Abstract
Alu sequences are the most abundant short interspersed repeated elements in the human genome. Here we show that in a cell culture model of colorectal cancer (CRC) progression, we observe accumulation of Alu RNA that is associated with reduced DICER1 levels. Alu RNA induces epithelial-to-mesenchymal transition (EMT) by acting as a molecular sponge of miR-566. Moreover, Alu RNA accumulates as consequence of DICER1 deficit in colorectal, ovarian, renal and breast cancer cell lines. Interestingly, Alu RNA knockdown prevents DICER1 depletion-induced EMT despite global microRNA (miRNA) downregulation. Alu RNA expression is also induced by transforming growth factor-β1, a major driver of EMT. Corroborating this data, we found that non-coding Alu RNA significantly correlates with tumor progression in human CRC patients. Together, these findings reveal an unexpected DICER1-dependent, miRNA-independent role of Alu RNA in cancer progression that could bring mobile element transcripts in the fields of cancer therapeutic and prognosis.
Introduction
About 50% of human genome consists of repetitive elements. These redundant sequences have been previously considered as ‘junk DNA’ based on their unknown function in physiological processes. However, recent findings revealed that DNA repeats could have a specific role on human genome integrity, gene regulation and developmental processes.1
Within the class of repetitive DNA elements, Alu sequences belong to short interspersed repeated elements dispersed throughout the whole primate genome. These sequences are the most abundant mobile elements in the human genome, representing 10.6% of nuclear DNA. They are so widely distributed because of their ability to undergo retrotransposition, which has long been considered the main fate of Alu repeats. These DNA elements can be transcribed by RNA polymerase III as ~300-nucleotide long non-coding RNAs (ncRNAs) with a polyA tail. They have a dimeric structure consisting of two monomers separated by a short A-rich linker.2 Free Alu transcripts are generally expressed at very low levels; it has been estimated that in physiological conditions there are only 102–103 molecules per cell because of their weak internal promoter. However, their abundance increases under various stresses, such as heat shock,3 hyperglycemia4 and viral infection.5 Their increased expression under stress conditions compared with the physiological presence of free Alu transcripts suggests that Alu transcription is strictly controlled and that these elements have a role in the regulation of vital cellular processes.6
We have recently shown a direct role of free Alu RNA in human disease. The accumulation of Alu RNA in the retinal-pigmented epithelium (RPE) of human eyes promotes geographic atrophy, an advanced form of age-related macular degeneration, causing blindness in millions of individuals.7 Alu RNA is enzymatically degraded by DICER1, an RNase III involved in microRNA (miRNA) biogenesis,8 and DICER1 deficiency leads to RPE degeneration not due to miRNA expression deficits but rather to accumulation of cytotoxic Alu RNA.7 Indeed, Alu transcripts can hijack innate immune machinery to induce RPE cell death by activating nucleotide-binding domain leucine-rich repeat containing 3 inflammasome—an innate immune complex that senses pattern/danger-associated molecular patterns—inducing RPE degeneration via interleukin-18.9
Until now, a specific role of free Alu RNA has also been demonstrated in systemic lupus erythematosus10 and there are several reports of a relationship between Alu expression and cancer. Increased levels of Alu RNA have been observed in hepatocellular carcinoma.11 Hypomethylation of Alu elements has also been reported in prostate adenocarcinomas,12 in non-small cell lung cancer13 and in gastric carcinomas.14 Another finding instead correlates mutations of the tumor suppressor gene p53 together with DNA methylation with an increase in the transcription of Alu repetitive elements and a variety of ncRNAs.15 Interestingly, downregulation of DICER1, the enzyme involved in Alu RNA processing, has also been identified and/or associated with poor prognosis in several cancers including colorectal,16 ovarian,17 renal,18 breast19 and hepatocellular carcinoma,20 the same cancer in which Alu RNA has been found to be upregulated.11 However, despite these findings, no correlation has been reported between increased Alu RNA levels and decreased expression of DICER1 in cancer.
Here we show that Alu expression correlates with colorectal cancer (CRC) progression and induces epithelial-to-mesenchymal transition (EMT). We also find that Alu RNA accumulation, due to DICER1 deficit, promotes EMT in colorectal, ovarian, renal and breast cancer cell lines. Both loss-of-function and gain-of-function experiments support the view that Alu transcripts regulate EMT acting as a molecular sponge for miR-566. Surprisingly, our data show that transforming growth factor-β1 (TGF-β1) upregulates Alu transcripts and their accumulation is correlated with tumor growth and metastasis in human CRC patients. Therefore, we provide novel insights into the molecular mechanisms of Pol III-derived Alu transcripts, which will enhance our understanding of the mechanisms underlying the most common interspersed repetitive elements in the human genome and into their role as a positive regulator of cancer progression.
Results
Alu RNA accumulation is a consequence of DICER1 deficit and correlates with CRC progression
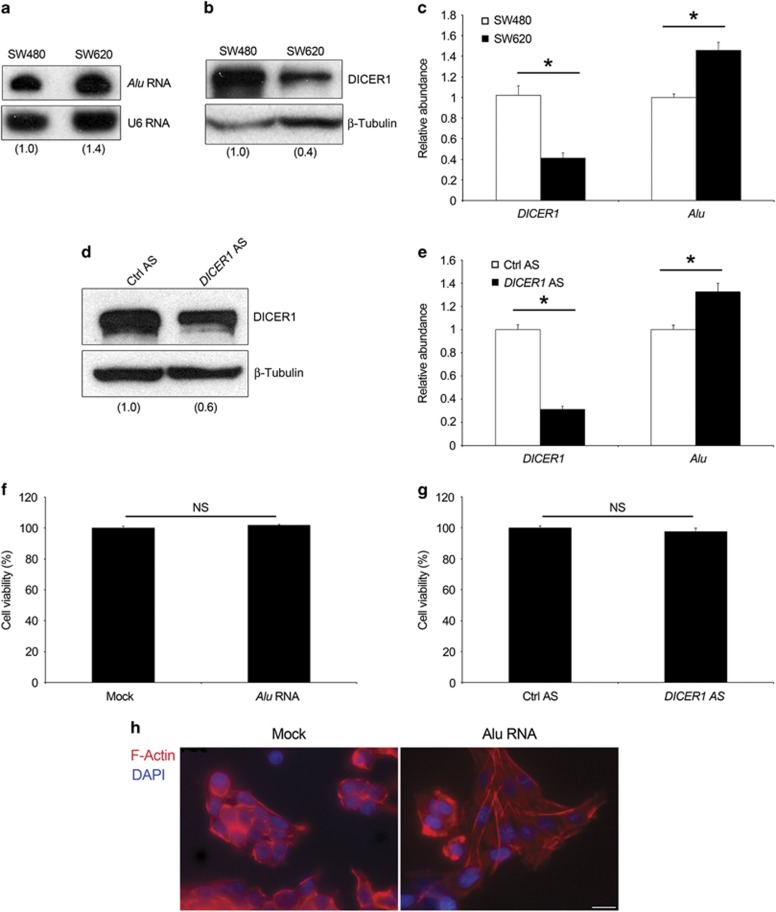
Recent experimental evidence supports a tumor suppressor role for Dicer1.21, 22 This effect has largely been attributed to global or specific imbalance of miRNA. However, the contribution of miRNA-independent Dicer1 activity into the clearance of Alu retroelements7 has not been elucidated in cancer thus far. Thus, we evaluated Alu RNA expression in two human CRC cell lines, SW480 and SW620, in which DICER1 protein levels have been previously shown to be decreased in the metastatic cell line (SW620) compared with primary tumor cell line (SW480).22 SW480 and SW620 cell lines are considered as a validated in vitro model for CRC that represents the progression from primary to lymph node metastatic stages.23 Indeed, these two cell lines are derived from the same cancer patient at two different stages: SW480 is derived from a primary colon carcinoma, whereas the SW620 from a lymph node metastasis resected from the same patient 6 months later. As shown in Figures 1b and c, a 60% of DICER1 reduction was observed in SW620 compared with SW480 cells. Interestingly, higher levels of Alu RNA in SW620 cells compared with SW480 cells corresponded to DICER1 reduction (Figures 1a and c). It is noteworthy that SW620 cells showed higher levels of Alu transcripts in comparison with SW480 cells in the cytoplasm as compared with that in the nucleus (Supplementary Figure S1a). Similarly, DICER1 knockdown in SW480 cells, achieved by transfecting an antisense oligonucleotide, increased Alu RNA accumulation (Figures 1d and e). Collectively, these results confirm our previous studies showing that Alu RNA accumulation is dependent on DICER1 deficit and suggest that Alu RNA expression correlates with cancer progression.
