Abstract
Mucin1 (MUC1), a heterodimeric oncoprotein, containing tandem repeat structures with a high proportion of threonine, is aberrantly overexpressed in many human cancers including pancreatic cancer. Since the overall survival rate of pancreatic cancer patients has remained low for several decades, novel therapeutic approaches are highly needed. Intestinal mucin has been known to be affected by dietary threonine supply since de novo synthesis of mucin proteins is sensitive to luminal threonine concentration. However, it is unknown whether biosynthesis of MUC1 is regulated by threonine in human cancers. In this study, data provided suggests that threonine starvation reduces the level of MUC1 and inhibits the migration of MUC1-expressing pancreatic cancer cells. Interestingly, knockdown of threonyl-tRNA synthetase (TRS), an enzyme that catalyzes the ligation of threonine to its cognate tRNA, also suppresses MUC1 levels but not mRNA levels. The inhibitors of TRS decrease the level of MUC1 protein and prohibit the migration of MUC1-expressing pancreatic cancer cells. In addition, a positive correlation between TRS and MUC1 levels is observed in human pancreatic cancer cells. Concurrent with these results, the bioinformatics data indicate that co-expression of both TRS and MUC1 is correlated with the poor survival of pancreatic cancer patients. Taken together, these findings suggest a role for TRS in controlling MUC1-mediated cancer cell migration and provide insight into targeting TRS as a novel therapeutic approach to pancreatic cancer treatment.
Introduction
Pancreatic cancer is one of the most aggressive human cancers. The lack of early diagnoses and effective treatment strategies are critical factors that can lead to rapid death and low survival rates of pancreatic cancer patients.1 Even after surgical resection with curative intent, the prognosis is very poor due to the high rate of metastasis.2 Hence, new strategies to find a novel therapeutic target are required to improve the treatment of pancreatic cancer.3
MUC1, a member of the mucin family and a heterogeneous glycoprotein, is normally expressed at the apical surface of polarized epithelial cells of the mammary gland, stomach, duodenum, pancreas, uterus, prostate and lungs.4 In malignancy, MUC1 is overexpressed and repositioned over the entire cell membrane of carcinoma cells and contributes to neoplastic transformation, tumor survival, angiogenesis, and metastasis.5 Additionally, the cytoplasmic tail of MUC1 (MUC1-CT) mediates intracellular signaling functions associated with cancer cell survival and metastasis.6 Aberrant overexpression of MUC1 is found in most human carcinomas including pancreatic cancer7 and often used as a diagnostic marker for metastatic progression.8
Mucins have a central backbone rich in threonine, proline, and serine residues that account for 20–55% of the total amino acid composition,9 with threonine alone constituting 28–35% of the total amino acids.10 In comparison with other essential amino acids, threonine is particularly important for the maintenance of the gut and a large proportion of threonine is retained in the intestines of piglets and humans.11, 12 Although previous reports have shown that mucin synthesis is sensitive to dietary threonine supply in the intestines of rats, pigs, piglets and mice,13, 14, 15, 16, 17 it is unknown whether mucins are affected by threonine in human cancer cells.
In this study, it is discovered that the levels of MUC1 are affected by threonine in human pancreatic cancer cells. The data presented has identified that the protein level of MUC1 is also affected by threonyl tRNA synthetase (TRS), which is one of the aminoacyl tRNA synthetases (ARSs), an essential enzyme transferring threonine to cognate tRNA for protein synthesis.18 In addition, it is demonstrated that TRS affects the migration of pancreatic cancer cells through MUC1 biosynthesis. Moreover, it appears that the expression of both TRS and MUC1 was positively correlated in pancreatic cancer cells, as well as associated with overall survival in the pancreatic cancer patients of the cancer genome atlas (TCGA) data set.
Materials and methods
Materials
Anti-MUC1 (cat# ab109185) was purchased from Abcam (Cambridge, UK), anti-MUC1 (cat# sc-7313), anti-ThrRS (cat# sc-166146), anti-c-Myc (cat# sc-40), and anti-AlaRS (cat# sc-98547) were purchased from Santa Cruz Biotechnology (Dallas, TX, USA), anti-alpha-tubulin (cat# T6074) was purchased from Sigma-Aldrich (St Louis, MO, USA), and anti-puromycin (cat# MABE343) was purchased from Millipore (Billerica, MA, USA). L-[35S]-Methionine (Met) (cat# NEG709A) was purchased from PerkinElmer (Waltham, MA, USA). Threonine, L-[3-3H] (cat# ART0330) was purchased from American Radiolabeled Chemicals (Saint Louis, MO, USA). Borrelidin (cat# ab144212) was purchased from Abcam. 5′-O-(N-(L-threonyl)-sulfamoyl) adenosine was purchased from Integrated DNA Technologies (Coralville, IA, USA). Lipofectamine 2000 (cat# 11668030) and puromycin (cat# A1113802) were purchased from Thermo Fisher (Waltham, MA, USA).
Cell culture
PANC-1, MIA PaCa-2 and HEK293T cells were cultured in high glucose Dulbecco’s Modified Eagle’s Medium (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) (HyClone) and 1% streptomycin and penicillin (S/P) (HyClone). AsPC-1, Panc 10.05 and BxPC-3 cells were cultured in Roswell Park Memorial Institute 1640 (HyClone) medium supplemented with 10% FBS and 1% S/P. HPAF-II cells were cultured in Eagle’s Minimum Essential Medium (HyClone) with 10% FBS and 1% S/P. CFPAC-1 cells were cultured in Iscove’s Modified Dulbecco’s Medium (HyClone) supplemented with 10% FBS and 1% S/P. Cells were incubated in a humidified atmosphere at 37 °C in 5% carbon dioxide (CO2).
Threonine starvation of cells
For threonine depletion, cells were rinsed twice with threonine-free medium (WelGENE, Daegu, Korea), and then incubated in threonine-free medium for the indicated times. For wound-healing assays, cells were rinsed with threonine-free medium twice and then incubated for the indicated times in threonine-free medium containing dialyzed FBS (Hyclone) after making scratches.
Preparation of cell lysates and immunoblotting
Cells were dissolved in lysis buffer containing 1% Triton X-100, 50 mM HEPES (pH 7.4), 2 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, and protease inhibitor cocktail (Calbiochem, San Diego, CA, USA), and lysates were centrifuged at 13 000 r.p.m. for 15 min. Then, 20 μg of the extracted proteins were fractionated by SDS–PAGE. Immunoblotting was performed according to previously described methods using the specified antibodies.19
Quantitative real-time PCR
Total RNA was extracted from cells using the RNeasy Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions and the purified RNAs were used for cDNA synthesis using a PrimeScript RT reagent kit (TaKaRa, Shiga, Japan). Quantitative polymerase chain reaction (PCR) was performed using gene-specific primer sets and SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). Real-time PCR was carried out in a 7500 real-time PCR system (Applied Biosystems, Foster city, CA, USA) according to the manufacturer’s instructions. Data were normalized against β-actin mRNA levels. Relative expression was calculated using the ΔΔCT method. PCR was performed using the human MUC1 primer set (forward primer: 5′-GAACTACGGGCAGCTGGACATC-3′, reverse primer: 5′-GCTCTCTGGGCCAGTCCTCCTG-3′), the human TRS primer set (forward primer: 5′-GTAAGCCATGATGGTG-3′, reverse primer: 5′-CTGCCTGTTTGCTGCGG-3′) and the β-actin primer set (forward primer: 5′-GAGCTGCCTGACGGCCAGG-3′, reverse primer: 5′-CATCTGCTGGAAGGTGGAC-3′).
[35S]-Met incorporation assay
Cells were seeded into 24-well plates, grown until subconfluence and incubated in Met-free media containing 10 μCi ml−1 [35S]-Met for 1 h. The cells were then washed twice with ice-cold PBS, treated with 5% trichloroacetic acid (TCA) for 30 min, washed twice with ice-cold PBS and solubilized in 0.5 N NaOH. An aliquot was analyzed with a liquid scintillation counter (Beckman Coulter, Brea, CA, USA).
