Dear Editor,
Hematopoietic stem and progenitor cells (HSPCs) have been documented to be specified from hemogenic endothelial (HE) cells in the ventral wall of the dorsal aorta (DA) through the endothelial-to-hematopoietic transition (EHT) during mouse embryogenesis1. N6-methyl-adenosine (m6A) is the most prevalent mRNA modification in eukaryotes. Although the function of m6A modification in cell fate determination of embryonic stem cells has been recently reported2, the physiological role and the underlying mechanism of m6A modification in definitive hematopoiesis during mouse embryogenesis have not been reported yet. Due to the early lethality of mutant mice with conventional knockout of methyltransferase like 3 (Mettl3)2, one of the most important m6A methyltransferase catalytic subunits identified so far3, here we utilized Vec-Cre mice crossed with Mettl3fl/fl mice to deplete the expression of Mettl3 specifically in endothelial cells of mouse aorta-gonad-mesonephros (AGM) region.
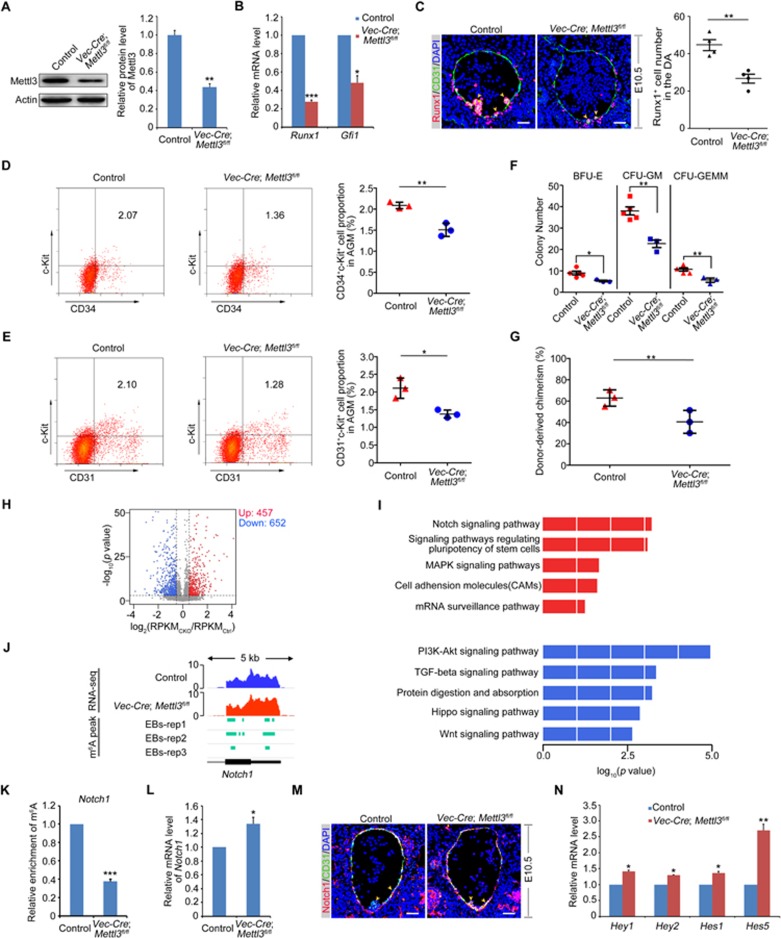
First, the knockout efficiency of Mettl3 in endothelial cells of Vec-Cre; Mettl3fl/fl AGM was validated by multiple approaches. The expression of Mettl3 in the whole AGM was significantly decreased at both protein and mRNA levels (Figure 1A; Supplementary information, Figure S1A). The obvious decrease of Mettl3 in endothelial cells of AGM was also confirmed by immunofluorescence (Supplementary information, Figure S1B). Then the developmental hematopoietic phenotype was examined. Interestingly, the expression of Runx1 and Gfi1, both specifically expressed in the intra-aortic hematopoietic cluster and essential for HSPC development4,5, was decreased in E10.5 Vec-Cre; Mettl3fl/fl AGM (Figure 1B). This is consistent with the reduced number of Runx1+ cells and hematopoietic clusters by immunofluorescence (Figure 1C). In addition, the proportion of HSPCs (CD34+c-Kit+) in the E10.5 Vec-Cre; Mettl3fl/fl AGM was apparently attenuated as compared to that in the control (Figure 1D). To further testify whether the