Abstract
Coactosin-like protein 1 (Cotl1), a member of the actin-depolymerizing factor (ADF)/cofilin family, was first purified from a soluble fraction of Dictyostelium discoideum cells. Neuronal migration requires cytoskeletal remodeling and actin regulation. Although Cotl1 strongly binds to F-actin, the role of Cotl1 in neuronal migration remains undescribed. In this study, we revealed that Cotl1 overexpression impaired migration of both early- and late-born neurons during mouse corticogenesis. Moreover, Cotl1 overexpression delayed, rather than blocked, neuronal migration in late-born neurons. Cotl1 expression disturbed the morphology of migrating neurons, lengthening the leading processes. This study is the first to investigate the function of Cotl1, and the results indicate that Cotl1 is involved in the regulation of neuronal migration and morphogenesis.
Keywords: Cotl1 protein, actins, in utero electroporation, neuronal migration
Introduction
Neuronal layers are formed during neuronal migration, a fundamental process in brain development. In the mammalian cerebral cortex, pyramidal neurons are generated in the ventricular zone (VZ) and migrate to the cortical plate (CP) via the subventricular zone (SVZ) and the intermediate zone (IZ) to form a six-layered structure [22,23]. Late-born neurons migrate past early-born neurons to superficial positions. This migration pattern produces an inside-first and outside-last gradient [1]. Early- and late-born cortical neurons display distinct migratory behaviors that are regulated by different molecular mechanisms [9]. Disturbing any process can impair neuronal migration and morphogenesis in the cerebral cortex, thereby causing neurological syndromes, such as epilepsy and Alzheimer's disease [15,21,25].
Neuronal migration is a coordinated process that includes extension of the leading process, translocation of the nucleus, and retraction of the trailing process. These processes are associated with cytoskeletal rearrangement [11,18]. Actin-associated proteins and microtubule-associated proteins regulate the reorganizations of microtubules and F-actin, which are essential for neuronal migration. The actin cytoskeleton is regulated by numerous actin-binding proteins, including the family of actin-depolymerizing factor homology (ADF-H) domains [8].
The ADF-H domains consist of three phylogenetically distinct classes: ADF/cofilins, twinfilins, and drebrin/Abp1s [14]. Coactosin-like protein 1 (Cotl1) is a member of the ADF/cofilin family and is highly related to coactosin, an actin-binding protein. Coactosin was first purified from a soluble fraction of Dictyostelium discoideum cells. In vitro, Cotl1 binds strongly to actin filaments [6] and to F-actin but not to G-actin [13]. In vitro studies have demonstrated that Cotl1 and cofilin compete with each other for binding to F-actin. Moreover, Cotl1 regulates lamellipodial dynamics by protecting F-actin from cofilin-mediated depolymerization [13]. A recent study showed that, in CHO cells, Cotl1 directly binds to F-actin without forming stable complexes with G-actin [19]. Chai et al. [4] have shown that cofilin and reelin cooperate in controlling cytoskeletal dynamics during neuronal migration. These results implicate Cotl1 in neuronal migration. However, the function of Cotl1 during neuronal migration remains undescribed.
In the present study, we examined the expression of Cotl1 mRNA in the developing cerebral cortex. Our results revealed that Cotl1 overexpression impaired the migration of both early-born and late-born neurons during mouse corticogenesis.
Materials and Methods
Animals
The ICR mice used in this study were purchased from Xi'an Jiao-tong University, China. Mice were fed and handled according to international guidelines. The animal protocols were approved by Northwest A&F University, China (approval No. 20111006-2).
Antibodies and agents
Antibodies included rabbit anti-green fluorescent protein (GFP), goat anti-myc, 4′,6-diamidino-2-phenylindole (DAPI; Millipore, USA), Alexa Fluor 488-conjugated donkey anti-rabbit IgG and Alexa Fluor 568-conjugated donkey anti-goat IgG (Life Technologies, USA). Bovine serum albumin, Triton X-100, and normal goat serum were purchased from Sigma-Aldrich (USA).
