Abstract
Most attempts at rational development of new analgesics have failed, in part because chronic pain involves multiple processes that remain poorly understood. To improve translational success, one strategy is to select novel targets for which there is proof of clinical relevance, either genetically through heritable traits, or pharmacologically. Such an approach by definition yields targets with high clinical validity. The biology of these targets can be elucidated in animal models before returning to the patients with a refined therapeutic. For optimal treatment, having biomarkers of drug action available is also a plus. Here we describe a case study in rational drug design: the use of controlled inhibition of peripheral tetrahydrobiopterin (BH4) synthesis to reduce abnormal chronic pain states without altering nociceptive-protective pain. Initially identified in a population of patients with low back pain, the association between BH4 production and chronic pain has been confirmed in more than 12 independent cohorts, through a common haplotype (present in 25% of Caucasians) of the rate-limiting enzyme for BH4 synthesis, GTP cyclohydrolase 1 (GCH1). Genetic tools in mice have demonstrated that both injured sensory neurons and activated macrophages engage increased BH4 synthesis to cause chronic pain. GCH1 is an obligate enzyme for de novo BH4 production. Therefore, inhibiting GCH1 activity eliminates all BH4 production, affecting the synthesis of multiple neurotransmitters and signaling molecules and interfering with physiological function. In contrast, targeting the last enzyme of the BH4 synthesis pathway, sepiapterin reductase (SPR), allows reduction of pathological BH4 production without completely blocking physiological BH4 synthesis. Systemic SPR inhibition in mice has not revealed any safety concerns to date, and available genetic and pharmacologic data suggest similar responses in humans. Finally, because it is present in vivo only when SPR is inhibited, sepiapterin serves as a reliable biomarker of target engagement, allowing potential quantification of drug efficacy. The emerging development of therapeutics that target BH4 synthesis to treat chronic pain illustrates the power of combining human and mouse genetics: human genetic studies for clinical selection of relevant targets, coupled with causality studies in mice, allowing the rational engineering of new analgesics.
Keywords: Chronic pain, Genetics, Mouse models, Tetrahydrobiopterin, Sepiapterin reductase, Analgesics
Introduction
Most attempts to develop new analgesics based on rational knowledge of pain pathways have failed, mainly due to lack of efficacy in phase II clinical trials, despite robust preclinical efficacy [1–3]. As a result, most analgesics currently prescribed for chronic pain were discovered by serendipity or empirical observation and consist of derivatives of ancient remedies such as the opiates (e.g. morphine), cannabinoids (e.g. sativex), cocaine analogues (e.g. lidocaine), and salicylates (e.g. aspirin). Further examples include compounds developed for other indications that patients have found to be efficacious analgesics, such as anticonvulsants (e.g. gabapentin and pregabalin) and tricyclic antidepressants (e.g. nortriptyline) [4–8]. Each of these treatments displays only moderate efficacy (30% improvement on average) with numerous side-effects (mostly due to disturbances in the central nervous system (CNS)) and these issues lead to a high level of patient dropout. In addition, some analgesics, notably opioid-based, exhibit strong tolerance and a risk of addiction, a particular worry in the control of chronic pain [9]. There is, therefore, a critical need to identify new biological targets to develop new classes of analgesics.
Why is Developing New Analgesics So Complex?
Although treatments conceived rationally have been very successful for a wide variety of diseases such as cancer or hypertension, most preclinical studies of pain in animals have not translated well into effective human drugs [10]. There are many possible explanations, including the following: chronic pain is caused by multiple processes, many of which remain poorly understood; in many cases, the mechanisms of drug action remain ambiguous; patient variability due to genetic and/or environmental factors results in differing drug responses; pain is notoriously difficult to quantify accurately across patients; and the source and/or original precipitating condition that results in chronic pain is often ambiguous. Comorbidities such as anxiety or depression further complicate chronic pain diagnosis.
This incomplete understanding at the clinical level has complicated preclinical studies, generating ambiguity with regard to critical design issues, including appropriate pain outcomes for predictive studies and choice of animal models for specific pain states. These factors are likely to have contributed to the fact that some therapies designed exclusively in pre-clinical models have subsequently shown no utility in patients [11, 12]. Such issues have greatly impeded rational pain drug design. Therefore, specifying and understanding the salient issues is a crucial first step towards future success.
How to Improve the Chance of Identifying a Target Relevant for Pain Patients
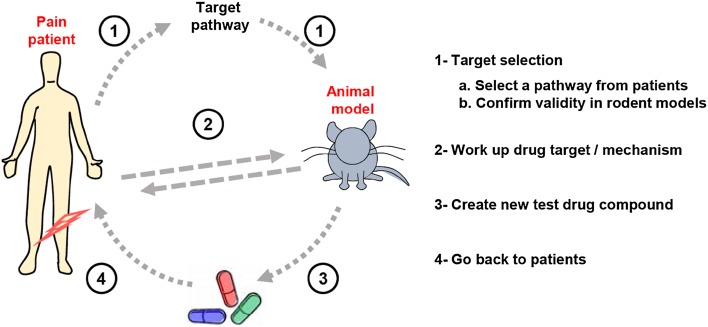
One strategy to improve translational success in developing drugs for pain is to select a target for which there is preexisting clinical support. This kind of support includes evidence from genetic association or congenic disease [13, 14], clues from naturally-occurring analgesic compounds in venoms or other sources [15, 16], and clinical experience with existing drugs with incomplete effects or that have been repositioned to treat pain [17–19]. Animal research is essential to understand biological pathways and formally demonstrate causality, but effective and efficient translation can only be achieved by regularly cycling model organism work with patient analysis, to keep the questions asked in the animal models relevant to the task at hand, namely developing new effective analgesics for humans (Fig. 1).
Fig. 1.
Reverse-engineering a relevant pathway from pain patients into rodents and back to patients. A complex interplay needs to be established between the patient and the experimental animal in order to achieve rational therapy design most efficiently.
Using Patient Genetics to Define New Rational Drug Targets
Currently, two major approaches in human genetics are used to define new molecular targets for pain drug development. The first is to screen for phenotypes with a strong effect size, but low population frequency, such as the damaging mutations responsible for congenital insensitivity to pain. These include recessive loss-of-function mutations for the Na+ channel Nav1.7 [20, 21] or nerve growth factor (NGF) signaling demonstrated by tropomyosin receptor kinase A and NGF-beta mutations [22–24] (Fig. 2A). Such drastic loss (or in some cases gain) of function mutations are extremely rare in the population, limiting the knowledge base that can be built from scientific investigation of their mechanisms and a full understanding of patient phenotype. However, modern technologies such as patient-derived stem cells [25] and CRISPR-mediated mutation modeling [26, 27] are beginning to aid this research area immensely and should allow high-throughput screening for mutations associated with hypo- or hyperexcitablity of nociceptors, both in mouse and human differentiated neurons (or other cell types of interest) in vitro [28].
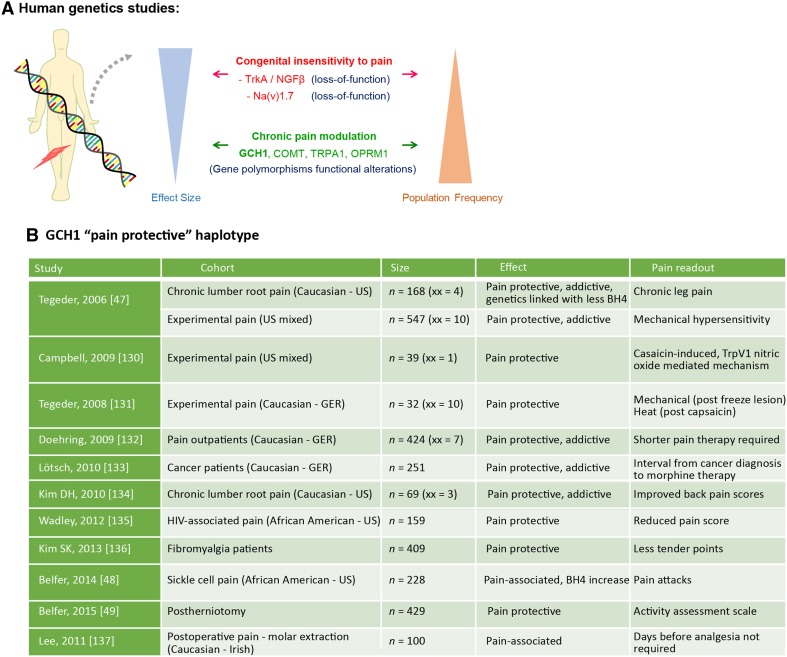
Fig. 2.
Human genetic studies for pain. A Strategies to identify pain-relevant genes by screening for rare congenic mutations with high effect size (e.g. congenital insensitivity to pain), or more common genetic variations with more modest effect size (e.g. polymorphisms). B Effects of GCH1 pain protective haplotype in various pain conditions. xx, homozygous carriers.
One potential confound of using highly penetrant genetic diseases with large effect sizes to define targets is that while the experimental aim is to reduce chronic pain hypersensitivity, these genes often play a fundamental role in protective, nociceptive pain transmission [14, 29, 30]. As a result, blocking these targets can block all nociceptive sensation completely, or paradoxically, can block nociceptive pain, with little or no effect on abnormal pathological pain, such as neuropathic pain [31, 32]. The goal is to develop medications for chronic pain that block abnormal pain hypersensitivity while maintaining nociceptive pain.
