Dear Editor,
N6-methyladenosine (m6A) is the most abundant mRNA chemical modification, and is modulated by m6A 'writers', 'erasers' and 'readers' proteins1,2,3. In vitro experiments suggest that m6A regulates several aspects of RNA metabolism, including RNA decay, splicing and translation1. Recent genetic analyses in vivo showed that m6A functions in sex determination in Drosophila4,5, in maternal-to-zygotic transition and haematopoietic stem cell specification during zebrafish embryogenesis6,7, in mouse spermatogenesis8,9,10, and in mouse brain development11. We recently discovered that lineage-specific deletion of the m6A 'writer' enzyme METTL3 in CD4+ T cells (Mettl3f/f; CD4-Cre) led to disruption of naïve T cell homeostasis12. CD4+ regulatory T cells (Tregs) comprise a critical subset of effector T cells, which are involved in resolution of inflammation and immunosuppression in tumor microenvironments13. However, the potential roles of m6A mRNA modification in Treg functions in vivo are unknown.
Mettl3f/f; CD4-Cre mice were normal without obvious defects during the first three months after birth. However, at the age of three months and older, the Mettl3f/f; CD4-Cre mice developed chronic inflammation in the intestine, evidenced by increased lymphocyte infiltration of the colon in Haematoxylin and eosin (H&E) staining analysis (Supplementary information, Figure S1). We hypothesized that the regulatory T cells might have lost their repressive functions in Mettl3f/f; CD4-Cre knockout mice.
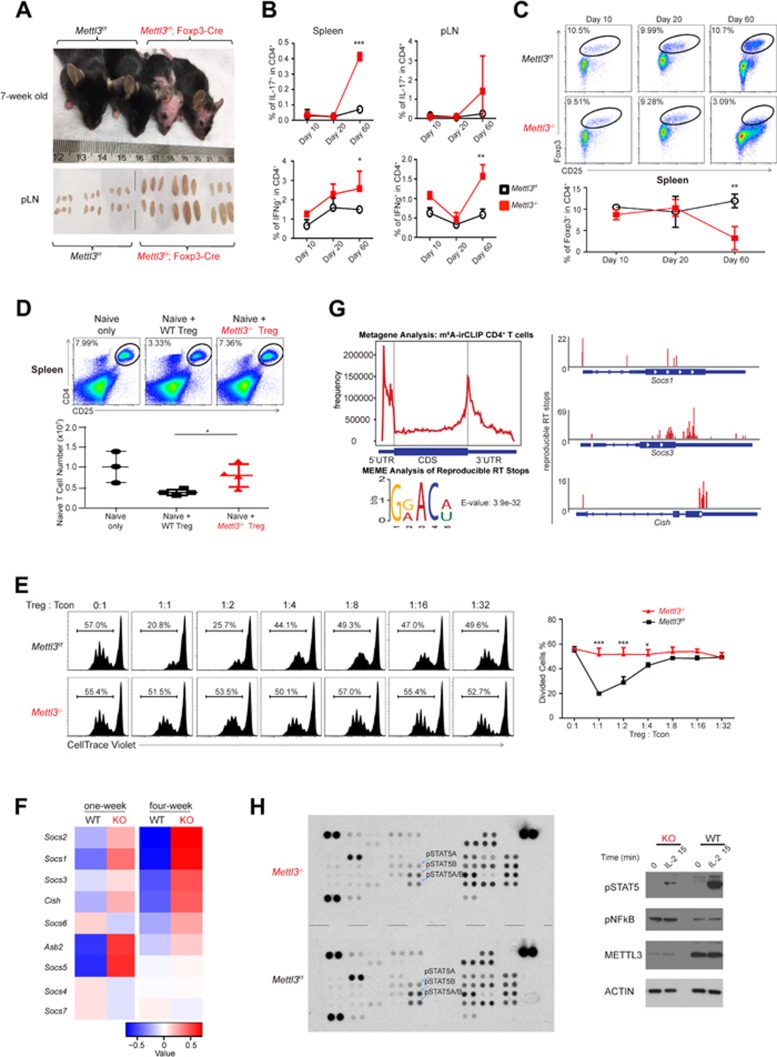
Therefore, we crossed Mettl3f/f mice with Foxp3Cre-YFP mice to specifically delete Mettl3 and consequently the m6A RNA modification in regulatory T cells. Because the Foxp3Cre-YFP transgene is on the X chromosome, we analyzed only male mice for all the following experiments in comparison to WT littermate controls. The Mettl3f/f; Foxp3Cre mice developed severe autoimmune diseases (for both male and female mice) and thus were infertile. Strikingly, the Mettl3f/f; Foxp3Cre mice had substantially larger peripheral lymph nodes and spleen, developed alopecia and severe systemic autoimmune disease after weaning, and started to die in 8∼9 weeks, indicating a systematic loss of suppressive function of Tregs without m6A RNA modification (Figure 1A).
Figure 1.
