Abstract
Milk microbiota has a great influence on the safety and quality of dairy products. However, few studies have investigated the variations of bacterial composition in raw milk. In this study, raw milk samples were collected in 12 successive months, and their bacterial compositions were determined by 16 S rRNA gene sequencing. The highest diversity of bacterial composition was detected in June, while the lowest was in December. Firmicutes, Proteobacteria and Actinobacteria were the most abundant phyla and exhibited a counter-balanced relationship. Pseudomonas, Lactococcus and Acinetobacter were the most prevalent genera (>1%), and a tiny core microbiota (Acinetobacter and Pseudomonas) was observed. Temperature and humidity were the determining factors for most variation in bacterial compositions at both the phylum and genus levels. Higher abundances of Pseudomonas, Propionibacterium and Flavobacterium were correlated with low temperature. Furthermore, Pseudomonas/Propionibacterium and Lactobacillus/Bifidobacterium were two pairs of genera that had synergistic effects. Associations between the microbiota and milk quality parameters were analyzed. The abundances of Propionibacterium and Pseudoalteromonas were negatively correlated to total bacterial count, which meant that they helped to maintain milk quality, while a series of environmental microorganisms contributed to the spoilage of raw milk.
Introduction
Milk and manufactured dairy products are important components of a healthy diet and are among the most frequently consumed foods. A highly nutritious food, milk is composed of proteins, fats, carbohydrates, vitamins and minerals, providing an ideal environment for the growth of microorganisms1. Currently, the demand for safe and high-quality food has become the focus of public attention. While milk used to be considered one of the healthiest foods with the highest quality, current food recalls and foodborne illness outbreaks have indicated the need for an in-depth investigation2,3.
Bacterial contamination of dairy products is a major concern throughout the world4, as dairy production and consumption have expanded extensively during the last decade. Raw milk is known to usually harbor complex microbial communities containing diverse microorganisms5,6. The indigenous microflora in raw milk has a direct impact on the subsequent development of dairy products7. Microorganisms could not only affect milk quality and shelf life but also produce extracellular lipases and proteases, which could result in spoilage8–10. In addition, abnormal microbial compositions in milk could cause health problems, in that the consumption of raw milk contaminated with pathogens can lead to severe illness11,12.
High-throughput sequencing has been revolutionarily developed for the analysis of microbial ecology and has provided detailed insights into a series of raw milks and dairy products, such as goat milk13, buffalo milk14, naturally fermented milk15, artisanal cheeses16 and breast milk17. In addition, factors related to raw milk processing, including storage tanks18, transportation vehicles19 and pasture altitude20, have been assessed. However, the role of weather conditions on the microbial communities in raw milk has not yet been studied.
On-farm knowledge of the bacterial communities in raw milk is important for the identification of bacterial composition and assessment of its regularity. Descriptions of microbial composition and distribution would provide necessary information for the risk assessment and quality control of raw milk throughout the year. In addition, it ensures the high performance and traceability of the subsequent pasteurization and processing. To get a better understanding of indigenous bacterial compositions and to guarantee the safety and quality of raw milk and dairy products, we investigated the bacterial communities of raw milk from ten dairy farms in Shanghai (China) with high-throughput sequencing technology. Specifically, we investigated the variation in the bacterial communities throughout one whole year. The effects of temperature and humidity were considered in a correlation analysis with the bacterial communities.
Results
Bacterial diversity in cow milk
From July 2015 to June 2016, we collected cow milk samples each month from 10 farms; a total of 112 samples were collected. A total of 2,503,353 (7,780~34,904) high-quality 16 S rRNA gene reads were obtained by high-throughput DNA sequencing that met quality-filtering requirements. To compare the samples without statistical bias, the minimal read number 7,780, as a normalization size for each sample, was chosen for calculation of the diversity indices. The operational taxonomic unit (OTU), or group of 16 S rRNA genes, was set at 97% similarity to estimate the diversity of the bacterial community. After all samples were grouped from the 12 months, a total of 12,898 OTUs (average 2,755, from 863 to 4,469) were obtained. Good’s coverage was an average of 98.2% (97.4%~98.7%, Table 1), which meant that the sequencing depth was sufficient for the microbiota investigation of cow milk over 12 months.
Table 1.
Bacterial richness and diversity estimates at genetic distances of 3%.
| Month | OTUs | Coverage | SEM | ACE | SEM | Chao | SEM | Shannon | SEM |
|---|---|---|---|---|---|---|---|---|---|
| Jan | 2570 | 0.982 | 0.003728 | 5424.179 | 151.447 | 4212.251 | 86.84022 | 3.261 | 0.285948 |
| Feb | 2183 | 0.984 | 0.005885 | 5123.543 | 200.1453 | 3818 | 151.0531 | 3.3 | 0.418796 |
| Mar | 3975 | 0.976 | 0.006953 | 9033.945* | 286.1278 | 6998.382* | 180.4165 | 4.843 | 0.301788 |
| Apr | 2611 | 0.983 | 0.004218 | 6585.588 | 178.1783 | 4788.6 | 95.91588 | 3.583 | 0.180692 |
| May | 1999 | 0.985 | 0.003139 | 6856.097 | 208.7313 | 4330.616 | 94.78375 | 3.015 | 0.089325 |
| June | 4469 | 0.977 | 0.004804 | 7941.831** | 205.4795 | 6749.65** | 130.5882 | 5.426 | 0.233656 |
| July | 3516 | 0.974 | 0.006174 | 8988.738** | 256.194 | 6607.327** | 114.5416 | 4.944 | 0.320875 |
| Aug | 2751 | 0.985 | 0.005982 | 5272.712 | 194.8767 | 4339.898 | 142.9341 | 4.245 | 0.256475 |
| Sept | 2720 | 0.984 | 0.00429 | 5251.533 | 143.3089 | 4236.942 | 107.8009 | 4.002 | 0.205467 |
| Oct | 2260 | 0.985 | 0.004677 | 5694.692 | 215.2667 | 4147.889 | 128.0317 | 3.206 | 0.140235 |
| Nov | 3139 | 0.984 | 0.003624 | 4722.395 | 220.7119 | 4703.58 | 107.3771 | 4.203 | 0.472844 |
| Dec | 863 | 0.987 | 0.004162 | 2841.298 | 210.8459 | 1818.37 | 110.98 | 2.961 | 0.432634 |
“*”indicated p < 0.05, “**”indicated p < 0.001.
