Abstract
An interaction between ribosomal protein S3 (rpS3) and nuclear factor kappa B or macrophage migration inhibitory factor in non-small-cell lung cancer is responsible for radioresistance. However, the role of rpS3 in glioblastoma (GBM) has not been investigated to date. Here we found that in irradiated GBM cells, rpS3 translocated into the nucleus and was subsequently ubiquitinated by ring finger protein 138 (RNF138). Ubiquitin-dependent degradation of rpS3 consequently led to radioresistance in GBM cells. To elucidate the apoptotic role of rpS3, we analyzed the interactome of rpS3 in ΔRNF138 GBM cells. Nuclear rpS3 interacted with DNA damage inducible transcript 3 (DDIT3), leading to DDIT3-induced apoptosis in irradiated ΔRNF138 GBM cells. These results were confirmed using in vivo orthotopic xenograft models and GBM patient tissues. This study aims to clarify the role of RNF138 in GBM cells and demonstrate that rpS3 may be a promising substrate of RNF138 for the induction of GBM radioresistance, indicating RNF138 as a potential target for GBM therapy.
Introduction
Glioblastoma (GBM), also known as glioblastoma multiforme and grade IV astrocytoma, is the most common and aggressive brain tumor.1 GBM carries a poor prognosis, with an ~15-month median survival time. Moreover, the 5-year survival rate following diagnosis in GBM patients is reported to be <5%.2 Because the presence of the blood–brain barrier limits the penetration of most chemotherapeutic drugs into the brain, the standard therapy for GBM is surgical resection followed by radiotherapy with adjuvant administration, such as temozolomide.3 Nevertheless, the overall outcome of GBM therapy has not been satisfactory, with frequent tumor relapse. The poor efficacy of the current therapeutic approaches for GBM is highly associated with the resistance of the tumor cell population based on their molecular and cellular characteristics.4, 5, 6 Overcoming this resistance of GBM to the current therapy is an ongoing challenge. Many researchers, to date, have put forth great efforts into the development of novel approaches to improve the sensitivity of GBM to current therapies and to identify specific factors that contribute to GBM aggressiveness.7
Ribosomal protein S3 (rpS3) is a member of the eukaryotic ribosome 40S subunit, which is responsible for the regulation of ribosome maturation and initiation of translation with the eukaryotic initiation factors elF2 and elF3.8, 9 Independent of ribosomal activities, rpS3 also plays multifunctional roles in DNA repair, apoptosis, survival and radioresistance via interactions with a variety of binding partners.10, 11, 12, 13, 14 RpS3 can be phosphorylated by PKCδ in response to DNA damage, resulting in the translocation of rpS3 to the nucleus and the functional switch of rpS3 from translation to DNA repair.12 In addition, rpS3 is reported to interact with the p65 subunit of nuclear factor kappa B (NF-κB) through the K homology domain (KH domain) of rpS3, which leads to NF-κB-induced transcriptional activation associated with cell survival and epithelial–mesenchymal transition.13, 14, 15 Another study demonstrated that rpS3 could interact with the TNF receptor type 1-associated DEATH domain protein in response to UV radiation, which consequently induces apoptosis through the activation of JNK/stress-activated protein kinase and caspase-3/8.16 Although the precise mechanism underlying the functional switch and regulation of rpS3 remains elusive, an investigation of rpS3-interacting partners might be a promising approach to clarify rpS3 functions.
Ring finger protein 138 (RNF138), also known as NEMO-like kinase-associated ring finger protein, has been characterized as an E3 ubiquitin-ligase that has several functional regions, including the ubiquitin-interacting motif, really interesting new gene (RING) domain, as well as C2HC and C2H2 zinc-binding motifs.17, 18, 19 RNF138 was initially identified as interacting with the NEMO-like kinase, leading to ubiquitination-mediated degradation of TCF/LEF and negative regulation of Wnt signaling.17 RNF138 has been shown to be involved in the regulation of secondary axis formation in the development of embryos and impairment of colonic mucosal regenerative capabilities in Crohn’s disease patients, indicating that RNF138 functions in embryo development, cell differentiation, cell proliferation and cell regeneration.17, 20 Interestingly, recent studies have suggested that RNF138 can be recruited to the regions of DNA double-strand breaks in order to participate in the DNA repair system by homologous recombination.18, 19 Moreover, the downregulation of RNF138 is associated with glioma cell apoptosis, suggesting tumorigenic activity of RNF138.21 Nevertheless, molecular and physiological roles of RNF138 in GBM currently remain unclear.
Herein, we demonstrated that rpS3 knockdown is associated with the induction of radioresistance in GBM cells. Interestingly, RNF138 led to the degradation of nuclear-translocating rpS3 in response to irradiation, consequently inhibiting rpS3-mediated apoptosis. We elucidate the role of RNF138 in GBM and identify rpS3 as a crucial substrate of ubiquitination by RNF138, which underlies the radioresistance of GBM.
Materials and methods
Chemicals, antibodies and reagents
Chemicals, antibodies, and reagents used are described in the Supplementary Materials and Methods.
Cell lines, cell culture and irradiation
Human GBM cell lines, U87MG, A172, U373 and T98G cells, were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA), authenticated and maintained in early passages for no more than 6 months after receipt from ATCC. The cells were grown in RPMI-1640, MEM or DMEM medium supplemented with 10% FBS, 100 U ml−1 penicillin, and 100 mg ml−1 streptomycin at 37 °C in 95% air/5% CO2. The cells were exposed to a single dose of γ-rays using a Gamma Cell 40 Exactor (Nordion International, Inc., Kanata, Ontario, Canada) at a dose of 0.81 Gy min−1. Flasks that contained control cells were placed in an irradiation chamber, but not exposed to radiation.
Western blotting and immunoprecipitation
The protein expression levels following treatments were measured by western blotting and immunohistochemistry (IHC). Detailed procedures are described in Supplementary Materials and Methods.
Cell viability assay
The effects of rpS3 knockdown on cell growth were measured by a cell viability assay using thiazolyl blue tetrazolium bromide solution. Detailed procedures are described in the Supplementary Materials and Methods.
Colony forming assay
The cells were plated at a density of 300 cells per well in six-well dishes. After 24 h, the cells were treated with the indicated siRNAs, were exposed to a specific dose of radiation, and subsequently grown for 14 days. Next, the cells were fixed with 10% methanol and 10% acetic acid, which were then stained with 1% crystal violet. Colonies containing more than 50 cells were identified using densitometry software and scored as survivors.22
Immunofluorescence staining
Immunofluorescence (IF) staining was performed as previously described.23 After the indicated treatments, the cells were fixed with 2% paraformaldehyde for 20 min and permeabilized in 0.5% Triton X-100 for 10 min. The cells and acini were washed three times with PBS and blocked in IF buffer (PBS, 0.1% BSA, 0.2% Triton X-100, and 0.05% Tween-20) containing 10% FBS at 37 °C for 30 min. The cells were stained with a primary antibody against rpS3 overnight at 4 °C, followed by three washes with IF buffer. After probing with DyLight 488-conjugated secondary antibodies (Thermo Scientific, Rockford, IL, USA) and counterstaining with 4′,6-diamidino-2-phenylindole, the slides were mounted with VECTASHIELD Hard-Set Mounting Medium (Vector Laboratories, Burlingame, CA). Fluorescent images were visualized with an Olympus IX71 fluorescence microscope (Olympus Optical Co. Ltd, Tokyo, Japan).
RNF138 gene knockout
We employed the Clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9 (CRISPR/Cas9) system with specific reagents for the RNF138 gene to generate RNF138-knockout U87MG cells. Detailed procedures are described in the Supplementary Materials and Methods.
Interactome analysis
We conducted the interactome analysis of rpS3 to identify the rpS3-interacting proteome in normal and ΔRNF138 U87MG cells. Detailed procedures are described in the Supplementary Materials and Methods.