RNA bioanalyzer
A bioanalyzer (Agilent Technologies, USA) was used to test Cotl1 expression in mouse cerebral cortex at different embryonic stages. Total RNA was extracted on ice by lysing cortical tissue in Trizol (Thermo Fisher Scientific, USA). Then, the cell lysate was centrifuged at 4℃, the supernatant removed, and isopropyl alcohol added. The solution was allowed to stand at room temperature for 10 min and then centrifuged at 4℃, discarded the supernatant. Then 75% alcohol was added to the solution and centrifuged at 4℃ to discard the supernatant. Diethylpyrocarbonate H2O was added to dissolve the precipitate. Finally, total RNA was transcribed to cDNA by using reverse transcripition kit (Thermo Fisher Scientific). We examined the cDNA sequences to determine the expression of Cotl1 in mouse cerebral cortex. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal reference. Forward 5′-AGCGAGACCCCACTAACA-3′ and reverse 5′-ATGAGCCCTTCCACAATG-3′ primers were used.
Plasmid construction
The full length of Cotl1 (GenBank: NM_028071.3) was constructed by performing reverse transcription polymerase chain reaction (RT-PCR) with total embryonic brain RNA and specific primers: forward 5′-GGAATTC ATGGCCACGAAGATCGACAAGGAGG-3′ and reverse 5′-GCGTCGACGCCTCTGACTGAGCGTCGTAGTTGGCC-3′. The enzyme sites (EcoR I and Sal I) were purchased from Fermentas (USA) and are indicated by the underlined sections of the primers. The Cotl1 fragment was inserted into pCAG-MCS-myc and pCAG-MCS-GFP was used as a control.
In utero electroporation
Plasmid DNA was prepared by using a QIAGEN Plasmid Plus Midi Kit (QIAGEN, Germany). Plasmids were transfected via trans-uterus pressure microinjection by using in utero electroporation (IUE) [4,10]. Briefly, a pregnant mouse was anesthetized with 0.56% sodium pentobarbitone (1 mL/100 g) and the uterus and embryos were exposed. The pCAG-Cotl1-myc, together with pCAG-MCS-GFP (pCAG-Cotl1-myc final concentration 3 µg/µL and pCAG-MCS-GFP final concentration 0.5 µg/µL), were mixed with Fast Green (1 mg/mL; Sigma-Aldrich) and injected into the embryo's lateral ventricle at embryonic days (E)12.5 and E15.5. The pCAG-MCS-GFP was used as the control (final concentration 3 µg/µL). For electroporation, electrical pulses were generated by a square wave electroporation system (ECM830; BTX, USA) and applied through five pulses (30 V; 50 msec duration; 950 msec interval). Subsequently, the embryos were replaced in the uterus to allow them to continue normal development. Embryo brains were harvested at specified time points.
Immunohistochemistry and confocal microscopy
Brains were harvested on E16.5 after electroporation at E12.5 and on postnatal days (P)1 and P7 after electroporation at E15.5 [7,9]. At least four embryonic mouse brains, regardless of sex, were harvested per group. Harvested brains were fixed in 4% paraformaldehyde for at least 48 h at 4℃. Subsequently, brains were sliced into 50 µm thick sections with a vibratome (VT 1000S; Leica Microsystems, Germany). The sections were incubated overnight at 4℃ with rabbit anti-GFP (1:1,000) or goat anti-myc (1:500). Sections were washed at least thrice with 0.1 M PBS and then incubated with Alexa Fluor 488-conjugated donkey anti-rabbit IgG (1:1,000) or Alexa Fluor 568-conjugated donkey anti-goat IgG (1:1,000) and DAPI at room temperature. The brain sections were then washed and covered with fluorescent mounting medium (Dako, Denmark) on glass and visualized by using a fluorescent microscope (Axio Observer Z1; Zeiss, Germany).
