Abstract
DNA methylation has been proposed to play critical roles in caste fate and behavioral plasticity in bumblebees, however, there is little information on its regulatory mechanisms. Here, we identified six important genes mediating the modification of DNA methylation and determined their expression patterns in the bumblebee Bombus terrestris. There is a complete functional DNA methylation system, including four DNA methyltransferases (DNMT1a, DNMT1b, DNMT2, and DNMT3), a DNA demethylase (Ten-eleven translocation), and a methyl-CpG-binding domain protein in B. terrestris. Most of these genes were highly expressed in fat bodies and gonads but lowly expressed in antennae and brains of bumblebee adults. Besides, these genes exhibited caste-specific expression patterns in bumblebees, with higher transcription levels in queens than workers and drones. Whereas their expression levels showed no remarkable difference in queenright and queenless workers. These results suggested potential roles of DNA methylation-related genes in caste differentiation in bumblebees.
Introduction
The bumblebee Bombus terrestris is one of the most effective plant pollinators featuring important economic and ecological values1. B. terrestris exhibits striking morphological and behavioral plasticities involved in unique caste differentiation2. Their morphology, physiology, behavior, longevity, and other life-history traits dramatically differ between queens and workers although they are derived from the same genome3. Reproduction plasticity also occurs between workers depending on social context in bumblebees4. In general, workers are sterile in the colony, but at the end of the colony, some workers become fertile and lay unfertilized eggs5. Besides, after removal of the queen, workers develop ovaries rapidly and are able to lay male destined eggs as early as the fifth day after emergence6. A series of studies suggested that several factors, such as nutrition, juvenile hormone, and social environments, play important roles in caste differentiation in B. terrestris7,8. However, the molecular mechanisms underlying caste differentiation and worker reproductive plasticity remain largely uncharacterized.
DNA methylation is a prominent epigenetic modification that plays essential roles in gene imprinting, X-chromosome inactivation, gene silencing and other significant biological processes9,10. Methylcytosine, a methyl group from S-adenosyl methionine is transferred to the fifth position on the pyrimidine group, which is one of the major DNA methylation modifications. Recently, DNA cytosine methylation has been suggested to be associated with caste differentiation in social insects11,12. After silencing the expression of the de novo methyltransferase DNMT3, the majority of worker-destined larvae emerged as queens with fully developed ovaries in the honeybee Apis mellifera13. Substantial differences in DNA methylation were identified between nurses and foragers, and methylation levels of a large number of genes showed great changes when reverting foragers back to nurses14. Over 500 different methylated genes have been identified in adult brains between queens and workers, and nearly all methylated cytosines are located in CpG dinucleotides in exons15. Another study indicated that there are 2399 genes showed significant differences of methylation in larval heads between queens and workers. Some of these genes are related to juvenile hormone and insulin pathways, which regulate caste determination in honeybees16. In addition, substantial differentially methylated genes were found between different castes in Zootermopsis nevadensis17 and Camponotus floridanus18.
Intricate cytosine methylation dynamics are achieved by methylation and demethylation processes catalyzed by methyltransferases and demethylases respectively. DNA methyltransferases (DNMTs) are divided into three types according to their functions. DNMT1 serves as maintenance methyltransferase, which is involved in the maintenance of cytosine methylation patterns during DNA replication19. DNMT3 is responsible for the establishment of DNA methylation during development, as the de novo methyltransferase20. DNMT2 also considered a methyltransferase because it features a conserved DNA methyltransferase catalytic domain. However, DNMT2 was proven to play an essential role in tRNA methylation21. Methyl-CpG-binding domain proteins (MBDs) can recognize and specifically bind to methylated sites, causing chromatin structural modification and remodeling by recruitment of repressive complexes, which are extremely imperative in DNA methylation mediated gene silencing22. Positive DNA demethylation mainly depends on the double oxygenase ten-eleven translocation protein (TET) through iterative oxidation reactions23.
Divergent levels of DNA methylation in B. terrestris have been reported at different developmental stages and genders24, and some DNA methylation-related genes have been mentioned in previous studies25,26, however, those genes involved in DNA methylation have not been characterised fully and the functional roles of these genes are not clear in the bumblebee. In this study, we characterized six key genes related to DNA methylation and determined their expression patterns in B. terrestris to lay the foundation for elucidating epigenetic mechanisms of caste differentiation in bumblebees.
Results
Sequence analysis of genes involved in DNA methylation from B. terrestris
Nucleotide sequences of six genes involved in DNA methylation in B. terrestris were obtained from the database of National Center for Biotechnology Information. Bioinformatically, we acquired four DNMTs (DNMT1a, DNMT1b, DNMT2, and DNMT3), a DNA demethylase (TET2), and a methyl-CpG-binding protein (MBD3). Supplementary Table S2 provides open reading frames (ORFs), numbers of amino acids, predicted relative molecular weights, and isoelectric points of these proteins.
By comparing domain structures of these genes, we found that DNMT1a and DNMT1b proteins had the same domain structures (Fig. 1). Both DNMT1s contain a replication foci (RFD) domain, a CXXC zinc finger (zf-C) domain, two bromo-adjacent homology (BAH) domains, and a site-specific DNA-cytosine methylase (MTase) domain. The MTase domain is responsible for cytosine methylation27. DNMT2 also contains a MTase domain (Fig. 1). Other than a MTase domain, two PWWP domains and a plant homeodomain (PHD) were also observed in DNMT3 (Fig. 1). The PHD domain serves as the catalytic center of DNMT3, which is essential for de novo methylation. MBD3 comprises a MeCP2 domain that can recognize methylated CpG dinucleotides specifically, and a MBD_C domain that is a typical characteristic of MBDs (Fig. 1). TET2 contains a zf-C domain and a double-stranded beta helix fold (DSΒH) domain (Fig. 1), the latter domain is involved in oxidizing reaction in DNA demethylation28. Above all, the bumblebee B. terrestris possesses a functional suite of DNA methylation-associated proteins, implying the possible important roles of DNA methylation in the bumblebee.