A fundamental advantage of harnessing the power of rare congenic disease is the relative ease of the clinical and molecular research involved, consisting of family identification, patient phenotyping, and exome sequencing. Such methods are straightforward relative to the complex analysis of large patient groups with complex phenotypes and mixed disease types. These extremely powerful congenic methods have led to great advances in understanding Na+ channel biology [33] and neuropathic pain therapeutics [34, 35], as well as in the action of NGF in pain mechanisms [36, 37]. However, targeting factors whose blockade can cause congenital insensitivity to pain may also affect other biological systems. For example, while pain therapies that inhibit NGF signaling have been shown to be effective analgesics, some strong side-effects in bone and cartilage have been reported and it remains to be determined how common these are across wide groups of patients [38, 39].
Although more complex, the second approach is to focus on genetic variations with more modest effect sizes but more commonly found in the general population (i.e. polymorphisms) that are directly related to chronic pain mechanisms (Fig. 2A). These genetic variations are usually not associated with any major defect in nociceptive pain, but instead with alterations of the risk of developing chronic pain and/or modulations of ongoing pain hypersensitivity. As such, characterizing them can identify specific molecular cascades responsible for inducing pathological pain hypersensitivity, ideal targets for developing new analgesics. With the advent of genome-wide-association studies in humans, several single nucleotide polymorphism (SNP) candidates have been identified in a non-biased manner for various chronic pain states [40]. Among these, many had already been implicated as pain-related genes in existing animal studies, including catechol-O-methyltransferase (COMT), opioid receptor mu-1 (OPRM1), and transient receptor potential cation channel A1 (TRPA1) [30, 41, 42], but several genes that were not previously associated with chronic pain states have also been discovered, such as K+ voltage-gated channel modifier subfamily S member 1 (KCNS1) and Ca2+ voltage-gated channel auxiliary subunit gamma 2 (CACNG2) [43–45] and others reviewed in [46].
One cascade that we have been interested in over the past decade is the pathway leading to pathological tetrahydrobiopterin (BH4) synthesis, which occurs in the damaged somatosensory system and can lead to chronic neuropathic and inflammatory pain. Through a combination of expression profiling and pharmacological analysis in the rat, we identified BH4 upregulation in injured sensory neurons and its rate-limiting enzyme GTP cyclohydrolase 1 (GTPCH1 or GCH1) as important in chronic pain signaling. Crucially, we also identified a specific haplotype of the human GCH1 gene among patients with low back pain which protected carriers from higher levels of chronic pain [47]. The mechanistic link to chronic pain in patients allowed us to define this cascade as potentially drugable. Further, since our initial description of the association of GCH1 with chronic pain in 2006, this link has been confirmed in more than twelve cohorts, demonstrating a robust link across multiple studies and populations [48–50] (Fig. 2B).
A Case Study in Rational Drug Design—Controlled Inhibition of Tetrahydrobiopterin Synthesis
GCH1 catalyzes the initial and rate-limiting step in the synthetic pathway of the pteridin (6R)-L-erythro-5,6,7,8-tetrahydrobiopterin (BH4) (Fig. 3A). BH4 is a critical co-factor for aromatic amino-acid hydroxylases such as phenylalanine hydroxylase [51–53], tyrosine hydroxylase [54], and tryptophan hydroxylases 1 and 2 [55, 56], as well as for all three isoforms of nitric oxide synthase [57–59], and alkylglycerol monooxygenase [60], making it indispensable for the synthesis of serotonin, epinephrine, norepinephrine, dopamine, and nitric oxide, and for adequate metabolism of phenylalanine and glycerolethers.
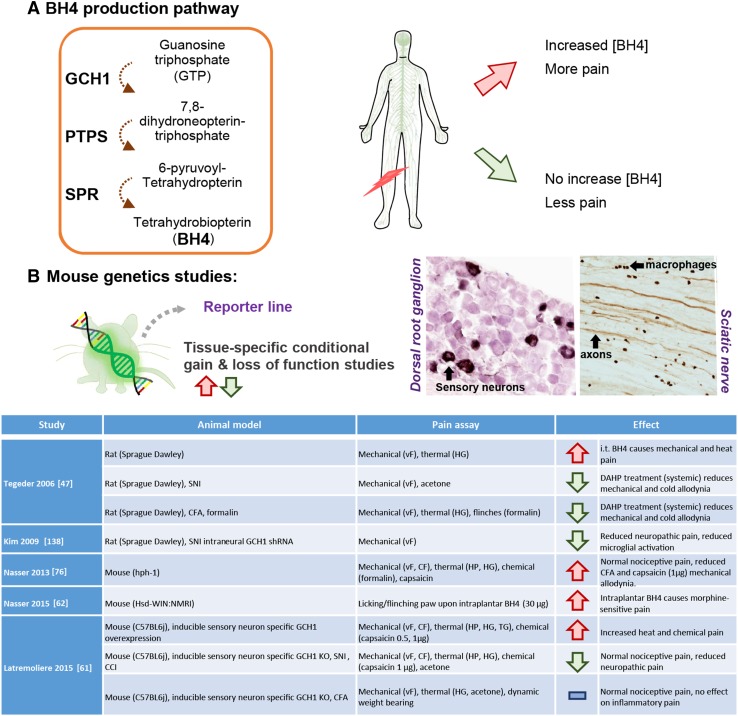
Fig. 3.
Tetrahydrobiopterin (BH4) is associated with pain hypersensitivity. A BH4 de novo production pathway. In patients carrying the pain-protective haplotype, stimulated lymphocytes produce less BH4 in patients who are less likely to develop chronic pain hypersensitivity. B Use of transgenic mouse models to specifically target the BH4 pathway reveals two major anatomical sites and cell types responsible for pain hypersensitivity: injured sensory neurons and macrophages within the sciatic nerve. Table describes existing rodent studies into BH4 and pain mechanisms. GCH1, GTP cyclohydrolase I; PTPS, pyruvoyl-tetrahydropterin synthase; SPR, sepiapterin reductase. vF, von Frey test; CF, calibrated forceps; HG, Hargreaves; HP, hot plate; TG, thermal gradient. Doses are indicated in brackets.
In defining the GCH1 pain-protective haplotype, we first linked (associated) a genetic version (haplotype) of GCH1 with less pain in a group of back pain patients. A major challenge is to move from an association of patient phenotype with their genotype to a functional relationship between a gene variant and its biological outcome. To achieve this in our study, we used immortalized leucocytes derived from the cohort participants as experimental proxies for the genetic response of each patient. Immortalized leucocytes from homozygous haplotype carriers (~2% of population) displayed reduced basal GCH1 transcriptional activity and decreased induction of BH4 upon stimulation. Furthermore, heterozygous carriers of the GCH1 haplotype (~22% of the population) produced intermediate levels of BH4 and GCH1 transcription, whereas those without the haplotype produced relatively high levels of BH4 and GCH1 expression. These results demonstrate a functional connection of BH4 synthesis upon cellular stimulation to the patient pain levels, a link previously only associated with the SNPs in the GCH1 gene [47] (Fig. 3A).
While congenic disease is usually caused by a damaging change in a gene’s protein coding sequence that directly confers the phenotype, differences spotted by association genetics can be two-fold. This method can not only identify differences in gene sequence, but it can also find changes in the genomic structure of the gene that alter gene expression and therefore gene function. Our functional data linking the level of BH4 production following patient leukocyte stimulation to patient pain phenotype strongly suggest that our haplotype is involved in GCH1 regulation, rather than damaging loss-of-function mutations, which would present as L-dopa-responsive dystonia (below) rather than lower levels of chronic pain. The existence of healthy GCH1 haplotype carriers [50], combined with the understanding that this haplotype acts specifically on new BH4 production, suggests that targeting BH4 synthesis in patients may reduce chronic pain without major side-effects.
From a Clinically Relevant Pathway to Mouse Genetic Studies
The specific contribution of excess BH4 production in pain hypersensitivity was confirmed in transgenic mice in which selective Cre recombinase expression allows specific overproduction or knockout of BH4 within a tissue of interest. Forced BH4 over-expression in all sensory neurons (Advillin lineage) or only in medium- and small-diameter sensory neurons (Na(V)1.8 lineage) is sufficient to heighten heat pain sensitivity in otherwise uninjured mice [61], supporting previous observations showing that intraplantar or intrathecal injection of BH4 increases pain sensitivity [47, 62, 63]. BH4 administered intraplantarly or intrathecally, however, is unstable and is very quickly reduced into BH2 [64]. BH2 and, to some limited extent, BH4 are transported into the cells through the nucleoside transporters ENT1 and ENT2 [65], where BH2 is then transformed back into BH4 under the action of dihydrofolate reductase or dihydropteridine reductase [66]. These multiple steps to function complicate interpretation of studies in which BH4 is injected directly into tissue in vivo.
