Abstract
Hepatocellular carcinoma (HCC) is one of the most common and aggressive cancers worldwide. HCC is the fifth common malignancy in the world and the second leading cause of cancer death in Asia. Long non-coding RNAs (lncRNAs) are RNAs with a length greater than 200 nucleotides that do not encode proteins. lncRNAs can regulate gene expression and protein synthesis in several ways by interacting with DNA, RNA and proteins in a sequence specific manner. They could regulate cellular and developmental processes through either gene inhibition or gene activation. Many studies have shown that dysregulation of lncRNAs is related to many human diseases such as cardiovascular diseases, genetic disorders, neurological diseases, immune mediated disorders and cancers. However, the study of lncRNAs is challenging as they are poorly conserved between species, their expression levels aren’t as high as that of mRNAs and have great interpatient variations. The study of lncRNAs expression in cancers have been a breakthrough as it unveils potential biomarkers and drug targets for cancer therapy and helps understand the mechanism of pathogenesis. This review discusses many long non-coding RNAs and their contribution in HCC, their role in development, metastasis, and prognosis of HCC and how to regulate and target these lncRNAs as a therapeutic tool in HCC treatment in the future.
Keywords: Tumor suppressor genes, Oncogenes, Long non-coding RNAs, Proliferation, Hepatocellular carcinoma, Metastasis
Core tip: Recent researches are focusing on targeting non-coding RNAs in an attempt to find therapeutic means for many health problems. Here, we are shedding the lights on the regulation of several proteins in hepatocellular carcinoma (HCC) by long non-coding RNAs (lncRNAs). lncRNAs try desperately to halt the aberrantly expressed oncogenic network by the fact that single lncRNA can have multiple downstream targets in one or more signaling pathway. This is an approach in an attempt to find an efficient radical cure for HCC.
INTRODUCTION
Epigenetic regulations
If we look up the word epigenetic in the dictionary, the result will be: the process by which the expression of genetic information is modified on a molecular level without alteration to the DNA sequence. The word epigenetic was also defined by previous researchers as “in addition to changes in genetic sequence”, “to act “on top of” or “in addition” to genetics” and “heritable changes in gene activity and expression that occur without alteration in DNA sequence”[1-4]. Various kinds of epigenetic processes have been discovered during the years, which include methylation, acetylation, phosphorylation, ubiquitylation and non-coding RNA. Such alterations can be transmitted to daughter cells or, as suggested in recent experiments, can be reversed. Epigenetic processes are significant to normal organism functions, however, if they develop incorrectly, severe unwanted health and mental effects could arise[4].
The most studied epigenetic process is DNA methylation. It’s the covalent addition or a methyl group (CH3) removal to the fifth position of the cytosine base within CpG dinucleotides. This modification is catalyzed by DNA methyltransferases (DNMTs), as DNMT1, DNMT3a and DNMT3b. DNMT3a and DNMT3b are considered de novo methyltransferases, initiating methylation to unmethylated CpGs during embryonic development or in cancer cells[5-7]. On the contrary, DNMT1 functions as the maintenance methyltransferase by methylating hemimethylated CpGs after mitosis, hence transmitting the methylation patterns to daughter strands, along with contributing to the de novo methylation process[8,9]. Both classes are said to function co-operatively to methylate DNA usually in regions known as CpG islands where the occurrence of CpG dinucleotides is high. DNA methylation causes gene silencing through two mechanisms; firstly, by decreasing the affinity of transcription factors to gene promoters through steric hindrance and the second mechanism is by the direct binding of methyl CpG binding domain (MBD)-containing proteins to the methylated DNA, leading to transcription repression through chromatin condensation[10]. This gene silencing could be reversed by active DNA demethylation which mainly happens by the removal of the methyl group from 5-methylcytosine via Methyl-CpG binding domain proteins[11].