Wound-healing assay
Wound-healing assays were performed with the IncuCyte Zoom according to the supplier’s protocols. Cells were seeded in 96-well ImageLock plates (Essen BioScience) and grown to 90–95% confluency. Subsequently, a WoundMaker (Essen Bioscience) was used to create scratches in all of the wells, after the media was replaced. Images of cell migration were recorded every 2 h for a total duration of 48 h with the IncuCyte Kinetic Live Cell Imaging System and analyzed with the IncuCyte Zoom software.
Cell proliferation analysis
Cells were seeded in triplicate using a 96-well plate at a density of 5000 cells per well and placed within a microplate tray on the IncuCyte ZOOM (Essen BioScience, Ann Arbor, MI, USA). Cells were monitored every 2 h for a total duration of 72 h using the IncuCyte Kinetic Live Cell Imaging System (Essen BioScience) and analyzed with the IncuCyte Zoom software (Essen BioScience).
RNA interference
Cells were transfected with duplex siRNA using the Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. ON-TARGETplus SMARTpool siRNAs against TRS (cat# L-011789) and AlaRS (cat# L-011565) were purchased from GE Healthcare Dharmacon (Lafayette, CO, USA). A non-targeting siRNA was used as a control (Dharmacon). Double-stranded siRNAs targeting human TRS (5′-TCGCTTTCGGGTTCTCTCATCGCTT-3′) and MUC1 (5′-GGUAAUGGUGGCAGCAGCCUCUCUU-3′) were purchased from Invitrogen. Cells were incubated with the siRNA for 36−72 h.
Puromycin incorporation assay
Surface sensing of translation (SUnSET) was performed as previously described.20 Briefly, cells were incubated with 1 μM puromycin (Thermo Fisher) for 30 min followed by washing with ice cold PBS and lysing with lysis buffer. Cell lysates were loaded onto SDS–PAGE, and western blotting was performed with a mouse anti-puromycin monoclonal antibody (Millipore), and normalized against Ponceau S staining (INtRON, Gyeongggi-do, Korea).
Aminoacylation assay
Aminoacylation assays were performed as previously described.21 Briefly, assays were carried out in a buffer containing 4 mM DTT, 50 mM HEPES-KOH (pH 7.6), 20 mM KCl, 10 mM MgCl2, 5 mM ATP, 2 mg ml−1 yeast tRNA (Roche, Indianapolis, IN, USA), threonine, L-[3-3H] (American Radiolabeled Chemicals), and cell lysates. Reactions were initiated with cell lysates and conducted at 37 °C. Aliquots (20 μl) were taken from the reactants after 30 min and quenched on Whatman filter pads that were presoaked with 5% TCA. Pads were washed three times for 10 min each time with cold 5% TCA, and once with cold 100% ethanol. Washed pads were then dried, and radioactivity was quantified using a scintillation counter (Beckman Coulter, Brea, CA, USA).
Establishment of a doxycycline-inducible cell line
Pancreatic cancer cells were seeded evenly in a 60 mm dish and incubated for 12 h to reach ~90% confluence. When the cells were ready for transfection, 2 μl of shTRS or myc-TRS lentiviral particles and 3 μl of 10 mg ml−1 polybrene were supplemented with 2 ml of complete media and added to the plate. After 16 h of incubation, the culture media was replaced with 3 ml of fresh complete media containing 1 μg ml−1 puromycin and incubated for an additional 48 h. The cells were gradually selected by treating with puromycin every 48 h. To check the efficiency of TRS knockdown or overexpression, cells were treated with 2.5 μg ml−1 of doxycycline (Dox) for the indicated times and immunoblotting was performed. shTRS was purchased from Dharmacon.
SynTarget software
Correlations between expression levels of MUC1 and TRS with the survival rates of pancreatic cancer patients were analyzed with SynTarget Software (http://www.chemoprofiling.org/cgi-bin/GEO/cancertarget/web_run_CT.V0.S1.pl) using the publicly available Gene Expression Omnibus (GEO) data set (GEO data set ID: TCGA_PAAD).
Tissue microarray
Commercially available tissue microarray sections of human PDACs (Cat# A307II AccuMax Array) were purchased from ISU ABXIS (Seoul, Korea) and used according to the manufacturer’s instructions. Slides were analyzed by bright-field microscopy using a Pannoramic 250 1.14 slide scanner and Pannoramic Viewer 1.15 (3DHISTECH, Budapest, Hungary).
Statistical analysis
Statistical analyses were performed using Prism6 (GraphPad Software, San Diego, CA, USA). All data values are represented as the mean±s.e.m., and statistical significance is denoted as follows: NS, Not significant, P⩾0.05; *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
Results
Threonine deprivation reduces MUC1 levels in pancreatic cancer cells
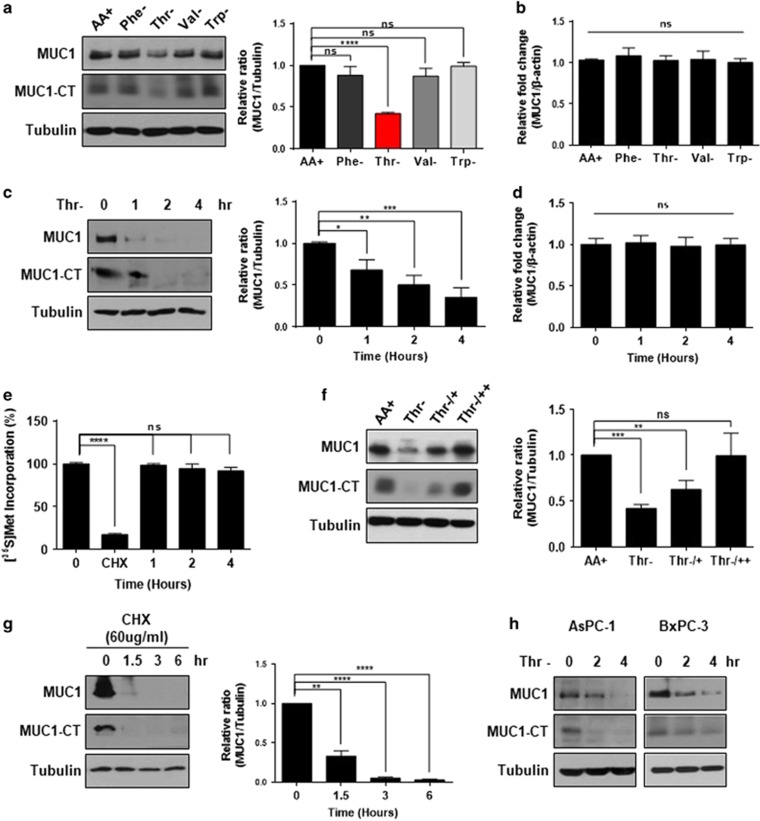
Since previous reports have shown that de novo synthesis of mucin is sensitive to threonine concentration,13, 14, 15, 16, 17 we hypothesized that MUC1 would be affected by threonine levels in pancreatic cancer cells. Thus, we examined whether MUC1 is more sensitive to the level of threonine than other amino acids in the media. The removal of threonine (Thr−), but not phenylalanine (Phe−), valine (Val−) or tryptophan (Trp−) significantly reduced MUC1 and MUC1-CT at the protein level (Figure 1a). However, no changes in MUC1 mRNA level were observed in Panc 10.05 cells in which threonine was deprived (Figure 1b). Next, the time-dependent change in MUC1 levels after the deprivation of threonine was investigated. When Panc 10.05 cells were incubated in threonine-free media, cellular levels of MUC1 and MUC1-CT were reduced in a time-dependent manner (Figure 1c) but mRNA levels of MUC1 were not changed under the same conditions (Figure 1d). To examine whether threonine depletion affects global translation or only the level of MUC1, a [35S]-Met incorporation assay was performed during threonine deprivation. Threonine deprivation had no time-dependent effect on global protein synthesis, whereas significant inhibition of MUC1 levels was observed under the same conditions. Cycloheximide (CHX), known as a protein translation inhibitor, completely blocked de novo protein synthesis22 (Figure 1e). After withdrawal of threonine for 2 h, threonine was re-added for 1 h (Thr−/+) or 2 h (Thr−/++) to Panc 10.05 cells, resulting in the restoration of the MUC1 and MUC1-CT level, further supporting the effect of threonine on MUC1 (Figure 1f). In the course of these experiments, MUC1 protein levels were rapidly reduced in the absence of threonine. To further characterize this observation, MUC1 protein stability was measured after blocking de novo protein synthesis with CHX, and found that the half-life of MUC1, as well as MUC1-CT, was shorter than 1.5 h (Figure 1g). These results suggest that MUC1 would need active biosynthesis to maintain its steady-state cellular level. In addition, the dependence of MUC1 on threonine in other pancreatic cancer cells was examined. The results showed a decrease in protein levels of MUC1 and MUC1-CT during the starvation of threonine in AsPC-1 and BxPC-3 cells (Figure 1h). Taken together, cancer-associated overexpression of MUC1 would highly increase the demands on the cellular supply of threonine.