The enhanced pain sensitivity caused by high intracellular levels of BH4 in uninjured sensory neurons is mostly mediated by an increase in nitric oxide (NO) production, as the administration of the NO synthase inhibitor L-NG-nitroarginine methyl ester can restore normal pain sensitivity in mice that overexpress BH4 within their sensory neurons specifically [61]. NO lowers the activation threshold for both TRPV1 and TRPA1 ion channels via nitrosylation [67], leading to increased Ca2+ influx into the sensory neurons following activation by various noxious stimuli (heat, low pH, or chemical irritants). Increased intracellular Ca2+ promotes cellular excitability and activates several pro-nociceptive cascades such as the phosphatidylinositol-3 kinases pathway [68]. In the injured peripheral nervous system, increased BH4 levels can also lead to the overproduction of serotonin, most likely produced by non-neuronal cells at the periphery, which can in turn sensitize nociceptors through activation of 5-HT2A [69] and 5-HT3 [70] receptors. In addition, overproduction of noradrenaline can increase the production of pro-inflammatory cytokines through α1-adrenergic receptors on macrophages [71]. Intracellular BH4 can affect macrophages by modulating the activity of the pro-inflammatory transcription factor nuclear factor erythroid 2 [72] as well as their lipidome through alkylglycerol monooxygenase [60]. Finally, a chronic increase in the cytoplasmic BH2:BH4 ratio can also cause the ‘uncoupling’ of NOS, leading to overproduction of reactive oxygen species, which can damage cells [73–75] and sensitize nociceptors.
Preventing BH4 production specifically in sensory neurons significantly attenuates the development of nerve injury-induced tactile hypersensitivity (allodynia) [61]. The ability to reduce these symptoms once they are fully established, however, is an essential prerequisite for effective therapeutic applications. To model this situation, we used inducible tissue-specific Cre-expressing transgenic mouse lines. We first established neuropathic pain symptoms in these mice and then blocked BH4 production specifically in sensory neurons. This also alleviated tactile pain hypersensitivity, confirming that targeting this pathway is a relevant therapeutic strategy (Fig. 3B). The ability to specifically modulate a target pathway in specific cell types and temporally regulate it relative to nerve injury or other experimental procedures, makes mouse genetic models invaluable to us in defining the link between a molecular pathway and specific pain phenotypes.
What is the Best Way to Systemically Reduce BH4 Production?
Since GCH1 is the rate-limiting enzyme for BH4 production, it is a powerful tool for gain- and loss-of-function studies allowing us to manipulate BH4 levels genetically in specific cell types, while leaving synthesis intact in other organs to limit confounding physiological factors. However, GCH1 is likely not the best target for reducing BH4 production systemically because it catalyzes an obligate step in synthesis, which no other enzyme can perform. Inhibiting GCH1 therefore carries a great risk of over-inhibiting the BH4 pathway, resulting in pathological rundown of BH4 in all cells, which will likely precipitate serious side-effects similar to those seen in global genetic BH4 deficiencies.
Inhibiting GCH1 systemically using 2,4 diamino-6-hydroxypyrimidine administration transiently reduces neuropathic and inflammatory pain hypersensitivity in rats [47] and mice [76]. Sustained reduction of GCH1 activity is associated with reduced inflammatory pain [76], as observed in HPH-1 mice, where damage to transcription control mechanisms involved in the basal expression of GCH1 leads to chronic reduction of GCH1 in many tissues [77–79]. However, it also leads to pulmonary hypertension, hyperphenylalaninemia, and severely reduced mono-amine production [79–82], which in turn dysregulates anxiety- and depression-like behaviors [83]. This profile is coherent with data from patients with a moderate reduction in global BH4 levels caused by a dominant-negative mutation of GCH1 (L-Dopa-responsive dystonia; also known as Segawa’s disease [84]). Here, BH4 synthesis is chronically inhibited but not prevented, causing relatively mild motor dysfunction that can be treated by L-dopa administration [85]. At the other end of the spectrum, recessive mutations within the GCH1 gene leading to an almost complete loss of function of the enzyme (and therefore a complete lack of BH4), are associated with severe systemic deficiencies including severe cognitive delays, dystonia and tremors (with diurnal fluctuation), and hyperphenylalaninemia [86–88]; problems that were likely created, if not seriously exacerbated, by developmental loss of GCH1 expression [89].
Alternative Ways to Modulate Pathological Increases in BH4 Synthesis
The evidence from the GCH1 pain-protective haplotype, where pathological BH4 synthesis is specifically absent only in chronic pain conditions, and from other genetic conditions that model systemic rundown of BH4, suggest that partial rather than complete inhibition is key to effective analgesic design. SPR is the last enzyme of the de novo BH4 synthesis pathway and carries out two reduction reactions. First, SPR converts 6-pyruvoyl-tetrahydropterin (produced by pyruvoyl-tetrahydropterin synthase) to 1’-hydroxy-2’-oxopropyltetrahydropterin (2’oxoPH4), then it catalyzes the reaction towards the formation of 1’-oxo-2’-hydroxypropyltetrahydropterin (1’oxoPH4; also known as 6-lactoyl tetrahydropterin) to produce BH4. Unlike the case for loss of GCH1 function, if SPR is absent or inhibited, 6-pyruvoyltetrahydrobiopterin can still be alternately reduced into BH4 by several enzymes that collectively form the BH4 salvage pathway [88, 90, 91] (Fig. 4A). This includes several independent enzymatic routes: the first pathway described, salvage pathway I, involves the successive action of an aldo-keto reductase, a carbonyl reductase, and dihydrofolate reductase (DHFR) [92–94]. Salvage pathway II relies on two specific isoforms of aldo-keto reductase (AKR1C3 and AKR1B1 in humans, respectively AKR1C18 and AKR1B3 in mice) to produce BH4 from 6-pyruvoyl-tetrahydropterin [91]. The tissue expression and enzymatic activity of each of these proteins are quite heterogeneous. As a result, it appears that overall, the CNS has a lower BH4 salvage pathway activity than peripheral organs, especially in humans [91, 92, 94, 95].
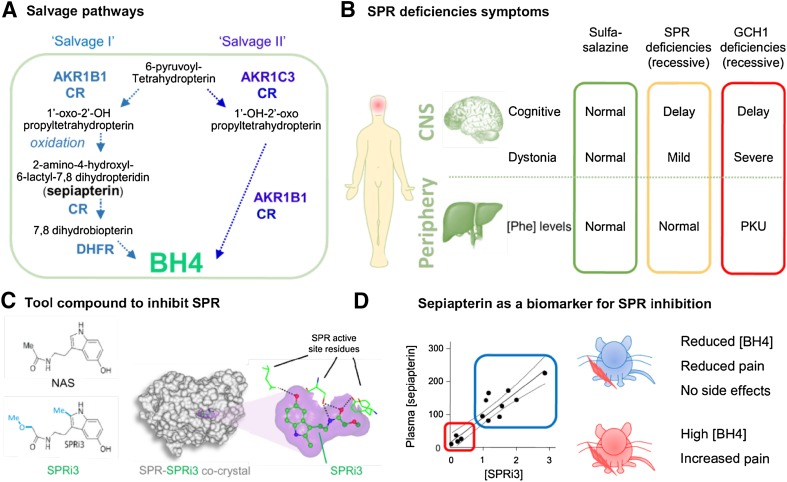
Fig. 4.
Targeting the BH4 pathway to reduce pain hypersensitivity. A Description of the salvage pathways that can produce essential cellular BH4 in the absence of SPR. Note that sepiapterin is not an endogenous SPR ligand and can only be produced when SPR is inactive. B Description of symptoms associated with recessive SPR and GCH1 deficiencies and after sulfasalazine treatment, a recently-identified SPR inhibitor. Most symptoms of SPR deficiency are central and happen during development. C A potent inhibitor was designed using a structure-based approach from N-acetyl-serotonin (NAS) to better fit the active pocket of SPR, and this was confirmed by co-crystal analysis. D Sepiapterin is a reliable biomarker for SPR inhibition that can be measured in plasma and confirms target engagement. Low levels of sepiapterin correspond to insufficient SPR inhibition, high BH4 levels, and pain hypersensitivity. High levels of sepiapterin indicate strong SPR inhibition, reduced BH4 levels, and reduced pain hypersensitivity. At these doses, no major side-effects were observed. PKU, phenylketonuria (= hyperphenylalaninemia).
For many years, autosomal recessive SPR deficiencies were not recognized. However, in 2001, two patients were characterized as homozygous carriers of loss-of-function mutations within the SPR gene and a more detailed clinical profile of the genetic effects of loss of this gene was defined (Fig. 4B) [94]. Since then, more than 50 patients with autosomal SPR deficiency have been documented, many having been initially misdiagnosed as suffering from cerebral palsy [96]. Common symptoms are mild motor defects (dystonia) with diurnal variation (symptoms are worse towards the end of the day) and oculogyric crises [94, 96–103]. Cognitive delay can occur, but most only have mild to moderate learning disabilities. Motor impairments can be substantially improved with L-Dopa. In some cases, patients also display mild hypersomnia and shortened ultradian cycles (12 h instead of 24 h) that can be treated by with 5-HT therapies [96, 98, 104, 105] (Fig. 4B).
The profile of SPR-deficient patients is encouraging with regard to the safety of therapeutic strategies targeting SPR in the periphery. Most of the side-effects of SPR deletion in patients are limited to the CNS, while peripheral function, such as phenylalanine metabolism and cardiovascular features, appear normal. Many of the cognitive impairments are developmental and can be mitigated when treated early enough [96], therefore these do not represent a strong risk for the population in need of chronic pain treatments. Later in life, if SPR-deficient patients stop their L-dopa/5-HTP treatment, they only exhibit a return of mild dystonia and hypersomnia, the latter being caused by a loss of melatonin tone (for which 5-HT is a precursor), and these symptoms disappear when treatment is reinitiated [98].
