu XY, Jeffry J, Karunarathne WK, Li JL, Munanairi A, et al. Descending control of itch transmission by the serotonergic system via 5-HT1A-facilitated GRP-GRPR signaling. Neuron. 2014;84:821–834. doi: 10.1016/j.neuron.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang FX, Liu XJ, Gong LQ, Yao JR, Li KC, Li ZY, et al. Inhibition of inflammatory pain by activating B-type natriuretic peptide signal pathway in nociceptive sensory neurons. J Neurosci. 2010;30:10927–10938. doi: 10.1523/JNEUROSCI.0657-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeong HJ, Mitchell VA, Vaughan CW. Role of 5-HT1 receptor subtypes in the modulation of pain and synaptic transmission in rat spinal superficial dorsal horn. Br J Pharmacol. 2012;165:1956–1965. doi: 10.1111/j.1476-5381.2011.01685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brumovsky P, Stanic D, Shuster S, Herzog H, Villar M, Hokfelt T. Neuropeptide Y2 receptor protein is present in peptidergic and nonpeptidergic primary sensory neurons of the mouse. J Comp Neurol. 2005;489:328–348. doi: 10.1002/cne.20639. [DOI] [PubMed] [Google Scholar]
- 46.Arcourt A, Gorham L, Dhandapani R, Prato V, Taberner FJ, Wende H, et al. Touch receptor-derived sensory information alleviates acute pain signaling and fine-tunes nociceptive reflex coordination. Neuron. 2017;93:179–193. doi: 10.1016/j.neuron.2016.11.027. [DOI] [PubMed] [Google Scholar]
- 47.Tetreault P, Beaudet N, Perron A, Belleville K, Rene A, Cavelier F, et al. Spinal NTS2 receptor activation reverses signs of neuropathic pain. FASEB J. 2013;27:3741–3752. doi: 10.1096/fj.12-225540. [DOI] [PubMed] [Google Scholar]
- 48.Guillemette A, Dansereau MA, Beaudet N, Richelson E, Sarret P. Intrathecal administration of NTS1 agonists reverses nociceptive behaviors in a rat model of neuropathic pain. Eur J Pain. 2012;16:473–484. doi: 10.1016/j.ejpain.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 49.Shi TJ, Xiang Q, Zhang MD, Barde S, Kai-Larsen Y, Fried K, et al. Somatostatin and its 2A receptor in dorsal root ganglia and dorsal horn of mouse and human: expression, trafficking and possible role in pain. Mol Pain. 2014;10:12. doi: 10.1186/1744-8069-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caterina MJ, Leffler A, Malmberg AB, Martin WJ, Trafton J, Petersen-Zeitz KR, et al. Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science. 2000;288:306–313. doi: 10.1126/science.288.5464.306. [DOI] [PubMed] [Google Scholar]
- 51.Yang L, Dong F, Yang Q, Yang PF, Wu R, Wu QF, et al. FGF13 Selectively Regulates Heat Nociception by Interacting with NaV1.7. Neuron. 2017;93(806–821):e9. doi: 10.1016/j.neuron.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 52.Dib-Hajj SD, Yang Y, Black JA, Waxman SG. The NaV1.7 sodium channel: from molecule to man. Nat Rev Neurosci. 2013;14:49–62. doi: 10.1038/nrn3404. [DOI] [PubMed] [Google Scholar]