Abstract
Our previous study has shown that genetic selection for susceptibility/resistance to diet-induced atherosclerosis has affected the Japanese quail’s cecal environment to accommodate distinctly different cecal microbiota. In this study, we fed the Atherosclerosis-resistant (RES) and -susceptable (SUS) quail a regular and a cholesterol enriched diet to examine the interaction of host genotype and diet on the diversity, composition, and metabolic functions of the duodenal and ileal microbiota with relations to atherosclerosis development. In the duodenal content, 9 OTUs (operational taxonomic units) were identified whose abundance had significant positive correlations with plasma total cholesterol, LDL level and/or LDL/HDL ratio. In the ileal content, 7 OTUs have significant correlation with plasma HDL. Cholesterol fed RES hosted significantly less Escherichia and unclassified Enterobacteriaceae (possibly pathogenic) in their duodenum than SUS fed the same diet. Dietary cholesterol significantly decreased the duodenal microbiome of SUS’s biosynthesis of Ubiquinone and other terpenoid-quinone. Cholesterol fed RES had significantly more microbiome genes for Vitamin B6, selenocompound, taurine and hypotaurine, and Linoleic acid metabolism; Bisphenol degradation; primary bile acid, and butirosin and neomycin biosynthesis than SUS on the same diet. Microbiome in the ileum and ceca of RES contributed significantly towards the resistance to diet induced atherosclerosis.
Introduction
Atherosclerosis is one of the leading causes of mortality in developed countries, and with increasing incidents in the developing countries1. It is a complex disease affected by the interaction of genetic and environmental risk factors2–4. In the past decade, research efforts in the biomedical field have focused on the implications of gut microbiome on human diseases. There is evidence that symptomatic atherosclerosis is associated with an altered gut microbial community in human and mice5–7. It has been demonstrated that cecal microbial transplantation from a susceptible strain to wild-type mice enhanced choline diet-dependent atheroslceorosis and TMAO (trimethylamine-N-oxide) levels in wild-type mice8. Examination of human oral and atherosclerotic plaque microbiota in patients with clinical atherosclerosis suggests that microbes from the oral cavity and perhaps the gut may be a reservoir for bacteria found in atherosclerotic plaques9. Our previous study has also shown that genetic selection for susceptibility/resistance to diet-induced atherosclerosis has affected the Japanese quail’s cecal environment to accommodate distinctly different cecal microbiota10. Most published studies have compared the fecal or cecal micriobial communities between atherosclerosis and healthy subjects, but there has been sparse information on the comparison of cecal and small intestinal microbiota in the same subject.
Mapping of bacterial communities in human, mouse and chicken intestinal tract11–14 revealed variations of microflora composition along the gastrointestinal tract relating to factors such as nutrient availability, pH and oxygen concentrations15. The small intestinal microbiota have the capacity to affect the absorption and metabolism of nutrients, the immune system, and defending the host from invasion of pathogenic bacteria16–18. While there have been studies on the intestinal microbiota metabolism of L-carnitine to TMAO19,20, the diversity and composition of microbes in the small intestine under healthy and disease states is still an area of ongoing research.
The atherosclerosis-resistant (RES) and -susceptible (SUS) Japanese quail21 is a proven animal model for studying atherosclerosis22,23. The two strains were developed by divergent selection from a common foundation population21. When challenged with a high cholesterol diet, about 80% of the SUS males will develop atherosclerosis whereas only about 4% of the RES males will. They host cecal microbial communities similar to that in mice and human24. In the current study, we fed the RES and SUS quail (from here on referred to as RES and SUS) a regular (control) and a cholesterol enriched diet, to examine the interaction of host genotype and diet on the diversity and composition of duodenal and ileal microbiota with relations to atherosclerosis development. We further included data from our cecal microbiota study24 for comparison and functional predictions.
Methods
Experimental Design
Duodenum and ileum contents were collected from the same individuals of RES and SUS whose cecal contents were analyzed in Liu et al.24. The experimental protocol has been described in Liu et al.24. Briefly, 80 RES and 80 SUS males were fed a semi-synthetic diet prepared according to the National Research Council (NRC) nutrient requirements standards recommended for Japanese quail (http://www.nap.edu/catalog/2114.html) from hatching to 6 weeks of age. At six weeks of age, they were divided into two dietary treatment groups and fed either the semi-synthetic diet (control) or the semi-synthetic diet with additional cholesterol (0.5% w/w) for another 6 weeks22. Individual birds were identified by numbered wing bands. Both RES and SUS fed the same diet were kept in the same pen. Birds on the alternative diet were kept in a neighbouring pen. The two side-by-side pens should have similar microbiological environment. At 12 weeks of age, six birds from each of the four treatment groups with body weight closest to the mean of the population were euthanized by decapitation and trunk blood was collected into Vacutainer tubes containing lithium heparin, and centrifuged at 4 °C for 10 min at 3,000 × g. Plasma was stored at −20 °C until it was later used for lipid analysis24. The aortic tree (the brachycephalic arteries to their bifurcations and the aorta to the iliac branching) of each bird was dissected out, opened longitudinally and examined under a 10–30X dissecting microscope for a semi-quantitative scoring23 of the seriousness of the atherosclerotic lesions on the interior wall. A score of 0 (normal) to 4 (presence of severe atherosclerotic lesions) was assigned by two independent scorers who were blind to the genetic and diet status of the bird. Segments of duodenum (The U-shape section at the beginning of small intestine) ileum (section between Jejunum and the large intestine), and ceca, including gut content were collected from each bird. All samples were quick frozen on dry ice immediately after collection and stored at −70 °C until processed for DNA extraction. The cecal microbiome was examined and reported in Liu et al.24.
DNA Extraction and Pyrosequencing
Dissected duodenum and ileum segments were thawed and the contents were gently scraped into sterile vials. Genomic DNA was extracted from samples (0.2–0.5g/sample) using the PowerMax Soil DNA Isolation Kit (MO BIO laboratories. Inc., Carlsbad, CA) according to the manufacture’s instructions. PCR amplifications were performed using the FastStart high fidelity PCR system (Roche Molecular Diagnostics, Branchburg, NJ, USA). The variable region 3–5(V3–V5) of the bacterial 16S rRNA gene was amplified with a primer set of 341F (5′-ACT CCT ACG GGA GGC AGC AG-3′) and 926R (5′-CCG TCA ATT CMT TTG AGT TT-3′) with the sample specific forward primer bearing a multiplex identifier (MID) sequences. All 341F and 926R primers were modified with adaptor A and B sequences respectively for pyrotag sequencing.The amplification program consisted of an initial denaturation step at 94 °C for 2 min; 32 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, and elongation at 72 °C for 30 s; and a final extension step at 72 °C for 7 min. The size of the PCR products was confirmed by gel electrophoresis.The PCR products was then purified using Gel extraction kit (Invitrogen) and were quantified using the NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA). The Amplicon libraries were subjected to pyrotag sequencing using a bench-top 454 GS Junior (454 Life Sciencesa Roche Company, Branford, CT, USA) with the GS Junior Titanium Sequencing Kit (https://lifescience.roche.com/shop/en/us/products/gs-junior-titanium-sequencing-kit).
Sequence Analysis
The raw 16s rRNA sequences were processed using Quantitative Insights Into Microbial Ecology (QIIME) software package25. Raw sequences were filtered to meet the following quality criteria: (1) complete barcode sequences followed by a forward primer sequence, with no mismatch in either barcode or primer sequence; (2) reads lengths between 150 and 900 bases; (3) average quality score >25; and (4) homophlymer run of 8 nt. De-noising of the dataset was performed using DENOISER v. 0.9.126 as implimented in QIIME. Chimeric sequences were removed using Chimera Slayer (http://microbiomeutil.sourceforge.net/). The filtered sequences were assigned to groups basing on their respective barcode sequences. Similar sequences were assigned into operational taxonomic units (OTUs) at a pairwise identify of 97% using UCLUST (http://www.drive5.com/usearch/). Representative sequence was the most abundant sequence in each OTU. Representative sequences (at 97% similarity) were then classified taxonomically using Ribosomal Database Project (RDP) classifier 2.0.127. The OTUs were aligned using PyNAST with a minimum alignment length of 150 bp and a minimum percent identity of 75%28. After alignment, PH LANE mask (http://greengenes.lbl.gov/) was conducted to screen out the hypervariable regions.
Statistical Analysis
Richness and Diversity Indices
Rarefaction Plots were constructed and Alpha-diversity (Chao1 Richness, Simpson’s and Shannon’s Diversity indices) were estimated as implemented in QIIME29. Beta-diversity among microbial communities was compared using Principal Component Analysis (PCA) in SIMCA-P (version 14.0) to visualize microbial community’s patterns caused by diet and host genotype. Results of the PCA were statistically tested by multivariate analysis of variance (MANOVA)30.
Comparison of Microbial Communities
Bacterial abundance difference at the genus level and plasma lipid parameters were examined by multivariate analysis, using Tukey’s HSD/ANOVA for mean separation (SPSS 13.0; SPSS Institute, 2001). P < 0.05 was considered significant,
Venn diagrams31 and “nearest-shrunken centroid” (NSC) classification32 were used to detect core microbiota community which best characterize each group. From each treatment group, the OTUs which were common in at least four of six samples were selected to generate OTUs files. For ileum segments, two SE samples were excluded because of low sequence reads, so ileal OTUs that were common in three of four samples in the group were selected. These newly generated OTU files were used to identify the key OTU communities. NSC analysis was performed on normalized Z-score profiles of OTUs, the amount of shrinkage was determined by cross-validation and test error was minimized. In duodenal NSC analysis, the training error was 0.46 and threshold was 0.43. In ileum NSC analysis, the training error was 0.36 and threshold was 0.63.