Statistical analysis
Eight slices were randomly selected from four transfected mouse brains per group. Slice selection was performed under fluorescent microscopy [7,27]. Quantification areas in the somatosensory cortex slices were selected by using a rectangular frame. All transfected (GFP-positive) cells in each layer of the frame were counted by using the ZEN 2012 program (Zeiss). Subsequently, the cell percentage in each layer was obtained to determine the effect of transfection on migration. The percentage of cells in each layer was calculated as the GFP-positive cells in each layer/GFP-positive cells in the frame. Statistical analysis was performed by using GraphPad Prism software (ver. 5.0; GraphPad Software, USA) to apply unpaired t-tests. A p value < 0.05 (*) was considered significant. A p value < 0.01 (**) was considered very significant, and a p value < 0.001 (***) was considered extremely significant.
Results
Cotl1 RNA expression was attenuated during specific developmental periods of mouse cerebral cortex
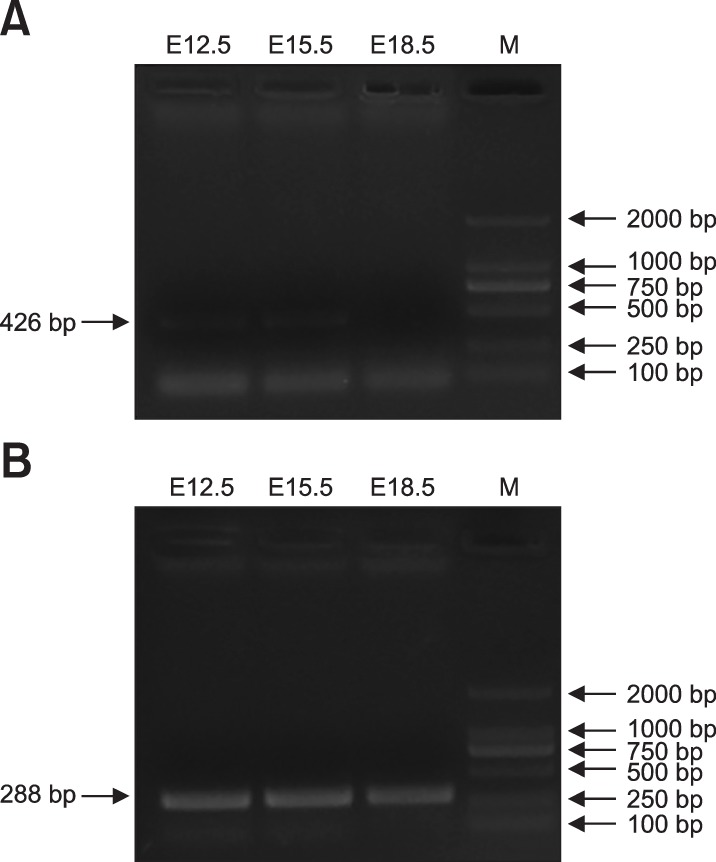
A previous study reported that Cotl1 mRNA transcripts are expressed in the human brain [19]. The results in the present study showed expression of Cotl1 mRNA in mouse embryonic brain, indicating its role in the development of the cerebral cortex. The results showed that Cotl1 mRNA was expressed at E12.5 and E15.5 at low levels but its expression could not be detected at E18.5 (Fig. 1).
Fig. 1. RNA bioanalyzer results for coactosin-like protein 1 (Cotl1) by the polymerase chain reaction. (A) Cotl1 RNA was expressed at a low level in specific periods in developing mouse cerebral cortex. (B) Internal reference (glyceraldehyde 3-phosphate dehydrogenase [GAPDH]) expression in mouse cerebral cortex. E, embryonic days; M, marker.
