Abstract
Sumoylation, the conjugation of a small ubiquitin-like modifier (SUMO) protein to a target, has diverse cellular effects. However, the functional roles of the SUMO modification during myogenesis have not been fully elucidated. Here, we report that basal sumoylation of histone deacetylase 1 (HDAC1) enhances the deacetylation of MyoD in undifferentiated myoblasts, whereas further sumoylation of HDAC1 contributes to switching its binding partners from MyoD to Rb to induce myocyte differentiation. Differentiation in C2C12 skeletal myoblasts induced new immunoblot bands above HDAC1 that were gradually enhanced during differentiation. Using SUMO inhibitors and sumoylation assays, we showed that the upper band was caused by sumoylation of HDAC1 during differentiation. Basal deacetylase activity was not altered in the SUMO modification-resistant mutant HDAC1 K444/476R (HDAC1 2R). Either differentiation or transfection of SUMO1 increased HDAC1 activity that was attenuated in HDAC1 2R. Furthermore, HDAC1 2R failed to deacetylate MyoD. Binding of HDAC1 to MyoD was attenuated by K444/476R. Binding of HDAC1 to MyoD was gradually reduced after 2 days of differentiation. Transfection of SUMO1 induced dissociation of HDAC1 from MyoD but potentiated its binding to Rb. SUMO1 transfection further attenuated HDAC1-induced inhibition of muscle creatine kinase luciferase activity that was reversed in HDAC1 2R. HDAC1 2R failed to inhibit myogenesis and muscle gene expression. In conclusion, HDAC1 sumoylation plays a dual role in MyoD signaling: enhancement of HDAC1 deacetylation of MyoD in the basally sumoylated state of undifferentiated myoblasts and dissociation of HDAC1 from MyoD during myogenesis.
Introduction
Similar to ubiquitination, sumoylation is a covalent binding of small ubiquitin-like modifiers (SUMOs) to specific lysine residues of target proteins. Since initially being discovered in 1997,1 sumoylation has been implicated in many biological functions and in disease status because it regulates diverse cellular processes. By identifying novel sumoylated proteins and evaluating the altered function of the sumoylated targets, we can extend the current understanding of the roles of sumoylation in cellular function.2 SUMOs comprise 92–97 amino acids, and sumoylation is different from classically characterized posttranslational modifications with relatively small chemical groups, such as acetylation, phosphorylation and methylation. Sumoylation has been well characterized at the molecular level, with a particular focus on site-specific conjugation of SUMO1, SUMO2 and SUMO3. Several hundred sumoylation target proteins are involved in various processes, such as chromatin organization, transcriptional regulation, DNA repair, macromolecular assembly, protein turnover, intracellular localization and signal transduction.3 Inversely, SUMO/sentrin-specific proteases remove SUMOs from target proteins, resulting in the tight balance of the sumoylation status.4
MyoD is a member of the myogenic basic helix-loop-helix (bHLH) family of transcription factors and functions as an initiator of the myogenic program.5 MyoD activation precedes sequential upregulation of other myogenic transcription factors, such as myogenin, myosin light chain and desmin, resulting in the eventual induction of genes that characterize terminal differentiation of skeletal muscle, such as muscle creatine kinase (MCK).6 Transcriptional activation of MyoD depends on its binding to a specific DNA sequence in the promoter, E-box (CNANNTG), that is present in the regulatory regions of muscle genes.5 Notably, however, MyoD is expressed, even in quiescent myoblasts, before the myogenic process, when it is unable to function as a transcriptional activator,7 suggesting that certain transcriptional inhibitors bind to MyoD before the start of the myogenic program. Indeed, MyoD is maintained in an inactivated status by binding to Id,8 Ezh2,9 ret finger protein,10 Suv39h1,11 Dec2,12 C/EBP homology protein,13 myb-binding protein 1a14 and NFATc1.15 In many cases, these repressors function as a complex and require transcriptional repression by removing an acetyl group from the histones of the MyoD target gene promoters that is primarily mediated by histone deacetylase 1 (HDAC1). Indeed, since HDAC1 was reported to inhibit MyoD-induced activation of the myogenic program,16, 17 many reports have elucidated that HDAC is required for repression of MyoD. In addition, during myogenesis, HDAC1 dissociates from MyoD and subsequently binds to retinoblastoma protein18 to form a complex with E2F to repress cell cycle progression.19 However, the mechanism of this complicated switching of binding partners has not been fully investigated.
HDAC1 contains many amino acid residues that undergo posttranslational modification.20 Considering that HDAC1 undergoes diverse posttranslational modifications19 that alter its activity and binding affinity, these modifications also affect the myogenic activity of MyoD in association with skeletal myoblasts. Similarly, HDAC1 sumoylation may also alter the myogenic activity of MyoD. However, the effects of HDAC1 sumoylation on MyoD and its following myogenic program have not been described. In the present study, we demonstrated HDAC1 sumoylation in myoblasts and subsequently explored the roles of HDAC1 sumoylation in MyoD-dependent myogenic differentiation.
Materials and methods
Cell culture, differentiation conditions and transfection
The C2C12 cell line has been previously described.21 Cells were maintained in Dulbecco’s modified Eagle’s medium containing 15% fetal bovine serum (Hyclone, Thermo Fisher Scientific, Waltham, MA, USA). Differentiation was induced in Dulbecco’s modified Eagle’s medium plus 2% horse serum (Hyclone). Cells were transfected using Lipofectamine LTX and plus reagent (Invitrogen Life Technologies, Carlsbad, CA, USA). HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Cells were transfected with Polyfect (QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions. All cells were cultured at 37 °C and 5% CO2.
Plasmids, antibodies and reagents
Both pcDNA3-HA-SUMO1 and pSG5-Flag-SUMO1 were kindly gifted by Professor Nacksung Kim (Department of Pharmacology, Chonnam National University Medical School, Hwasun-gun, South Korea). pcDNA6-SUMO1-myc was constructed by subcloning, utilizing pSG5-Flga-SUMO1 (CosmoGeneTech, Dagjeon, Republic of Korea). pHM6-HA-MyoD was obtained from Professor Young-Gyu Ko (College of Life Sciences and Biotechnology, Korea University, Seoul, South Korea). pGL3-Basic-myogenin-luciferase and pGL3-Mck-luciferase were kind gifts from Professor Da-Zhi Wang (Department of Cardiology, Harvard Medical School, Boston, MA, USA). pBJ5.1-Flag-HDAC1 (HDAC1 wild-type (WT)) was a kind gift from Professor Jonathan A Epstein (University of Pennsylvania, Philadelphia, PA, USA). pBJ5.1-Flag-HDAC1 K444/476R mutant (HDAC1 2R) was constructed by site-directed mutagenesis based on pBJ5.1-Flag-HDAC1 WT (CosmoGeneTech).
The following antibodies were used: anti-HDAC1 (H-51; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-HDAC2 (ab12169; Abcam, San Francisco, CA, USA), anti-SUMO1 (D-11; Santa Cruz Biotechnology), anti-MyoD (C-20; Santa Cruz Biotechnology), anti-MCK (C-14; Santa Cruz Biotechnology), anti-myogenin (M-225; Santa Cruz Biotechnology), α-myosin heavy chain antibody (MF20, Developmental Studies Hybridoma Bank, Iowa city, IA, USA), anti-Rb (sc-102; Santa Cruz Biotechnology), anti-actin (Alpha Sr-1; Santa Cruz Biotechnology), anti-GAPDH (H-12; Santa Cruz Biotechnology), anti-lamin B1 (ab16048; Abcam), anti-acetyl lysine (ab21623; Abcam), anti-Flag (F7425; Sigma-Aldrich, St Louis, MO, USA) and anti-HA (H9658; Sigma-Aldrich).
The chemical 2-D08 was purchased from Sigma-Aldrich (St. Louis, MO, USA), and ginkgolic acid was obtained from Abcam. MG132 was purchased from Cayman Chemical (Ann Arbor, MI, USA). Texas Red-X-phalloidin was purchased from Invitrogen.