Figure 1.
MUC1 protein levels in pancreatic cancer cells after threonine deprivation. (a) Panc 10.05 cells were maintained in DMEM (AA+) or were deprived of all amino acids (AA−), phenylalanine (Phe−), Threonine (Thr−), Valine (Val−) or Tryptophan (Trp−) for 2 h. MUC1 and MUC1-CT protein levels were evaluated by immunoblotting (left) and are displayed relative to tubulin levels as a bar graph (right). (b) The effects of amino acid deprivation on MUC1 transcription were evaluated by quantitative RT-PCR analysis in Panc 10.05 cells. (c) Panc 10.05 cells were deprived of threonine (Thr−) for 1 to 4 h. MUC1 and MUC1-CT protein levels were evaluated by immunoblotting (left) and are displayed relative to tubulin levels as a bar graph (right). (d) The effects of threonine starvation on MUC1 transcription were evaluated by qRT–PCR analysis in Panc 10.05 cells. (e) Panc 10.05 cells were incubated with [35S]-Met with or without threonine for the indicated times. Protein synthesis was monitored by [35S]-Met incorporation assay and displayed as a bar graph. (f) Panc 10.05 cells were cultured in media containing (AA+) or lacking threonine (Thr−) for 2 h. After withdrawal for 2 h, threonine-containing media was added for 1 or 2 h (Thr−/+ or Thr−/++). MUC1 and MUC1-CT protein levels were evaluated by immunoblotting (left) and are displayed as a bar graph (right). (g) Panc 10.05 cells were treated with 60 μg ml−1 cycloheximide (CHX) for 1 to 4 h and protein levels of MUC1 and MUC1-CT levels were evaluated by immunoblotting with anti-MUC1 antibody (left) and displayed as a bar graph (right). (h) Immunoblotting analysis of the MUC1 and MUC1-CT in lysates of AsPC-1 and BxPC-3 cells that were deprived threonine (Thr−) for 2 to 4 h. (a–g) *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001; NS, not significant vs control group. Values represent means±s.e.m. of three independent experiments.
Threonine deprivation suppresses pancreatic cancer cell migration
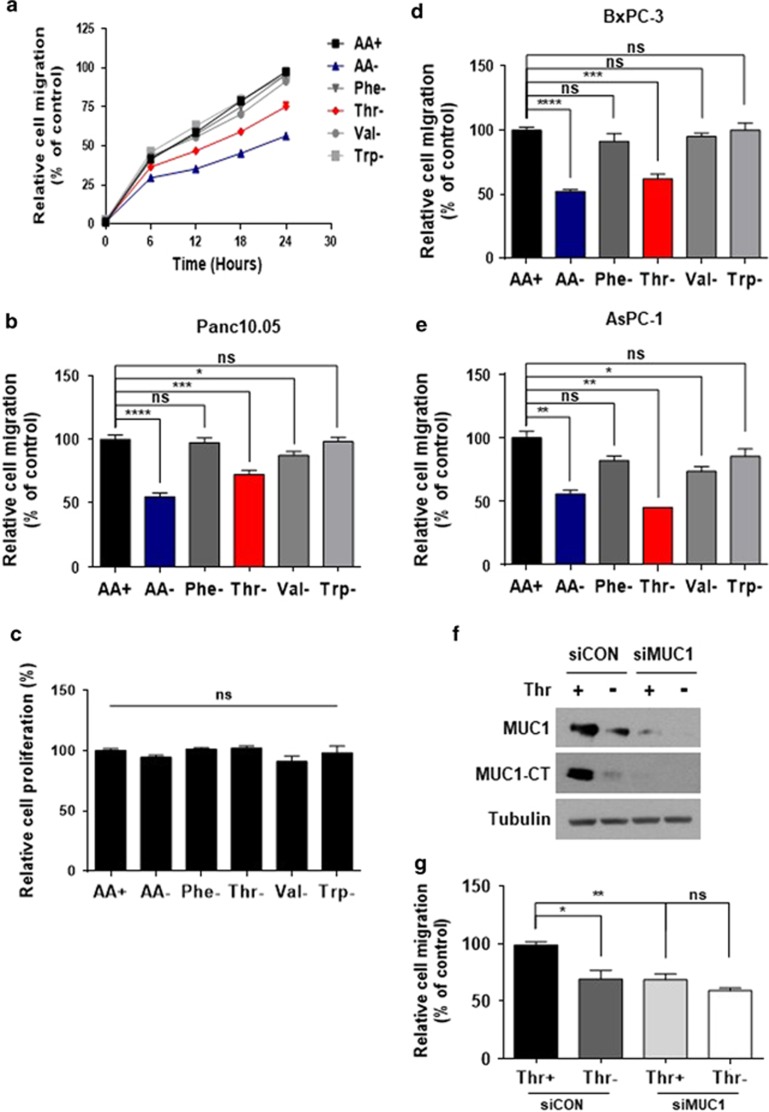
It is known that MUC1 contributes to cancer cell migration.23, 24, 25 To verify the effect of threonine in MUC1-expressing pancreatic cancer cells, experiments that assessed the migration of pancreatic cancer cells following starvation of amino acids were performed. While migration of Panc 10.05 cells was significantly diminished by starvation of total amino acids, threonine displayed the highest sensitivity among single amino acid depletion (Figures 2a and b). In this experimental condition, cell growth was minimally affected (Figure 2d). Migration of other MUC1-expressing pancreatic cancer cells, AsPC-1 (Figure 2d) and BxPC-3 (Figure 2e) was also reduced in the absence of threonine. To evaluate whether the reduction in migration by threonine deprivation is dependent on MUC1 level, Panc 10.05 cells were transfected with siMUC1, and incubated in threonine-free media. The protein levels of MUC1 and MUC1CT were decreased by threonine deprivation or siMUC1 treatment (Figure 2f). Knockdown of MUC1 decreased migration of Panc 10.05 cells and showed little effect on migration in the absence of threonine (Figure 2g), indicating that the threonine-mediated decrease in migration is dependent on MUC1. Taken together, these results demonstrate that threonine deprivation suppresses the migration of pancreatic cancer cells expressing MUC1.
Figure 2.
Effects of threonine deprivation on pancreatic cancer cell migration. (a) Panc 10.05 cells were seeded at equal densities into 96-well plates, cultured to confluency, mechanically wounded by scratching, incubated in each medium and then monitored for 24 h by IncuCyte Zoom. The data is displayed as line graphs. (b) Quantification of migration of Panc 10.05 cells treated as described in (a) for 24 h. Migration distances were measured using the IncuCyte software and displayed as a bar graph. (c) Panc 10.05 cells were seeded at equal densities into 96-well plates, imaged within the IncuCyte Zoom, and analyzed as percentage confluent. The data is displayed as a bar graph. (d and e) Quantification of the migration of AsPC-1 (d) and BxPC-3 (e) cells treated as described in a for 24 h. Migration distance was measured using the IncuCyte software and displayed as a bar graph. (f) Panc 10.05 cells were transfected with non-targeting or MUC1 targeting siRNAs for 24 h and then incubated in media with or without threonine for 24 h. Protein levels of MUC1 and TRS were monitored by immunoblotting with the respective antibody. (g) Panc 10.05 cells were seeded at equal densities into 96-well plates, cultured to confluency, mechanically wounded by scratching, incubated in medium with or without threonine and then monitored for 24 h by IncuCyte Zoom. Cell migration was quantified based on the distance separating wound images and displayed as a bar graph. (a–e and g) *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001; NS, not significant vs control group. Values represent means±s.e.m. of three independent experiments.