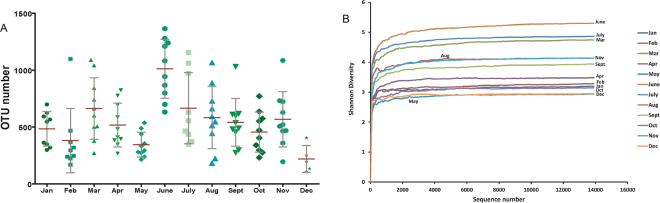
Phylogenetic and taxonomic assessments of the 16 S rRNA V3–4 regions revealed that the diversity of bacterial populations in the raw milk greatly varied among different months. Based on the OTU evaluation, the total estimated bacterial richness per sample differed between months. The milk collected in March, June and July contained higher species richness according to the ACE and Chao index (Table 1). Similarly, a higher number of OTUs was observed in milk examined in these three months (Fig. 1A). There was an average of 29.3% of OTUs (1,158 OTUs) uniquely present in March, June and July, and other months had an average of 19.0% total unique OTUs. The highest number of OTUs was detected in June, while the lowest was in December.
Figure 1.
(A) Scatter plots showing OTU numbers with respect to total sequences obtained from 16 S rRNA genes associated with months. The error bars were the mean with SD. (B) Shannon diversity curve of bacterial community derived from 12 months.
The Shannon index showed that the samples from December had the lowest diversity, while the highest diversity was from the June samples (Shannon: December/June = 2.96/5.43, p < 0.01, Fig. 1B).
Variable richness and high diversity of bacterial populations in cow milk depending on month
An RDP (Ribosomal Database Project) analysis of 112 samples assigned 98.3% (SD = 0.014) of the 16 S rRNA genes to 33 phyla and 91.0% (SD = 0.039) of the 16 S rRNA genes to 785 genera, respectively. A total of 70% or more of the taxa detected represented less than 1% of the abundance in all OTUs. At the phylum level, 4 of 33 phyla were abundant in cow milk, i.e., 40.8% (SD = 0.217) of 16 S rRNA genes were assigned to Firmicutes, 39.0% (SD = 0.141) to Proteobacteria, 9.40% (SD = 0.058) to Actinobacteria and 7.47% (SD = 0.032) to Bacteroidetes. Among the 785 genera, four were abundant, i.e., 19.6% (SD = 0.122) of 16 S rRNA genes were Pseudomonas, 13.8% (SD = 0.096) were Bacillus, 11.7% (SD = 0.078) were Lactococcus and 10.2% (SD = 0.088) were Acinetobacter (Fig. 2). The remaining 35.6% of the 16 S rRNA genes were assigned to 781 other bacterial genera, demonstrating the high diversity of the cow milk bacterial community.
Figure 2.
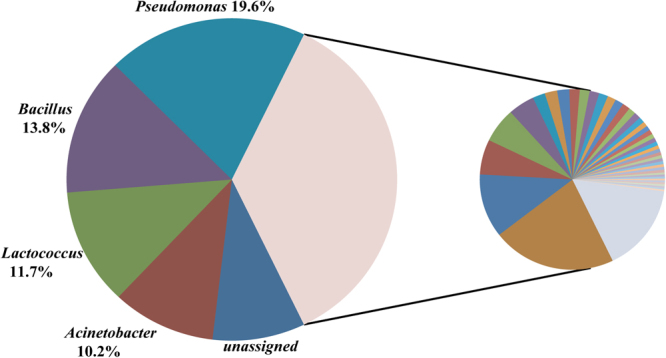
Illustration for four major abundant genera. All genera were showed in the pie chart. The left pie showed four major abundant genera and unsigned sequences which were not classified to genus; the right pie showed other 781 genera.
Based on phylum information (Fig. 3), which was derived from the classification of OTU representative sequences by the RDP classifier, Firmicutes, Proteobacteria and Actinobacteria showed large differences (p < 1E-20) by month. In January, February, March and November, the abundance of Actinobacteria was over 10% of all phyla and was obviously greater (p < 1E-50) than that in other months. In April, May, June, August, September and October, the abundance of Firmicutes was up to 40%~70% of all phyla. In addition, the abundance of Proteobacteria was almost 60% of all phyla in July, November and December. The total proportion of these three phyla was up to 80%~95%.
Figure 3.
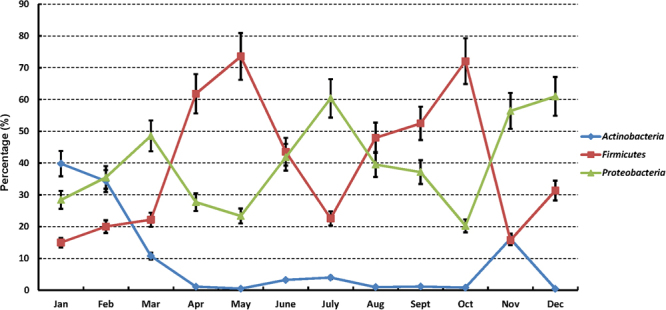
Microbiota variation of three phyla in 12 months. These three phyla showed the difference by the months which might be caused by the temperature and humidity changes. The three phyla were Actinobacteria (0.4%~39.8%), Firmicutes (15.0%~73.6%) and Proteobacteria (20.3%~61.0%).