Luciferase reporter assay, RNA isolation, quantitative real-time PCR, western blotting and immunoprecipitation analyses
These procedures have been previously described.21, 22, 23 The primer sequences for quantitative real-time PCR will be provided upon request. The real-time PCR amplification products were confirmed by sequencing. For western blotting, primary antibodies against HDAC1 (1:1000), HDAC2 (1:5000), MCK (1:1000), MyoD (1:500), Myogenin (1:1000), SUMO1 (1:1000), Rb (1:1000), actin (1:1000), GAPDH (1:1000), Lamin B1 (1:1000), Flag (1:4000) and HA (1:20,000) were used.
For detection of sumoylated protein by immunoprecipitation analysis, 2 mM N-ethylmaleimide was included in the lysis buffer to block isopeptidases and deubiquitinating enzymes. Immunoprecipitated SUMO1 complexes were resolved using sodium dodecyl sulfate–polyacrylamide gel electrophoresis and detected by western blotting.
Nuclear fractionation, in vitro sumoylation assay and in vitro translation
To prepare the nuclear fractions, NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Fischer Scientific) were used according to the manufacturer’s instructions. The protein levels of the nuclear fractions were normalized to Lamin B1. The in vitro sumoylation assay was conducted using a commercial assay kit from Abcam (ab139470). The mixtures were set up in 20 μl of reaction buffer and incubated at 37 °C for 90 min. The reaction mixtures were subsequently separated using sodium dodecyl sulfate–polyacrylamide gel electrophoresis. For in vitro translation, protein synthesis was achieved using the TNT T7/T3 Coupled Reticulocyte Lysate System (Promega, Fitchburg, WI, USA) according to the manufacturer’s instructions. In vitro translation was performed in a 25 μl reaction volume with 1 μg of construct. The reaction was incubated at 30 °C for 90 min.
Chromatin immunoprecipitation
The chromatin immunoprecipitation assay was performed as previously described,22 with slight modifications. Briefly, cells were treated with 1% formaldehyde (Sigma) for 10 min to induce crosslinking at room temperature and neutralized with 0.125 M glycine. The assay was conducted using the EpiQuik Chromatin Immunoprecipitation kit (EpiGentek, Farmingdale, NY, USA). The procedure was performed according to the manufacturer’s instructions. PCR was performed as previously described using specific primers for either MCK or myogenin promoter.
HDAC activity measurement
HDAC activity assays were performed as previously described.24, 25 HDAC activity was determined using a commercial assay kit from Enzo Life Sciences (HDAC Fluorometric Assay/Drug Discovery kit, Farmingdale, NY, USA). For the HDAC1 activity assay, cells were transfected with HDAC1 WT or the HDAC1 2R mutant and the SUMO1 construct, followed by immunoprecipitation using an anti-Flag antibody.
Immunocytochemistry and multinucleated cell count
C2C12 cells were fixed with 4% paraformaldehyde and permeabilized with Triton X-100 (0.1%). Cells were incubated overnight with anti-Flag and anti-Fast skeletal myosin heavy chain antibodies in 1% bovine serum albumin and detected using an Alex Fluor 568-conjugated goat anti-mouse IgG secondary antibody (Invitrogen). Before mounting on slides, cells were stained with 4',6-diamidino-2-phenylindole (DAPI, Molecular Probes, Eugene, OR, USA). Cells were visualized using a Nikon Zoom stereomicroscope (SMZ1000, Tokyo, Japan). Cells with more than 2 nuclei per cell were counted as multinucleated cells after DAPI nuclear staining. Multinucleated cell numbers were divided by the total cell count to obtain percentage values. For each experimental condition, 10 fields were randomly selected per well, averaged and considered as one case.
Statistics
The data are presented as the means±s.e.m. Statistical significance was determined using Student’s t-tests or one-way analysis of variance, followed by Tukey’s honest significant difference multiple comparison post hoc test. PASW Statistics 19 (SPSS, an IBM Company, Chicago, IL, USA) was used for statistical analysis.
Results
Additional immunoreactivity with HDAC1 during myoblast differentiation
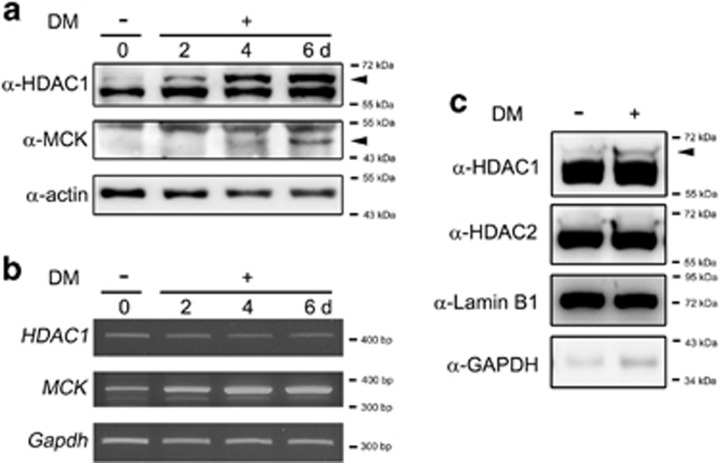
Because HDAC1 interacts with MyoD,17 a key myogenic transcription factor, we examined whether either the protein level or the posttranslational modification of HDAC1 is altered during myogenesis. We induced C2C12 cells to differentiate by switching the culture media from growth media (GM) to differentiation media (DM) and subsequently examined the expression of HDAC1. Although we did not observe apparent changes in the protein levels of HDAC1, those of MCK, a terminal differentiation marker of myogenesis,6 were increased, particularly in the late phase of differentiation. Interestingly, we observed an additional faint upper band of HDAC1 in GM, and the intensity of the upper band gradually increased when differentiation was induced (DM 2–6 days, Figure 1a, arrowhead). DM did not alter the mRNA level of HDAC1, but induced MCK mRNA expression (Figure 1b). We next assessed whether DM induces the additional band of HDAC2 and HDAC1 in the nuclear fractions of 2-day DM-treated C2C12 cells; the HDAC1-associated upper bands appeared, although no clear upper band was observed with HDAC2 (Figure 1c).
Figure 1.
Differentiation of myoblasts showed an additional band that appeared above histone deacetylase 1 (HDAC1). (a) Switching the culture media to differentiation media (DM) resulted in additional immunoblot bands above HDAC1. C2C12 cells were treated and maintained in DM for the indicated duration. Arrowhead shows new bands that were upregulated during myoblast differentiation. Muscle creatine kinase (MCK) expression was increased during myogenesis. (b) HDAC1 mRNA level was not changed after DM treatment. Real-time PCR (RT-PCR) was performed with C2C12 cells. (c) Treatment with DM for 2 days was sufficient to enhance the additional band of HDAC1 in the nuclear fraction of C2C12 cells. However, DM failed to induce an additional band of HDAC2.
HDAC1 is sumoylated during myogenesis
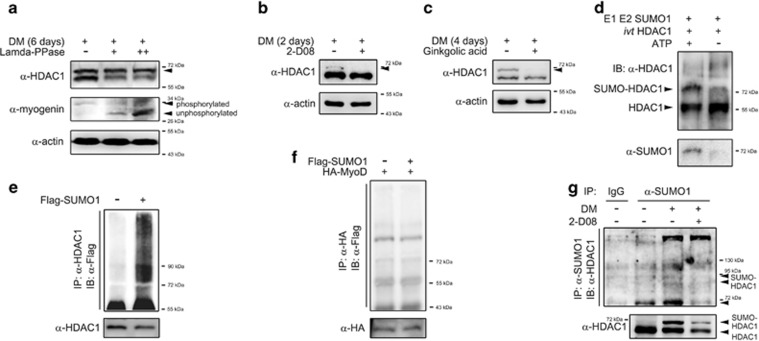
We next examined which modification of HDAC1 is involved in the differentiation process. We first examined the possibility of phosphorylation. However, treatment with lamda-protein phosphatase (Lamda-PPase) did not affect the upper band (Figure 2a, upper panel). Myogenin is known to be phosphorylated during myoblast differentiation.26 Indeed, in our experimental model, treatment with Lamda-PPase attenuated the upper phosphorylated band of myogenin and the amount of the lower unphosphorylated band was increased in a Lamda-PPase dose-dependent manner (Figure 2a, middle panel). Sumoylation is mediated by three different SUMO ligases, E1, E2 and E3.27 We observed that a series of sumoylation inhibitors blocked immunoreactivity; specifically, treatment with 2-D08, a SUMO E1/E2 ligase inhibitor,28 completely blocked the upper band (Figure 2b). Similarly, ginkgolic acid, a SUMO E1 ligase inhibitor,29 also blocked the band (Figure 2c).