Figure 1.
Alu RNA levels in CRC cells lines. (a) Northern blot analysis shows Alu RNA abundance in SW480 and SW620 cells. Densitometric values normalized against U6 small nucleolar RNA are shown in parentheses. (b) Western blot analysis shows DICER1 protein abundance in SW480 and SW620 cells. Densitometric values normalized against β-tubulin are shown in parentheses. (c) Levels of DICER1 mRNA and Alu transcripts in SW480 and SW620 as evaluated by quantitative reverse transcriptase (qRT–PCR). Transfection of DICER1 antisense oligonucleotide (DICER1 AS) downregulates DICER1 protein (d) and mRNA, and increases the abundance of Alu RNA transcripts (e) in SW480 cells as compared with control antisense oligonucleotide (Ctrl AS). Alu RNA overexpression (f) and DICER1 reduction (g) do not alter SW480 cell viability. (h) Alu RNA induces a reorganization of the cytoskeleton. Representative images of Alu RNA- (right) or vehicle- (Mock, left) transfected SW480 cells stained for F-actin with rhodamine-phalloidin (red). Nuclei are counterstained with 4',6-diamidino-2-phenylindole (DAPI, blue). Scale bar: 25 μm. For all panels: n=3; *P<0.05. Error bars denote s.e.m.
We previously showed that Alu RNA induces cell death in RPE cells.7 Therefore, we tested whether a similar mechanism also occurs in SW480 cells. We found that Alu RNA overexpression, through the transfection of a synthetic transcribed RNA, did not affect SW480 cell viability as well as the cell cycle distribution (Figure 1f and Supplementary Figure S1b). In line with this result, DICER1 knockdown using an antisense oligonucleotide did not alter SW480 cell viability (Figure 1g), showing that Alu RNA accumulation, even if induced by DICER1 deficit, does not affect SW480 cell proliferation.
Alu RNA has been shown to activate the nucleotide-binding domain leucine-rich repeat containing 3 inflammasome.9 Thus, we checked whether Alu RNA induces this inflammatory machinery in SW480 cells. However, we did not detect either cleavage of Caspase-1 or secretion of interleukins-1β/18 (data not shown), suggesting that this pathway is not activated in SW480 cells. Interestingly, we observed a substantial morphological change in Alu RNA-transfected SW480 cells, the cobblestone-like appearance of epithelial cells feature changed to a fibroblast-like morphology with long tubular shape, a characteristic of mesenchymal cells (Figure 1h).
Collectively, these data point to an unforeseen mechanism of Alu RNA signaling not mediated by nucleotide-binding domain leucine-rich repeat containing 3 inflammasome activation in this cancer context.
Alu RNA induces EMT transition
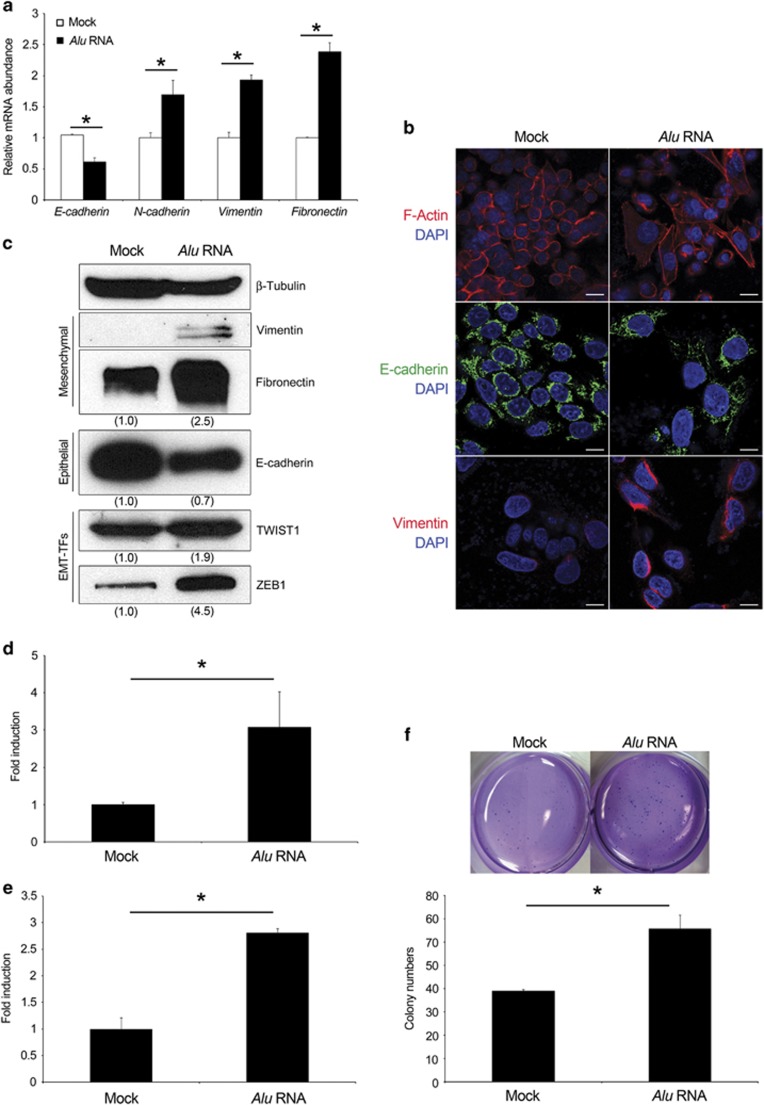
EMT is associated with cancer initiation, progression and metastasis.24 During this progress, endothelial cells acquire fibroblast-like features, reducing the expression of epithelial markers and increasing that of mesenchymal genes. Therefore, we investigated the expression of mesenchymal and epithelial markers in SW480 cells 72 h after Alu RNA transfection. As shown in Figure 2a, epithelial marker E-cadherin (encoded by Cdh1) decreased in Alu RNA-transfected SW480 cells. In contrast, the mesenchymal markers Vimentin, Fibronectin and N-cadherin were increased. Western blotting and immunofluorescence analyses confirmed increased expression of the mesenchymal markers and the reduction of E-cadherin (Figures 2b and c).