Bimolecular fluorescence complementation assay
In response to radiation, the interaction between rpS3 and DDIT3 in live ΔRNF138 U87MG cells was monitored by a bimolecular fluorescence complementation (BiFC) assay, as previously described.24 The N-terminal Venus-conjugated EGFR (pBiFC-EGFR-VN) and C-terminal Venus-conjugated EGFR (pBiFC-EGFR-VC) constructs were kindly provided by Dr Chang-Deng Hu (Purdue University, West Lafayette, IN, USA) and Dr Ichi N. Maruyama (Okinawa Institute of Science and Technology Graduate University, Okinawa, Japan). To construct pBiFC-rpS3-VN, an rpS3-encoding DNA fragment was amplified by PCR and then inserted into the KpnI site of pBiFC-EGFR-VN, where the region encoding EGFR had been located. To construct pBiFC-DDIT3-VC, a DNA fragment encoding DDIT3 was amplified and inserted between the SalI and XhoI sites of pBiFC-EGFR-VC, thus eliminating the EGFR DNA fragment. The ΔRNF138 U87MG cells were transiently transfected with pBiFC-rpS3-VN and pBiFC-DDIT3-VC, and fluorescence was measured with an Olympus IX71 fluorescence microscope (Olympus Optical Co. Ltd).
Luciferase reporter gene assay
A luciferase assay was performed to measure the transcriptional activity of DDIT3. Detailed procedures are described in the Supplementary Materials and Methods.
Chromatin immunoprecipitation assay
We conducted a chromatin immunoprecipitation (ChIP) assay to measure the transcriptional activity of DDIT3 on the GADD34 promoter. Detailed procedures are described in the Supplementary Materials and Methods.
Apoptosis assay
Apoptosis was assessed by measuring Caspase 3/7 activity using the Caspase-Glo 3/7 assay kit (Promega, Madison, WI, USA) in accordance with the manufacturer’s protocol.25 In brief, the treated cells (104 cells per ml) in 100 μl of culture medium were transferred to a 96-well microplate. Subsequently, 100 μl of Caspase-Glo 3/7 reagent containing Caspase 3/7 substrate was added to each well. After mixing the contents of the wells gently at 300–500 rpm for 30 s, the plate was incubated at room temperature for 1 h. Luminescence of each sample was measured using a Glomax multi-detection system (Promega).
Apoptosis induction was also assessed by analyzing the cytoplasmic histone-associated DNA fragmentation.26 In brief, the cells were placed in 96-well plates and allowed to attach overnight. The cells were then subjected to transfection, irradiation and/or drug treatment. Cytoplasmic histone-associated DNA fragmentation was monitored using a cell death detection kit (Roche Applied Science, Indianapolis, IN, USA) in accordance with the manufacturer’s instructions.
Orthotopic xenograft mouse model
Six-week-old male BALB/c athymic nude mice purchased from Central Lab Animals Inc., Seoul, Republic of Korea were used in a xenograft mouse model. U87MG cells were harvested through trypsinization and suspended at a density of 1 × 105 cells per μl in serum-free media. Then, 5 × 105 cells were stereotactically injected into the brain of mice (n=5 for each group, weight: 18±2 g). Twelve days after the injection date, the brains of the mice were irradiated with 2 Gy for 5 days (2 Gy × 5 times). Mice with 20% body weight loss were killed by CO2 asphyxiation. Brain tissues were embedded in a frozen block, and the sections were prepared by Leica Biosystems CM3050 S Research Cryostat (Leica, Deerfield, IL, USA) for immunostaining. The survival of mice is presented in Kaplan–Meier survival curves. P-values were calculated by Mantel–Cox testing. The GBM cells that were irradiated were done so as single cells using Gamma Cell 40 Exactor (Nordion International, Inc.).
Hematoxylin and eosin staining and IHC
The mice were killed after the experimental treatments. Soon thereafter, 4-mm-thick sections were cut, fixed in formalin and embedded in paraffin before being dewaxed and rehydrated as previously described.27 For hematoxylin and eosin (H&E) staining, the sections were incubated with H&E. For IHC, the sections were incubated in a 3% hydrogen peroxide/methanol solution. Antigen retrieval was performed by incubation with 0.25% pepsin (Dako, Carpinteria, CA, USA). The sections were then blocked in a blocking solution (Dako) and incubated overnight with the indicated primary antibody (ABcam, Cambridge, UK) at 4 °C. After washing with TBST, the sections were incubated with a polymer-HRP-conjugated secondary antibody (Dako). Following hematoxylin staining and bluing (1% HCl in 70% ethanol), a 3,3′-diaminobenzidine Substrate Chromogen System (Dako) was used to detect antibody binding, and the stained sections were examined with an Olympus IX71 inverted microscope (Olympus Optical Co. Ltd.).
Patient tissue samples
Specimens and data used in this study, which included frozen tissue samples from six GBM patients and paired normal adjacent tissue samples, were provided by the Biobank of Pusan National University Hospital (Busan, Republic of Korea and a member of the Korea Biobank Network). Informed consent was obtained from all individuals participants in this study. The use of these archival tissues for gene expression analysis, western blotting and immunohistochemistry was approved by the Ethics Committee of Pusan National University.
Real-time quantitative RT-PCR
The levels of gene expression and miRNAs were measured by real-time quantitative RT-PCR (qRT-PCR) as previously described.28 Total RNA from the cells with the indicated treatments (transfection of gene constructs or siRNAs, reagents or irradiation) or tissue samples from GBM patients was isolated using TRIzol (15596-026, Invitrogen, Carlsbad, CA, USA). To obtain cDNA for the analysis of mRNA levels, the isolated RNA (100 ng μl−1 final concentration) was reverse-transcribed by the ImProm-II reverse transcription system (A3800, Promega, Madison, WI, USA) in accordance with the manufacturer’s protocol. The cycle parameters for the RT reaction were 25 °C for 5 min, 42 °C for 60 min, 85 °C for 5 min, and a 4 °C hold. The aliquots of a master mix containing all of the reaction components with the primers (Supplementary Table S1) were dispensed into a real-time PCR plate (Applied Biosystems, Foster City, CA, USA). All of the PCR reagents were from the SYBR Green core reagent kit (Applied Biosystems). The expression of all genes evaluated was measured in triplicate in the reaction plate. The qRT-PCR was performed with the Applied Biosystems-7900 HT qRT-PCR instrument. PCR was performed for 15 s at 95 °C and 1 min at 60 °C for 40 cycles, followed by thermal denaturation. The expression of each gene relative to GAPDH mRNA was determined using the 2−ΔΔCT method.29 To simplify the presentation of the data, the relative expression values were multiplied by 102.
Statistical analysis
All numeric data are presented as the means±standard deviation (s.d.) of at least three independent experiments. The experimental results were analyzed by one-way ANOVA for ranked data, followed by the Tukey’s honestly significant difference test. Prism 5 software (GraphPad Software, SanDiego, CA, USA) was used to perform all statistical analyses. A P-value<0.05 was considered statistically significant.