Another strong argument in favor of the safety profile expected from systemic SPR inhibition comes from the recent discovery that sulfasalazine (SSZ), a Food and Drug Administration-approved drug used to treat various inflammatory diseases such as rheumatoid arthritis (RA) [106, 107], and sulfapyridine, its metabolite, are SPR inhibitors [108, 109] (Fig. 4B). Administration of extremely high doses of SSZ can in some rare cases lead to some motor impairments (mostly dystonia) with concentrations of SSZ and sulfapyridine in the cerebrospinal fluid (CSF) consistent with central SPR inhibition [108]. At much lower doses than those required to reach pathological exposure in the CNS, SSZ administration can reduce chronic pain symptoms in RA patients in the clinic [110] and diabetic neuropathic pain in rodents [111]. Since (i) SSZ-related CNS side-effects are extremely rare, (ii) motor symptoms caused by loss of SPR can be easily treated by L-dopa administration, and (iii) the involvement of BH4 in chronic pain states seems mostly limited to peripheral tissues (sensory neurons and macrophages), SPR inhibition appears to be a good logical approach to reduce pathological BH4 production after nerve injury or during chronic inflammation without causing systemic pathological BH4 deficiency.
We used a structure-based approach to find small molecule compounds with improved affinity and potency relative to the endogenous SPR inhibitor, N-acetyl-serotonin, and identified SPRi3. Systemic administration of this compound reduced pain hypersensitivity in mouse models of neuropathic and chronic inflammatory pain [61] (Fig. 4C). Systemic administration of SPRi3 to mice did not alter motor, exploratory, or depression-like behaviors, which are especially sensitive to BH4 deficiencies. In addition, sub-chronic treatment (twice daily injections for 3 days) did not affect amine levels in the brain, or phenylalanine metabolism [61]. SPR activity was reduced in tissues of interest (dorsal root ganglia and sciatic nerve for neuropathic pain, hindpaw skin for inflammatory pain caused by intraplantar injection of complete Freund’s adjuvant), and led to decreased BH4 levels [61]. SPRi3 administration significantly reduced the swelling caused by ongoing inflammation as well as NO production in macrophages, suggesting a possible disease-modifying role of BH4 in inflammatory processes, similar to those seen for SSZ [61]. Interestingly, methotrexate, which inhibits the BH4 salvage pathway enzyme dihydrofolate reductase and consequently reduces cellular BH4 levels, is a first-line medication for inflammatory states such as RA [112, 113]. It is therefore tempting to speculate that BH4 production could play a more pivotal role in chronic inflammation than initially thought.
Sepiapterin: A Biomarker for Personalized Medicine?
Since the two reactions that convert 6-pyruvoyl-tetrahydropterin into BH4 occur within SPR’s active pocket, the intermediates 1’oxoPH4 and 2’oxoPH4 are normally not present in the cytosol. In contrast, when SPR is blocked, aldo-keto-reductases and carbonyl reductases in the salvage pathways can produce these two compounds. However, 1’oxoPH4 and 2’oxoPH4 are very unstable and, in the absence of enzymatic activity, 1’oxoPH4 oxidizes into 2-amino-4-hydroxyl-6-lactyl-7,8 dihydropteridin (also known as sepiapterin), which can be further metabolized into BH2 by carbonyl reductase, and then to BH4 by DHFR through salvage pathway I. The name “sepiapterin” is derived from the fact that it is the biochemical structure found in the eye pigments of the mutant line of Drosophila melanogaster ‘sepia’ [114–117]. The enzyme capable of reducing sepiapterin in vitro was purified a decade later and named sepiapterin reductase [118]. Sepiapterin is therefore not the endogenous substrate for SPR [119] and actually only accumulates when SPR is absent or blocked. While carbonyl reductase and DHFR can carry out the reactions from sepiapterin to BH4, their efficiency is much lower than that of SPR. As a result, there is an accumulation of sepiapterin, which increases in the cytosol, but is also found extracellularly [61] (Fig. 3A). Extremely stable, sepiapterin can be detected in CSF or urine from patients with SPR deficiencies [94, 120], and in plasma from mice after acute SPR inhibition [61]. Since sepiapterin is not detectable when SPR is active, and its production is strongly correlated with the amount of SPR inhibition, it represents a reliable biomarker for target engagement by an SPR-inhibiting compound. Such a biomarker could be extremely helpful to titrate the degree of SPR inhibition required for an adequate decrease in BH4 production without over-inhibition (which would manifest with mild motor defects) (Fig. 3D). Over time, it may be possible to use the sepiapterin levels in blood or urine to correlate levels of SPR inhibition in patients with effective pain relief to allow tailored dosing of an SPR-inhibiting drug. Indeed, we used this biomarker to demonstrate that the SPRi3 compound is a more effective SPR inhibitor than SZZ at the maximum dose deliverable in mice, and this difference correlates well with analgesic efficacy [61].
Combining Human and Rodent Genetics to Perform Personalized Medicine–A Path Forward
Chronic pain conditions have for too long been diagnosed as essentially a single disorder, but many distinct diseases exist within the ‘chronic pain’ diagnosis. To develop effective treatments, each of these must be clearly defined and studied so that clinicians can readily differentiate them and achieve more efficient, personalized medicine [121]. While human genetics can identify potentially relevant targets in patients, and preclinical studies can formally demonstrate their action in animal models, the drugs developed from these studies are not likely to work for all manifestations of chronic pain. As new mechanistic subtypes of ongoing pain are identified, further animal model work will be required to understand the mechanisms at play and how the drugs developed to target each new cascade act. The use of treatment-specific biomarkers should help in both the study and application of this rational mechanism-based approach by identifying responders versus non-responders for each therapeutic class. Genetic screening tools, behavioral evaluation, or ideally both, will help improve patient phenotyping [122]. Improving our understanding of how individual mechanisms relate to pain symptoms will likely require a more detailed quantitative analysis of individual pain modalities, via a more comprehensive use of quantitative sensory testing across all pain patients [123–125].
To precisely phenotype individuals, we should consider whole-genome sequencing for all patients as soon as it is practically possible. Currently, our understanding of how the structure of a genome is translated into a phenotype is miniscule [126]. However, this knowledge will increase with time, and the more people sequenced, the quicker we will learn [127, 128]. Using genome-editing tools such as CRISPR to alter in vitro reprogrammed cells derived from pain patients could represent a breakthrough for mechanism-based medicine, where screening to assess genetic susceptibilities to abnormal pain development will be coupled with functional genomics to formally confirm the involvement of a specific pathway, such as pathological BH4 synthesis [129]. However, while such approaches would give access to human-derived cells which closely mimic their endogenous counterparts, they will remain in vitro assessments, which do not take into account the physiological interactions that occur in vivo. Such strategies will still need to be coupled with animal model studies to understand how identified defects affect pain pathways and the resultant symptoms in vivo.
Using genetic material from patients raises multiple legal and ethical concerns that will need to be considered before implementing these approaches systematically, including privacy rights, the right to abstain, and life and health insurance liabilities [128]. Environmental risk factors are clearly key to triggering some types of chronic pain, such as that induced by chemotherapeutic agents, alcohol abuse, and diabetes, and for each we need an in-depth knowledge of the precipitating mechanism. Again, this will only be possible through collaborative efforts in the laboratory and clinic. As our understanding develops, we can look forward to the day when those at risk of certain chronic conditions can be advised or treated ahead of time. Until that point, we must push to develop specific strategies to offer individualized treatment for those diseases we truly understand at the molecular level. While still a formidable task, the emergent success of controlled inhibition of excess BH4 synthesis in chronic pain helps demonstrate how the advent of molecular genetic medicine is making this ever more achievable.
Acknowledgements
We thank Drs Susan E. Lewis and Nick Andrews for helpful comments and suggestions and careful reading. AL was supported by NIH grant DE022912. MC was supported by NIH grant NS074430.
Complaince with Ethical Standards
Conflict of interest
AL and MC are founding share owners in Quartet Medicine, a company created to produce clinically relevant inhibitors of SPR. These inhibitors do not include SPRi3 which is available as a research tool on request.
Contributor Information
Alban Latremoliere, Email: alban.latremoliere@childrens.harvard.edu, Email: allodynie@hotmail.com.
Michael Costigan, Email: michael.costigan@childrens.harvard.edu.