Specific deletion of Mettl3 in Tregs led to loss of Treg suppression function and severe autoimmune diseases. (A) 7-week-old Mettl3f/f; Foxp3Cre mice developed severe autoimmune disease, lost hair, and had enlarged periphery lymph nodes (pLN). (B) Th1 and Th17 cytokine analysis by FACS of the spleen and periphery lymph nodes from Mettl3f/f; Foxp3Cre and WT control mice. (C) FACS analysis of Treg population (Foxp3+CD25+) at different days after birth, and the statistics is shown below (n = 7). (D) In vivo suppression assay by co-transferring naïve T cells only, naïve T cells with WT or Mettl3−/− Tregs into TCRβ−/− mice (n = 4). (E) In vitro suppression assay by co-culturing the WT naïve T cells with either KO Tregs or WT Treg cells at different ratios. The mixing ratio is shown above the graph, and the percentage of divided cells and statistics are shown on the right graph (n = 3). (F) RNA-Seq analysis of WT and Mettl3 KO Tregs isolated from spleen of one-week-old and four-week-old mice, and the Socs genes are among the most up-regulated genes. (G) m6A irCLIP-Seq analysis of CD4+ T cells shows m6A enrichment at 3′ UTR and 5′ UTR regions and confirms Socs genes are m6A targets (n = 2). (H) Phospho-kinase array with lysates of FACS-sorted WT or Mettl3−/− Tregs from 4-week-old mice; pSTAT5 signals decrease in the Mettl3 KO Tregs and are highlighted. (I) Signaling assay of the IL-2-STAT5 pathway with FACS-sorted pure WT and Mettl3 KO Tregs from 6-week-old mice (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
To explore the cellular mechanisms of the severe autoimmunity of Mettl3f/f; Foxp3Cre mice, we analyzed T cell effector cytokines in spleen and peripheral lymph nodes. The inflammatory Th1 and Th17 responses in Mettl3f/f; Foxp3Cre were at basal levels 10 days after birth, and were still under control before weaning (day 20), but became significantly elevated compared to WT at day 60, which was consistent with the observed autoimmune phenotypes (Figure 1B). Uncontrolled inflammation in Mettl3f/f; Foxp3Cre mice was not due to Treg differentiation defects, as the percentage and the number of Tregs in spleen and thymus at day 10 and day 20 were similar to WT littermate controls, but Mettl3−/− Tregs were exhausted at day 60 in the spleen due to excessive inflammation (Figure 1C and Supplementary information, Figures S2, S3, S4). Because we previously showed that in vitro differentiation of Mettl3f/f; CD4-Cre naïve T cells to Tregs was similar to littermate WT naïve T cells12, these results suggest a potential role for m6A in Treg function.
We surmise that Mettl3−/− Tregs lost their suppressive function over the effector T cells. To test this hypothesis, we first set up an in vivo co-suppression assay by co-transferring naïve T cells without Treg cells, or with WT Tregs, or with Mettl3−/− Tregs in different ratios into TCRβ−/− mice. The results showed that Mettl3−/− Tregs completely lost the ability to suppress naïve T cell in vivo proliferation comparing to WT Tregs (Figure 1D and Supplementary information, Figure S5). Second, we also performed the standard in vitro suppression assay by mixing Tregs isolated from Mettl3f/f; Foxp3Cre mice or WT littermate control mice with WT naïve T cells labeled by CellTrace at different ratios. We measured the cell proliferation by FACS at day 4 after co-culture, and the results showed that naïve T cells co-cultured with m6A KO Tregs proliferated much faster (Figure 1E). These data confirmed that Mettl3f/f; Foxp3Cre Tregs lost their suppressive function over T cell proliferation.
We next sought to understand the molecular mechanism of the loss of suppressive function of Tregs from Mettl3 conditional knockout mice. We used FACS to sort Tregs from spleen by the Foxp3Cre-YFP marker from Mettl3f/f; Foxp3Cre-YFP and littermate Foxp3Cre-YFP mice at one week and four weeks of age, and subjected the Tregs to RNA sequencing analysis. Consistent with our previous observation in naïve T cells from Mettl3f/f; CD4-Cre mice12, members of the inhibitory Socs gene family, including Cish, Socs1, Socs2, Socs3, Asb2, were among the top up-regulated genes in Mettl3f/f; Foxp3Cre CD4+ Tregs comparing to WT CD4+ Tregs. Elevation of Socs gene family mRNA levels in Mettl3−/− Tregs was evident at the very early age of one week, and further increased by four weeks of age (Figure 1F). RT-qPCR analysis confirmed the increase of Socs family mRNA levels without changes in levels of transcripts of other genes in the IL-2-STAT5 pathway (Supplementary information, Figure S6). Furthermore, we applied m6A irCLIP-Seq, a newly developed and sensitive method to map m6A modification at near nucleotide resolution, in CD4+ T cells. We confirmed that m6A modification was enriched at 3′ UTR and 5′ UTR regions, with a consensus sequence of GG/AACA/U (Figure 1G). In addition, Socs gene mRNAs, including Socs3 and Cish, were indeed modified by m6A (Figure 1G). We have previously shown that these Socs gene mRNAs are m6A targets in CD4+ T cells, and deletion of Mettl3 led to a global decrease of mRNA m6A levels and loss of m6A modification in specific Socs gene transcripts. Decreased m6A modification led to enhanced Socs mRNA stability, increased levels of SOCS proteins, and blockage in cytokine signal transduction12.