Results
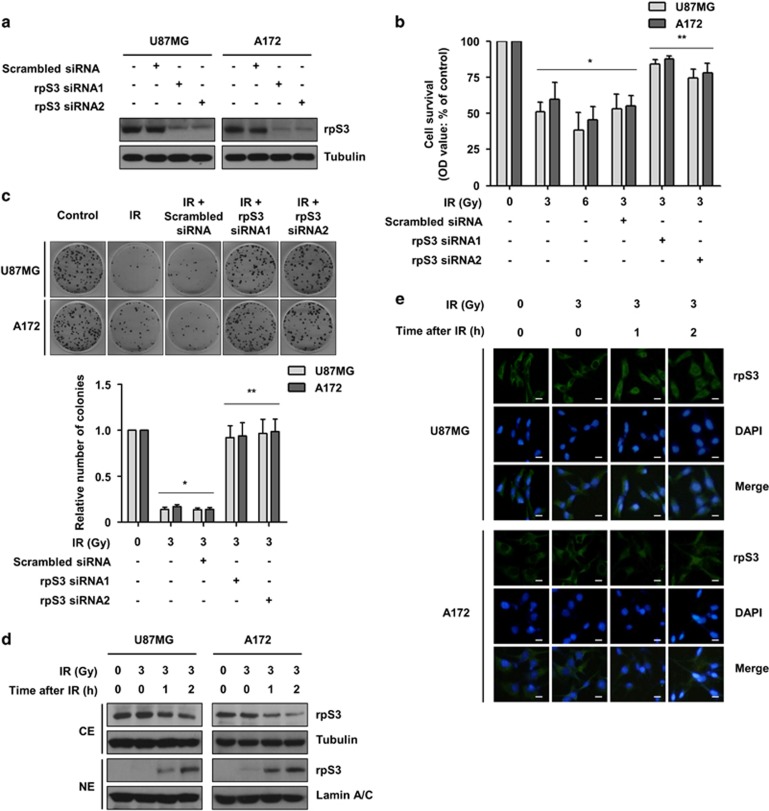
RpS3 knockdown increased resistance of GBM cells to ionizing radiation
Our previous study demonstrated that rpS3 is involved in radioresistance via its interaction with NF-κB or macrophage migration inhibitory factor in non-small-cell lung cancer (NSCLC).13, 14 To investigate whether rpS3 increases the radioresistance of GBM cells, cell survival and proliferation were measured. After the inhibition of rpS3 expression by treatment with rpS3-specific siRNAs (Figure 1a), the short-term effects of rpS3 knockdown on cell survival after irradiation were evaluated with a cell viability assay in the GBM U87MG and A172 cell lines. Unlike the function in NSCLC cells, rpS3 knockdown significantly increased the growth of irradiated GBM cells, suggesting that rpS3 sensitizes GBM cells to irradiation (Figure 1b). To evaluate the long-term effect of rpS3 knockdown on cell proliferation after irradiation, a colony formation assay was conducted. Colony formation in the presence of radiation was greatly increased by rpS3 knockdown in both U87MG and A172 cells (Figure 1c). The possible radioresistant effects of rpS3 knockdown were further verified in additional GBM cell lines, U373 and T98G cells (Supplementary Figure S1a and b). Next, radiation-dependent subcellular localization of rpS3 in GBM cells was investigated since rpS3 is known to be translocated by irradiation in NSCLC cells.13, 14 As shown in Figure 1d, irradiation caused a marked reduction of cytosolic rpS3 and a concomitant increase of nuclear rpS3 in both U87MG and A172 cells. Irradiation led to an increase in nuclear staining of rpS3, with fluorescence intensity evident by 2 h and maintained until 4 h (Figure 1e and Supplementary Figure S1c). Taken together, nuclear localization of rpS3 may be radiation-dependent and rpS3 knockdown may increase the resistance of GBM cells to radiation.
Figure 1.
Knockdown of rpS3 enhances the resistance of GBM cells to irradiation. (a) SiRNA knockdown efficiency of rpS3 in U87MG and A172 cells was analyzed by western blotting. (b) Short-term effects of rpS3 knockdown on cell growth in both U87MG and A172 cells following irradiation (IR) were assessed via MTT assay. The data are represented as the mean±s.e.m. (n=3); *P<0.05 compared with non-irradiated cells, **P<0.05 compared with cells treated with irradiation-alone or cells treated with irradiation and scrambled siRNA. (c) Long-term effects of rpS3 knockdown on cell growth in both U87MG and A172 cells following radiation exposure were assessed via colony forming assay. The data are represented as the mean±s.e.m. (n=3); *P<0.05 compared with non-irradiated cells, **P<0.05 compared with cells treated with irradiation-alone or cells treated with irradiation and scrambled siRNA. (d) Radiation-induced translocation of rpS3 from the cytoplasm to the nucleus was assessed by western blotting after cytoplasmic or nuclear fractionation (CE, cytoplasmic extract; NE, nuclear extract). Tubulin and Lamin A/C were used as markers for the CE and NE, respectively. (e) Radiation-induced translocation of rpS3 from the cytoplasm to the nucleus was assessed by IF staining. Scale bars, 25 μm.
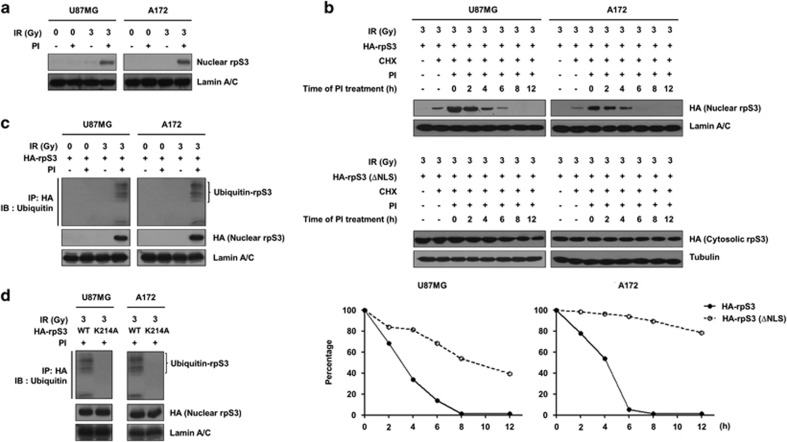
Protein stability of rpS3 in irradiated GBM cells is negatively regulated in a ubiquitination-dependent manner
In microscopy analysis, the fluorescence intensity of nuclear rpS3 was diminished after 6 h of irradiation (Supplementary Figure S1c). To determine whether the reduction of rpS3 was due to proteasomal degradation, the protein levels of nuclear rpS3 were monitored after treatment with proteasomal inhibitors. As shown in Figure 2a, irradiation caused a marked decrease in the levels of nuclear rpS3 in both U87MG and A172 cells, while treatment with proteasomal inhibitors restored the nuclear rpS3 protein levels. These results were additionally verified in U373 and T89G cells (Supplementary Figure S2a). Indeed, the estimated half-life of nuclear rpS3 (4 h) was significantly shorter than that of NLS (nuclear localization signal, amino acids 7–10)-deleted cytoplasmic rpS3 (Figure 2b). These results indicated that radiation-induced nuclear localization and proteasomal degradation of rpS3 in the nucleus caused a reduction in nuclear rpS3 protein stability. In line with these findings, proteasome inhibitors caused a pronounced accumulation of poly-ubiquitinated nuclear rpS3 (Figure 2c and Supplementary Figure S2b). RpS3 consists of 243 amino acids in humans and contains 20 Lys residues (Supplementary Figure S2c). To determine which among these Lys are ubiquitination sites, based on the three-dimensional structure of rpS3 (Protein Data Bank ID: 3KC4) and prediction results from the ubiquitination prediction database (PhosphoSitePlus, UbPred, iUbiq-Lys and UbiProber), we mutated six Lys residues (K10, K90, K151, K202, K214 and K227) to Ala and measured radiation-induced poly-ubiquitination (Supplementary Figure S2d). This analysis revealed a ubiquitin modification of rpS3 at a single site, K214, which is conserved among several other species (Figure 2d and Supplementary Figure S2e). According to these data, we proposed that radiation-induced rpS3 ubiquitination in the nucleus is directly associated with the destabilization of rpS3 protein in GBM cells.
Figure 2.
Poly-ubiquitination is required for degradation of rpS3 in irradiated GBM cells. (a) The effects of proteasome inhibition on the protein stability of nuclear rpS3 were measured by western blotting (proteasome inhibitors, PI; a mix of 5 mmol l−1 MG115 and 5 mmol l−1 MG262, 24 h incubation). (b) The half-life of rpS3 WT or mut (ΔNLS) in response to radiation with cycloheximide (CHX; 50 mmol l−1) in the presence of PI was estimated by western blotting. The cells were treated with PI at different time points after irradiation (0–12 h). The protein levels of HA-rpS3 WT or ΔNLS at each time point measured in b were analyzed by densitometric analysis. The expression level of HA-rpS3 WT or ΔNLS at time point 0 (lane 3) was set at 100%. (c) The accumulation of poly-ubiquitinated nuclear rpS3 in response to radiation was measured. U87MG and A172 cells expressing HA-rpS3 were treated with or without PI. HA-rpS3 protein was immunoprecipitated with an anti-HA antibody, and western blotting was performed with an anti-ubiquitin antibody. (d) The effects of the Lys214 residue on the poly-ubiquitination of rpS3 were measured. U87MG and A172 cells expressing rpS3 WT or mut (K214A) were treated with PI. HA-rpS3 protein was immunoprecipitated with an anti-HA antibody, and western blotting was performed with an anti-ubiquitin antibody.