Microbial metabolic function prediction
The PICRUSt (phylogenetic investigation of communities by reconstruction of unobserved states)33 was employed to predict functional genes of the classified members of the microbiome (including Cecal OTU data (SRE accession number SRR2537231) obtained from Liu et al.24) through closed-reference based OTU mapping against the Greengenes database33. Mapped closed-reference OTUs are normalized based on the copies of 16S rRNA gene within the known bacterial genomes in Integrated Microbial Genomes (IMG). Predicted genes were clustered hierarchically and categorised on the basis of KEGG34 orthologues (KO’s) and pathways (level -3). Using STAMP software35. To compare differences in predicted metagenomic functions among different treatment groups, Welch’s t-test was applied on the predicted microbiome functions determined by KEGG functional modules (level-3) under various microbiome metabolism36.
Availability of data and supporting materials
The Cecal, duodenal and ileal microbiota OTU sequences have been submitted to Sequence Read Archive (SRA); Cecal OTU sequences with accession number SRR2537231, Duodenal OTU sequences with accession number SRR6312041, and Ileal OTU sequences with accession number SRR6312040.
Ethics approval
All experiments were performed in accordance with protocols reviewed and approved by the UBC Animal Care Committee (Certificate # A12-0087).
Results
Atherosclerotic Lesions on the Intimal Surface of the Aortae
Atherosclerotic lesions of all SUS and RES fed the control diet (SC and RC, respectively) scored 0. Four of six SUS on cholesterol diet (SE) scored 4, the remaining two scored 3+. Three of six RES on the cholesterol diet (RE) scored 0 and the other three scored 1.
Richness of Small Intestinal Microbiota
Duodenum
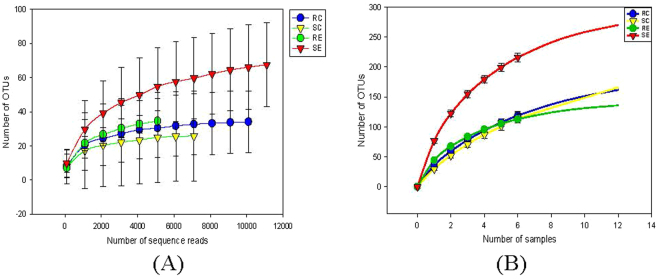
After quality filteration and trimming, a total of 450,576 sequences were generated with a mean length of 534bp and 18,774 ± 1,633.68 sequences/sample. Rarefaction analysis (Fig. 1) of the sample sizes in the four groups covered most of the OTUs in the sampled population (RC 28.00 ± 4.46; SC 19.40 ± 5.71; RE 44.17 ± 4.78; SE 68.60 ± 8.27). The sequences were classified into 360 species-level operational taxonomic units (OTUs) belong to 15 microbial phyla. Phyla of Firmicutes (95.49%), Proteobacteria (2.67%) and Bacteroidetes (0.83%) dominated in the duodenum where the remaining sequences were identified as the phyla Cyanobacteria, Tenericutes, TM6, Actinobacteria, Chloroflexi, Chlorobi, Acidobacteria, Armatimonadetes, Fusobacteria, Gemmatimonadetes, and Synergistetes, Spirochaetes (together no more than 2% of total sequences).
Figure 1.
Rarefraction analysis, calculated at 97% dissimilarity, for the assessment of operational taxonomic units (OTU) coverage within the 16S rRNA gene-based duodenal bacterial communities in the RES and SUS quail fed the control or cholesterol diets. (A) The number of OTUs as a function of the number of sequence reads. (B) The number of OTUs as a function of the number of individual quail sampled.
Ileum
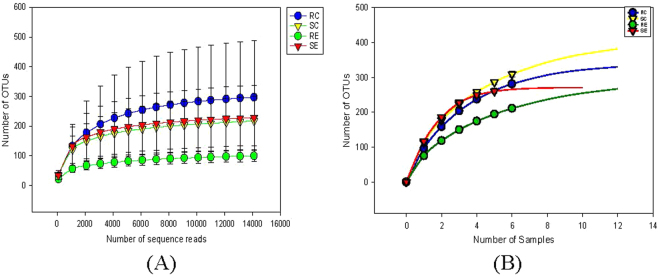
The ileum segment yielded a total of 515,982 sequences with a mean length of 475bp and 23453.73 ± 1104.55 sequences/sample. Rarefaction curve (Fig. 2) reached plateau, showing that the sample size would include most OTUs present in the sampled populations (RC 90.17 ± 19.77; SC 121.50 ± 6.84; RE 105.33 ± 18.18; SE 108.00 ± 30.57). The sequences were classified into 381 OTUs belonging to 8 microbial phyla. The most abundant sequences belonged to four microbial phyla: Firmicutes (73.94%), Proteobacteria (16.28%), Actinobacteria (8.57%) and Bacteroidetes (0.37%). The remaining sequences were identified as Thermi, Acidobacteria, Chlorobi, Chloroflexi, Cyanobacteria, Tenericutes (together less than 2% of total sequences).
Figure 2.
Rarefraction analysis, calculated at 97% dissimilarity, for the assessment of operational taxonomic units (OUT) coverage within the 16S rRNA gene-based ileal bacterial communities in the RES and SUS quail fed the control or cholesterol diets. (A) The number of OTUs as a function of the number of sequence reads. (B) The number of OTUs as a function of the number of individual quail sampled.
A comparison of Small Intestinal Microbiota Diversity
Duodenum
Chao1 species richness was significantly (P < 0.05) affected by diet X host genotype interaction. SUS on the cholesterol diet (SE) had the highest OTU richness than the other three treatment groups (SC, RE, and RC) in which no significant difference was detected. There were no significant differences (P > 0.05) in Simpson and Shannon estimators among the four treatment groups.
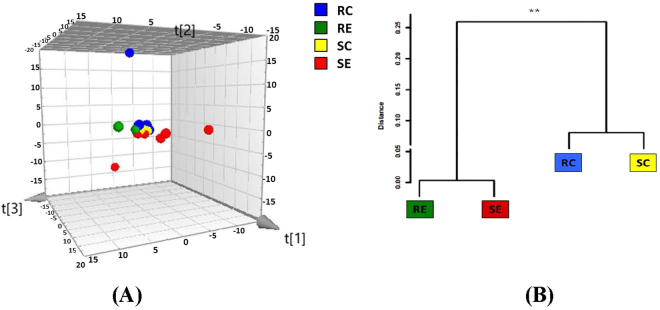
Although PCA plot of β-diversity showed that the four treatment groups were partly overlapped (Fig. 3A), MANOVA analysis of the duodenal microbial community indicated significant (P < 0.01) difference among four treatment groups. A Mahalanobis Distances dendrogram was developed to provide a better visualization of the relationships among the four treatment groups (Fig. 3B). Duodenum samples from birds fed high-cholesterol diet formed a close cluster whereas those from the control diet groups formed another close cluster that was significantly (P < 0.048) distant from the first.
Figure 3.
(A) Three-dimensional projection of PCA of whole duodenal microbial community. Each symbol represents a single sample. PC1 = 0.249; PC2 = 0.186; PC3 = 0.146 Ellipse: Hotellings’ T2 = 0.97. (B) Clustering of duodenal microbiota based on distances between different groups calculated with multivariate analysis of variance test of the first six PCs of the OTU data. The Mahalanobis distances between group means are shown. **P < 0.01.
At the genus level, the abundance of Escherichia (Enterobacteriaceae), Streptococcus (Streptococcaceae), and Staphylococcus (Staphylococcaceae) was significantly affected by both diet and host genotype (Table 1). Escherichia was most abundant in RC and not different among the other three groups. Streptococcus was most abundant in SE but not detectable in SC. Staphylococcus was most abundant in SE but not detectable in SC and RE. Unclassified Enterobacteriaceae was also found in significant abundance in RC and SE but not detected in SC and RE. There was significantly more unclassified Rikenellaceae, unclassified Coprobacillaceae, Blautia, and Collinsella in birds fed the cholesterol diet (SE and RE) than in birds on the control diet (RC and SC). The abundance of Bacteroides and Enterococcus was significantly affected by host genotype. RES (both RC and RE) had more Bacteroides in their duodenum than SUS (SC and SE), while the opposite was true for Enterococcus (Table 1).
Table 1.
Genus level differences in abundance of duodenal microbiota among treatment groups.
| RC | SC | RE | SE | P | |
|---|---|---|---|---|---|
| Escherichia § | 4.03 ± 1.05A | 0.33 ± 0.33B | 0.63 ± 0.17B | 0.82 ± 0.17B | 0.003 |
| Streptococcus § | 0.64 ± 0.49AB | 0.00 ± 0.00B | 0.29 ± 0.21AB | 1.66 ± 0.60A | 0.021 |
| Staphylococcus § | 0.96 ± 0.50AB | 0.00 ± 0.00B | 0.00 ± 0.00B | 2.10 ± 1.15A | 0.025 |
| Uncl. Enterobacteriaceae§ | 0.31 ± 0.19A | 0.00 ± 0.00B | 0.00 ± 0.00B | 0.25 ± 0.16AB | 0.038 |
| Alicyclobacillus † | 0.58 ± 0.28 | 0.00 ± 0.00 | 0.07 ± 0.07 | 0.12 ± 0.12 | 0.059 |
| Control diet | Cholesterol diet | P | |||
| Uncl. Rikenellaceae* | 0.00 ± 0.00 | 0.92 ± 0.36 | 0.023 | ||
| Uncl. Coprobacillaceae* | 0.51 ± 0.21 | 3.65 ± 1.49 | 0.044 | ||
| Blautia* | 0.30 ± 0.16 | 1.94 ± 0.77 | 0.045 | ||
| Collinsella* | 0.29 ± 0.21 | 1.09 ± 0.32 | 0.050 | ||
| RES | SUS | P | |||
| Bacteroides ‡ | 3.00 ± 0.91 | 0.20 ± 0.11 | 0.007 | ||
| Enterococcus ‡ | 0.28 ± 0.17 | 2.79 ± 1.13 | 0.047 | ||
§Significant diet X host genotype interaction.