hematopoietic phenotype was due to endothelial cell-specific depletion of Mettl3, Vav-Cre; Mettl3fl/fl embryo, in which the expression of Mettl3 was specifically deleted in hematopoietic cells, was used as a control. Immunofluorescence results showed that there were no obvious changes in the Runx1+ cell number and hematopoietic cluster formation in the E10.5 Vav-Cre; Mettl3fl/fl AGM (Supplementary information, Figure S1C). Moreover, flow cytometry analysis also indicated that the proportion of HSPCs (CD34+c-Kit+) in the Vav-Cre; Mettl3fl/fl AGM was similar to that in the control (Supplementary information, Figure S1D). Meanwhile, we also examined the percentage of pre-HSCs, the maturation of which is required for the formation of definitive HSCs6. Intriguingly, the proportion of pre-HSCs (CD144+CD45+c-Kit+) was dramatically reduced in the E10.5 Vec-Cre; Mettl3fl/fl AGM but was unaltered in the E10.5 Vav-Cre; Mettl3fl/fl AGM as compared with that in controls (Supplementary information, Figure S1E and S1F). Since the earliest HSPCs are derived from HE cells in the AGM region, we then examined the development of HE cells. As expected, the proportion of HE (CD31+c-Kit+) cells was decreased in the E10.5 Vec-Cre; Mettl3fl/fl AGM, but not in the E10.5 Vav-Cre; Mettl3fl/fl AGM and controls (Figure 1E; Supplementary information, Figure S1G). Finally, to determine whether endothelial-specific loss of Mettl3 affects the function of HSPC, we performed the colony-forming unit culture (CFU-C) assay as well as transplantation assay. As shown in Figure 1F and 1G, the colony formation ability and the short-term reconstitution capability of HSPCs in the E10.5 Vec-Cre; Mettl3fl/fl AGM were diminished, suggesting that the function of HSPC was also impaired. Nevertheless, HSPCs functioned normally in the E10.5 Vav-Cre; Mettl3fl/fl AGM (Supplementary information, Figure S1H and S1I). Taken together, these data demonstrate that Mettl3 in endothelial cells of AGM plays a pivotal role in HSPC development during mouse definitive hematopoiesis.
Figure 1.