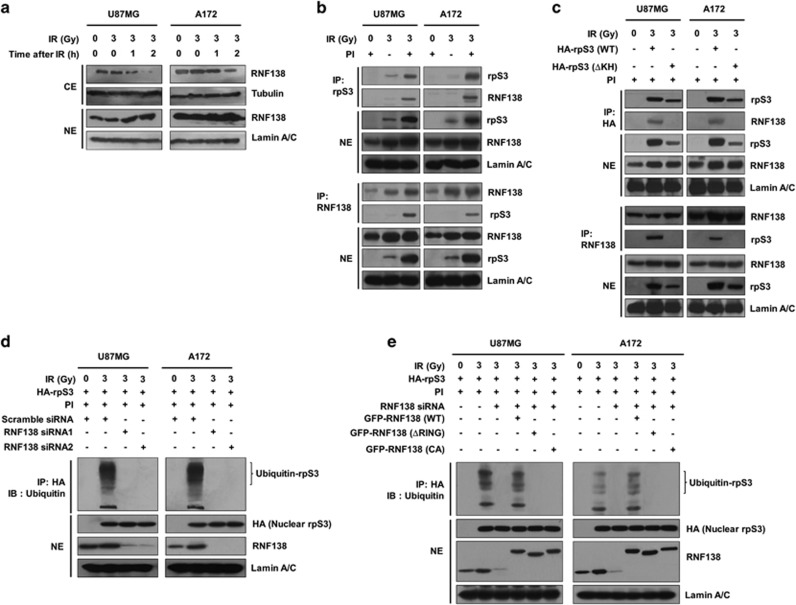
RNF138 ubiquitinates rpS3 in a radiation-stimulated manner
To explore how rpS3 is ubiquitinated in irradiated GMB cells, we revisited our previous yeast two-hybrid results for the identification of potential rpS3-interacting E3 ligases and focused on RNF138 among these rpS3-associated proteins.13, 14, 30 RNF138 has a ubiquitin-interacting motif for ubiquitin binding that is commonly found in damage-responsive nuclear proteins (Supplementary Figure S3a).31 In addition, RNF138 might interact with the Qki protein, which has a KH domain found in rpS3 (Supplementary Figure S2c).32 Furthermore, in comparison with normal brain tissue, RNF138 was reported to be mislocalized from the ER to the nucleus in glioma.33 Thus, we set out to investigate the physiological relevance of these findings. Although RNF138 was present in both the cytosol and nucleus, we were able to detect a radiation-dependent translocation of RNF138 from the cytosol to the nucleus in GBM cells (Figure 3a and Supplementary Figure S3b). We assessed the interaction between RNF138 and rpS3 in GBM cells. As shown in Figure 3b and Supplementary Figure S3c, rpS3 specifically interacted with RNF138 in radiation-treated U87MG, A172, U373 and T98G cells. Under the same conditions, reciprocally, RNF138 also specifically interacted with rpS3. In addition, KH domain-deleted rpS3 did not interact with RNF138 (Figure 3c). These results indicated that RNF138 interacted with radiation-activated nuclear rpS3 through the KH domain of rpS3 in GBM cells. Next, we examined the RNF138-dependent rpS3 ubiquitination following irradiation in GBM cells. We found that rpS3 was ubiquitinated in an RNF138-dependent manner (Figure 3d and Supplementary Figure S3d). After siRNA-mediated depletion of RNF138, we found a markedly reduced rpS3 ubiquitination following irradiation. Moreover, a catalytically inactive mutant (C18A/C54A, CA) and RING domain deletion of RNF138 abrogated the ability of RNF138 to ubiquitinate rpS3 (Figure 3e). These results are consistent with RNF138 regulating the ubiquitination of rpS3. Taken together, we can infer that RNF138 ubiquitinates rpS3 in a radiation-stimulated manner.
Figure 3.
RpS3 is ubiquitinated by RNF138 in response to radiation. (a) Radiation-induced translocation of RNF138 from the cytoplasm to the nucleus was assessed by western blotting after cytoplasmic or nuclear fractionation. (b) RpS3–RNF138 interaction in U87MG and A172 cells after treatment with PI and irradiation was monitored by reciprocal IP assay. After preparation of NE, lysates were subjected to IP assay with an anti-rpS3 antibody or anti-RNF138 antibody followed by western blotting. (c) The effects of rpS3 WT or mut (ΔKH) on the interaction with RNF138 were measured by reciprocal IP assay. (d) The effects of RNF138 knockdown on the ubiquitination of rpS3 were measured. (e) The compensatory effects of RNF138 WT or mut (ΔRING or C18A/C54A) overexpression on the effects of RNF138 knockdown were determined.
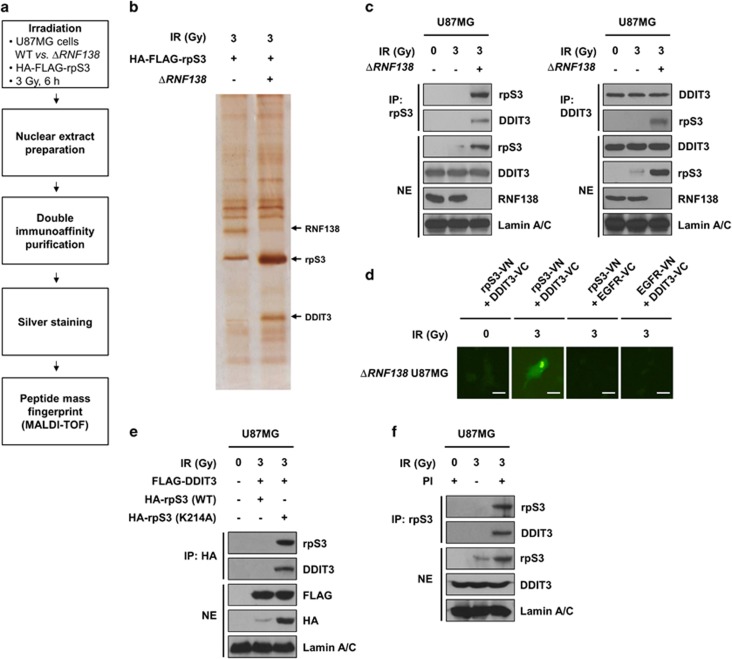
RpS3 ubiquitination is required for downregulation of radiation-induced DDIT3 signaling in GBM
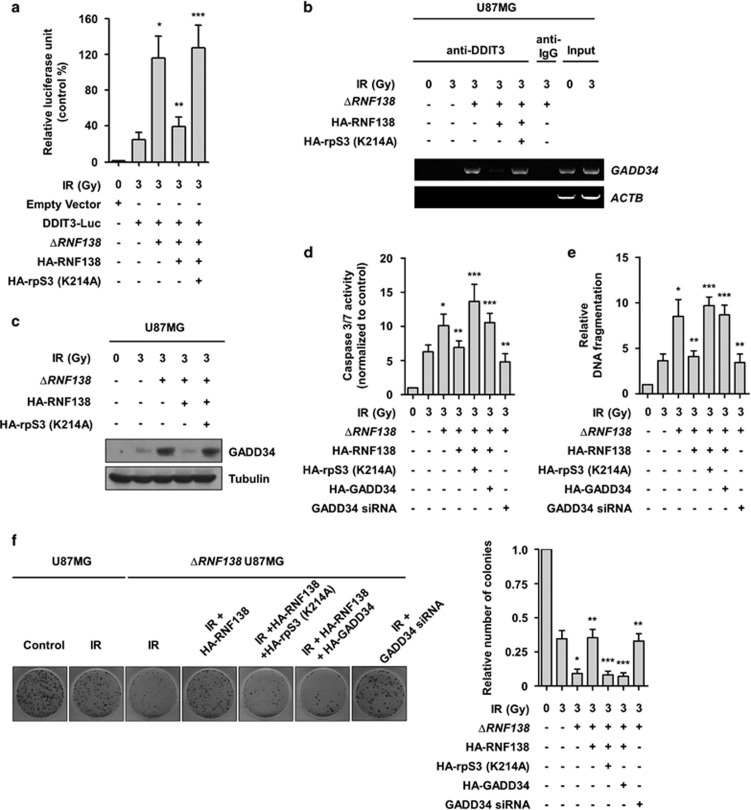
We observed that RNF138-mediated rpS3 ubiquitination is highly associated with radioresistance in GBM cells, indicating that RNF138 knockdown could be a promising strategy for improving the radiotherapeutic efficacy of GBM treatment. To further investigate the effects of RNF138 knockdown on nuclear rpS3 in response to radiation, we established ΔRNF138 GBM cells using the CRISPR/Cas9 system and confirmed the knockout efficacy at the mRNA and protein levels of RNF138 (Supplementary Figure S4a and b). Based on previous studies reporting that rpS3 could mediate its associated functions by protein–protein interactions,13, 14, 15, 34 we focused on the interactors of rpS3 responsible for radiation-mediated apoptotic signaling. We further performed an interactome analysis of rpS3 to identify a rpS3-interacting proteome in the irradiated GBM cells to elucidate the molecular functions of rpS3 in the nucleus. The strategy used to identify the nuclear proteins associated with nuclear rpS3 after ectopic overexpression of rpS3 WT in normal U87MG or ΔRNF138 U87MG cells is presented in Figure 4a. Nuclear extracts were prepared from normal U87MG or ΔRNF138 U87MG cells treated with radiation, and soluble nuclear protein complexes were separated to reduce sample complexity. The complexes containing rpS3 were purified using a double immunoaffinity purification, and the identity of rpS3 interactors was determined by repeated rounds of peptide mass fingerprinting.35 Several radiation-dependent rpS3-interacting proteins were identified with high confidence. On the basis of the information from published databases of physical and functional interactions, we focused on DNA damage inducible transcript 3 (DDIT3) (Figure 4b), which is a key regulator of ER stress-induced cell death.36, 37, 38 We subsequently confirmed the interaction between rpS3 and DDIT3 via reciprocal IP assays (Figure 4c). We observed the radiation-dependent interaction between rpS3 and DDIT3 in live ΔRNF138 U87MG cells by BiFC assay (Figure 4d). Unlike rpS3 WT, rpS3 ub-mut (K214A) interacted with DDIT3 after radiation exposure (Figure 4e), indicating that rpS3 WT might be degraded in a ubiquitination-dependent manner and rpS3 ub-mut might not undergo degradation and therefore still interact with DDIT3. It was confirmed that rpS3 interacted with DDIT3 in the nucleus after inhibition of proteasome activity in a radiation-dependent manner (Figure 4f). According to these results, it is possible for rpS3 to interact with DDIT3 in irradiated GBM cells, and this interaction can be negatively regulated by RNF138-mediated rpS3 ubiquitination.