Figure 2.
New bands are produced by sumoylation. (a) The upper bands did not result from phosphorylation in C2C12 cells. Induction of upper bands by differentiation media (DM; 6 days) was not blocked by treatment with lamda-protein phosphatase (Lamda-PPase, 800 units for 2 h at 37 °C) that reduces phosphorylation (upper panel). In contrast, DM-induced formation of the myogenin upper bands was dose-dependently blocked and shifted to the lower band by Lamda-PPase. (b) The upper band disappeared in C2C12 cells after treatment with 2-D08 (25 μM, 24 h), a small ubiquitin-like modifier (SUMO) E1/E2 ligase inhibitor. DM was administered for 2 days. (c) The upper band induced after treatment with DM for 4 days disappeared in C2C12 cells after treatment with ginkgolic acid (25 μM, 24 h), a SUMO E1 ligase inhibitor. (d) The in vitro sumoylation assay was performed with a commercially available SUMO kit as described in the Materials and Methods. We used in vitro translated (ivt) histone deacetylase 1 (HDAC1) and synthetic proteins of E1 and E2 ligases. The sumoylated bands disappeared in the absence of adenosine triphosphate (ATP). (e) Forced transfection of SUMO1 induces sumoylation of HDAC1 in C2C12 cells. At 2 days after transfection with Flag-SUMO1, endogenous HDAC1 was immunoprecipitated using an anti-HDAC1 antibody, and subsequently the precipitates were separated on the gel for the immunoblot analysis using an anti-Flag antibody (SUMO1). A smearing pattern was observed by forced expression of SUMO1. (f) Transfection of SUMO1 did not induce sumoylation of MyoD. Either Flag-SUMO1 or HA-MyoD was transfected into 293T cells and immunoprecipitated with an anti-HA antibody. No increase in the smearing pattern was observed. (g) Differentiation of C2C12 cells induces sumoylation of HDAC1. Endogenous SUMO1 was immunoprecipitated and HDAC1 was detected. Compared with growth media (GM, second lane), DM enhanced the smearing pattern of HDAC1 (third lane) that was attenuated by simultaneous treatment with 2-D08 (fourth lane).
SUMO ligation requires adenosine triphosphate (ATP).30, 31, 32, 33 We examined sumoylation of HDAC1 in the presence or absence of ATP by utilizing the SUMO assay kit34 in the cell-free condition. We used both synthetic E1 and E2 proteins and synthetic in vitro transcribed/translated HDAC1. In the presence of ATP, we observed the sumoylated HDAC1 band that was detected by an anti-HDAC1 antibody. However, this sumoylated band was attenuated in the absence of ATP, suggesting that this sumoylation of HDAC1 is ATP dependent (Figure 2d, upper panel). Utilization of an anti-SUMO1 antibody also detected a 72 kDa sumoylated band (Figure 2d, lower panel), and SUMO-HDAC1 was detected with an anti-HDAC1 antibody (upper panel).
Sumoylation of certain proteins can also be demonstrated by immunoprecipitation between SUMO1 protein and the target molecule.34 We also performed an immunoprecipitation assay using mammalian expression vectors of SUMO1. Forced expression of SUMO1 protein induced a smearing pattern of HDAC1 in either C2C12 cells (Figure 2e) or 293T cells (data not shown), suggesting successful conjugation of SUMO1 to HDAC1. In the following studies to induce forced sumoylation, transfection with either Flag-SUMO1 or HA-SUMO1 to cells was performed. In our preliminary study utilizing C2C12 cells, transfection of SUMO2 did not induce a clear sumoylation pattern of HDAC1 as SUMO1 did in our experimental models (data not shown). To rule out the argument that MyoD can be directly sumoylated, we assessed whether MyoD is sumoylated by SUMO1 transfection in 293T cells. However, we could not detect sumoylation of MyoD under our experimental conditions (Figure 2f). Using immunoprecipitation assay with endogenous SUMO1 and HDAC1, we observed that HDAC1 was slightly sumoylated in C2C12 cells in GM (Figure 2g, second lane). Interestingly, the smearing pattern of HDAC1 was enhanced when DM was added to C2C12 cells (third lane), whereas the enhancement was not observed with treatment with 2-D08 (fourth lane).
HDAC1 sumoylation affects enzymatic activity during differentiation
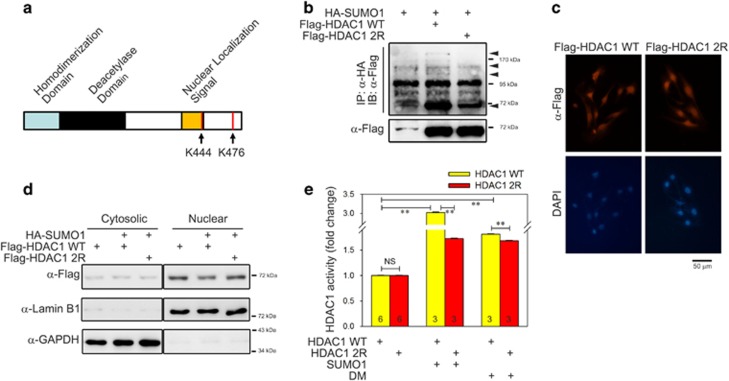
The K444 and K476 residues of HDAC1 are known to be sumoylated.35, 36 Those lysine residues together with the whole structure of HDAC1 are shown in Figure 3a. We generated a double mutant construct to express sumoylation-dead HDAC1 by substituting those lysines with arginines (HDAC1 K444/476R), designated as HDAC1 2R. To confirm that those lysines are responsible for the sumoylation of HDAC1, we subsequently performed an immunoprecipitation-based sumoylation assay with the mutant protein. Transfection of HDAC1 WT generated additional bands (Figure 3b, second lane upper panel) compared with the nontransfected group (arrowheads at right side). However, those additional bands disappeared when HDAC1 2R was transfected (third lane upper panel). The expression levels of the WT and mutant forms were not different (Figure 3b, lower panel).
Figure 3.
Histone deacetylase 1 (HDAC1) sumoylation is required for deacetylase activity. (a) Molecular structure of HDAC1 and the lysine residues responsible for sumoylation. The predicted sumoylation residues are indicated with arrows. (b) Sumoylation of HDAC1 is dependent on lysines 444 and 476. HDAC1 K444/476R was constructed by site-directed mutagenesis and utilized for sumoylation assay in C2C12 cells. Note that the increase in sumoylation pattern in HDAC1 wild-type (WT) was not observed in HDAC1 2R, indicating that HDAC1 K444/476 are the main sumoylation residues, and HDAC1 2R can be used as a sumoylation-resistant mutant in the following studies. (c) HDAC1 sumoylation did not induce nuclear-to-cytosolic shift of HDAC1 in C2C12 cells. To determine the intracellular localization of HDAC1, 2 days after the transfection of either Flag-HDAC1 WT or Flag-HDAC1 2R, immunocytochemical analysis was performed using an anti-Flag antibody. Note that HDAC1 2R was still abundant in the nucleus. (d) Western blot analysis with fractionated C2C12 cells further demonstrated that no cytosolic-nuclear shuttling was observed. C2C12 cells were fractionated at 2 days after the transfection of either Flag-HDAC1 WT or Flag-HDAC1 2R as indicated. Forced expression of small ubiquitin-like modifier 1 (SUMO1) did not result in an increase in HDAC1 WT in the cytosol. (e) Deacetylase activity of HDAC1 is dependent on sumoylation under either forced sumoylation or with differentiation media (DM; 2 days). Basal HDAC1 WT activity was calculated as 1 and the fold changes over basal activity are displayed. The activity of HDAC1 2R was similar to that of HDAC1 WT. Forced sumoylation significantly increased the HDAC1 WT enzymatic activity and the increase was dramatically attenuated in HDAC1 2R. Although the magnitude was less than that of forced sumoylation, treatment with DM also significantly increased the deacetylase activity of HDAC1 WT. However, the increase was significantly reduced in HDAC1 2R. Error bars represent s.e.m. NS, not significant, **P<0.01. Numerals in bar graphs are the numbers of samples.