TRS regulates MUC1 biosynthesis
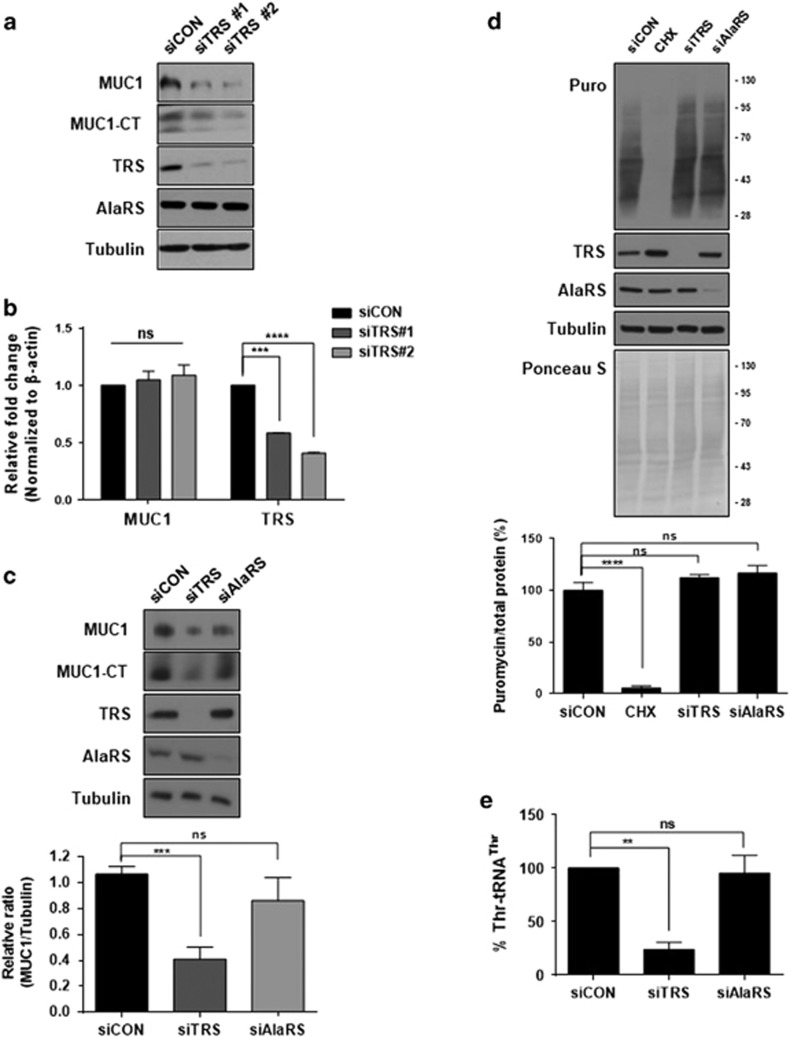
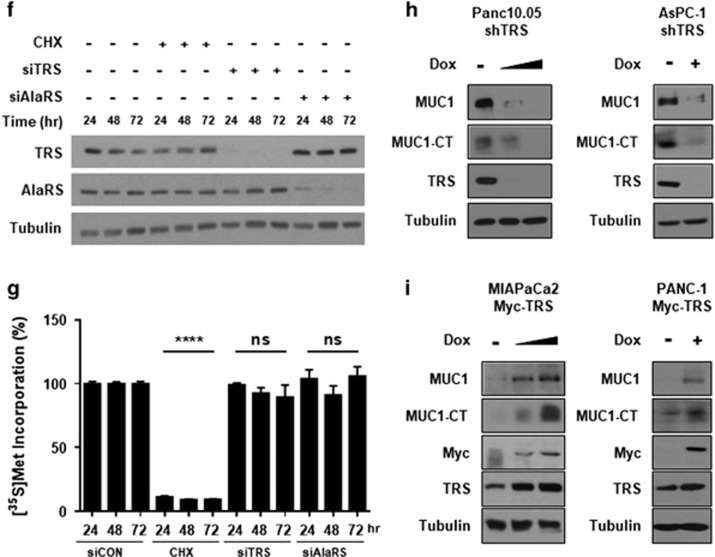
Since TRS catalyzes the ligation of threonine to its substrate tRNA for incorporation into nascently synthesized proteins, the inhibition of TRS catalytic activity or its expression can reduce MUC1 biosynthesis. To assess this possibility, TRS expression was suppressed using its specific siRNAs and checked whether it would affect the cellular level of MUC1. To improve experimental validation, two different siRNAs targeting different regions of human TRS were utilized. Both of the siRNAs targeting TRS appeared to significantly reduce MUC1 and MUC1-CT levels when compared to non-targeting siRNA (Figure 3a). There were no changes observed in MUC1 mRNA under the same conditions (Figure 3b). To determine the TRS dependence on MUC1 levels, we transfected siRNA targeting TRS or alanyl-tRNA synthetase (AlaRS) into Panc 10.05 cells and found a specific reduction of MUC1 and MUC1-CT levels in cells treated with siTRS, but not with siAlaRS (Figure 3c). The next step was to examine how the suppression of TRS or AlaRS would affect global protein synthesis using a SUnSET technique20 since they are protein synthesis enzymes. For this approach, Panc 10.05 cells transfected with siRNA were incubated with a low dose of puromycin (1 μM), which was incorporated into newly synthesized polypeptide chains. Newly synthesized proteins labelled with puromycin were quantified by immunoblotting of the whole cell lysates. The results showed no changes among siCON-, siTRS- and siAlaRS-treated cells in the amounts of newly synthesized proteins while CHX treatment completely blocked de novo protein synthesis (Figure 3d). In this condition, threonylation activity was only decreased in the cells transfected with siTRS, but not with siAlaRS (Figure 3e). It was also important to evaluate total protein synthesis at different times (24, 48 and 72 h) after the transfection of siRNAs using the [35S]-Met incorporation assay. The protein levels of TRS and AlaRS were decreased in the siRNA-treated cells (Figure 3f), but global protein synthesis was not significantly impacted despite the reduction in TRS or AlaRS protein levels (Figure 3g). These results indicate that suppression of TRS specifically reduced biosynthesis of threonine-rich MUC1 without affecting global protein synthesis. To evaluate the effects of TRS on the MUC1 protein level, two types of pancreatic cancer cells were generated expressing TRS shRNA or Myc-tagged TRS induced by doxycycline (Dox). To induce the target genes, pancreatic cancer cells were incubated with Dox (10 ng ml−1) for 24 h. The silencing of TRS following Dox treatment resulted in a marked decrease in the levels of MUC1 and MUC1-CT in Panc 10.05 and AsPC-1 cells (Figure 3h). On the other hand, overexpression of TRS by Dox induction resulted in a significant increase in the level of MUC1 and MUC1-CT in MIA PaCa2 and PANC-1 cells (Figure 3i). These data suggest that TRS specifically regulates the biosynthesis of MUC1.
Figure 3.