Figure 4.
RpS3 ubiquitination negatively affects radiation-induced DDIT3 signaling in GBM cells. (a) Schematic presentation of the strategy used to identify the rpS3 nuclear interactome in U87MG cells. To generate ΔRNF138 U87MG cells, the CRISPR/Cas9 system was employed. (b) After irradiation, double-immuno-purified rpS3 complexes in normal U87MG or ΔRNF138 U87MG cells were separated by PAGE and visualized by silver staining. The silver-stained gel was analyzed by mass spectrometry (PMF). It was shown that three bands were particularly increased by irradiation. The bands indicated by arrowheads correspond to RNF138, rpS3, and DDIT3, respectively. (c) RpS3–DDIT3 interaction in normal U87MG and ΔRNF138 U87MG cells in response to irradiation was measured by reciprocal IP assay. (d) A BiFC assay was performed to evaluate the interaction of rpS3-DDIT3 in live cells. The ΔRNF138 U87MG cells were transiently transfected with pBiFC-rpS3-VN, pBiFC-DDIT3-VC, pBiFC-EGFR-VN and/or pBiFC-EGFR-VC. Fluorescence indicative of rpS3-DDIT3 binding was measured in irradiated ΔRNF138 U87MG cells. Scale bars, 25 μm. (e) The effects of rpS3 WT or mut (K214A) in response to irradiation on the interaction with DDIT3 were measured. (f) The effects of rpS3 degradation on the interaction with DDIT3 were determined.
Downregulation of DDIT3-mediated apoptosis confers radioresistance in GBM cells
A previous report has shown that DDIT3 functions as a transcription factor in promoting apoptosis under stressful conditions by expressing pro-apoptotic target genes, including GADD34.39, 40 GADD34 is one of the major DDIT3 effectors that positively correlates with apoptosis under conditions of DNA damage and cellular stress.40 To explore the potential effect of radiation-mediated rpS3 signaling on the functions of DDIT3, we next examined the influence of rpS3 on the ability of DDIT3 to increase the promoter activity of GADD34. The results from a luciferase assay demonstrated that DDIT3 showed an increased effect on the promoter activity of the GADD34 gene with radiation exposure in ΔRNF138 U87MG cells (Figure 5a). Moreover, the increase in luciferase activity in ΔRNF138 U87MG cells was reduced by transient transfection of RNF183 WT constructs, but this effect was reversed by a further overexpression of rpS3 ub-mut protein. To further verify the effects of rpS3 on transcriptional activation of DDIT3, we performed ChIP assays by focusing on DDIT3 binding sites within the GADD34 promoter (Supplementary Table S2). Compared with the normal U87MG cells, ΔRNF138 U87MG cells had to a significant increase in the recruitment of DDIT3 to the promoter regions of the GADD34 gene in response to irradiation (Figure 5b). The binding of DDIT3 to the GADD34 promoter was markedly reduced by the overexpression of RNF138 WT, while this effect was rescued by further overexpression of rpS3 ub-mut. Concomitantly, the expression level of GADD34 was increased in ΔRNF138 U87MG cells (Figure 5c). The overexpression of rpS3 ub-mut in ΔRNF138 U87MG cells with recovery of RNF138 WT also increased the expression of GADD34 protein. These results demonstrated a significant reduction in DDIT3 transcriptional activity and DDIT3-target gene expression in RNF138-positive GBM cells as a response to irradiation. This could be caused by a decrease in the DNA binding property of DDIT3 through radiation-induced rpS3 ubiquitination and degradation. To further determine whether the downregulation of DDIT3 signaling and its target gene expression had functional involvement with respect to radioresistance in GBM cells, an apoptosis assay was conducted. It was observed that radiation-induced caspase 3/7 activation and consequent cytoplasmic histone-associated DNA fragmentation in GBM cells were further increased by RNF138 knockdown or transient transfection of rpS3 ub-mut and GADD34 (Figure 5d and e and Supplementary Figure S5). The effects of caspase 3/7 activation and consequent DNA fragmentation observed in ΔRNF138 GBM cells were further decreased by GADD34 knockdown. In addition, we confirmed that RNF138 knockout or overexpression of rpS3 ub-mut and GADD34 induced lower colony-forming ability in the irradiated GBM cells (Figure 5f). The effects of colony formation in the irradiated ΔRNF138 GBM cells were further restored by GADD34 knockdown. These results indicate that RNF138-mediated rpS3 ubiquitination, inactivation of DDIT3, and subsequent downregulation of GADD34 in response to the irradiation could inhibit the activation of apoptotic signaling leading to the radioresistance in GBM cells.
Figure 5.