Another epigenetic regulation is histone modification which encounters any post translational modification regulating chromatin structure and function. Chromatin consists of DNA and proteins bundled together in a compact way to fit inside the nucleus[4]. These complexes can be modified mainly through acetylation and methylation of the histone lysine residues. The resultant effect differs according to the type of modification and its location on the histone. The lysine residues at the histone terminals are subject to acetylation or deacetylation by histone acetyltransferases or histone deacetylases. Acetylation decreases the positive charges of lysine residues and decreases the affinity between histones and DNA which results in decondensation of the chromatin hence, disrupting the chromatin structure. Moreover, acetylated residues act as binding sites for histone modifying enzymes or chromatin remodeling factors that facilitate gene expression[12,13]. Methylation of histone occurs on different lysine residues with different degree of methylation thus giving a wide variety of results either repressive or activating depending on the combination of factors[14]. Methylation of the lysine at the fourth residue of histone H3 (H3K4Me) promotes a transcriptionally active conformation, as H3K9Me which promotes a transcriptionally repressive conformation. H3K36Me can be activating or repressive, depending upon proximity to a gene promoter region[15].
Non-coding RNAs
RNA species beyond mRNA which lack clear potential to encode proteins or peptides, and they include intronic RNAs, microRNAs (miRNAs), circular RNAs (circRNAs), extracellular RNAs and long non-coding RNAs (lncRNAs), that will be our main focus in this review[16].
lncRNAs
LncRNAs are a diverse group of transcripts whose natural functions and potential as drug targets remain largely undefined. These RNA species are greater than 200 nucleotides in length and do not encode protein. lncRNAs are thought to involve nearly 30000 different transcripts in humans, lncRNA transcripts account for the major part of the non-coding transcriptome. LncRNA discovery remains at a primary stage.
LncRNAs biogenesis
Several long non-coding RNAs (lncRNAs) or classes of lncRNAs are differentially regulated at different levels of their biogenesis, maturation and degradation. At the chromatin level state, lncRNAs and mRNAs exhibit similar properties, For example, an enrichment of H3K4me3 at promoters; however, lncRNA genes have a higher enrichment of H3K27ac and are definitely more repressed by certain chromatin remodelling complexes, such as Swr1, Isw2, Rsc and Ino80. Transcription starts from divergent promoters differs for the sense (mRNA) and the antisense (lncRNA) directions; divergent antisense transcription is enriched for H3K56ac and phosphorylation of RNA polymerase II (Pol II) Tyr1. Transcription in the divergent direction is additionally increased by the SWI/SNF proteins and repressed by CAF-1. Transcriptional elongation is more strongly regulated by DICER1 and MYC for lncRNAs than for mRNAs. The occurrence of U1 and polyadenylation signals differs on either side of bidirectional promoters (along the U1–PAS axis), favoring the splicing of mRNAs in the sense direction and the cleavage and polyadenylation in the divergent, antisense direction. Despite the the fact that mRNAs localize very specifically to ribosomes in the cytoplasm, lncRNA localization is substantially more differed, as certain lncRNAs can occupy the chromatin, subnuclear domains, the nucleoplasm or the cytoplasm. Finally, whereas mRNAs are primarily degraded in the cytoplasm by decapping and 5′-to-3′ exonuclease digestion, numerous unstable lncRNA transcripts are liable to the nuclear exosome or to cytosolic nonsense-mediated decay (NMD)[17].
LncRNAs functions and mechanism of action
LncRNAs can regulate gene expression and the synthesis of protein in various ways. Some lncRNAs are relatively highly expressed, and functions as scaffolds for particular subnuclear domains. Carrying on, lncRNA have secondary structures which facilitate their interactions with DNA, RNA and proteins. Also, its binding to DNA or RNA is sequence-specific. Gene regulation may take place in cis (in close proximity to the transcribed lncRNA) or in trans (at a distance from the transcription site). In the function of chromatin modulation, the effect of lncRNA is usually gene-specific, exerted at a local level (in cis) but the regulation of chromatin can also take place in trans[18].
Some lncRNAs already had their functions experimentally elucidated and have proved to be involved in gene regulation processes including: Chromatin modification and structure, direct transcriptional regulation, regulation of RNA processing such as splicing, editing, localization, translation and degradation, post-translational regulation of protein activity and localization, facilitation of ribonucleoprotein (RNP) complex formation, modulation of microRNA regulation, in addition to gene silencing through endogenous siRNA (endo-siRNA) and regulation of genomic imprinting.
Recently, lncRNA functions have been classified into main different molecular mechanisms. One lncRNA may be described as one or more of the following five models: First, the Signal model that functions as a molecular signal or indicator of transcriptional activity. The Decoy model in which it binds and titrates away other regulatory RNAs (e.g., microRNAs) or proteins (e.g., transcription factors). Third type is the Guide model in which it directs the localization of ribonucleoprotein complexes to specific targets (e.g., chromatin modification enzymes are recruited to DNA). Last but not least the Scaffold model that has a structural role as platform upon which relevant molecular components (proteins and or RNA) can be assembled into a complex or spatial proximity. And the last type is the Enhancer model which controls higher order chromosomal looping in an enhancer-like model[19].
