Abstract
Gram-positive bacteria use peptides as auto-inducing (AI) signals to regulate the production of extracellular enzymes (e.g., proteases). ComX is an AI peptide, mostly known for its role in the regulation of bacterial competence and surfactant production in Bacillus subtilis. These two traits are regulated accordingly to the bacterial population size, thus classifying ComX as a quorum sensing signal. ComX also indirectly regulates exoprotease production through the intermediate transcriptional regulator DegQ. We here use this peptide-based AI system (the ComQXPA system) as a model to address exoprotease regulation by ComX in biofilms. We also investigate the potential of ComX regulated proteases to degrade the ComX AI peptide. Results indicate that ComX indeed induces the expression of aprE, the gene for the major serine protease subtilisin, and stimulates overall exoprotease production in biofilms of B. subtilis PS-216 and several other B. subtilis soil isolates. We also provide evidence that these exoproteases can degrade ComX. The ComX biological activity decay is reduced in the spent media of floating biofilms with low proteolytic activity found in the comP and degQ mutants. ComX biological activity decay can be restored by the addition of subtilisin to such media. In contrast, inhibition of metalloproteases by EDTA reduces ComX biological activity decay. This suggests that both serine and metalloproteases, which are induced by ComX, are ultimately capable of degrading this signaling peptide. This work brings novel information on regulation of exoproteases in B. subtilis floating biofilms and reveals that these proteolytic enzymes degrade the AI signaling peptide ComX, which is also a major determinant of their expression in biofilms.
Keywords: cell signaling, protease, quorum quenching, biofilms, degradative enzymes, quorum sensing, auto inducing signal, pellicles
Introduction
Auto-inducing (AI) signaling provides the means to integrate information on cell density, mass transfer and other environmental parameters by sensing secreted signaling molecules (Redfield, 2002; Bassler and Losick, 2006; Hense and Schuster, 2015). AI is especially relevant in biofilms, where cell density is very high (Nadell et al., 2008; Hense et al., 2012; Estrela and Brown, 2013). Signaling molecules bind to specific receptors, which then induce transcription and consequently the synthesis of beneficial and secreted products termed public goods (e.g., proteases) (Brown and Taddei, 2007; Diggle et al., 2007; Hense and Schuster, 2015; Schuster et al., 2017). The AI regulation of extracellular proteases (exoproteases) has been most extensively studied in Gram-negative bacteria, especially Pseudomonas aeruginosa (Brint and Ohman, 1995; Redfield, 2002; Hense et al., 2007; Hense and Schuster, 2015). These bacteria usually use lactonases for self-degradation of AI signals, acyl homo-serine lactones (Fekete et al., 2010; Terwagne et al., 2013).
In Gram-positive bacteria AI systems employ peptide pheromones as signaling molecules (Kleerebezem et al., 1997), which induce many adaptive processes at critical cell concentrations including the production of exoprotease in Staphylococcus aureus (Tegmark et al., 1998; Boles and Horswill, 2008); or in Bacillus subtilis (Msadek et al., 1991; Stanley and Lazazzera, 2005). In B. subtilis, the comQXPA gene cluster encodes the major peptide based AI system (Weinrauch et al., 1990), which is wide spread in the phylum Firmicutes (Dogsa et al., 2014). Existing studies focus on the regulatory role of ComX in the expression of the srf operon, responsible for the synthesis of surfactin (lipopetide antibiotic) and on the development of genetic competence for transformation (Nakano et al., 1991; D’Souza et al., 1994; Lazazzera et al., 1997; Ansaldi et al., 2002; Stefanic and Mandic-Mulec, 2009; Oslizlo et al., 2014, 2015; Stefanic et al., 2015; Aleti et al., 2016; Pollak et al., 2016). The observations that the former traits are regulated mostly in a cell density dependent manner (Ansaldi et al., 2002; Oslizlo et al., 2015) made the ComQXPA system widely known as the quorum sensing (QS) system of B. subtilis. In the ComQXPA system, the activity of the histidine kinase response regulator pair, ComP – ComA, is modulated by a signaling peptide, ComX (Magnuson et al., 1994; Ansaldi et al., 2002; Okada et al., 2005). ComX is modified by the isoprenyl transferase ComQ (Tortosa et al., 2001; Ansaldi et al., 2002; Schneider et al., 2002; Okada et al., 2005). Extracellular accumulation of the modified ComX leads to phosphorylation of ComA and subsequent induction of the ComA regulon (Ogura et al., 2001; Comella and Grossman, 2005). The degQ gene is also part of this regulon (Msadek et al., 1991; Stanley and Lazazzera, 2005). DegQ enhances phosphorylation of the response regulator DegU (Kobayashi, 2007) by DegS (Dahl et al., 1992; Jers et al., 2011). The level of phosphorylated DegU is a key information needed for proper expression of the DegU regulon (Murray et al., 2009). High DegU-P positively regulates production of extracellular enzymes including exoproteases (Verhamme et al., 2007; Veening et al., 2008). The aprE gene encodes the major serine exoprotease of B. subtilis that is under direct DegU-P control (Kawamura and Doi, 1984; Veening et al., 2008). AprE together with the metalloprotease NprE accounts for 95% of all extracellular proteolytic activity in B. subtilis (Kawamura and Doi, 1984), which are also under negative control of several repressors (Barbieri et al., 2016). B. subtilis encodes four other minor exoproteases that are either metalloproteases or serine type proteases (Wu et al., 1991). Dependence of DegQ synthesis on ComA (Msadek et al., 1991) strongly suggests that ComX indirectly controls aprE transcription. Additionally, aprE is known to be expressed in biofilms (Marlow et al., 2014) and therefore it is of interest to study the link between the ComQXPA system and proteases in a biofilm setting, which has not been attempted yet to our knowledge.
Overall, we still lack a deeper understanding of the importance of the ComQXPA system in exoprotease production in B. subtilis. Therefore, the first part of this study will be concerned in establishing that ComX is indeed crucial for the production of exoproteases. Furthermore, since ComX is a signaling peptide, it should be susceptible to degradation in highly proteolytic environments. Therefore, we investigate this question in the second part of our study, by testing in vitro degradation of ComX by Bacillus exoproteases.
With the latter points in mind we present the two main hypotheses:
-
simple (1)
Exoprotease production in B. subtilis populations with a non-functioning ComQXPA system will be severely diminished, because ComX plays a major role in the induction of exoprotease production.
-
simple (2)
ComX will be degraded by the induced exoproteases, because as a peptide it should be subject to degradation in highly proteolytic environments.
The results section is organized in two parts, in the first part we present evidence supporting the first hypothesis, and in the second part we present evidence supporting the second hypothesis.
Materials and Methods
Bacterial Strains, Growth Media, and Growth Conditions
Bacterial strains used in this study are listed in Table 1. Overnight cultures were incubated at 37°C and shaken at 200 rpm in LB medium with the appropriate antibiotics. To express heterologous ComX, a 2% (v/v) of overnight culture of Escherichia coli ED367 grown in LB medium supplemented with 100 μg/ml ampicillin was inoculated into the fresh M9 minimal medium (Ansaldi et al., 2002). Cells were grown to OD650 of ∼0.7 a.u. at 37°C and 200 rpm. Then IPTG was added in final concentration 0.4 mM. Cells were incubated further for 4 h at 200 rpm and 37°C and then centrifuged at 8,000 g for 10 min. M9 spent medium was sterilized through filters with 0.2 μm pores and stored at 4°C until use.
Table 1.
