Abstract
MicroRNAs (miRNAs) are a family of short noncoding RNAs that posttranscriptionally regulate gene expression and play an important role in multiple cellular processes. A significant percentage of miRNAs are intragenic, which is often functionally related to their host genes playing either antagonistic or synergistic roles. In this study, we constructed and analyzed the entire network of intergenic interactions induced by intragenic miRNAs. We further focused on the core of this network, which was defined as a union of nontrivial strongly connected components, i.e., sets of nodes (genes) mutually connected via directed paths. Both the entire network and its core possessed statistically significant non-random properties. Specifically, genes forming the core had high expression levels and low expression variance. Furthermore, the network core did not split into separate components corresponding to individual signalling or metabolic pathways, but integrated genes involved in key cellular processes, including DNA replication, transcription, protein homeostasis and cell metabolism. We suggest that the network core, consisting of genes mutually regulated by their intragenic miRNAs, could coordinate adjacent pathways or homeostatic control circuits, serving as a horizontal inter-circuit link. Notably, expression patterns of these genes had an efficient prognostic potential for breast and colorectal cancer patients.
Introduction
MicroRNAs (miRNAs) are a family of short (~22 nt) noncoding RNAs that posttranscriptionally regulate gene expression and play an important role in various cellular processes, including oncogenesis, epithelial–mesenchymal transition, regeneration, embryogenesis, and cellular differentiation1–5. Furthermore, miRNAs can function in coordination with various epigenetic regulators6 and transcription factors7. The miRNA concentration in a cell can rapidly change8, and therefore, miRNA expression is considered an element of early genetic response to external stimuli9,10. Finally, miRNAs also regulate cellular homeostasis by serving as nodes of signalling networks11.
A significant percentage of miRNAs are intragenic, i.e., located within intronic or exonic regions of coding genes (host genes)12. In humans, more than half of miRNAs are intragenic13. At the same time, the majority of intragenic miRNAs are located within introns14; specifically, humans intronic miRNAs constitute more than 85% of all intragenic miRNAs13. Moreover, intronic miRNAs are usually transcribed in the same direction as their host genes14. In humans, more than 80% of intronic miRNA genes have a sense orientation with respect to their host genes13. Therefore, most human intragenic miRNAs are co-transcribed with their host genes and subsequently released at the splicing stage15,16.
In addition to having overlapping genome locations, intragenic miRNAs and their host genes can be functionally connected; however, in studies demonstrating these links, the pairs of {host gene – intragenic miRNA} were analyzed independently from each other. Specifically, a number of theoretical and experimental studies have shown that the host genes can be the direct targets of their intragenic miRNAs. Targeting of a host gene by its intragenic miRNA was observed not only for exonic antisense miRNAs (which are complementary to a gene region but transcribed independently of their host genes)17,18 but also for multiple intronic and exonic sense miRNAs19–23. Furthermore, computational analysis demonstrated that depending on the parameters applied in the model, self-regulation of genes via their intragenic miRNAs can have various biological roles, including buffering of “expression noise”, conferring expression robustness, regulating the timing of responses to external signals, and adapting gene expression to persistent stimuli, thus providing responses conforming to Weber’s law24. Notably, a non-canonical mechanism of self-regulation of genes, where intragenic miRNA enhances transcription instead of mediating posttranscriptional repression, was also reported25. Co-transcription of a host gene and its targeting miRNA (followed by subsequent release of the miRNA precursor during splicing and its processing into a mature miRNA via a common well-described mechanism16,26,27) can be regarded as a negative (in most cases) or a positive (occasionally) feedback loop28 (Fig. 1a,b). This self-regulation of gene expression may be regarded as a specific case of regulatory network motifs that include both transcription factors and miRNAs7,11,29,30.
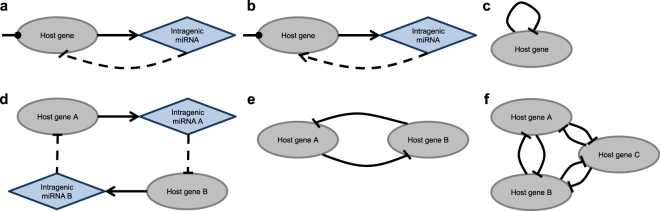
Figure 1.
Regulatory network motifs involving intragenic miRNAs. (а) A self-regulatory negative feedback loop. (b) A self-regulatory positive feedback loop. (c) Representation of a self-regulatory feedback loop in the constructed network of intergenic interactions induced by intragenic miRNAs (note that loops are removed prior to the analysis of the network). (d) A pair of genes mutually targeting each other via their intragenic miRNAs. (e) Representation of miRNA-induced intergenic interactions shown in panel (d) in the constructed network. (f) A three-node sub-network in which each pair of nodes is mutually (bidirectionally) connected.
Other reported types of functional relations between host genes and their intragenic miRNAs include miRNA-targeting of genes whose products are downstream effectors of host gene products, genes antagonistic to a host gene, or genes belonging to the same pathway as a host gene14,31–35.
We hypothesized that regulatory network motifs involving intragenic miRNAs could simultaneously function in a cell as interconnected parts of the entire mechanism of gene expression regulation. Therefore, in the present study, we did not focus on individual regulatory motifs but, for the first time, constructed and analyzed the entire network of intergenic interactions induced by intragenic miRNAs. It is a gene oriented network which edges correspond to miRNAs and represent targeting of one gene by a miRNA hosted in the other gene. In particular, we identified condensed core of the constructed network. The core contained 21 or 12 genes (for all intragenic miRNAs or only intronic sense miRNAs, respectively) involved in key cellular processes, including DNA replication, transcription, protein homeostasis and cell metabolism. Intragenic miRNAs located in the core genes are likely to confer the robustness of the core by buffering internal and external noises30,36–38, fine-tune the expression of the core genes39 and, more generally, coordinate adjacent pathways or homeostatic control circuits by serving as a horizontal inter-circuit link. We further hypothesized that regulation of expression of the core genes mediated by their intragenic miRNAs could be important for normal cell functioning, and distortion of the expression patterns of these genes could have a significant diagnostic potential. As proof of concept, we identified gene expression signatures consisting solely of core genes for highly efficient recurrence prognosis of breast and colorectal cancer. Remarkably, these expression signatures were as efficient as ones identified from a genome-wide transcriptome analysis40,41.
Results
The network design
The construction and analysis of the network of intergenic interactions induced by intragenic miRNAs were performed on the levels of (1) all intragenic miRNAs and (2) only intronic sense miRNAs. For network construction we used only validated targets of miRNAs. Therefore, we focused on humans since human databases of validated miRNA targets are currently the most comprehensive.
The designed network is gene oriented and has the following structure. Its nodes are genes, and nodes (genes) A and B are connected by a directed edge (A –┤ B) if gene A is a host for an intragenic miRNA that targets gene B. We use bar-headed arrows to represent edges as miRNAs generally suppress their targets.