†Effect of diet X host genotype interaction tends to be significant (P < 0.059).
*Significant diet effect.
‡Significant host genotype effect.
Combining the results of the Venn Diagram/NSC analyses (Table 2), we found that Unclassified Streptophyta (ID 4420570) and Unclassified Lachnospiraceae (ID 158971) were overabundant in RC but rare in SC. Unclassified Lactobacillaceae (ID 292057) was overabundant in RE but rare in SE, while Unclassified Methylobacterium (ID 105470) was overabundant in SC but became rare when SUS was fed the cholesterol diet. The abundance of Unclassified Streptococcaceae (ID 255359), Unclassified Lachnospiraceae (ID 130773), and Unclassified Sinobacteraceae (ID 100307) were affected by both the host genotype and dietary cholesterol.
Table 2.
Summary of key duodenal OTUs characteristics generated by Venn diagram and NSC analysis.
| RC | SC | |
|---|---|---|
| Over-abundant | Uncl. Streptococcaceae (255359) Uncl. Streptophyta (4420570) Uncl. Escherichia (114510) Bacteroides fragilis (3474081) Uncl. Lachnospiraceae (158971)** |
Uncl. Enterococcus (839152) Uncl. Sinobacteraceae (100307) Uncl. Lactobacillus (137580) Uncl. Sphingomonas (4449609) Uncl. Caulobacteraceae (4353264) Uncl. Afipia (92573) |
| Unique |
Bacteroides ovatus (2563561) Pseudomonas stutzeri (4312845) Uncl. Ruminococcaceae (134174) Uncl. Ruminococcus (136507)† |
Lactobacillus reuteri (137043)†* |
| RE | SE | |
| Over-Abundant | Uncl. Lactobacillaceae (292057)ठ| Uncl. Lachnospiraceae (130773) Uncl. Coprobacillaceae (592616) Uncl. Ruminococcaceae (306633) Uncl. Ruminococcus (182245) |
| Unique | Uncl. RF39 (235065) Uncl. Sphingomonas (582921)† Uncl. Ruminococcaceae (157546) |
Uncl. Coprobacillaceae (364722) Uncl. Ruminococcus (3438642)† Uncl. Ruminococcus (191273)† Uncl. Ruminococcus (548503)† Uncl. Lachnospiraceae (NCR37)† |
Ileum
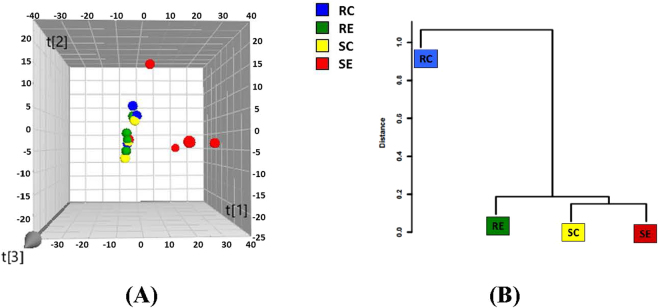
There were no significant difference (P > 0.05) in α-diversity among the 4 treatment groups according to the Chao1, Simpson and Shannon indices. PCA plot indicated no detectable differences in microbial composition among treatment groups (Fig. 4). MANOVA analysis of variance derived from PCA scores showed there was no statistically significant (P > 0.05) separation among the microbiota of the four treatment groups.
Figure 4.
(A) Three-dimensional projection of PCA of whole ileal microbial community. Each symbol represents a single sample. PC1 = 0.173; PC2 = 0.112; PC3 = 0.105 Ellipse: Hotellings’ T2 = 0.97. (B) Clustering of duodenal microbiota based on distances between different groups calculated with multivariate analysis of variance test of the first six PCs of the OTU data.
At the genus level, only Lactobacillus (Lactobacillaceae) was significantly (P < 0.013) more abundant in RES (51.24 ± 7.01) than in SUS (27.51 ± 2.47).
Summarizing the results of the Venn Diagram/NSC analyses (Table 3), we found that Unclassified Lactobacillus (ID 192832) was overabundant in RC but rare in SC. Unclassified Lactobacillus (ID 292057) was overabundant in RE but rare in SE. Unclassified Lactobacillus (ID 137580) was overabundant in RC but became rare when RES was fed the cholesterol diet.
Table 3.
Summary of key ileal OTUs characteristics generated by Venn diagram and NSC analysis.
| RC | SC | |
|---|---|---|
| Over-abundant | Uncl. Lactobacillus (137580)* Uncl. Lactobacillus (958496) Uncl. Lactobacillus (192832) Uncl. Lactobacillus (242917) |
Streptococcus alactolyticus (4473883) Uncl. Streptococcus (237444) Uncl. Acinetobacter (4482598) |
| Unique | Lactobacillus reuteri (354256) |
Propionibacterium acnes (933896) Uncl. Hydrogenophilus (104987) Uncl. Hydrogenophilus (575143) Uncl. Acinetobacter (4482598) Uncl. Acinetobacter (4431922) |
| RE | SE | |
| Over-abundant | Uncl. Lactobacillus (292057)ठUncl. Lactobacillus (1142657) Uncl. Lactobacillus (823916) Uncl. Lactobacillus (574102) Uncl. Lactobacillus (4361528) Acinetobacter guillouiae (4449456) |
|
| Unique | Uncl. Lachnospiraceae (158971)** Uncl. Streptococcus (15440) Uncl. Lactobacillus (332718) Uncl. Flectobacillus (539293) Uncl. Microbacterium (215095) |
Escherichia coli (656881) Uncl. Peptostreptococcaceae (4409730)† Uncl. Enterobacteriaceae (4457268) Uncl. Enterobacteriaceae (782953) Uncl. Enterobacteriaceae (4454531) |
Association of key bacteria species with plasma lipid parameters
There was a significant diet × genotype interaction in plasma Total Cholesterol (TC) (P < 0.0003), LDL (P < 0.0002) levels, and LDL/HDL ratio (P < 0.0001) (Table 4). SE was significantly higher in these parameters than the other three treatment groups.
Table 4.
Significant Diet X Host Genotype interaction in plasma lipids level.
| Plasma lipids (mmol/L) | N | RES/CON | SUS/CON | RES/CHOL | SUS/CHOL |
|---|---|---|---|---|---|
| Total Cholesterol1 | 24 | 4.49 ± 0.46a | 5.23 ± 0.31a | 13.90 ± 1.84a | 35.84 ± 4.37b |
| Plasma LDL2 | 24 | 1.04 ± 0.08a | 1.34 ± 0.08a | 8.56 ± 2.11a | 31.39 ± 4.26b |
| Plasma LDL/HDL ratio3 | 24 | 0.37 ± 0.03a | 0.42 ± 0.04a | 2.60 ± 0.74a | 9.05 ± 1.10b |
1P < 0.0003 Diet X Genotype interaction.
2P < 0.0002 Diet X Genotype interaction.
3P < 0.0001 Diet X Genotype interaction.
In the duodenum, we have identified 13 bacteria species whose abundance had significant correlation with plasma lipid parameters (Table 5). Nine species have positive correlations with plasma total cholesterol, LDL level or LDL/HDL ratio. Three species showed negative correlation with LDL/HDL ratio and only one species, Unclassified Lachnospiraceae (158971), showed positive correlation with HDL. In the ileum, 7 bacteria species have significant correlation with HDL and none showed significant correlation with plasma total cholesterol, LDL level or LDL/HDL ratio (Table 6). Only Unclassified Lactobacillus (4414257) showed a negative correlation. None of the bacteria species in the duodenum or ileum had significant correlation with plasma triglycerides level.
Table 5.
Significant Pearson’s Correlations§ between the abundance of duodenum bacteria species and plasma lipid parameters†.
| OTUs | Total Chol | HDL | LDL | LDL/HDL |
|---|---|---|---|---|
| Firmicutes | ||||
| Uncl. Lactobacillus (NCR39) | 0.608* | |||
| Uncl. Ruminococcus (130103) | 0.592* | |||
| Blautia producta (158211) | 0.609* | 0.601* | ||
| Uncl. Lachnospiraceae (158971) | 0.578* | |||
| Uncl. Ruminococcaceae (157804) | 0.675* | |||
| Uncl. Ruminococcaceae (158217) | 0.647* | |||
| Uncl. Ruminococcaceae (157546) | −0.588* | |||
| Uncl. Ruminococcaceae (313037) | 0.766** | 0.744** | 0.603* | |
| Uncl. Collinsella (NCR69) | 0.769** | 0.748** | 0.620* | |
| Uncl. Coprobacillaceae (592616) | 0.681* | |||
| Proteobacteria | ||||
| Uncl. Afipia (92573) | −0.653* | |||
| Uncl. Escherichia (114510) | 0.683* | 0.693* | 0.725** | |
| Tenericutes | ||||
| Uncl. RF39 (235065) | −0.609* | |||
†RES and SUS fed the cholesterol diet; N = 12.