Strains used in this study.
| Strain name | Background | Genome description | Reference |
|---|---|---|---|
| Bacillus subtilis strains | |||
| PS-31 | Undomesticated strain | Stefanic and Mandic-Mulec, 2009 | |
| PS-53 | Undomesticated strain | Stefanic and Mandic-Mulec, 2009 | |
| PS-196 | Undomesticated strain | Stefanic and Mandic-Mulec, 2009 | |
| PS-216 | Undomesticated strain | Stefanic and Mandic-Mulec, 2009 | |
| PS-218 | Undomesticated strain | Stefanic and Mandic-Mulec, 2009 | |
| BD2876 | 168 | his leu met srfA-lacZ (tet) comQ::kan | Tortosa et al., 2001 |
| BD2962 | 168 | his met srfA-lacZ (tet) amyE::xylR Pxyl-comK (cat) (comQ::pED345 comX comP replaced by genes from B. mojavensis RO-H-1) | Tortosa et al., 2001 |
| BD3019 | 168 | his leu met srfA-lacZ (tet) amyE::xylR Pxyl-comK (cat) (comQ::pED375 comX comP replaced by genes from B. subtilis RS-D-)2 | Ansaldi et al., 2002 |
| BM1289 | PS-31 | comQ::pED345 (spec) | This work |
| BM1290 | PS-53 | comQ::pED345 (spec) | This work |
| BM1291 | PS-196 | comQ::pED375 (spec) | This work |
| BM1292 | PS-218 | comQ::pED375 (spec) | This work |
| BM1400 | PS-216 | comQ::kan | This work |
| BM1402 | PS-216 | comP::cat | Oslizlo et al., 2014 |
| BM1127 | PS-216 | ΔcomQ marker less | This work |
| BM1133 | PS-216 | degQ::tet | This work |
| BM1445 | PS-216 | ΔcomQ degQ::tet | This work |
| BM1142 | PS-216 | PaprE-gfp (cm) | This work |
| BM1144 | PS-216 | degQ::tet PaprE-gfp (cm) | This work |
| O8G57 | 168 | PaprE-gfp (cm) | Veening et al., 2008 |
| BM1443 | PS-216 | ΔcomQ PaprE-gfp (cm) | This work |
| BM1448 | PS-216 | ΔcomQ degQ::tet PaprE-gfp (cm) | This work |
| DL722 | 3610 | amyE::PsrfAA-yfp (spec) | López et al., 2009 |
| BM1456 | PS-216 | comQ::kan amyE::PsrfAA-yfp (spec) | This work |
| BD7123 | 168 | degQ::tet PcomGA-luc (cm) | Miras and Dubnau, 2016 |
| Escherichia coli strains | |||
| ED367 | BL21 (DE3) | pET22(b) – comQ comX from B. subtilis 168 (amp) | Ansaldi et al., 2002 |
| DE553 | pMiniMAD2 oriBsTs amp mls | Parashar et al., 2013 | |
To prepare B. subtilis spores, the overnight culture [1% (v/v)] was inoculated into sporulation medium (SM) (Warriner and Waites, 1999). After 5 days of incubation, the culture was exposed to 80°C for 30 min and then centrifuged at 10,000 g for 10 min. The pellet was re-suspended and washed three times with physiological saline. Before freezing at -20°C 10% (v/v) glycerol was added to the spore suspension. Spores were enumerated using the MPN method (data not shown). A 1% (v/v) solution of triphenyltetrazolium chloride (TTC; BioLife, Italy) was sterilized through filters with 0.2 μm pores. 180 μl of the liquid LB medium with TTC [0.01% (v/v) final concentration] was dispensed in standard 96-well sterile microtiter plates. A 10-fold serial dilution of the spore suspension was prepared in microtiter plates with eight technical replicates for each 10-fold dilution down to 10-11. Microtiter plates were then incubated at 37°C overnight. Positive wells were identified by red color development due to the bacterial growth. Spore suspensions with MPN count in the 108 MPN/mL range were used in the experiments.
To measure gene expression in floating biofilms (pellicles) a spore suspension 1% (v/v) was inoculated into the liquid MSgg medium. In some experiments the MSgg medium was supplemented with 20% (v/v) of the spent medium containing ComX, which was heterologously produced by E. coli ED367 (Ansaldi et al., 2002). The spent M9 medium of E. coli ED367 that was not induced by IPTG was used as a negative control.
To harvest pellicle spent media 1% (v/v) of B. subtilis spores were inoculated in 4 ml of MSgg medium (Branda et al., 2001), in some cases complemented with 20% (v/v) spent M9 minimal medium and incubated in sterile 12-well microtiter plates in static conditions at 37°C.
Strain Construction
Construction of the mutant strains was performed by transformation of specific markers into competent B. subtilis strains grown in competence medium (CM) at 37°C (Albano et al., 1987). Antibiotic selections were carried out on LB agar plates at 37°C containing chloramphenicol (Cm) 5 μg/ml, kanamycin (Kan) 50 μg/ml, spectinomycin (Spec) 100 μg/ml, tetracycline (Tet) 10 μg/ml, erythromycin 0.5 μg/ml, and lincomycin 12.5 μg/ml (mls). The comQ::spec mutants were constructed by transforming the B. subtilis PS-31, PS-53 with the DNA isolated from the strain BD2962 (Tortosa et al., 2001) and by transforming PS-196 and PS-218 with the DNA isolated from the strain BD3019 (Ansaldi et al., 2002). The comQ::kan mutant (BM1400) was constructed by transforming the B. subtilis PS-216 with the DNA isolated from the strain BD2876 (Tortosa et al., 2001). The degQ::tet mutant was constructed by transforming the genomic DNA from the strain BD7123 (Miras and Dubnau, 2016) to appropriate PS-216 strains. Mutants with PaprE-gfp were constructed by transforming O8G57 genomic DNA (Veening et al., 2008) into appropriate PS-216 strains. When constructing the amyE::PsrfAA-yfp mutants, the genomic DNA from the strain DL722 (López et al., 2009) was transformed into the strain BM1400. When transforming DNA into comQ mutants, the competence was achieved by the addition of exogenous ComX in the form of spent medium [5% (v/v) of the E. coli ED367 M9, which was grown in the presence of IPTG].
To construct the ΔcomQ marker less deletion strain, the region upstream of the comQ gene was PCR amplified using the primer pair Up-F/Up-R (Table 2) and digested with EcoRI and BamHI. Also the region downstream of comQ gene was PCR amplified using the primer pair Down-F/Down-R (Table 2) and digested with BamHI and SalI. The two fragments were then simultaneously ligated into the EcoRI and SalI sites of pMiniMAD2 (Parashar et al., 2013), which carries a temperature-sensitive origin of replication and an erythromycin resistance cassette to generate pMiniMAD2-updowncomQ. The constructed plasmid was transformed into B. subtilis PS-216 at the restrictive temperature for plasmid replication (37°C) using 0.5 μg/ml erythromycin and 12.5 μg/ml lincomycin (mls) as a selection. To evict the plasmid, the strain was harvested according to an established protocol (Patrick and Kearns, 2008). Chromosomal DNA from colonies that had excised the plasmid was isolated and screened by PCR using primers Up-F/Down-R to determine which isolates carried a deletion in comQ gene.
Table 2.