Cotl1 overexpression impaired the migration of early-born neurons in a dose-dependent manner
To investigate the function of Cotl1 in neuronal migration, we transfected neural progenitor cells with the Cotl1 expression vector and the GFP plasmid via IUE. Given that early- and late-born neurons display distinct migratory behaviors, electroporation was first performed at E12.5 to investigate the role of Cotl1 in the migration of early-born neurons. The distribution of the transfected cells was analyzed at E16.5. We found that approximately 72% of the transfected neurons reached the CP in the control group, whereas only approximately 14% of the cells migrated to the IZ. Moreover, only approximately 59% of the Cotl1-transfected cells reached the CP, while more than 30% of the neurons clustered in the IZ (panels A and B in Fig. 2). These results indicated that Cotl1 overexpression impaired the migration of early-born neurons.
Fig. 2. Coactosin-like protein 1 (Cotl1) overexpression impaired the migration of early-born neurons in a dose-dependent manner. (A) Cotl1 impeded neuronal migration at embryonic days (E)16.5. (B) Quantification of transfected neurons in each layer at E16.5. Significance of differences between the Cotl1 and green fluorescent protein (GFP) control groups for each layer were determined by using t-tests. (C) Average fluorescence intensity in intermediate zone (IZ) and cortical plate (CP) layers at E16.5 were significantly different (***p<0.001). Bars indicate mean±SEM. Scale bar = 100 µm. DAPI, 4′,6-diamidino-2-phenylindole; VZ/SVZ, ventricular zone/subventricular zone.
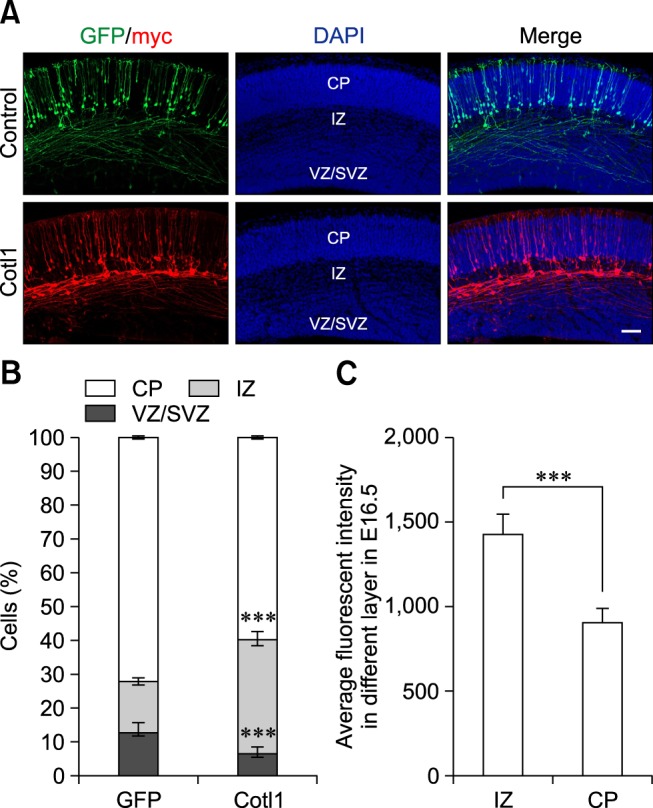
We further analyzed Cotl1 expression in the CP and IZ. The fluorescence intensity of the transfected cells was measured by using ZEN 2012 software. The average fluorescence intensity of the neurons in the IZ (1,430.0±128.2) was significantly higher than that in the CP (907.5±81.8) (panel C in Fig. 2). This result indicated that Cotl1 impaired the migration of early-born neurons in a dose-dependent manner.
Cotl1 overexpression slowed down the neuronal migration of late-born neurons
To investigate the function of Cotl1 in late-born neurons, E15.5 neural progenitor cells were transfected with the Cotl1 expression vector and the GFP plasmid. In the control group, nearly 80% of the transfected neurons reached the upper CP (UCP), whereas only approximately 8% of the transfected neurons resided in the IZ in the control group at P1, and approximately 6.7% of transfected neurons resided in the VZ. However, the distribution of the transfected cells was significantly different in the Cotl1-transfected group: 27% (p<0.0001) of Cotl1-transfected neurons reached the UCP, 32% (p<0.0001) of neurons had arrested in the IZ, and 30% of the neurons (p=0.0097) remained in the VZ (panels A and C in Fig. 3). These results indicated that Cotl1 overexpression impaired the migration of late-born neurons.