Specifically, a trivial cycle (loop) A –┤ A (Fig. 1c) in this network represents self-regulation of gene A by its intragenic miRNA. In addition to the loops, the resulting network contains nontrivial cycles, including pairs of mutually connected genes (Fig. 1d,e). To the best of our knowledge, this type of regulatory network pattern was not previously reported and analyzed for miRNA-induced intergenic interactions. At the same time, there are no triples in which each pair of genes is mutually (bidirectionally) connected. In other words, the network does not contain a sub-network depicted in Fig. 1f.
However, if not confined to one-edge paths (i.e., to direct miRNA-induced interactions) and considering longer paths with intermediate nodes, two relatively small components comprising nodes mutually connected by directed paths can be revealed. In other words, the network contains exactly two nontrivial (i.e., containing at least two nodes) strongly connected components. These components can be considered analogues of the host genes that are self-regulated by their intragenic miRNAs. Each node of a component is a host gene whose intragenic miRNA targets the members of the same component (although a direct miRNA-induced interaction exists not for all pairs of genes). And vice versa, each node is a gene targeted by a miRNA hosted in another gene from this component.
The union of the nontrivial strongly connected components will be further referred to as a “network core”. Section Network core: a simple example of Supplementary Information illustrates the definition of the network core.
Quantitative characteristics of networks
A full network (F-network) of miRNA-induced intergenic interactions in humans was designed based on the lists of experimentally validated miRNA targets. A target was considered validated if it was present simultaneously in two databases of validated miRNA targets — DIANA-TarBase v7.042 and miRTarBase (Release 6.1)43.
The miRNA–target interactions were validated in total for 842 mature miRNAs, 445 of which were intragenic. Furthermore, these 445 mature miRNAs originated from 389 pre-miRNAs located in 305 different host genes and targeted 8,416 genes. The targeted genes included 176 out of 305 host genes. Consequently, the F-network contained 176 nodes with both in- and out-edges, 129 nodes with only out-edges and 8,240 nodes with only in-edges (see Supplementary Fig. 1 as an illustration): 8,545 non-isolated nodes in total. The number of edges in the F-network (after removing loops and merging multiple edges) was 19,081. These numbers are summarized in Table 1.
Table 1.
Quantitative characteristics of the F-network and IS-network.
| TotalN | BothDirN | OutN | InN | TotalE | Mat-mi | Pre-mi | HostGenes | |
|---|---|---|---|---|---|---|---|---|
| F-network | 8,545 | 176 | 129 | 8,240 | 19,081 | 445 | 389 | 305 |
| IS-network | 8,307 | 140 | 106 | 8,061 | 16,913 | 364 | 277 | 246 |
TotalN – total number of non-isolated nodes. BothDirN – number of nodes with both in- and out-edges. OutN – number of nodes with out-edges but no in-edges. InN – number of nodes with in-edges but no out-edges. TotalE – total number of edges after removing loops and merging multiple edges. Mat-mi – number of mature miRNAs that induce intergenic interactions represented as network edges. Pre-mi – number of pre-miRNAs for these mature miRNAs. HostGenes – number of genes hosting these pre-miRNAs. Clearly, TotalN = BothDirN + OutN + InN, HostGenes = BothDirN + OutN.
Along with the F-network, which was based on all intragenic miRNAs, we constructed an IS-network based solely on intronic sense miRNAs. In the IS-network, the number of different nodes and edges was evidently lower compared to that of the F-network; however, this difference was minor since intronic sense miRNAs constitute the majority of all intragenic miRNAs (Table 1).
Formal description of the constructed networks through the listing of their edges is presented in Supplementary Data (worksheets F-network formal descr. and IS-network formal descr.).
Nodes with the highest in-degree
The analysis of lists of nodes with the highest in-degree (i.e., lists of genes having the highest regulation by intragenic miRNAs) revealed a moderate enrichment for several major functional categories. Specifically, for both networks, the enrichment was evident for such categories as cell cycle, p53 signalling pathway, oncogenic pathway, focal adhesion, apoptosis, and transcriptional and posttranscriptional regulation of gene expression (Table 2). Additional data are presented in Supplementary Information (section The enrichment analysis of lists of nodes with the highest in-degree). Histograms of in-degree for the F-network and the IS-network are presented in Supplementary Data (worksheets F-network in-degree hist. and IS-network in-degree hist).
Table 2.
Major functional categories overrepresented in lists of genes with the highest in-degree.
| Term | F-network adj. p-val | IS-network adj. p-val |
|---|---|---|
| Cell cycle | 1.09 × 10−5 | 1.16 × 10−5 |
| p53 signalling pathway | 3.11 × 10−5 | 3.62 × 10−5 |
| Pathways in cancer | 2.15 × 10−5 | 4.24 × 10−4 |
| Focal adhesion | 6.54 × 10−4 | 5.25 × 10−4 |
| Apoptosis | 1.17 × 10−3 | 1.21 × 10−3 |
| Posttranscriptional regulation of gene expression | 3.32 × 10−5 | 1.63 × 10−3 |
| Transcription regulation | 4.12 × 10−3 | 1.19 × 10−2 |
F-network adj. p-val – Benjamini-corrected p-value for the F-network list reported by DAVID; IS-network adj. p-val – Benjamini-corrected p-value for the IS-network list reported by DAVID.
Overrepresented three-node network motifs
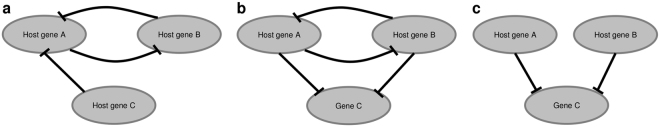
Both F- and IS-networks had properties different from those of random graphs with the same number of vertices and edges. Notably, the number of occurrences of the three-node network motif presented in Fig. 2a (41 for the F-network and 18 for the IS-network) was significantly higher than the expected number for this motif in a random graph. For random graphs generated using a model with a fixed set of out-degrees of vertices (which is equivalent to the randomization of lists of miRNA targets preserving sizes of the lists), p-values did not exceed 10−4 for the F-network and 0.017 for the IS-network (see Supplementary Table 5 – sub-network type 6). For random graphs generated using the Erdős–Rényi model44, the p-value was 2.6 × 10−3 for the F-network, but for the IS-network the statistical significance was lower, with a p-value of 0.1. However, a clear difference between the constructed networks and random graphs generated with the Erdős–Rényi model was revealed by the analysis of other three-node motifs. E.g., an evident overrepresentation in comparison with Erdős–Rényi– random graphs was observed for a motif shown in Fig. 2b (p-value < 10−4 for both the F-network and the IS-network; see Supplementary Table 5 – sub-network type 14).
Figure 2.
Three-node network motifs overrepresented in the F-network and IS-network in comparison with random graphs (a) generated using a model with a fixed set of out-degrees of vertices; (b) generated with the Erdős–Rényi model; (c) generated using any of these two models.
Furthermore, a three-node network motif presented in Fig. 2c was also overrepresented in the constructed networks in comparison with random graphs. The number of its occurrence in the F-network and in the IS-network was 30757 and 22913, respectively, which resulted in p-values < 10−4 for both the Erdős–Rényi model and the model with a fixed set of out-degrees (see Supplementary Table 5 – sub-network type 3).