Oligonucleotides used in this study.
| Name | Sequence 5′–3′ |
|---|---|
| Up-F | CCGGAATTCATGACAAAGCGAAAAGGCCAC |
| Up-R | CGCGGATCCCTCCTTCATTTTCTCCTTGATCCGGAC |
| Down-F | CGCGGATCCACAAGATGCAAGACCTAATTAACTAC |
| Down-R | ACGCGTCGACCCTATTTCTCCAAGGTATCTTTGTATA |
Estimation of the Proteolytic Activity Produced by a B. subtilis Colony
Skim milk powder 20% (w/v) was reconstituted with distilled water and autoclaved at 110°C. An agar solution 3% (w/v) was also prepared and autoclaved at 121°C. After autoclaving, the suspensions were carefully mixed in a 1:1 ratio. Finally 20 ml of the mixture was poured into 9 cm diameter Petri dishes, yielding 10% skim milk (w/v) and 1.5% (w/v) agar. For overnight cultures B. subtilis strains were incubated in LB medium supplemented with appropriate antibiotics at 37°C and 200 rpm. Cultures were then diluted 100-fold in physiological saline and a 5 μl droplet of diluted culture was surface spotted in the center of the skim milk agar plate. Photos of the proteolytic clearing zones were taken after 16 h of incubation at 37°C.
Preparation of Casein-Gelatin Plates
To prepare casein gelatin agar plates 1% (w/v) casein sodium salt from bovine milk (Sigma–Aldrich, United States) and 1% (w/v) gelatin from porcine skin (Sigma–Aldrich, United States) were thoroughly dissolved in 0.02 M NaOH and the pH was equilibrated to 7 ± 0.2 as described by Montville (1983). Wherever, the proteolytic inhibition by ethylenediaminetetraacetic acid (EDTA) was tested, EDTA was added to the casein gelatin medium in 1 mM final concentration. Finally, 1.5% (w/v) of agar was added to the casein gelatin medium. The casein gelatin medium was autoclaved at 110°C. 40 ml of the medium was poured into Petri dishes with 9 cm diameter. Wells (6 mm diameter) were cut into the casein gelatin agar using an agar punch cutter.
Determination of Proteolytic Activity in Spent Media of Floating Biofilms
Proteolytic activity was determined in B. subtilis spent medium at different time points. The spent medium below the biofilm was centrifuged for 5 min at 8000 g to remove remaining planktonic cells and sterilized through filters with 0.2 μm pores. Spent medium was diluted in 10 mM sodium acetate buffer with 5 mM calcium acetate (pH 7.5). If EDTA was added to experimental samples, spent medium was diluted in PBS buffer (10 mM sodium phosphate dibasic, 1 mM potassium phosphate monobasic, 137 mM NaCl, and 2.7 mM KCl pH 7). The diluted spent medium (100 μl) was dispensed into the wells punched into the casein gelatin agar. The plates were incubated for 16 h at 37°C when the proteolytic zones were measured. To estimate the proteolytic activity of spent media in U/ml of subtilisin, the proteolytic zone diameters of the spent media were compared to the proteolytic zone diameters obtained by different concentrations of commercial subtilisin (Sigma–Aldrich). One such subtilisin dose-response curve is shown in Supplementary Figure 1. The subtilisin concentration equivalents were displayed as units where 1 unit of subtilisin hydrolyzes casein to produce color equivalent to 1.0 μmol (181 μg) of tyrosine per minute at pH 7.5 at 37°C using Folin-Ciocalteu reagent. According to Sigma–Aldrich and to our in-house control of subtilisin proteolytic activity following the Sigma’s protease activity assay (Cupp-Enyard, 2008), 1 mg of subtilisin corresponds to roughly 13 units. Protease activity estimation was done routinely after storage of subtilisin for over 6 months at -20°C, to determine that the storage conditions did not lower the subtilisin standard proteolytic activity.
Expression of PaprE-gfp during Formation of Floating Biofilm
Briefly, 200 μl aliquots of inoculated MSgg medium, sometimes supplemented with M9 spent medium were dispensed in a sterile 96-well black transparent bottom microtiter plate in four technical replicates. The lid was sealed with micropore tape and the space between wells was filled with sterile distilled water to minimize the effect of medium evaporation. The microtiter plate was incubated in the Cytation 3 imaging reader (BioTek, United States) at 37°C without shaking. Optical density at 650 nm and fluorescence intensity were measured in half hour intervals for up to 60 h. Fluorescence intensity of GFP (green fluorescent protein) was used to monitor PaprE-gfp expression with excitation at 480 nm and emission at 510 nm. The gain was set on 50. In a parallel experimental setup the same strains without the fluorescent marker were always cultured. To calculate the final expression the autofluorescence of unmarked strains was deducted from the fluorescence of the marked strains. Fluorescence intensity was normalized per OD650 of fluorescently labeled strains at each time point.
The ComX Biological Activity Assay
The ComX biological activity was quantified in spent media of various B. subtilis strains harvested after 36 and 48 h of static growth, using biosensor strains: B. subtilis BM1456 and BM1400. We chose the 48 h time point because then all strains have comparable floating biofilm (pellicle) thickness, estimated by measuring OD650 (Supplementary Figure 2B). The BM1400 strain only served to determine background fluorescence, since it carries no fluorescent marker. Both biosensor strains do not produce their own ComX. The BM1456 biosenor responds to ComX in the spent medium by inducing the PsrfAA-yfp reporter (Oslizlo et al., 2014). Since the induction of PsrfAA is proportional to the quantity of ComX (Ansaldi et al., 2002) we used this assay to estimate the quantity of biologically active ComX in the spent media of various pellicles.
Both biosensor strains were grown in 5 ml of CM medium supplemented with 1% (v/v) of filtered spent media of selected strains for 6 h at 37°C with shaking (200 rpm). After 6 h of incubation 200 μl aliquots were dispensed in a sterile 96-well black transparent bottom microtiter plate. The PsrfAA-yfp expression was quantified by YFP (yellow fluorescent protein) fluorescence intensity. YFP was excited at 510 nm and emissions were measured at 530 nm. The gain was set to 100. Each PsrfAA-yfp reading was normalized to OD650. The normalized autofluorescence background of BM1400 grown in the same experimental conditions was subtracted. Additionally, we subtracted the PsrfAA-yfp expression of cells exposed to ΔcomQ spent medium (lacks ComX), to account for ComX independent YFP expression.
ComX biological activity was determined in the spent media immediately after harvest (T0) and after a 24 h (T24) incubation at 37°C. We reasoned that if exoproteases in the spent medium degrade ComX the ComX biological activity at T0 will be greater than at T24. To calculate the ComX biological activity decay we used the following equation:
The same approach was used to determine ComX biological activity decay in the spent media supplemented with subtilisin (0.1 mg/ml) or metalloprotease inhibitor EDTA (1 mM) after harvesting or to estimate the effect of commercially available subtilisin on ComX heterologously produced in E. coli ED367.
HPLC Purification of Exoprotease Treated Spent Media
ComX was purified as described previously (Ansaldi et al., 2002; Oslizlo et al., 2014). Briefly, 5 ml of ED367 spent medium was acidified and separated with a C-18 reverse phase HPLC column. ComX presence was determined by measuring the absorbance at 214 nm, where it appears as two chromatographic peaks, both of which exhibit biological activity (Oslizlo et al., 2014). The medium was purified after adding subtilisin (0.1 mg/ml) and incubating it for 24 h at 37°C. To ensure heterologous ComX stability over time bovine serum albumin (BSA, 50 μg/ml) was added to the spent media prior to tests. The resulting chromatogram was compared to a control experiment, where subtilisin was not added. To control for the presence of other signals potentially produced by E. coli ED367, the spent medium of the non-induced E. coli was subjected to the same procedure.
Statistical Analysis
All results were statistically analyzed using the non-parametric Mann–Whitney U-test. The threshold level for accepting significant differences was p = 0.05. All of the error bars show the standard error of mean (SEM).