The IL2-STAT5 signaling axis is critical for Treg function13. The increased Socs function in Tregs in Mettl3f/f; Foxp3Cre mice is likely to inhibit the IL2-STAT5 pathway, similar to the scenario that depletion of m6A leads to increased Socs mRNAs and inhibition of IL7-STAT5 signaling in CD4+ naïve T cells12. To test this hypothesis, we FACS-sorted YFP+ Tregs from Mettl3f/f; Foxp3Cre-YFP mice and Foxp3Cre-YFP control mice, and stimulated these cells in vitro with IL-2. 15 minutes after stimulation, we applied the cell lysates to the phospho-kinase array to interrogate multiple signaling pathways. As shown in Figure 1H, phosphorylation of STAT5A and STAT5B decreased in Mettl3 KO Tregs comparing to WT Tregs (Quantification shown in Supplementary information, Figure S7). Western blots confirmed that the phosphorylation levels of STAT5 in m6A KO Tregs were severely diminished comparing to those in littermate control WT cells isolated from 6-week-old mice (Figure 1I). Taken together, we conclude that depletion of Mettl3/m6A in Tregs leads to increased Socs mRNA levels, thus suppresses the IL-2-STAT5 signaling pathway which is essential to Treg functions and stability.
We reported in our previous work that m6A controlled the rapid degradation of transcripts of early inducible genes in naïve T cells, and here again we observed similar elevated Socs mRNA levels in Mettl3f/f; Foxp3Cre Tregs. But in the latter case of Tregs, elevated Socs function targeted the IL-2-STAT5 signaling pathway, which critically controls Treg cell functions. Our results show that the m6A RNA modification specifically targets the same class of genes encoding components of essential signaling pathways in different T cell subtypes, thereby controlling the differentiation of naïve T cells12 and also sustaining the suppressive functions of Tregs. Since Tregs in the tumor microenvironment suppress the tumor-killing functions of CD8 T cells, it is possible that selective depletion of m6A in tumor-infiltrated Tregs may be beneficial in combination with other forms of cancer immunotherapy.
Acknowledgments
We thank W Bailis, W Li, Kroehling L, J Alderman, C Lieber, J Stein and other members of the Flavell laboratory and the Li laboratory for discussions and technical support. This work was supported by Howard Hughes Medical Institute (RAF), the start-up fund from the Shanghai Jiao Tong University School of Medicine (H-BL), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (H-BL), the National Natural Science Foundation of China (No. 91753141 to H-BL, No. 81725004 to HL, No. 31470845 and 81430033 to BS, No. 81788104 and 31770990 to SZ), the US NIH R01-HG004361 and R35-CA209919 (HYC).
Footnotes
(Supplementary Information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Supplementary information, Figures S1–S7
References
- Cao G, Li HB, Yin Z, Flavell RA. Open Biol 2016; 6:160003. [DOI] [PMC free article] [PubMed]
- Liu J, Yue Y, Han D, et al. Nat Chem Biol 2014; 10:93–95. [DOI] [PMC free article] [PubMed]
- Ping XL, Sun BF, Wang L, et al. Cell Res 2014; 24:177–189. [DOI] [PMC free article] [PubMed]
- Haussmann IU, Bodi Z, Sanchez-Moran E, et al. Nature 2016; 540:301–304. [DOI] [PubMed]
- Lence T, Akhtar J, Bayer M, et al. Nature 2016; 540:242–247. [DOI] [PubMed]
- Zhang C, Chen Y, Sun B, et al. Nature 2017; 549:273–276. [DOI] [PubMed]
- Zhao BS, Wang X, Beadell AV, et al. Nature 2017; 542:475–478. [DOI] [PMC free article] [PubMed]
- Lin Z, Hsu PJ, Xing X, et al. Cell Res 2017; 27:1216–1230. [DOI] [PMC free article] [PubMed]
- Xu K, Yang Y, Feng GH, et al. Cell Res 2017; 27:1100–1114. [DOI] [PMC free article] [PubMed]
- Hsu PJ, Zhu Y, Ma H, et al. Cell Res 2017; 27:1115–1127. [DOI] [PMC free article] [PubMed]
- Yoon KJ, Ringeling FR, Vissers C, et al. Cell 2017; 171:877–889 [DOI] [PMC free article] [PubMed]
- Li HB, Tong J, Zhu S, et al. Nature 2017; 548:338–342. [DOI] [PMC free article] [PubMed]
- Li MO, Rudensky AY. Nat Rev Immunol 2016; 16:220–233. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information, Figures S1–S7