Epigenetic regulation and lncRNAs in hepatocellular carcinoma
The lncRNA X-inactive specific transcript (XIST) recruits EZH2 which is a component of polycomb repressive complex “PRC2”, to the X chromosome, leading to histone methylation and silencing of one X chromosome[20]. Also ANRIL, a lncRNA which recruits polycomb repressive complexes “PRC1 and PRC2” to the locus, causing the reduction transcription of ARF, CDKN2B, and CDKN2A[21,22]. Another lncRNA “ TUG1” can regulate KLF2 expression in the epigenetic level by binding to PRC2. And it was found that those lncRNAs are deregulated in hepatocellular carcinoma (HCC) beside other lncRNAs that will be mentioned below.
LncRNAs in cancer
Numerous previous studies concluded that long non-coding RNAs play an important role in regulating many biological processes including gene expression, cell cycle regulation and cellular differentiation[23]. Hence, deregulated lncRNA expression may contribute to disease pathogenesis and pathophysiology such as cancers, cardiovascular disorders and metabolic disorders. This relation unveils the role lncRNAs could have as therapeutic targets or biomarkers for diseases[24]. LncRNAs are usually observed in cancer cases due to the expression difference between cancerous cells and normal cells[25]. Moreover, they fall under two categories; either oncogenic, promoting cancer or tumor suppressor, inhibiting cancer progression and both have equal importance in cancer therapeutic treatment. Long non-coding RNAs exert their effect through various mechanisms for example, modulating epigenetic regulations including imprinting as X-inactivation in females, controlling cell apoptosis signaling as well as cellular proliferation pathways. Sometimes, they interact with RNA-binding proteins mediating transcriptional processes, bind to their intracellular steroid receptors activating or inhibiting downstream targets or transcription factors. Furthermore, lncRNAs could induce chromatin modification through recruiting chromatin remodeling complex to specific loci and regulate gene expression at the post transcriptional level as well (as shown in Figure 1)[26].
Figure 1.
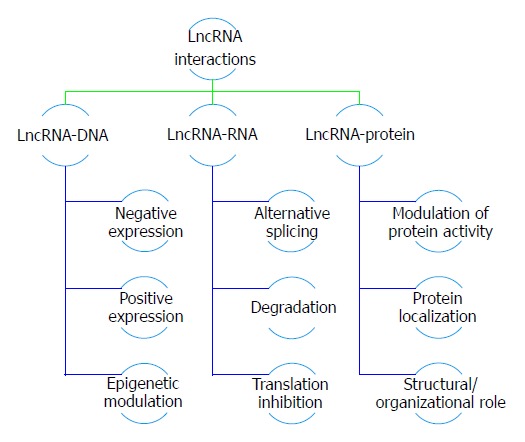
Long non-coding RNAs actions and interaction in cancer development; a schematic representation to the role of deregulated long non-coding RNAs in various types of cancers. lncRNAs: Long non-coding RNAs.
As an example to the function of deregulated lncRNAs in various types of cancers, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is reported overexpressed in breast cancer, prostate cancer, uterine cancer, colon cancer, pancreatic cancer, osteosarcoma, NSCLC and other cancers[26-28]. Additionally, HOX transcript antisense RNA (HOTAIR) is found upregulated in breast cancer and prostate cancer aiding in tumor invasion and metastasis[29,30]. Furthermore, H19 present on chromosome 11, is reported crucial in bladder, lung, breast, cervix, prostate and colorectal cancers. Another famous lncRNA is urothelial carcinoma-associated 1 (UCA1) which serves as an oncogene in colon, cervix, lung, bladder, breast and stomach carcinomas through enhancing cellular proliferation and migration[27]. ANRIL, one of the antisense lncRNAs is found in high levels in prostate cancers and leukemia[26,27]. BC200, an intergenic gene on chromosome 2 modulates translational processes in breast, cervix, esophagus, lung, ovary and tongue cancers[26]. On the contrary, MEG3 has proved to be one of the tumor suppressor genes in cancers like pituitary adenomas as it inhibits cell proliferation and modulates angiogenesis through regulating p53[31]. Another tumor suppressor gene is growth arrest-specific transcript 5 (GAS5) that induces apoptosis in breast and prostate cancers by inhibiting several responsive genes leading to altering various cellular processes[26].