RNF138 is responsible for downregulation of DDIT3-mediated apoptosis leading to radioresistance in GBM cells. (a) The effects of RNF138 WT or rpS3 mut (K214A) overexpression on the transcriptional activities of DDIT3 in normal U87MG and ΔRNF138 U87MG cells were compared by luciferase reporter gene assay. The data are represented as the mean±s.e.m. (n=3); *P<0.05 compared to normal U87MG cells treated with irradiation-alone, **P<0.05 compared with ΔRNF138 U87MG cells treated with irradiation-alone, ***P<0.05 compared with ΔRNF138 U87MG cells treated with RNF138 WT overexpression and irradiation. (b) Binding of DDIT3 to the promoter of GADD34 in normal U87MG and ΔRNF138 U87MG cells was measured by ChIP analysis. (c) The expression levels of GADD34 protein in normal U87MG and ΔRNF138 U87MG cells were measured by western blotting. (d, e) Functional involvement of RNF138, rpS3, DDIT3 and GADD34 in radiation-induced apoptosis was analyzed by caspase assay (d) and cytoplasmic histone-associated DNA fragmentation assay (e). The data are represented as the mean±s.e.m. (n=3); *P<0.05 compared with normal U87MG cells treated with irradiation-alone, **P<0.05 compared with ΔRNF138 U87MG cells treated with irradiation-alone, ***P<0.05 compared with ΔRNF138 U87MG cells treated with RNF138 WT overexpression and irradiation. (f) Functional involvement of RNF138, rpS3, DDIT3 and GADD34 in normal U87MG and ΔRNF138 U87MG cells following radiation exposure was assessed by colony forming assay. The data are represented as the mean±s.e.m. (n=3); *P<0.05 compared with normal U87MG cells treated with irradiation-alone, **P<0.05 compared with ΔRNF138 U87MG cells treated with irradiation-alone, ***P<0.05 compared with ΔRNF138 U87MG cells treated with RNF138 WT overexpression and irradiation.
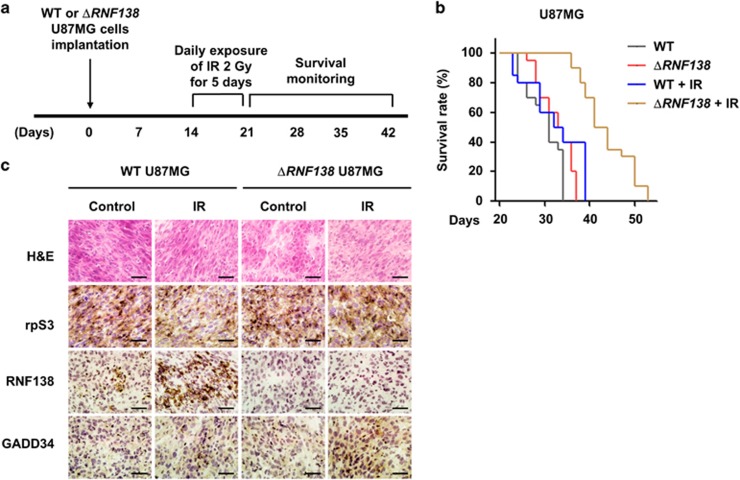
RNF138 knockdown combined with irradiation impeded tumor growth in GBM orthotopic xenograft tumor models
The above in vitro data collectively suggest that RNF138 targeting can be an effective anti-GBM therapy, especially in combination with irradiation. To assess this issue in a pre-clinical animal model, we developed an in vivo tumor model (Figure 6a). Normal U87MG or ΔRNF138 U87MG cells were injected into the brains of immune-deficient nude mice. Once tumors were formed, fractionated cranial irradiation was applied to these mice. Animal survival, tumor histology and immunostaining analyses were evaluated in these mice. Normal U87MG cell-transplanted mice treated with irradiation exhibited a slightly longer median survival (by 8%) compared with those mice that did not undergo radiation (33.5 vs 31 days; P-value=0.040) (Figure 6b and Table 1). In the non-irradiated condition, ΔRNF138 U87MG cell-transplanted mice also showed a slightly longer median survival by 6% compared with their normal U87MG cell-transplanted counterparts (33 vs 31 days; P-value=0.016). Importantly, ΔRNF138 U87MG cell-transplanted mice treated with irradiation showed a significantly prolonged median survival (42.5 days; P<0.0001). We further analyzed rpS3, RNF138 and GADD34 expression levels in brain tissues obtained from the xenograft mice (Figure 6c). The group with transplantation of ΔRNF138 U87MG cells and treatment with irradiation exhibited higher levels of GADD34 compared with the other groups. Together, these in vivo results strongly support the efficacy of RNF138 knockdown combined with irradiation for anti-GBM therapy.
Figure 6.
Combination treatment of RNF138 knockdown and irradiation reduced tumor growth in GBM orthotopic xenograft mice models. (a) An experimental protocol to determine whether RNF138 knockdown increases in vivo radiosensitization in GBM orthotopic xenograft mice models. (b) Kaplan–Meier survival curves of orthotopic tumor-bearing mice (n=5 for each group) after implantation of normal U87MG or ΔRNF138 U87MG cells and radiation exposure. (c) Representative H&E staining images and IHC images for rpS3, RNF138 and GADD34 on tumor sections. Scale bars, 25 μm.
Table 1. Summary of survival and statistical significance in orthotopic tumor-bearing mice.
| Group | Median survival (days) | ISRa (%) | P-value |
|||
|---|---|---|---|---|---|---|
| Wild type | Wild type+IR | ΔRNF138 | ΔRNF138+IR | |||
| Wild type | 31 | 0.040 | 0.016 | <0.0001 | ||
| Wild type+IR | 33.5 | 8 | 0.040 | 0.103 | <0.0001 | |
| ΔRNF138 | 33 | 6 | 0.016 | 0.103 | <0.0001 | |
| ΔRNF138+IR | 42.5 | 35 | <0.0001 | <0.0001 | <0.0001 | |
ISR, increasing survival rate.
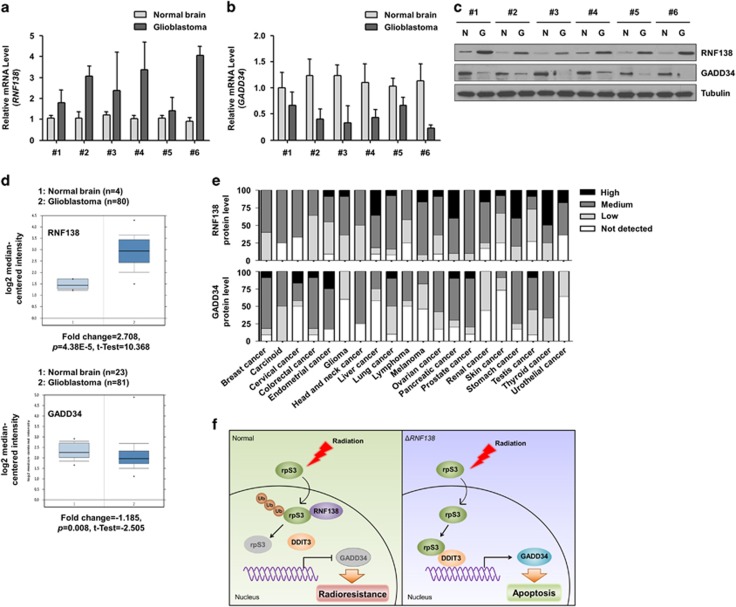
Negative correlation between RNF138 and GADD34 expression in GBM patients
We next analyzed the expression of RNF138 and GADD34 in six sets of GBM patient samples and normal brain tissues (Table 2). As seen in Figure 7a and b, RNF138 mRNA was highly expressed in the GBM samples, but was negligible in normal brain tissues. Conversely, GADD34 mRNA expression was negligible in brain tumor samples, but highly expressed in normal counterparts. The differential expression levels of RNF138 and GADD34 proteins in these tumor sets were confirmed by western blotting (Figure 7c). Upon examination of the database on Oncomine (www.oncomine.org), we found several datasets indicating that RNF138 expression is significantly upregulated and GADD34 expression is slightly downregulated in brain tumors compared with corresponding normal tissues (Figure 7d).41, 42 Moreover, GADD34 showed considerably lower levels of protein expression, while RNF138 was expressed moderately (or highly) in gliomas compared with other cancer types in accordance with the database from the Human Protein Atlas (www.proteinatlas.org) (Figure 7e). The Betastasis database (www.betastasis.com) showed that RNF138-positive (high expression) patients demonstrated a poorer prognosis compared to RNF138-negative (low expression) patients (Supplementary Figure S6). These in vivo data substantiated our in vitro studies regarding the importance of RNF138 as a prognostic marker for GBM malignancy.