Figure 2.
Alu RNA induces EMT. (a) Alu RNA reduces E-cadherin and increase N-cadherin, Vimentin and Fibronectin mRNAs as evaluated by quantitative reverse transcriptase (qRT–PCR). (b) Representative images of Alu RNA- (right) or vehicle- (Mock, left) transfected SW480 cells stained for F-actin (red), E-cadherin (green) and Vimentin (red). Nuclei are counterstained with 4',6-diamidino-2-phenylindole (DAPI, blue). Scale bar: 50 μm. (c) Alu RNA increases the level of the mesenchymal markers (Fibronectin and Vimentin) and of the EMT-TFs TWIST1 and ZEB1, and reduces the epithelial marker E-cadherin, as evaluated by western blot. Densitometric values normalized against β-Tubulin are shown in parentheses. (d and e) Alu RNA increases SW480 cell migration and invasion. Data are expressed as fold of induction with respect to vehicle-transfected cells (Mock). (f) Alu RNA increases SW480 ability to grow in anchorage-dependent manner. The bars indicate the average of colony numbers in each well. Representative pictures of two conditions are shown. For all panels: n=3; *P<0.05. Error bars denote s.e.m.
EMT is orchestrated by several transcription factors (TFs) mainly involved in the repression of Cdh1.24 Quantitative reverse transcriptase–PCR showed that Alu RNA increases the expression of Snail, Slug, TWIST1, ZEB1 and ZEB2 mRNAs (Supplementary Figure S2b). Among these, we confirmed the ability of Alu RNA to increase the protein abundance of TWIST1 and ZEB1, which have been reported to be highly expressed in CRC specimens25, 26 (Figure 2c). Collectively, these data suggest that Alu RNA induces EMT by modulating these transcription factors.
In contrast, the knockdown of Alu RNA achieved transfecting an antisense oligonucleotide (Alu AS) decreases the expression of Fibronectin and of the EMT-TFs (TWIST1 and ZEB1), whereas it increases the expression of E-cadherin compared with cell transfected with a scramble oligonucleotide (Ctrl AS) (Supplementary Figure S2a), further confirming the ability of Alu RNA to induce EMT.
A hallmark of cells undergoing EMT is the acquisition of migratory and invasion ability due to the changes in their shape and polarity, and the ability to grow in an anchorage-independent manner. We examined whether Alu RNA is able to enhance cell migration, invasion and colony formation. As shown in Figures 2d and e, Alu RNA transfection induced a significant increase of both migratory and invasion ability of SW480 cells. More interestingly, it increased the ability of SW480 cells to grow in soft agar (Figure 2f), although Alu RNA was transiently transfected, further showing that these cells had gained functional mesenchymal characteristics.
Collectively, these data demonstrate that Alu RNA induces EMT in SW480 CRC cells.
Alu RNA mediates DICER1 deficit-induced EMT
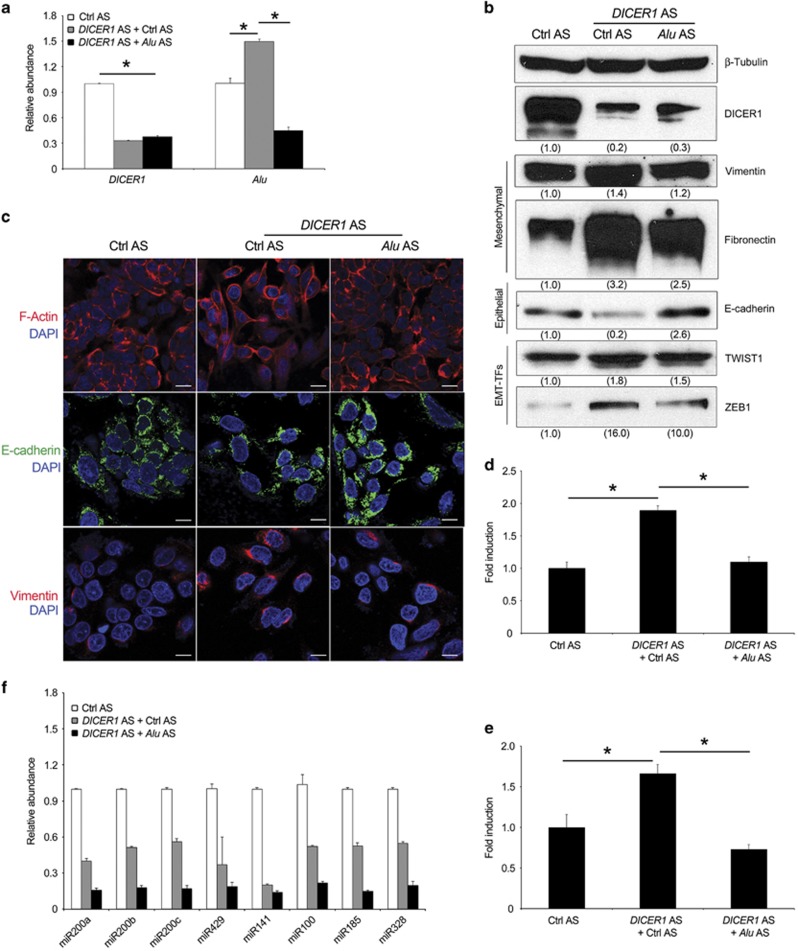
DICER1 reduction has been shown to induce EMT by reducing the abundance of the miR-200 family involved in the degradation of ZEB1 and ZEB2 transcripts.22, 27 We hypothesized that Alu RNA accumulation induced by DICER1 reduction is involved in this process. To verify this hypothesis, we performed a rescue experiment transfecting Alu AS following DICER1 knockdown. As shown in Figure 3a, Alu AS efficiently reduced Alu RNA accumulation induced by DICER1 reduction. Next, we monitored the expression levels of the EMT markers. Consonant with previous reports, our results confirmed that DICER1 knockdown (central lane, Figure 3b) induces EMT by reducing E-cadherin levels through regulation of EMT-TFs (Supplementary Figure S3a) and increasing the expression of mesenchymal markers (Fibronectin and Vimentin), compared with SW480 cells transfected with control antisense oligonucleotides (left lane, Figure 3b). Surprisingly, transfection of Alu AS in DICER1 AS-SW480 transfected cells rescued EMT induced by DICER1 deficit (right lane, Figure 3b). These data were further confirmed by immunofluorescence analyses (Figure 3c). Corroborating this data, the co-transfection of Alu AS prevented the ability of DICER1 knockdown SW480 cells to migrate and to invade (Figures 3d and e).
Figure 3.
Antisense oligonucleotide (Alu AS) targeting Alu RNA sequences rescues the DICER1 deficit-induced EMT. (a) Alu AS reduces Alu RNA accumulation induced by DICER1 deficit in SW480 cells as evaluated by quantitative reverse transcriptase (qRT–PCR). (b) Evaluation of DICER1, Vimentin, Fibronectin, E-cadherin, TWIST1 and ZEB1 protein levels by western blot analysis. Densitometric values normalized against β-Tubulin are shown in parentheses. (c) Representative images of F-actin (red), E-cadherin (green) and Vimentin (red) staining. Nuclei are counterstained with 4',6-diamidino-2-phenylindole (DAPI, blue). Scale bar: 50 μm. (d and e) Alu AS inhibits DICER1-deficit induced SW480 cell migration and invasion. Data are expressed as fold of induction with respect to Ctrl AS. (f) DICER1 AS induced global miRNA expression deficits in SW480 cells compared with Ctrl AS. No significant difference in miRNA abundance between Alu AS and Ctrl AS-treated DICER1-depleted cells as evaluated by qRT–PCR. For all panels: n=3; *P<0.05. Error bars denote s.e.m.