Endothelial-specific deletion of Mettl3 impairs definitive hematopoiesis via Notch signaling during mouse embryogenesis. (A) Expression of Mettl3 in the E10.5 control and Vec-Cre; Mettl3fl/fl AGM detected by western blotting. The right panel is the quantification of the western blotting results. (B) qPCR analysis of mRNA levels of Runx1 and Gfi1 in the E10.5 control and Vec-Cre; Mettl3fl/fl AGM. (C) Immunofluorescence on the sections of the control and Vec-Cre; Mettl3fl/fl AGM at E10.5 with anti-Runx1 and anti-CD31 antibodies. Yellow arrowheads mark hematopoietic clusters. The right panel is the quantification of Runx1 positive cells in the DA. (D, E) Flow cytometry analysis of the percentages of HSPCs (CD34+c-Kit+) and HE (CD31+c-Kit+) cells in the E10.5 AGM of Vec-Cre; Mettl3fl/fl embryos compared with control. (F) CFU-C assays to detect the colony-forming ability of HSPC in the control and Vec-Cre; Mettl3fl/fl embryos. (G) Donor-derived chimerism in peripheral blood (PB) of recipients 4 weeks post transplantation (n = 3 for each group). (H) Volcano plot showing differentially expressed genes upon Mettl3 deficiency. Genes with significantly increased and decreased expression levels were highlighted using red and blue. (I) Representative Kyoto Encyclopedia of Genes and Genomes (KEGG) of upregulated (top) or downregulated (bottom) genes in the signaling pathway upon Mettl3 deficiency. (J) Integrative Genomics Viewer (IGV) tracks displaying RNA-seq reads distribution in the E10.5 control (blue) and Vec-Cre; Mettl3fl/fl (red) AGM and m6A peaks (green) (m6A peak data were downloaded from GSE619952) at the Notch1 last exon. (K) m6A-RIP-qPCR assay to show the enrichment of m6A on Notch1 mRNA in the AGM of Vec-Cre; Mettl3fl/fl embryo at E10.5 compared with control. (L) The mRNA level of Notch1 in the E10.5 control and Vec-Cre; Mettl3fl/fl AGM by qPCR. (M) Expression of Notch1 in the E10.5 control and Vec-Cre; Mettl3fl/fl AGM by immunofluorescence assay. (N) Expression of Notch signaling target genes Hey1, Hey2, Hes1 and Hes5 in the E10.5 control and Vec-Cre; Mettl3fl/fl AGM. Student's t-test: *P < 0.05; **P < 0.01; ***P < 0.001. Scale bar, 10 μm. The statistical results are presented as mean ± SEM. BFU-E, burst-forming unit-erythroid; CFU-GEMM, CFU-granulocyte, erythroid, macrophage, megakaryocyte; CFU-GM, CFU-granulocyte, macrophage. CKO, conditional knockout (Vec-Cre; Mettl3fl/fl); Ctrl, control.
Our recent data showed that Mettl3-mediated m6A modification on notch1a mRNA could repress the arterial-endothelial Notch activity to promote HSPC specification in zebrafish7. Furthermore, previous studies in zebrafish and mouse showed that the inhibition of Notch signaling in HE cells facilitated EHT and enhanced the generation of HSPCs8,9. To determine whether Notch signaling pathway is regulated by Mettl3 during mouse definitive hematopoiesis, we performed RNA-seq using the E10.5 control and Vec-Cre; Mettl3fl/fl AGM. In total, 457 upregulated and 652 downregulated genes were identified in the Vec-Cre; Mettl3fl/fl AGM (Figure 1H). As expected, among the upregulated ones, genes that encode components of Notch signaling pathway were highly enriched (Figure 1I). The gene-set enrichment analysis (GSEA) also revealed a marked upregulation of genes in Notch signaling pathway (Supplementary information, Figure S2A). Furthermore, the majority of mRNA transcripts of these genes involved in Notch signaling pathway contained m6A modifications (Supplementary information, Figure S2B), suggesting that m6A might play an important regulatory role in Notch signaling activity. Among these m6A-modified genes, we found that Hes5, Dtx4, Dll3, Dll1, Mfng and Nocth1 were remarkably upregulated upon Mettl3 deficiency (Supplementary information, Figure S2C). Similar to the m6A methylation of notch1a mRNA in zebrafish7, we also identified conserved m6A peak near the stop codon of Notch1 mRNA (Figure 1J). In addition, m6A-RIP-qPCR assay showed that the enrichment of m6A on Notch1 mRNA was notably decreased in the E10.5 Vec-Cre; Mettl3fl/fl AGM (Figure 1K). Subsequently, the increased expression of Notch1 was observed at mRNA level (Figure 1L). Consistently, immunofluorescence results further showed that in wild-type embryos, Notch1 was only enriched in the endothelial cells but not in the emerging hematopoietic clusters, whereas the increased expression of Notch1 was observed in both endothelial cells and poorly formed hematopoietic clusters in the E10.5 Vec-Cre; Mettl3fl/fl AGM (Figure 1M). A list of Notch signaling downstream targets including Hey1, Hey2, Hes1 and Hes5 were significantly increased in the E10.5 Vec-Cre; Mettl3fl/fl AGM detected by qPCR (Figure 1N). In addition, both the mRNA and protein levels of Dll4 were increased in the AGM of Vec-Cre; Mettl3fl/fl embryo (Supplementary information, Figure S2D-S2F). Dll4 is not only a downstream target gene of Notch signaling, but also a well-established arterial marker. The increased expression of Dll4 and Notch1 in the AGM of Vec-Cre; Mettl3fl/fl embryo indicated that Notch signaling was over-activated and the enhanced arterial-endothelial identity inhibited the transition from HE cells to HSPCs. Taken together, endothelial-specific Mettl3 facilitates m6A methylation on Notch1 mRNA to inhibit Notch activity in HE cells, thereby promoting HSPC generation through EHT.