Results
ComX Induces Exoproteases
The Regulation of PaprE-gfp Expression in Floating Biofilms Is ComQ and DegQ Dependent
Previous work suggested that ComA positively regulates expression of degQ that is needed for exoprotease production (Msadek et al., 1991). Furthermore, the DegU system was shown to be important for exoprotease activity during biofilm growth (Verhamme et al., 2007). Based on this knowledge we predicted that disruptions of comQ, required for ComX maturation, will cause a decrease in the transcriptional activity of the major protease gene in B. subtilis pellicles grown in MSgg medium.
In order to test this hypothesis, we monitored the population expression of PaprE-gfp of the wild type PS-216 strain and ΔcomQ mutant grown as floating biofilms. PaprE-gfp expression is indeed higher in the wt strain compared to the ΔcomQ mutant at all time points (Figure 1A). This is very similar to the PaprE-gfp expression in the degQ::tet mutant and ΔcomQ degQ::tet double mutant. In all tested mutants PaprE-gfp expression is very low as compared to the expression in the wt strain (Supplementary Figure 2A). These results support the hypothesis 1 that ComQXPA AI system contributes to regulation of exoproteases. Also, expression of PaprE-gfp in the wt strain reaches maximal levels at 36 h of incubation (Figure 1A and Supplementary Figure 2A). This is the approximate time point where OD650 of wt strain and all mutant floating biofilms are comparable, despite their difference in surface morphology (Supplementary Figures 2B, 3). Nevertheless, PaprE-gfp expression remains significantly higher in the wt strain than in ΔcomQ or degQ::tet mutants at early time points and even after 60 h of floating biofilm growth.
FIGURE 1.
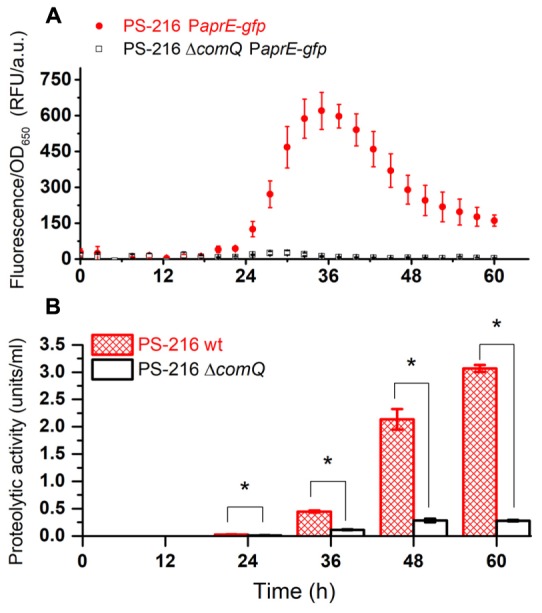
PaprE-gfp expression of floating biofilms grown in MSgg medium (A) and the proteolytic activity of harvested floating biofilm spent media (B). (A) PaprE-gfp expression of floating biofilms grown in MSgg media was evaluated by measuring fluorescence. Measurements were performed every half an hour (only every fifth measured data point is shown for clarity). Averages and SEM (standard error of means) of four biological replicates are shown. (B) The proteolytic activity of floating biofilm spent media grown in MSgg media at different time points. Averages and SEM of three biological replicates are shown. A Mann–Whitney U-test was performed to determine statistical significance of discussed differences (∗p < 0.05).
The Proteolytic Activity during Floating Biofilm Growth Is ComQ and DegQ Dependent
Then we measured the overall protease activity in the wt and ΔcomQ mutant floating biofilm spent media (Figure 1B). Again, the exoprotease activity in the spent medium of the ΔcomQ mutant is drastically lower than in the wt strain at all the measured time points. A significantly higher proteolytic activity of the wt strain than of the ΔcomQ mutant spent medium is detectable already at 24 h and the difference increases with time. However, PaprE-gfp expression and proteolytic activity (Figure 1) do not correlate, especially after the 36 h incubation time point, but results further corroborate, that proteolytic activity in floating biofilm spent medium is ComQ dependent.
This conclusion is further supported by experiments where we addressed the role of ComQ and DegQ in production of exoprotease during growth on skim milk agar. As predicted, wt colonies produced clearly visible proteolytic zones which were hardly visible in ΔcomQ, degQ::tet and ΔcomQ degQ::tet mutants (Figure 2). Also, different wt soil isolates of B. subtilis (PS-31, PS-53, PS-196, and PS-218) produce strong clearing zones around colonies grown on skim milk agar but lack them in ΔcomQ isogenic mutants (Supplementary Figure 4). Therefore, the results support predictions made in the first hypothesis and reinforce the assumption of the importance of ComX in positive regulation of exoprotease production.
FIGURE 2.
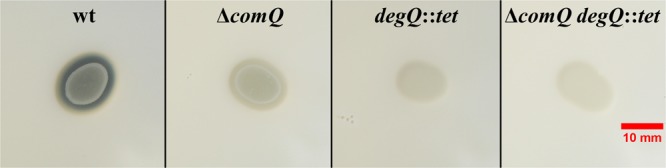
Proteolytic activity of Bacillus subtilis PS-216 wild type and ΔcomQ, degQ::tet, and ΔcomQ degQ::tet mutants on skim milk agar. Photos were taken on a dark blue background after 16 h of incubation at 37°C. Note the clearing zone around the wt colony, indicating the proteolysis of skim milk. Colonies of the mutant strains did not degrade skim milk as much and thus the clearing zones are less apparent.
ComX Complements the Defect in PaprE-gfp Expression and Protease Activity in the ΔcomQ Mutant
Next, we tested whether addition of ComX complements PaprE-gfp expression in the ΔcomQ mutant. We added ComX in the form of a spent M9 minimal medium [20% (v/v)] of E. coli ED367, which heterologously produces ComX upon induction with IPTG. The M9 spent medium without IPTG induction was used as a control. Thus growth conditions for the complementation assay with M9 spent medium were slightly different as for results presented in Figure 1 and Supplementary Figure 2. For example, OD650 measurements indicate that in MSgg + M9 medium floating biofilms start forming after 10 h (Supplementary Figure 5), while in MSgg medium this occurs after 20 h (Supplementary Figure 2B). This may explain different values for PaprE-gfp expression in MSgg + M9 medium (Figures 3A,B) compared to the ones presented in Figure 1A. Nevertheless, the addition of ComX complements PaprE-gfp expression and protease activity in the ΔcomQ mutant (Figures 3A,B) and even restores the ΔcomQ mutant floating biofilm morphology to the wt morphology at 24 h (Supplementary Figure 6). Although complementation was partial for PaprE-gfp expression, the spent media proteolytic activity at 24 h was also complemented by ComX (Figures 3C,D). In contrast and as expected, ComX does not complement PaprE-gfp expression in degQ::tet mutant and ΔcomQ degQ::tet double mutant (Supplementary Figure 7), which is consistent with published results indicating that ComX works upstream of degQ (Msadek et al., 1991).
FIGURE 3.