References
- 1.van der Worp HB, Howells DW, Sena ES, Porritt MJ, Rewell S, O’Collins V, et al. Can animal models of disease reliably inform human studies? PLoS Med. 2010;7:e1000245. doi: 10.1371/journal.pmed.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook D, Brown D, Alexander R, March R, Morgan P, Satterthwaite G, et al. Lessons learned from the fate of AstraZeneca’s drug pipeline: a five-dimensional framework. Nat Rev Drug Discov. 2014;13:419–431. doi: 10.1038/nrd4309. [DOI] [PubMed] [Google Scholar]
- 3.Kissin I. Scientometrics of drug discovery efforts: pain-related molecular targets. Drug Des Devel Ther. 2015;9:3393–3404. doi: 10.2147/DDDT.S85633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dworkin RH. Introduction: Recommendations for the diagnosis, assessment, and treatment of neuropathic pain. Am J Med. 2009;122:S1–S2. doi: 10.1016/j.amjmed.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Dworkin RH, Backonja M, Rowbotham MC, Allen RR, Argoff CR, Bennett GJ, et al. Advances in neuropathic pain: diagnosis, mechanisms, and treatment recommendations. Arch Neurol. 2003;60:1524–1534. doi: 10.1001/archneur.60.11.1524. [DOI] [PubMed] [Google Scholar]
- 6.Finnerup NB, Otto M, McQuay HJ, Jensen TS, Sindrup SH. Algorithm for neuropathic pain treatment: an evidence based proposal. Pain. 2005;118:289–305. doi: 10.1016/j.pain.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Finnerup NB, Sindrup SH, Jensen TS. The evidence for pharmacological treatment of neuropathic pain. Pain. 2010;150:573–581. doi: 10.1016/j.pain.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 8.O’Connor AB, Dworkin RH. Treatment of neuropathic pain: an overview of recent guidelines. Am J Med. 2009;122:S22–S32. doi: 10.1016/j.amjmed.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Corder G, Tawfik VL, Wang D, Sypek EI, Low SA, Dickinson JR, et al. Loss of mu opioid receptor signaling in nociceptors, but not microglia, abrogates morphine tolerance without disrupting analgesia. Nat Med. 2017;23:164–173. doi: 10.1038/nm.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill R. NK1 (substance P) receptor antagonists–why are they not analgesic in humans? Trends Pharmacol Sci. 2000;21:244–246. doi: 10.1016/s0165-6147(00)01502-9. [DOI] [PubMed] [Google Scholar]
- 12.Yaksh TL, Woller SA, Ramachandran R, Sorkin LS. The search for novel analgesics: targets and mechanisms. F1000Prime Rep 2015, 7: 56. [DOI] [PMC free article] [PubMed]
- 13.Dib-Hajj SD, Waxman SG. Translational pain research: Lessons from genetics and genomics. Sci Transl Med 2014, 6: 249sr244. [DOI] [PubMed]
- 14.Nahorski MS, Chen YC, Woods CG. New mendelian disorders of painlessness. Trends Neurosci. 2015;38:712–724. doi: 10.1016/j.tins.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Gazerani P, Cairns BE. Venom-based biotoxins as potential analgesics. Expert Rev Neurother. 2014;14:1261–1274. doi: 10.1586/14737175.2014.962518. [DOI] [PubMed] [Google Scholar]
- 16.Serhan CN, Arita M, Hong S, Gotlinger K. Resolvins, docosatrienes, and neuroprotectins, novel omega-3-derived mediators, and their endogenous aspirin-triggered epimers. Lipids. 2004;39:1125–1132. doi: 10.1007/s11745-004-1339-7. [DOI] [PubMed] [Google Scholar]
- 17.Ai N, Wood RD, Welsh WJ. Identification of Nitazoxanide as a Group I Metabotropic Glutamate Receptor Negative Modulator for the Treatment of Neuropathic Pain: An In Silico Drug Repositioning Study. Pharm Res. 2015;32:2798–2807. doi: 10.1007/s11095-015-1665-7. [DOI] [PubMed] [Google Scholar]
- 18.Sisignano M, Parnham MJ, Geisslinger G. Drug repurposing for the development of novel analgesics. Trends Pharmacol Sci. 2016;37:172–183. doi: 10.1016/j.tips.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Lotsch J, Doehring A, Mogil JS, Arndt T, Geisslinger G, Ultsch A. Functional genomics of pain in analgesic drug development and therapy. Pharmacol Ther. 2013;139:60–70. doi: 10.1016/j.pharmthera.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Dib-Hajj SD, Yang Y, Black JA, Waxman SG. The Na(V)1.7 sodium channel: from molecule to man. Nat Rev Neurosci. 2013;14:49–62. doi: 10.1038/nrn3404. [DOI] [PubMed] [Google Scholar]
- 21.Zakrzewska JM, Palmer J, Morisset V, Giblin GM, Obermann M, Ettlin DA, et al. Safety and efficacy of a Nav1.7 selective sodium channel blocker in patients with trigeminal neuralgia: a double-blind, placebo-controlled, randomised withdrawal phase 2a trial. Lancet Neurol. 2017;16:291–300. doi: 10.1016/S1474-4422(17)30005-4. [DOI] [PubMed] [Google Scholar]
- 22.Indo Y. Molecular basis of congenital insensitivity to pain with anhidrosis (CIPA): mutations and polymorphisms in TRKA (NTRK1) gene encoding the receptor tyrosine kinase for nerve growth factor. Hum Mutat. 2001;18:462–471. doi: 10.1002/humu.1224. [DOI] [PubMed] [Google Scholar]
- 23.Einarsdottir E, Carlsson A, Minde J, Toolanen G, Svensson O, Solders G, et al. A mutation in the nerve growth factor beta gene (NGFB) causes loss of pain perception. Hum Mol Genet. 2004;13:799–805. doi: 10.1093/hmg/ddh096. [DOI] [PubMed] [Google Scholar]
- 24.Indo Y. Nerve growth factor and the physiology of pain: lessons from congenital insensitivity to pain with anhidrosis. Clin Genet. 2012;82:341–350. doi: 10.1111/j.1399-0004.2012.01943.x. [DOI] [PubMed] [Google Scholar]
- 25.McNeish J, Gardner JP, Wainger BJ, Woolf CJ, Eggan K. From dish to bedside: lessons learned while translating findings from a stem cell model of disease to a clinical trial. Cell Stem Cell. 2015;17:8–10. doi: 10.1016/j.stem.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 26.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 27.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wainger BJ, Buttermore ED, Oliveira JT, Mellin C, Lee S, Saber WA, et al. Modeling pain in vitro using nociceptor neurons reprogrammed from fibroblasts. Nat Neurosci. 2015;18:17–24. doi: 10.1038/nn.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JH, Park CK, Chen G, Han Q, Xie RG, Liu T, et al. A monoclonal antibody that targets a NaV1.7 channel voltage sensor for pain and itch relief. Cell. 2014;157:1393–1404. doi: 10.1016/j.cell.2014.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Bennett DL, Woods CG. Painful and painless channelopathies. Lancet Neurol. 2014;13:587–599. doi: 10.1016/S1474-4422(14)70024-9. [DOI] [PubMed] [Google Scholar]
- 31.Hockley JR, Gonzalez-Cano R, McMurray S, Tejada-Giraldez MA, McGuire C, Torres A, et al. Visceral and somatic pain modalities reveal NaV 1.7-independent visceral nociceptive pathways. J Physiol. 2017;595:2661–2679. doi: 10.1113/JP272837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minett MS, Falk S, Santana-Varela S, Bogdanov YD, Nassar MA, Heegaard AM, et al. Pain without nociceptors? Nav1.7-independent pain mechanisms. Cell Rep 2014, 6: 301–312. [DOI] [PMC free article] [PubMed]
- 33.Waxman SG, Merkies IS, Gerrits MM, Dib-Hajj SD, Lauria G, Cox JJ, et al. Sodium channel genes in pain-related disorders: phenotype-genotype associations and recommendations for clinical use. Lancet Neurol. 2014;13:1152–1160. doi: 10.1016/S1474-4422(14)70150-4. [DOI] [PubMed] [Google Scholar]
- 34.Goldberg YP, Pimstone SN, Namdari R, Price N, Cohen C, Sherrington RP, et al. Human Mendelian pain disorders: a key to discovery and validation of novel analgesics. Clin Genet. 2012;82:367–373. doi: 10.1111/j.1399-0004.2012.01942.x. [DOI] [PubMed] [Google Scholar]
- 35.Rivara M, Zuliani V. Novel sodium channel antagonists in the treatment of neuropathic pain. Expert Opin Investig Drugs. 2016;25:215–226. doi: 10.1517/13543784.2016.1121992. [DOI] [PubMed] [Google Scholar]
- 36.Lewin GR, Lechner SG, Smith ES. Nerve growth factor and nociception: from experimental embryology to new analgesic therapy. Handb Exp Pharmacol. 2014;220:251–282. doi: 10.1007/978-3-642-45106-5_10. [DOI] [PubMed] [Google Scholar]
- 37.Chang DS, Hsu E, Hottinger DG, Cohen SP. Anti-nerve growth factor in pain management: current evidence. J Pain Res. 2016;9:373–383. doi: 10.2147/JPR.S89061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller RE, Block JA, Malfait AM. Nerve growth factor blockade for the management of osteoarthritis pain: what can we learn from clinical trials and preclinical models? Curr Opin Rheumatol. 2017;29:110–118. doi: 10.1097/BOR.0000000000000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mullard A. Drug developers reboot anti-NGF pain programmes. Nat Rev Drug Discov. 2015;14:297–298. doi: 10.1038/nrd4612. [DOI] [PubMed] [Google Scholar]