Aggregate information on the number of non-equivalent (non-isomorphic) three-node sub-networks in the constructed networks and in random graphs is presented in Supplementary Information (section Three-node sub-networks). Histograms supporting the specified p-values are also presented in Supplementary Information (section Histograms supporting p-values).
The network core
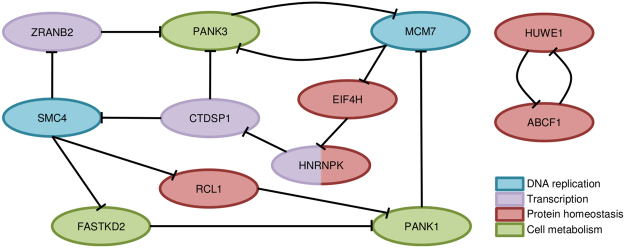
For the IS-network, the core consisted of two strongly connected components that included 10 and 2 genes (Fig. 3; Supplementary Data – worksheet IS-network core formal descr.). The core of the F-network contained the IS-network core as a subgraph and also consisted of two components, with 18 and 3 genes (see Supplementary Fig. 6 and Supplementary Data – worksheet IS-network core formal descr.).
Figure 3.
The core of the IS-network. The nodes are genes, and nodes (genes) A and B are connected by a directed edge (A –┤ B) if gene A is a host for an intronic sense miRNA that targets gene B.
Similarly to the three-node network motifs discussed above, network core size clearly differentiated both the F-network and the IS-network from random graphs generated using the Erdős–Rényi model (for random graphs, the size of the core was substantially higher, p-value < 10−4). Applying the model with a fixed set of out-degrees of vertices resulted in the reduction of the core size in random graphs: the core size for the F- and IS-networks was still smaller than a median core size for random graphs, but statistical significance was violated (p-value was close to 0.17 for both the F- and IS-networks). Histograms supporting the specified p-values are presented in Supplementary Information (subsection Histograms supporting p-values).
The network core comprised genes that are essential for key cellular processes. For example, MCM7 and SMC4 are indispensable for DNA replication: MCM7 promotes unwinding of dsDNA in the replication fork45 and SMC4 is involved in condensation of mitotic chromosomes46. Furthermore the CTDSP1 and ZRANB2 genes are important transcriptional regulators responsible for gene silencing and alternative splicing, respectively47,48. Four other genes from the IS-network core are involved in translation control and maintenance of protein homeostasis. EIF4H and ABCF1 are both involved in translation initiation49,50. RCL1 encodes a nuclease that cleaves pre-rRNA at the A2 site, thus separating rRNA destined for small and large ribosomal subunits51. The product of the HUWE1 gene is a E3 ubiquitin-protein ligase that mediates ubiquitination and subsequent proteasomal degradation of target proteins, including the anti-apoptotic protein Mcl1, p53, c-Myc and core histones52. The protein encoded by HNRNPK is a key regulator of transcription and translation53. The PANK1, PANK3 and FASTKD2 genes are essential for basic cellular metabolism. PANK1 and PANK3 genes encode isoforms of pantothenate kinase, which catalyzes the first rate-limiting step of panthotenate biosynthesis. At the same time, pantothenate is the precursor of coenzyme A — a key cellular metabolite that serves as a cofactor for multiple enzymes and is involved in various processes, including fatty and amino acid biosynthesis, cell signalling and regulation of gene expression54. Finally, the FASTKD2 protein is localized in the mitochondria, and its knockdown leads to the impairment of cellular respiration55.
To summarize, the network core connects the genes involved in basic cellular processes: DNA replication, transcription, protein homeostasis and underlying metabolic activity.
High expression levels and low expression variance of the core genes
The analysis of genome-wide transcriptome profiles of 675 cell lines56 revealed that genes belonging to the cores of the IS-network and F-network have generally high expression levels. More specifically, after ranking the transcripts present in the dataset (ca. 26 thousand transcripts) by the median expression level, 10 out of 12 core genes from the IS-network were in the upper quarter, while two remaining genes were still in the upper half (binomial test p-value < 4 × 10−5). Similar ranking by the third (higher) or the first (lower) quartile led to similar results. Specifically, the distribution among quarters was exactly the same after ranking by the third quartile, while after the first quartile ranking, 11 out of 12 genes were located in the upper quarter, and the remaining gene was still found in the upper half. For the core of the F-network, statistically significant enrichment of highly expressed transcripts was even more remarkable. For example, after ranking by the median expression levels, 16 out of 21 genes comprising the core of the F-network resided in the upper quarter, and the remaining 5 genes were still in the upper half, with the binomial test p-value less than 10−5.
At the same time, genes from the core were not the most highly expressed ones: after ranking of transcripts by a median expression level, as well as by the first quartile, only one gene from the core (HNRNPK) appeared in the top 1%. After ranking by the third quartile, HNRNPK moved down (but remained in the top 2% zone), and no genes from the core were present in the top 1%.
Along with high expression, genes from the core have generally low variance in expression levels. Even after excluding low expression transcripts from the analysis and retaining the upper half of the transcripts (with respect to the ranking by the first quartile of expression levels), most genes from the core were in the half with a lower variance — 10 genes for the core of the IS-network (binomial test p-value 0.019) and 16 genes for the core of the F-network (binomial test p-value 0.013). Moreover, 8 genes were in the quarter of transcripts with the lowest variance for the core of the IS-network (binomial test p-value 0.0028), and for the core of the F-network, the number of such genes was 12 (binomial test p-value 0.0017).
Boxplots illustrating the described data are presented in Supplementary Information (Supplementary Figs 7, 8). Details of the analysis, including measures used to quantify expression levels and expression variance, are described in Methods section (subsection Analysis of expression levels).
Further analysis of expression levels of genes comprising the core in tissue specimens from 298 breast cancer patients and 519 colorectal patients confirmed these results. Details are presented in Supplementary Information (section Expression levels of core genes in tissue specimens).
Therefore, our data strongly indicate that the genes comprising the network core of intragenic miRNA-induced gene-gene interactions can be characterized as generally having higher expression levels and lower variance compared to the median values.
The prognostic power of genes belonging to the core
Analysis of expression pattern of genes within the core led to clear conclusions about a cell and even a whole organism. More specifically, highly reliable prognostic gene signatures were derived for breast and colorectal cancer using only expression patterns of the genes from the network core.
These prognostic gene signatures were constructed separately for breast cancer and colorectal cancer using an SVM-based exhaustive search strategy described previously40. Microarray data utilized for this construction are specified in Methods section (subsection Construction of prognostic gene signatures). Both for breast cancer and for colorectal cancer these data included one training, two filtration and one validation (testing) dataset. For breast cancer the datasets reported expression levels for 17 genes from the core of the F-network, including 10 genes from the core of the IS-network; for colorectal cancer the datasets reported expression levels of all genes comprising the core of the F-network. Log-scaled expression levels were used.