§After elimination of significant correlations due to a single outlier.
*P < 0.05; **P < 0.01.
Table 6.
Significant Pearson’s Correlations§ between the abundance of ileum bacteria species and plasma lipid parameters†.
| OTUs | HDL |
|---|---|
| Firmicutes | |
| Uncl. Lactobacillus (4414257) | −0.654* |
| Proteobacteria | |
| Uncl. Gluconacetobacter (656881) | 0.690* |
| Uncl. Acinetobacter (4361528) | 0.748* |
| Uncl. Acinetobacter (1085703) | 0.717* |
| Acinetobacter guillouiae (4449456) | 0.655* |
| Acinetobacter johnsonii (4333705) | 0.782** |
| Uncl. Enhydrobacter (235065) | 0.643* |
†RES and SUS fed the cholesterol diet; N = 10.
§After elimination of significant correlations due to a single outlier.
*P < 0.05; **P < 0.01.
PICRUSt predictions
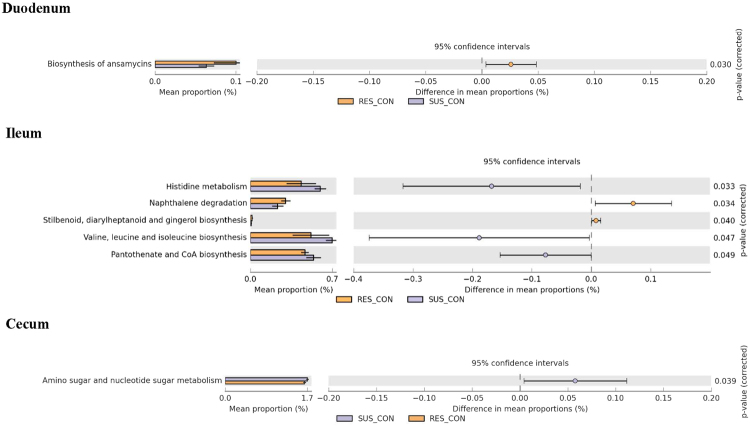
When fed the control diet, the RES duodenal microbiome has significantly more functional genes for anasmycinins biosynthesis, and RC has significantly more ileal microbiome genes for naphthalene degradation, stilbenoid, diarylheptanoid and gingerol biosynthesis than SC (Fig. 5). SC, on the other hand, has more Ileal microbiome genes that metabolize Histidine, biosynthesize Valine, leucine, isoleucine, pentothinate and CoA, and a more cecum microbiome genes for amino sugar and nucleotide sugar metabolism.
Figure 5.
Differences in intestinal microbiome metabolic functions between RC and SC.
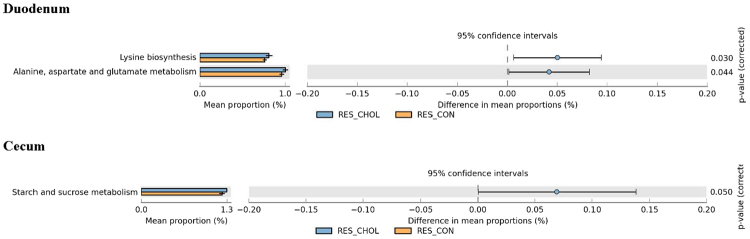
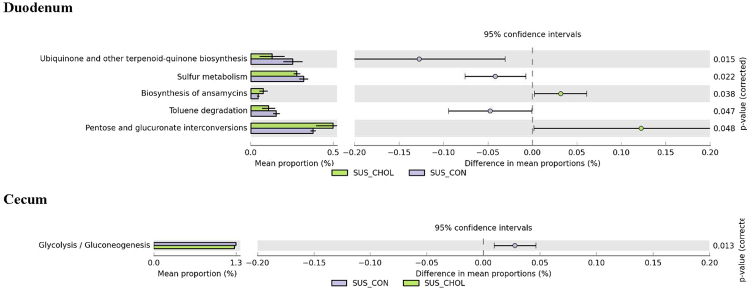
In response to the cholesterol diet, there were more duodenal microbiome genes in RE for Lysine biosynthesis, Alanine, aspartate, and glutamate metabolism than in RC (Fig. 6). SUS responded to the cholesterol diet in a different manner (Fig. 7). While there were more duodenal microbiome genes in SE for ansamycines biosynthesis, and pentose and gluconate interconversions than in SC, there were significantly less duodenal microbiome genes in SE for biqinone and other terpenoid-quinone biosynthesis, sulfur metabolism, and toluene degradation than in SC. Note that all these changes in the microbiome genome of the RES and SUS in response to the cholesterol diet were in the duodenum.
Figure 6.
Differences in intestinal microbiome metabolic functions between RC and RE.
Figure 7.
Differences in intestinal microbiome metabolic functions between SC and SE.
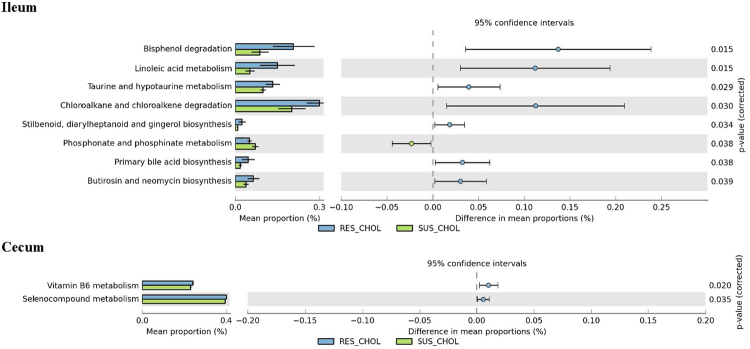
When the intestinal microbiome genome of RE and SE were compared, RE had significantly more cecum microbiome genes for Vitamin B6, and selenocompounds metabolism than SE (Fig. 8). RE also had significantly more ileal microbiome genes for bisphenol, chloroalkane and choloroalkene degradation, linoleic acid, taurine and hypotaurine metabolism, Stilbenoid, Diarylheptanoid and gingerol, primary bile acids, butirosin and neomycine biosynthesis compared with SE. On the other hand, SE had significantly more ileum microbiome genes for phosphonate and phosphinate metabolism than RE.
Figure 8.
Differences in intestinal microbiome metabolic functions between RE and SE.
Discussions
We examined the duodenal and ileal microbiota of 12 week old Japanese quail that had been fed their respective diets for at least 6 weeks. Taxonomic analysis showed that composition of the quail’s duodenal and ileal microbiota at various levels is similar to that of human, mice, hamsters, chickens, emu, and Bobwhite quail5,37–42.
Duodenal microbiota
Although there were 15 microbial phyla found in the quail duodenum, 95% of the sequences detected were Firmicutes. While there was no significant difference in Chao1 richness and Simpson & Shannon diversity between RES and SUS duodenum microbiota when they were fed the regular diet, the cholesterol diet had significantly increased the Chao1 richness of SUS but not the RES quail. The exposure to the cholesterol diet has significantly increased the abundance of Unclassified Rikenellaceae, Unclassified Coprobacillaceae, Blautia, and Collinsella in both RES and SUS.
In the human gut, Collinsella was enriched in patients with symptomatic atherosclerosis5. Rikenellaceae are bacteria commensal to the gut and these bacteria thrive on high-fat diets and are enriched in gut microbiomes of obese human. In laboratory mice, a high fat diet also induced a significant increase in Ruminococcaceae and Rikenellaceae43. Blautia digest complex carbohydrates. Blautia levels were decreased in patients with liver disease and colorectal cancer and children with diabetes. The increased abundance of commensal bacteria belonging to the Blautia genus is associated with reduced lethal Graft-versus-Host Disease and improved overall survival44.
It is interesting that RES hosted significantly less Escherichia and unclassified Enterobacteriaceae in their duodenum in response to the cholesterol diet but the SUS was not affected. Many species of Escherichia and Enterobacteriaceae are known to be pathogenic in human. In mice fed a high-fat diet, there was a significantly higher abundance of Enterobacteriaceae and the abundance of Enterobacteriaceae correlated with an increase in endotoxin levels in the gut43. In chickens, inclusion of probiotic (a mixture of Bacillus, Lactobacillus, Streptococcus, and Clostridium) in a high cholesterol diet significantly reduced the number of cecal Enterobacteriaceae species45. RE hosted an overabundance of Unclassified Lactobacillaceae (292057) in the ileum and this may have an effect on combating Enterobacteriaceae. Furthermore, RE also uniquely hosted with abundance a species of Unclassified Sphingomonas (582921) in the duodenum. Ryu et al.46 reported isolating many strains of Sphingomonas sanguis from wild pheasant GI tracts that could produce antagonistic substance against Salmonella gallinarum, the cause of fowl typhoid.