MUC1 biosynthesis via TRS in pancreatic cancer cells. (a) TRS was suppressed with two different siRNAs for 48 h in Panc 10.05 cells and protein levels of MUC1 and MUC1-CT were evaluated by immunoblotting with anti-MUC1 antibody. AlaRS and tubulin were used as controls. siCON, non-targeting control siRNA. (b) Effects of TRS suppression on the transcription levels of MUC1 and TRS were analyzed by qRT–PCR with primers targeting TRS and MUC1. (c) TRS and AlaRS were suppressed with their specific siRNAs for 48 h in Panc 10.05 cells, and protein levels of MUC1 and MUC1-CT were evaluated by immunoblotting with an anti-MUC1 antibody (top) and displayed as a bar graph (bottom). (d) TRS and AlaRS were suppressed with their specific siRNAs for 48 h in Panc 10.05 cells and treated with puromycin (1 μℳ) for 30 min. De novo global protein synthesis was then monitored by immunoblotting of cell lysates with an anti-puromycin antibody. The protein levels were quantified by Ponceau S staining. CHX was employed as a positive control to block de novo protein synthesis (top).The ratio of de novo protein synthesis versus global protein synthesis was determined as a percentage and shown as a bar graph (bottom). (e) Panc 10.05 cells were transfected with siRNAs against TRS, AlaRS, or a non-targeting control, and the catalytic activities of cell lysates were determined by threonylation of tRNAThr as described in the Methods. The amount of threonine-charged tRNAs was measured by scintillation counting and the data was displayed as a bar graph. (f) Panc 10.05 cells were transfected with siTRS or siAlaRS for the indicated times, and the protein levels of TRS and AlaRS were evaluated by immunoblotting using anti-TRS or anti-AlaRS antibodies. CHX was used as a positive control and treated for 30 min. (g) Protein synthesis was measured in Panc 10.05 cells transfected with siCON, siTRS, or siAlaRS for the indicated times by [35S]-Met incorporation assay. Data is displayed as a bar graph. CHX was used as a positive control and treated for 30 min. (h) Panc 10.05 and AsPC-1 cells that expressed inducible shRNA-targeting TRS were cultured in the presence or in the absence of Doxycycline (Dox) for 24 to 48 h. Expression levels of MUC1, MUC1-CT and TRS were determined by immunoblotting using anti-MUC1 or anti-TRS antibodies. (i) MIA PaCa-2 and PANC-1 cells that expressed inducible myc-tagged TRS were cultured in the presence or absence of Dox for 24 to 48 h. Expression of MUC1, MUC1-CT, and myc-tagged TRS were detected by immunoblotting with each respective antibody. (b–e and g) **P<0.01; ***P<0.001; ****P<0.0001; NS, not significant vs control group. Values represent means±s.e.m. of three independent experiments.
TRS inhibitors suppress pancreatic cancer cell migration
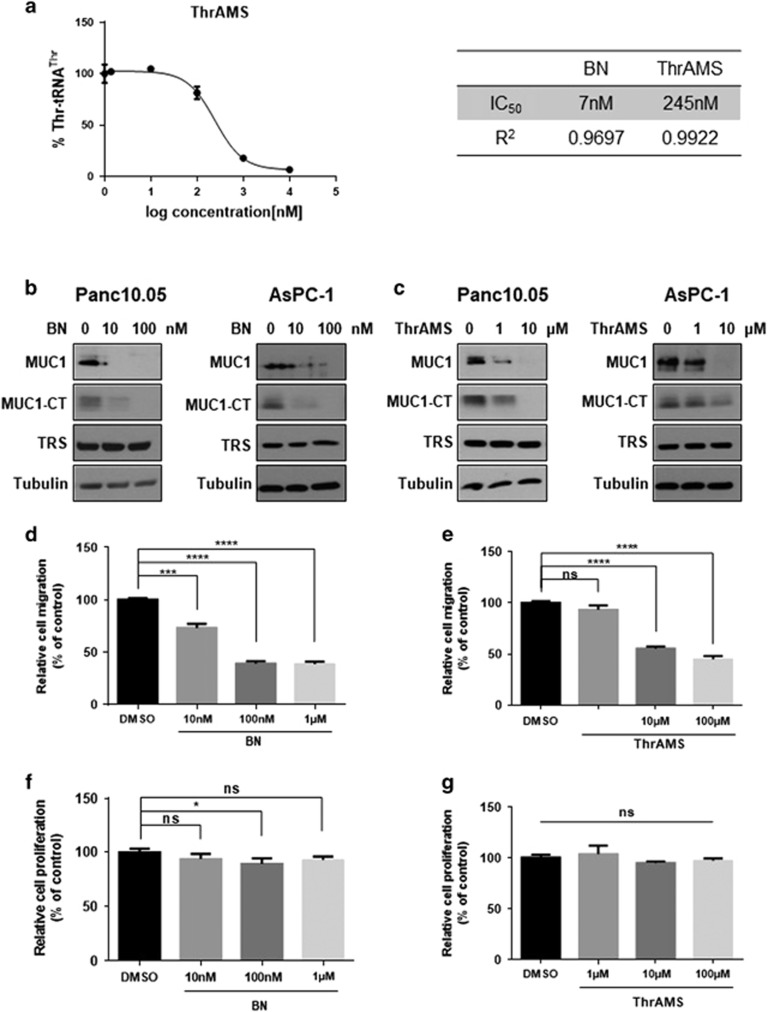
Next, we investigated whether the inhibition of TRS catalytic activity would also affect MUC1 level. For this study, we inhibited the TRS activity with the macrolide antibiotic, borrelidin (BN), a potent non-competitive inhibitor of TRS, and 5′-O-(N-(L-threonyl)-sulfamoyl)-adenosine (ThrAMS), a non-hydrolyzable analog of the reaction intermediate, threonyl-adenylate. In the aminoacylation assay with human TRS, activity was decreased by ThrAMS treatment in a dose-dependent manner with an estimated IC50 of 245 nM (Figure 4a, left). Previous studies have shown that BN inhibits the threonine activation step of human TRS with an inhibition constant (Ki) value of ~7 nM (Figure 4a, right).21 Panc 10.05 and AsPC-1 cells were then treated with different doses of BN and ThrAMS to investigate the effects on MUC1 levels by western blotting. The results showed a dose-dependent decrease of MUC1 and MUC1-CT levels in these cells (Figures 4b and c). To see whether TRS inhibitors would affect the growth and migration of pancreatic cancer cells, cell growth and wound-healing assays were performed. When Panc 10.05 cells were treated with different doses of BN or ThrAMS for 12 h, both BN and ThrAMS dramatically inhibited cell migration (Figures 4d and e), with little effect on cell growth (Figures 4f and g). Collectively, these results suggest the significance of TRS activity for biosynthesis of the protein MUC1.
Figure 4.
Effects of TRS inhibitors on pancreatic cancer cell migration. (a) Aminoacylation activity of a recombinant human TRS protein was assayed with ThrAMS titration (10–10 000 nM), with an estimated IC50 of 245 nM. The activity was normalized to DMSO, which was used as the solvent for ThrAMS. ThrAMS, 5′-O-(N-(L-threonyl)-sulfamoyl) adenosine; BN, Borrelidin. (b) Panc 10.05 and AsPc-1 cells were treated with the indicated concentration (10 and 100 nM) of BN for 4 h. Protein levels of MUC1, MUC1-CT and TRS were monitored by immunoblotting with each respective antibody. (c) Panc 10.05 and AsPC-1 cells were treated with the indicated concentration (1 and 10 μM) of ThrAMS for 4 h. Protein levels of MUC1, MUC1-CT and TRS were monitored by immunoblotting with each antibody. (d) Panc 10.05 cells were seeded at equal densities into 96-well plates, cultured to confluency, mechanically wounded by scratching, incubated in medium with the indicated concentrations (10 nM to 1 μM) of BN and then monitored for 12 h by IncuCyte Zoom. Cell migration was quantified based on the distance separating wound images and displayed as a bar graph. (e) Panc 10.05 cells were seeded at equal densities into 96-well plates, cultured to confluency, mechanically wounded by scratching, incubated in medium with the indicated concentrations (1 to 100 μM) of ThrAMS and then monitored for 12 h by IncuCyte Zoom. Cell migration was quantified based on the distance separating wound images and displayed as a bar graph. (f) Panc 10.05 cells were seeded at equal densities into 96-well plates with the indicated concentrations (10 nM to 1 μM) of BN and imaged within the IncuCyte Zoom. Proliferation was analyzed as the percentage confluent and displayed as a bar graph. (g) Panc 10.05 cells were seeded at equal densities into 96-well plates with the indicated concentrations (1 to 100 μM) of ThrAMS and imaged within the IncuCyte Zoom. Proliferation was analyzed as the percentage confluent and displayed as a bar graph. (d–g) *P<0.05; ***P<0.001; ****P<0.0001; NS, not significant vs control group. Values represent means±s.e.m. of three independent experiments.
