Abstract
The large expansion of GGGGCC (G4C2) repeats of the C9orf72 gene have been found to lead to the pathogenesis of devastating neurological diseases, amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). The structural polymorphisms of C9orf72 HRE DNA and RNA may cause aberrant transcription and contribute to the development of ALS and FTD. Here we showed that the two-repeat G4C2 DNA, d(G4C2)2, simultaneously formed parallel and antiparallel G-quadruplex conformations in the potassium solution. We separated different folds of d(G4C2)2 by anion exchange chromatography, followed with characterizations by circular dichroism and nuclear magnetic resonance spectroscopy. The parallel d(G4C2)2 G-quadruplex folded as a symmetric tetramer, while the antiparallel d(G4C2)2 adopted the topology of an asymmetric dimer. These folds are distinct from the antiparallel chair-type conformation we previously identified for the d(G4C2)4 G-quadruplex. Our findings have demonstrated the conformational heterogeneity of the C9orf72 HRE DNA, and provided new insights into the d(G4C2)n folding. Meanwhile, the purified d(G4C2)2 G-quadruplex samples are suitable for further three-dimensional structure characterizations, which are required for the structure-based design of small molecules targeting ALS and FTD related C9orf72 HRE.
Introduction
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are severe neurological diseases1–3. The abnormal expansion of a GGGGCC (G4C2) hexanucleotide repeat (HRE) in a noncoding region of the C9orf72 gene has been found to be a single genetic cause of ALS and FTD4–6. Fewer than 25 G4C2 repeats are present in the healthy control population7, whereas more than 700 G4C2 repeats exist in the ALS and FTD patients4,5. In patient cells, the G4C2 HRE transcripts accumulate as nuclear RNA foci, which may sequester and inactivate RNA binding proteins8,9. The transcribed G4C2 expansion repeats can also undergo non-ATG initiated translation, which result in toxic dipeptide accumulation10–12. Despite these findings, the detailed disease mechanism remains to be elucidated. Recent studies have demonstrated that the structure polymorphisms of both C9orf72 HRE DNA and RNA may contribute to the pathogenesis of ALS and FTD diseases13. They can form complex structures, including DNA or RNA G-quadruplexes and DNA∙RNA hybrids, which may cause transcriptional abortion leading to the loss of full-length RNA transcripts and accumulation of abortive RNA transcripts9,14,15.
The G-quadruplex is a stable four-stranded structure formed by guanosine-rich DNA and RNA sequences. Hoogsteen hydrogen bonds connect four guanine bases to form a square planar structure called guanine tetrad. Two or more guanine tetrad layers stack to form a G-quadruplex and monovalent cations can stabilize the G-quadruplex formation. Various cations affect the topology and stability of G-quadruplexes differently16. G-quadruplexes can adopt parallel, antiparallel, or mixed topology, depending on the orientation of the strand loops17,18. In recent years, accumulating evidence point to the existence of G-quadruplex structures under physiological conditions, which may have critical functional roles in many cellular processes. G-quadruplex and other peptides, like β-amyloid, are highly attractive targets for therapeutic interventions for neurological disorders15,19–21. G-quadruplex-based assays and probes are also used to detect exonuclease activity or gene deletion22,23.
Several studies have investigated C9orf72 HRE DNA G-quadruplexes9,24–27. Circular dichroism (CD) spectroscopy showed that the C9orf72 HRE DNA with different lengths adopted distinct G-quadruplex topologies. The d(G4C2)G4 formed parallel G-quadruplex; d(G4C2)4 formed antiparallel G-quaduplex; d(G4C2)2, d(G4C2)3 and d(G4C2)5 formed mixed G-quadruplexes with mixed topologies9,25. Using NMR approaches, we previously solved the topology of d(G4C2)4, which exhibited a chair-type G-quadruplex with a four-layer antiparallel G-tetra core and three edgewise loops25. In another study, the solution structure of a modified G4C2 DNA, d[(G4C2)3GGBrGG], was determined to adopted an antiparallel topology with four G-quartets and three edgewise C-C loops26.