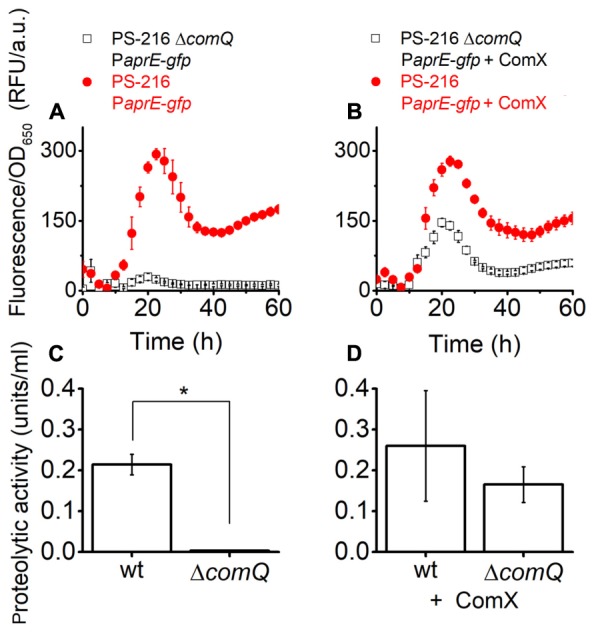
PaprE-gfp expression (A,B) and the proteolytic activity (C,D) of 24 h floating biofilm spent media of strains grown in MSgg medium complemented with spent M9 minimal medium. Cells were grown in MSgg medium with the addition of M9 Escherichia coli ED367 spent medium [20% (v/v)]. (A,C) Were complemented with spent M9 medium where ED367 heterologous expression of ComX was not induced with IPTG and therefore the spent medium lacked ComX. (B,D) Were complemented with M9 spent medium, where heterologous expression of ComX in ED367 was induced with IPTG, and therefore the spent medium contained heterologous ComX. Measurements on (A,B) were made every half an hour, only every fifth measured data point is shown for clarity. (C,D) Show the proteolytic activity of spent medium from floating biofilms of different PS-216 strains after 24 h growth in the same media as on (A,B) but in 12-well microtiter plates. A Mann–Whitney U-test was performed to determine statistical significance of discussed differences (∗p < 0.05). Averages and SEM (standard error of means) of three independent biological replicates are shown.
Exoproteases Degrade ComX
Exoproteases in the Floating Biofilm Spent Media Contribute to the ComX Biological Activity Decay
ComX is a small peptide (Magnuson et al., 1994; Ansaldi et al., 2002), which implies that it might be susceptible to proteolytic degradation. If this is the case, we expect that the ComX biological activity decay will be high in exoprotease rich spent media of floating biofilms that produce exoproteases but low in the spent media of mutants that have low exoprotease activity (hypothesis 2).
Floating biofilms of different strains were grown in 12-well microtiter plates and imaged at different time points. At 36 and 48 h the ΔcomQ and comP::cat mutants form floating biofilms with a more structured surface than the wt strain and degQ::tet mutant, however, floating biofilm biomass appears similar (Supplementary Figure 3). Therefore, we chose these two time points (36 and 48 h) to measure the proteolytic activity and the ComX biological activity decay in the spent media of floating biofilms. Results show that at both time points proteolytic activity and ComX biological activity decay is high in spent media of the wt strain, but low in the spent media of the mutant strains (Figure 4 and Supplementary Figure 8). This supports the second hypothesis. Furthermore, metalloprotease specific inhibitor EDTA (Ellaiah et al., 2002), added to the spent medium of the wt strain decreases the proteolytic activity (Figure 4A) and consequently the ComX biological activity decay (Figure 4B). This suggests that metaloproteases influence ComX stability. Finally, we tested whether subtilisin, which is the major serine protease produced by B. subtilis (Kawamura and Doi, 1984), can also degrade ComX. We added exogenous commercially available subtilisin to spent media harvested from comP::cat or degQ::tet mutants, which have otherwise very low proteolytic activities. Subtilisin increased proteolytic activity of mutant’s spent media to the levels of the wt strain (Figure 4A) and consequently also the ComX biological activity decay (Figure 4B). This supports the prediction that subtilisin, which expression is under ComX control, also modulates the ComX biological activity decay.
FIGURE 4.
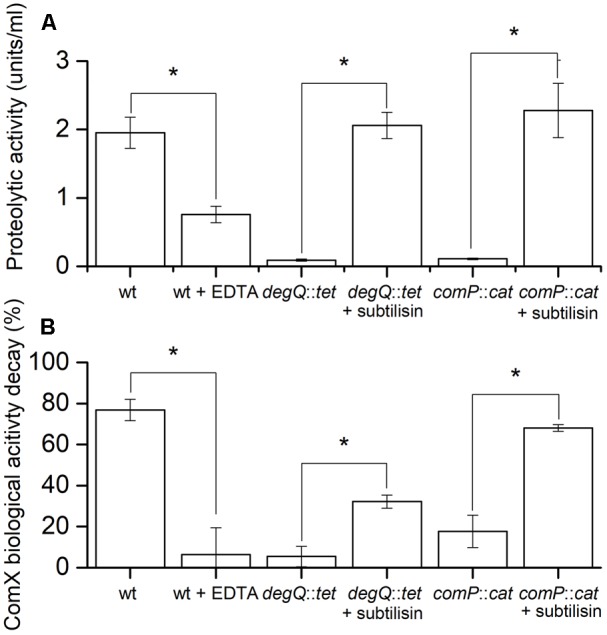
Comparisons of the proteolytic activity (A) and ComX biological activity decay (B) of different B. subtilis 48 h floating biofilm spent media. (A) Proteolytic activity of the spent medium from floating biofilms. Different B. subtilis PS-216 strains (wt, degQ:tet, comP::cat) were grown for 48 h at 37°C in MSgg medium. Where indicated, subtilisin or EDTA was added to spent media after harvest in order to increase or inhibit the proteolytic activity of the media, respectively. Averages and SEM of three independent biological replicates are shown. A Mann–Whitney U-test was performed to determine statistical significance of discussed measurements (∗p < 0.05). (B) The ComX biological activity decay of the same harvested floating biofilm spent medium as above using a signal deficient biosensor strain BM1456. Averages and SEM of three independent biological replicates are shown. A Mann–Whitney U-test was performed to determine statistical significance of discussed measurements (∗p < 0.05).
Summing up the results presented in Figure 4 and Supplementary Figure 8, we conclude that the ComX biological activity decay in spent medium (i) coincides with the proteolytic activity of the media; (ii) is inhibited by metalloprotease inhibitor EDTA and (iii) is promoted by subtilisin. This speaks strongly in favor of our second hypothesis and implies that both, metalloproteases and subtilisin, degrade ComX.
The Protease Subtilisin Is Sufficient to Degrade ComX
One might still argue, that exoproteases degrade ComX indirectly through some unknown intermediate factor, present in the native B. subtilis floating biofilm spent media. Therefore, to provide more direct experimental evidence of ComX degradation by exoproteases, we treated the E. coli ED367 M9 spent medium that contains heterologously expressed ComX with subtilisin (Figure 5). In the spent M9 minimal media from E. coli, such a confounding factor is less likely to be present. To test this, we incubated spent M9 media with heterologous ComX with or without subtilisin (0.1 mg/ml) for 24 h at 37°C. The results confirm that subtilisin increases the decay of the ComX biological activity as compared to the subtilisin untreated control (Figure 5A). Finally, one might argue that in all cases above no degradation of ComX occurs, only a modulation of the sensitivity of the ComX biosensor assay, since this is the only measure we used in order to quantify ComX so far. Therefore, in order to provide more direct proof of ComX degradation, we present the HPLC chromatograms of E. coli M9 spent media with heterologously expressed ComX, which were either treated with subtilisin or not (Figure 5B). We see that the chromatogram of the M9 minimal spent medium with ComX treated with subtilisin lacks the two distinct peaks (Figure 5B), which are indicative of ComX (Oslizlo et al., 2014). These two peaks are visible on the chromatograms of the M9 spent medium containing heterologous ComX induced by IPTG, but untreated with subtilisin. The chromatograms of additional negative controls, namely the ED367 spent media that were not induced by IPTG also lack the two ComX indicative peaks.