Figure 1.
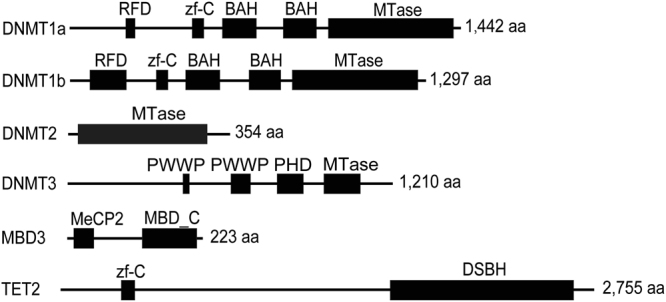
Predicted protein domains encoded by DNA methylation-related genes from B. terrestris. RFD: cytosine specific DNA methyltransferase replication foci domain; zf-C: CXXC zinc finger domain; BAH: bromo-adjacent homology domain; MTase: site-specific DNA-cytosine methylase domain; PWWP: domain with conserved PWWP motif; PHD: plant homeodomain; MeCP2: methyl-CpG-binding domain; MBD_C: C-terminal domain of methyl-CpG-binding protein; DSΒH: double-stranded beta helix fold domain.
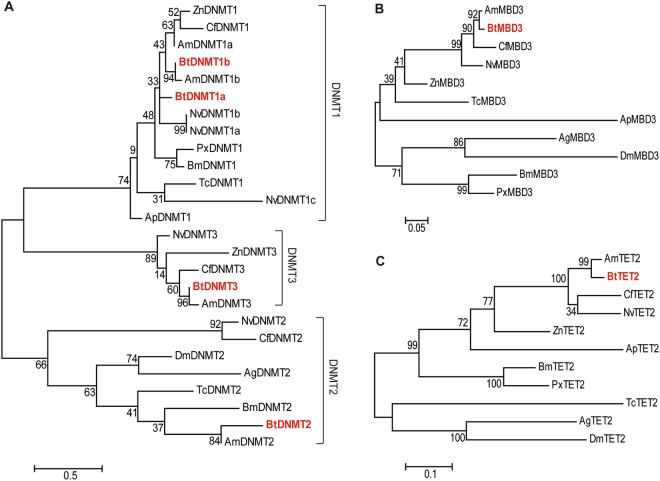
Phylogenetic analysis of genes involved in DNA methylation from B. terrestris and other insects
The homologs of methylation-related genes in all insect species were firstly searched by using these sequences from the mice as seed sequences in NCBI database with a Blast tool. And then, we selected 11 insect species with the available whole genome sequences from six orders, including A. mellifera, Anopheles gambiae, Acyrthosiphon pisum, Bombyx mori, B. terrestris, Camponotus floridanus, Drosophila melanogaster, Plutella xylostella, Tribolium castaneum, Nasonia vitripennis and Z. nevadensis to constructed the phylogenetic trees. Nevertheless, some genes were lost in the genomes from several species or were not found based on the gene database of NCBI. Such as, D. melanogaster genome lacks DNMT1 and DNMT329. Although DNMT1, DNMT2 and DNMT3 were reported in A. pisum genome from previous study30, we have not found the sequences of DNMT2 and DNMT3 of A. pisum according to the NCBI gene database. Thus, not all of DNA methylation-related genes from these species were included in phylogenetic analysis.
DNMT1a, DNMT1b, DNMT2, and DNMT3 from different species were grouped to form a monophyletic clade respectively (Fig. 2A). Two members of DNMT1 from B. terrestris were clustered with the homologous proteins of other insects. The relationship of DNMT1a between B. terrestris and A. mellifera was closer. DNMT1b in B. terrestris showed close evolutionary relationship with DNMT1a in B. terestris and the homologous proteins in A. mellifera, N. vitripennis, Z. nevadensis and C. floridanus (Fig. 2A). DNMT2 and DNMT3 in B. terrestris presented closer relationships with these of A. mellifera (Fig. 2A). Similarly, MBD3 from B. terrestris showed a closer relationship with that of A. mellifera (Fig. 2B). And phylogenetic analysis suggested TET2 from B. terrestris and A. mellifera shared closer relationship (Fig. 2C). Above all, DNA methylation-related genes in B. terrestris displayed closer relationships with those of other Hymenoptera insects. Hence, the obtained sequences in B. terrestris are reliable.
Figure 2.
Maximum Likelihood-based phylogenetic trees of DNA methylation-related proteins. (A) Phylogenic tree based on amino acid sequences of DNMT1, DNMT2, and DNMT3 from B. terrestris and other insects. (B) Phylogenic tree based on amino acid sequences of MBD3 from B. terrestris and other insects. (C) Phylogenic tree based on amino acid sequences of TET2 from B. terrestris and other insects. Genes from B. terrestris are highlighted in bold red. Taxon abbreviations: Bt, B. terrestris; Am, A. mellifera; Ag, A. gambiae; Ap, A. pisum; Bm, B. mori; Cf, C. floridanus; Dm, D. melanogaster; Px, P. xylostella; Tc, T. castaneum; Nv, N. vitripennis; Zn, Z. nevadensis. Gene names, species names and GenBank accession numbers were shown in Supplementary Table S3.