- 40.Hoofwijk DM, van Reij RR, Rutten BP, Kenis G, Buhre WF, Joosten EA. Genetic polymorphisms and their association with the prevalence and severity of chronic postsurgical pain: a systematic review. Br J Anaesth. 2016;117:708–719. doi: 10.1093/bja/aew378. [DOI] [PubMed] [Google Scholar]
- 41.Segall SK, Maixner W, Belfer I, Wiltshire T, Seltzer Z, Diatchenko L. Janus molecule I: dichotomous effects of COMT in neuropathic vs nociceptive pain modalities. CNS Neurol Disord Drug Targets. 2012;11:222–235. doi: 10.2174/187152712800672490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crist RC, Berrettini WH. Pharmacogenetics of OPRM1. Pharmacol Biochem Behav. 2014;123:25–33. doi: 10.1016/j.pbb.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costigan M, Belfer I, Griffin RS, Dai F, Barrett LB, Coppola G, et al. Multiple chronic pain states are associated with a common amino acid-changing allele in KCNS1. Brain. 2010;133:2519–2527. doi: 10.1093/brain/awq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsantoulas C, Zhu L, Shaifta Y, Grist J, Ward JP, Raouf R, et al. Sensory neuron downregulation of the Kv9.1 potassium channel subunit mediates neuropathic pain following nerve injury. J Neurosci. 2012;32:17502–17513. doi: 10.1523/JNEUROSCI.3561-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nissenbaum J. From mouse to humans: discovery of the CACNG2 pain susceptibility gene. Clin Genet. 2012;82:311–320. doi: 10.1111/j.1399-0004.2012.01924.x. [DOI] [PubMed] [Google Scholar]
- 46.Mogil JS. Pain genetics: past, present and future. Trends Genet. 2012;28:258–266. doi: 10.1016/j.tig.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 47.Tegeder I, Costigan M, Griffin RS, Abele A, Belfer I, Schmidt H, et al. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nat Med. 2006;12:1269–1277. doi: 10.1038/nm1490. [DOI] [PubMed] [Google Scholar]
- 48.Belfer I, Youngblood V, Darbari DS, Wang Z, Diaw L, Freeman L, et al. A GCH1 haplotype confers sex-specific susceptibility to pain crises and altered endothelial function in adults with sickle cell anemia. Am J Hematol. 2014;89:187–193. doi: 10.1002/ajh.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belfer I, Dai F, Kehlet H, Finelli P, Qin L, Bittner R, et al. Association of functional variations in COMT and GCH1 genes with postherniotomy pain and related impairment. Pain. 2015;156:273–279. doi: 10.1097/01.j.pain.0000460307.48701.b0. [DOI] [PubMed] [Google Scholar]
- 50.Latremoliere A, Costigan M. GCH1, BH4 and pain. Curr Pharm Biotechnol. 2011;12:1728–1741. doi: 10.2174/138920111798357393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaufman S. A new cofactor required for the enzymatic conversion of phenylalanine to tyrosine. J Biol Chem. 1958;230:931–939. [PubMed] [Google Scholar]
- 52.Kaufman S. Phenylalanine hydroxylation cofactor in phenylketonuria. Science. 1958;128:1506–1508. doi: 10.1126/science.128.3337.1506. [DOI] [PubMed] [Google Scholar]
- 53.Kaufman S, Levenberg B. Further studies on the phenylalanine-hydroxylation cofactor. J Biol Chem. 1959;234:2683–2688. [PubMed] [Google Scholar]
- 54.Kettler R, Bartholini G, Pletscher A. In vivo enhancement of tyrosine hydroxylation in rat striatum by tetrahydrobiopterin. Nature. 1974;249:476–478. doi: 10.1038/249476a0. [DOI] [PubMed] [Google Scholar]
- 55.Friedman PA, Kappelman AH, Kaufman S. Partial purification and characterization of tryptophan hydroxylase from rabbit hindbrain. J Biol Chem. 1972;247:4165–4173. [PubMed] [Google Scholar]
- 56.Sawada M, Sugimoto T, Matsuura S, Nagatsu T. (6R)-tetrahydrobiopterin increases the activity of tryptophan hydroxylase in rat raphe slices. J Neurochem. 1986;47:1544–1547. doi: 10.1111/j.1471-4159.1986.tb00792.x. [DOI] [PubMed] [Google Scholar]
- 57.Gorren AC, Bec N, Lange R, Mayer B. Redox role for tetrahydrobiopterin in nitric oxide synthase catalysis: low-temperature optical absorption spectral detection. Methods Enzymol. 2002;353:114–121. doi: 10.1016/s0076-6879(02)53041-x. [DOI] [PubMed] [Google Scholar]
- 58.Gorren AC, Mayer B. Tetrahydrobiopterin in nitric oxide synthesis: a novel biological role for pteridines. Curr Drug Metab. 2002;3:133–157. doi: 10.2174/1389200024605154. [DOI] [PubMed] [Google Scholar]
- 59.Tayeh MA, Marletta MA. Macrophage oxidation of L-arginine to nitric oxide, nitrite, and nitrate. Tetrahydrobiopterin is required as a cofactor. J Biol Chem. 1989;264:19654–19658. [PubMed] [Google Scholar]
- 60.Watschinger K, Keller MA, McNeill E, Alam MT, Lai S, Sailer S, et al. Tetrahydrobiopterin and alkylglycerol monooxygenase substantially alter the murine macrophage lipidome. Proc Natl Acad Sci U S A. 2015;112:2431–2436. doi: 10.1073/pnas.1414887112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Latremoliere A, Latini A, Andrews N, Cronin SJ, Fujita M, Gorska K, et al. Reduction of neuropathic and inflammatory pain through inhibition of the tetrahydrobiopterin pathway. Neuron. 2015;86:1393–1406. doi: 10.1016/j.neuron.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nasser A, Ali S, Wilsbech S, Bjerrum OJ, Moller LB. Intraplantar injection of tetrahydrobiopterin induces nociception in mice. Neurosci Lett. 2015;584:247–252. doi: 10.1016/j.neulet.2014.10.050. [DOI] [PubMed] [Google Scholar]
- 63.Nasser A, Birk Moller L. GCH1 variants, tetrahydrobiopterin and their effects on pain sensitivity. Scandinavian Journal of Pain. 2014;5:121–128. doi: 10.1016/j.sjpain.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 64.Hasegawa H, Sawabe K, Nakanishi N, Wakasugi OK. Delivery of exogenous tetrahydrobiopterin (BH4) to cells of target organs: role of salvage pathway and uptake of its precursor in effective elevation of tissue BH4. Mol Genet Metab. 2005;86(Suppl 1):S2–S10. doi: 10.1016/j.ymgme.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 65.Ohashi A, Sugawara Y, Mamada K, Harada Y, Sumi T, Anzai N, et al. Membrane transport of sepiapterin and dihydrobiopterin by equilibrative nucleoside transporters: a plausible gateway for the salvage pathway of tetrahydrobiopterin biosynthesis. Mol Genet Metab. 2011;102:18–28. doi: 10.1016/j.ymgme.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 66.Werner ER, Blau N, Thony B. Tetrahydrobiopterin: biochemistry and pathophysiology. Biochem J. 2011;438:397–414. doi: 10.1042/BJ20110293. [DOI] [PubMed] [Google Scholar]
- 67.Miyamoto T, Dubin AE, Petrus MJ, Patapoutian A. TRPV1 and TRPA1 mediate peripheral nitric oxide-induced nociception in mice. PLoS One. 2009;4:e7596. doi: 10.1371/journal.pone.0007596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhuang ZY, Xu H, Clapham DE, Ji RR. Phosphatidylinositol 3-kinase activates ERK in primary sensory neurons and mediates inflammatory heat hyperalgesia through TRPV1 sensitization. J Neurosci. 2004;24:8300–8309. doi: 10.1523/JNEUROSCI.2893-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okamoto K, Imbe H, Morikawa Y, Itoh M, Sekimoto M, Nemoto K, et al. 5-HT2A receptor subtype in the peripheral branch of sensory fibers is involved in the potentiation of inflammatory pain in rats. Pain. 2002;99:133–143. doi: 10.1016/s0304-3959(02)00070-2. [DOI] [PubMed] [Google Scholar]
- 70.Kayser V, Elfassi IE, Aubel B, Melfort M, Julius D, Gingrich JA, et al. Mechanical, thermal and formalin-induced nociception is differentially altered in 5-HT1A-/-, 5-HT1B-/-, 5-HT2A-/-, 5-HT3A-/- and 5-HTT-/- knock-out male mice. Pain. 2007;130:235–248. doi: 10.1016/j.pain.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 71.Grisanti LA, Woster AP, Dahlman J, Sauter ER, Combs CK, Porter JE. alpha1-adrenergic receptors positively regulate Toll-like receptor cytokine production from human monocytes and macrophages. J Pharmacol Exp Ther. 2011;338:648–657. doi: 10.1124/jpet.110.178012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McNeill E, Crabtree MJ, Sahgal N, Patel J, Chuaiphichai S, Iqbal AJ, et al. Regulation of iNOS function and cellular redox state by macrophage Gch1 reveals specific requirements for tetrahydrobiopterin in NRF2 activation. Free Radic Biol Med. 2015;79:206–216. doi: 10.1016/j.freeradbiomed.2014.10.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Crabtree MJ, Tatham AL, Al-Wakeel Y, Warrick N, Hale AB, Cai S, et al. Quantitative regulation of intracellular endothelial nitric-oxide synthase (eNOS) coupling by both tetrahydrobiopterin-eNOS stoichiometry and biopterin redox status: insights from cells with tet-regulated GTP cyclohydrolase I expression. J Biol Chem. 2009;284:1136–1144. doi: 10.1074/jbc.M805403200. [DOI] [PubMed] [Google Scholar]