Ability to suppress Escherichia and Enterobacteriaceae abundance when exposed to a high cholesterol diet may be a defence strategy of RES that was modified in SUS by selective breeding for susceptibility to diet induced atherosclerosis47–49. SUS hosted significantly more duodenal Streptococcus and Staphylococcus in response to the high cholesterol diet whereas RES did not. Many species of Staphylococcus are pathogenic in human. A high fat diet will induce the increase of these bacteria in the upper gut50,51. We have also demonstrated that the harboring of these bacteria is also affected by the host’s genotype. The difference in defense strategy employed by the two quail strains is also supported by our finding that, regardless of diets, SUS had significantly more Enterococcus (pathogenic in human) than RES while RES had significantly more Bacteroides than SUS. The inability of SUS to suppress the increase of Enterobacteriaceae, Escherichia, and Staphylococcus may partially facilitate the development of atherosclerosis when exposed to a high cholesterol diet.
Ileal microbiota
Of the 8 microbial Phyla found in the ileum, 74% were Firmicutes, 16% were Proteobacteria and 8.5% were Actinobacteria. There were no significant difference among the 4 treatment groups in Chao1 richness and Simpson and Shannon diversity. Regardless of diets, RES has significantly more Lactobacillus (Lactobacillaceae) then SUS. Earlier research on Lactobacillus found L. acidophilus improved egg production, food conversion and reduced the cholesterol concentration in the eggs in chickens52. Supplementation with L. rhamnosus GG decreased serum total cholesterol by 32% in rats with induced hypercholesterolemia53. However, the administration of L. fermentum did not appear to produce a major change in serum lipid fractions in human subjects with elevated serum cholesterol54. Recent research has gathered more evidence to show that Lactobacillus can be an effective probiotic to lower cholesterol both in vivo and in vitro55. Administration of L. reuteri NCIMB 30242 significantly reduced serum LDL-C, total cholesterol, and non-HDL cholesterol but not triglycerides and HDL-C in healthy hypercholesterolemic human subjects56. It was suggested that L. reuteri facilitated the deconjugation of intraluminal bile acids to inhibit absorption of non-cholesterol sterols as well as a novel cholesterol-reducing mechanism56. It was also demonstrated57 that soluble effector molecules produced by L. acidophilus down regulate the expression of NPC1L, a gene in the small intestine that regulates cholesterol absorption, thus inhibiting the cellular uptake of micellar cholesterol. Fuentes et al.58 administered a mixture of three strains of L. plantarum to hypercholesterolemic adults and found significant reduction of plasma total cholesterol and LDL-C. In the ceca, RC also hosted 2 unique species of Lactobacillus24. Lactobacillus species has been commonly used as a probiotic to suppress GI tract inflammation in human59. The selection for resistance to diet induced atherosclerosis have also changed the RES gut microbiome when RES is not exposed to dietary cholesterol and may have improved the general gut health of RES59.
Our present study found that when exposed to similar diet and microbiological environment, RES was able to harbor significantly more Lactobacillus than SUS and thus provided evidence to support the contention that host genotype can affect the gut microbiome60.
Gut Microbiota and Atherosclerosis
A high level of serum total cholesterol is considered to be a risk factor for atherosclerosis. The abundance of Unclassified Ruminococcaceae (OTU313037), Unclassified Collinsella (NCR69), Blautia producta (OTU158211), Unclassified Escherichia (OTU114510) in the duodenum is significantly correlated with plasma total cholesterol, LDL levels and LDL/HDL ratio. Only Unclassified Escherichia (OTU114510) was found in abundance in the RC duodenum. Liu et al.24 reported that the abundance of four species of Lachnospiraceae, three species of Ruminococcaceae and one species of Coprobacillaceae in the ceca was positively correlated with plasma Total Cholesterol, plasma LDL, and LDL/HDL ratio. Among these, Unclassified Ruminococcus (OTU548503) was found in overabundance in the SE ceca and unique and abundant in the SE duodenum. Another Ruminococcus species (OTU191273) was not abundant in the ceca but unique and abundant in the SE duodenum. Interestingly, the abundance of these two species in the ceca was significantly correlated with the plasma lipids level but not significantly so when they were in the duodenum. None of the microbiota species in the ileum was significantly correlated with TC, LDL, or LDL/HDL. The abundance of 5 Proteobacteria species in the ileum was significantly correlated with plasma HDL level but none of these species were found in over-abundance in the ileum. Further research on the functionality of these bacteria with relationship to their abundance and the intestinal environment is warranted61–63 to better our understanding of gut microbial activity.
RES has higher abundance of Lactobacillus in the ileum than SUS. The administration of L. acidophilus ATCC 4356 was found to protect against atherosclerosis in ApoE knock-out mice through the inhibition of intestinal cholesterol absorption64. The administration of L. plantarum 299v to smokers also resulted in the reduction of several cardiovascular disease risk factors65.
In comparison with SE, RE has significantly more microbiome genes for Vitamin B6 metabolism in their ceca. Pyridoxal 5′ phosphate (PLP), the active form of vitamin B6, is involved in a wide variety of physiologic processes including gluconeogenesis and the synthesis of sphingolipids and neurotransmitters. It also functions as a cofactor for many enzymes required for amino acid metabolism66–68. Complex sphingolipids are essential structural components of intestinal membranes, providing protection and integrity to the intestinal mucosa and regulating intestinal absorption processes. The role of sphingolipid signaling has been established in numerous cellular events, including intestinal cell survival, growth, differentiation, and apoptosis68,69. An important biochemical change associated with vitamin B6 deficiency is that of hyperhomocysteinemia which was recognized as a cardiovascular event risk factor67. The oxidative stress due to low level of vitamin B6 accelerates the development of homocysteine-induced atherosclerosis in rats70.
RE had significantly more microbiome genes than SE in the ceca that are involved with selenocompound metabolism. Selenomethionine is the major selenocompound in cereal grains, grassland legumes and soybeans. Methylseleninic acid (MASIV) has been proposed to be a nutritional selenium source71. Nutritional selenocompounds are considered to be transformed into the common intermediate selenide for utilization as selenoenzymes. Selenium affects expression of 15% of the genes that participate in lipid metabolism, especially in lipid transport, including apolipoprotein. It is a major determinant of the capacity of HDL to promote cholesterol efflux. Plasma HDL levels are inversely correlated with atherosclerosis72.
In the ileum, the RE microbiome is significantly better in bisphenol degradation than the SE microbiome. Bisphenol A (BPA) has been reported to cause hypercholesterolemia. BPA caused an overexpression of key genes (Mvd, Lss, Hmgcr, and Sqle) involved in cholesterol biosynthesis. BPA also induced the expression of the sterol regulatory element-binding proteins 2, a master regulator of hepatic cholesterol biosynthesis73.
Another microbiome metabolic function that was significantly better in the RE ileum than the SE ileum was Linoleic acid metabolism. Intestinal bacteria can metabolize linoleic acid to form vaccenic acid or 10-hydroxy-18:1. Both of these compounds are precursors of conjugated linoleic acid (CLA, also known as Omega-6)74. CLA improves blood lipids by lowering triglycerides and cholesterol levels. Deficiency in dietary Linoleic acids increased the risk of Coronary Heart Disease and obesity75,76.
Another two related microbiome metabolic functions that were performed significantly better in the RE ileum than in the SE ileum were primary bile acid biosynthesis, and taurine and hypotaurine metabolism. The synthesis of bile acids is the major pathway of cholesterol catabolism in mammals and most vertebrate. Although several of the enzymes involved in bile acid synthesis are active in many different cell types, the liver is the only organ in mammals where complete biosynthesis of primary bile acids (cholic acid, chenodeoxycholic acid) can occur. Secondary bile acids (e.g. lithocholic acid, deoxycholic acid), on the other hand, can be biosynthesized by intestinal microbiota. The prediction by PICRUSt that the ileal microbiota in quail can biosynthesize primary bile acids seems to contradict current knowledge. However, Kim et al.77 reported biosynthesis of primary bile acids in a variety of marine bacterial taxa. Therefore, it may be that excessive dietary cholesterol can stimulate some ileal microbiota to completely or incompletely biosynthesize primary bile acids. Bile acids have long been known to facilitate digestion and absorption of lipids in the small intestine as well as regulate cholesterol homeostasis78. It has now become clear that bile acids are also hormones involved in the regulation of various metabolic processes. Through activation of various signaling pathways, bile acids regulate not only their own synthesis and enterohepatic circulation, but also triglyceride, cholesterol, glucose, and energy homeostasis79,80.
Taurine is a major constituent of bile and can be found in the large intestine. It has many fundamental biological roles, such as conjugation of bile acids, antioxidation, osmoregulation, membrane stabilization, and modulation of calcium signaling. It is essential for cardiovascular function, and development and function of skeletal muscle, the retina, and the central nervous system. Taurine has been shown to reduce the secretion of apolipoprotein B100 and lipids in HepG2 cells. High concentrations of serum lipids and apolipoprotein B100 (essential structural component of VLDL and LDL) are major risk factors of atherosclerosis and coronary heart disease81.
Finally, the RE ileal microbiome was significantly better than the SE ileal microbiome in butirosin and neomycin biosynthesis. Both butirosin and neomycin are antibiotics. Neomycin is not adsorbed through the gastrointestinal wall and has been used as a preventive measure for hypercholesterolemia in human82.
Thus it seems that microbiome in the ileum and ceca of RES contributed significantly towards the resistance to diet induced atherosclerosis.