Next, we sought to determine whether this pathway has broad implication in cancer. Alu transcripts accumulate as consequence of DICER1 deficit in different cancer cell lines, independently from the tissues of origin, such as ovarian (A2780), kidney (A498) and breast (MCF-7). Furthermore, the co-transfection of Alu AS is able to inhibit their accumulation and to rescue the EMT induced by DICER deficit (Supplementary Figures S3b, c and d).
Then we evaluated whether Alu inhibition recovered the miRNA deficits induced by DICER1 reduction could explain the functional rescue of DICER1 deficit-induced EMT. As expected, DICER1 knockdown in SW480 cells markedly reduced the abundance of numerous miRNAs, including those of miR-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429). However, the inhibition of Alu RNA did not recover the miRNA deficits (Figure 3f), indicating that DICER1 deficit-induced EMT is mediated by Alu RNA accumulation and not due to miRNA downregulation.
Overall, these data show that Alu RNA mediates DICER1 deficit-induced EMT in cancer cells, and that DICER1 function in this context should be reconsidered in view of its role in the clearance of Alu RNA transcripts.
Alu RNA induces EMT by acting as a molecular decoy for miR-566
miRNAs are involved in regulating numerous biological processes, including EMT, and their expression is altered during cancer progression.28 Recently, the competing endogenous RNA hypothesis proposed that a large number of ncRNAs might function as molecular sponges for miRNA.29 As Pol III-derived Alu transcripts can also be considered ncRNAs, we investigated whether these sequences can act as a molecular decoy for miRNA. To identify the potential target miRNAs, we used bioinformatics tools (miRbase) to predict potential miRNA binding sites in the Alu RNA sequence. Among the predicted miRNAs, we focused on miR-566 because its downregulation is associated with CRC progression.30 It is important to underscore that the miRNA binding site for miR-566 is conserved among the Alu sequences. Supplementary Figure S4a shows the base pairing between miR-566 and the sequence of human Alu primary transcript used to transfect in SW480 cells.
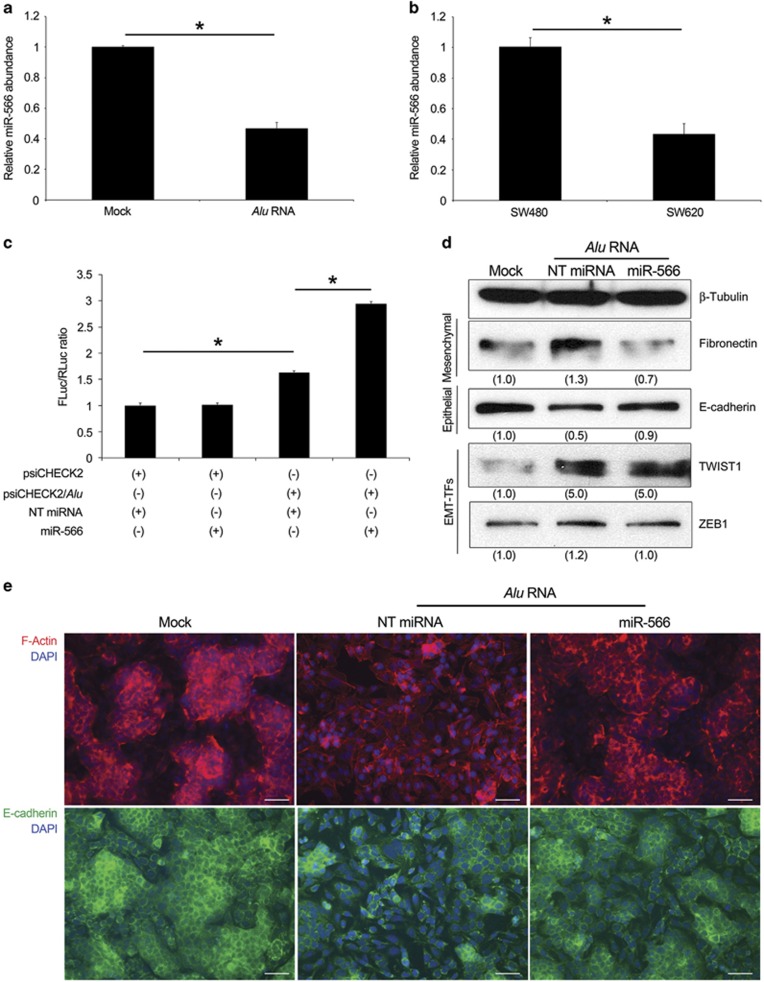
The level of miR-566 was first evaluated in SW480 cells transiently transfected with Alu RNA. As shown in Figure 4a, the level of miR-566 is much lower in transfected cells compared with control cells. Moreover, SW620 cells, which express a higher level of Alu sequences compared with SW480 cells (Figure 1a), also had reduced abundance of miR-566 (Figure 4b), indicating that the interaction of Alu/miR-566 occurs endogenously and does not depend on the Alu sequence that we transfected into SW480 cells.
Figure 4.
Alu RNA acts as a molecular decoy for miR-566 inducing EMT. (a) Alu RNA transfection in SW480 cells reduces miR-566 level as evaluated by quantitative reverse transcriptase (qRT–PCR). (b) SW620 cells show less miR-566 level compared with SW480 cells, as evaluated by qRT–PCR. (c) The activity of Renilla luciferase (RLuc) was downregulated by miR-566 and not by NT miRNA. SW480 cells were co-transfected with psiCHECK2 and/or psiCHECK2/Alu, and miR-566 and/or NT miRNA. The bars indicate Firefly luciferase (FLuc) activities normalized against Renilla. Each experiment was repeated at least three times and each sample was assayed in triplicate. (d) miR-566 prevents the Alu-induced EMT, as shown by Fibronectin, E-cadherin, TWIST1 and ZEB1 levels evaluated by western blot. Densitometric values normalized against β-Tubulin are shown in parentheses. (e) Representative images of F-actin (red) and E-cadherin (green) staining. Nuclei are counterstained with 4',6-diamidino-2-phenylindole (DAPI, blue). Scale bar: 75 μm.
To investigate whether Alu can interact with miR-566, we performed a dual-luciferase reporter assay. We cloned Alu RNA in psiCHECK2 vector downstream of Renilla luciferase sequence (psiCHECK2/Alu) that was co-transfected into SW480 cells with a miR-566 mimic or with a non-targeting (NT) miRNA. We assayed Renilla Luciferase 48 h after transfection. Data, normalized to Firefly luciferase expression, indicated that miR-566 decreases the activity of Renilla luciferase by directly binding to the Alu sequence downstream of the Renilla luciferase gene, whereas the luciferases activity in the cells co-transfected with psiCHECK2 vector and miR-566 mimic or with NT miRNA were not altered. Furthermore, the significant increased signal in cells co-transfected with psiCHECK2/Alu and NT miRNA compared with cells transfected with empty vector is possibly due to the interaction with endogenous miR-566 (Figure 4c).