For each gene its mean expression level and standard deviation of expression level were estimated based on the training dataset, and expressions in all datasets were (0, 1)-scaled by subtracting these means and dividing by these deviations.
Then for every studied subset of genes from the core of the F-network, ranging from individual genes to the whole core of the F-network, a binary classifier was constructed using Support Vector Machine57 (SVM) with the linear kernel and class weights inversely proportional to the number of patients in each class. This construction utilized only the training dataset. For triples of genes the classifier had a form
where e1, e2, e3 were (0, 1)-scaled expressions (with scaling parameters derived from the training dataset), and weights w1, w2, w3 as well as ρ were computed using a standard SVM construction algorithm. Taking expression levels e1, e2, e3 associated with a patient, this classifier attributed the patient to the high risk group if R (e1, e2, e3) was nonnegative, and to a low-risk group otherwise. For sets with another number of genes the difference in the classifier forms was only in the number of terms in a linear combination.
If the resulting classifier did not pass thresholds on sensitivity and specificity for the training dataset (the utilized values are specified in Methods section, subsection Construction of prognostic gene signatures), it was excluded from the further analysis. Otherwise, in order to avoid overfitting, this classifier was additionally applied to each of the filtration datasets. If sensitivity or specificity for at least one filtration dataset was lower than a filtration threshold, the classifier was also filtered out. For classifiers that passed the filtration their prognostic characteristics were estimated using the validation dataset. Samples from the validation dataset were utilized neither for classifier construction, nor for filtration.
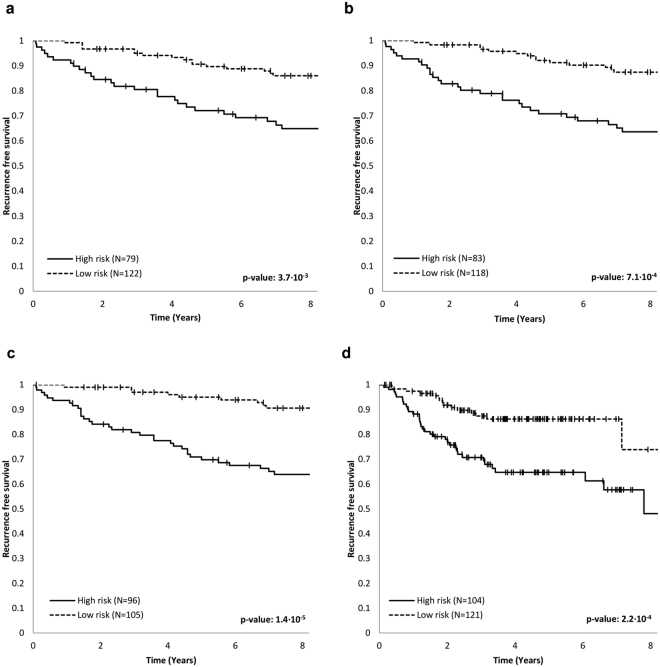
Limiting the analysis to the triples comprising the genes from the core of the IS-network, we identified HNRNPK, PANK3 and SMC4 gene triple as having the highest prognostic power for estrogen receptor-positive (ER-positive) breast cancer (with respect to the training and filtration datasets – see Methods, subsection Construction of prognostic gene signatures). Furthermore, on the validation dataset this triple provided prediction of breast cancer recurrence with a sensitivity and specificity of 63.6% and 66.4%, respectively. The AUC value for the validation dataset was 0.662, while its mean value for the training and filtration datasets was equal to 0.708. The Kaplan-Meier curves for the validation dataset are presented in Fig. 4a.
Figure 4.
Kaplan-Meier curves for prognostic three-gene signatures. Log-rank p-values are reported. (a) Breast cancer, genes HNRNPK, PANK3, SMC4. (b) Breast cancer, genes MCM7, PANK3, SMC4. (c) Breast cancer, genes MAP2K4, PANK3, SMC4. (d) Colorectal cancer, genes FASTKD2, PANK1, HUWE1.
The genes HNRNPK, PANK3 and SMC4 are not involved in one cellular process but contribute to different pathways related to DNA replication (SMC4), regulation of transcription and translation (HNRNPK) and cellular metabolism (PANK3). Notably, SMC4 and PANK3 genes had the highest degree (i.e., total number of in- and out-edges for a node) in the subgraph of the IS-network induced by its core (Fig. 3). Interestingly, the substitution of HNRNPK for MCM7 (the remaining gene with degree 4) yielded a three-gene signature with a similar prognostic power for the training and filtration datasets (mean AUC value 0.693) and even higher characteristics for the validation dataset (sensitivity 69.7%, specificity 66.4%, AUC 0.670; Fig. 4b).
Extension of the SMC4-PANK3 pair to the triple with MAP2K4, which belongs to the core of the F-network, slightly enhanced the mean AUC value for the training and filtration datasets and increased the sensitivity, specificity and AUC value for the validation dataset to 84.8%, 60.2% and 0.728, respectively (Fig. 4c).
Therefore, the analysis limited to the core of the network of intragenic miRNA-induced gene-gene interactions allowed construction of three-gene signatures for the prognosis of ER-positive breast cancer recurrence with sensitivity and specificity comparable to the most reliable three-gene signatures identified by extensive genome-wide analysis40 as well as the reliability of multigene commercial prognostic signatures, such as OncotypeDX58,59 and MammaPrint59,60.
For colorectal cancer, an exhaustive search revealed the FASTKD2, PANK1, and HUWE1 gene triple from the core to have the highest prognostic power (with respect to the training and filtration datasets). All these genes belong to the core of the IS-network, but this triple was also optimal for the core of the F-network. The sensitivity and specificity of this triple in predicting the recurrence for colorectal cancer on the validation dataset were 68.3% and 60.0%, respectively, while the AUC value was 0.659 (Fig. 4d). These values of sensitivity and specificity were similar to the ones for the OncotypeDX for Colon Cancer41, which was designed based on a large-scale transcriptome analysis (expression levels were measured for 761 preselected candidate genes)41,61.
It is worth mentioning that the prognostic power of the above described triples of genes from the network core was provided by the totality of all genes in a triple. For gene pairs (and, obviously, for individual genes), the prognostic power was much lower. Moreover, none of the pairs of genes from the core passed the filtration.
Individual genes in each triple had different weights w (which characterize prognostic potential) in the resulting classifier value. The SMC4 gene had the highest weight in all the above described triples for breast cancer, and increased expression of this gene (in the case of unaltered expression levels of the other two genes in the triple) was associated with a higher risk of recurrence. These data are consistent with emerging reports on the oncogenic potential of the SMC4 gene62,63. Similarly, elevated expression of the MCM7 gene, whose oncogenic potential has been widely discussed64–66, was also associated with a higher risk of cancer recurrence. For PANK3, HNRNPK and MAP2K4 genes, this correlation was opposite. Two of these genes (HNRNPK and MAP2K4) were previously identified as tumor suppressors67–69; however, a pro metastatic-role of these genes was also reported70,71. To the best of our knowledge, the role of PANK3 in oncogenesis has not yet been described.