Excessive dietary cholesterol has significantly affected the duodenal microbiome of SUS. In comparison with SC, duodenal microbiome of SE had significantly less Ubiquinone and other terpenoid-quinone biosynthesis. Ubiquinone, or Coenzyme Q10, shares a biosynthetic pathway with cholesterol83. In human, CoQ10 deficiency may be associated with a multitude of diseases including heart failure84. The severity of heart failure correlates with the severity of CoQ10 deficiency. Emerging data suggest that the harmful effects of reactive oxygen species are increased in patients with heart failure and CoQ10 may help to reduce these toxic effects because of its antioxidant activity. However, only a few randomized controlled trials have compared CoQ10 to other therapeutic modalities, and no systematic review of existing randomized trials has been conducted. There was “no conclusive evidence to support or refute” the use of ubiquinone for the treatment of heart failure84. A 2009 Cochrane review concluded that studies looking at the effects of CoQ10 on blood pressure were unreliable, and therefore no conclusions could be made regarding its effectiveness in lowering blood pressure. A couple of Terpenoid-Quinones isolated from plants have found to possess significant antihyperglycemic activity85.
The duodenal microbiome of SE also had significantly less sulfur metabolism ability. Sulfur is generally acquired in the diet through protein86. Gut microbes require sulfur inputs and, because of their active metabolism and tremendous number, are likely to play a major role in the metabolism of luminal sulfur. Sulfur is either converted to sulfated compounds, assimilated by host cells or excreted. The importance of the microbiota and the metabolism of sulfated bile acids are now established, but further work is needed to understand how dietary fat intake influences these pathways. In addition, one essential and another conditionally essential amino acid (methionine and cysteine, respectively) are sulfated. Cysteine and methionine are used by the body to make glutathione (GSH). GSH is an important antioxidant, preventing damage to important cellular components caused by reactive oxygen species. Hoekstra et al.87 examined cultured aortic endothelial cells from RES and SUS and found that SUS cells were more susceptible to oxidative stress than RES cells. In particular, SUS cells had significantly lower level of GSH.
Thus, in terms of metabolic functions, the selective breeding for susceptibility to diet induced atherosclerosis has mainly affected the microbiome in the duodenum while selection for resistance has mainly affected the microbiome in the ileum and ceca.
Gut microbiome of RES and SUS on control diet
RES and SUS were developed by divergent selective breeding and the selection criteria were based on the atherosclerotic plaque score of males fed a high cholesterol diet21. Males with the highest plaque scores were selected as breeders to establish the SUS strain and males with the lowest scores were selected to be breeders to establish the RES strain. After this first generation, selection was done within strain and there was no crossing back to the foundation population. SE males in the SUS strain with the highest plaque scores were selected as breeders to propagate the next generation of SUS. RE males in RES strain with the lowest plaque scores were used for breeding in the RES. The change in gut microbiome in the males fed a high cholesterol diet is considered to be a correlated response as selection was not done on the microbes. The change in gut microbiome in males not exposed to a high cholesterol diet is also a correlated trait as selection was not done on birds fed the regular diet.
In our study, RC duodenal microbiome was predicted to be significantly better than the SC duodenal microbiome in the biosynthesis of ansamycins. Ansamycin antibiotics are a class of microbial metabolites that exhibit an array of biological activities. It has been postulated that Ansamycins can be used to control microbes that are involved in systemic inflammatory processes88. Ansamycins are highly effective against Chlamydia pneumoniae which plays important role in the occurrence and development of coronary heart disease89. The microbiome in the RC ileum also has more genes than SC for naphthalene degradation. Naphthalene has serious deleterious effects on bird health90,91. Strains of Pseudomonas putida and Escherichia coli carry the genes for naphthalene degradation on a recombinant plasmid pRKJ392,93. In general, gut microbiome of RES (both RE and RC) facilitated better health of their host more so than their SUS counterparts.
The ileal microbiome of SC, on the other hand, has significantly more genes for histidine metabolism, valine/leucine/isoleucine biosynthesis, and pantothenate/CoA biosynthesis than RC. Many gut bacteria, especially those in Clostridia and Fuscobacteria, have been identified to metabolize histidine94. Histidine can be metabolized through 5 different pathways which give rise to several important metabolic products such as histamine and formiminoglutamic acid (FIGLU). The N10-formyl tetrahydrofolic acid (THF) is essential in protein synthesis in microorganisms94,95. Two-component signal transduction systems are the major routes bacteria use to detect environmental signals that mediate changes in cellular behavior or biological processes. These systems consist typically of two proteins—a sensory histidine kinase and a response regulator96. Pantothenic acid (Vitamin B5) is an essential vitamin required for the biosynthesis of Coenzyme A (CoA) and CoA functions as a carrier of acyl groups in enzymatic reactions involved in synthesis of fatty acids, cholesterol, and sterols. CoA also has an essential function in lipid metabolism97,98. The gene clusters involved in amino acid biosynthetic pathways have been described in lactobacillus, and systems controlling gene expression have been identified96. Leucine, isoleucine and valine are branched-chain amino acids (BCAA). There is an increasing body of evidence that the functional output of the gut microbiota, in particular bacterial metabolites like amino acids, are important modulators of host physiology97. BCAA leucine, isoleucine and valine are preferred substrates of gut bacteria to generate a complex mixture of metabolic end products such as short-chain fatty acids (SCFA). Some of these have been shown to modulate bacterial gene expression for amino acid metabolism and also to modulating the mucosal immune system of the host98,99. The most abundant SCFA are acetate, propionate and butyrate. Once absorbed, butyrate is used by liver cells for gluconeogenesis and cholesterol synthesis100. Furthermore, it has been postulated that chronic elevation of systemic BCAA levels, as seen in obesity, impair transport of these BCAA from the intestine lumen into systemic circulation.
Thus leading to more amino acid catabolism in the lumen and more SCFA formation. Thus, it seems that selection for susceptibility to atherosclerosis in SUS has made them more efficient in the absorption and synthesis of fatty acids, cholesterol, and sterols.
Conclusions
Our study has well controlled independent variables to provide conclusive evidence on the interaction of host genotype and diet on gut microbiota. The SUS and RES quail strains are a result of divergent genetic selection from a common foundation population21. The eggs of the four treatment groups were artificially incubated at the same time in close proximity in the same incubator. Birds of the two strains fed the same diet were raised in the same pen. Blood and tissue sampling was done at the same age for all the birds24. The difference in gut microbiota in the two quail strains fed the same diet can therefore be attributed to host genotype. The gut microbiota difference in birds of the same strain fed different diets can be attributed to dietary effect. The difference in the diets is only in the level of cholesterol and so the dietary effect can be attributed to the effect of cholesterol. Difference among the 4 treatment groups can be attributed to the interaction between host genotype and diet.
The interaction between host genotype and diet provided possible explanations for the varying efficacy of probiotic treatments101,102. Of particular interest is the overabundance of Lactobacillus in the ileum of RES. With the help of PICRUSt predictions, we can relate gut microbiota with the host’s resistance and susceptibility to diet induced atherosclerosis. Atherosclerosis is a complex disease affected by the interaction of genetic and environmental risk factors. To understand better the role that gut microbiota play in the development of atherosclerosis, it will be worthwhile to examine the gene expression of the intestinal wall and the liver in association with these microbiome genes.
Electronic supplementary material
Acknowledgements
This paper was submitted by SL as part of a thesis for the partial fulfillment of the requirements for a PhD degree, Chinese Academy of Agricultural Sciences/China Agricultural University. The study was carried out while SL was a visiting student at the UBC Avian Research Centre. The feeding trial was carried out at the Agriculture and Agri-Food Canada Research Station at Agassiz, British Columbia, Canada, under the supervision of Dr. Frederick Silversides. The NGS analyses was carried out at the School of Biological Science, University of Hong Kong. We would like to thank Dr. Raymond Kin-hi Hui, Dr. Tae-Jin Park, Ms. Charis Chan and Ms Angel Ma at The University of Hong Kong for technical assistance; Dr. June Kim (UBC Avian Research Centre) supervised the sample preparation for NGS analysis. The funding for this research was provided by the BC Ministry of Agriculture (funds administered by the UBC Specialty Birds Research Committee) to KMC, a grant from the China Agriculture Research System (CARS-42-G13) to SL, and by the Strategic Initiative (Biomedical Engineering) Fund to FCL. SL was also supported by a scholarship from the China Scholarship Council.
Author Contributions
This manuscript is an extension of the thesis research carried out by S.L. D.C.B. provided expertise in quail nutrition and gut microbiota. H.M.T. provided expertise in Bioinformatics and assisted in manuscript preparation. F.L. provided expertise and laboratory facilities for pyrosequencing and microbiome analysis. H.Z. was S.L.’s thesis supervisor at CAAS and contributed to the early planning and development of the research project. K.M.C. was the thesis research supervisor and provided expertise in avian genetics, development of the RES/SUS quail model, cholesterol metabolism and atherosclerosis, experimental design, and manuscript preparation.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Shasha Liu and Hein Min Tun contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20508-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hongfu Zhang, Email: zhanghf6565@vip.sina.com.
Kimberly M. Cheng, Email: kmtc@mail.ubc.ca
References
- 1.World Health Organization. The top 10 causes of death. http://who.int/mediacentre/factsheets/fs310/en/ accessed: 25 Sept (2016).