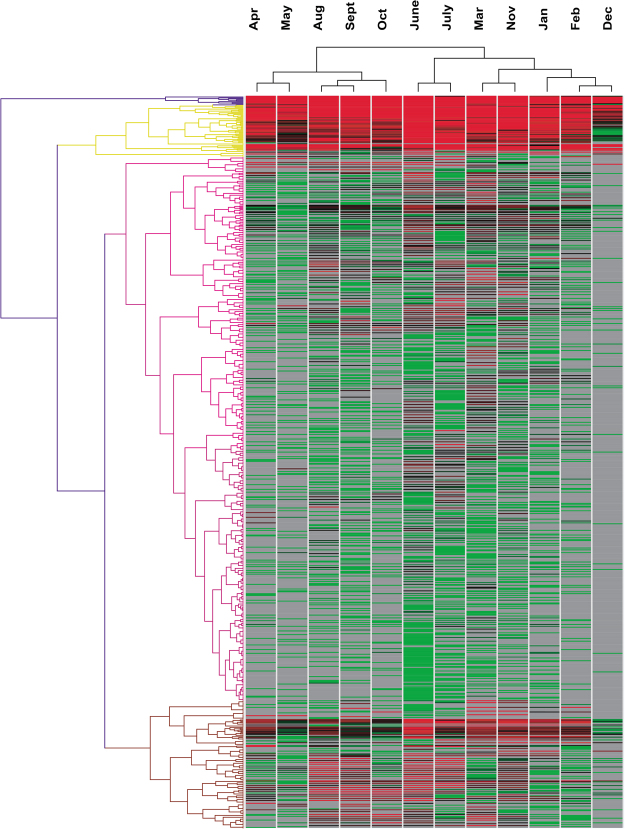
Information regarding genus prevalence for the samples from each month was illustrated with a heatmap by hierarchical cluster analysis (Fig. 4), which showed a similar topological structure of monthly distribution with the results based on a sequence matrix (Fig. 5). Adjacent months were usually located in the same clade, such as July and June, September and August, January and February, which meant that the microbiota compositions were similar between adjacent months. It was also shown that the samples could be grouped into two clades according to the season. Spring and autumn formed one clade, while summer and winter belonged to the other clade. This effect might be partly attributed to temperature and humidity changes (Table S1).
Figure 4.
Heatmap illustrating different genera percentage by 12 months. Hierarchical cluster was calculated for the month and genera. More details of different genera were listed on the Supplementary Tables S2–S12.
Figure 5.
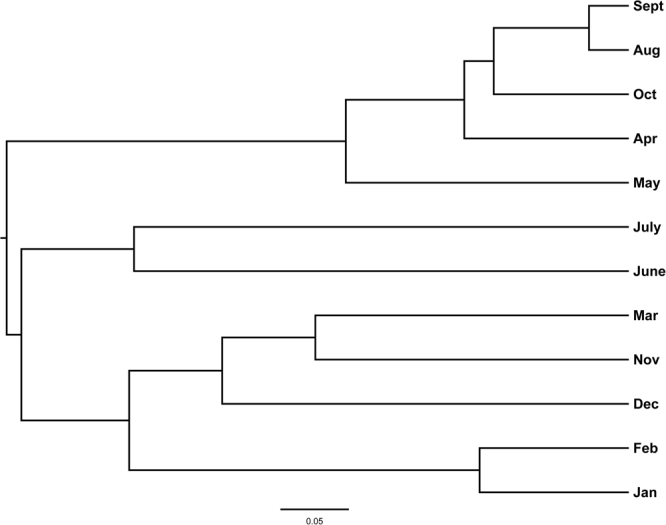
The tree showing the cluster of bacterial community using the Yue & Clayton theta similarity coefficient based on the sequences matrix.
An analysis of the 16 S rRNA gene sequences showed that three genera were most prevalent (>1%) in all 12 months, which were Pseudomonas, Lactococcus and Acinetobacter. Pseudomonas represented an average of 12.0% (SD = 0.039) in April-October (average temperature of 22.8 °C) and 32.4% (SD = 0.090) in November-March (average temperature of 8.8 °C). Acinetobacter represented an average of 19.4% (SD = 0.079) in June-September (average temperature of 25.7 °C) and 6.0% (SD = 0.031) in October-May (average temperature of 12.6 °C). Lactococcus was abundant throughout the year and varied irregularly between adjacent months. Many other significant differences were present in the bacterial communities between months (Fig. 6, Tables S2–S12, p < 1E-5). Considering only the genera whose proportion was greater than 0.1% in one of two compared months, we could reveal a total of 122 genera showing significant differences between months. That meant most genera (84.5%) showed no difference between months, and these genera were a low proportion of the total (summing up to average of 10.7%, 2.7%~18.5%). Therefore, the differences were caused by the high abundance genera. When the criterion for genus proportion was raised to 1%, only 27 genera had significant differences, and those genera occupied an average of 81.8% (68.1%~95.8%) of the total bacterial population (Table S13). Thus, the subsequent analysis was focused on these 27 major genera.
Figure 6.
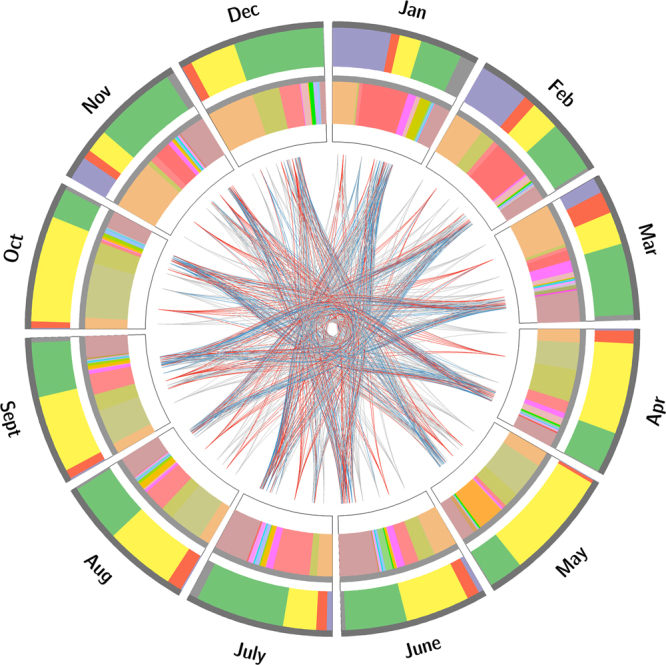
The complex illustrations showing the major phyla and genera in 12 months and the significant differences between major genera among 12 months. (1) The outer circle showed the phylum level and percentage of major phyla, purple: Actinobacteria, red: Bacteroidetes, yellow: Firmicutes, green: Proteobacteria, grey: other phyla. (2) The inner circle showed the genus level and percentage of major genera which accounting for at least 1% and had significant difference (Q-value < 1E-5) between months. (3) The lines linked the different months showed the major genera difference, red line: fold change >50, blue line: 50≥ fold change >10, grey line: 10≥ fold change >2, and more details were showed in the supplementary Tables S2–S12.