Protein sumoylation has diverse biological functions, one of which is the regulation of intracellular localization.37, 38 Thus, we first examined whether HDAC1 sumoylation affects the localization of protein after fractionation of the cellular components. We examined the intracellular localization of HDAC1 by immunocytochemistry analysis and probed HDAC1 with an anti-Flag antibody. Both HDAC1 WT and HDAC1 2R were abundant in the nucleus, and no clear nuclear-to-cytosolic shift of HDAC1 2R localization was observed (Figure 3c). We next examined whether forced sumoylation of HDAC1 affected its localization. Interestingly, forced sumoylation slightly decreased the amount of HDAC1 WT in the nucleus (Figure 3d, second lane right upper panel). However, the decrease in HDAC1 WT was not accompanied by an increase in cytosolic HDAC1 (Figure 3d, second lane left upper panel), suggesting that no clear nuclear-to-cytosolic shift of HDAC1 protein occurred. However, the decrease in the nucleus was not observed with HDAC2 2R (Figure 3d, third lane right upper panel). It has been reported that HDAC1 sumoylation induces proteasomal degradation.
We next examined whether HDAC1 sumoylation is required for the deacetylase activity of HDAC1 and compared the activity of WT with 2R. HDAC1 2R basal activity was not different from that of WT (Figure 3e, left two columns). Interestingly, forced sumoylation by transfection of SUMO1 significantly increased the activity of HDAC1 by threefold. Surprisingly, the SUMO1-induced increase in HDAC1 activity was halved when HDAC2 2R was transfected (middle two columns). DM also significantly increased HDAC1 WT intrinsic activity; however, the increase was blunted in HDAC1 2R. These results suggest that although basal HDAC1 activity was not altered, when the sumoylation status is induced by either forced expression of SUMO1 or DM, deacetylase activity is upregulated and the increase in activity is sumoylation dependent.
HDAC1 dissociates from MyoD to derepress acetylation of MyoD
It is known that MyoD activity and protein amount remain silent during the proliferative phase of the myoblast that is in part mediated by the binding of MyoD to HDAC1.16, 17, 39 During myoblast differentiation into myotubes, however, HDAC1 dissociates from MyoD, thus freeing up space for binding by pCAF.40 Dissociated HDAC1 subsequently interacts with other transcription factors, including the pRb/E2F complex, to induce myoblast proliferation.18 However, the detailed mechanism of this switching of binding partners is not known.
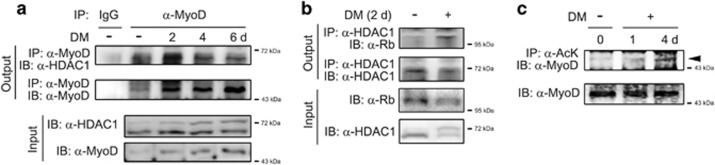
First, we assessed whether the binding of HDAC1 with MyoD is altered during myoblast differentiation in our experimental models. The binding of MyoD to HDAC1 seemed to be enhanced after 2 days of treatment with DM compared with the untreated group (Figure 4a, second vs third lanes in uppermost panel). However, considering that MyoD expression is dramatically increased by even 2 days of DM (second vs third lanes in the second panel and first vs second lanes in the lowermost panel), the enhancement of binding might be caused by the simple increase in the amount of MyoD protein, rather than by the enhancement of protein affinity between MyoD and HDAC1. Interestingly, however, the binding affinity was reduced at days 4 and 6 of DM treatment; the binding of HDAC1 and MyoD was attenuated (third–fifth lanes in uppermost panel), although MyoD protein expression was relatively maintained (second panel) or even increased (Figure 4a, lowermost panel). In contrast, DM induced the association of HDAC1 with Rb, suggesting switching of binding partners during myocyte differentiation, as previously described (Figure 4b).18
Figure 4.
Histone deacetylase 1 (HDAC1) dissociates from MyoD in myogenesis. (a) HDAC1 dissociates from MyoD during the differentiation media (DM)-induced differentiation of C2C12 cells. Immunoprecipitation assay was performed using an anti-MyoD antibody, and subsequently an anti-HDAC1 antibody was used for the immunoblot analysis. Treatment with DM for 2 days tended to increase the binding of MyoD and HDAC1 (uppermost panel). However, this would be caused by the increase in MyoD during myoblast differentiation (second and fourth panels). However, with further differentiation after 2 days, the association was gradually decreased, suggesting that HDAC1 dissociates from MyoD by differentiation. (b) HDAC1 associates with Rb in DM. Binding of HDAC1 to Rb was enhanced even though the protein amount of Rb was reduced after treatment with DM for 2 days. (c) MyoD is acetylated during C2C12 differentiation. The acetylation of MyoD was examined by immunoprecipitation assay using anti-acetyl lysine antibody, and subsequently an anti-MyoD antibody was used for the immunoblot analysis.
Acetylation of MyoD is important for its transcriptional activity, and the acetylation status of MyoD is balanced through the opposing actions of two enzymes, pCAF (or p300) and HDAC1, that function as an acetyltransferase and deacetylase, respectively.17 Similarly, in our experimental model, acetylation of MyoD was increased in DM at day 4 compared with GM or DM at day 1 (Figure 4c).
HDAC1 sumoylation is required for its dissociation from MyoD during myogenesis
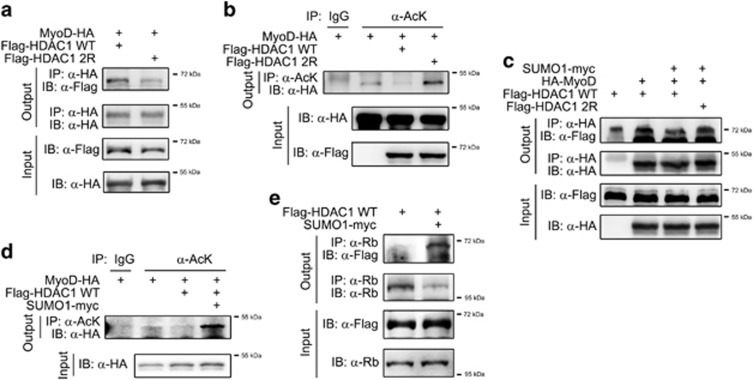
We examined whether the binding of MyoD to HDAC1 2R is altered compared with HDAC1 WT. Compared with the HDAC1 input control detected using an anti-Flag antibody, substitution of lysines (HDAC1 WT) with arginines (HDAC1 2R) slightly reduced the binding of HDAC1 to MyoD (Figure 5a). We next examined whether sumoylation of HDAC1 affects the acetylation status of MyoD using an immunoprecipitation-based acetylation assay with an anti-acetyl lysine antibody. Transfection of HDAC1 WT decreased the acetylation of MyoD (Figure 5b, second vs third lanes in upper panel), and this reduction was attenuated when HDAC1 2R was transfected (fourth lane). These results suggest that the reduction of the MyoD deacetylation activity of HDAC1 2R (Figure 5b) results from both the reduction of its deacetylase activity (Figure 3e) and its dissociation from MyoD (Figure 5a).
Figure 5.