Fig. 3. (A) Coactosin-like protein 1 (Cotl1) overexpression slowed the migration of late-born neurons at P1. (B) Neurons reached the upper cortical plate (UCP) at P7. (C) Quantification showed that neuron numbers were significantly decreased in the CP layers in the Cotl1-transfected group than in the green fluorescent protein (GFP) control group, but were significantly increased in the intermediate zone (IZ) and ventricular zone (VZ). Scale bars = 100 µm. *p<0.05, **p<0.01, ***p<0.001. E, embryonic days; P, postnatal days; DAPI, 4′,6-diamidino-2-phenylindole; DCP, deeper cortical plate.
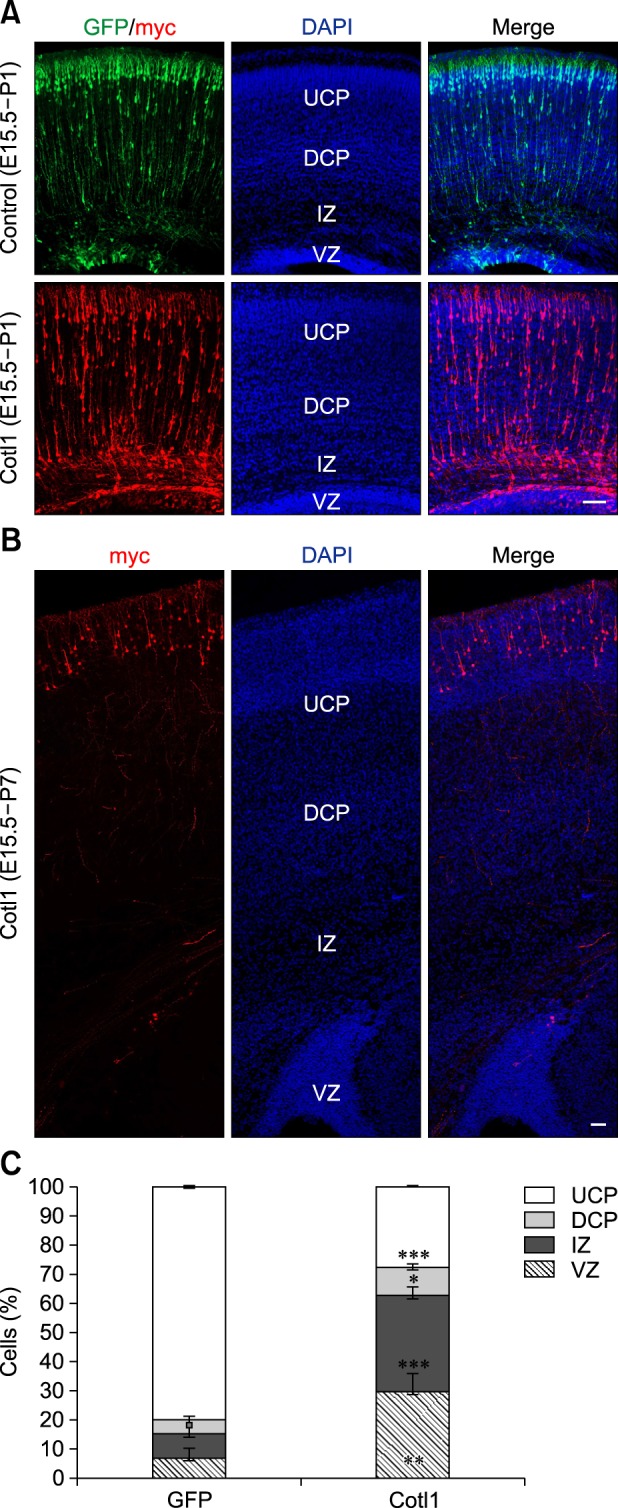
The distribution of the transfected neurons was analyzed at P7 to investigate whether Cotl1 expression blocked or delayed neuronal migration. We observed that nearly all transfected neurons reached the UCP (panel B in Fig. 3), suggesting that Cotl1 overexpression delayed, rather than blocked, the migration of late-born neurons.