Previous findings have suggested that the parallel topology is adopted by most C9orf72 (G4C2)n DNAs9,24,25. However, to date, no parallel topology of the C9orf72 HRE DNA has been determined, largely due to the difficulty imposed by the heterogeneous conformations of G-rich oligonucleotides for structural determination. Commonly, the adjustment of length of oligonucleotide repeats, or mutation and modification of specific nucleotides could be employed to stabilize a particular structural fold of G-quadruplexes. The experimental conditions, including pH, temperature and type of cations in the buffers, could also profoundly impact the G-quadruplex folding.
To obtain a C9orf72 HRE DNA sample with the parallel topology and of sufficient quality for structural determination, we carefully examined our previous data, including CD and NMR spectra of G4C2 repeats of different lengths25. The 1D 1H NMR spectra suggested that d(G4C2)2 was composed of less heterogeneous G-quadruplex conformations compared to other tested d(G4C2)n DNAs. Therefore, in this study, we employed d(G4C2)2 for further purifications, and obtained several distinct fractions by anion exchange chromatography. The native polyacrylamide gel electrophoresis (PAGE) and CD spectroscopy were performed to investigate all the purified fractions, which demonstrated that homogeneous samples each composed of a distinct parallel or antiparallel G-quadruplex fold were obtained. Using NMR approaches, we further characterized the parallel and antiparallel G-quadruplex conformations. The parallel d(G4C2)2 G-quadruplex was found to fold as a symmetric tetramer, while the antiparallel d(G4C2)2 G-quadruplex adopted the topology of an asymmetric dimer.
Results
d(G4C2)2 formed mixed parallel and antiparallel topologies
During the preliminary screening of d(G4C2)n samples for structural studies, the CD spectra of d(G4C2)2, d(G4C2)3 and d(G4C2)5 showed a negative peak at 240 nm and positive peaks at 260 and 290 nm25. As previous reports presented, characteristic peaks for parallel quadruplexes include a positive maximum at ∼260 nm and a negative minimum at ∼240 nm. Meanwhile, the positive maximum for an antiparallel quadruplex is typically at around 290 nm28. Thus, the characteristic peaks of both parallel and antiparallel G-quadruplexes existed in the above CD spectra, indicating mixed topologies in these samples. The NMR spectroscopy was also employed to investigate their folds. The 1D 1H spectra of d(G4C2)n except d(G4C2)2 showed broaden peaks in the imino region, suggesting the formation of mixtures with multiple G-quadruplex conformations25. In contrast, the 1D 1H spectrum of d(G4C2)2 showed sharp peaks with narrow line widths at 10–12 ppm (Fig. 1a), indicating that its conformation was less heterogeneous. Therefore, we focused on d(G4C2)2 in further efforts to obtain a G4C2 DNA G-quadruplex sample with the parallel topology.
Figure 1.
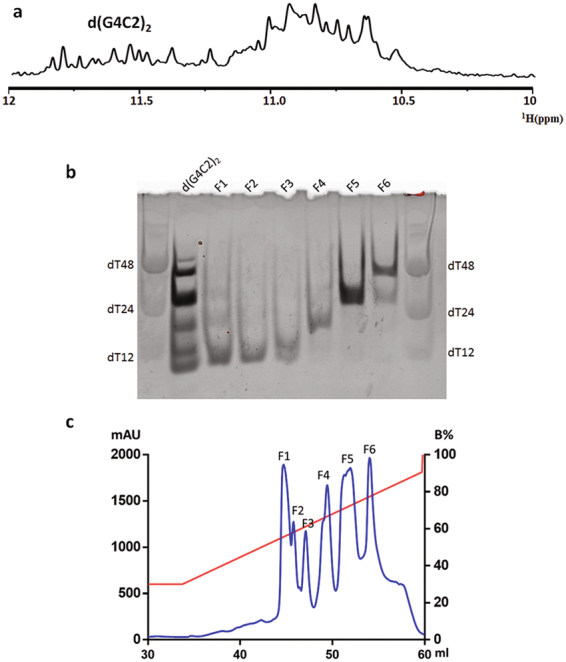
Anion exchange chromatography purification of d(G4C2)2. (a) The imino region of 1D 1H spectrum of d(G4C2)2. (b) Gel electrophoresis of d(G4C2)2 and anion exchange chromatography fractions monitored by the staining method. (c) Elution profile of d(G4C2)2 from Mono-Q column.