- 74.Vasquez-Vivar J, Martasek P, Whitsett J, Joseph J, Kalyanaraman B. The ratio between tetrahydrobiopterin and oxidized tetrahydrobiopterin analogues controls superoxide release from endothelial nitric oxide synthase: an EPR spin trapping study. Biochem J. 2002;362:733–739. doi: 10.1042/0264-6021:3620733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vasquez-Vivar J, Kalyanaraman B, Martasek P. The role of tetrahydrobiopterin in superoxide generation from eNOS: enzymology and physiological implications. Free Radic Res. 2003;37:121–127. doi: 10.1080/1071576021000040655. [DOI] [PubMed] [Google Scholar]
- 76.Nasser A, Bjerrum OJ, Heegaard AM, Moller AT, Larsen M, Dalboge LS, et al. Impaired behavioural pain responses in hph-1 mice with inherited deficiency in GTP cyclohydrolase 1 in models of inflammatory pain. Mol Pain. 2013;9:5. doi: 10.1186/1744-8069-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khoo JP, Nicoli T, Alp NJ, Fullerton J, Flint J, Channon KM. Congenic mapping and genotyping of the tetrahydrobiopterin-deficient hph-1 mouse. Mol Genet Metab. 2004;82:251–254. doi: 10.1016/j.ymgme.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 78.McDonald JD, Cotton RG, Jennings I, Ledley FD, Woo SL, Bode VC. Biochemical defect of the hph-1 mouse mutant is a deficiency in GTP-cyclohydrolase activity. J Neurochem. 1988;50:655–657. doi: 10.1111/j.1471-4159.1988.tb02961.x. [DOI] [PubMed] [Google Scholar]
- 79.Bode VC, McDonald JD, Guenet JL, Simon D. hph-1: a mouse mutant with hereditary hyperphenylalaninemia induced by ethylnitrosourea mutagenesis. Genetics. 1988;118:299–305. doi: 10.1093/genetics/118.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Belik J, McIntyre BA, Enomoto M, Pan J, Grasemann H, Vasquez-Vivar J. Pulmonary hypertension in the newborn GTP cyclohydrolase I-deficient mouse. Free Radic Biol Med. 2011;51:2227–2233. doi: 10.1016/j.freeradbiomed.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hyland K, Gunasekera RS, Engle T, Arnold LA. Tetrahydrobiopterin and biogenic amine metabolism in the hph-1 mouse. J Neurochem. 1996;67:752–759. doi: 10.1046/j.1471-4159.1996.67020752.x. [DOI] [PubMed] [Google Scholar]
- 82.Nandi M, Miller A, Stidwill R, Jacques TS, Lam AA, Haworth S, et al. Pulmonary hypertension in a GTP-cyclohydrolase 1-deficient mouse. Circulation. 2005;111:2086–2090. doi: 10.1161/01.CIR.0000163268.32638.F4. [DOI] [PubMed] [Google Scholar]
- 83.Nasser A, Moller LB, Olesen JH, Konradsen Refsgaard L, Andreasen JT. Anxiety- and depression-like phenotype of hph-1 mice deficient in tetrahydrobiopterin. Neurosci Res. 2014;89:44–53. doi: 10.1016/j.neures.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 84.Segawa M, Hosaka A, Miyagawa F, Nomura Y, Imai H. Hereditary progressive dystonia with marked diurnal fluctuation. Adv Neurol. 1976;14:215–233. [PubMed] [Google Scholar]
- 85.Segawa M, Nomura Y, Nishiyama N. Autosomal dominant guanosine triphosphate cyclohydrolase I deficiency (Segawa disease) Ann Neurol. 2003;54(Suppl 6):S32–S45. doi: 10.1002/ana.10630. [DOI] [PubMed] [Google Scholar]
- 86.Longo N. Disorders of biopterin metabolism. J Inherit Metab Dis. 2009;32:333–342. doi: 10.1007/s10545-009-1067-2. [DOI] [PubMed] [Google Scholar]
- 87.Opladen T, Hoffmann GF, Blau N. An international survey of patients with tetrahydrobiopterin deficiencies presenting with hyperphenylalaninaemia. J Inherit Metab Dis. 2012;35:963–973. doi: 10.1007/s10545-012-9506-x. [DOI] [PubMed] [Google Scholar]
- 88.Thony B, Blau N. Mutations in the BH4-metabolizing genes GTP cyclohydrolase I, 6-pyruvoyl-tetrahydropterin synthase, sepiapterin reductase, carbinolamine-4a-dehydratase, and dihydropteridine reductase. Hum Mutat. 2006;27:870–878. doi: 10.1002/humu.20366. [DOI] [PubMed] [Google Scholar]
- 89.Douglas G, Hale AB, Crabtree MJ, Ryan BJ, Hansler A, Watschinger K, et al. A requirement for Gch1 and tetrahydrobiopterin in embryonic development. Dev Biol. 2015;399:129–138. doi: 10.1016/j.ydbio.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bonafe L, Thony B, Leimbacher W, Kierat L, Blau N. Diagnosis of dopa-responsive dystonia and other tetrahydrobiopterin disorders by the study of biopterin metabolism in fibroblasts. Clin Chem. 2001;47:477–485. [PubMed] [Google Scholar]
- 91.Hirakawa H, Sawada H, Yamahama Y, Takikawa S, Shintaku H, Hara A, et al. Expression analysis of the aldo-keto reductases involved in the novel biosynthetic pathway of tetrahydrobiopterin in human and mouse tissues. J Biochem. 2009;146:51–60. doi: 10.1093/jb/mvp042. [DOI] [PubMed] [Google Scholar]
- 92.Blau N, Bonafe L, Thony B. Tetrahydrobiopterin deficiencies without hyperphenylalaninemia: diagnosis and genetics of dopa-responsive dystonia and sepiapterin reductase deficiency. Mol Genet Metab. 2001;74:172–185. doi: 10.1006/mgme.2001.3213. [DOI] [PubMed] [Google Scholar]
- 93.Thony B, Auerbach G, Blau N. Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem J. 2000;347(Pt 1):1–16. [PMC free article] [PubMed] [Google Scholar]
- 94.Bonafe L, Thony B, Penzien JM, Czarnecki B, Blau N. Mutations in the sepiapterin reductase gene cause a novel tetrahydrobiopterin-dependent monoamine-neurotransmitter deficiency without hyperphenylalaninemia. Am J Hum Genet. 2001;69:269–277. doi: 10.1086/321970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Milstien S, Kaufman S. Immunological studies on the participation of 6-pyruvoyl tetrahydropterin (2’-oxo) reductase, an aldose reductase, in tetrahydrobiopterin biosynthesis. Biochem Biophys Res Commun. 1989;165:845–850. doi: 10.1016/s0006-291x(89)80043-9. [DOI] [PubMed] [Google Scholar]
- 96.Friedman J, Roze E, Abdenur JE, Chang R, Gasperini S, Saletti V, et al. Sepiapterin reductase deficiency: a treatable mimic of cerebral palsy. Ann Neurol. 2012;71:520–530. doi: 10.1002/ana.22685. [DOI] [PubMed] [Google Scholar]
- 97.Abeling NG, Duran M, Bakker HD, Stroomer L, Thony B, Blau N, et al. Sepiapterin reductase deficiency an autosomal recessive DOPA-responsive dystonia. Mol Genet Metab. 2006;89:116–120. doi: 10.1016/j.ymgme.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 98.Leu-Semenescu S, Karroum E, Brion A, Konofal E, Arnulf I. Dopamine dysregulation syndrome in a patient with restless legs syndrome. Sleep Med. 2009;10:494–496. doi: 10.1016/j.sleep.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 99.Lohmann E, Koroglu C, Hanagasi HA, Dursun B, Tasan E, Tolun A. A homozygous frameshift mutation of sepiapterin reductase gene causing parkinsonism with onset in childhood. Parkinsonism Relat Disord. 2012;18:191–193. doi: 10.1016/j.parkreldis.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 100.Neville BG, Parascandalo R, Farrugia R, Felice A. Sepiapterin reductase deficiency: a congenital dopa-responsive motor and cognitive disorder. Brain. 2005;128:2291–2296. doi: 10.1093/brain/awh603. [DOI] [PubMed] [Google Scholar]
- 101.Verbeek MM, Willemsen MA, Wevers RA, Lagerwerf AJ, Abeling NG, Blau N, et al. Two Greek siblings with sepiapterin reductase deficiency. Mol Genet Metab. 2008;94:403–409. doi: 10.1016/j.ymgme.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 102.Wali GM, Thony B, Blau N. Sepiapterin reductase deficiency: two Indian siblings with unusual clinical features. Mov Disord. 2010;25:954–955. doi: 10.1002/mds.23032. [DOI] [PubMed] [Google Scholar]