Despite these differences of bacterial communities in cow milk samples, a tiny core milk microbiome was still present at the genus level. Only two genera (Acinetobacter and Pseudomonas) belonging to the Proteobacteria phylum were detected in all 112 cow milk samples examined. Notably, Lactococcus was not a part of the core microbiome, although it was present in relatively high proportions in some of the cow milk tested; Lactococcus was entirely absent from one sample and was therefore not included in the core. When focusing on the OTU (species) level, no core species was found in all samples, reflecting the high diversity of microorganisms in cow milk.
Correlations between bacterial composition in cow milk and weather conditions
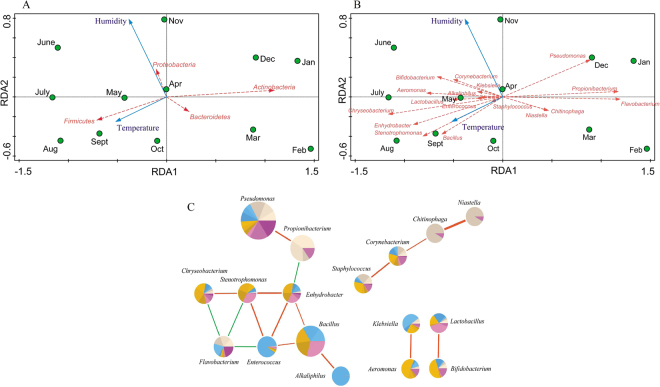
Redundancy analysis (RDA) indicated that temperature and humidity (Table S1) were the determining factors for most of the variation in the bacterial compositions of the samples over 12 months (Fig. 7A,B). Higher abundance of phylum Actinobacteria was correlated with low temperature, while higher abundance of Firmicutes was correlated with high temperature. Higher abundance of phylum Bacteroidetes was correlated with low humidity, while higher abundance of Proteobacteria was correlated with high humidity. At the genus level, higher abundances of Pseudomonas, Propionibacterium and Flavobacterium were correlated with low temperature, while the abundances of Niastella and Chitinophaga were correlated with low humidity. In addition, higher abundances of most genera were correlated with high temperature and high humidity. Meanwhile, correlation patterns were also analyzed among the 785 genera. A Spearman algorithm analysis validated that 17 of the 27 major genera had strong correlations (Fig. 7C, Table S14, p < 0.01), with parameters set as coefficient >0.68 or <−0.68, which were considered to represent strong or high correlations21. Genus Flavobacterium was negatively correlated with Enterococcus, Chryseobacterium and Stenotrophomonas, while genus Propionibacterium was negatively correlated with Enhydrobacter. Other genera exhibited positive correlations, such as Pseudomonas and Propionibacterium, Lactobacillus and Bifidobacterium.
Figure 7.
(A) Redundancy analysis (RDA) of the relationship between the environment factors and the relative abundance of bacterial phylum of 12 months’ samples. (B) Redundancy analysis (RDA) of the relationship between the environment factors and the relative abundance of bacterial genus of 12 months’ samples. (C) Correlation of 17 major genera. Line: orange-red (positive relation) and green (negative relation). Line weight: correlation strength. Circle size represented the reads numbers. The pie chart presented relative proportion in 12 months, seashell series: 1–3 months, blue series: 4–6 months, orange series: 7–9 months, orchid series: 10–12 months.
Correlations between microflora composition and raw milk quality parameters
According to the correlation analysis among 785 genera, many genera exhibited significant correlations with raw milk quality parameters (Table S15), such as somatic cell count (SCC), total bacterial count (TBC), heat-resisting spores, psychrotrophic bacteria, milk protein content and milk fat content.
There were five genera (Azospira, Brachymonas, Chryseobacterium, Cloacibacterium and Sulfurospirillum) showing extremely significant correlations with SCC (p < 0.001), indicating that these bacteria had a great influence on the quality of raw milk and the health status of milking cows.
Stackebrandtia, Teredinibacter and Gemella were the top three genera positively correlated to TBC (p < 0.001), coinciding with the factor that they were also negatively correlated with milk fat content (p < 0.05).
Dozens of genera were demonstrated to be significantly correlated with heat-resisting spores (p < 0.001), while Methylonatrum showed extreme significance.
Only Brochothrix was found to be positively correlated with psychrotrophic bacteria (p < 0.01). To our surprise, the correlation between Pseudomonas and psychrotrophic bacteria was not significant (p = 0.22).
Quite a few bacteria were negatively correlated with protein content, such as Bacteroides, Streptococcus, Agrococcus, and Ornithinimicrobium (p < 0.05), which meant that milk protein decreased with the increased abundance of these bacteria.
In addition, Atopobium was found to be the most significantly contributor to the degradation of milk fat (p < 0.01).
Microbial function prediction
The microbial functions of each month were predicted by PICRUSt software. Using strict parameters, we found no functional difference among some consecutive months (Tables S16–S25), especially in two groups of consecutive months (one group included November, December, January, February and March, and the other group included August, September and October). However, comparing these two groups of consecutive months, we found that some pathways showed significant differences, including “endocytosis”, “dioxin degradation”, “D-Arginine and D-ornithine metabolism”, and “isoflavonoid biosynthesis”. The OTU abundances involved in these four different pathways were increased in the “August, September and October” group.