FIGURE 5.
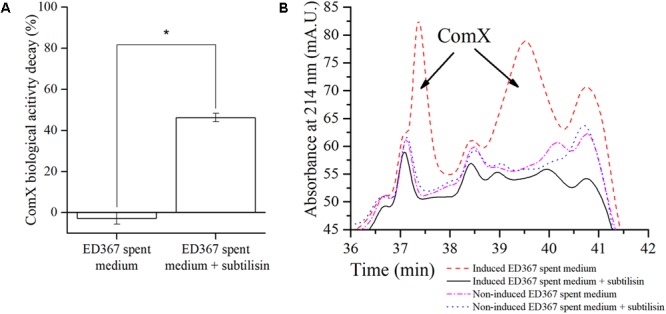
ED367 heterologous ComX biological activity decay (A) and the purification of heterologous ComX on HPLC (B). (A) The ComX biological activity decay of the E. coli ED367 spent medium containing heterologously expressed ComX, using a signal deficient biosensor strain BM1456 with a PsrfAA-yfp fluorescent reporter fusion. The spent medium was incubated for 24 h at 37°C with and without subtilisin. When adding subtilisin, the ComX biological activity decay increases. Averages and SEM of three independent biological replicates are shown. A Mann–Whitney U-test was performed to determine statistical significance of discussed measurements (∗p < 0.05). (B) Chromatograms of the E. coli ED367 M9 spent media that were incubated for 24 h at 37°C with and without subtilisin. After treating the medium with subtilisin, the two peaks containing ComX are no longer present in the medium that contained heterologously expressed ComX. As a control a parallel experiments were made where non-IPTG induced E. coli ED367 spent medium was incubated for 24 h at 37°C with and without subtilisin.
Overall these results speak strongly in favor of our second hypothesis and show that native exoproteases, which are according to hypothesis 1 induced by ComX, create a proteolytic environment that leads to the degradation of this AI signaling peptide.
Discussion
We here report that the ComQXPA AI system of B. subtilis positively controls the transcription of the subtilisin gene aprE and exoprotease production during biofilm growth at liquid–air interface or on agar surface. Moreover, we provide evidence, that exoproteases, which are induced by ComX in a DegQ dependent manner, can degrade ComX AI signaling peptide (Figure 6).
FIGURE 6.
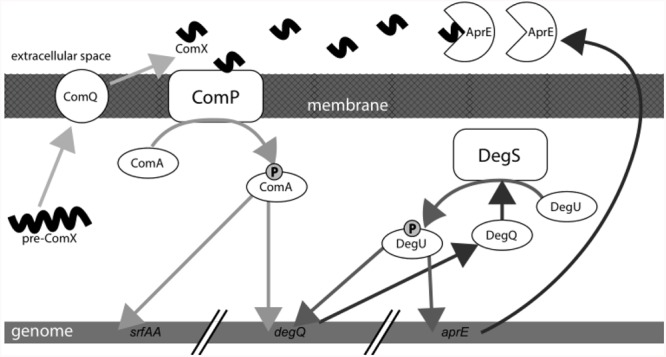
Schematic presentation of the quorum sensing (QS) ComQXPA system. ComX induces exoprotease production. Exoproteases ultimately degrade ComX in the extracellular milieu.
Many bacterial auto-inducing systems regulate the production of exoproteases (Diggle et al., 2007; Hense and Schuster, 2015; Schuster et al., 2017) and this regulation has mostly been studied in Gram-negative Pseudomonas aeruginosa (Brint and Ohman, 1995), but also in Gram-positive Staphylococcus aureus (Tegmark et al., 1998). We know less how AI systems regulate exoproteases in Gram-positive model organism, B. subtilis. The ComQXPA system has been implicated in expression of DegU regulon previously (Msadek et al., 1991; Stanley and Lazazzera, 2005) but the work was focused on planktonic cultures. We here investigate the role of ComX in exoprotease production during biofilm growth. Results confirm that ComX positively influences PaprE-gfp expression and exoproteases production in the model PS-216 strain and in a set of B. subtilis undomesticated wt strains, that were isolated from soil microscale (Stefanic and Mandic-Mulec, 2009) and differ in their physiology (Stefanic et al., 2012).
The ΔcomQ mutant shows lower PaprE-gfp expression and proteolytic activity in floating biofilms (pellicles) at all time points compared to the wt strain. In this strain PaprE-gfp expression, but not proteolytic activity, starts decreasing in intensity after 36 h (Figure 1A). This drop is unexpected, because fluorescence measurements of promoter labeled strains usually indicate cumulative transcription (Miras and Dubnau, 2016). We also observe that the ΔcomQ mutant forms pellicles at a faster rate than the wt strain up to 36 h (Supplementary Figure 2B). As the ΔcomQ mutant is expected to have low levels of DegU-P leading to a more active primary metabolism (Tanaka et al., 2015) and faster growth. In addition, we note that after 36 h floating biofilms show indication of brown coloration (Supplementary Figure 3) indicative of sporulation (Koo et al., 2017). Changed physiological conditions may affect the stability of GFP. Nevertheless, this discrepancy does not compromise our main conclusion of the ComX AI system playing a significant positive role in expression/activity of exoproteases.
To our knowledge, this work also provides the first evidence that AI regulated exoproteases degrade the peptide auto-inducer ComX in B. subtilis. This is evident through experiments in vitro in which we show that ComX biological activity decay correlates with proteolytic activity of spent media. Both, serine proteases (e.g., subtilisin) and metalloproteases influence the ComX biological activity decay. Gram-negative bacteria use AHLs (acyl homoserine lactones) auto-inducing molecules for QS (Papenfort and Bassler, 2016). Quorum quenchers, like lactonases, degrade the signaling molecules in bacteria and often act as weapons in intra-species competition (Kalia, 2013; Hense and Schuster, 2015). Also in Gram-negative bacteria auto-degradation of AHLs is not well-understood. In Brucella melitensis a protein was identified, which has a potential for AHL degradation (Terwagne et al., 2013). Similarly, Pseudomonas putida accumulates AHL degradation by products during growth, indicating potential AHL degradation (Fekete et al., 2010). However, these findings, like ours for B. subtilis, show only the degradation of the AI molecule. The effect of such degradation on the microbial physiology is, to our knowledge, unknown.
Future experiments will show whether the self-directed AI signal degrading strategy is potentially a negative feedback regulatory loop of the ComQXPA auto-inducing response. The fact that the two most studied ComQXPA regulated traits, namely competence and surfactant production (Nakano et al., 1991; D’Souza et al., 1994; Lazazzera et al., 1997; Ansaldi et al., 2002; Stefanic and Mandic-Mulec, 2009; Oslizlo et al., 2014, 2015; Stefanic et al., 2015; Aleti et al., 2016; Pollak et al., 2016), are regulated in a cell density dependent manner (Ansaldi et al., 2002; Oslizlo et al., 2015) made the ComQXPA system widely known as the QS system of B. subtilis. However, if the proteolytic degradation of ComX has physiological consequences then this would imply that these traits may not be regulated in density dependent manner in conditions that support synthesis of exoproteases. This provides further support for the validity of the efficiency sensing theory (Hense et al., 2007; Hense and Schuster, 2015). This work describes two different types of regulation that involve the signaling peptide ComX: a positive genetic regulation and a negative biochemical regulation. The latter may represent a negative feedback loop, which could elegantly link the bacterial supply and demand for beneficial public goods. The bacterial population will increase public good supply via ComX. This will also increase proteolytic activity, which will decrease ComX and thus potentially further supply of public goods. If ComX is produced and degraded simultaneously, this may prevent cells to overinvest in protease production, which is more costly than the synthesis of a small peptide. Therefore, cells could potentially obtain information on the demand for exoproteases through ComX but this remains to be demonstrated.