In the case of the FASTKD2-PANK1-HUWE1 triple, a higher risk of cancer recurrence was associated with increased expression of the HUWE1 gene and lower expression levels of FASTKD2 and PANK1. The FASTKD2 gene, which has the greatest weight in the classifier for this triple, has been previously shown to promote apoptosis of cancer cells72. Furthermore, both oncogenic and tumor suppression roles were previously reported for the HUWE1 gene73–76. In contrast, as for the PANK3 gene, the role of the PANK1 gene in the oncogenesis remains unknown.
Interestingly, larger gene sets (e.g., quadruples) did not provide a substantial increase in prognostic power. Moreover, the reliability of cancer prognosis based on the set consisting of all genes in the core was even lower in comparison with the most reliable triples due to overfitting of the classifier.
Weights of genes in the above classifiers and additional details of triples filtration are presented in Supplementary Information (section Prognostic gene triples).
A connection between the IS-network core and the Myc protein
Notably, a striking functional connection between the core of the IS-network and the Myc protein has been found. The transcription factor Myc is known to have a high number of different targets and has a pronounced oncogenic potential77–79. Analysis of the published data revealed that 9 out of 12 genes comprising the core were either regulated by Myc directly or shown to conversely interact with Myc at the protein level, regulating its activity. See Supplementary Information for details (section Interplay of the IS-network core and Myc).
Discussion
Despite the fact that a typical miRNA-target interaction mediates only a subtle reduction in protein levels36,80, miRNAs play an important physiological role by conferring robustness of biological processes36,37, fine-tuning essential cellular pathways, controlling signal transduction11,39,81,82 and maintaining metabolic homeostasis83,84. In particular, one report showed that aberrant levels of miR-103 and miR-107, intronic sense miRNAs located in the pantothenate kinase genes PANK3 and PANK1, respectively, induced impaired glucose homeostasis85.
Over the last years, miRNAs have been shown to have significant impact in cancer, playing a role in each step of cancer biogenesis and progression86,87. Numerous networks involved in different cancer types and comprising miRNAs as well as the target genes have been identified88,89. The reported networks proved that miRNAs can generate connected graphs and revealed cancer type-specific patterns, pointing out to diagnostic, prognostic and therapeutic potential of miRNAs and miRNA-based networks90.
Intragenic miRNAs constitute a significant percentage of total miRNAs12,13 and are often functionally related with their host genes14, playing either antagonistic or synergistic roles31,33,35. In the present study, instead of focusing on individual regulatory network motifs or disease-specific networks involving miRNAs, we analyzed the entire network of intergenic interactions induced by either intronic sense miRNAs or all intragenic miRNAs. Furthermore, in these networks, we focused on the core, which has been defined as a union of nontrivial strongly connected components, i.e., sets of genes mutually connected by directed paths.
Generally, the genes belonging to the core had high expression levels and low expression variance. The latter is consistent with the concept of the miRNome as an instrument for conferring robustness of gene expression30,36,37 and maintaining cellular homeostasis83,84. Interestingly, the core could not be split into the components corresponding to individual signalling or metabolic pathways, but it integrated genes involved in the key cellular processes, including DNA replication, transcription, protein homeostasis and cell metabolism. Thus, interactions between genes from the core mediated by intragenic miRNAs could be involved not only in independent pathways or homeostatic control circuits91 but also in a horizontal regulation that subtly coordinates adjacent circuits.
Remarkably, the expression pattern of genes forming the core was shown to be an efficient prognostic marker in breast and colorectal cancer patients. While individual genes, as well as gene pairs, had relatively low prognostic power, a number of triples of genes from the network core provided a prognostic reliability comparable to that of the gene expression signatures identified by large-scale transcriptome analyses, including common commercial prognostic gene signatures. Furthermore, the higher risk of cancer recurrence was associated with differently directed changes in expression of genes for all identified triples, i.e., higher imbalance in the expression patterns of core genes.
The impact of expression changes of individual genes on the values of constructed prognostic classifiers was consistent with the previous reports on oncogenic or tumor suppressive roles of these genes. However, in the prognostic classifier for breast cancer, the weight of the well-studied MCM7 oncogene64–66 was essentially lower than the weight of the SMC4 gene with less reported oncogenic potential62,63. Similarly, there is limited evidence for the tumor suppressor potential of the FASTK2 gene, which had the highest weight in the prognostic three-gene expression signature for colorectal cancer72.
To the best of our knowledge, the roles of PANK1 and PANK3 in oncogenesis have not been yet confirmed. However, transcription of PANK1 has been previously reported to be directly targeted by p5392.
To summarize, we constructed a network of intergenic interactions induced by intragenic miRNAs and identified its core, which consists of genes involved in the key cellular processes. Both the network and its core possessed statistically significant non-random properties. Specifically, genes forming the core generally had high expression levels and low expression variance. Importantly, our data indicate that the expression pattern of these genes could be used for a reliable prognosis of recurrence for breast and colorectal cancer patients. Finally, we hypothesized that miRNA-induced intergenic interactions represented by directed edges in the network core could orchestrate subtle coordination of adjacent pathways and/or homeostatic control circuits by serving as horizontal inter-circuit regulatory links.
Methods
Network construction and analysis
The list of identifiers for intragenic pre-miRNAs and their respective host genes was downloaded from the miRIAD intragenic microRNA database13. The information for the classification of intragenic miRNAs into intronic and exonic, as well as their sorting into sense and antisense miRNAs, was also derived from this database. The identifiers of mature miRNAs corresponding to pre-miRNAs were obtained from the miRBase database93,94 (Release 21). The lists of validated targets of mature miRNAs were downloaded from the DIANA-TarBase v7.042 and miRTarBase (Release 6.1)43 databases. An mRNA target was considered validated if both databases confirmed the targeting by a corresponding miRNA.
The analysis of the resulting networks was performed using the igraph library95. Multiple edges and loops, as well as isolated nodes, were removed prior to the analysis. This graph simplification used igraph R-package command simple.
The network core was defined as the union of nontrivial (i.e., containing at least two nodes) strongly connected components. These components were found using igraph command components with parameter mode = “strong”.
The comparison of properties of the constructed networks with the properties of random graphs was performed using the Erdős–Rényi model44 and a model based on fixed out-degrees. For both models, the number of vertices and edges coincided with the number of nodes and edges in the analyzed networks of miRNA-induced intergenic interactions. For the Erdős–Rényi model, the presence of an edge with given starting and ending vertices was equiprobable for all ordered pairs of vertices. For the second model, a number of outgoing edges Ng was fixed for each vertex g, and all possible sets of ending vertices for these edges (i.e., sets consisting of exactly Ng vertices and not containing g) were equiprobable.
Random graph generation under the Erdős–Rényi model was performed using igraph R-package command erdos.renyi.game with parameters type = “gnm”, directed = TRUE, loops = FALSE.