Table 2. Patient characteristics.
| Characteristics | No. of patients (n=6) |
|---|---|
| Sex | |
| Male | 3 |
| Female | 3 |
| Age (year) | |
| Median | 60.5 |
| Range | 28–77 |
| Diagnosis | |
| Glioblastoma (primary) | 6 |
Figure 7.
RNF138 expression shows a negative correlation with GADD34 expression in GBM patients. (a, b) The expression levels of RNF138 (a) and GADD34 (b) mRNA in tumor tissues of GBM patients were quantified by real-time qRT-PCR. (c) The expression levels of RNF138 and GADD34 protein in tumor tissues of GBM patients were determined by western blotting. (d) Data sets obtained from Oncomine demonstrated that glioblastoma expressed higher RNF138 mRNA levels (fold change=2.708, p=4.38E-5, t-test=10.368) but lower GADD34 mRNA levels (fold change=−1.185, p=0.008, t-test=−2.505) than normal brain tissues. (e) Data sets obtained from Protein Atlas demonstrated the protein levels of RNF138 were moderately expressed but those of GADD34 were rarely expressed in glioma. (f) Schematic diagram illustrating that RNF138-mediated rpS3 ubiquitination is responsible for radioresistance in GBM cells through down-regulation of rpS3/DDIT3-mediated apoptosis.
Discussion
In this study, we investigated the contributions and the clinical relevance of rpS3 and RNF138 in the context of GBM radioresistance. The current study provides evidence suggesting that RNF138-mediated rpS3 degradation allows GBM to resist radiation-induced cell death via mechanisms involving DDIT3 and GADD34 (Figure 7f). To the best of our knowledge, we report for the first time that RNF138 binds to rpS3 in the nucleus for nuclear rpS3 ubiquitination and degradation. Our findings demonstrate that nuclear rpS3 is accumulated through the depletion of RNF138 (via the CRISPR/Cas9 system) in response to radiation exposure leading to apoptosis via interaction with DDIT3 and subsequent expression of the pro-apoptotic protein GADD34. Moreover, the RNF138 protein levels were highly expressed in GBM patients, and its levels were negatively correlated with the GADD34 protein levels. Taken together, our study suggests that RNF138 can be a potent target for the activation of rpS3/DDIT3-mediated radiosensitizing signaling in GBM cells.
We previously reported that rpS3 promotes cell survival in response to irradiation through interactions with NF-κB or macrophage migration inhibitory factor, leading to radioresistance in NSCLC cells.13, 14 However, the current study demonstrated that rpS3 plays a pro-apoptotic role via its interaction with DDIT3 in GBM cells undergoing radiation exposure. This discrepancy can be partially explained by multi-functional proteins in differential cellular contexts. For instance, ERK might be involved in a PAI-1/AKT axis for pro-survival signaling leading to the radioresistance in NSCLC.25 However, ERK could interact with death-associated protein kinase 1 to promote p53-dependent apoptosis under radiation exposure.24 GBM cells might suffer from oxidative stress induced by aerobic glycolysis associated with tumor aggressiveness,4, 43 and this oxidative stress could affect several cellular compartments resulting in ER stress and DNA damage responses, including DDIT3-mediated apoptosis as an ER stress-responsive signaling.39, 40 In the GBM cellular context, rpS3 could translocate to the nucleus in response to irradiation, enhancing the transcriptional activity of DDIT3 to promote the induction of apoptotic signaling against the radiation response. However, this does not mean that rpS3 might only be involved in the induction of cell death in GBM. We could not rule out the possibility of other functions of rpS3, as a pro-survival factor that can interact with the NF-κB subunit, that may lead to cell survival and radioresistance.
We observed that RNF138 mainly resides in the nucleus, although it also translocated to the nucleus in response to irradiation (Figure 3). RNF138 consequently inhibits rpS3/DDIT3-dependent apoptotic signaling, leading to the radioresistance in GBM cells. It has been reported that mislocalization of many proteins occurs in glioma due to intracellular contexts formed by oxidative stress or the accumulation of misfolded proteins.33 One of those proteins, RNF138, was mislocalized from the ER to the nucleus. Moreover, a recently reported study demonstrated that RNF138 is a major ubiquitin E3 ligase playing a role in the DNA damage response in the nucleus.44 After laser microirradiation, RNF138 was activated by ATM-induced phosphorylation that resulted in recruitment of RNF138 to the DNA damage site and the interaction between RNF138 and RAD51D in order to mediate a homologous recombination repair pathway. Once in the nucleus, RNF138, as an E3 ligase, might have the opportunity to interact with its targets. In our study, we found that rpS3 localizing to the nucleus in response to irradiation could interact with RNF138 and be degraded in a ubiquitin-dependent manner. Based on previously reported studies and our results, we hypothesized that RNF138 is involved in an important signaling axis for cellular protection against severe ER stress and DNA damage induced by radiotherapy through participation in the DNA repair system and inhibition of apoptotic signaling, which consequently leads to radiotherapy resistance in GBM.
The molecular mechanism underlying the acquisition of tumor aggressiveness and radioresistance in GBM remains elusive. The current study suggests that RNF138 might be a critical factor responsible for conferring the resistance to radiotherapy and tumor malignancy. For the first time, we provide evidence that RNF138 promotes the ubiquitin-dependent degradation of rpS3 and subsequently results in the inhibition of rpS3/DDIT3-mediated apoptosis signaling in GBM cells. We propose that RNF138 might be a promising prognostic indicator in GBM and that targeting RNF138 in combination with chemotherapy could enhance the radiotherapeutic efficacy of GBM treatment.
Acknowledgments
This work was supported by the Radiation Technology R&D Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning (2017M2A2A7A01019304), the Ministry of Education (2016R1D1A1B03932822 to WK and 2016R1D1A1B03931405 to HY) and the Korea Institute of Energy Technology Evaluation and Planning (KETEP) and the Ministry of Trade, Industry & Energy (MOTIE) of the Republic of Korea (20131610101840). The biospecimens and data used for this study were provided by the Biobank of Pusan National University Hospital, a member of the Korea Biobank Network.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors declare no conflict of interest.