To our knowledge this is the first report which links peptide-based AI system to production of exoproteases and subsequent self-degradation of the signaling molecules. We envision that protease dependent control of signaling peptides may be more wide spread among Gram-positive bacteria and hope that this work will initiate new research in this direction.
Author Contributions
All authors (MS, TD, and IM-M) were involved in experimental design and the writing of the manuscript. MS and TD performed the experiments.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Roberto Kolter’s lab for providing the DL722 strain, Daniel B. Kearns’ lab for providing the pMiniMAD2 plasmid, Oscar Kuipers’ lab for providing the O8G57 strain and Dave Dubnau’s lab for providing all the BD strains. We thank Sam Brown, Nicola Stanley-Wall, and Ákos Kovács for insightful comments on this work. We thank our colleagues working at the Chair of Microbiology for all the discussions and advice; especially our lab technician Simona Leskovec for general assistance. We thank Marija Kralj for help in the graphical design of Figure 6. We thank both reviewers for their insightful comments, which improved the manuscript.
Footnotes
Funding. The authors acknowledge the financial support from the Slovenian Research Agency, namely the program financing no. P4-0116 and the ARRS Young Researcher grant.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00105/full#supplementary-material
References
- Albano M., Hahn J., Dubnau D. (1987). Expression of competence genes in Bacillus subtilis. J. Bacteriol. 169 3110–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleti G., Lehner S., Bacher M., Compant S., Nikolic B., Plesko M., et al. (2016). Surfactin variants mediate species-specific biofilm formation and root colonization in Bacillus. Environ. Microbiol. 18 2634–2645. 10.1111/1462-2920.13405 [DOI] [PubMed] [Google Scholar]
- Ansaldi M., Marolt D., Stebe T., Mandic-Mulec I., Dubnau D. (2002). Specific activation of the Bacillus quorum-sensing systems by isoprenylated pheromone variants. Mol. Microbiol. 44 1561–1573. 10.1046/j.1365-2958.2002.02977.x [DOI] [PubMed] [Google Scholar]
- Barbieri G., Albertini A. M., Ferrari E., Sonenshein A. L., Belitsky B. R. (2016). Interplay of CodY and ScoC in the regulation of major extracellular protease genes of Bacillus subtilis. J. Bacteriol. 198 907–920. 10.1128/JB.00894-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler B. L., Losick R. (2006). Bacterially speaking. Cell 125 237–246. 10.1016/j.cell.2006.04.001 [DOI] [PubMed] [Google Scholar]
- Boles B. R., Horswill A. R. (2008). agr-mediated dispersal of Staphylococcus aureus biofilms. PLOS Pathog. 4:e1000052. 10.1371/journal.ppat.1000052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda S. S., González-Pastor J. E., Ben-Yehuda S., Losick R., Kolter R. (2001). Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 98 11621–11626. 10.1073/pnas.191384198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brint J. M., Ohman D. E. (1995). Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177 7155–7163. 10.1128/JB.177.24.7155-7163.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. P., Taddei F. (2007). The durability of public goods changes the dynamics and nature of social dilemmas. PLOS ONE 2:e593. 10.1371/journal.pone.0000593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comella N., Grossman A. D. (2005). Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol. Microbiol. 57 1159–1174. 10.1111/j.1365-2958.2005.04749.x [DOI] [PubMed] [Google Scholar]
- Cupp-Enyard C. (2008). Sigma’s non-specific protease activity assay-casein as a substrate. J. Vis. Exp. 19:899. 10.3791/899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl M. K., Msadek T., Kunst F., Rapoport G. (1992). The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J. Biol. Chem. 267 14509–14514. [PubMed] [Google Scholar]
- Diggle S. P., Griffin A. S., Campbell G. S., West S. A. (2007). Cooperation and conflict in quorum-sensing bacterial populations. Nature 450 411–414. 10.1038/nature06279 [DOI] [PubMed] [Google Scholar]
- Dogsa I., Choudhary K. S., Marsetic Z., Hudaiberdiev S., Vera R., Pongor S. (2014). ComQXPA quorum sensing systems may not be unique to Bacillus subtilis: a census in prokaryotic genomes. PLOS ONE 9:e96122. 10.1371/journal.pone.0096122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza C., Nakano M. M., Zuber P. (1994). Identification of comS, a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 91 9397–9401. 10.1073/pnas.92.2.646e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellaiah P., Srinivasulu B., Adinarayana K. (2002). A review on microbial alkaline proteases. J. Sci. Ind. Res. 61 690–704. [Google Scholar]
- Estrela S., Brown S. P. (2013). Metabolic and demographic feedbacks shape the emergent spatial structure and function of microbial communities. PLOS Comput. Biol. 9:e1003398. 10.1371/journal.pcbi.1003398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete A., Kuttler C., Rothballer M., Hense B. A., Fischer D., Lucio M., et al. (2010). Dynamic regulation of N-acyl-homoserine lactone production and degradation in Pseudomonas putida IsoF. FEMS Microbiol. Ecol. 72 22–34. 10.1111/j.1574-6941.2009.00828.x [DOI] [PubMed] [Google Scholar]
- Hense B. A., Kuttler C., Müller J., Rothballer M. (2007). Does efficiency sensing unify diffusion and quorum sensing? Nat. Rev. Microbiol. 5 230–239. [DOI] [PubMed] [Google Scholar]
- Hense B. A., Müller J., Kuttler C., Hartmann A. (2012). Spatial heterogeneity of autoinducer regulation systems. Sensors 12 4156–4171. 10.3390/s120404156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hense B. A., Schuster M. (2015). Core principles of bacterial autoinducer systems. Microbiol. Mol. Biol. Rev. 79 153–169. 10.1128/MMBR.00024-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jers C., Kobir A., Søndergaard E. O., Jensen P. R., Mijakovic I. (2011). Bacillus subtilis two-component system sensory kinase DegS is regulated by serine phosphorylation in its input domain. PLOS ONE 6:e14653. 10.1371/journal.pone.0014653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia V. C. (2013). Quorum sensing inhibitors: an overview. Biotechnol. Adv. 31 224–245. 10.1016/j.biotechadv.2012.10.004 [DOI] [PubMed] [Google Scholar]
- Kawamura F., Doi R. H. (1984). Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J. Bacteriol. 160 442–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M., Quadri L. E., Kuipers O. P., de Vos W. M. (1997). Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24 895–904. 10.1046/j.1365-2958.1997.4251782.x [DOI] [PubMed] [Google Scholar]
- Kobayashi K. (2007). Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol. Microbiol. 66 395–409. 10.1111/j.1365-2958.2007.05923.x [DOI] [PubMed] [Google Scholar]
- Koo B. M., Kritikos G., Farelli J. D., Todor H., Tong K., Kimsey H., et al. (2017). Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst. 4 291–305. 10.1016/j.cels.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazazzera B. A., Solomon J. M., Grossman A. D. (1997). An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell 89 917–925. 10.1016/S0092-8674(00)80277-9 [DOI] [PubMed] [Google Scholar]
- López D., Vlamakis H., Losick R., Kolter R. (2009). Paracrine signaling in a bacterium. Genes Dev. 23 1631–1638. 10.1101/gad.1813709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson R., Solomon J., Grossman A. D. (1994). Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell 77 207–216. 10.1016/0092-8674(94)90313-1 [DOI] [PubMed] [Google Scholar]