Generation of random graphs under the second model was performed as follows. Let |V| be the number of nodes (genes) in the analyzed networks of miRNA-induced intergenic interactions, and let (N1, N2, …, N|V|) be a vector of out-degrees. We obtained this vector using igraph command degree with parameter mode = “out”. Random graph generation started from a graph with |V| nodes {1, 2, …, |V|} and no edges. Then for each k from 1 to |V| a random permutation on {1, 2, …, |V| − 1} was generated using the Fisher-Yates shuffle96, first Nk elements of the generated permutation were selected, and for each selected element l a directed edge from node k to node l (if l < k) or to node (l + 1) (if l ≥ k) was added to the graph.
Distribution of properties of random graphs was assessed using the standard Monte Carlo approach after generating 10,000 graphs for each model.
The enrichment analysis was performed using DAVID 6.7 online service97 with the default background setting (i.e., with a background automatically selected by DAVID).
Construction of prognostic gene signatures
The prognostic gene signatures were constructed using SVM57 with the linear kernel as described previously40. The triples with the highest prognostic power were identified by an exhaustive search with the threshold for sensitivity and specificity equal to 60% and 50% for the training and the filtration datasets, respectively. The triples that passed the filtration were further sorted in a descending order with respect to the mean value of AUC for the training and filtration datasets. The same procedure was used for the analysis of signatures with a different size (pairs, quadruples).
Construction of signatures for patients with ER-positive breast cancer utilized the collection of microarray datasets identical to the one we used previously40, as well as the previously used division into groups (recurrence within 5 years, no recurrence and follow-up of at least 7 years, and the remaining “gray zone”).
Construction of signatures for patients with colorectal cancer utilized the GSE39582 series98 as a training dataset, GSE3789299 and GSE17536100,101 as filtration datasets, and GSE14333102 as a validation (testing) dataset. Datasets were jointly preprocessed using RMA method103. Numbers of patients in these datasets are presented in Supplementary Table 6. At the training and filtration stages, only patients with recurrence within the first three years and patients without recurrence and follow-up of for at least four years were considered; other patients (comprising the so-called “grey zone”) were excluded from the analysis. Patients with an unknown recurrence status and patients with a recurrence within the first month were excluded from the analysis as well.
For the identified triples of genes with the highest prognostic power a 3-fold and 10-fold cross validation on the training dataset was performed as an additional test that proved an absence of overfitting. The results of cross-validation are presented in Supplementary Information (section Cross-validation of the identified triples of genes on the training datasets).
Analysis of expression levels
Analysis of expression levels and expression variance of transcripts in the cell lines was performed using information from the RNA-Seq dataset E-MTAB-270656, which contains genome-wide transcriptome profiles of 675 cancer cell lines. Reads per kilobase per million mapped reads (RPKM) metrics was used to represent the expression levels of genes; values of RPKM reported in the dataset file 140625_Klijn_RPKM_coding.txt were used. The variance in expression level for a given transcript was quantified as the third quartile to the first quartile ratio of expression levels for this transcript. With respect to the gene ranking, this quantification was equivalent to the quantification based on the interquartile range in log-scale. Transcripts that remained in the upper half after ranking by the first quartile of expression level had positive values of this quartile, and hence, the above quantification of expression variance was applicable for these transcripts.
Analysis of expression levels of genes in tissue specimens obtained from breast and colorectal cancer patients was performed similarly and utilized the following microarray datasets: GSE17705104 for breast cancer patients and GSE3958298 for colorectal cancer patients. These datasets were preprocessed using RMA method103 as parts of larger sets utilized for construction of prognostic gene signatures as described in the previous subsection. The analysis used expression levels reported by the RMA preprocessing algorithm.
Statistical analysis
The significance level was set to 0.05. For Monte Carlo hypothesis testing, a conventional estimate of the p-value was used105. Log-rank tests were utilized to compare survival distribution between groups of patients; two-tailed p-values are reported. Binomial tests were used to analyze the size of intersection between halves or quarters of genes (with respect to a certain ranking) with the lists of genes comprising the network core.
Data availability
The authors declare that the data supporting the findings of this study are available within the article and Supplementary Information or available from the authors upon request. All utilized datasets are properly referenced in the article and can be downloaded from publicly available databases.
Electronic supplementary material
Acknowledgements
We thank Prof. U. Schumacher, Prof. M.S. Gelfand, Prof. B.D. Zhivotovsky and Dr. L. Brodsky for valuable discussions, comments and suggestions. The research was supported by the Russian Science Foundation (project 17-14-01338).
Author Contributions
V.V.G., A.V.G., M.Y.S. and T.R.S. conceived and designed the research. V.V.G. and A.V.G. integrated data and conducted in silico experiments. V.V.G., J.A.M., A.A.T., M.Y.S., T.R.S. and A.G.T. interpreted the results. V.V.G., A.V.G., A.A.T. and T.R.S. wrote the manuscript. All the authors edited, discussed and approved the whole paper.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Vladimir V. Galatenko and Alexey V. Galatenko contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20215-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vladimir V. Galatenko, Email: vgalat@imscs.msu.ru
Alexander G. Tonevitsky, Email: tonevitsky@mail.ru
References
- 1.Papagiannakopoulos T, Kosik KS. MicroRNAs: regulators of oncogenesis and stemness. BMC Med. 2008;6:15. doi: 10.1186/1741-7015-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samatov TR, Tonevitsky AG, Schumacher U. Epithelial-mesenchymal transition: focus on metastatic cascade, alternative splicing, non-coding RNAs and modulating compounds. Mol. Cancer. 2013;12:107. doi: 10.1186/1476-4598-12-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eulalio A, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 4.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 2011;12:136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Osella M, Riba A, Testori A, Corà D, Caselle M. Interplay of microRNA and epigenetic regulation in the human regulatory network. Front. Genet. 2014;5:345. doi: 10.3389/fgene.2014.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwama H. Coordinated networks of microRNAs and transcription factors with evolutionary perspectives. Adv. Exp. Med. Biol. 2013;774:169–187. doi: 10.1007/978-94-007-5590-1_10. [DOI] [PubMed] [Google Scholar]
- 8.Tonevitsky AG, et al. Dynamically regulated miRNA-mRNA networks revealed by exercise. BMC Physiol. 2013;13:9. doi: 10.1186/1472-6793-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avraham R, et al. EGF decreases the abundance of microRNAs that restrain oncogenic transcription factors. Sci. Signal. 2010;3:ra43. doi: 10.1126/scisignal.2000876. [DOI] [PubMed] [Google Scholar]