TRS affects pancreatic cancer cell migration
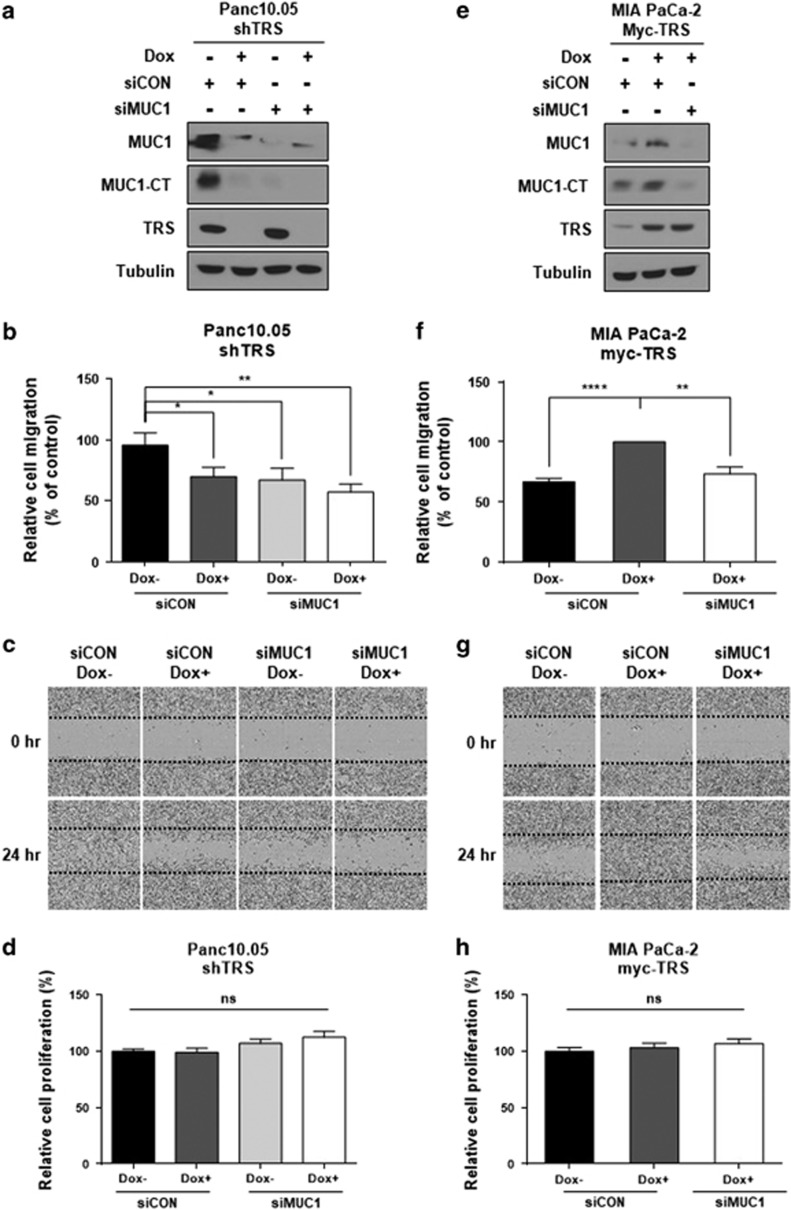
We next sought to utilize the two Dox-inducible TRS knockdown and overexpression cell lines that were used above to confirm that TRS-dependent MUC1 synthesis would affect pancreatic cancer cell migration. MUC1 and MUC1-CT protein levels were significantly reduced 24 h after TRS suppression (Figure 5a). Unsurprisingly, treatment of siRNA targeting MUC1 in shTRS-induced Panc 10.05 cells also decreased MUC1 and MUC1-CT protein levels (Figure 5a). Interestingly, suppression of either TRS or MUC1 decreased Panc 10.05 cell migration to similar extents (Figures 5b and c), with little effect on cell growth (Figure 5f). On the contrary, there was an observed increase in MUC1 and MUC1-CT levels, following induction of Myc-TRS but not when MUC1 was suppressed with its specific siRNA (Figure 5e). MIA PaCa-2 cell migration was increased when Myc-TRS was induced, but decreased after treatment with siMUC1 (Figures 5f and g) without affecting cell growth (Figure 5h). Taken together, our data indicate that TRS could affect pancreatic cancer cell migration by regulating protein levels of MUC1.
Figure 5.
Effects of TRS on pancreatic cancer cell migration. (a–d) Panc 10.05 cells were transfected with non-targeting or MUC1 targeting siRNAs for 24 h and then treated with Dox for 24 h to induce shRNA specific to TRS. (a) Protein levels of MUC1 and TRS were monitored by immunoblotting with each respective antibody. (b) Panc 10.05 cells were seeded at equal densities into 96-well plates, cultured to confluency, mechanically wounded by scratching, incubated in medium and then monitored for 24 h by IncuCyte Zoom. Cell migration was quantified based on the distance separating wound images and displayed as a bar graph. (c) Light microscopy images depicting migration in wound-healing assays as described in (b). Images were captured at 0 h (top) and 24 h (bottom), and showed the wound gap filled by cells after scratching. (d) Panc 10.05 cells were seeded at equal densities into 96-well plates and imaged within the IncuCyte Zoom. Proliferation was analyzed as the percentage confluent and displayed as a bar graph. (e–h) MIA PaCa-2 cells were transfected with non-targeting or MUC1 targeting siRNAs for 24 h and then treated with Dox for 24 h to induce myc-tagged TRS. (e) Protein levels of MUC1 and TRS were monitored by immunoblotting with each respective antibody. (f) MIA PaCa-2 cells were seeded at equal densities into 96-well plates, cultured to confluency, mechanically wounded by scratching, incubated in medium and then monitored for 24 h by IncuCyte Zoom. Cell migration was quantified based on the distance separating wound images and displayed as a bar graph. (g) Light microscopy images depicting migration in wound-healing assays as described in (f). Images were captured at 0 h (top) and 24 h (bottom), and showed the wound gap filled by cells after scratching. (h) MIA PaCa-2 cells were seeded at equal densities into 96-well plates and imaged within the IncuCyte Zoom. Proliferation was analyzed as the percentage confluent and displayed as a bar graph. (b–d and f–h) *P<0.05; **P<0.01; ****P<0.0001; NS, not significant vs control group. Values represent means±s.e.m. of three independent experiments. All images were captured by IncuCyte Zoom.
TRS and MUC1 levels are positively correlated with pancreatic cancer
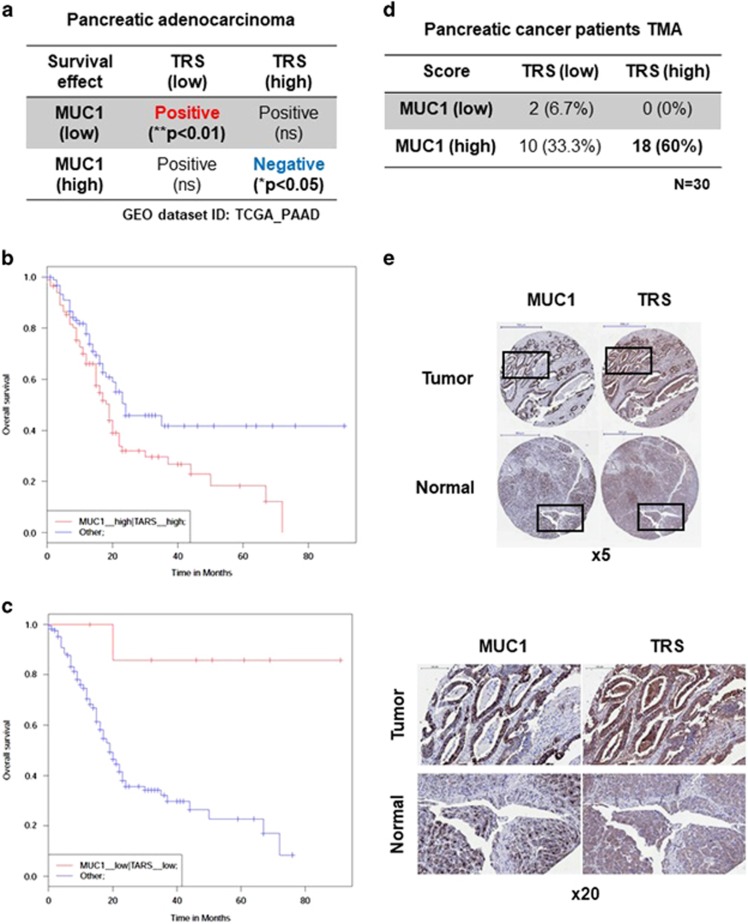
The next step was to understand whether expression of TRS and MUC1 would show any relevance to the survival of pancreatic cancer patients. This required bioinformatics analysis using SynTarget, a new online tool that investigates the expression of multiples genes from microarray datasets for synergistic survival relationships (Figure 6a).26, 27 Interestingly, co-expression of TRS and MUC1 at high levels showed a positive correlation with poor survival outcomes of the patients (Figure 6b). On the contrary, when the expression levels of both TRS and MUC1 were low, patient survival was significantly improved (Figure 6c). To further assess the correlation between MUC1 and TRS levels in clinical pancreatic tumors, it was important to analyze the tissue levels of the two proteins using a tumor microarray (TMA) including 30 clinical pancreatic tumor samples. Immunohistochemical signals of MUC1 and TRS were scored according to staining intensity, and it was found that tissue samples with high levels of both TRS and MUC1 accounted for 60% of the total tissue samples. (Figure 6d). Moreover, strong staining of MUC1 and TRS was observed in the duct region of tumor samples, whereas no staining was observed in the duct region of normal samples (Figure 6e). These results support the hypothesis that higher expression of TRS would increase the MUC1 level, contributing to the poor survival of pancreatic cancer patients.