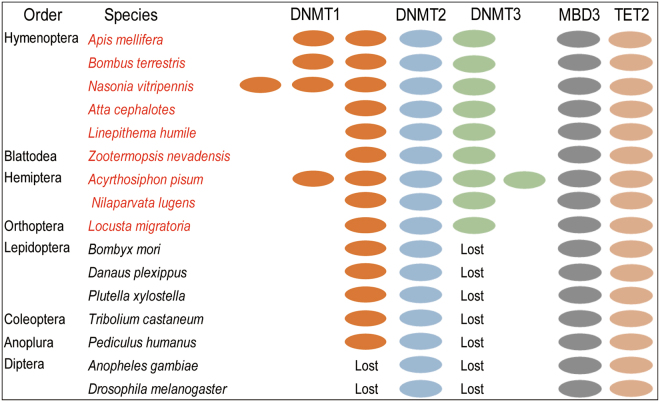
The comparison of DNA methylation systems between B. terrestris and other insects
The DNA methylation process needs a set of essential enzymes including DNA methyltransferases and demethylases31. However, DNA methylation-related proteins were selectively lost in insect species. Based on previous researches25,32, we revised the patterns of DNA methylation-related proteins in several insects from eight orders, including Hymenoptera, Blattodea, Hemiptera, Orthoptera, Lepidoptera, Coleoptera, Anoplura, and Diptera (Fig. 3). Bumblebees, honeybees, ants and wasps contain full DNA methylation toolkits with different duplications of DNMT133–35. Likewise, Z. nevadensis was found to have a full DNA methylation toolkit36. Aphides and brown planthoppers also harbor the full functional DNA methylation systems30,32. Besides, the locust genome possesses a complete DNA methylation toolkit37 (Fig. 3). Nevertheless, DNMT3 was lost in some Lepidoptera, Coleoptera and Anoplura insects as shown in Fig. 338–41. Both DNMT1 and DNMT3 were lost in flies and mosquitos, only DNMT2 was found in their genomes42 (Fig. 3). These results suggested that B. terrestris has a complete DNA methylation system, providing new insights into the important roles of DNA methylation in social insects.
Figure 3.
Comparison of DNA methylation systems in insect species. Different insect species display varied DNA methylation systems. The numbers of ellipses of each color indicate the numbers of gene copies found in each species. Lost in figure means that DNMTs are absent in insect genomes. The red marked species in the figure contain a complete functional DNA methylation toolkit.
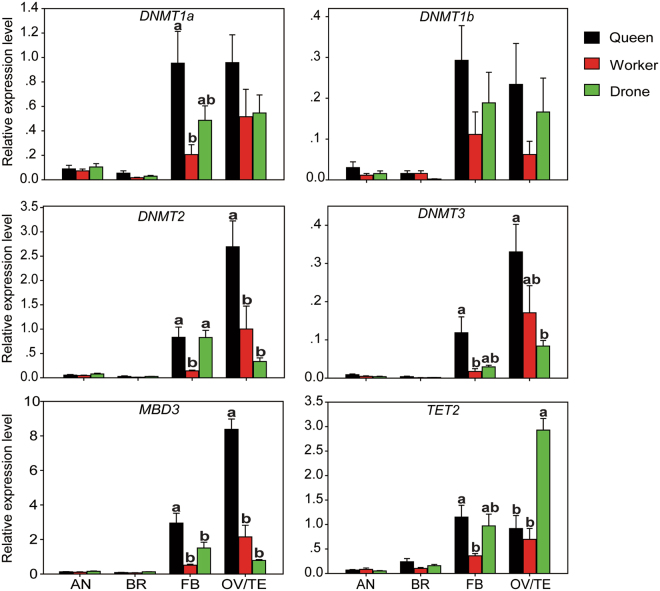
Caste-specific expression patterns of DNA methylation-related genes in B. terrestris
qRT-PCR was performed to investigate the expression patterns of DNA methylation-related genes in different tissues from queen, worker, and drone adults. The results showed that most of these genes were highly expressed in fat bodies, ovaries and testes in bumblebee adults (Fig. 4). In queens, DNMT1a showed higher expression levels in fat bodies and ovaries than antennae and brains (ANOVA, F3,12 = 11.072, P = 0.0012), whereas there was no significant difference in the expression levels of DNMT1b among different tissues (Fig. 4A). The expression levels of DNMT2, DNMT3 and MBD3 were significantly higher in ovaries than other tissues (ANOVA, DNMT2: F3,12 = 19.097, P < 0.0001; DNMT3: F3,12 = 13.393, P < 0.001; MBD3: F3,12 = 88.284, P < 0.0001) (Fig. 4A). TET2 showed higher expression levels in fat bodies and ovaries than antennae and brains (ANOVA, F3,12 = 7.809, P = 0.004) (Fig. 4A).
Figure 4.