Supplementary Material
References
- Nehoff H, Parayath NN, McConnell MJ, Taurin S, Greish K. A combination of tyrosine kinase inhibitors, crizotinib and dasatinib for the treatment of glioblastoma multiforme. Oncotarget 2015; 6: 37948–37964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 2007; 21: 2683–2710. [DOI] [PubMed] [Google Scholar]
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 2009; 10: 459–466. [DOI] [PubMed] [Google Scholar]
- Wolf A, Agnihotri S, Guha A. Targeting metabolic remodeling in glioblastoma multiforme. Oncotarget 2010; 1: 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR et al. The somatic genomic landscape of glioblastoma. Cell 2013; 155: 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eramo A, Ricci-Vitiani L, Zeuner A, Pallini R, Lotti F, Sette G et al. Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ 2006; 13: 1238–1241. [DOI] [PubMed] [Google Scholar]
- Chakravarti A, Palanichamy K. Overcoming therapeutic resistance in malignant gliomas: current practices and future directions. Cancer Treat Res 2008; 139: 173–189. [PubMed] [Google Scholar]
- Westermann P, Heumann W, Bommer UA, Bielka H, Nygard O, Hultin T. Crosslinking of initiation factor eIF-2 to proteins of the small subunit of rat liver ribosomes. FEBS Lett 1979; 97: 101–104. [DOI] [PubMed] [Google Scholar]
- Tolan DR, Hershey JW, Traut RT. Crosslinking of eukaryotic initiation factor eIF3 to the 40S ribosomal subunit from rabbit reticulocytes. Biochimie 1983; 65: 427–436. [DOI] [PubMed] [Google Scholar]
- Graifer D, Malygin A, Zharkov DO, Karpova G. Eukaryotic ribosomal protein S3: a constituent of translational machinery and an extraribosomal player in various cellular processes. Biochimie 2014; 99: 8–18. [DOI] [PubMed] [Google Scholar]
- Lee SB, Kwon IS, Park J, Lee KH, Ahn Y, Lee C et al. Ribosomal protein S3, a new substrate of Akt, serves as a signal mediator between neuronal apoptosis and DNA repair. J Biol Chem 2010; 285: 29457–29468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Kim HD, Kim J. PKCdelta-dependent functional switch of rpS3 between translation and DNA repair. Biochim Biophys Acta 2009; 1793: 395–405. [DOI] [PubMed] [Google Scholar]
- Youn H, Son B, Kim W, Jun SY, Lee JS, Lee JM et al. Dissociation of MIF-rpS3 complex and sequential NF-kappaB activation is involved in IR-induced metastatic conversion of NSCLC. J Cell Biochem 2015; 116: 2504–2516. [DOI] [PubMed] [Google Scholar]
- Yang HJ, Youn H, Seong KM, Jin YW, Kim J, Youn B. Phosphorylation of ribosomal protein S3 and antiapoptotic TRAF2 protein mediates radioresistance in non-small cell lung cancer cells. J Biol Chem 2013; 288: 2965–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L et al. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell 2007; 131: 927–939. [DOI] [PubMed] [Google Scholar]
- Jang CY, Kim HD, Kim J. Ribosomal protein S3 interacts with TRADD to induce apoptosis through caspase dependent JNK activation. Biochem Biophys Res Commun 2012; 421: 474–478. [DOI] [PubMed] [Google Scholar]
- Yamada M, Ohnishi J, Ohkawara B, Iemura S, Satoh K, Hyodo-Miura J et al. NARF, an nemo-like kinase (NLK)-associated ring finger protein regulates the ubiquitylation and degradation of T cell factor/lymphoid enhancer factor (TCF/LEF). J Biol Chem 2006; 281: 20749–20760. [DOI] [PubMed] [Google Scholar]
- Ismail IH, Gagne JP, Genois MM, Strickfaden H, McDonald D, Xu Z et al. The RNF138 E3 ligase displaces Ku to promote DNA end resection and regulate DNA repair pathway choice. Nat Cell Biol 2015; 17: 1446–1457. [DOI] [PubMed] [Google Scholar]
- Schmidt CK, Galanty Y, Sczaniecka-Clift M, Coates J, Jhujh S, Demir M et al. Systematic E2 screening reveals a UBE2D-RNF138-CtIP axis promoting DNA repair. Nat Cell Biol 2015; 17: 1458–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen OH, Bjerrum JT, Csillag C, Nielsen FC, Olsen J. Influence of smoking on colonic gene expression profile in Crohn's disease. PLoS ONE 2009; 4: e6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YX, Chen SS, Wu TF, Ding DD, Chen XH, Chen JM et al. A novel gene RNF138 expressed in human gliomas and its function in the glioma cell line U251. Anal Cell Pathol 2012; 35: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Youn H, Kang C, Youn B. Inflammation-induced radioresistance is mediated by ROS-dependent inactivation of protein phosphatase 1 in non-small cell lung cancer cells. Apoptosis 2015; 20: 1242–1252. [DOI] [PubMed] [Google Scholar]
- Son B, Jun SY, Seo H, Youn H, Yang HJ, Kim W et al. Inhibitory effect of traditional oriental medicine-derived monoamine oxidase B inhibitor on radioresistance of non-small cell lung cancer. Sci Rep 2016; 6: 21986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T, Youn H, Son B, Kim D, Seong KM, Park S et al. DANGER is involved in high glucose-induced radioresistance through inhibiting DAPK-mediated anoikis in non-small cell lung cancer. Oncotarget 2016; 7: 7193–7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Kim W, Kwon T, Youn H, Kim JS, Youn B. Plasminogen activator inhibitor-1 enhances radioresistance and aggressiveness of non-small cell lung cancer cells. Oncotarget 2016; 7: 23961–23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Youn H, Kwon T, Kang J, Kim E, Son B et al. PIM1 kinase inhibitors induce radiosensitization in non-small cell lung cancer cells. Pharmacol Res 2013; 70: 90–101. [DOI] [PubMed] [Google Scholar]
- Kim W, Kim E, Yang HJ, Kwon T, Han S, Lee S et al. Inhibition of hedgehog signalling attenuates UVB-induced skin photoageing. Exp Dermatol 2015; 24: 611–617. [DOI] [PubMed] [Google Scholar]
- Kim W, Kim E, Lee S, Kim D, Chun J, Park KH et al. TFAP2C-mediated upregulation of TGFBR1 promotes lung tumorigenesis and epithelial-mesenchymal transition. Exp Mol Med 2016; 48: e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Jang CY, Shin HS, Kim HD, Kim JW, Choi SY, Kim J. Ribosomal protein S3 is stabilized by sumoylation. Biochem Biophys Res Commun 2011; 414: 523–527. [DOI] [PubMed] [Google Scholar]
- Giannini AL, Gao Y, Bijlmakers MJ. T-cell regulator RNF125/TRAC-1 belongs to a novel family of ubiquitin ligases with zinc fingers and a ubiquitin-binding domain. Biochem J 2008; 410: 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 2006; 125: 801–814. [DOI] [PubMed] [Google Scholar]
- Lee K, Byun K, Hong W, Chuang HY, Pack CG, Bayarsaikhan E et al. Proteome-wide discovery of mislocated proteins in cancer. Genome Res 2013; 23: 1283–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan F, Weaver A, Gao X, Bern M, Hardwidge PR, Lenardo MJ. IKKbeta phosphorylation regulates RPS3 nuclear translocation and NF-kappaB function during infection with Escherichia coli strain O157:H7. Nat Immunol 2011; 12: 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol 2003; 370: 430–444. [DOI] [PubMed] [Google Scholar]
- Liu X, Yue P, Zhou Z, Khuri FR, Sun SY. Death receptor regulation and celecoxib-induced apoptosis in human lung cancer cells. J Natl Cancer Inst 2004; 96: 1769–1780. [DOI] [PubMed] [Google Scholar]
- Xie M, Sun M, Zhu YN, Xia R, Liu YW, Ding J et al. Long noncoding RNA HOXA-AS2 promotes gastric cancer proliferation by epigenetically silencing P21/PLK3/DDIT3 expression. Oncotarget 2015; 6: 33587–33601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iurlaro R, Munoz-Pinedo C. Cell death induced by endoplasmic reticulum stress. FEBS J 2016; 283: 2640–2652. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 2004; 18: 3066–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Qiu Y, Yang L, Peng L, Xia Z, Hou LN et al. Desipramine induces apoptosis in rat glioma cells via endoplasmic reticulum stress-dependent CHOP pathway. J Neurooncol 2011; 101: 41–48. [DOI] [PubMed] [Google Scholar]
- Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF et al. Stem cell-related ‘self-renewal’ signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol 2008; 26: 3015–3024. [DOI] [PubMed] [Google Scholar]
- Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 2006; 9: 287–300. [DOI] [PubMed] [Google Scholar]
- Rinaldi M, Caffo M, Minutoli L, Marini H, Abbritti RV, Squadrito F et al. ROS and brain gliomas: an overview of potential and innovative therapeutic strategies. Int J Mol Sci 2016; 17: pii: E984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Liang J, Lu Y, Xu L, Miao S, Lu LY et al. Ubiquitylation of Rad51d mediated by E3 ligase Rnf138 promotes the homologous recombination repair pathway. PLoS ONE 2016; 11: e0155476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.