Discussion
In the present study, it was demonstrated that bacterial composition and structure were highly diverse across the 12 months, indicating that environmental factors play crucial roles in shaping the composition of the milk microbiota.
The microbiota composition in March, June and July contained higher species richness, indicating that raw milk collected in these months was exposed to a greater risk of bacterial contamination (Fig. 1A). Coinciding with the temperature, it was shown that bacterial richness was generally low in winter and high in summer. However, the clusters of bacterial communities could be classified into two categories. It is easy to understand that the situations in spring and autumn are similar, but the bacterial structures in winter and summer were unexpectedly grouped together despite their large differences in bacterial richness (Fig. 4).
The phylum-level structure of the bacterial communities detected in the present study was similar to those found in previous studies1. However, an interesting phenomenon was observed that Actinobacteria, Firmicutes and Proteobacteria had counter-balanced relationships. Although the richnesses of these bacteria fluctuated over the year, the total proportion remained at a certain level (80%~95%), indicating that these microorganisms competed with each other for ecological niches.
Furthermore, the common genera previously detected in raw milk, such as Pseudomonas22, Bacillus1, Lactococcus and Acinetobacter23, were also identified in our samples. Pseudomonas was abundant in winter (November-March, average temperature 8.8 °C), which might be due to its psychrotrophic features. Acinetobacter was abundant in summer (June-September, average temperature 25.7 °C), since most species are widely distributed in nature and can survive at a broad range of temperatures. There was no obvious trend observed in Lactococcus, the richness of which changed randomly in adjacent months. It is probable that Lactococcus in raw milk might be affected by other environmental factors than temperature and humidity.
According to the alignment at the genus level, a tiny core microbiota (two genera, Acinetobacter and Pseudomonas) was observed. Only Acinetobacter was found to be consistent with the previously reported milk core microbiota (29 genera) after storage and transportation in tanker trucks19, hinting that the raw milk microbiota is easily subject to change by environmental factors rather than the consistent bacterial composition in tanker trucks or silos. Four phyla were abundant in all cow milk samples, including Firmicutes, Proteobacteria, Actinobacteria and Bacteroidetes. The four phyla showed more or less significant differences between months, which could be interpreted according to temperature and humidity changes. Firmicutes and Proteobacteria were very consistently correlated with high temperature and high humidity, respectively, while Actinobacteria and Bacteroidetes were the opposite (Fig. 7A).
Considering genus-genus correlations (Fig. 7C, Table S14, p < 0.01), Lactobacillus and Bifidobacterium had a strong positive correlation, which might be because both of them are main sources of probiotics and lactic acid producing strains. In addition, there might be a synergistic fermentation effect between these two genera. Meanwhile, both genera had positive correlations with high temperature and humidity. Pseudomonas (one of the four most abundant genera) had strong positive correlations with Propionibacterium and Flavobacterium (Fig. 7B), which could be due to their psychrotrophic characteristics. Bacillus (one of the four most abundant genera) had strong positive correlations with Enhydrobacter, Enterococcus and Alkaliphilus. Neither Pseudomonas nor Bacillus was correlated with humidity, but Bacillus had a positive correlation with temperature, while Pseudomonas had a negative correlation with temperature (Fig. 7B). Therefore, the moderate negative correlation (coefficient = −0.55, p = 0.06) between Pseudomonas and Bacillus might be mediated by temperature. Figure 7C also revealed positive correlations among the four genera Niastella, Chitinophaga, Corynebacterium and Staphylococcus, but no obvious consistency of these genera with temperature or humidity was observed (Fig. 7B). This result meant that some other environmental factors might affect the composition of bacterial populations, such as microbiota from teat apex24, milking equipment25, air, water26, etc.
With respect to the roles of the microorganisms in milk1, four kinds of microorganisms were confirmed, including genera that facilitate dairy fermentation (F), cause spoilage (S), promote health (H) and cause disease (D). In this study, genera with three of these roles (F, S and H) were abundant (Table S13). The genera Salmonella, Campylobacter and Listeria, which could cause disease, were identified in only five raw milk samples, and their proportion of the total microbiota was very low (<1E-4), coinciding with the strict quality control of cow breed and raw milk production toward opportunistic pathogens.
Since the genera Pseudomonas and Bacillus are known to play important roles in causing spoilage (S) after long periods of cold incubation27, we could easily conclude that raw milk from April, May, August, September and October would be subject to Bacillus spoilage, while the risk from Pseudomonas existed throughout the year (Table S13). The genera Pseudomonas/Propionibacterium and Lactobacillus/Bifidobacterium had strong positive correlations with each other, which could facilitate dairy fermentation (F)1. According to these results, we speculated that Pseudomonas/Propionibacterium and Lactobacillus/Bifidobacterium were two pairs of genera with synergistic effects in raw milk.
The interactive relationships of milk bacteria with raw milk parameters were elucidated. According to the correlation analysis, many genera significantly influenced raw milk quality, such as milk protein content, milk fat content, SCC, TBC, thermophilic spores, and psychrotrophic bacteria number.
A high SCC in raw milk usually indicated mastitis occurrence and problems in milking or general hygiene. It was reported that Acinetobacter, Aeromonas and Brevundimonas were persistent members of genera occurring in tap water28. This information provides an explanation for the implication of tap water bacteria in raw milk contamination. Meanwhile, Aeromonas, Chryseobacterium and Stenotrophomonas are opportunistic pathogens causing a broad spectrum of illnesses29–31. Therefore, the presence of these pathogens from udder, teats or wound infections might lead to an abnormal increase in SCC. The positive correlation between these genera and SCC (Table S15) suggested that there might be important pathogens within these genera.