Histone deacetylase 1 (HDAC1) sumoylation is required for its deacetylase activity of MyoD during myoblast differentiation. (a) The association between HDAC1 and MyoD is slightly reduced in HDAC1 2R. Immunoprecipitation was performed with either HDAC1 WT or HDAC2 2R in 293T cells. The association between HDAC1 2R and MyoD was reduced (first panel). Notably, the input amount of HDAC1 2R is also less than that of HDAC1 WT (third panel). (b) MyoD deacetylation is dependent on HDAC1 sumoylation. Acetylation of MyoD was measured by immunoprecipitation-based analysis by utilizing anti-acetyl lysine antibody (α-AcK) in 293T cells. Transfection of HDAC1 WT decreased the acetylation of MyoD (third lane), suggesting the intact deacetylase activity of HDAC1. HDAC1 2R failed to deacetylate MyoD (fourth lane). (c) Forced sumoylation of HDAC1 caused its dissociation from MyoD. We transfected Flag-HDAC1 WT, MyoD-HA and SUMO1-Flag into 293T cells as indicated and immunoprecipitation assay was performed. Note that forced sumoylation reduced the interaction between MyoD and HDAC1 (second vs third lanes in the uppermost panel). (d) Forced sumoylation enhanced acetylation of MyoD. Transfection of HDAC1 WT induced deacetylation of MyoD (second vs third lanes), whereas co-transfection of small ubiquitin-like modifier 1 (SUMO1) restored the acetylation of MyoD in 293T cells. Acetylation of MyoD was measured by immunoprecipitation. (e) Forced sumoylation enhanced the binding of HDAC1 to Rb in C2C12 cells.
Figure 1a showed that sumoylation of HDAC1 was enhanced during myoblast differentiation. Thus, we examined whether forced sumoylation mimicked HDAC1 under the differentiated conditions by assessing whether forced sumoylation of HDAC1 affects its binding to MyoD. When Flag-SUMO1 was co-transfected, the binding of MyoD to HDAC1 WT was reduced (Figure 5c, second vs third lanes). However, binding of HDAC1 2R to MyoD was not significantly affected (Figure 5c, second vs fourth lanes). Considering that SUMO1 transfection failed to inhibit the interaction between MyoD and HDAC1 2R, the reduction of HDAC1 WT binding to MyoD must be dependent on the K444 and K476 residues of HDAC1 (Figure 5c). We extended this analysis by assessing whether forced sumoylation and subsequent dissociation of HDAC1 from MyoD (Figure 5c) would result in the recovery of the acetylation of MyoD. As expected, HDAC1 WT-induced deacetylation of MyoD (second vs third lanes) was recovered when SUMO1 was transfected (Figure 5d, third vs fourth lanes). Interestingly, forced sumoylation of HDAC1 induced the enhancement of its binding to Rb in C2C12 cells (Figure 5e) and in 293T cells (data not shown). These results suggest that HDAC1 binding to MyoD and the subsequent deacetylation of MyoD are reduced either through forced sumoylation of HDAC1 (Figures 5c and d), which simulates the late phase of differentiation (Figure 1a), or the loss of the sumoylation residue (Figures 5a and b). In addition, sumoylation of HDAC1 induces its dissociation from MyoD and association with Rb (Figure 5e).
HDAC1 sumoylation attenuates MyoD-induced transcriptional activity of MCK and myogenin promoter
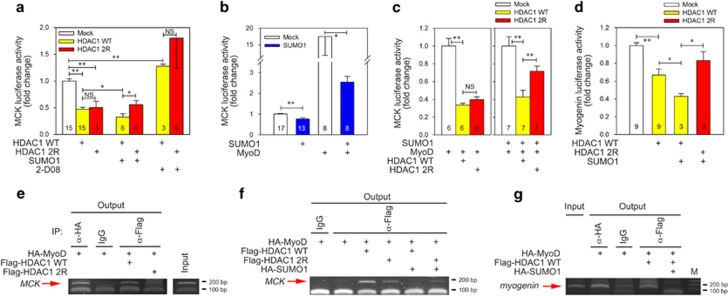
As a key myogenic transcription factor, MyoD induces the transcription of downstream targets, such as MCK and myogenin, through direct association with MyoD binding elements containing an E-box in the promoter.41 We assumed that the HDAC1 sumoylation-dependent alteration of MyoD acetylation and HDAC1 binding to MyoD might result in the regulation of the transcriptional activity of MyoD targets. After transfection of HDAC1 WT, MCK promoter luciferase activity was halved compared with the mock-transfected control (Figure 6a, first vs second columns). HDAC1 2R also decreased MCK luciferase activity, and the magnitude of the decrease was similar to that of WT (second vs third columns), further suggesting that the basal activity of HDAC1 2R is the same as that of WT (Figure 3e, left two columns). Forced sumoylation further decreased MCK promoter activity (Figure 6a, second vs fourth columns), suggesting an enhancement of the deacetylation activity, as shown in the first vs third columns in Figure 3e. Interestingly, this potentiation of reduction was not observed when HDAC1 2R was used (Figure 6a, third vs fifth columns), suggesting that the forced sumoylation-dependent enhancement of the reduction of the MCK promoter is dependent on K444 and K476. The E1/E2 ligase inhibitor, 2-D08, slightly but significantly increased basal MCK promoter activity (Figure 6a, first vs sixth columns).
Figure 6.
Histone deacetylase 1 (HDAC1)-mediated inhibition of MyoD transactivation is inhibited by HDAC1 2R. (a) Forced sumoylation enhances HDAC1 action on luciferase activity of the muscle creatine kinase (MCK) promoter in C2C12 cells. HDAC1 wild type (WT) repressed MCK-luciferase activity (second column). The magnitude of repression of HDAC1 2R (third column) was the same as that of HDAC1 WT. Forced sumoylation by transfection of small ubiquitin-like modifier 1 (SUMO1) further enhanced HDAC1 WT-induced repression (fourth column). However, forced sumoylation failed to enhance the HDAC1 2R-induced repression (fifth column). 2-D08 (25 μM, 24 h)-induced inhibition of sumoylation prevented HDAC1 from its repression of MCK promoter activity. Rather, the MCK promoter activity was increased by HDAC1 WT (sixth column) or HDAC 2R (seventh column). (b) Forced sumoylation repressed MCK promoter activity in C2C12 cells. Forced sumoylation depressed the basal MCK promoter activity (first vs second columns). MyoD significantly increased the MCK promoter activity (third column), but the increase was significantly attenuated by co-transfection of SUMO1 (third vs fourth columns). (c) Sumoylation is required for the full repressive effect of HDAC1 in C2C12 cells. In the absence of SUMO1, HDAC1-induced repression of MCK promoter activity was not affected by 2R (second and third columns in left graph). However, in the presence of forced sumoylation, HDAC1-induced repression was attenuated when HDAC1 2R was used (second and third columns in right graph), suggesting that sumoylation is required for the full repressive activity of HDAC1. (d) Sumoylation further enhanced the HDAC1-mediated repressive action of myogenin promoter in C2C12 cells. Forced sumoylation potentiated HDAC1 WT-induced depression of myogenin promoter (second vs third columns). However, this depression was not observed with HDAC1 2R. (e) Chromatin immunoprecipitation analysis revealed that HDAC1 2R lost the binding of HDAC1 to E-box-containing MyoD-responsive element of MCK promoter. Either anti-MyoD or anti-Flag antibody was utilized for the immunoprecipitation of MyoD-transfected C2C12 cell lysates. Compared with HDAC1 WT, HDAC1 2R lost its binding affinity to MyoD-binding element in MCK promoter region. (f) Chromatin immunoprecipitation analysis showed that forced sumoylation blocked binding of HDAC1 to MCK promoter. Again, binding of HDAC1 2R was less than that of HDAC1 WT (third vs fourth lanes). Forced sumoylation completely blocked the binding of HDAC1 WT to MCK promoter (third vs fifth lanes). (g) Forced sumoylation decreased the binding of HDAC1 WT to MyoD-binding element of myogenin promoter region (fourth vs fifth lanes). M: size marker. Error bars represent s.e.m. NS, not significant, *P<0.05, **P<0.01. Numerals in bar graphs represent the numbers of samples.
In the absence of HDAC1 overexpression, transfection of Flag-SUMO1 decreased basal MCK promoter activity (Figure 6b, first vs second columns). Forced expression of SUMO1 also blunted MyoD-induced transactivation of MCK promoter (Figure 6b, third vs fourth columns). MyoD-induced transactivation of MCK promoter was reduced to 30% after co-transfection of HDAC1 WT, but the magnitude of the reduction was similar to that of HDAC1 2R (Figure 6c, left graph). However, in the presence of forced sumoylation through transfection of Flag-SUMO1, the HDAC1 WT-induced reduction of promoter activity was blunted with HDAC1 2R (right graph). HDAC1 WT-induced reduction of myogenin promoter activity was also enhanced after forced sumoylation (Figure 6d, second vs third columns) that was recovered by HDAC1 2R (fourth column).