LncRNA IN HCC
Tumor suppressor lncRNA
lncRNA SVUGP2 is downregulated in HCC cells and its overexpression inhibits cellular proliferation and invasion through the inhibition of several mRNA and proteins of growth markers and invasion markers; hence acting as a tumor suppressor[32]. AF113014 acts as a tumor suppressor by regulating the expression of the tumor suppressor gene, Egr2 through interaction with miR-20a causing a decrease in cellular proliferation[33]. Uc.134 is reported to be significantly downregulated in HCC cells; its overexpression suppresses HCC by inhibiting cellular invasion and proliferation via repression of CUL4-mediated ubiquitination of LATS1 and activation of YAPS127 phosphorylation[34]. SchLAH is a lncRNA that is downregulated in HCC via possible histone deacetylation; its overexpression inhibits cell migration and metastasis through interaction with FUS protein and modulation of RhoA and Rac1 downstream targets[35]. SRA1 is one of the downregulated RNAs in HCC cells that serves as a tumor suppressive through its regulation of tumor size and serum glucose levels[36]. DGCR5 is one of the downregulated lncRNAs that can acts as a tumor suppressor in HCC patients however its molecular mechanism is yet to be studied[37]. Also, the lncRNA ZNF674-AS1 is found to be downregulated in HCC patients with significant relation to poor prognosis in patients; however, no significant molecular mechanism is established yet[38]. STARD13 is a tumor suppressor as its expression levels in HCC is low and its upregulation enhances cellular apoptosis by acting as a ceRNA for Fas and increases tumor sensitivity to chemotherapy[39].
Linc-cdh4-2 is downregulated in HCC and its overexpression causes a significant decrease in migration and invasion with no effect on cell viability and proliferation. The decrease in migration could be through an increase in the protein levels of R-cadherin and decrease in the protein levels of small GTPase RAC1, as an increase in RAC1 protein levels enhances cell invasion abilities[40]. Low levels of LINC RP1130-1 are significant in HCC patients compared to normal patients. This decrease is associated with number of tumors, portal vein tumor thrombus, microvascular invasion and most importantly a shorter recurrence-free survival hence, classifying it as a tumor suppressor lncRNA[41]. ZNFX1-AS1 is significantly downregulated in HCC and its overexpression suppressed cell proliferation, colony formation and promoted cell apoptosis. miR-9 exhibits a positive correlation with ZNFX1-AS1 hence an upregulation of miR-9 and a downregulation of its promoter methylation is observed with ZNFX1-AS1 overexpression[42]. LINC00052 is one of the tumor suppressor lncRNAs as its upregulation inhibit cell invasion and proliferation abilities of HCC cells which is achieved through regulating NTRK3 expression by establishing complementary base pairing with miR-128 ad miR-485-3p. Downregulation of NTRK3 promotes cellular invasion and proliferation abilities just like the low levels of LINC00052[43]. PRAL is a tumor suppressor lncRNA present on chromosome 17p13.1 usually in low levels in HCC patients and its overexpression inhibits tumor growth. This occurs due to the binding of PRAL to HSP90 which enhances its binding to p53, opposes MDM2 induced p53 degradation hence promoting HCC cell apoptosis[44]. Moreover, studying several deregulated genes in HCC revealed the downregulation of AF070632 associated with various cellular processes such as oxidation-reduction, cofactor binding in catabolic processes[45]. The expression of the lnc-Dreh was evaluated and found to be downregulated in HCC tissues proving to be a tumor suppressor as it binds to vimentin, decreasing its expression and disrupting the normal cytoskeleton structure required for growth and invasion hence inhibiting metastasis in HCC or HBV related HCC patients[46]. LET is considered as one of the lncRNAs that function as a tumor suppressor as it inhibits HCC metastasis and invasion when overexpressed on chromosome 15. This occur through a complex positive feedback loop of HIF-1α/HDAC3/lncRNA-LET/NF90 that modulates HIF-1α response and gives the tumor suppressive effect[47].