Excessive bacterial count usually results from internal or external contamination32. The genus Enterococcus is an important contamination source and has been known in the dairy industry for decades33. As a risk or a foreign intrusive flora, the positive correlation between Enterococcus and TBC is suggestive of fecal contamination, indicating poor hygiene during milk handling and processing. However, Propionibacterium was found to be negatively correlated to TBC, which meant that it was helpful for maintaining the quality of raw milk. Dairy Propionibacterium was reported to be a candidate non-lactic acid probiotic, which predominantly ferments lactate to short-chain fatty acid (SCFA)34. Acetate and propionate have been shown to modulate immunity, reduce inflammation and protect against pathogens, and more importantly, to create an acid environment that inhibits the growth of other bacteria. This might be an explanation for its inhibitory effect.
Pseudoalteromonas was another genus negatively correlated to TBC. This genus of bacteria was reported to produce a broad range of anti-bacterial products, which have been found to aid them in colonization35. This might be the reason that it was able to limit TBC. It was suggested that opportunistic pathogens are inhibited by a cooperative network of bacterial communities, which are important in the maintenance of milk ecology. These findings led us to hypothesize that distinct interactive relationships existed in the milk microbiota.
Some genera were correlated with milk protein or milk fat, which are important nutritional parameters. Bacteroides and Streptococcus exist widely in the environment. In the raw milk production process, the lipolytic and proteolytic activities of these genera greatly contributed to the spoilage of raw milk.
Another important group of bacteria related to food safety and spoilage are the psychrotrophic bacteria, which could be detected in almost all natural environments. Due to their psychrotrophic nature and adaptability to different substrates and growth conditions, they can grow during refrigeration. It was estimated that psychrotrophic bacteria could cause a 10% loss of milk fats and proteins36. Because of their effects on milk quality, it is important that we develop sensitive and efficient tools to monitor the presence of these psychrotrophic bacteria. To our surprise, Pseudomonas was not found to be the major microorganism responsible for the degradation of milk properties, although it was one of the main genera identified in the raw milk microbiota. This might be another piece of evidence for the diversity of bacterial compositions in raw milk.
More genera were positively correlated to heat-resisting spore number, indicating that these genera themselves might be the main contributors of heat-resisting spores in raw milk, such as Methylonatrum, Cloacibacillus, and Novispirillum. In addition, the appearance of Cloacibacterium might indicate contamination with raw sewage.
Based on the microbial function predictions, some pathways differed between months, and some differences existed in adjacent months, such as March and April (Table S18), June and July (Table S21). On the other hand, some consecutive months showed no differences due to similar weather conditions, e.g., consecutive months November-March (average temperature 8.8 °C and humidity 71.8%) and consecutive months August-October (average temperature 23.9 °C and humidity 73.3%). A comparison between these two groups of consecutive months revealed that the OTU abundances involved in four pathways were highly increased in the high-temperature months (August-October), reflecting a more active metabolism in the microbiota during summer.
In most cases, management practices have a significant influence on milk quality. The importance of cow and stall hygiene and equipment hygiene for the quality of raw milk should be highlighted. More preventive actions related to environmental factors should be implemented to reduce the incidence of contamination.
Materials and Methods
Milk sources and sampling
In this study, 112 raw milk samples were aseptically collected from 10 different farms in Shanghai, China. All methods were carried out in accordance with the approved guidelines and regulations (GB 4789.1–2010, National Health and Family Planning Commission of the People’s Republic of China). Ten-milliliter milk samples were collected from the storage tanks of each farm and kept in sterile plastic tubes without preservative (Corning Life Sciences, USA). Then, the samples were immediately refrigerated at 4 °C, transported to the laboratory on ice, and frozen at −80 °C prior to DNA extraction. Samples were collected every month between July 2015 and June 2016.
Genomic DNA extraction, PCR and 16 S rRNA gene sequencing
Bacterial genomic DNA was extracted by a Milk Bacterial DNA Isolation Kit (Norgen, Thorold, ON, Canada). The V3–4 region of the 16 S rRNA genes was amplified with primers 338 F and 806R37 with a random 8-bp bar code separately on the 5′ ends of 338 F and 806 R. PCR amplification was performed using TransStart FastPfu DNA Polymerase (TransGen, Beijing, China). The thermocycling steps were as follows: 5 min at 95 °C; 20 cycles of 45 sec at 95 °C, 30 sec at 55 °C, and 30 sec at 72 °C; and a final extension step for 10 min at 72 °C. Each sample was PCR amplified in triplicate. Then, the PCR products were purified with an AxyPrep DNA Gel Extraction kit (Axygen, Hangzhou, China) and pooled in equivalent amounts. The 16 S rRNA gene amplicons were sequenced on an Illumina MiSeq instrument38 at Shanghai Yuanxu Biotechnology Co., Ltd (China).
16 S rRNA gene sequence bioinformatics and statistical analysis
16 S rRNA gene bioinformatic analysis
The raw data were separated by barcode using the MiSeq system program, and each sample was assigned paired FASTQ files. The paired FASTQ files were assembled using Mothur39 (version 1.37.0) with the command “make.contigs”. After removing sequences with ambiguous bases and shorter sequences (<350 bp), sequences were also excluded if they were identified as chimeric sequences or contaminants. The high-quality DNA sequences were grouped into OTUs with 97% by comparing them to the SILVA reference database (V119)40. Data normalization was carried out with the command “sub.sample” based on the minimum sample size (7,780). Community richness and diversity analysis (Shannon, ACE, Chao1 and Good’s coverage) were performed using the command “single.summary”. Taxonomy was assigned using the online software RDP classifier41 at an 80% threshold based on the Ribosomal Database Project42. The functional predictions based on the 16 S rRNA genes were performed using PICRUSt software.
16 S rRNA gene statistical analysis
A t-test with 2 tails was used to determine whether the means of the Shannon diversity indices between June and Dec were significantly different using an R package, and p < 0.01 was considered a significant difference.