Using the chromatin immunoprecipitation assay, we further examined whether HDAC1 sumoylation-dependent regulation of MyoD transcriptional activity is accompanied by binding of the HDAC1/MyoD complex to the MyoD binding element of the MCK promoter. After transfection of either Flag-HDAC1 WT or Flag-HDAC1 2R, an anti-Flag antibody was used to precipitate HDAC1 to assess the association of HDAC1 with the MyoD-binding element of the MCK promoter. Compared with WT, HDAC1 2R lost its binding affinity to the MCK promoter (Figure 6e, third vs fourth lanes). We repeated the chromatin immunoprecipitation assay under forced sumoylation conditions. Similarly, the binding of HDAC1 2R to the MyoD-binding element was reduced compared with that of HDAC1 WT (Figure 6f, third vs fourth lanes). Interestingly, forced sumoylation completely abolished the binding of HDAC1 WT to the MyoD element (third vs fifth lanes). Moreover, the binding of HDAC1 2R to the MyoD element was further blocked by forced sumoylation (fourth vs sixth lanes). However, compared with WT, HDAC1 2R slightly restored its binding to the MyoD element under forced sumoylation (fifth vs sixth lanes). We performed a similar experiment using the myogenin promoter spanning the MyoD-binding element E-box (Figure 6g). Binding of HDAC1 WT to the myogenin promoter was dramatically reduced through forced expression of HA-SUMO1 (fourth vs fifth lanes).
HDAC1 sumoylation is required for HDAC1-mediated inhibition of myocyte differentiation
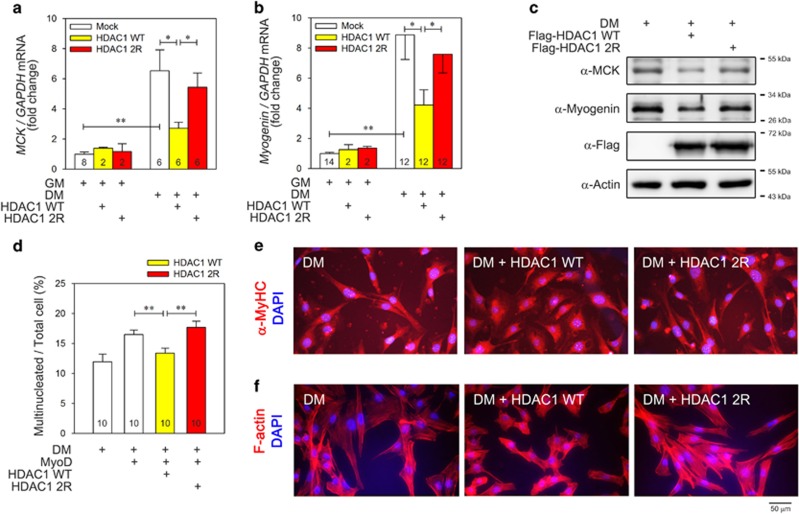
We next examined whether sumoylation of HDAC1 affects the overall differentiation of myoblasts and myotube formation. Switching the culture media from GM to DM for 1 day significantly increased the mRNA amount of both MCK (Figure 7a) and myogenin (Figure 7b) that was dramatically attenuated by transfection of HDAC1 WT. Compared with HDAC1 WT, however, transfection of HDAC1 2R relieved the repression (Figures 7a and b). Similarly, after treatment with DM for 1 day, the protein levels of MCK and myogenin were reduced by transfection of HDAC1 WT that was not observed after transfection of HDAC1 2R (Figure 7c).
Figure 7.
Histone deacetylase 1 (HDAC1)-induced depression of myogenic differentiation is dependent on sumoylation of HDAC1 in C2C12 cells. (a) Repression of muscle creatine kinase (MCK) mRNA is dependent on sumoylation of HDAC1. Switching from growth media (GM) to differentiation media (DM) significantly increased MCK mRNA level (first vs fourth columns). The increase was significantly blocked when HDAC1 wild type (WT) was co-transfected (fourth vs fifth columns). However, the inhibition was not observed when HDAC1 2R was used (fourth to sixth columns). (b) Sumoylation of HDAC1 also affects the repression of myogenin mRNA level. HDAC1 WT significantly blocked the DM-induced increase in myogenin mRNA (fourth vs fifth columns). However, this was relieved when HDAC1 2R was transfected (sixth column). (c) Reductions of protein levels of MCK and myogenin were also blunted when HDAC1 2R was transfected. (d) Transfection of MyoD further increased DM-induced multinucleation of C2C12 cells (first vs second columns). However, this increase was completely blocked when HDAC1 WT was transfected (third column). The inhibition was not observed with HDAC1 2R (fourth column). After the transfection of plasmids as indicated, the C2C12 cells were treated with DM for 4 days. The nuclei were visualized using 4',6-diamidino-2-phenylindole (DAPI) staining, and C2C12 cells containing more than 2 nuclei were counted as multinucleated cells. A total of 10 randomly selected fields per well were evaluated, averaged and calculated as one case. (e) Immunocytochemical analysis was performed with an anti-α-myosin heave chain (α-MyHC) antibody to visualize the cellular shapes. DM (2 days) induced changes of the morphology of C2C12 cells to an elongated or polygonal shape. The α-MyHC was evenly distributed in the entire cellular spaces (left image). Transfection of HDAC1 WT prevented this change and cells formed a spindle shape with fibroblast-like features. α-MyHC was localized in the perinuclear space. HDAC1 2R failed to induce the changes in cell shape. Red color represents α-MyHC, whereas blue indicates nucleus. (f) For further visualization of the C2C12 cell shape changes, phalloidin was utilized to stain F-actin. After treatment with DM for 4 days, the C2C12 cells were fixed and stained with Texas Red-X-phalloidin and DAPI. DM-induced polygonal changes of the C2C12 shape were blocked by the transfection of HDAC1 WT (middle image). These changes were not observed when HDAC1 2R was transfected (right image). *P<0.05, **P<0.01.
Multinucleation is a key marker of sarcomeric muscle differentiation and myotube formation. We thus examined multinucleation after transfection of either HDAC1 WT or 2R. Switching to DM increased the multinucleated cell numbers that were significantly reduced by transfection of HDAC1 WT (Figure 7d, second vs third columns). However, HDAC1 2R failed to inhibit multinucleation of C2C12 cells (third vs fourth columns). Cellular shapes were observed after immunocytochemical analysis using an anti α-MyHC antibody (Figure 7e). DM induced fibroblast-like polygonal shapes of C2C12 cells that was blocked by transfection with HDAC1 WT. However, these changes were not observed when HDAC1 2R was transfected (Figure 7e). Cellular shape was also assessed after staining of F-actin with Texas Red-X-phalloidin (Figure 7f). The HDAC1-induced impairment of differentiation was not observed in HDAC1 2R. These results suggest that sumoylation of HDAC1 is required for HDAC1-mediated inhibition of myotube differentiation.
Discussion
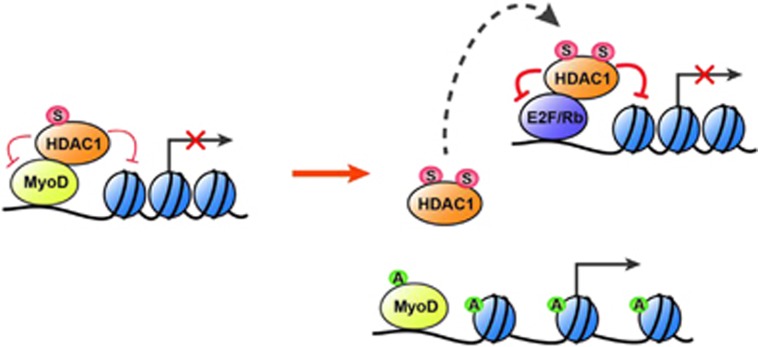
In the present study, we identified a novel sumoylation signaling pathway for HDAC1 in MyoD-mediated myogenesis. Specifically, basal sumoylation of HDAC1 in undifferentiated myoblasts enhances deacetylation of MyoD, whereas a further increase in sumoylation of HDAC1 during differentiation may contribute to the switching of HDAC1 binding from MyoD to Rb (Figure 8).