An interesting mechanism of tumor suppression is encountered with PTENP1, a pseudogene of PTEN, where it induces autophagy of HCC cell inhibiting its survival. This is achieved as PTENP1 acts as an endogenous sponge for miR-17, miR-19b and miR-20a abolishing their inhibitory ability of PTEN hence inhibiting the PIK3T/AKT pathway and inducing pro-death autophagy[48]. CPS1-IT1 acts as tumor suppressor in HCC by inactivation of HIF-1α which results in decreased the expression of epithelia-mesenchymal transition (EMT) related proteins[49]. TUSC7 acts as tumor suppressor through the TUSC7-miR-10a-EphA4 axis leading to EMT suppression[50]. WT1-AS may function as a tumor suppressor in HCC by reversing the oncogenic effects of WT1. WT1-AS downregulates WT1 expression in HCC tumors and promotes apoptosis by binding to the promoter region of WT1[51]. AOC4P lncRNA also plays role as an HCC tumor suppressor by promoting vimentin degradation and suppressing the EMT[52]. SRHC is noncoding tumor suppressor that can inhibit HCC cell proliferation and promote its differentiation[53]. CASC2 inhibits cell proliferation, migration and invasion and promotes apoptosis by inactivation of the MAPK pathway this indicates that CASC2 may act as a tumour suppressor in HCC[54]. lncRNA-AK058003 can inhibit HCC proliferation and metastasis acting as a tumor suppressor by suppressing the SNCG in a HuR-dependent manner[55]. Suppression of treRNA by miR-190a significantly inhibited migration of HCC cells. This lead to reduced expression of Vimentin and SNAI1 (mesenchymal markers), as well as induced expression of E-cadherin and Claudin-1 (epithelial markers)[56].
Oncogenic lncRNA
HCG11 acts as an oncogenic lncRNA as it induces cell proliferation and invasion of HCC tumor cells, suppresses apoptotic indicators, such as: p21 and caspase-3, promotes anti-apoptotic factors including ERK, JNK and p38 via the regulation of IGF2BP1 protein and activation of MAPK pathway which consequently promoted HCC progression[57]. LOC90784 long non-coding RNA functions as an oncogene as its knockdown promotes Bax and represses CDK4 and Cyclin D protein expression which induces apoptosis and suppresses proliferation. Moreover, its downregulation inhibits cellular invasion and migration by repressing MMP2 and MMP9 expression in HCC cells[58]. Another lncRNA that is found to be upregulated in HCC cells is UBE2CP3; it induces the process of cell invasion and migration in HCC by increasing the expression of Snail-1 and N-cadherin while decreasing the expression of E-cadherin hence, promoting the EMT leading to tumor metastasis[59]. Linc00462 is reported to be upregulated in HCC tumors promoting tumor progression through the regulation of PI3K/AKT pathway, inducing cellular proliferation and migration[60]. TINCR lncRNA is found to be upregulated in HCC cells acting as an oncogene regulating differentiation, invasion and metastasis of tumor cells[61]. AB019562 is reported to be upregulated in HCC cells; its knockdown inhibits cell proliferation, arrests cell cycle at G0/G1 phase, suppresses cellular metastasis, induces apoptosis and activates caspase-3 activity[62]. LncARSR is a long non-coding RNA that regulates the sensitivity of the tumor cells to doxorubicin. Upregulation of lncARSR in HCC cells induces its resistance to doxorubicin, indicating poor prognosis through PTEN mRNA depletion and PI3k/Akt activation[63]. AWPPH serves as an oncogene as its overexpression in HCC promotes cell proliferation, migration and metastasis. This is achieved through YBX1 interaction and its mediated activation of downstream transcription factors such as SNAIL1 and PIK3CA and their pathways[64].