Recently, miR-566 has been shown to target Von Hippel–Lindau (VHL) mRNA in glioblastoma cells.31 Therefore, we explored whether Alu RNA, by acting as a molecular sponge of miR-566, could regulate VHL mRNA. miR-566 reduced the abundance of VHL mRNA in SW480 cells, whereas Alu RNA transfection led to an increase in VHL transcript abundance (Supplementary Figure S4b), further confirming that Alu RNA acts as molecular sponge for miR-566.
Next, we explored whether miR-566 was involved in Alu RNA-induced EMT in SW480 cells. Co-transfection of miR-566, but not of NT miRNA, with Alu RNA restored the Alu RNA-induced EMT (Figure 4d). F-Actin and E-cadherin staining confirmed that miR-566 prevented the mesenchymal phenotype induced by Alu RNA (Figure 4e). As we demonstrated that Alu RNA mediates DICER1 deficit-induced EMT (Figure 3), we tested whether miR-566 is also involved in this process. DICER1 AS transfection induced EMT in SW480 cells, which is blocked by co-transfecting miR-566 but not by NT miRNA (Supplementary Figure S4c). Corroborating these data, miR-566 is able to revert the mesenchymal properties of SW620 cells decreasing the level of Fibronectin and reducing those of E-cadherin (Supplementary Figure S4d).
Collectively, these data demonstrate that miR-566 is an essential modulator of Alu RNA-induced EMT.
TGF-β1-mediated EMT is driving Alu RNA accumulation
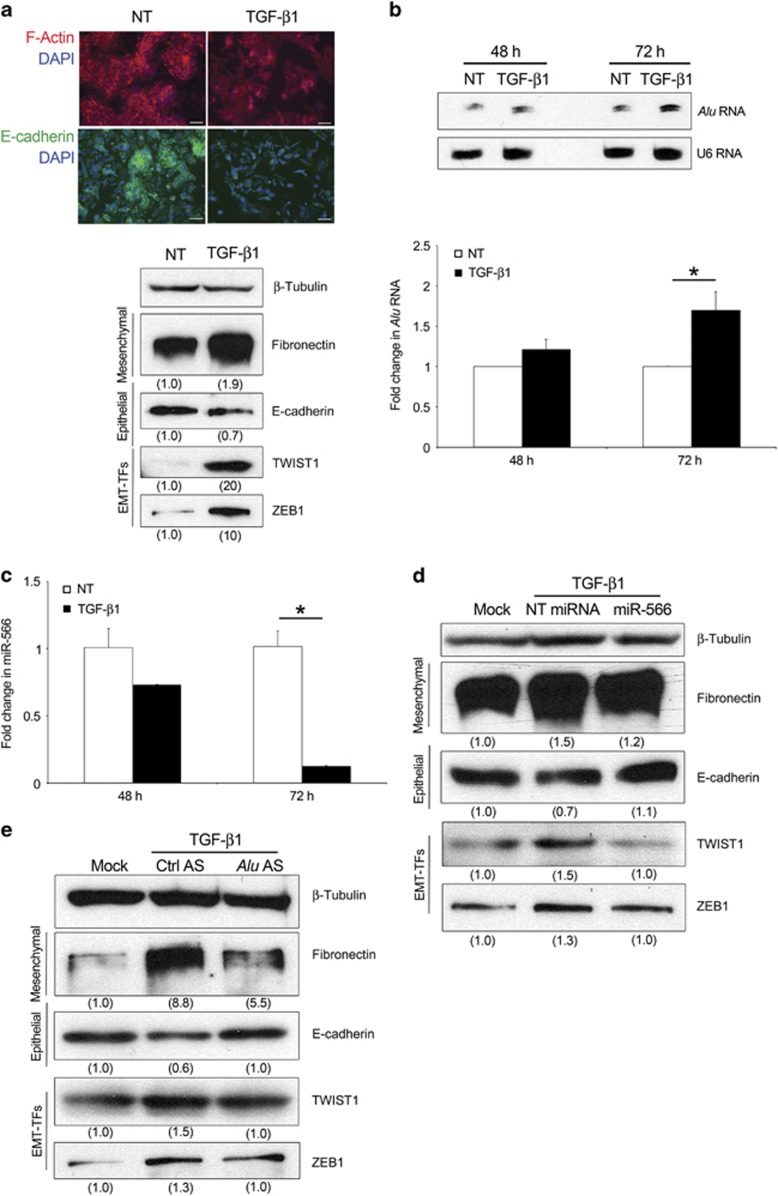
In order to further assess the role of Alu RNA in EMT, we used TGF-β1, the major inducer of EMT in cancer metastasis,32 to establish a canonical EMT model in our system. As shown in Figure 5a, TGF-β1 is able to induce EMT in SW480 cells. Next, we monitored the expression of Alu transcripts after TGF-β1 stimulation. TGF-β1 treatment induced accumulation of Alu RNA over time (Figure 5b). We then tested the effect of TGF-β1 on miR-566 expression: consonant with our previous results, miR-566 was dramatically reduced (Figure 5c) and miR-566 transfection is able to inhibit the TGF-β1-induced EMT (Figure 5d and Supplementary Figure S5a). Moreover, Alu AS transfection after TGF-β1 treatment is able to rescue the TGF-β1-induced EMT, indicating that TGF-β1 induces a reduction of miR-566 in an Alu-dependent manner (Figure 5e and Supplementary Figure S5b). Interestingly, the effect of TGF-β1 treatment on Alu RNA accumulation was similar in A2780, A498 and MCF-7 (Supplementary Figure S5c), indicating that this signaling has broad implication in cancer.
Figure 5.
TGF-β1 stimulation induces Alu RNA accumulation. (a) The stimulation of SW480 cells with 5 ng/ml TGF-β1 induces EMT. Representative images (upper) of TGF-β1 (right) or not- (NT, left) treated SW480 cells stained for F-actin (red) and E-cadherin (green). Nuclei are counterstained with 4',6-diamidino-2-phenylindole (DAPI, blue). Scale bar: 75 μm. Western blot (lower) shows abundance of Fibronectin, E-cadherin, TWIST1 and ZEB1. Densitometric values normalized against β-Tubulin are shown in parentheses. (b) TGF-β1 increases Alu RNA accumulation as evaluated by northern blot (upper) and quantitative reverse transcriptase (qRT–PCR) (lower). (c) TGF-β1 decreases miR-566 in SW480 cells, as evaluated by qRT–PCR. (d and e) miR-566 and Alu AS rescues the TGF-β1-induced EMT. Western blots show abundance of Fibronectin, E-cadherin, TWIST1 and ZEB1. Densitometric values normalized against β-Tubulin are shown in parentheses. For all panels: n=3; *P<0.05. Error bars denote s.e.m.
Collectively, these findings demonstrate that Alu RNA is involved in TGF-β1-mediated EMT through miR-566 in cancer.