Figure 8.
The effect of sumoylation of histone deacetylase 1 (HDAC1). (Left) In the quiescent phase of undifferentiated myoblasts, basally sumoylated HDAC1 with basal deacetylase activity binds to MyoD and thereby inhibits the transactivation of MyoD. HDAC1 inhibits acetylation of both MyoD and histones associated with promoters of MyoD target genes. (Right) During myoblast differentiation, enhancement of sumoylation of HDAC1 induces its dissociation from MyoD. This dissociation relieves the repression of MyoD and thereby induces the transactivation of MyoD target genes. A, acetylation; S, sumoylation.
Recent evidence suggests that the activities of transcription factors can be altered or even redirected by posttranslational modifications. Unlike modifications with relatively small chemical moieties, such as acetylation, phosphorylation and methylation, conjugation of a large protein to the lysine residues of a target molecule is involved in both ubiquitination and sumoylation. Polyubiquitination-dependent proteasomal degradation of a target protein is an example of this type of protein modification. Other recent evidence suggests that monoubiquitination and conjugation of ubiquitin-like proteins may also play direct roles in modulating protein function.42 Generally, sumoylation of nuclear receptors is associated with transcriptional repression, particularly via transrepression, and both ligand-operated sumoylation and signal-dependent crosstalk may further contribute to modulate transcriptional regulation of nuclear receptors.43
Notably, sumoylation was initially described as a regulator of intranuclear localization of nuclear receptors and an inducer of the translocation of target proteins into promyelocytic leukemia protein bodies or nuclear speckles.38, 44, 45 Although the present study did not reveal the intranuclear localization of sumoylated HDAC1, the localization issue may also contribute to the regulation of MyoD by sumoylated HDAC1. Indeed, HDAC1 sumoylation might contribute to the dissociation of HDAC1 from MyoD in the relatively late phase of myocyte differentiation, as observed in Figures 4a and 5c.
Two independent groups demonstrated that the sumoylation sites of HDAC1 are K444 and K476, although a consensus has not been reached on the function of sumoylation. For example, Colombo et al.35 did not observe changes in the enzymatic activity of HDAC1 2R compared with WT. In contrast, David et al.36 reported a 60% reduction of activity of the mutant. We also observed that mutation of HDAC1 did not impair the basal activity (Figure 3f, left two columns); interestingly, however, under forced sumoylation conditions through transfection of SUMO1, the activity of HDAC1 2R was significantly reduced compared with WT (Figure 3f, middle two columns), suggesting that the discrepancies between the two groups reflects basal sumoylation conditions. In the present study, we observed that DM induces sumoylation of HDAC1 that may increase HDAC1 activity, and this increase was slightly but significantly attenuated with HDAC1 2R (Figure 3e, right two columns).
Direct binding of MyoD with HDAC1 in association with other transcriptional repressors has been well demonstrated, as described in the Introduction. Indeed, the association of HDAC1 with MyoD causes the deacetylation of histones in the promoter region of MyoD target genes. However, as described by two independent groups,46, 47 MyoD itself can be an acetylation target. Acetylation of MyoD transactivates downstream target myogenic genes and thereby initiates the muscle program. As for acetyltransferases, both pCAF46 and p30048 have been demonstrated, whereas deacetylation is primarily mediated by HDAC1.17 In the present study, we investigated sumoylation of HDAC1 in muscle cells, and the results successfully extended the mechanism of HDAC1/MyoD signaling in myogenesis.
So what is the biological implication of HDAC1 sumoylation and its role in the MyoD pathway during myogenesis? Herein, we showed that differentiation enhanced sumoylation of HDAC1 and further activated its enzymatic activity (Figure 3e). However, sustained HDAC1 activation may not result in persistent inhibition of MyoD; rather, sumoylation of HDAC1 during myoblast differentiation (Figure 1a) may contribute to the dissociation of HDAC1 from MyoD (Figure 4a) following acetylation of MyoD (Figure 4c), resulting in the failure of further inhibition, although HDAC1 is activated by enhanced sumoylation during myogenic differentiation. Indeed, considering that forced sumoylation reduces the binding of HDAC1 WT to MyoD (Figure 5c) and thereby to MyoD-binding elements in the target gene promoters (Figures 6f and g), sumoylation itself might induce dissociation of HDAC1 from MyoD during differentiation. Notably, there might be targets of forced sumoylation other than HDAC1 that indirectly affect the binding of MyoD and HDAC1. However, considering that forced sumoylation failed to block the binding of HDAC1 2R to MyoD (Figure 5c) and HDAC1 2R restored binding to the MyoD element under forced sumoylation (Figure 6f), it is highly plausible that the SUMO-mediated interference of binding of HDAC1 and MyoD is dependent on sumoylation of HDAC1 itself. Thus, HDAC1 sumoylation plays dual roles: (1) inhibition of MyoD transactivation by basally sumoylated HDAC1 in quiescent myoblasts (left drawing in Figure 8) and (2) switching of binding partners of HDAC1 from MyoD to Rb and following derepression of MyoD transactivation during differentiation where further enhancement of sumoylation occurs (right in Figure 8).
We observed that both forced sumoylation of HDAC1 WT (Figure 5c) and HDAC1 2R (Figure 5a) induce its dissociation from MyoD and from the MyoD-binding element in the MCK promoter (Figure 6e). Considering that HDAC1 2R, a sumo-resistant mutant, was utilized to simulate the ‘unsumoylated condition’ of HDAC1, these two observations somewhat contradict each other concerning the ultimate function of sumoylation in the association of HDAC1 with MyoD. However, considering that in the basal quiescent condition without further sumoylation stimuli in GM, HDAC1 WT can interact well with MyoD, it is plausible that the dissociation of HDAC1 2R from MyoD might reflect alterations of the molecular structure. Together with the reduction of intrinsic deacetylation activity, this dissociation of HDAC1 2R may explain its function in the recovery of MyoD acetylation (Figure 5b) and thereby derepression of the myogenic program in DM (Figure 7). If HDAC1 2R has two functions of ‘sumo-dead’ and ‘structural alteration’, then HDAC1 2R might not simulate the ‘unsumoylated’ condition per se that may limit the interpretation of these experimental results.
Although SUMO2 can induce sumoylation of HDAC1 in breast cancer cells,49 SUMO1 predominates in myoblast cells over SUMO2. In the present study, we did not examine a specific E3 ligase for HDAC1 sumoylation during myogenesis. However, further studies are required to determine the E3 ligase that induces sumoylation of HDAC1 in skeletal muscle and to establish the implication of persistent sumoylation of HDAC1, even after dissociation from MyoD during myogenesis. Nonetheless, this study may have implications as the first demonstration of HDAC1 sumoylation-mediated regulation of the MyoD signaling pathway.
Acknowledgments
We thank Jennifer Holmes at Medical Editing Services for language editing and careful reading of the manuscript. This work was financially supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science, ICT & Future Planning (2016R1A4A1009895), by the NRF grant by the Ministry of Education (2016R1A6A3A01010622) and by the NRF grant funded by the Korean government (2015R1A2A1A05001708).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors declare no conflict of interest.