To analyze the molecular size of d(G4C2)2, the native PAGE was performed and DNA oligonucleotides dT12, dT24 and dT48 were employed as size indicators (Fig. 1b; Supplementary Fig. S1). The d(G4C2)2 DNA migrated as multiple bands, in consistency with a mixture of topologies observed in CD spectroscopy.
Purification of d(G4C2)2 by anion exchange chromatography
To separate different conformations of d(G4C2)2 in the original sample, anion exchange chromatography was performed using a high resolution Mono-Q column. We generated a potassium chloride gradient to elute DNA fractions from the column using buffer A (20 mM Tris, 50 mM KCl, pH7) and buffer B (20 mM Tris, 1 M KCl, pH7). Twenty-five column volumes of potassium chloride concentration from 30% B to 90% B produced six distinct peaks during the elution process (Fig. 1c). Native PAGE demonstrated that each fraction contained particular components of d(G4C2)2 G-quadruplex folds (Fig. 1b). The d(G4C2)2 component with lower stoichiometry was eluted at lower salt concentrations. The elution peaks of fractions 1 and 2 (F1 and F2) were closely adjacent and each contained a single band that migrated faster than dT12. Since the monomeric d(G4C2)2 could not form a G-quadruplex, the result suggested that F1 and F2 were consisted of homogeneous dimeric G-quadruplexes of similar conformations. Several bands were observed in F3 or F4, which migrated between dT12 and dT24, indicating the composition of different low-order G-quadruplexes. F5 migrated as a single band near dT24, which was likely a homogeneous tetrameric G-quadruplex. In F6, several bands migrated slower than dT24, indicating the composition of high-order structural forms. To further validate the oligomeric status, we compared the electrophoretic mobility of F1 and F5 with monomeric 23-nt human telomere h-telo d[TAGGG(TTAGGG)3]29 and dimeric 16-nt 93del d[GGGGTGGGAGGAGGGT]30 (Supplementary Fig. S2). F1 migrated similarly as h-telo, supporting the dimeric form of d(G4C2)2. F5 migrated slower than 93del, which agreed with the tetrameric composition of d(G4C2)2.
CD spectroscopy characterizations of purified d(G4C2)2 fractions
To investigate the conformation of d(G4C2)2 in different fractions, CD spectroscopy was employed to analyze the topology of each fraction (Fig. 2). In addition to characteristic peaks for parallel and antiparallel quadruplexes as described above, the 290-nm peak indicates that two adjacent G-tetrads have opposite hydrogen-bond directionalities, whereas the 260-nm peak is characteristic of a G-quadruplex in which all the G-tetrads have the same polarity31. F1 and F2 fractions displayed two positive maxima at around 260 and 290 nm, and a negative minimum at around 240 nm. Therefore, based on their CD spectra, F1 and F2 adopted a mixed parallel/antiparallel topology with G-tetrad neighbors of mixed relative polarities.
Figure 2.
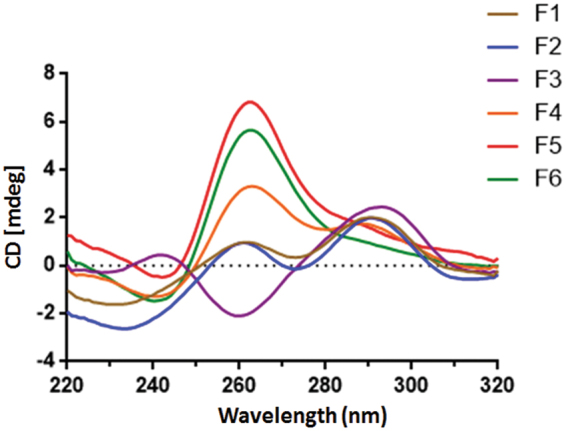
CD spectra of anion exchange chromatography fractions of d(G4C2)2. The concentration of each fraction was 20 μM.
In contrast, F3 displayed a single positive absorption peak and a negative peak at 290 nm and 260 nm, respectively, suggesting the formation of a predominantly antiparallel G-quadruplex fold. The spectrum of F4 exhibited two positive peaks at 260 and 290 nm, which was probably resulted from its mixed conformations as observed in the native PAGE. The CD spectra of F5 and F6 displayed a positive peak at 260 nm and a negative peak at 240 nm, respectively, indicating parallel G-quadruplex folds. These results also demonstrated that the lower stoichiometry of d(G4C2)2 was prone to adopt antiparallel topology and the higher stoichiometry of d(G4C2)2 favored parallel topologies.