- Marlow V. L., Cianfanelli F. R., Porter M., Cairns L. S., Dale J. K., Stanley-Wall N. R. (2014). The prevalence and origin of exoprotease-producing cells in the Bacillus subtilis biofilm. Microbiology 160 56–66. 10.1099/mic.0.072389-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miras M., Dubnau D. (2016). A DegU-P and DegQ-dependent regulatory pathway for the K-state in Bacillus subtilis. Front. Microbiol. 7:1868. 10.3389/fmicb.2016.01868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montville T. J. (1983). Dual-substrate plate diffusion assay for proteases. Appl. Environ. Microbiol. 45 200–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msadek T., Kunst F., Klier A., Rapoport G. (1991). DegS-DegU and ComP-ComA modulator-effector pairs control expression of the Bacillus subtilis pleiotropic regulatory gene degQ. J. Bacteriol. 173 2366–2377. 10.1128/jb.173.7.2366-2377.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray E. J., Kiley T. B., Stanley-Wall N. R. (2009). A pivotal role for the response regulator DegU in controlling multicellular behaviour. Microbiology 155 1–8. 10.1099/mic.0.023903-0 [DOI] [PubMed] [Google Scholar]
- Nadell C. D., Xavier J. B., Levin S. A., Foster K. R. (2008). The evolution of quorum sensing in bacterial biofilms. PLOS Biol. 6:e14. 10.1371/journal.pbio.0060014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M. M., Xia L., Zuber P. (1991). Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis. J. Bacteriol. 173 5487–5493. 10.1128/jb.173.17.5487-5493.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M., Yamaguchi H., Ki Y., Fujita Y., Tanaka T. (2001). DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29 3804–3813. 10.1093/nar/29.18.3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Sato I., Cho S. J., Iwata H., Nishio T., Dubnau D., et al. (2005). Structure of the Bacillus subtilis quorum-sensing peptide pheromone ComX. Nat. Chem. Biol. 1 23–24. 10.1038/nchembio709 [DOI] [PubMed] [Google Scholar]
- Oslizlo A., Stefanic P., Dogsa I., Mandic-Mulec I. (2014). Private link between signal and response in Bacillus subtilis quorum sensing. Proc. Natl. Acad. Sci. U.S.A. 111 1586–1591. 10.1073/pnas.1316283111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oslizlo A., Stefanic P., Vatovec S., Beigot Glaser S., Rupnik M., Mandic-Mulec I. (2015). Exploring ComQXPA quorum-sensing diversity and biocontrol potential of Bacillus spp. isolates from tomato rhizoplane. Microb. Biotechnol. 8 527–540. 10.1111/1751-7915.12258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K., Bassler B. L. (2016). Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14 576–588. 10.1038/nrmicro.2016.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar V., Konkol M. A., Kearns D. B., Neiditch M. B. (2013). A plasmid-encoded phosphatase regulates Bacillus subtilis biofilm architecture, sporulation, and genetic competence. J. Bacteriol. 195 2437–2448. 10.1128/JB.02030-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick J. E., Kearns D. B. (2008). MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol. Microbiol. 70 1166–1179. 10.1111/j.1365-2958.2008.06469.x [DOI] [PubMed] [Google Scholar]
- Pollak S., Omer-Bendori S., Even-Tov E., Lipsman V., Bareia T., Ben-Zion I., et al. (2016). Facultative cheating supports the coexistence of diverse quorum-sensing alleles. Proc. Natl. Acad. Sci. U.S.A. 113 2152–2157. 10.1073/pnas.1520615113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield R. J. (2002). Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10 365–370. 10.1016/S0966-842X(02)02400-9 [DOI] [PubMed] [Google Scholar]
- Schneider B. K., Palmer T. M., Grossman A. D. (2002). Characterization of comQ and comX, two genes required for production of ComX pheromone in Bacillus subtilis. J. Bacteriol. 184 410–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Sexton D. J., Hense B. A. (2017). Why quorum sensing controls private goods. Front. Microbiol. 8:885 10.3389/fmicb.2017.00885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley N. R., Lazazzera B. A. (2005). Defining the genetic differences between wild and domestic strains of Bacillus subtilis that affect poly-γ-dl-glutamic acid production and biofilm formation. Mol. Microbiol. 57 1143–1158. 10.1111/j.1365-2958.2005.04746.x [DOI] [PubMed] [Google Scholar]
- Stefanic P., Decorosi F., Viti C., Petito J., Cohan F. M., Mandic-Mulec I. (2012). The quorum sensing diversity within and between ecotypes of Bacillus subtilis. Environ. Microbiol. 14 1378–1389. 10.1111/j.1462-2920.2012.02717.x [DOI] [PubMed] [Google Scholar]
- Stefanic P., Kraigher B., Lyons N. A., Kolter R., Mandic-Mulec I. (2015). Kin discrimination between sympatric Bacillus subtilis isolates. Proc. Natl. Acad. Sci. U.S.A. 112 14042–14047. 10.1073/pnas.1512671112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanic P., Mandic-Mulec I. (2009). Social interactions and distribution of Bacillus subtilis pherotypes at microscale. J. Bacteriol. 191 1756–1764. 10.1128/JB.01290-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Iwasaki K., Morimoto T., Matsuse T., Hasunuma T., Takenaka S., et al. (2015). Hyperphosphorylation of DegU cancels CcpA-dependent catabolite repression of rocG in Bacillus subtilis. BMC Microbiol. 15:43. 10.1186/s12866-015-0373-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegmark K., Morfeldt E., Arvidson S. (1998). Regulation of agr -dependent virulence genes in Staphylococcus aureus by RNAIII from coagulase-negative staphylococci. J. Bacteriol. 180 3181–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwagne M., Mirabella A., Lemaire J., Deschamps C., De Bolle X., Letesson J. J. (2013). Quorum sensing and self-quorum quenching in the intracellular pathogen Brucella melitensis. PLOS ONE 8:e82514. 10.1371/journal.pone.0082514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortosa P., Logsdon L., Kraigher B., Itoh Y., Mandic-Mulec I., Dubnau D. (2001). Specificity and genetic polymorphism of the Bacillus competence quorum-sensing system. J. Bacteriol. 183 451–460. 10.1128/JB.183.2.451-460.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening J., Igoshin O. A., Eijlander R. T., Nijland R., Hamoen L. W., Kuipers O. P. (2008). Transient heterogeneity in extracellular protease production by Bacillus subtilis. Mol. Syst. Biol. 4:184. 10.1038/msb.2008.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhamme D. T., Kiley T. B., Stanley-Wall N. R. (2007). DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol. Microbiol. 65 554–568. 10.1111/j.1365-2958.2007.05810.x [DOI] [PubMed] [Google Scholar]
- Warriner K., Waites W. M. (1999). Enhanced sporulation in Bacillus subtilis grown on medium containing glucose:ribose. Lett. Appl. Microbiol. 29 97–102. 10.1046/j.1365-2672.1999.00593.x [DOI] [Google Scholar]
- Weinrauch Y., Penchev R., Dubnau E., Smith I., Dubnau D. (1990). A Bacillus subtilis regulatory gene product for genetic competence and sporulation resembles sensor protein members of the bacterial two-component signal-transduction systems. Genes Dev. 4 860–872. 10.1101/gad.4.5.860 [DOI] [PubMed] [Google Scholar]
- Wu X. C., Lee W., Tran L., Wong S. L. (1991). Engineering a Bacillus subtilis expression-secretion system with a strain deficient in six extracellular proteases. J. Bacteriol. 173 4952–4958. 10.1128/jb.173.16.4952-4958.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