Abundance differences in genera between months were analyzed by Metastats43. Differences with q-values <1E-5 and fold changes >2 between months were considered significant, and the detailed results are listed in Tables S2–S12. The complex relations among the different genera were drawn using Circos software44 based on the major genera (percentage >1% in at least one month) and the phylum percentage. The coefficients of the relationships among genus abundances and between genera and milk quality parameters were analyzed using the non-parametric Spearman rank correlation with an R package. The p-value and adjusted p-value (q-value) were both calculated for finding relationships with a multiple test procedure45. Because most q-values were over 0.05 (some even 0.5), we chose p-value (p < 0.01) as a less stringent cutoff. The software Genesis (version 1.7.7)46 was used to draw heatmaps using the taxonomy information (genus information) of the representative OTU sequences, and hierarchical clustering was calculated for the monthly distributions. More differences between major genera are listed in Tables S2–S12. An RDA analysis based on the value of “axis lengths” (<3) was performed using the Vegan package in R. The identification of species was determined through BlastN based on the SILVA database and the NCBI NT database, and the parameters were set as identity ≥99% and alignment ≥97% based on a previous study38. To analyze the functional differences between months, a two-sided Welch’s test was performed using STAMP software, and the resultant P-values were multiply adjusted by the Benjamini-Hochberg procedure (q < 0.05); then, ratios of proportions higher than 2 were retained.
Accession numbers
The sequence data have been submitted to the GenBank Sequence Read Archive (Accession number PRJNA389757).
Electronic supplementary material
Acknowledgements
We thank Shanghai Denuo Product Testing Service Co., Ltd. for providing the raw milk samples, and Shanghai Meteorological Bureau for providing weather data necessary for this study. This work was supported by National Science and Technology Support Program of China during the 12th Five-Year Plan Period (Grant No. 2012BAD12B08) and Shanghai Science and Technology Committee Engineering Center Capacity Improvement Project (Grant No. 16DZ2280600).
Author Contributions
C.Y., J.R. and Z.L. conceived and designed the experiments. N.L., Y.W. and W.C. performed the experiments. N.L., Y.W., C.Y. and H.Z. analyzed the data. N.L. and Y.W. contributed to the writing of the manuscript. All authors reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Nan Li, Yuezhu Wang and Chunping You contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20862-8.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Huajun Zheng, Email: zhenghj@chgc.sh.cn.
Zhenmin Liu, Email: liuzhenmin@brightdairy.com.
References
- 1.Quigley L, et al. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013;37:664–698. doi: 10.1111/1574-6976.12030. [DOI] [PubMed] [Google Scholar]
- 2.LeJeune JT, Rajala-Schultz PJ. Unpasteurized Milk: A Continued Public Health Threat. Food Safety. 2009;48:93–100. doi: 10.1086/595007. [DOI] [PubMed] [Google Scholar]
- 3.Buehner KP, Anand S, Garcia A. Prevalence of thermoduric bacteria and spores on 10 Midwest dairy farms. J. Dairy Sci. 2014;97:6777–6784. doi: 10.3168/jds.2014-8342. [DOI] [PubMed] [Google Scholar]
- 4.Gopal N, et al. The prevalence and control of Bacillus and related spore-forming bacteria in the dairy industry. Front. Microbiol. 2015;6:1418. doi: 10.3389/fmicb.2015.01418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quigley L, et al. Molecular approaches to analysing the microbial composition of raw milk and raw milk cheese. Int. J. Food Microbiol. 2011;150:81–94. doi: 10.1016/j.ijfoodmicro.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Vacheyrou M, et al. Cultivable microbial communities in raw cow milk and potential transfers from stables of sixteen French farms. Int. J. Food Microbiol. 2011;146:253–262. doi: 10.1016/j.ijfoodmicro.2011.02.033. [DOI] [PubMed] [Google Scholar]
- 7.Schornsteiner E, Mann E, Bereuter O, Wagner M, Schmitz-Esser S. Cultivation-independent analysis of microbial communities on Austrian raw milk hard cheese rinds. Int. J. Food Microbiol. 2014;180:88–97. doi: 10.1016/j.ijfoodmicro.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Julien M, Dion P, Lafreniere C, Antoun H, Drouin P. Sources of Clostridia in raw milk on farms. Appl. Environ. Microbiol. 2008;74:6348–6357. doi: 10.1128/AEM.00913-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huck JR, Hammond BH, Murphy SC, Woodcock NH, Boor KJ. Tracking spore-forming bacterial contaminants in fluid milk-processing systems. J. Dairy Sci. 2007;90:4872–4883. doi: 10.3168/jds.2007-0196. [DOI] [PubMed] [Google Scholar]
- 10.Raats D, Offek M, Minz D, Halpern M. Molecular analysis of bacterial communities in raw cow milk and the impact of refrigeration on its structure and dynamics. Food Microbiol. 2011;28:465–471. doi: 10.1016/j.fm.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 11.McAuley CM, Gobius KS, Britz ML, Craven HM. Heat resistance of thermoduric enterococci isolated from milk. Int. J. Food Microbiol. 2012;154:162–168. doi: 10.1016/j.ijfoodmicro.2011.12.033. [DOI] [PubMed] [Google Scholar]
- 12.Hou QC, et al. Evaluation of bacterial contamination in raw milk, ultra-high temperature milk and infant formula using single molecule, real-time sequencing technology. J. Dairy Sci. 2015;98:1–9. doi: 10.3168/jds.2015-9886. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F, et al. Bacterial diversity in goat milk from the Guanzhong area of China. J. Dairy Sci. 2017;100:1–13. doi: 10.3168/jds.2016-11302. [DOI] [PubMed] [Google Scholar]
- 14.Li L, et al. Characterization of the indigenous microflora in raw and pasteurized buffalo milk during storage at refrigeration temperature by high-throughput sequencing. J. Dairy Sci. 2016;99:1–9. doi: 10.3168/jds.2015-9445. [DOI] [PubMed] [Google Scholar]