Stability of d(G4C2)2 fractions investigated by CD melting experiments
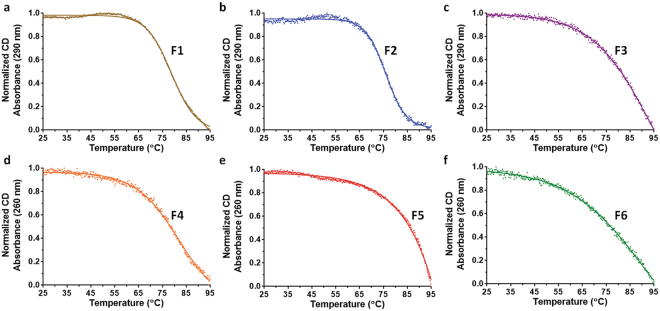
The thermal stability of the purified d(G4C2)2 fractions were examined by CD melting experiments, which were performed with a temperature range of 25 °C to 95 °C at 1 °C/min. The CD absorbance was measured at a single wavelength (290 nm for antiparallel G-quadruplexes F1, F2 and F3, and 260 nm for parallel G-quadruplexes F4, F5 and F6). The normalized CD absorbance was fitted by the Boltzmann sigmoid equation (Fig. 3). The melting temperatures of F1 and F2 were 78.79 °C and 75.96 °C, respectively. Both their melting curves exhibited apparent transitions, suggesting the stable G-quadruplex structure formation by these DNA samples. In contrast, the melting profiles of F3, F4, F5 and F6 showed no obvious transitions, indicating that these DNA samples formed less stable G-quadruplex structures or adopted multiple conformations. As such, the d(G4C2)2 G-quadruplexes in F1 and F2 adopted the most stable conformations among the purified samples.
Figure 3.
CD melting curves of anion exchange chromatography fractions of d(G4C2)2. The melting experiments were performed with a temperature range of 25 °C to 95 °C at 1 °C/min. The CD absorbance was measured at a single wavelength (290 nm for F1, F2 and F3, and 260 nm for F4, F5 and F6). Data were fit by the Boltzmann sigmoid equation (Prism).
Detailed G-quadruplex topologies revealed by NMR spectroscopy
To obtain more structural insights into d(G4C2)2 G-quadruplex conformations, 1D 1H NMR spectra were examined for the unpurified d(G4C2)2 and the purified d(G4C2)2 fractions (Fig. 4). The imino region of 1H NMR spectrum of unpurified d(G4C2)2 exhibited signals between 10 and 12 ppm, corresponding to imino protons of guanines involved in G-tetrads. Interestingly, the imino region of the antiparallel-dominant G-quadruplex fractions (F1 and F2) showed peaks between 11 and 12 ppm, while that of the parallel-dominant G-quadruplex fractions (F5 and F6) lay between 10.5 and 11 ppm. Thus, the location of the imino peaks may be indicative of the topology of d(G4C2)n G-quadruplexes. The 1H NMR spectra of F3 and F4 contained peaks in both antiparallel and parallel regions, indicating the formation of heterogeneous mixtures. The spectrum of F6 showed broaden peaks, which was consistent with the high-order structures determined by the native PAGE experiment.
Figure 4.
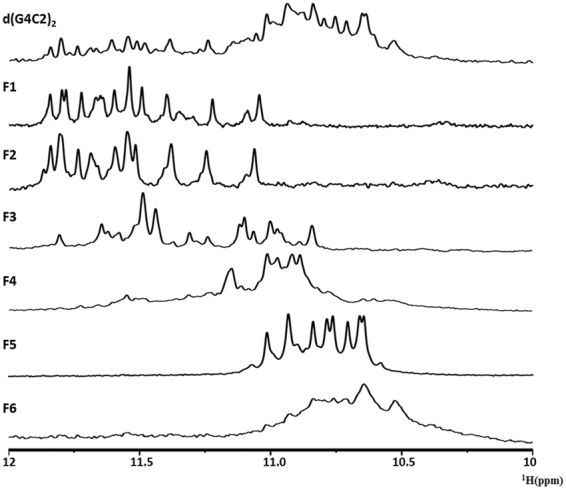
1D 1H spectra showed the imino region of d(G4C2)2 and anion exchange chromatography fractions. The spectra were recorded in 90% H2O, 10% D2O at 25 °C.