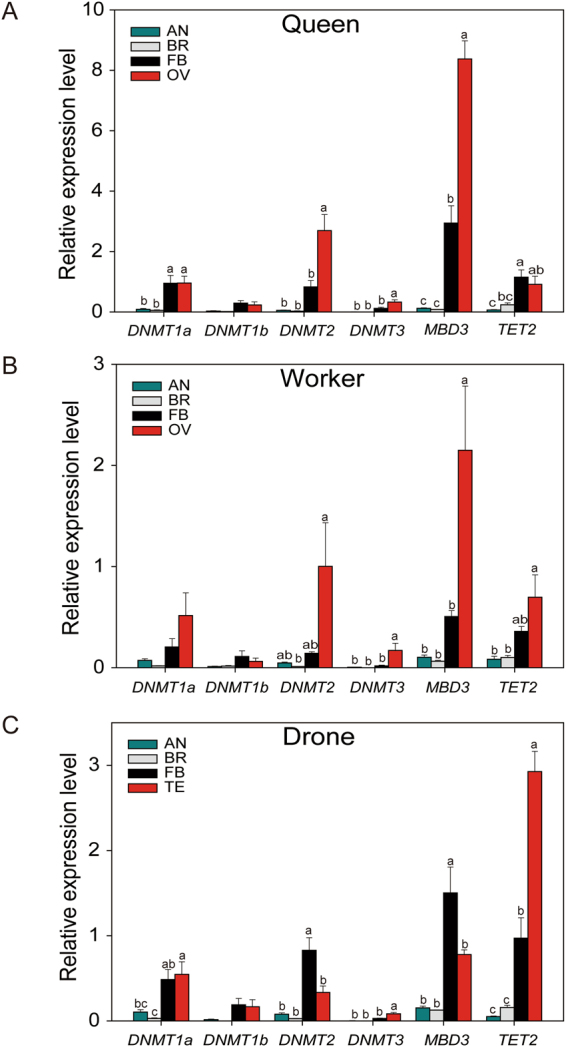
The expression patterns of DNA methylation-related genes of B. terrestris between different tissues in queen (A), worker (B), and drone (C) adults respectively. AN: antenna; BR: brain; FB: fat body; OV: ovary; TE: testis. Data in the figure represent mean ± SM of four biological replicates, and the different letters indicate significant difference among different castes by multiple testing in one-way ANOVA model (P < 0.05).
In workers, the expression levels of DNMT1a and DNMT1b showed no remarkable difference among four tissues (Fig. 4B). DNMT2, DNMT3 and MBD3 exhibited higher expression levels in ovaries than other three tissues (ANOVA, DNMT2: F3,12 = 3.996, P = 0.035; DNMT3: F3,12 = 5.289, P = 0.015; MBD3: F3,12 = 8.405, P = 0.003) (Fig. 4B). TET2 displayed higher expression levels in fat bodies and ovaries than antennae and brains (ANOVA, F3,12 = 6.226, P = 0.009) (Fig. 4B). In drones, DNMT1a was highly expressed in testis and fat body tissues (ANOVA, F3,12 = 6.838, P = 0.007) (Fig. 4C). While the expression levels of DNMT1b showed no obvious distinction in all tissues. Both DNMT2 and MBD3 displayed higher expression levels in fat bodies than other three tissues (ANOVA, DNMT2: F3,12 = 18.972, P < 0.0001; MBD3: F3,12 = 14.594, P = 0.0003) (Fig. 4C). Relative expression levels of DNMT3 and TET2 were significantly higher in testes than other tissues (ANOVA, DNMT3: F3,12 = 24.640, P < 0.0001; TET2: F3,12 = 61.465, P < 0.0001) (Fig. 4C).
In different castes, the expression levels of these genes showed significant differences in fat bodies and gonads but not in antennae and brains (Fig. 5). The expression levels of most genes were significantly higher in fat bodies of queen adults than in those of worker adults (ANOVA, DNMT1a: F2,9 = 4.839, P = 0.037; DNMT2: F2,9 = 7.086, P = 0.014; DNMT3: F2,9 = 5.071, P = 0.034; MBD3: F2,9 = 10.177, P = 0.005; TET2: F2,9 = 4.435, P = 0.046). For ovaries, DNMT2 and MBD3 were significantly higher expressed in queens than workers (ANOVA, DNMT2: F2,9 = 8.689, P = 0.008; MBD3: F2,9 = 59.993, P < 0.0001). There was no significant difference in the expression levels of most genes between females and males in fat bodies (Fig. 5). However, TET2 exhibited much higher expression levels in male testes than female ovaries, and no obvious difference occurred between ovaries of queens and workers (TET2: F2,9 = 25.778, P = 0.0002) (Fig. 5). These results indicated that these DNA methylation-related genes displayed apparent caste-dependent expression patterns.
Figure 5.
The expression patterns of DNA methylation-related genes of B. terrestris in different tissues among queen, worker, and drone adults. AN: antenna; BR: brain; FB: fat body; OV: ovary; TE: testis. Data in the figure represent mean ± SM of four biological replicates, and the different letters indicate significant difference among different castes by multiple testing in one-way ANOVA model (P < 0.05).
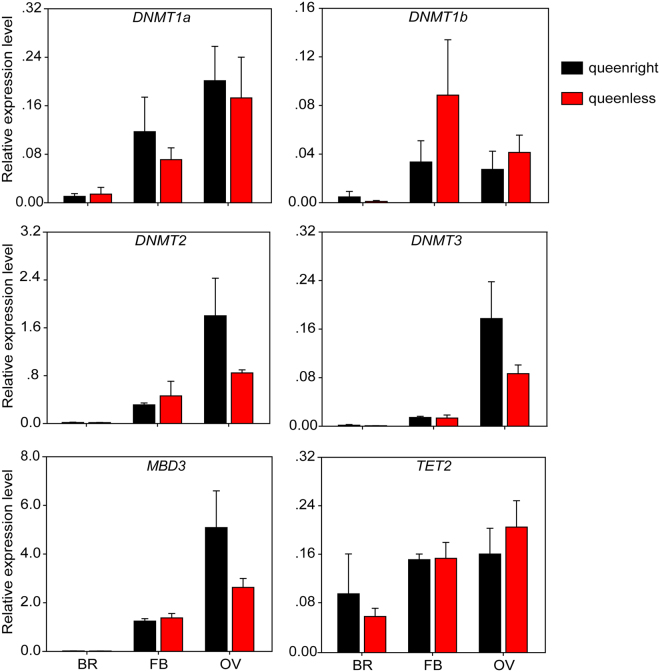
Expression patterns of DNA methylation-related genes in queenright and queenless workers
In bumblebees, workers are able to activate their ovaries and lay haploid eggs after the death of the queen or the removal of the queen in a colony43. We investigated whether DNA methylation is associated with queen-repressed worker fertility in B. terrestris. After measuring the length of terminal oocytes of workers in queenless and queenright colonies, we found that the ovaries in queenless workers were significantly larger than those in queenright workers (Supplementary Fig. S6). The expression patterns of these genes were determined in different tissues, including brains, fat bodies, and ovaries from queenright and queenless workers (Fig. 6). In brains, TET2 showed a scarcely higher expression level in queenright workers than in queenless workers. In fat bodies, DNMT1a displayed a barely higher expression level in queenright workers than in queenless workers. Whereas the expression levels of DNMT1b and DNMT2 were slightly higher in fat bodies of queenless workers than in those of queenright workers (Fig. 6). For ovaries, DNMT1a, DNMT2, DNMT3, and MBD3 showed marginally decreased expression levels in queenless workers compared with those in queenright workers. In contrary, the expression levels of DNMT1b and TET2 were slightly higher in ovaries of queenless workers than in those of queenright workers. However, no significant difference was observed in the expression levels of these genes between queenright and queenless workers (Fig. 6).