- 15.Zhong Z, et al. Bacterial microbiota compositions of naturally fermented milk are shaped by both geographic origin and sample type. J. Dairy Sci. 2016;99:1–10. doi: 10.3168/jds.2015-10825. [DOI] [PubMed] [Google Scholar]
- 16.Quigley L, et al. High-throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl. Environ. Microbiol. 2012;78:5717–5723. doi: 10.1128/AEM.00918-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakwinska O, et al. Microbiota in breast milk of chinese lactating mothers. Plos One. 2016;11:e0160856. doi: 10.1371/journal.pone.0160856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillespie BE, et al. Short communication: Evaluation of bulk tank milk microbiological quality of nine dairy farms in Tennessee. J. Dairy Sci. 2012;95:4275–4279. doi: 10.3168/jds.2011-4881. [DOI] [PubMed] [Google Scholar]
- 19.Kable ME, et al. The core and seasonal microbiota of raw bovine milk in tanker trucks and the impact of transfer to a milk processing facility. mBio. 2016;7:e00836–16. doi: 10.1128/mBio.00836-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonizzi I, Buffoni JN, Feligini M, Enne G. Investigating the relationship between raw milk bacterial composition, as described by intergenic transcribed spacer-PCR fingerprinting, and pasture altitude. J. Appl. Microbiol. 2009;107:1319–1329. doi: 10.1111/j.1365-2672.2009.04311.x. [DOI] [PubMed] [Google Scholar]
- 21.Taylor R. Interpretation of the correlation coefficient: A Basic Review. J. Diagn. Med. Sonogr. 1990;6:35–39. doi: 10.1177/875647939000600106. [DOI] [Google Scholar]
- 22.von Neubeck M, et al. Biodiversity of refrigerated raw milk microbiota and their enzymatic spoilage potential. Int. J. Food Microbiol. 2015;211:57–65. doi: 10.1016/j.ijfoodmicro.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Gurung M, et al. Prevalence and antimicrobial susceptibility of Acinetobacter from raw bulk tank milk in Korea. J. Dairy Sci. 2013;96:1997–2002. doi: 10.3168/jds.2012-5965. [DOI] [PubMed] [Google Scholar]
- 24.Verdier-Metz I, et al. Cow teat skin, a potential source of diverse microbial populations for cheese production. Appl. Environ. Microbiol. 2012;78:326–333. doi: 10.1128/AEM.06229-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostrov I, Harel A, Bernstein S, Steinberg D, Shemesh M. Development of a method to determine the effectiveness of cleaning agents in removal of biofilm derived spores in milking system. Front. Microbiol. 2016;7:1498. doi: 10.3389/fmicb.2016.01498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michel AL, et al. Longevity of Mycobacterium bovis in raw and traditional souring milk as a function of storage temperature and dose. PloS one. 2015;10:e0129926. doi: 10.1371/journal.pone.0129926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Champagne CP, Laing RR, Roy D, Mafu AA, Griffiths MW. Psychrotrophs in dairy products: their effects and their control. Crit. Rev. Food Sci. Nutr. 1994;34:1–30. doi: 10.1080/10408399409527648. [DOI] [PubMed] [Google Scholar]
- 28.Vaz-Moreira I, Nunes OC, Manaia CM. Ubiquitous and persistent Proteobacteria and other Gram-negative bacteria in drinking water. Sci. Total Environ. 2017;586:1141–1149. doi: 10.1016/j.scitotenv.2017.02.104. [DOI] [PubMed] [Google Scholar]
- 29.Stratev D, Odeyemi OA. 2016. Antimicrobial resistance of Aeromonas hydrophila isolated from different food sources: A mini-review. J. Infect. Public Health. 2016;9:535–544. doi: 10.1016/j.jiph.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 30.Hantsis-Zacharov E, Shakéd T, Senderovich Y, Halpern M. Chryseobacterium oranimense sp. nov., a psychrotolerant, proteolytic and lipolytic bacterium isolated from raw cow’s milk. Int. J. Syst. Evol. Microbiol. 2008;58:2635–2639. doi: 10.1099/ijs.0.65819-0. [DOI] [PubMed] [Google Scholar]
- 31.Ohnishi M, et al. Antimicrobial susceptibility and genetic relatedness of bovine Stenotrophomonas maltophilia isolates from a mastitis outbreak. Lett. Appl. Microbiol. 2012;54:572–576. doi: 10.1111/j.1472-765X.2012.03246.x. [DOI] [PubMed] [Google Scholar]
- 32.Lan XY, et al. Short communication: Microbiological quality of raw cow milk and its association with herd management practices in Northern China. J. Dairy Sci. 2017;100:4294–4299. doi: 10.3168/jds.2016-11631. [DOI] [PubMed] [Google Scholar]
- 33.Rodrigues MX, Lima SF, Canniatti-Brazaca SG, Bicalho RC. The microbiome of bulk tank milk: Characterization and associations with somatic cell count and bacterial count. J. Dairy Sci. 2017;100:2536–2552. doi: 10.3168/jds.2016-11540. [DOI] [PubMed] [Google Scholar]
- 34.Kwon G, Lee J, Lim YH. Dairy Propionibacterium extends the mean lifespan of Caenorhabditis elegans via activation of the innate immune system. Sci. Rep. 2016;6:31713. doi: 10.1038/srep31713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmstrom C, et al. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active cellular agents. FEMS Microbiol. Ecol. 1999;30:285–293. doi: 10.1016/S0168-6496(99)00063-X. [DOI] [PubMed] [Google Scholar]
- 36.Hantsis-Zacharov E, Halpern M. 2007. Culturable psychrotrophic bacterial communities in raw milk and their proteolytic and lipolytic traits. Appl. Environ. Microbiol. 2007;73:7162–7168. doi: 10.1128/AEM.00866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome biology. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng H, et al. Altered gut microbiota composition associated with Eczema in Infants. PloS one. 2016;11:e0166026. doi: 10.1371/journal.pone.0166026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PloS one. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cole JR, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. Plos Comput. Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krzywinski M, et al. Circos: an Information Aesthetic for Comparative Genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–230. [Google Scholar]
- 46.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.