Alu RNA accumulation correlates with human CRC progression
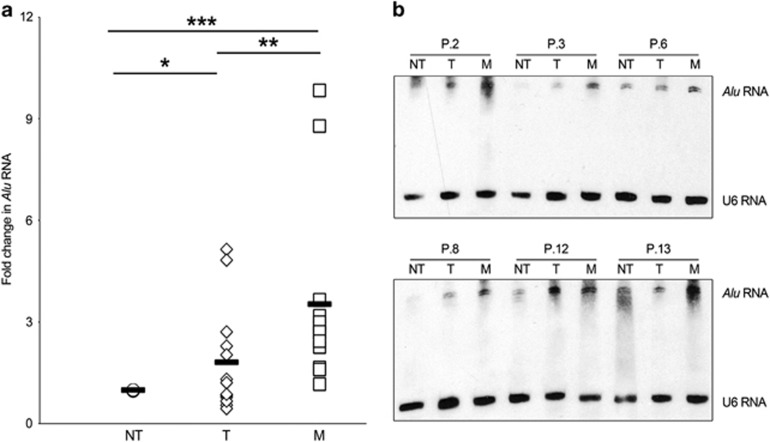
Next, we examined whether Alu RNA accumulation occurs in human CRC tumors and whether it increases with progression and malignancy. We quantified Alu RNA in 13 matched non-tumoral tissues (NT), primary colon tumor (T) and liver metastasis (M) derived from CRC patients. We found that Alu RNA transcripts are significantly upregulated in primary colon tumor (T) compared with non-tumoral tissues (NT) (P=0.05, Figure 6a). Furthermore, Alu RNA is significantly increased in liver metastasis (M) compared with normal (NT, P=0.001) and tumoral (T, P=0.049) tissues (Figure 6a) showing that the expression of Alu RNA correlates with CRC progression. To further validate the accumulation of Pol III-derived Alu transcripts (roughly 300 nucleotides long) in human samples, we performed northern blot analyses on total RNA of six out of thirteen patients analyzed. As shown in Figure 6b, CRC (T) and liver metastasis (M) tissues showed a robust accumulation of Alu transcripts compared with normal tissues, strengthening the data obtained by quantitative reverse transcriptase–PCR. These data provide evidence of Alu RNA accumulation in primary and metastatic human CRC, mirroring the functional data obtained in cell culture studies.
Figure 6.
Alu RNA accumulation in CRC patients. (a) Alu RNA expression in non-tumoral tissues (NT), primary colon tumor (T) and liver metastasis (M) of 13 matched patients, as evaluated by quantitative reverse transcriptase (qRT–PCR) and normalized them to 18S mRNA. *P=0.05; **P=0.049; ***P=0.001. (b) Northern blot analysis shows Alu RNA abundance in six patients. U6 small nucleolar RNA was used as loading control.
Discussion
Our study establishes a novel functional role for Alu elements, the most common small interspersed repetitive elements in the human genome, in cancer progression. Collectively, our findings demonstrate that Alu RNA promotes EMT by acting as molecular sponge for miR-566. Furthermore, Alu RNA accumulation is downstream of TGF-β1 signaling and correlates with CRC progression in patients. As EMT is closely related to epithelial plasticity and malignant transformation,24 this is the first study, to our knowledge, that demonstrates the active role of Alu RNA retroelements in cancer pathogenesis.
The initial observation that Pol III-derived Alu transcripts are increased and DICER1 is reduced in the metastatic CRC cell line SW620, with respect to the SW480 cell line derived from the primary tumor of the same patient, suggested that Alu RNA accumulation could have a role in cancer progression. In contrast to other studies that showed a cytotoxic role of Alu RNA,7, 33, 34 we found that the transfection of an in vitro transcribed Alu RNA does not alter cell viability and cell cycle distribution, but induces EMT in SW480 cells.
Furthermore, we have demonstrated that Alu RNA induces cell death by activating the nucleotide-binding domain leucine-rich repeat containing 3 inflammasome, inducing interleukin-18 secretion, and consequent MyD88 signaling in RPE cells.9 Interestingly, Alu RNA activates this pathway in a broad range of cell types including primary cells, such as RPE cells, and immortalized cells such as human monocyte leukemia cells (THP-1) and HeLa.9 However, Alu RNA does not induce inflammasome activation in SW480 cells. This could be due to differences in malignancies or to different mutations on tumor suppressor genes in the cancer cell lines analyzed. Alternatively, it could be due to the lack of the expression of inflammasome components necessary for Alu-induced cytotoxicity in SW480 cells.
In addition, our study confirms the role of DICER1 in the clearance of Alu RNA sequences previously reported.7 Indeed, we showed that DICER1 deficiency induces Alu RNA accumulation in all the cancer cell lines analyzed (SW480 colorectal, A2780 ovarian, A498 renal and MCF7 breast). Although we observed this close association between DICER1 deficit and Alu RNA accumulation in vitro, we did not find the same correlation in human CRC samples, where high variation of the abundance of DICER1 transcript was found (data not shown). However, this does not exclude that in other tissues this correlation could be maintained.
As TGF-β1 has been found to increase during CRC progression,35 we hypothesized that the abundance of Alu RNA in CRC human samples could be mainly due to TGF-β1 signaling. Indeed, we demonstrated that in cultured cells Alu accumulation is downstream to TGF-β1. However, we cannot exclude that the triggering event of Alu RNA accumulation in cancer cells could be DICER1-deficit independent. Indeed, a potential culprit could include hypoxia, which is involved not only in cancer pathogenesis but also in the epigenetic reduction of DICER1 transcription.36 It is noteworthy that we found that hypoxia induces Alu RNA accumulation together with a robust downregulation of DICER1 mRNA (manuscript in preparation). However, future studies should determine the nature of the Alu RNA sequences that accumulate and how their transcription is regulated in cancer pathogenesis.
Furthermore, it has been reported that DICER1 knockdown promotes EMT through reduction of miRNAs belonging to miR-200 family.22, 27 In this scenario, our novel finding that Alu knockdown prevents DICER1 deficit-induced EMT in different cancer cell lines without affecting the miRNA imbalance can open new research avenues. Indeed, this study introduces the concept that a miRNA-independent activity of DICER1, that is, degradation of Alu RNA, is involved in cancer. Furthermore, the recognition of the DICER1/Alu RNA axis as a more general mechanism of pathogenesis could inspire a new view in other pathologic conditions in which DICER1 and/or Alu RNA are involved. Indeed, DICER1 deficit has also been observed in Parkinson’s disease,37 in rheumatoid arthritis38 and, recently, increase of Alu like elements was found to be associated with a Dicer1 decrease in a murine model of proliferative retinopathy.39
According to the competing endogenous RNA hypothesis, ncRNAs communicate with other RNA transcripts through miRNA response elements as the letters of a novel RNA language.29 In addition to ncRNAs, the idea that Alu RNA might serve as a competing endogenous RNA by sequestering miRNA opens new and interesting perspectives on the study of the role of these transcripts. Our findings are consistent with the study providing the first miRNA signature able to discriminate CRC metastasis: together with five miRNAs found to be upregulated, it identified miR-566 significantly downregulated in colon metastatic cancer.30 Based on these findings, future studies will aim to determine how the downregulation of miR-566 mediates the Alu-induced EMT. Preliminary bioinformatics analyses have revealed that miR-566 has different potential targets, whose increased expression could enhance the EMT process, for example, β-parvin that has been shown to be a key regulator of filopodium-like protrusion.40
In conclusion, these findings reveal an unexpected role of Alu RNAs, whose accumulation correlates with DICER1 deficit, EMT, TGFβ-1 signaling and cancer progression, providing new insights into the importance of Alu RNA sequences and non-canonical DICER1 signaling, which could open new horizons for alternative prognostic and therapeutic strategies.