- 2.Nabel EG. Lessons learned from monogenic cardiovascular disorders. N Engl J Med. 2003;349:60–72. doi: 10.1056/NEJMra035098. [DOI] [PubMed] [Google Scholar]
- 3.Garrido J, Garcés C, De Oya M. Diet and atherosclerosis. Rev Espan de Card. 1997;51:36–44. [PubMed] [Google Scholar]
- 4.Watts G, et al. Effects on coronary artery disease of lipid-lowering diet, or diet plus cholestyramine, in the St Thomas’ Atherosclerosis Regression Study (STARS) Lancet. 1992;339:563–569. doi: 10.1016/0140-6736(92)90863-X. [DOI] [PubMed] [Google Scholar]
- 5.Karlsson FH, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nature Comm. 2012;3:1245. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martínez I, et al. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl Env Micro. 2009;75:4175–4184. doi: 10.1128/AEM.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavallini D, et al. Influence of a probiotic soy product on fecal microbiota and its association with cardiovascular risk factors in an animal model. Lipids Health Dis. 2011;10:126. doi: 10.1186/1476-511X-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregory JC, et al. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J Biol Chem. 2015;290:5647–5660. doi: 10.1074/jbc.M114.618249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koren O, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci. 2011;108:4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu S, et al. The effect of diet and host genotype on ceca microbiota of Japanese quail fed a cholesterol enriched diet. Front Microb. 2015;6:1092. doi: 10.3389/fmicb.2015.01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savage DC. Microbial ecology of the gastrointestinal tract. Ann Rev Microb. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 12.Gong J, et al. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS Microb Ecol. 2007;59:147–157. doi: 10.1111/j.1574-6941.2006.00193.x. [DOI] [PubMed] [Google Scholar]
- 13.Gu S, et al. Bacterial community mapping of the mouse gastrointestinal tract. PloS one. 2013;8:e74957. doi: 10.1371/journal.pone.0074957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Nat Acad Sci. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci trans med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sommer F, Bäckhed F. The gut microbiota—masters of host development and physiology. Nat Rev Microb. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 17.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 18.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 19.Koeth RA, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koeth RA, et al. γ-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-Carnitine to TMAO. Cell metab. 2014;20:799–812. doi: 10.1016/j.cmet.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shih JC, Pullman E, Kao K. Genetic selection, general characterization, and histology of atherosclerosis-susceptible and-resistant Japanese quail. Atherosclerosis. 1983;49:41–53. doi: 10.1016/0021-9150(83)90006-0. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Schulte P, Godin DV, Cheng KM. Differential mRNA expression of seven genes involved in cholesterol metabolism and transport in the liver of atherosclerosis-susceptible and-resistant Japanese quail strains. Gen Sel Evol. 2012;44:20. doi: 10.1186/1297-9686-44-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godin DV, Garnett ME, Cheng KM, Nichols CR. Sex-related alterations in antioxidant status and susceptibility to atherosclerosis in Japanese quail. Can J Cardiol. 1995;11:945–951. [PubMed] [Google Scholar]
- 24.Liu S, et al. The effect of diet and host genotype on ceca microbiota of Japanese quail fed a cholesterol enriched diet. Front Microb. 2015;6:1092. doi: 10.3389/fmicb.2015.01092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinf. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole JR, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucl Acids Res. 2009;37(Suppl 1):D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caporaso G, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colwell RK, et al. Models and estimators linking individual-based and sample-based rarefaction, extrapolation and comparison of assemblages. J Plant Eco. 2012;5:3–21. doi: 10.1093/jpe/rtr044. [DOI] [Google Scholar]
- 30.Weinfurt, K. P. Multivariate analysis of variance in Reading and understanding multivariate statistics (eds Grimm, L. G. and Yarnold, P. R.) 245–276 (Amer Psyc Assoc 1995).
- 31.Oliveros, J. C. VENNY: An interactive tool for comparing lists with Venn Diagrams; http://bioinfogp.cnb.csic.es/tools/venny/index.html. accessed: Sept 2014 (2007).
- 32.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Nat Acad Sci. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucl Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucl Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ley RE, et al. Obesity alters gut microbial ecology. Proc Nat Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turnbaugh PJ, Bäckhed F, Fulton. L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bennett DC, Tun HM, Kim JE, Leung FC, Cheng KM. Characterization of cecal microbiota of the emu (Dromaius novaehollandiae) Vet Microb. 2013;166:304–310. doi: 10.1016/j.vetmic.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 41.Su H, et al. Cultivable bacterial microbiota of northernbobwhite (Colinus virginianus): a new reservoir of antimicrobial resistance? PLoS one. 2014;9:e99826. doi: 10.1371/journal.pone.0099826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Videnska, P. et al. Succession and replacement of bacterial populations in the caecum of egg laying hens over their whole life. PLoS one, 10.1371/journal.pone.0115142 (2014). [DOI] [PMC free article] [PubMed]
- 43.Kim K-A, Gu W, Lee I-A, Joh E-H, Kim D-H. High fat diet-Induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS one. 2012 doi: 10.1371/journal.pone.0047713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenq RR, et al. Intestinal Blautia is associated with reduced death from Graft-versus-Host Disease. Biol Blood Marrow Transp. 2015;21:373–383. doi: 10.1016/j.bbmt.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Endo T, Nakano M, Shimizu S, Fukushima M, Miyoshi S. Effects of a Probiotic on the Lipid Metabolism of Cocks Fed on a Cholesterol-enriched Diet. Biosci Biotech Biochem. 1999;63:1569–1575. doi: 10.1271/bbb.63.1569. [DOI] [PubMed] [Google Scholar]
- 46.Ryu H-S, Lee H-S, Lim J-H, Kim J-R, Kim S-D. Isolation and identification of Sphingomonas sanguis from wild pheasant and production of antagonistic substance against fowl Typhoid causing Salmonella gallinarum. Appl Biol Chem. 2004;47:27–32. [Google Scholar]
- 47.Clarke SF, et al. The gut microbiota and its relationship to diet and obesity. Gut Microbes. 2012;3:186–202. doi: 10.4161/gmic.20168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heinritz SN, et al. Impact of a high-fat or high fibre diet on intestinal microbiota and metabolic markers in a pig model. Nutrients. 2016;8:317. doi: 10.3390/nu8050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown K, DeCoffe D, Molcan E, Gibson DL. Diet-Induced Dysbiosis of the Intestinal Microbiota and the Effects on Immunity and Disease. Nutrients. 2012;4:1095–1119. doi: 10.3390/nu4081095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graber CD, et al. Autochthonous intestinal bacterial flora and cholesterol levels in specific pathogen-free swine fed high-lipid and high-sucrose diets. J Bacteriol. 1966;92:1290–1297. doi: 10.1128/jb.92.5.1290-1297.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conlon MA, Bird AR. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2015;7:17–44. doi: 10.3390/nu7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abdulrahim SM, Haddadin MSY, Hashlamoun EAR, Robinson RK. The influence of lactobacillus acidophilus and bacitracin on layer performance of chickens and cholesterol content of plasma and egg yolk. Brit Poult Sci. 1996;37:341–346. doi: 10.1080/00071669608417865. [DOI] [PubMed] [Google Scholar]
- 53.Kumar L, et al. Probiotic Lactobacillus rhamnosus GG and Aloe vera gel improve lipid profiles in hypercholesterolemic rats. J Nutr. 2013;29:574–579. doi: 10.1016/j.nut.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 54.Simons LA, Amansec SG, Conway P. Effect of Lactobacillus fermentum on serum lipids in subjects with elevated serum cholesterol. Nutr Met Card Dis. 2006;16:531–535. doi: 10.1016/j.numecd.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 55.Tomaro-Duchesneau C, et al. Cholesterol assimilation by Lactobacillus probiotic bacteria: An in vitro investigation. BioMed Res Intern. 2014;2014:Article ID 380316. doi: 10.1155/2014/380316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones ML, Martoni CJ, Prakash S. Cholesterol lowering and inhibition of sterol absorption by Lactobacillus reuteri NCIMB 30242: a randomized controlled trial. Eur J Clin Nutr. 2012;66:1234–1241. doi: 10.1038/ejcn.2012.126. [DOI] [PubMed] [Google Scholar]
- 57.Huang Y, Zheng Y. The probiotic Lactobacillus acidophilus reduces cholesterol absorption through the down-regulation of Niemann-Pick C1-like 1 in Caco-2 cells. Brit J Nut. 2010;103:473–478. doi: 10.1017/S0007114509991991. [DOI] [PubMed] [Google Scholar]
- 58.Fuentes MC, Lajo T, Carrio JM, Cuñé J. Cholesterol-lowering efficacy of Lactobacillus plantarum CECT 7527, 7528 and 7529 in hypercholesterolaemic adults. Brit J Nut. 2013;109:1866–1872. doi: 10.1017/S000711451200373X. [DOI] [PubMed] [Google Scholar]