Figure 6.
Positive correlation between MUC1 and TRS in pancreatic cancer. (a) Positive correlation between MUC1/TRS co-expression and reduced survival of pancreatic cancer patients based on the dataset from TCGA. (GEO dataset ID: TCGA_PAAD). (b and c) The bioinformatics analysis of synergistic effects of MUC1 and TRS co-expression on overall survival in months within human pancreatic adenocarcinoma patients with (b) high MUC1 and high TRS expression (**P<0.01) or (c) with low MUC1 and low TRS expression (*P<0.05). (d) Summary table of the quantitative expression of MUC1 and TRS with the score of the respective pancreatic adenocarcinoma tissue microarray (TMA). A TMA of pancreatic adenocarcinomas was immunostained with antibodies against MUC1 and TRS, respectively. Immunohistochemical (IHC) signals were scored by the staining intensity (0: no signal, 1: weak signal, 2: intermediate signal and 3: strong signal). Low, 0–1; high, 2–3. (e) Representative examples of patient biopsies of pancreatic adenocarcinoma assembled into a TMA stained for MUC1 and TRS, depicting the reciprocal expression pattern (× 5 magnification) (top). Boxed areas are shown at higher magnification (× 20) (bottom). Scale bar (× 5), 500 μm; scale bar (× 20), 100 μm.
Discussion
The oncoprotein MUC1 is overexpressed in 90% of pancreatic cancers and associated with a short survival rate.28, 29 MUC1 also induces epithelial-mesenchymal transition, as well as increased cell invasion.30 For these reasons, MUC1 is considered an attractive therapeutic target to moderate pancreatic cancer progression. In this study, deprivation of threonine (Figure 1), and reduction of TRS expression (Figure 3) and TRS catalytic activity (Figure 4) were associated with decreased MUC1 levels, and consequently, the suppression of pancreatic cancer cell migration. A number of preclinical studies suggest that decreasing mucin expression in tumors has potential as a novel molecular approach for the treatment of pancreatic cancer.31, 32, 33, 34 Since MUC4 and MUC16 are also threonine-rich glycoproteins, their cellular levels are also presumably regulated by TRS in pancreatic cancer cells.35, 36, 37, 38
Previous reports showed that threonine is considered an essential amino acid that is important for mucin synthesis.13, 14, 15, 16, 17 In line with these previous studies, the work presented here indicates that threonine supplementation is critical for MUC1 oncoprotein biosynthesis in pancreatic cancer cells (Figure 1).
Many reports have shown the functional implications of human aminoacyl-tRNA synthetases (ARSs) in cancer growth and progression via their unique activities other than their catalytic activities.39, 40, 41, 42 However, it is unknown whether the increased expression of ARSs would catalytically contribute to tumorigenesis. Presented in this study is the first report showing that the biosynthesis of mucin can be controlled either by the inhibition of TRS activity or by the suppression of TRS expression without affecting global protein synthesis. TRS expression has been shown to be increased in ovarian cancer and was suggested to be associated with angiogenesis.43 However, it may be more related to the unique activity of the secreted TRS in the extracellular space, as it was previously shown that TRS can be secreted to promote angiogenesis.44 Considering that TRS specifically regulates the biosynthesis of the threonine-rich protein MUC1, it would be interesting to investigate whether specific amino acid-rich proteins can also be specifically regulated by their cognate ARSs.
ARSs are validated drug targets against pathogens.45, 46 Recently, there have been increasing efforts to target human ARSs for various pharmacological indications.47, 48 For instance, halofuginone (HF), the halogenated derivative of febrifugine, has been tested for fibrosis treatment.49 The accumulation of extra-cellular matrix proteins, especially collagen, is a major feature of fibrotic disease. Since collagen is a proline-rich protein, prolyl-tRNA synthetase (PRS) could serve as a preferred target to block collagen biosynthesis. In fact, HF, a specific PRS inhibitor, showed high efficacy in decreasing collagen levels.50, 51
In summary, this study provides evidence illuminating the potential role of TRS in the migration of human pancreatic cancer cells by enhancing MUC1 biosynthesis. The effect of TRS inhibitors on MUC1 levels and cancer cell migration suggests the possibility targeting of TRS as a novel therapeutic avenue against pancreatic cancer. Further validation studies are currently under way using in vivo models and patient analysis.
Acknowledgments
This work was supported by a Global Frontier Project grant (NRF-M3A6A4-2010-0029785) of the National Research Foundation funded by the Ministry of Science, ICT & Future Planning (MSIP) of Korea.
Footnotes
The authors declare no conflict of interest.
References
- Schneider G, Siveke JT, Eckel F, Schmid RM. Pancreatic cancer: basic and clinical aspects. Gastroenterology 2005; 128: 1606–1625. [DOI] [PubMed] [Google Scholar]
- Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet 2011; 378: 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HH, Lemoine NR. Pancreatic cancer: molecular pathogenesis and new therapeutic targets. Nat Rev Gastroenterol Hepatol 2009; 6: 412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendler SJ. MUC1, the renaissance molecule. J Mammary Gland Biol Neoplasia 2001; 6: 339–353. [DOI] [PubMed] [Google Scholar]
- Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer 2009; 9: 874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PK, Hollingsworth MA. Cell surface-associated mucins in signal transduction. Trends Cell Biol 2006; 16: 467–476. [DOI] [PubMed] [Google Scholar]
- Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med 2014; 20: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horm TM, Schroeder JA. MUC1 and metastatic cancer: expression, function and therapeutic targeting. Cell Adh Migr 2013; 7: 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Klinken BJ, Dekker J, Buller HA, Einerhand AW. Mucin gene structure and expression: protection vs. adhesion. Am J Physiol 1995; 269: G613–G627. [DOI] [PubMed] [Google Scholar]
- Mantle M, Allen A. Isolation and characterization of the native glycoprotein from pig small-intestinal mucus. Biochem J 1981; 195: 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaart MW, Schierbeek H, van der Schoor SR, Stoll B, Burrin DG, Reeds PJ et al. Threonine utilization is high in the intestine of piglets. J Nutr 2005; 135: 765–770. [DOI] [PubMed] [Google Scholar]
- van der Schoor SR, Wattimena DL, Huijmans J, Vermes A, van Goudoever JB. The gut takes nearly all: threonine kinetics in infants. Am J Clin Nutr 2007; 86: 1132–1138. [DOI] [PubMed] [Google Scholar]
- Faure M, Moennoz D, Montigon F, Mettraux C, Breuille D, Ballevre O. Dietary threonine restriction specifically reduces intestinal mucin synthesis in rats. J Nutr 2005; 135: 486–491. [DOI] [PubMed] [Google Scholar]
- Law GK, Bertolo RF, Adjiri-Awere A, Pencharz PB, Ball RO. Adequate oral threonine is critical for mucin production and gut function in neonatal piglets. Am J Physiol Gastrointest Liver Physiol 2007; 292: G1293–G1301. [DOI] [PubMed] [Google Scholar]
- Wang X, Qiao S, Yin Y, Yue L, Wang Z, Wu G. A deficiency or excess of dietary threonine reduces protein synthesis in jejunum and skeletal muscle of young pigs. J Nutr 2007; 137: 1442–1446. [DOI] [PubMed] [Google Scholar]