Materials and methods
Cell lines
All cancer cell lines were purchased from ATCC (Manassas, VA, USA). The human colon cancer (SW480 and SW620), the human breast cancer (MCF-7) and the human kidney carcinoma (A498) cell lines were grown in Dulbecco’s modified Eagle’s medium medium supplemented with 10% fetal bovine serum, 2 mM glutamine and standard concentration of antibiotics. Human ovarian cancer cell line (A2780) was cultured in RPMI, supplemented with 10% heat-inactivated fetal bovine serum, 2 mM L-glutamine and standard concentration of antibiotics (all reagents from Euroclone). For TGF-β1 activation, the cells were treated with 5 ng/ml TGF-β1 in serum-free medium for the indicated times.
Human tissues
Human primary CRC, CRC liver metastasis and their corresponding adjacent normal snap-frozen tissue samples were obtained from the Biological Resources Center (CRB, Centre de Resource Biologique) of Centre Léon Bérard (protocol number: BB-0033-00050) (Lyon, France). Protocol using human material was approved by the CRB medical and scientific committee. All patients signed informed consent to participate to research according to the French laws. RNA from human tissues was extracted QIACUBE kit (Qiagen, Hilden, Germany), following the manufacturer’s protocol. RNA concentration was measured by Nanodrop system (Thermo Scientific, Waltham, MA, USA) and RNA quality was evaluated by TapeStation system (Life Technologies, Carlsbad, CA, USA).
In Vitro Transcription of Alu RNA
We synthesized a 281 nucleotides Alu RNA sequence originating from the CDNA clone TS 103, which is known to be expressed in human cell.41 The pT7/Alu plasmid was linearized with DraI and subjected to HiScribe T7 Quick High Yield RNA Synthesis Kit (BioLabs, Ipswich, MA, USA) according to the manufacturer’s instructions. After the T7 transcription reaction, RNA was treated with DNase and purified following phenol–chlorophorm extraction and ethanol precipitation. RNA was quantified at Nanodrop and the integrity was monitored by gel electrophoresis. This yields single-stranded RNA that fold into a defined secondary structure identical to Pol III derived transcripts.
Transient transfection
Cells were transfected with Alu RNA, DICER1 antisense oligonucleotide (as), control (for DICER1) as, and miR-566- and miRNA-negative control mimics (Thermo Scientific) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), and Alu as and control (for Alu) as using Oligofectamine (Invitrogen) according to the manufacturer’s instructions.
Cell viability
Cell viability was assessed using CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA) according to manufacturer’s instructions.
Quantitative reverse transcriptase–PCR and western blotting
Total, cytoplasmic and nuclear RNA were isolated from cells by TRIzol (Thermo Fisher Scientific, Waltham, MA, USA) and by Cytoplasmic and Nuclear RNA Purification kit (Norgen, Thorold, ON, Canada) according to the manufacturer’s protocol and complementary DNA was generated using QuantiTect reagents (Qiagen). For miRNA quantification, RNA was reverse transcribed using universal primer using Universal cDNA synthesis Kit (Exiqon, Vedbaek, Denmark) according to the manufacturer’s specifications. The primers for the target miRNAs and the reference U6 small nucleolar RNA were also purchased from Exiqon. Quantitative reverse transcriptase–PCR assays were performed using SYBR Green PCR Master Mix (BioRad, Hercules, CA, USA) along with primers listed in the Supplementary Methods and normalized to the 18S gene. Cell lysates were prepared with lysis buffer (Tris-HCl 20 mM pH 8, NaCl 150 mM, Triton X-100 1%, EDTA 10 mM, Glycerol 10%, ZnAc 1 mM), separated by SDS–polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes and probed with antibodies listed in the Supplementary Methods.
Northern blot analysis
Total RNA (100 ng) was denatured for 10 min at 70 °C in a Tris-Borate-EDTA buffer–urea 2 × loading buffer (BioRad) and then run on a 10% denaturing Tris-Borate-EDTA buffer–urea–polyacrylamide gel. Then the RNA was transferred to a positively charged nylon membrane (Amersham, Little Chalfont, UK). The membrane was cross-linked by UV irradiation and saturated with a pre-hybridization solution (formamide 50%, sodium phosphate buffer 0.12 M pH 7.2, NaCl 0.25 M, EDTA 1 mM, SDS 7%) at 42 °C for 1 h. One hundred and twenty-five nanograms of denatured Alu probe was added and incubated for 16 h at 42 °C. Alu probe was obtained by PCR reaction using as template the plasmid pAlu, the specific primers (Alu probe F: 5′-GGGCCGGGCGCGGTG-3′ and Alu probe R 5′-GTACCTTTAAAGAGACAGAGTCTCGC-3′, biotinylated on the 5′-side) and the following cycle: 5 min at 95 °C, 1 min at 95 °C, 30 s 58 °C, 1 min at 72 °C for 40 cycles and 8 min at 72 °C. The reaction product was then purified using ProbeQuant G-50 Micro Columns (GE Healthcare, Little Chalfont, UK) following manufacturer instructions. For the normalization we used a 5′-biotinylated probe against U6 RNA (5′-AGCCTACAGCACCCGGTATT-3′) at final concentration of 10 ng/μl. The membrane was washed twice with SSC 2 × SDS 0.2% at 65 °C. For the detection we used Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific) following manufacturer instructions.
Transwell cell migration and invasion assay
Cells (1 × 105) were then seeded into the upper chamber of a 24-multiwell insert system with 8 μm pore size polycarbonate filter (Corning, Corning, NY, USA) coated or not-coated with 200 μg/ml of Matrigel. After 30 h, the cells on the top of the filter were removed and those on the bottom side were stained with 4',6-diamidino-2-phenylindole. Images were recorded on Leica DM6000 (Leica, Wetzlar, Germany) fluorescence microscope. Single cells were counted using ImageJ (NIH, Bethesda, MD, USA).
Soft agar assay
After 24 h from transfection cells were trypsinized and then subjected to soft agar assay. Six-well plates were pre-coated with 0.75% basal agar layer with culture media. Cells were re-suspended in 0.32% upper agar layer and seeded at the density of 1 × 104 cells per well. After 12 days, cell colonies were visualized by 0.2% crystal violet (Sigma-Aldrich, St Louis, MO, USA) staining. Images were captured and the colony number was counted by Image J software (NIH).
Luciferase reporter assay
We cloned Alu sequence downstream of the luciferase gene in the psiCHECK-2 luciferase vector (Promega) in the XhoI and NotI sites. SW480 cells were co-transfected in 96-well with 20 ng of psiCHECK-2 or psiCHECK-2/Alu and 1 μM of miR-566 or miRNA-negative control mimics for each well. After 48 h, the luciferase activity was measured using the Dual-Glo(R) Luciferase Assay System (Promega). Renilla luciferase activity was normalized against the firefly luciferase activity.
Statistical analysis
Results are expressed as mean±s.e.m., with P-values<0.05 considered statistically significant. Differences among groups were compared by the Student’s t-test (two-tailed) or one-way analysis of variance.