Figure 6.
The expression patterns of DNA methylation-related genes in different tissues of workers in queenright and queenless colonies. AN: antenna; BR: brain; FB: fat body; OV: ovary; TE: testis. Data in the figure represent mean ± SM of four biological repetitions. The values between groups were analyzed by independent sample T-test (P < 0.05).
Discussion
In this study, we identified six genes involved in DNA methylation in bumblebees. These genes exhibited caste-specific expression patterns in B. terrestris. No remarkable difference in the expression levels of these genes was observed between queenright workers and queenless workers. This study provides molecular insights into the DNA methylation-related gene system of B. terrestris.
Our results revealed that there is a complete DNA methylation system in B. terrestris, similar with other social insects, aphids, and locusts (Fig. 3). Actually, DNA methylation systems display diverse patterns among insects. For example, D. melanogaster lacks two key DNMTs (DNMT1 and DNMT3), and a very low level of cytosine methylation exists at the early embryonic stage44. Homologous genes of DNMT1 and DNMT2 were discovered in B. mori genome, but no homologous gene of DNMT3 was detected38. Similarly, T. castaneum lacks DNMT3, and it contains a significantly lower DNA methylation level45. However, the complete functional DNA methylation toolkit was observed in most of social insects. Social insects usually exhibit a striking environment-driven plasticities including morphological, reproductive and behavioral diversities from the same genome46. Epigenetic mechanisms are widely considered additional aspects of genome regulation for increasing flexibility of organisms47. Therefore, the conservation of functional DNA methylation systems in these social insects suggests that DNA methylation may be associated with social evolution.
DNA methylation-related genes displayed obvious caste-dependent expression patterns in B. terrestris. A previous large-scale RNA-seq analysis from 11 different combinations of caste and developmental stages in B. terrestris48, also implied that possible differential expression patterns of DNA cytosine methyltransferase genes in different castes. And DNMT3 shows different expression patterns between queens and workers in the honeybee Apis cerana cerana49. Besides, a number of documents have reported that DNA methylation is widespread and associated with caste differences in many social insects, including Z. nevadensis17, C. floridanus50, Harpegnathos saltator50 and A. mellifera15. We also found that most DNA methylation-related genes were higher expressed in fat bodies and ovaries of queen adults than in those of workers and drones in B. terrestris, implying that DNA methylation processes bears important functions in caste determination.
In mammals, relative mRNA expressions of DNMTs have been reported to be associated with the global dynamic of DNA methylation status51. DNA methylation, as an important epigenetic factor in regulations of gene expression and transcriptional status52, may plays vital roles in physiological processes relies on the participation of DNMTs. Higher DNMTs expression levels existed in fat bodies and ovaries of adult queens imply that more active methylation processes may occur in these two tissues of queens than in those of workers, which is consistent with a large demand of energy substances to be accumulated in fat bodies and ovaries for ovary development and egg laying in the queen53. These results suggest that DNA methylation may be closely related to the development of ovary and reproduction of the queen in the bumblebee.
In the present study, most genes in drones displayed intermediate expression levels compared to the two female castes. Although females and males differ in their ploidies, they share the same genome. The differential expression patterns of these genes implied distinct DNA methylation status in these three castes in the bumblebee. These wide difference of DNA methylation may be associated with unique roles of specific castes played in the bumblebee colony. However, more research is needed to understand the mechanisms underlying the result.
Our results showed that TET displayed the highest expression level in drone testes. In mammals, three members of TET proteins have been reported to be involved in the demethylation process28. Until now, only one member of TET enzymes has been identified in insect genomes31. In the honeybee A. mellifera, TET dioxygenase has been reported to possess catalytic activity to convert 5-methylcytosine to 5-hydroxymethylcytosine54, suggesting similar functions of insect TET enzymes with that of mammals. It was reported that DNA methylation dynamics are crucial for the differentiation of spermatogenic cells in mammals55. During spermatogenesis, TET proteins are involved in the regulation of maintenance of pluripotency and proliferation of male germline stem cells56. The highest expression level of TET2 in drone testes of bumblebees implied potentially active demethylation in male gonads. Therefore, TET-mediated demethylation may be necessary for spermatogenesis in bumblebees, however, the detailed functions of TET should be further determined.