In summary, we utilized Cre/LoxP system to generate the endothelial-specific knockout mice of Mettl3, and identified the indispensable function of Mettl3-mediated m6A modification in mouse definitive hematopoiesis. Mechanistically, endothelial expression of Mettl3 can methylate Notch1 mRNA and then lead to the repression of Notch activity during EHT. Interestingly, bioinformatics analysis showed that m6A peak near the stop codon of Notch1 mRNA can be recognized by Ythdf2 (Supplementary information, Figure S2G), suggesting that Ythdf2-mediated Notch1 mRNA decay is responsible for the observed HSPC phenotype10. These results are consistent with our recent findings in zebrafish7, supporting an evolutionally conserved function of m6A in HSPC specification in mammals. In addition, three recent studies have demonstrated that mRNA m6A modification mediated by Mettl3/Mettl14 plays essential roles during spermatogenesis in mice11,12,13, further confirming a general regulatory role of Mettl3-mediated m6A modification in multiple tissues.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31425016 and 81530004), the Ministry of Science and Technology of China (2016YFA0100500) and the Strategic Priority Research Program of the Chinese Academy of Sciences, China (XDA01010110).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
Supplementary Methods, Table, and Figures
References
- Boisset JC, van Cappellen W, Andrieu-Soler C, et al. Nature 2010; 464:116–120. [DOI] [PubMed]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. Science 2015; 347:1002–1006. [DOI] [PubMed]
- Ping XL, Sun BF, Wang L, et al. Cell Res 2014; 24:177–189. [DOI] [PMC free article] [PubMed]
- Chen MJ, Yokomizo T, Zeigler BM, et al. Nature 2009; 457:887–891. [DOI] [PMC free article] [PubMed]
- Thambyrajah R, Mazan M, Patel R, et al. Nat Cell Biol 2016; 18:21–32. [DOI] [PubMed]
- Taoudi S, Gonneau C, Moore K, et al. Cell Stem Cell 2008; 3:99–108. [DOI] [PubMed]
- Zhang C, Chen Y, Sun B, et al. Nature 2017; 549:273–276. [DOI] [PubMed]
- Zhang P, He Q, Chen D, et al. Cell Res 2015; 25:1093–1107. [DOI] [PMC free article] [PubMed]
- Lizama CO, Hawkins JS, Schmitt CE, et al. Nat Commun 2015; 6:7739. [DOI] [PMC free article] [PubMed]
- Wang X, Lu Z, Gomez A, et al. Nature 2014; 505:117–120. [DOI] [PMC free article] [PubMed]
- Hsu PJ, Zhu Y, Ma H, et al. Cell Res 2017; 27:1115–1127. [DOI] [PMC free article] [PubMed]
- Lin Z, Hsu PJ, Xing X, et al. Cell Res 2017; 27:1216–1230. [DOI] [PMC free article] [PubMed]
- Xu K, Yang Y, Feng GH, et al. Cell Res 2017; 27:1100–1114. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods, Table, and Figures