- 59.Walter J. Ecological role of Lactobacillus in the gastrointestinal tract: Implications for fundamental and biomedical research. Appl Environ Microbiol. 2008;74:4985. doi: 10.1128/AEM.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spor. A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microb. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 61.Tsai K-N, Lin S-H, Liu W-C, Wang D. Inferring microbial interaction network from microbiome data using RMN algorithm. BMC Sys Biol. 2015;9:54. doi: 10.1186/s12918-015-0199-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steinway, S. N., Biggs, M. B., Loughran, T. P. Jr., Papin, J. A. & Albert, R. Inference of network dynamics and metabolic interactions in the gut microbiome. PLoS Comp Biol, 10.1371/journal.pcbi.1004338 (2015). [DOI] [PMC free article] [PubMed]
- 63.Moya A, Ferre RM. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microb. 2016;24:402–413. doi: 10.1016/j.tim.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 64.Huang Y, et al. Lactobacillus acidophilus ATCC 4356 prevents atherosclerosis via inhibition of intestinal cholesterol absorption in apolipoprotein E knockout mice. Appl Environ Microb. 2014;80:7496–7506. doi: 10.1128/AEM.02926-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Naruszewicz M, Johansson M-L, Zapolska-Downar D, Bukowska H. Effect of Lactobacillus plantarum 299v on cardiovascular disease risk factors in smokers. Am J Clin Nutr. 2002;76:1249–1255. doi: 10.1093/ajcn/76.6.1249. [DOI] [PubMed] [Google Scholar]
- 66.Brigelius-Flohé R, Banning A, Schnurr K. Selenium-dependent enzymes in endothelial cell function. Antioxid Redox Signal. 2003;5:205–215. doi: 10.1089/152308603764816569. [DOI] [PubMed] [Google Scholar]
- 67.Ciorba MA. Kynurenine pathway metabolites: relevant to vitamin B-6 deficiency and beyond. Am J Clin Nutr. 2013;98:863–864. doi: 10.3945/ajcn.113.072025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hadi LA, Di Vito C, Riboni L. Fostering inflammatory bowel disease: sphingolipid strategies to join forces. Media Inflam. 2016;2016:Article ID 3827684. doi: 10.1155/2016/3827684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duthie SJ, et al. Nutritional B vitamin deficiency alters the expression of key proteins associated with vascular smooth muscle cell proliferation and migration in the aorta of atherosclerotic apolipoprotein E null mice. Genes Nutr. 2015;10:446. doi: 10.1007/s12263-014-0446-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Endo N, et al. Antioxidant activity of vitamin B6 delays homocysteine-induced atherosclerosis in rats. Brit J Nutri. 2006;95:1088–1093. doi: 10.1079/BJN20061764. [DOI] [PubMed] [Google Scholar]
- 71.Sinha R, Unni E, Ganther HE, Medina D. Methylseleninic acid, a potent growth inhibitor of synchronized mouse mammary epithelial tumor cells in vitro. Biochem Pharmacol. 2001;61:311–317. doi: 10.1016/S0006-2952(00)00545-1. [DOI] [PubMed] [Google Scholar]
- 72.Kaur HD, Bansal MP. Studies on HDL associated enzymes under experimental hypercholesterolemia: possible modulation on selenium supplementation. Lipids Health Dis. 2009;8:55. doi: 10.1186/1476-511X-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marmugi A, et al. Adverse effects of long-term exposure to bisphenol A during adulthood leading to hyperglycaemia and hypercholesterolemia in mice. Toxicology. 2014;325:133–143. doi: 10.1016/j.tox.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 74.Devillard E, McIntosh FM, Duncan SH, Wallace R. Metabolism of linoleic acid by human gut bacteria: different routes for biosynthesis of conjugated linoleic acid. J bacteriol. 2007;189:2566–2570. doi: 10.1128/JB.01359-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benito P, et al. Effect of conjugated linoleic acid on platelet function, platelet fatty acid composition, and blood coagulation in humans. Lipids. 2001;36:221. doi: 10.1007/s11745-001-0711-y. [DOI] [PubMed] [Google Scholar]
- 76.Farvid MS, et al. Dietary linoleic acid and risk of coronary heart disease: A systematic review and meta-analysis of prospective cohort studies. Circulation. 2016;130:1568–1578. doi: 10.1161/CIRCULATIONAHA.114.010236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim D, et al. Biosynthesis of bile acids in a variety of marine bacterial taxa. J Microbiol Biotech. 2007;17:403–407. [PubMed] [Google Scholar]
- 78.Hagey LR, Schteingart CD, Ton-Nu H-T, Hofmann AF. A novel primary bile acid in the Shoebill stork and herons and its phylogenetic significance. J Lipid Res. 2002;43:685–690. [PubMed] [Google Scholar]
- 79.Staels B, Fonseca VA. Bile acids and metabolic regulation - Mechanisms and clinical responses to bile acid sequestration. Diabetes Care. 2009;32(Suppl 2):S237–245. doi: 10.2337/dc09-S355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 81.Ridker PM, Rifai N, Cook NR, Bradwin G, Buring JE. Non–HDL cholesterol, apolipoproteins a-i and b100, standard lipid measures, lipid ratios, and CRP as risk factors for cardiovascular disease in women. JAMA. 2005;294:325–333. doi: 10.1001/jama.294.3.326. [DOI] [PubMed] [Google Scholar]
- 82.Meisel, S., Rate, R. Neomycin for hypercholesterolemia. N Engl J Med302, 233–234 PMID: 7350468, 10.1056/NEJM198001243020414. (1980). [DOI] [PubMed]
- 83.Meganathan R. Ubiquinone biosynthesis in microorganisms. FEMS Microb Letters. 2001;203:131–139. doi: 10.1111/j.1574-6968.2001.tb10831.x. [DOI] [PubMed] [Google Scholar]
- 84.Madmani, M. E. et al. Coenzyme Q10 for heart failure. Cochrane Data Sys Rev6, Art. No: CD008684, 10.1002/14651858.CD008684.pub2 (2014). [DOI] [PubMed]
- 85.Fort DM, et al. Novel Antihyperglycemic Terpenoid-Quinones from Pycnanthus angolensis. J Org Chem. 2000;65:6534–6539. doi: 10.1021/jo000568q. [DOI] [PubMed] [Google Scholar]
- 86.Stipanuk MH. Sulfur amino acid metabolism: pathways for production and removal of homocysteine and cysteine. Ann Rev Nutr. 2004;24:539–577. doi: 10.1146/annurev.nutr.24.012003.132418. [DOI] [PubMed] [Google Scholar]
- 87.Hoekstra KA, Godin DV, Kurtu J, Cheng KM. Heme oxygenase and antioxidant status in cultured aortic endothelial cells isolated from atherosclerosis-susceptible and –resistant Japanese quail. Mol Cell Biochem. 2003;252:253–262. doi: 10.1023/A:1025555525661. [DOI] [PubMed] [Google Scholar]
- 88.Curry K, Lawson L. Links between infectious diseases and cardiovascular disease: A growing body of evidence. J Nurse Prac. 2009;5:733–741. doi: 10.1016/j.nurpra.2008.12.006. [DOI] [Google Scholar]
- 89.Suchland RJ, Bourillon A, Denamur E, Stamm WE, Rothstein DM. Rifampin-resistant RNA polymerase mutants of Chlamydia trachomatis remain susceptible to the Ansamycin Rifalazil. Anti Agents Chemo. 2005;49:1120–1126. doi: 10.1128/AAC.49.3.1120-1126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.De Witt JB. Pesticide Toxicity, Effects of Chlorinated Hydrocarbon Insecticides upon Quail and Pheasants. J Agric Food Chem. 1955;3:672–676. doi: 10.1021/jf60054a003. [DOI] [Google Scholar]
- 91.Gorsline J, Holmes WN, Cronshaw J. The effects of ingested petroleum on the naphthalene-metabolizing properties of liver tissue in seawater-adapted mallard ducks (Anas platyrhynchos) Enviro Res. 1981;24:377–390. doi: 10.1016/0013-9351(81)90167-5. [DOI] [Google Scholar]
- 92.Bhushan B, Samanta SK, Jain RK. Indigo production by naphthalene-degrading bacteria. Lett Appl Microb. 2000;31:5–9. doi: 10.1046/j.1472-765x.2000.00754.x. [DOI] [PubMed] [Google Scholar]
- 93.Dennis JJ, Zylstra GJ. Complete sequence and genetic organization of pDTG1, the 83 kilobase Naphthalene Degradation Plasmid from Pseudomonas putida strain NCIB 9816-4. J Mol Biol. 2004;341:753–768. doi: 10.1016/j.jmb.2004.06.034. [DOI] [PubMed] [Google Scholar]
- 94.Dai Z-L, Wu G, Zhu W-Y. Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front in Biosci. 2011;16:1768–1786. doi: 10.2741/3820. [DOI] [PubMed] [Google Scholar]
- 95.Stiefel FB, Herman RH. Histidine metabolism. Am J Clin Nutr. 1971;24:207–217. doi: 10.1093/ajcn/24.2.207. [DOI] [PubMed] [Google Scholar]
- 96.Lau P, et al. A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. Proc Natl Acad Sci. USA. 1997;94:1453–1458. doi: 10.1073/pnas.94.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chopin A. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microb Rev. 1993;12:21–37. doi: 10.1111/j.1574-6976.1993.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 98.Sridharan GV, et al. Prediction and quantification of bioactive microbiota metabolites in the mouse gut. Nat Commun. 2014;5:5492. doi: 10.1038/ncomms6492. [DOI] [PubMed] [Google Scholar]
- 99.Blachier F, Mariotti F, Huneau JF, Tome D. Effects of amino acid-derived luminal metabolites on the colonic epithelium and physiopathological consequences. Amino Acids. 2007;33:547–562. doi: 10.1007/s00726-006-0477-9. [DOI] [PubMed] [Google Scholar]
- 100.Neis PJG, Dejong CHC, Rensen SS. The role of microbial amino acid metabolism in host metabolism. Nutrients. 2015;7:2930–2946. doi: 10.3390/nu7042930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ooi L-G, Liong M-T. Cholesterol-lowering effects of probiotics and prebiotics: a review of in vivo and in vitro findings. Int J Mol Sci. 2010;11:2499–2522. doi: 10.3390/ijms11062499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ng. SC, Hart AL, Kamm MA, Stagg AJ, Knight SC. Mechanisms of action of probiotics: Recent advances. Inf Bowel Dis. 2009;15:300–310. doi: 10.1002/ibd.20602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.