Cotl1 overexpression lengthened the leading process
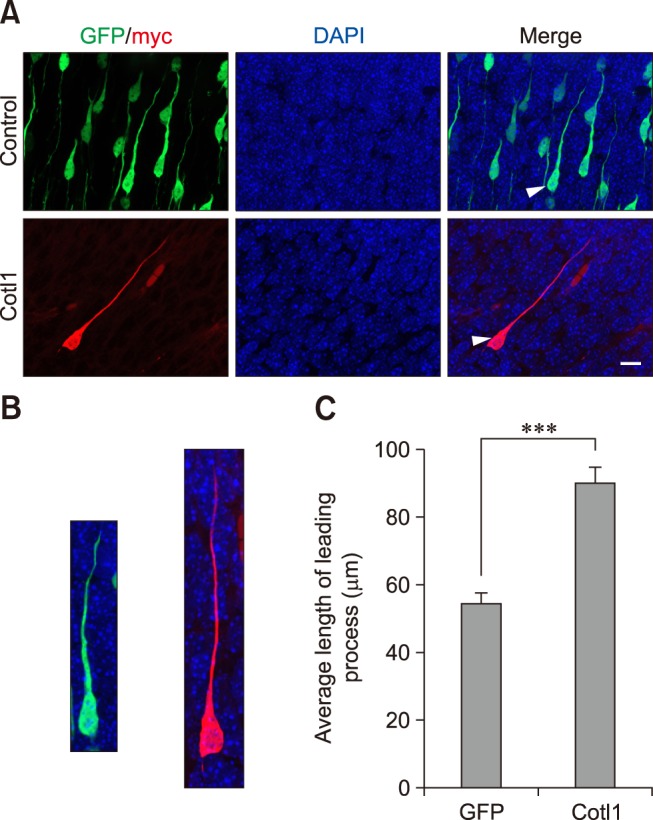
We examined the morphology of Cotl1-transfected neurons in the CP to investigate the mechanisms contributing to the Cotl1 effect on late-born neuronal migration. The Cotl1-transfected neurons exhibited a typical bipolar morphology with a leading process toward the pial surface. However, the leading processes in the Cotl1-transfected group were longer than those in the control group (panels A and B in Fig. 4). The average length of the leading processes in the control group was 54.41±2.91 µm, whereas it was 89.6± 4.45 µm in the Cotl1-transfected group, an extremely significant difference (Fig. 4). Thus, Cotl1 overexpression modified the morphology of the migrating neurons, which delayed neuronal migration.
Fig. 4. Coactosin-like protein 1 (Cotl1) induced an abnormal morphology in neurons of the cerebral cortex. (A) The Cotl1-transfected neurons that migrated into the cortical plate were different from those in the green fluorescent protein (GFP) control group. The average length of the leading process in the Cotl1-transfected group was significantly greater than that in the GFP control group. The arrow refers to a single neuron as shown in panel B. (B) Single neuron transfected with plasmid; left image is a representative neuron from the control group and the right image is from the Cotl1-transfected group. (C) The statistic of the length of neuronal leading process. Scale bar = 10 µm. ***p<0.001. DAPI, 4′,6-diamidino-2-phenylindole.
Discussion
We observed that Cotl1 RNA was expressed during specific periods in the course of development of mouse cerebral cortex. Moreover, Cotl1 overexpression impaired both early- and late-born neuronal migration in a dose-dependent manner. In addition, neuronal leading processes were elongated when late-born neurons migrated to the CP. Overall, these results suggested that Cotl1 overexpression impaired late-born neuronal migration partly because of the longer leading process in the CP.