We found that no difference was observed in the expression levels of DNA methylation-related genes between queenless and queenright workers in B. terrestris. Bumblebee workers are well acknowledged to show remarkable reproductive plasticity, depending on social context. After the competition point in queenright colony, workers were not inhibited by the queen and then became fertile57. Influence of queen on worker reproduction constantly has been an interesting question, which presents great significance to evolution of social insects. Many studies suggested that chemical pheromones and direct contact with the queen play important roles in regulating reproductive dominance in bumblebees58,59. Several molecular mechanisms, such as epigenetic regulations, also were proposed to determine plasticity of worker reproduction60,61. However, differential expression levels of genes related to DNA methylation were not observed between queenright workers and queenless workers, implying that DNA methylation may not be directly involved in queen-inhibited worker reproductions. The result is in agreement with a recent study which reported that levels of DNA methylation exhibited no difference between queenright nonreproductive workers and queenless reproductive workers62. Therefore, differential expressed genes involved in DNA methylation may mainly be involved in caste differentiation, but not in worker reproductive plasticity depending on the social context.
DNA methylation serves as a complex dynamic system involved in regulating many important biological processes in insects. In this study, we focused on the expression patterns of genes related to DNA methylation in bumblebee adults. Functional studies on DNA methylation system in different developmental stages should provide significant discoveries for exploring mechanisms of caste differentiation in bumblebees.
Methods
Insects
B. terrestris was bought from the breeding base of pollinating bees, Information Institute of Beijing Academy of Agriculture and Forestry Sciences, and reared with pollen and 50% sucrose water at 28 °C and relative humidity of 50–60% under red light in well-ventilated plastic boxes.
Sequence analysis
Nucleotide sequences of genes were obtained from the National Center for Biotechnology Information with blast tool. Putative ORFs and amino acid sequences of DNA methylation-associated genes in B. terrestris were predicted using DNAMAN software. Protein properties were analyzed with EXPASy server, and protein domains were predicted with SMART software63. Sequences used in phylogenetic analysis were downloaded from the National Center for Biotechnology Information gene database (http://www.ncbi.nlm.nih.gov). DNA methylation-associated genes of B. terrestris and other selected insects were subjected to multiple sequence alignments using Clustal W program64 and edited with GeneDoc software65 (Supplementary Figs S1–S5). The phylogenetic trees were constructed using a Maximum Likelihood method with a Poisson model and 1000 Bootstrap replications of MEGA5.0 software66.
Sample preparation
Different tissues (antenna, brain, fat body, ovary, and testis) of B. terrestris were dissected from one-day-old adults that from queens, workers and drones. Wherein adult workers, virgin queens and drones were obtained from four colonies separately. Four biological replicates were prepared for each experiment, and each biological replicate included eight individuals. Collected samples were immediately stored at −80 °C for further total RNA extraction.
Five newly emerged workers were placed in a new nest containing the mother queen from the native colony, as a queenright treatment. In queenless treatment, five newly emerged workers were selected and reared in a new colony without the queen. There were four biological replicates in each treatment. Five days later, different tissues (brain, fat body, and ovary) were dissected from workers in queenright and queenless colonies. Harvested samples were kept in −80 °C at once to extract total RNA.
RNA extraction and cDNA synthesis
Total RNAs were isolated using a TRIzol kit (Life Technologies) following the manufacturer’s instructions. Quantity and quality of total RNA were determined by ultraviolet spectrophotometry. Before first-stand cDNA synthesis, DNase was used to treat total RNA to prevent genomic DNA contamination. First-strand cDNA was synthesized from 2 μg total RNA using the M-MLV Reverse Transcriptase and oligo (dT)-primer (Promega) according to the manufacturer’s instructions. Synthesized first-stand cDNA was stored at −80 °C immediately for subsequent use.
Quantitative RT-PCR (qRT-PCR)
Primers used for qRT-PCR were designed by Primer 5.0 software, and are listed in Supplementary Table S1. The gene Elongation Factor 1a (EF1a) in B. terrestris was used as a reference gene for normalization in accordance with the previous study67. The cDNA was subjected to qRT-PCR by using the SYPR Green RealMasterMix (Roche) according to the manufacturer’s instructions on a LightCycler 480 instrument (Roche). Melting curve analysis was used to confirm unique amplification. Data were analyzed by 2−ΔΔCT method to quantify relative mRNA expression levels68.
Statistical analysis
Data in figures are expressed as means ± standard error (M ± SM), and qRT-PCR results were analyzed using multiple comparison in one-way ANOVA and independent sample T-test by SPSS17.0 software.
Electronic supplementary material
Acknowledgements
We express sincere gratitude to Le Kang (Institute of Zoology, Chinese Academy of Sciences) for comments and supports for this work. We are grateful to Qing Liu (Institute of Zoology, Chinese Academy of Sciences) for revising the sequences of genes in this study. We also thank MaoWu Luo (Institute of Zoology, Chinese Academy of Sciences), Minmin Cao (Institute of Zoology, Chinese Academy of Sciences) and Xiaoming Liu (College of Plant Protection, China Agricultural University) for comments on the manuscript. This study was supported by the National Natural Science Foundation of China (No. 31472047) and the Open Funding of State Key Laboratory of Integrated Management of Pest Insects and Rodents (ChineseIPM1507).
Author Contributions
X.H.W. and S.H.A. designed and conceived the study; B.B.L., L.H. and D.Z. performed the experiments; B.B.L. analysed data and wrote the manuscript; X.H.W., S.H.A. and X.L.X. reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20831-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shiheng An, Email: anshiheng@aliyun.com.
Xianhui Wang, Email: wangxh@ioz.ac.cn.
References
- 1.Goulson, D. Bumblebees Behavior and Ecology. (Oxford University Press, 2003).