In 1D 1H NMR spectra of F1 and F2, at least 12 imino proton resonances were observed at 10–12 ppm with narrow line widths, indicating the formation of an asymmetric dimeric G-quadruplex fold with three or four G-tetrads. The hydrogen-deuterium exchange (HDX) experiment showed that many imino protons were exposed and subjected to exchange with solvent after 30 min in D2O (Supplementary Fig. S3a,b), in consistency with a dimeric G-quadruplex conformation. Meanwhile, the 1D 1H NMR spectrum of F5 exhibited 8 well-resolved peaks at 10–12 ppm, indicating a symmetric tetrameric conformation with eight G-tetrads. Around six peaks were well-protected from the exchange with solvent in the HDX experiment, which was consistent with the G-quadruplex fold containing eight tetrad planes (Supplementary Fig. S3c).
No obvious signals were detected in the Watson-Crick base pairing region, implicating that the cytosines were located in the loop region of the d(G4C2)2 conformation. The detailed guanine compositions and loop orientations of the asymmetric dimeric and symmetric tetrameric G-quadruplexes remain unclear and need further experiments to resolve.
Purified d(G4C2)2 G-quadruplex fractions are suitable for investigations by crystallography
To evaluate the sample quality of these anion exchange fractions for further structure determination, we employed the X-ray crystallography approach. We screened various crystallization conditions for fractions F1 and F5. In initial trials we obtained crystals of fraction F5 (Supplementary Fig. S4), and a diffraction data set was collected to 1.8 Å resolution. This suggests that the purified samples are suitable for high resolution structural investigations. Since this set of diffraction data is still not sufficient to determine the F5 structure by the molecular replacement method, we will optimize crystal conditions or use bases with heavy-atom substitutions to solve the phase problem in the future study.
Discussion
The G-quadruplexes formed by the C9orf72 (G4C2)n DNA or RNA have been found to lead to the pathogenesis of devastating neurological diseases ALS/FTD. The detailed structure information of these G-quadruplexes is critical for understanding the related disease etiology. Previous studies have demonstrated that the C9orf72 (G4C2)n DNA formed parallel and/or antiparallel topologies9,24,25. The d(G4C2)4 formed 100% antiparallel topology, while all the other tested d(G4C2)n contained the parallel G-quadruplex topology. Despite topologies resolved for the antiparallel G-quadruplex folds of d(G4C2)425,26, the knowledge of the parallel G-quadruplex conformation adopted by the (G4C2)n HRE DNA was limited to indications from CD spectroscopy9,24,25.
The structure polymorphisms of G-rich oligonucleotides create the obstacle for obtaining homogenous parallel G-quadruplex folds for the 3-dimensional structure determination. Some modification tools, such as the 8-bromo guanine substitution favoring syn conformation, have been employed to improve the homogeneity of the samples. However, using these approaches, many mutational combinations need to be tested to find the optimal one, which is labor-intensive, tedious and time consuming. In some cases, no guanine with syn conformation in the G-quadruplex is available for chemical modifications. Therefore, other approaches need to be considered for purification of homogeneous G-quadruplex samples for structural determination.
Although anion exchange chromatography has not been frequently used in the structure study of G-quadruplexes, it represents one of the most common methods for the purification of oligonucleotide samples. It is widely used to purify DNA or RNA before crystallization trials32. Here we demonstrated that using the method of anion exchange chromatography, we could isolate homogeneous d(G4C2)2 samples with either the parallel or antiparallel G-quadruplex conformation. Therefore, the approach of anion exchange chromatography could be routinely adopted to purify distinct conformations of G-quadruplexes for structural characterizations.
In summary, we presented the first report of the purification and characterization of the parallel topology adopted by the G4C2 repeat DNA. Using the method of anion exchange chromatography, we isolated homogeneous d(G4C2)2 samples with either parallel or antiparallel G-quadruplex conformation. Further CD and NMR spectroscopy characterizations suggested that the parallel d(G4C2)2 G-quadruplex formed a symmetric tetramer while the antiparallel G-quadruplex folded into an asymmetric dimer. Moreover, the purified d(G4C2)2 G-quadruplex fractions provided suitable samples for further 3-dimensional structure determinations, which may build the necessary structural basis for designing small molecules targeting ALS and FTD related C9orf72 HRE.