- Nichols NL, Bertolo RF. Luminal threonine concentration acutely affects intestinal mucosal protein and mucin synthesis in piglets. J Nutr 2008; 138: 1298–1303. [DOI] [PubMed] [Google Scholar]
- van der Sluis M, Schaart MW, de Koning BA, Schierbeek H, Velcich A, Renes IB et al. Threonine metabolism in the intestine of mice: loss of mucin 2 induces the threonine catabolic pathway. J Pediatr Gastroenterol Nutr 2009; 49: 99–107. [DOI] [PubMed] [Google Scholar]
- Park SG, Ewalt KL, Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem Sci 2005; 30: 569–574. [DOI] [PubMed] [Google Scholar]
- Lee JY, Kim DG, Kim BG, Yang WS, Hong J, Kang T et al. Promiscuous methionyl-tRNA synthetase mediates adaptive mistranslation to protect cells against oxidative stress. J Cell Sci 2014; 127: 4234–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 2009; 6: 275–277. [DOI] [PubMed] [Google Scholar]
- Fang P, Yu X, Jeong SJ, Mirando A, Chen K, Chen X et al. Structural basis for full-spectrum inhibition of translational functions on a tRNA synthetase. Nat Commun 2015; 6: 6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga BS, Pronczuk AW, Munro HN. Mechanism of cycloheximide inhibition of protein synthesis in a cell-free system prepared from rat liver. J Biol Chem 1969; 244: 4480–4489. [PubMed] [Google Scholar]
- Besmer DM, Curry JM, Roy LD, Tinder TL, Sahraei M, Schettini J et al. Pancreatic ductal adenocarcinoma mice lacking mucin 1 have a profound defect in tumor growth and metastasis. Cancer Res 2011; 71: 4432–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto S, Yokoyama S, Higashi M, Yamada N, Takao S, Yonezawa S. MUC1 enhances hypoxia-driven angiogenesis through the regulation of multiple proangiogenic factors. Oncogene 2013; 32: 4614–4621. [DOI] [PubMed] [Google Scholar]
- Liu X, Yi C, Wen Y, Radhakrishnan P, Tremayne JR, Dao T et al. Interactions between MUC1 and p120 catenin regulate dynamic features of cell adhesion, motility, and metastasis. Cancer Res 2014; 74: 1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonov AV. BioProfiling.de: analytical web portal for high-throughput cell biology. Nucleic Acids Res 2011; 39: W323–W327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio I, Tsvetkov PO, Knight RA, Lisitsa A, Melino G, Antonov AV. SynTarget: an online tool to test the synergetic effect of genes on survival outcome in cancer. Cell Death Differ 2016; 23: 912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MP, Chakraborty S, Souchek J, Batra SK. Mucin-based targeted pancreatic cancer therapy. Curr Pharm Des 2012; 18: 2472–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Kumar S, Momi N, Sasson AR, Batra SK. Mucins in pancreatic cancer and its microenvironment. Nat Rev Gastroenterol Hepatol 2013; 10: 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy LD, Sahraei M, Subramani DB, Besmer D, Nath S, Tinder TL et al. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene 2011; 30: 1449–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP, Moniaux N, Chauhan SC, Meza JL, Batra SK. Inhibition of MUC4 expression suppresses pancreatic tumor cell growth and metastasis. Cancer Res 2004; 64: 622–630. [DOI] [PubMed] [Google Scholar]
- Tsutsumida H, Swanson BJ, Singh PK, Caffrey TC, Kitajima S, Goto M et al. RNA interference suppression of MUC1 reduces the growth rate and metastatic phenotype of human pancreatic cancer cells. Clin Cancer Res 2006; 12: 2976–2987. [DOI] [PubMed] [Google Scholar]
- Chaturvedi P, Singh AP, Moniaux N, Senapati S, Chakraborty S, Meza JL et al. MUC4 mucin potentiates pancreatic tumor cell proliferation, survival, and invasive properties and interferes with its interaction to extracellular matrix proteins. Mol Cancer Res 2007; 5: 309–320. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Wong S, Borrelli A, Chung MA. Down-regulation of MUC1 in cancer cells inhibits cell migration by promoting E-cadherin/catenin complex formation. Biochem Biophys Res Commun 2007; 362: 740–746. [DOI] [PubMed] [Google Scholar]
- Felder M, Kapur A, Gonzalez-Bosquet J, Horibata S, Heintz J, Albrecht R et al. MUC16 (CA125): tumor biomarker to cancer therapy, a work in progress. Mol Cancer 2014; 13: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg G, Gibbs J, Belt B, Powell MA, Mutch DG, Goedegebuure P et al. Novel treatment option for MUC16-positive malignancies with the targeted TRAIL-based fusion protein Meso-TR3. BMC Cancer 2014; 14: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam SK, Kumar S, Cannon A, Hall B, Bhatia R, Nasser MW et al. MUC4 mucin- A therapeutic target for pancreatic ductal adenocarcinoma. Expert Opin Ther Targets 2017; 21: 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachagani S, Macha MA, Ponnusamy MP, Haridas D, Kaur S, Jain M et al. MUC4 potentiates invasion and metastasis of pancreatic cancer cells through stabilization of fibroblast growth factor receptor 1. Carcinogenesis 2012; 33: 1953–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SG, Schimmel P, Kim S. Aminoacyl tRNA synthetases and their connections to disease. Proc Natl Acad Sci USA 2008; 105: 11043–11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Kwon NH, Kim S. Association of aminoacyl-tRNA synthetases with cancer. Top Curr Chem 2014; 344: 207–245. [DOI] [PubMed] [Google Scholar]
- Kim S, You S, Hwang D. Aminoacyl-tRNA synthetases and tumorigenesis: more than housekeeping. Nat Rev Cancer 2011; 11: 708–718. [DOI] [PubMed] [Google Scholar]
- Kim YW, Kwon C, Liu JL, Kim SH, Kim S. Cancer association study of aminoacyl-tRNA synthetase signaling network in glioblastoma. PLoS ONE 2012; 7: e40960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellman TL, Eckenstein M, Wong C, Rincon M, Ashikaga T, Mount SL et al. Threonyl-tRNA synthetase overexpression correlates with angiogenic markers and progression of human ovarian cancer. BMC Cancer 2014; 14: 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TF, Mirando AC, Wilkinson B, Francklyn CS, Lounsbury KM. Secreted Threonyl-tRNA synthetase stimulates endothelial cell migration and angiogenesis. Sci Rep 2013; 3: 1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvian LF, Wang J, Steitz TA. Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin. Science 1999; 285: 1074–1077. [PubMed] [Google Scholar]
- Rock FL, Mao W, Yaremchuk A, Tukalo M, Crepin T, Zhou H et al. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 2007; 316: 1759–1761. [DOI] [PubMed] [Google Scholar]
- Gadakh B, Van Aerschot A. Aminoacyl-tRNA synthetase inhibitors as antimicrobial agents: a patent review from 2006 till present. Expert Opin Ther Pat 2012; 22: 1453–1465. [DOI] [PubMed] [Google Scholar]
- Zhou H, Sun L, Yang XL, Schimmel P. ATP-directed capture of bioactive herbal-based medicine on human tRNA synthetase. Nature 2013; 494: 121–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines M, Snyder D, Yarkoni S, Nagler A. Halofuginone to treat fibrosis in chronic graft-versus-host disease and scleroderma. Biol Blood Marrow Transplant 2003; 9: 417–425. [DOI] [PubMed] [Google Scholar]
- Pines M, Knopov V, Genina O, Lavelin I, Nagler A. Halofuginone, a specific inhibitor of collagen type I synthesis, prevents dimethylnitrosamine-induced liver cirrhosis. J Hepatol 1997; 27: 391–398. [DOI] [PubMed] [Google Scholar]
- Gnainsky Y, Spira G, Paizi M, Bruck R, Nagler A, Abu-Amara SN et al. Halofuginone, an inhibitor of collagen synthesis by rat stellate cells, stimulates insulin-like growth factor binding protein-1 synthesis by hepatocytes. J Hepatol 2004; 40: 269–277. [DOI] [PubMed] [Google Scholar]