- 2.Yagound B, Blacher P, Chameron S, Chaline N. Social context and reproductive potential affect worker reproductive decisions in a eusocial insect. PloS ONE. 2012;7:e52217. doi: 10.1371/journal.pone.0052217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiner SA, Toth AL. Epigenetics in social insects: a new direction for understanding the evolution of castes. Genet res int. 2012;2012:609810. doi: 10.1155/2012/609810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardinal, S. & Danforth, B. N. The Antiquity and Evolutionary History of Social Behavior in Bees. PloS ONE6, doi:ARTN e21086/journal.pone.0021086 (2011). [DOI] [PMC free article] [PubMed]
- 5.Alaux C, Boutot M, Jaisson P, Hefetz A. Reproductive plasticity in bumblebee workers (Bombus terrestris)—reversion from fertility to sterility under queen influence. Behav Ecol Sociobiol. 2007;62:213–222. doi: 10.1007/s00265-007-0455-6. [DOI] [Google Scholar]
- 6.Roseler PF, Roseler I, Vanhonk CGJ. Evidence for Inhibition of Corpora Allata Activity in Workers of Bombus Terrestris by a Pheromone from the Queens Mandibular Glands. Experientia. 1981;37:348–351. doi: 10.1007/BF01959856. [DOI] [Google Scholar]
- 7.Bortolotti L, Duchateau MJ, Sbrenna G. Effect of juvenile hormone on caste determination and colony processes in the bumblebee Bombus terrestris. Entomol Exp Appl. 2001;101:143–158. doi: 10.1046/j.1570-7458.2001.00899.x. [DOI] [Google Scholar]
- 8.Pereboom JJM, Velthuis HHW, Duchateau MJ. The organisation of larval feeding in bumblebees (Hymenoptera, Apidae) and its significance to caste differentiation. Insect Soc. 2003;50:127–133. doi: 10.1007/s00040-003-0639-7. [DOI] [Google Scholar]
- 9.Paulsen M, Ferguson-Smith AC. DNA methylation in genomic imprinting, development, and disease. J Pathol. 2001;195:97–110. doi: 10.1002/path.890. [DOI] [PubMed] [Google Scholar]
- 10.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 11.Elango N, Hunt BG, Goodisman MA, Yi SV. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee. Apis mellifera. Proc Natl Acad Sci USA. 2009;106:11206–11211. doi: 10.1073/pnas.0900301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foret S, Kucharski R, Pittelkow Y, Lockett GA, Maleszka R. Epigenetic regulation of the honey bee transcriptome: unravelling the nature of methylated genes. BMC genom. 2009;10:472. doi: 10.1186/1471-2164-10-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 14.Herb BR, et al. Reversible switching between epigenetic states in honeybee behavioral subcastes. Nat Neurosci. 2012;15:1371–1373. doi: 10.1038/nn.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyko, F. et al. The Honey Bee Epigenomes: Differential Methylation of Brain DNA in Queens and Workers. PloS Biol. 8, 10.1371/journal.pbio.1000506 (2010). [DOI] [PMC free article] [PubMed]
- 16.Foret S, et al. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc Natl Acad Sci USA. 2012;109:4968–4973. doi: 10.1073/pnas.1202392109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glastad, K. M., Gokhale, K., Liebig, J. & Goodisman, M. A. D. The caste- and sex-specific DNA methylome of the termite Zootermopsis nevadensis. Sci Rep6, doi:Artn 37110/Srep37110 (2016). [DOI] [PMC free article] [PubMed]
- 18.Alvarado, S., Rajakumar, R., Abouheif, E. & Szyf, M. Epigenetic variation in the Egfr gene generates quantitative variation in a complex trait in ants. Nat Commun6, doi:Artn 6513/Ncomms7513 (2015). [DOI] [PubMed]
- 19.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 20.Chen T, Tsujimoto N, Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol Cell Biol. 2004;24:9048–9058. doi: 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goll MG, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 22.Bogdanovic O, Veenstra GJ. DNA methylation and methyl-CpG binding proteins: developmental requirements and function. Chromosoma. 2009;118:549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He YF, et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bigot Y, Jegot G, Casteret S, Aupinel P, Tasei JN. DNA modifications and genome rearrangements during the development and sex differentiation of the bumble bee Bombus terrestris. Insect Mol Biol. 2011;20:165–175. doi: 10.1111/j.1365-2583.2010.01052.x. [DOI] [PubMed] [Google Scholar]
- 25.Li-Byarlay H. The Function of DNA Methylation Marks in Social Insects. Front. Ecol. Evol. 2016;4:1–8. doi: 10.3389/fevo.2016.00057. [DOI] [Google Scholar]
- 26.Sadd BM, et al. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 2015;16:76. doi: 10.1186/s13059-015-0623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raddatz G, et al. Dnmt2-dependent methylomes lack defined DNA methylation patterns. Proc Natl Acad Sci USA. 2013;110:8627–8631. doi: 10.1073/pnas.1306723110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh TK, et al. A functional DNA methylation system in the pea aphid, Acyrthosiphon pisum. Insect Mol Biol. 2010;19(Suppl 2):215–228. doi: 10.1111/j.1365-2583.2009.00974.x. [DOI] [PubMed] [Google Scholar]
- 31.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu rev biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, et al. DNA methyltransferases have an essential role in female fecundity in brown planthopper. Nilaparvata lugens. Biochem biophys res commun. 2015;464:83–88. doi: 10.1016/j.bbrc.2015.05.114. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, et al. Functional CpG methylation system in a social insect. Science. 2006;314:645–647. doi: 10.1126/science.1135213. [DOI] [PubMed] [Google Scholar]