Neuronal migration requires the balance of many molecules in multiple steps [3,7,24]. Postmitotic neurons that are generated in the VZ/SVZ migrate toward the pial surface and experience various morphological changes and neuronal maturation events [12,20]. There are two modes of neuronal migration in the cerebral cortex: radial and tangential [2,5,17,26]. Radial migration is crucial in the establishment of layered structures. Previous reports have suggested that both early-born neurons (the earliest emigrants from the E12.5 VZ) and late-born neurons (those derived from the E15.5 VZ) transform from a stellate to a bipolar shape. Neurons show migratory behavior that is regulated by distinct molecular mechanisms [9]. Somal translocation occurs during relatively early stages of corticogenesis in the E13 to E14 mouse [17], whereas locomotion subsequently dominates [16]. In the present study, Cotl1 overexpression impaired the neuronal migration of both early- and late-born neurons. This result suggested that Cotl1 is required for both somal translocation and locomotion. Neuronal migration requires precisely coordinated cytoskeletal dynamics. Cotl1 competes with cofilin to bind with F-actin, and previous studies have shown that cofilin is important for neuronal migration. Moreover, cofilin overexpression has been reported to lengthen leading processes, indicating that Cotl1 may cooperate with cofilin in controlling cytoskeletal dynamics during neuronal migration.
In summary, our study showed that Cotl1 is involved in neuronal migration and morphology. The results further support the hypothesis that Cotl1 is important for cell architecture and motility in actin regulation.
Acknowledgments
This work was supported by the the National Natural Science Foundation of China (31301970), the Postdoctoral Science Foundation of China (2015M570858), Fundamental Research Funds for the Central Universities (2452015039), the Natural Science Foundation of Shaanxi (2016JM3033), and the Resource-based Industry Key Technology Project of Shaanxi Province (2016KTCL02-19).
Footnotes
Conflict of Interest: The authors declare no conflicts of interest.
References
- 1.Angevine JB, Jr, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- 2.Austin CP, Cepko CL. Cellular migration patterns in the developing mouse cerebral cortex. Development. 1990;110:713–732. doi: 10.1242/dev.110.3.713. [DOI] [PubMed] [Google Scholar]
- 3.Ayala R, Shu T, Tsai LH. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 4.Chai X, Zhao S, Fan L, Zhang W, Lu X, Shao H, Wang S, Song L, Failla AV, Zobiak B, Mannherz HG, Frotscher M. Reelin and cofilin cooperate during the migration of cortical neurons: a quantitative morphological analysis. Development. 2016;143:1029–1040. doi: 10.1242/dev.134163. [DOI] [PubMed] [Google Scholar]
- 5.de Carlos JA, López-Mascaraque L, Valverde F. Dynamics of cell migration from the lateral ganglionic eminence in the rat. J Neurosci. 1996;16:6146–6156. doi: 10.1523/JNEUROSCI.16-19-06146.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Hostos EL, Bradtke B, Lottspeich F, Gerisch G. Coactosin, a 17 kDa F-actin binding protein from Dictyostelium discoideum. Cell Motil Cytoskeleton. 1993;26:181–191. doi: 10.1002/cm.970260302. [DOI] [PubMed] [Google Scholar]
- 7.Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Müller U. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron. 2011;69:482–497. doi: 10.1016/j.neuron.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goroncy AK, Koshiba S, Tochio N, Tomizawa T, Sato M, Inoue M, Watanabe S, Hayashizaki Y, Tanaka A, Kigawa T, Yokoyama S. NMR solution structures of actin depolymerizing factor homology domains. Protein Sci. 2009;18:2384–2392. doi: 10.1002/pro.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatanaka Y, Hisanaga S, Heizmann CW, Murakami F. Distinct migratory behavior of early- and late-born neurons derived from the cortical ventricular zone. J Comp Neurol. 2004;479:1–14. doi: 10.1002/cne.20256. [DOI] [PubMed] [Google Scholar]