Methods
Sample preparation
DNA oligonucleotides were chemically synthesized by IDT. The synthesized DNA was solved at 100 μM concentration in 20 mM potassium phosphate (pH 7.0) and 70 mM KCl. The solved DNA was heated to 95 °C for 15 min, and slowly cooled to room temperature. The NMR sample was concentrated with the use of a Centricon 3kD ultrafiltration column (Millipore, MA).
Circular dichroism spectroscopy
Circular dichroism spectra were recorded with 400 μl SNA samples at room temperature on an Applied Photophysics Chirascan CD spectrometer using a 1 mm path length quartz cell. DNA concentration was 20 μM. The DNA samples were prepared in the buffer containing 20 mM potassium phosphate (pH 7.0) and 70 mM KCl.
CD melting
The CD melting experiments were performed with a temperature range from 25 °C to 95 °C at 1 °C/min. The CD absorbance were measured at a single wavelength and then normalized by using the equation (Abst − min)/(max − min), in which Abst is the absorbance at a given temperature, max is the maximum absorbance at 260 nm (parallel G-quadruplexes) and at 290 nm (antiparallel G-quadruplexes), and min is the minimum value. Data were fit by the Boltzmann sigmoid equation (Prism).
Polyacrylamide gel electrophoresis (PAGE)
The 18% TBE polyacrylamide gel (acrylamide:bis-acrylamide 29:1) was used to run non-denaturing PAGE. 20 mM KCl was supplemented in both gel and running buffer (0.5× TBE). The annealed samples were prepared at a strand concentration of 100 μM. For the anion exchange chromatography fractions, 5 μl of each fraction was combined and loaded in the gel. Red-safe staining was employed to reveal gel bands. The images were acquired by the Molecular Imager Gel Doc XR system (Bio-Rad).
NMR spectroscopy
The 1D 1H NMR experiments were performed on 750 MHz and 800 MHz Varian spectrometers. Watergate or Jump-and-Return water suppression techniques were employed for samples in water solution. NMRPipe was used to process NMR spetra. For the HDX exchange experiments, the oligonucleotides samples were annealed and lyophilized. The lyophilized oligonucleotides were resuspended in 99% D2O before NMR measurements.
DNA crystallization
DNA crystals of fraction F1 and F5 were grown by vapor diffusion from hanging drops at 289 K using Natrix HT (Hampton research). The diffraction data were collected on beamline BL-17U of the Shanghai Synchrotron Radiation Facility (SSRF)33.
Electronic supplementary material
Acknowledgements
We thank the staff at the SSRF for help with collection of diffraction data. We thank Dr. Xin Dai and Prof. Zhihong Guo at Department of chemistry at HKUST for help with the CD melting experiment. This work described here was supported by the Research Grants Council of the Hong Kong Special Administrative Region, China to B.Z. (Project No. 16101615 and 16118416) and G.Z. (Project No.16104315, 16103714and AoE/M-06/08), the National Natural Science Foundation of China to B.Z. (No. 31500604) and the Health and Medical Research Fund of Food and Health Bureau of the Hong Kong Special Administrative Region Government to G.Z. (Ref. No: 02133056).
Author Contributions
B.Z., Y.G., C.L. and G.Z. designed the study. B.Z., Y.G. and C.L. mainly performed the experiments and analyzed data. B.Z., Y.G., C.L. and G.Z. wrote the manuscript. H.M., Y.R., N.X., X.S., Y.Y. and T.L. provided help with the experiments.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Bo Zhou, Yanyan Geng and Changdong Liu contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20852-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Bo Zhou, Email: bzhou@ust.hk.
Guang Zhu, Email: gzhu@ust.hk.