- 34.Suen G, et al. The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLoS Genet. 2011;7:e1002007. doi: 10.1371/journal.pgen.1002007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith CD, et al. Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile) Proc Natl Acad Sci USA. 2011;108:5673–5678. doi: 10.1073/pnas.1008617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terrapon N, et al. Molecular traces of alternative social organization in a termite genome. Nat commun. 2014;5:3636. doi: 10.1038/ncomms4636. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, et al. The locust genome provides insight into swarm formation and long-distance flight. Nat commun. 2014;5:2957. doi: 10.1038/ncomms3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang H, et al. Single base-resolution methylome of the silkworm reveals a sparse epigenomic map (vol 28, pg 516, 2010) Nat Biotechnol. 2010;28:756–756. doi: 10.1038/nbt0710-756d. [DOI] [PubMed] [Google Scholar]
- 39.You M, et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat genet. 2013;45:220–225. doi: 10.1038/ng.2524. [DOI] [PubMed] [Google Scholar]
- 40.Kirkness EF, et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci USA. 2010;107:12168–12173. doi: 10.1073/pnas.1003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glastad, K. M., Hunt, B. G. & Goodisman, M. A. Evidence of a conserved functional role for DNA methylation in termites. Insect Mol Biol, 10.1111/imb.12010 (2012). [DOI] [PubMed]
- 42.Marhold J, et al. Conservation of DNA methylation in dipteran insects. Insect Mol Biol. 2004;13:117–123. doi: 10.1111/j.0962-1075.2004.00466.x. [DOI] [PubMed] [Google Scholar]
- 43.Alaux C, Jaisson P, Hefetz A. Queen influence on worker reproduction in bumblebees (Bombus terrestris) colonies. Insect Soc. 2004;51:287–293. doi: 10.1007/s00040-004-0741-5. [DOI] [Google Scholar]
- 44.Boffelli D, Takayama S, Martin DI. Now you see it: genome methylation makes a comeback in Drosophila. Bioessays. 2014;36:1138–1144. doi: 10.1002/bies.201400097. [DOI] [PubMed] [Google Scholar]
- 45.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 46.Keller L, Jemielity S. Social insects as a model to study the molecular basis of ageing. Exp Gerontol. 2006;41:553–556. doi: 10.1016/j.exger.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 47.Feinberg AP. Epigenomics reveals a functional genome anatomy and a new approach to common disease. Nat Biotechnol. 2010;28:1049–1052. doi: 10.1038/nbt1010-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harrison MC, Hammond RL, Mallon EB. Reproductive workers show queenlike gene expression in an intermediately eusocial insect, the buff-tailed bumble bee Bombus terrestris. Mol Ecol. 2015;24:3043–3063. doi: 10.1111/mec.13215. [DOI] [PubMed] [Google Scholar]
- 49.Liu TT, et al. Cloning and expression profiling of the DNA methyltransferase Dnmt3 gene in the Chinese honeybee, Apis cerana cerana (Hymenoptera: Apidae) Acta Entomol Sinica. 2012;55:284–290. [Google Scholar]
- 50.Bonasio R, et al. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr biol. 2012;22:1755–1764. doi: 10.1016/j.cub.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oh BK, et al. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. Int J Mol Med. 2007;20:65–73. [PubMed] [Google Scholar]
- 52.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 53.Arrese EL, Soulages JL. Insect Fat Body: Energy, Metabolism, and Regulation. Annu Rev Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wojciechowski, M. et al. Insights into DNA hydroxymethylation in the honeybee from in-depth analyses of TET dioxygenase. Open Biol4, doi:Unsp 140110/Rsob.140110 (2014). [DOI] [PMC free article] [PubMed]
- 55.Gan, H. Y. et al. Dynamics of 5-hydroxymethylcytosine during mouse spermatogenesis. Nat Commun4, doi:Artn 1995/Ncomms2995 (2013). [DOI] [PubMed]
- 56.Zheng L, et al. Modification of Tet1 and histone methylation dynamics in dairy goat male germline stem cells. Cell Prolif. 2016;49:163–172. doi: 10.1111/cpr.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plowright RC, Laverty TM. The Ecology and Sociobiology of Bumble Bees. Annu Rev Entomol. 1984;29:175–199. doi: 10.1146/annurev.en.29.010184.001135. [DOI] [Google Scholar]
- 58.Holman, L. Bumblebee size polymorphism and worker response to queen pheromone. Peerj2, doi:ARTN e604/peerj.604 (2014). [DOI] [PMC free article] [PubMed]
- 59.Padilla M, Amsalem E, Altman N, Hefetz A, Grozinger CM. Chemical communication is not sufficient to explain reproductive inhibition in the bumblebee Bombus impatiens. R Soc Open Sci. 2016;3:160576. doi: 10.1098/rsos.160576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amarasinghe HE, Toghill BJ, Nathanael D, Mallon EB. Allele specific expression in worker reproduction genes in the bumblebee Bombus terrestris. Peerj. 2015;3:e1079. doi: 10.7717/peerj.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson GJ, Kucharski R, Maleszka R, Oldroyd BP. Genome-wide analysis of genes related to ovary activation in worker honey bees. Insect Mol Biol. 2008;17:657–665. doi: 10.1111/j.1365-2583.2008.00838.x. [DOI] [PubMed] [Google Scholar]
- 62.Amarasinghe HE, Clayton CI, Mallon EB. Methylation and worker reproduction in the bumble-bee (Bombus terrestris) P Roy Soc B-Biol Sci. 2014;281:20132502. doi: 10.1098/rspb.2013.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nicholas KB, Nicholas HB, Jr, Deerfield DW. GeneDoc: Analysis and Visualization of Genetic Variation. EMBNET News. 1997;4:14. [Google Scholar]
- 66.Tamura K, et al. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shpigler, H. et al. Gonadotropic and Physiological Functions of Juvenile Hormone in Bumblebee (Bombus terrestris) Workers. PloS ONE9, doi:ARTN e100650/journal.pone.0100650 (2014). [DOI] [PMC free article] [PubMed]
- 68.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.