References
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 1997; 88: 97–107. [DOI] [PubMed] [Google Scholar]
- Sarge KD. Analysis of protein sumoylation. Curr Protoc Protein Sci 2016; 83: 14.8.1–8. [DOI] [PubMed] [Google Scholar]
- Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 2013; 82: 357–385. [DOI] [PubMed] [Google Scholar]
- Hay RT. SUMO-specific proteases: a twist in the tail. Trends Cell Biol 2007; 17: 370–376. [DOI] [PubMed] [Google Scholar]
- Puri PL, Sartorelli V. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J Cell Physiol 2000; 185: 155–173. [DOI] [PubMed] [Google Scholar]
- Sabourin LA, Rudnicki MA. The molecular regulation of myogenesis. Clin Genet 2000; 57: 16–25. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 1993; 75: 1351–1359. [DOI] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell 1990; 61: 49–59. [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev 2004; 18: 2627–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloor A, Kotsopoulou E, Hayward P, Champion B, Green A. RFP represses transcriptional activation by bHLH transcription factors. Oncogene 2005; 24: 6729–6736. [DOI] [PubMed] [Google Scholar]
- Mal AK. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J 2006; 25: 3323–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto K, Hamaguchi H, Hashiba T, Nakamura T, Kawamoto T, Sato F et al. Transcriptional repression by the basic helix-loop-helix protein Dec2: multiple mechanisms through E-box elements. Int J Mol Med 2007; 19: 925–932. [PubMed] [Google Scholar]
- Alter J, Bengal E. Stress-induced C/EBP homology protein (CHOP) represses MyoD transcription to delay myoblast differentiation. PLoS ONE 2011; 6: e29498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CC, Liu H, Chen SL, Wang TH, Hsieh CL, Huang Y et al. Epigenetic silencing of myogenic gene program by Myb-binding protein 1a suppresses myogenesis. EMBO J 2012; 31: 1739–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers ML, Celona B, Black BL. NFATc1 controls skeletal muscle fiber type and is a negative regulator of MyoD activity. Cell Rep 2014; 8: 1639–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbac OC, Wolffe AP, Rupp RA. Histone deacetylase activity is required for the induction of the MyoD muscle cell lineage in Xenopus. Biol Chem 2000; 381: 1013–1016. [DOI] [PubMed] [Google Scholar]
- Mal A, Sturniolo M, Schiltz RL, Ghosh MK, Harter ML. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J 2001; 20: 1739–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Iezzi S, Stiegler P, Chen TT, Schiltz RL, Muscat GE et al. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol Cell 2001; 8: 885–897. [DOI] [PubMed] [Google Scholar]
- Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev 2000; 14: 2393–2409. [DOI] [PubMed] [Google Scholar]
- Eom GH, Kook H. Posttranslational modifications of histone deacetylases: implications for cardiovascular diseases. Pharmacol Ther 2014; 143: 168–180. [DOI] [PubMed] [Google Scholar]
- Joung H, Eom GH, Choe N, Lee HM, Ko JH, Kwon DH et al. Ret finger protein mediates Pax7-induced ubiquitination of MyoD in skeletal muscle atrophy. Cell Signal 2014; 26: 2240–2248. [DOI] [PubMed] [Google Scholar]
- Kim J-R, Kee HJ, Kim J-Y, Joung H, Nam K-I, Eom GH et al. Enhancer of polycomb1 acts on serum response factor to regulate skeletal muscle differentiation. J Biol Chem 2009; 284: 16308–16316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee H, Kim J, Joung H, Choe N, Lee S, Eom G et al. Ret finger protein inhibits muscle differentiation by modulating serum response factor and enhancer of polycomb1. Cell Death Differ 2012; 19: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom GH, Cho YK, Ko JH, Shin S, Choe N, Kim Y et al. Casein kinase-2alpha1 induces hypertrophic response by phosphorylation of histone deacetylase 2 S394 and its activation in the heart. Circulation 2011; 123: 2392–2403. [DOI] [PubMed] [Google Scholar]
- Kee HJ, Eom GH, Joung H, Shin S, Kim JR, Cho YK et al. Activation of histone deacetylase 2 by inducible heat shock protein 70 in cardiac hypertrophy. Circ Res 2008; 103: 1259–1269. [DOI] [PubMed] [Google Scholar]
- Zhou J, Olson EN. Dimerization through the helix-loop-helix motif enhances phosphorylation of the transcription activation domains of myogenin. Mol Cell Biol 1994; 14: 6232–6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J 2010; 428: 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Keyser SG, Schneekloth JS Jr. Synthesis of 2',3',4'-trihydroxyflavone (2-D08), an inhibitor of protein sumoylation. Bioorg Med Chem Lett 2014; 24: 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda I, Ito A, Hirai G, Nishimura S, Kawasaki H, Saitoh H et al. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem Biol 2009; 16: 133–140. [DOI] [PubMed] [Google Scholar]
- Alontaga AY, Bobkova E, Chen Y. Biochemical analysis of protein SUMOylation. Curr Protoc Mol Biol 2012; Chapter 10: Unit10.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Kemp GD, Hay RT. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J Biol Chem 1999; 274: 10618–10624. [DOI] [PubMed] [Google Scholar]
- Johnson PR, Hochstrasser M. SUMO-1: ubiquitin gains weight. Trends Cell Biol 1997; 7: 408–413. [DOI] [PubMed] [Google Scholar]
- Okuma T, Honda R, Ichikawa G, Tsumagari N, Yasuda H. In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem Biophys Res Commun 1999; 254: 693–698. [DOI] [PubMed] [Google Scholar]
- Sarge KD, Park-Sarge OK. Detection of proteins sumoylated in vivo and in vitro. Methods Mol Biol 2009; 590: 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo R, Boggio R, Seiser C, Draetta GF, Chiocca S. The adenovirus protein Gam1 interferes with sumoylation of histone deacetylase 1. EMBO Rep 2002; 3: 1062–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G, Neptune MA, DePinho RA. SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J Biol Chem 2002; 277: 23658–23663. [DOI] [PubMed] [Google Scholar]
- Chen WY, Lee WC, Hsu NC, Huang F, Chung BC. SUMO modification of repression domains modulates function of nuclear receptor 5A1 (steroidogenic factor-1). J Biol Chem 2004; 279: 38730–38735. [DOI] [PubMed] [Google Scholar]
- Chalkiadaki A, Talianidis I. SUMO-dependent compartmentalization in promyelocytic leukemia protein nuclear bodies prevents the access of LRH-1 to chromatin. Mol Cell Biol 2005; 25: 5095–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey P, Downes M, Lau P, Harris J, Chen SL, Hamamori Y et al. The nuclear receptor corepressor N-CoR regulates differentiation: N-CoR directly interacts with MyoD. Mol Endocrinol 1999; 13: 1155–1168. [DOI] [PubMed] [Google Scholar]
- Mal A, Harter ML. MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc Natl Acad Sci USA 2003; 100: 1735–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi TM, Walsh K, Schimmel P. Rabbit muscle creatine kinase: genomic cloning, sequencing, and analysis of upstream sequences important for expression in myocytes. Nucleic Acids Res 1991; 19: 3027–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature 2009; 458: 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuter E, Venteclef N. Transcriptional control of metabolic and inflammatory pathways by nuclear receptor SUMOylation. Biochim Biophys Acta 2011; 1812: 909–918. [DOI] [PubMed] [Google Scholar]
- Lee MB, Lebedeva LA, Suzawa M, Wadekar SA, Desclozeaux M, Ingraham HA. The DEAD-box protein DP103 (Ddx20 or Gemin-3) represses orphan nuclear receptor activity via SUMO modification. Mol Cell Biol 2005; 25: 1879–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Hu X, Gupta P, Lin YP, Ha SG, Wei LN. SUMOylation of Tr2 orphan receptor involves Pml and fine-tunes Oct4 expression in stem cells. Nat Struct Mol Biol 2007; 14: 68–75. [DOI] [PubMed] [Google Scholar]
- Sartorelli V, Puri PL, Hamamori Y, Ogryzko V, Chung G, Nakatani Y et al. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell 1999; 4: 725–734. [DOI] [PubMed] [Google Scholar]
- Polesskaya A, Duquet A, Naguibneva I, Weise C, Vervisch A, Bengal E et al. CREB-binding protein/p300 activates MyoD by acetylation. J Biol Chem 2000; 275: 34359–34364. [DOI] [PubMed] [Google Scholar]
- Polesskaya A, Harel-Bellan A. Acetylation of MyoD by p300 requires more than its histone acetyltransferase domain. J Biol Chem 2001; 276: 44502–44503. [DOI] [PubMed] [Google Scholar]
- Citro S, Jaffray E, Hay RT, Seiser C, Chiocca S. A role for paralog-specific sumoylation in histone deacetylase 1 stability. J Mol Cell Biol 2013; 5: 416–427. [DOI] [PubMed] [Google Scholar]