- 103.Zielonka M, Makhseed N, Blau N, Bettendorf M, Hoffmann GF, Opladen T. Dopamine-responsive growth-hormone deficiency and central hypothyroidism in sepiapterin reductase deficiency. JIMD Rep. 2015;24:109–113. doi: 10.1007/8904_2015_450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koht J, Rengmark A, Opladen T, Bjornara KA, Selberg T, Tallaksen CM, et al. Clinical and genetic studies in a family with a novel mutation in the sepiapterin reductase gene. Acta Neurol Scand Suppl 2014: 7–12. [DOI] [PubMed]
- 105.Friedman J, Hyland K, Blau N, MacCollin M. Dopa-responsive hypersomnia and mixed movement disorder due to sepiapterin reductase deficiency. Neurology. 2006;67:2032–2035. doi: 10.1212/01.wnl.0000247274.21261.b4. [DOI] [PubMed] [Google Scholar]
- 106.Plosker GL, Croom KF. Sulfasalazine: a review of its use in the management of rheumatoid arthritis. Drugs. 2005;65:1825–1849. doi: 10.2165/00003495-200565130-00008. [DOI] [PubMed] [Google Scholar]
- 107.Suarez-Almazor ME, Belseck E, Shea B, Wells G, Tugwell P. Sulfasalazine for rheumatoid arthritis. Cochrane Database Syst Rev 2000: CD000958. [DOI] [PMC free article] [PubMed]
- 108.Haruki H, Pedersen MG, Gorska KI, Pojer F, Johnsson K. Tetrahydrobiopterin biosynthesis as an off-target of sulfa drugs. Science. 2013;340:987–991. doi: 10.1126/science.1232972. [DOI] [PubMed] [Google Scholar]
- 109.Chidley C, Haruki H, Pedersen MG, Muller E, Johnsson K. A yeast-based screen reveals that sulfasalazine inhibits tetrahydrobiopterin biosynthesis. Nat Chem Biol. 2011;7:375–383. doi: 10.1038/nchembio.557. [DOI] [PubMed] [Google Scholar]
- 110.Costigan M, Latremoliere A, Woolf CJ. Analgesia by inhibiting tetrahydrobiopterin synthesis. Curr Opin Pharmacol. 2012;12:92–99. doi: 10.1016/j.coph.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Berti-Mattera LN, Kern TS, Siegel RE, Nemet I, Mitchell R. Sulfasalazine blocks the development of tactile allodynia in diabetic rats. Diabetes. 2008;57:2801–2808. doi: 10.2337/db07-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lopez-Olivo MA, Siddhanamatha HR, Shea B, Tugwell P, Wells GA, Suarez-Almazor ME. Methotrexate for treating rheumatoid arthritis. Cochrane Database Syst Rev 2014: CD000957. [DOI] [PMC free article] [PubMed]
- 113.Zhu H, Deng FY, Mo XB, Qiu YH, Lei SF. Pharmacogenetics and pharmacogenomics for rheumatoid arthritis responsiveness to methotrexate treatment: the 2013 update. Pharmacogenomics. 2014;15:551–566. doi: 10.2217/pgs.14.25. [DOI] [PubMed] [Google Scholar]
- 114.Forrest HS, Mitchell HK. H.K. Pteridines from Drosophila. II. Structure of the Yellow Pigment. J. Am. Chem. Soc. 1954;76:5658–5662. [Google Scholar]
- 115.Forrest HS, Mitchell HK. Pteridines from Drosophila. I. Isolation of a Yellow Pigment. J Am Chem Soc. 1954;76:5656–5658. [Google Scholar]
- 116.Matsubara M, Akino M. On the presence of sepiapterin reductase different from folate and dihydrofolate reductase in chicken liver. Experientia. 1964;20:574–575. doi: 10.1007/BF02150303. [DOI] [PubMed] [Google Scholar]
- 117.Nawa S, Forrest HS. Synthesis of the yellow pteirdine, isosepiapterin. Nature. 1962;196:169–170. doi: 10.1038/196169a0. [DOI] [PubMed] [Google Scholar]
- 118.Taira T. Enzymatic reduction of the yellow pigment of Drosophila. Nature. 1961;189:231–232. doi: 10.1038/189231a0. [DOI] [PubMed] [Google Scholar]
- 119.Nichol CA, Lee CL, Edelstein MP, Chao JY, Duch DS. Biosynthesis of tetrahydrobiopterin by de novo and salvage pathways in adrenal medulla extracts, mammalian cell cultures, and rat brain in vivo. Proc Natl Acad Sci U S A. 1983;80:1546–1550. doi: 10.1073/pnas.80.6.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Carducci C, Santagata S, Friedman J, Pasquini E, Carducci C, Tolve M, et al. Urine sepiapterin excretion as a new diagnostic marker for sepiapterin reductase deficiency. Mol Genet Metab. 2015;115:157–160. doi: 10.1016/j.ymgme.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 121.Vardeh D, Mannion RJ, Woolf CJ. Toward a mechanism-based approach to pain diagnosis. J Pain. 2016;17:T50–T69. doi: 10.1016/j.jpain.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Edwards RR, Dworkin RH, Turk DC, Angst MS, Dionne R, Freeman R, et al. Patient phenotyping in clinical trials of chronic pain treatments: IMMPACT recommendations. Pain. 2016;157:1851–1871. doi: 10.1097/j.pain.0000000000000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Neuropathy Eisenstein M. A name for their pain. Nature. 2016;535:S10–S11. doi: 10.1038/535S10a. [DOI] [PubMed] [Google Scholar]
- 124.Finnerup NB, Haroutounian S, Kamerman P, Baron R, Bennett DL, Bouhassira D, et al. Neuropathic pain: an updated grading system for research and clinical practice. Pain. 2016;157:1599–1606. doi: 10.1097/j.pain.0000000000000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Baron R, Maier C, Attal N, Binder A, Bouhassira D, Cruccu G, et al. Peripheral neuropathic pain: a mechanism-related organizing principle based on sensory profiles. Pain. 2017;158:261–272. doi: 10.1097/j.pain.0000000000000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guttmacher AE, Collins FS. Welcome to the genomic era. N Engl J Med. 2003;349:996–998. doi: 10.1056/NEJMe038132. [DOI] [PubMed] [Google Scholar]
- 127.Klein CJ, Foroud TM. Neurology individualized medicine: when to use next-generation sequencing panels. Mayo Clin Proc. 2017;92:292–305. doi: 10.1016/j.mayocp.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 128.Lindor NM, Thibodeau SN, Burke W. Whole-genome sequencing in healthy people. Mayo Clin Proc. 2017;92:159–172. doi: 10.1016/j.mayocp.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 129.Jung-Klawitter S, Ebersold J, Gohring G, Blau N, Opladen T. Generation of an iPSC line from a patient with GTP cyclohydrolase 1 (GCH1) deficiency: HDMC0061i-GCH1. Stem Cell Res. 2017;20:38–41. doi: 10.1016/j.scr.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 130.Campbell CM, Edwards RR, Carmona C, Uhart M, Wand G, Carteret A, et al. Polymorphisms in the GTP cyclohydrolase gene (GCH1) are associated with ratings of capsaicin pain. Pain. 2009;141:114–118. doi: 10.1016/j.pain.2008.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tegeder I, Adolph J, Schmidt H, Woolf CJ, Geisslinger G, Lotsch J. Reduced hyperalgesia in homozygous carriers of a GTP cyclohydrolase 1 haplotype. Eur J Pain. 2008;12:1069–1077. doi: 10.1016/j.ejpain.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 132.Doehring A, Freynhagen R, Griessinger N, Zimmermann M, Sittl R, Nv Hentig, et al. Cross-sectional assessment of the consequences of a GTP cyclohydrolase 1 haplotype for specialized tertiary outpatient pain care. Clin J Pain. 2009;25:781–785. doi: 10.1097/AJP.0b013e3181b43e12. [DOI] [PubMed] [Google Scholar]
- 133.Lotsch J, Klepstad P, Doehring A, Dale O. A GTP cyclohydrolase 1 genetic variant delays cancer pain. Pain. 2010;148:103–106. doi: 10.1016/j.pain.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 134.Kim DH, Dai F, Belfer I, Banco RJ, Martha JF, Tighiouart H, et al. Polymorphic variation of the guanosine triphosphate cyclohydrolase 1 gene predicts outcome in patients undergoing surgical treatment for lumbar degenerative disc disease. Spine (Phila Pa 1976) 2010, 35: 1909–1914. [DOI] [PubMed]
- 135.Wadley AL, Lombard Z, Cherry CL, Price P, Kamerman PR. Analysis of a previously identified “pain-protective” haplotype and individual polymorphisms in the GCH1 gene in Africans with HIV-associated sensory neuropathy: a genetic association study. J Acquir Immune Defic Syndr. 2012;60:20–23. doi: 10.1097/QAI.0b013e31824bcc17. [DOI] [PubMed] [Google Scholar]
- 136.Kim SK, Kim SH, Nah SS, Lee JH, Hong SJ, Kim HS, et al. Association of guanosine triphosphate cyclohydrolase 1 gene polymorphisms with fibromyalgia syndrome in a Korean population. J Rheumatol. 2013;40:316–322. doi: 10.3899/jrheum.120929. [DOI] [PubMed] [Google Scholar]
- 137.Lee PJ, Delaney P, Keogh J, Sleeman D, Shorten GD. Catecholamine-o-methyltransferase polymorphisms are associated with postoperative pain intensity. Clin J Pain. 2011;27:93–101. doi: 10.1097/AJP.0b013e3181f15885. [DOI] [PubMed] [Google Scholar]
- 138.Kim SJ, Lee WI, Lee YS, Kim DH, Chang JW, Kim SW, et al. Effective relief of neuropathic pain by adeno-associated virus-mediated expression of a small hairpin RNA against GTP cyclohydrolase 1. Mol Pain. 2009;5:67. doi: 10.1186/1744-8069-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]