Acknowledgments
We thank the IGB integrated microscopy and FACS facilities and Grazia Mercadante for technical assistance, and Ivana Apicella, Valeria Cicatiello, Nagaraj Kerur and Monica Autiero for discussions. This work was supported by Marie Curie Action (FP7-PEOPLE-2013-CIG 631311 CanAlu) to VT, Italian Ministry of Scientific Research (PON 01_01434 REACT) and EU Horizon2020 (TRANSCAN-2, project BeFIT) to SDF, and Nuovo-Soldati Cancer Research Foundation to VB.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
The authors declare no conflict of interest.
Supplementary Material
References
- Cordaux R, Sen SK, Konkel MK, Batzer MA. Computational methods for the analysis of primate mobile elements. Methods Mol Biol 2010; 628: 137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. Alu elements: know the SINEs. Genome Biol 2011; 12: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, Mandal AK, Jha V, Mukerji M. Heat shock factor binding in Alu repeats expands its involvement in stress through an antisense mechanism. Genome Biol 2011; 12: R117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wang WH, Azadzoi KM, Dai P, Wang Q, Sun JB et al. Alu RNA accumulation in hyperglycemia augments oxidative stress and impairs eNOS and SOD2 expression in endothelial cells. Mol Cell Endocrinol 2016; 426: 91–100. [DOI] [PubMed] [Google Scholar]
- Panning B, Smiley JR. Activation of RNA polymerase III transcription of human Alu elements by herpes simplex virus. Virology 1994; 202: 408–417. [DOI] [PubMed] [Google Scholar]
- Hasler J, Samuelsson T, Strub K. Useful 'junk': Alu RNAs in the human transcriptome. Cell Mol Life Sci 2007; 64: 1793–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 2011; 471: 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001; 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 2012; 149: 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Pratt GA, Sundararaman B, Townsend MJ, Chaivorapol C, Bhangale T et al. The Ro60 autoantigen binds endogenous retroelements and regulates inflammatory gene expression. Science 2015; 350: 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang RB, Wang HY, Lu HY, Xiong J, Li HH, Qiu XH et al. Increased level of polymerase III transcribed Alu RNA in hepatocellular carcinoma tissue. Mol Carcinog 2005; 42: 93–96. [DOI] [PubMed] [Google Scholar]
- Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC, Cho YM et al. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol 2007; 211: 269–277. [DOI] [PubMed] [Google Scholar]
- Daskalos A, Nikolaidis G, Xinarianos G, Savvari P, Cassidy A, Zakopoulou R et al. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer 2009; 124: 81–87. [DOI] [PubMed] [Google Scholar]
- Xiang S, Liu Z, Zhang B, Zhou J, Zhu BD, Ji J et al. Methylation status of individual CpG sites within Alu elements in the human genome and Alu hypomethylation in gastric carcinomas. BMC Cancer 2010; 10: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonova KI, Brodsky L, Lipchick B, Pal M, Novototskaya L, Chenchik AA et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc Natl Acad Sci USA 2013; 110: E89–E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faggad A, Kasajima A, Weichert W, Stenzinger A, Elwali NE, Dietel M et al. Down-regulation of the microRNA processing enzyme Dicer is a prognostic factor in human colorectal cancer. Histopathology 2012; 61: 552–561. [DOI] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med 2008; 359: 2641–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Fan Y, Gao Y, Zhang Y, Huang Q, Ai Q et al. Dicer is down-regulated in clear cell renal cell carcinoma and in vitro Dicer knockdown enhances malignant phenotype transformation. Urol Oncol 2014; 32: 46 e9–46 e17. [DOI] [PubMed] [Google Scholar]
- Grelier G, Voirin N, Ay AS, Cox DG, Chabaud S, Treilleux I et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer 2009; 101: 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa N, Ojima H, Shirakihara T, Shimizu H, Kokubu A, Urushidate T et al. Downregulation of the microRNA biogenesis components and its association with poor prognosis in hepatocellular carcinoma. Cancer Sci 2013; 104: 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev 2009; 23: 2700–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S et al. A MicroRNA targeting dicer for metastasis control. Cell 2010; 141: 1195–1207. [DOI] [PubMed] [Google Scholar]
- Hewitt RE, McMarlin A, Kleiner D, Wersto R, Martin P, Tsokos M et al. Validation of a model of colon cancer progression. J Pathol 2000; 192: 446–454. [DOI] [PubMed] [Google Scholar]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013; 13: 97–110. [DOI] [PubMed] [Google Scholar]
- Gomez I, Pena C, Herrera M, Munoz C, Larriba MJ, Garcia V et al. TWIST1 is expressed in colorectal carcinomas and predicts patient survival. PLoS One 2011; 6: e18023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer Res 2008; 68: 537–544. [DOI] [PubMed] [Google Scholar]
- van den Beucken T, Koch E, Chu K, Rupaimoole R, Prickaerts P, Adriaens M et al. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat Commun 2014; 5: 5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009; 10: 704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011; 146: 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drusco A, Nuovo GJ, Zanesi N, Di Leva G, Pichiorri F, Volinia S et al. MicroRNA profiles discriminate among colon cancer metastasis. PLoS One 2014; 9: e96670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KL, Zhou X, Han L, Chen LY, Chen LC, Shi ZD et al. MicroRNA-566 activates EGFR signaling and its inhibition sensitizes glioblastoma cells to nimotuzumab. Mol Cancer 2014; 13: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15: 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Fordis CM, Corsico CD, Howard TH, Howard BH. Modulation of HeLa cell growth by transfected 7SL RNA and Alu gene sequences. J Biol Chem 1991; 266: 3031–3038. [PubMed] [Google Scholar]
- Baryakin DN, Semenov DV, Savelyeva AV, Koval OA, Rabinov IV, Kuligina EV et al. Alu- and 7SL RNA analogues suppress MCF-7 cell viability through modulating the transcription of endoplasmic reticulum stress response genes. Acta Natur 2013; 5: 83–93. [PMC free article] [PubMed] [Google Scholar]
- Tsushima H, Ito N, Tamura S, Matsuda Y, Inada M, Yabuuchi I et al. Circulating transforming growth factor beta 1 as a predictor of liver metastasis after resection in colorectal cancer. Clin Cancer Res 2001; 7: 1258–1262. [PubMed] [Google Scholar]
- Rupaimoole R, Wu SY, Pradeep S, Ivan C, Pecot CV, Gharpure KM et al. Hypoxia-mediated downregulation of miRNA biogenesis promotes tumour progression. Nat Commun 2014; 5: 5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E et al. A microRNA feedback circuit in midbrain dopamine neurons. Science 2007; 317: 1220–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsaleh G, Nehmar R, Bluml S, Schleiss C, Ostermann E, Dillenseger JP et al. Reduced DICER1 expression bestows rheumatoid arthritis synoviocytes proinflammatory properties and resistance to apoptotic stimuli. Arthritis Rheumatol 2016; 68: 1839–1848. [DOI] [PubMed] [Google Scholar]
- Liu CH, Wang Z, Sun Y, SanGiovanni JP, Chen J. Retinal expression of small non-coding RNAs in a murine model of proliferative retinopathy. Sci Rep 2016; 6: 33947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibue T, Brooks MW, Weinberg RA. An integrin-linked machinery of cytoskeletal regulation that enables experimental tumor initiation and metastatic colonization. Cancer Cell 2013; 24: 481–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TH, Roy AM, Kim J, Batzer MA, Deininger PL. cDNAs derived from primary and small cytoplasmic Alu (scAlu) transcripts. J Mol Biol 1997; 271: 222–234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.