- 10.He M, Zhang ZH, Guan CB, Xia D, Yuan XB. Leading tip drives soma translocation via forward F-actin flow during neuronal migration. J Neurosci. 2010;30:10885–10898. doi: 10.1523/JNEUROSCI.0240-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jovceva E, Larsen MR, Waterfield MD, Baum B, Timms JF. Dynamic cofilin phosphorylation in the control of lamellipodial actin homeostasis. J Cell Sci. 2007;120:1888–1897. doi: 10.1242/jcs.004366. [DOI] [PubMed] [Google Scholar]
- 12.Kawauchi T, Hoshino M. Molecular pathways regulating cytoskeletal organization and morphological changes in migrating neurons. Dev Neurosci. 2008;30:36–46. doi: 10.1159/000109850. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Shapiro MJ, Bamidele AO, Gurel P, Thapa P, Higgs HN, Hedin KE, Shapiro VS, Billadeau DD. Coactosin-like 1 antagonizes cofilin to promote lamellipodial protrusion at the immune synapse. PLoS One. 2014;9:e85090. doi: 10.1371/journal.pone.0085090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lappalainen P, Kessels MM, Cope MJ, Drubin DG. The ADF homology (ADF-H) domain: a highly exploited actin-binding module. Mol Biol Cell. 1998;9:1951–1959. doi: 10.1091/mbc.9.8.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marín O, Rubenstein JL. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 16.Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 17.Nadarajah B, Brunstrom JE, Grutzendler J, Wong RO, Pearlman AL. Two modes of radial migration in early development of the cerebral cortex. Nat Neurosci. 2001;4:143–150. doi: 10.1038/83967. [DOI] [PubMed] [Google Scholar]
- 18.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 19.Provost P, Doucet J, Stock A, Gerisch G, Samuelsson B, Rådmark O. Coactosin-like protein, a human F-actin-binding protein: critical role of lysine-75. Biochem J. 2001;359:255–263. doi: 10.1042/0264-6021:3590255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rakic P. A century of progress in corticoneurogenesis: from silver impregnation to genetic engineering. Cereb Cortex. 2006;16(Suppl 1):i3–i17. doi: 10.1093/cercor/bhk036. [DOI] [PubMed] [Google Scholar]
- 21.Rakic P. Developmental and evolutionary adaptations of cortical radial glia. Cereb Cortex. 2003;13:541–549. doi: 10.1093/cercor/13.6.541. [DOI] [PubMed] [Google Scholar]
- 22.Rakic P. Guidance of neurons migrating to the fetal monkey neocortex. Brain Res. 1971;33:471–476. doi: 10.1016/0006-8993(71)90119-3. [DOI] [PubMed] [Google Scholar]
- 23.Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol. 1972;145:61–83. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- 24.Sekine K, Kawauchi T, Kubo K, Honda T, Herz J, Hattori M, Kinashi T, Nakajima K. Reelin controls neuronal positioning by promoting cell-matrix adhesion via inside-out activation of integrin α5β1. Neuron. 2012;76:353–369. doi: 10.1016/j.neuron.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valiente M, Marín O. Neuronal migration mechanisms in development and disease. Curr Opin Neurobiol. 2010;20:68–78. doi: 10.1016/j.conb.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Walsh C, Cepko CL. Clonally related cortical cells show several migration patterns. Science. 1988;241:1342–1345. doi: 10.1126/science.3137660. [DOI] [PubMed] [Google Scholar]
- 27.Xie J, Li X, Zhang W, Chai X, Huang Y, Li K, Cheng X, Zhao S. Aberrant expression of LIMK1 impairs neuronal migration during neocortex development. Histochem Cell Biol. 2017;147:471–479. doi: 10.1007/s00418-016-1514-8. [DOI] [PubMed] [Google Scholar]