References
- 1.Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. N. Engl. J. Med. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 2.Kiernan MC, et al. Amyotrophic lateral sclerosis. Lancet. 2011;377:942–955. doi: 10.1016/S0140-6736(10)61156-7. [DOI] [PubMed] [Google Scholar]
- 3.Rademakers R, Neumann M, Mackenzie IR. Advances in understanding the molecular basis of frontotemporal dementia. Nat. Rev. Neurol. 2012;8:423–434. doi: 10.1038/nrneurol.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majounie E, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rutherford NJ, et al. Length of normal alleles of C9ORF72 GGGGCC repeat do not influence disease phenotype. Neurobiol. Aging. 2012;33(2950):e2955–2957. doi: 10.1016/j.neurobiolaging.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee YB, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haeusler AR, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mori K, et al. The C9orf72GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 11.Ash PE, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zu T, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. USA. 2013;110:E4968–4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vatovec S, Kovanda A, Rogelj B. Unconventional features of C9ORF72 expanded repeat in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Neurobiol. Aging. 2014;35:2421 e2421–2421 e2412. doi: 10.1016/j.neurobiolaging.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 14.Kumar V, Kashav T, Islam A, Ahmad F, Hassan MI. Structural insight into C9orf72 hexanucleotide repeat expansions: Towards new therapeutic targets in FTD-ALS. Neurochem. Int. 2016;100:11–20. doi: 10.1016/j.neuint.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Cammas A, Millevoi S. RNA G-quadruplexes: emerging mechanisms in disease. Nucleic Acids Res. 2017;45:1584–1595. doi: 10.1093/nar/gkw1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huppert JL. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chemical Society Reviews. 2008;37:1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- 17.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simone R, Fratta P, Neidle S, Parkinson GN, Isaacs AM. G-quadruplexes: Emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett. 2015;589:1653–1668. doi: 10.1016/j.febslet.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Crenshaw E, et al. Amyloid Precursor Protein Translation Is Regulated by a 3’UTR Guanine Quadruplex. PLoS One. 2015;10:e0143160. doi: 10.1371/journal.pone.0143160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Man BY-W, et al. Group 9 metal-based inhibitors of β-amyloid (1-40) fibrillation as potential therapeutic agents for Alzheimer’s disease. Chem. Sci. 2011;2:917–921. doi: 10.1039/c0sc00636j. [DOI] [Google Scholar]
- 22.Leung CH, et al. Simple and convenient G-quadruplex-based turn-on fluorescence assay for 3′– > 5′ exonuclease activity. Anal. Chem. 2011;83:463–466. doi: 10.1021/ac1025896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He HZ, Chan DS, Leung CH, Ma DL. A highly selective G-quadruplex-based luminescent switch-on probe for the detection of gene deletion. Chem. Commun. (Camb) 2012;48:9462–9464. doi: 10.1039/c2cc32253f. [DOI] [PubMed] [Google Scholar]
- 24.Sket P, et al. Characterization of DNA G-quadruplex species forming from C9ORF72 G4C2-expanded repeats associated with amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Neurobiol. Aging. 2015;36:1091–1096. doi: 10.1016/j.neurobiolaging.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 25.Zhou B, Liu C, Geng Y, Zhu G. Topology of a G-quadruplex DNA formed by C9orf72 hexanucleotide repeats associated with ALS and FTD. Sci. Rep. 2015;5:16673. doi: 10.1038/srep16673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brcic J, Plavec J. Solution structure of a DNA quadruplex containing ALS and FTD related GGGGCC repeat stabilized by 8-bromodeoxyguanosine substitution. Nucleic Acids Res. 2015;43:8590–8600. doi: 10.1093/nar/gkv815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zamiri B, et al. Quadruplex formation by both G-rich and C-rich DNA strands of the C9orf72 (GGGGCC)8*(GGCCCC)8 repeat: effect of CpG methylation. Nucleic Acids Res. 2015;43:10055–10064. doi: 10.1093/nar/gkv1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paramasivan S, Rujan I, Bolton PH. Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding. Methods. 2007;43:324–331. doi: 10.1016/j.ymeth.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 29.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K + solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phan AT, et al. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc. Natl. Acad. Sci. USA. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray DM, et al. Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality. 2008;20:431–440. doi: 10.1002/chir.20455. [DOI] [PubMed] [Google Scholar]
- 32.Hollis T. Crystallization of protein-DNA complexes. Methods Mol. Biol. 2007;363:225–237. doi: 10.1007/978-1-59745-209-0_11. [DOI] [PubMed] [Google Scholar]
- 33.Wang QS, et al. The macromolecular crystallography beamline of SSRF. Nucl. Sci. Tech. 2015;26:12–17. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.