- 10.Emmerling VV, et al. miR-483 is a self-regulating microRNA and can activate its own expression via USF1 in HeLa cells. Int. J. Biochem. Cell Biol. 2016;80:81–86. doi: 10.1016/j.biocel.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 11.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 12.França GS, Vibranovski MD, Galante PA. Host gene constraints and genomic context impact the expression and evolution of human microRNAs. Nat. Commun. 2016;7:11438. doi: 10.1038/ncomms11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinske LC, et al. miRIAD – integrating microRNA inter- and intragenic data. Database (Oxford) 2014;2014:bau099. doi: 10.1093/database/bau099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinske LC, Galante PA, Kuo WP, Ohno-Machado L. A potential role for intragenic miRNAs on their hosts’ interactome. BMC Genomics. 2010;11:533. doi: 10.1186/1471-2164-11-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monteys AM, et al. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, et al. Systematic study of cis-antisense miRNAs in animal species reveals miR-3661 to target PPP2CA in human cells. RNA. 2016;22:87–95. doi: 10.1261/rna.052894.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito M, et al. A trans-homologue interaction between reciprocally imprinted miR-127 and Rtl1 regulates placenta development. Development. 2015;142:2425–2430. doi: 10.1242/dev.121996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Megraw M, et al. Isoform specific gene auto-regulation via miRNAs: a case study on miR-128b and ARPP-21. Theor. Chem. Acc. 2009;125:593–598. doi: 10.1007/s00214-009-0647-4. [DOI] [Google Scholar]
- 20.Nikolic I, Plate KH, Schmidt MH. EGFL7 meets miRNA-126: an angiogenesis alliance. J. Angiogenes. Res. 2010;2:9. doi: 10.1186/2040-2384-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kos A, et al. A potential regulatory role for intronic microRNA-338-3p for its host gene encoding apoptosis-associated tyrosine kinase. PLoS One. 2012;7:e31022. doi: 10.1371/journal.pone.0031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dill H, Linder B, Fehr A, Fischer U. Intronic miR-26b controls neuronal differentiation by repressing its host transcript, ctdsp2. Genes Dev. 2012;26:25–30. doi: 10.1101/gad.177774.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sundaram GM, et al. ‘See-saw’ expression of microRNA-198 and FSTL1 from a single transcript in wound healing. Nature. 2013;495:103–106. doi: 10.1038/nature11890. [DOI] [PubMed] [Google Scholar]
- 24.Bosia C, Osella M, Baroudi ME, Corà D, Caselle M. Gene autoregulation via intronic microRNAs and its functions. BMC Syst. Biol. 2012;6:131. doi: 10.1186/1752-0509-6-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu M, et al. The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev. 2013;27:2543–2548. doi: 10.1101/gad.224170.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corcoran DL, et al. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One. 2009;4:e5279. doi: 10.1371/journal.pone.0005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramalingam P, et al. Biogenesis of intronic miRNAs located in clusters by independent transcription and alternative splicing. RNA. 2014;20:76–87. doi: 10.1261/rna.041814.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez NJ, et al. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao X, Qiao Y, Han D, Zhang Y, Ma N. Enemy or partner: relationship between intronic micrornas and their host genes. IUBMB Life. 2012;64:835–840. doi: 10.1002/iub.1079. [DOI] [PubMed] [Google Scholar]
- 32.Schmitt DC, Tan M. The Enemy within: regulation of host genes by intronic miRNAs. Chemotherapy. 2014;3:e126. [Google Scholar]
- 33.Lutter D, Marr C, Krumsiek J, Lang EW, Theis FJ. Intronic microRNAs support their host genes by mediating synergistic and antagonistic regulatory effects. BMC Genomics. 2010;11:224. doi: 10.1186/1471-2164-11-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandemakers W, et al. Co-regulation of intragenic microRNA miR-153 and its host gene Ia-2 β: identification of miR-153 target genes with functions related to IA-2β in pancreas and brain. Diabetologia. 2013;56:1547–1556. doi: 10.1007/s00125-013-2901-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barik S. An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res. 2008;36:5232–5241. doi: 10.1093/nar/gkn513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siciliano V, et al. miRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat. Commun. 2013;4:2364. doi: 10.1038/ncomms3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat. Genet. 2006;38:S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 39.Del Rosario RC, Damasco JR, Aguda BD. MicroRNA inhibition fine-tunes and provides robustness to the restriction point switch of the cell cycle. Sci. Rep. 2016;6:32823. doi: 10.1038/srep32823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galatenko VV, et al. Highly informative marker sets consisting of genes with low individual degree of differential expression. Sci. Rep. 2015;5:14967. doi: 10.1038/srep14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gray RG, et al. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J. Clin. Oncol. 2011;29:4611–4619. doi: 10.1200/JCO.2010.32.8732. [DOI] [PubMed] [Google Scholar]
- 42.Vlachos, I. S. et al. DIANA-TarBasev7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 43(Database issue), D153–D159 (2015). [DOI] [PMC free article] [PubMed]
- 43.Chou CH, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44(D1):D239–D247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erdős P, Rényi A. On Random Graphs I. (PDF) Publ. Math. 1959;6:290–297. [Google Scholar]
- 45.Costa A, Onesti S. The MCM complex: (just) a replicative helicase? Biochem. Soc. Trans. 2008;36(Pt 1):136–140. doi: 10.1042/BST0360136. [DOI] [PubMed] [Google Scholar]
- 46.Jeppsson K, Kanno T, Shirahige K, Sjögren C1. The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nat. Rev. Mol. Cell. Biol. 2014;15:601–614. doi: 10.1038/nrm3857. [DOI] [PubMed] [Google Scholar]
- 47.R HR, Kim H, Noh K, Kim YJ. The diverse roles of RNA polymerase II C-terminal domain phosphatase SCP1. BMB Rep. 2014;47:192–196. doi: 10.5483/BMBRep.2014.47.4.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mangs AH, Morris BJ. ZRANB2: structural and functional insights into a novel splicing protein. Int. J. Biochem. Cell Biol. 2008;40:2353–2357. doi: 10.1016/j.biocel.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 49.Chu J, Cargnello M, Topisirovic I, Pelletier J. Translation Initiation Factors: Reprogramming Protein Synthesis in Cancer. Trends Cell Biol. 2016;26:918–933. doi: 10.1016/j.tcb.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 50.Paytubi S, et al. ABC50 promotes translation initiation in mammalian cells. J. Biol. Chem. 2009;284:24061–24073. doi: 10.1074/jbc.M109.031625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Horn DM, Mason SL, Karbstein K. Rcl1 protein, a novel nuclease for 18 S ribosomal RNA production. J. Biol. Chem. 2011;286:34082–34087. doi: 10.1074/jbc.M111.268649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheffner M, Kumar S. Mammalian HECT ubiquitin-protein ligases: biological and pathophysiological aspects. Biochim. Biophys. Acta. 2014;1843:61–74. doi: 10.1016/j.bbamcr.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 53.Barboro P, Ferrari N, Balbi C. Emerging roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in cancer progression. Cancer Lett. 2014;352:152–159. doi: 10.1016/j.canlet.2014.06.019. [DOI] [PubMed] [Google Scholar]
- 54.Dansie LE, et al. Physiological roles of the pantothenate kinases. Biochem. Soc. Trans. 2014;42:1033–1036. doi: 10.1042/BST20140096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Popow J, et al. FASTKD2 is an RNA-binding protein required for mitochondrial RNA processing and translation. RNA. 2015;21:1873–1884. doi: 10.1261/rna.052365.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klijn C, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 2015;33:306–312. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 57.Cortes C, Vapnik V. Support-Vector Networks. Mach. Learn. 1995;20:273–297. [Google Scholar]
- 58.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 59.EGAPP Working Group. Recommendations from the EGAPP Working Group: can tumor gene expression profiling improve outcomes in patients with breast cancer? Genet. Med. 11, 66–73 (2009) [DOI] [PMC free article] [PubMed]
- 60.Van’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 61.O’Connell MJ, et al. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J. Clin. Oncol. 2010;28:3937–3944. doi: 10.1200/JCO.2010.28.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou B, et al. A novel miR-219-SMC4-JAK2/Stat3 regulatory pathway in human hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2014;33:55. doi: 10.1186/1756-9966-33-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang C, et al. SMC4, which is essentially involved in lung development, is associated with lung adenocarcinoma progression. Sci. Rep. 2016;6:34508. doi: 10.1038/srep34508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ren B, et al. MCM7 amplification and overexpression are associated with prostate cancer progression. Oncogene. 2006;25:1090–1098. doi: 10.1038/sj.onc.1209134. [DOI] [PubMed] [Google Scholar]
- 65.Erkan EP, et al. Depletion of minichromosome maintenance protein 7 inhibits glioblastoma multiforme tumor growth in vivo. Oncogene. 2014;33:4778–4785. doi: 10.1038/onc.2013.423. [DOI] [PubMed] [Google Scholar]
- 66.Qu K, et al. MCM7 promotes cancer progression through cyclin D1-dependent signaling and serves as a prognostic marker for patients with hepatocellular carcinoma. Cell Death Dis. 2017;8:e2603. doi: 10.1038/cddis.2016.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gallardo M, et al. hnRNP K is a haploinsufficient tumor tuppressor that regulates proliferation and differentiation programs in hematologic malignancies. Cancer Cell. 2015;28:486–499. doi: 10.1016/j.ccell.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ahn YH, et al. Map2k4 functions as a tumor suppressor in lung adenocarcinoma and inhibits tumor cell invasion by decreasing peroxisome proliferator-activated receptor γ2 expression. Mol. Cell. Biol. 2011;31:4270–4285. doi: 10.1128/MCB.05562-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davis SJ, et al. Analysis of the mitogen-activated protein kinase kinase 4 (MAP2K4) tumor suppressor gene in ovarian cancer. BMC Cancer. 2011;11:173. doi: 10.1186/1471-2407-11-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao R, et al. Heterogeneous nuclear ribonucleoprotein K (hnRNP-K) promotes tumor metastasis by induction of genes involved in extracellular matrix, cell movement, and angiogenesis. J. Biol. Chem. 2013;288:15046–15056. doi: 10.1074/jbc.M113.466136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pavese JM, et al. Mitogen-activated protein kinase kinase 4 (MAP2K4) promotes human prostate cancer metastasis. PLoS One. 2014;9:e102289. doi: 10.1371/journal.pone.0102289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Das S, Yeung KT, Mahajan MA, Samuels HH. Fas Activated Serine-Threonine Kinase Domains 2 (FASTKD2) mediates apoptosis of breast and prostate cancer cells through its novel FAST2 domain. BMC Cancer. 2014;14:852. doi: 10.1186/1471-2407-14-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adhikary S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 74.Inoue S, et al. Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15. Genes Dev. 2013;27:1101–1114. doi: 10.1101/gad.214577.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vaughan L, et al. HUWE1 ubiquitylates and degrades the RAC activator TIAM1 promoting cell-cell adhesion disassembly, migration, and invasion. Cell Rep. 2015;10:88–102. doi: 10.1016/j.celrep.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Myant KB, et al. HUWE1 is a critical colonic tumour suppressor gene that prevents MYC signalling, DNA damage accumulation and tumour initiation. EMBO Mol. Med. 2017;9:181–197. doi: 10.15252/emmm.201606684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, Metabolism, and Cancer. Cancer Discov. 2015;5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kress TR, Sabò A, Amati B. MYC: connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer. 2015;15:593–607. doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- 80.Baek J, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 2011;11:163–175. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- 82.Kurtz, C. L. et al. MicroRNA-29 fine-tunes the expression of key FOXA2-activated lipid metabolism genes and is dysregulated in animal models of insulin resistance and diabetes. Diabetes63, 3141–3148 (2014). [DOI] [PMC free article] [PubMed]
- 83.Rottiers V, Näär AM. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hartig SM, Hamilton MP, Bader DA, McGuire SE. The miRNA Interactome in metabolic homeostasis. Trends Endocrinol Metab. 2015;26:733–745. doi: 10.1016/j.tem.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Trajkovski M, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–663. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 86.Lee YS, Dutta A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15:38. doi: 10.1186/s12935-015-0185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O’Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010;12:201. doi: 10.1186/bcr2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet. 2016;17:719–732. doi: 10.1038/nrg.2016.134. [DOI] [PubMed] [Google Scholar]
- 90.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kotas ME, Medzhitov R. Homeostasis, inflammation, and disease susceptibility. Cell. 2015;160:816–827. doi: 10.1016/j.cell.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang SJ, et al. p53-Dependent regulation of metabolic function through transcriptional activation of pantothenate kinase-1 gene. Cell Cycle. 2013;12:753–761. doi: 10.4161/cc.23597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Griffiths-Jones, S., Grocock, R. J., van Dongen, S., Bateman, A. & Enright, A. J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34(Database issue), D140–D144 (2006). [DOI] [PMC free article] [PubMed]
- 94.Kozomara, A. & Griffiths-Jones, S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42(Database issue), D68–D73 (2014). [DOI] [PMC free article] [PubMed]
- 95.Csardi G, Nepusz T. The igraph software package for complex network research. InterJ. Complex Syst. 2006;1695:1–9. [Google Scholar]
- 96.Knuth, D. E. The Art of Computer Programming, volume 2: Seminumerical Algorithms (Addison-Wesley, 1969).
- 97.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 98.Marisa L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Laibe S, et al. A seven-gene signature aggregates a subgroup of stage II colon cancers with stage III. OMICS. 2012;16:560–565. doi: 10.1089/omi.2012.0039. [DOI] [PubMed] [Google Scholar]
- 100.Smith JJ, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Freeman TJ, et al. Smad4-mediated signaling inhibits intestinal neoplasia by inhibiting expression of β-catenin. Gastroenterology. 2012;142:562–571.e2. doi: 10.1053/j.gastro.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jorissen RN, et al. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clin. Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 104.Symmans WF, et al. Genomic index of sensitivity to endocrine therapy for breast cancer. J. Clin. Oncol. 2010;28:4111–4119. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ewens WJ. On estimating P values by the Monte Carlo method. Am. J. Hum. Genet. 2003;72:496–498. doi: 10.1086/346174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the article and Supplementary Information or available from the authors upon request. All utilized datasets are properly referenced in the article and can be downloaded from publicly available databases.