Prostate cancer-associated transcript (PCAT)-14 is overexpressed in HCC cells; it induces proliferation, migration and cell cycle arrest. Moreover, it regulates the expression of ATAD2 and Hedgehog pathway via its inhibitory effect on miR-372 that’s achieved by methylation of miR-372’s promoter region. Conversely, miR-372 eliminates PCAT-14’s effects on proliferation and invasion of HCC cells[65]. Another crucial lncRNA is P73 antisense 1 T that is upregulated in HCC cell lines; its knockdown resulted in reduced cellular proliferation, invasion and downregulated HMGB1/ RAGE signaling pathway that also correlates with dysregulated cell death and survival. miR-200a has a negative correlation with TP73-AS1 and HMGB1 expressions and studies revealed that both compete for the same binding site on miR-200a concluding that TP73-AS1 could be oncogenic through miR-200a dependent HMGB1/RAGE regulation[66]. Linnc00441 is an oncogene that’s upregulated in HCC promoting cell proliferation, tumor growth and downregulating RB1 tumor suppressor gene. Transcription factor (TCF)-4 and H3K27 acetylation were found to contribute to Linc00441 upregulation which in turn downregulated RB1 expression through possible enhancement of CpG islands methylation in its promoter region via DNMT3A Linc00441-dependent recruitment[67]. HOST2 is reported to be upregulated in various HCC cell lines and its silencing results in decreased cell proliferation, blocked cell cycle at G0/Gi phase to S phase and reduced cell migration and invasion. Therefore, its marked as an oncogene that supports cellular proliferation, metastasis and inhibits apoptosis in human HCC cells[68]. UC001kfo is an overexpressed lncRNA in HCC cells that positively regulates cellular proliferation and migration through the modulation of its target gene, α-SMA, in a positive manner. α-SMA also serves as a crucial marker for EMT which in turn regulates cellular migration and invasion via its effect on UC001kfo function[69]. LncBRM is an upregulated RNA in HCC and liver CSCs; it may contribute to the recurrence and self-renewal of the tumor cells. LncBRM modulates BRG1/BRM switch that in turn activates YAP1 signaling pathway via a KLF4-dependent manner giving a result that’s positively correlated with tumor severity and self-renewal ability of the cells[70]. HOX Antisense lincRNA HOXA-AS2 is an oncogene that’s overexpressed in HCC cells and its upregulation correlates with poor prognosis, increased proliferation and metastasis. The knockdown of HOXA-AS2 significantly suppressed proliferation, migration and promoted apoptosis[71]. CRNDE is reported to be overexpressed in HCC cells inducing cellular proliferation, migration and invasion. It is found that CRNDE inhibits the expression of miR-384 that’s usually downregulated in HCC while enhancing the expression of NF-κB and p-AKT, suggesting that CRNDE-miR-384 axis is promising for treatment of HCC[72].
One of the antisense lncRNAs investigated is ZEB2-AS1; it is reported to be overexpressed in HCC cells especially Huh7. The study confirmed the that changes in ZEB2-AS1 levels fluctuates EMT-induced markers and the silencing of the RNA decreases cellular proliferation, viability and invasion[73]. Unigene56159 is an HBV related lncRNA that could be induced by HBV infection aiding in HCC development, migration and EMT. This happens because Unigene56159 directly binds to miR-140-5p acting as a competing endogenous RNA (ceRNA) for the miRNA, abolishing its inhibitory effect on its other target Slug hence, promoting EMT in HCC cells[74]. CCHE1 is an oncogene that’s noted to be upregulated in HCC and its knockdown enhanced growth arrest and cell apoptosis. The knockdown also reduced tumorigenicity and inhibited ERK/MAPK pathway, with a function in cellular proliferation, differentiation and survival, hence the decreased cellular viability[75]. GPC3-AS1 is suggested to be upregulated in HCC due to possible increase in histone acetylation in the promoter region. GPC-AS1 upregulation recruits PCAF to the GPC3 body upregulating its transcription and hence promoting cellular proliferation and migration[76]. GIHCG is upregulated in HCC cells and significantly promotes tumor growth and metastasis through association with EZH2. This upregulates H3K27me3 and DNA methylation levels by DNMT1 of miR-200b/a/429 promoter hence epigenetically silencing miR-200b/a/429 expression in HCC cells[77]. LncCAMTA1 is overexpressed in HCC and is associated with poor prognosis, increased HCC proliferation and tumorigenesis. LncCAMTA1 is noted to inhibit CAMTA1 transcription by promoting a repressive chromatin structure of the gene and this relation is essential for lncCAMTA1 proliferative effect on HCC cells[78]. LncSox4 is upregulated in HCC and tumor initiating cells (TIC) that has proved essential for liver TIC self-renewal and tumor initiation. LncSox4 interacts with and recruits Stat3 to the Sox4 promoter enhancing Sox4 expression which is also required for TIC self-renewal[79]. Small nucleolar rna host gene (SNHG)3 shows high expression levels in HCC patients and is significantly correlated with tumor size, overall survival, poor prognosis and malignant status[80].
SNHG12 is overexpressed in HCC tissues promoting cellular proliferation and inhibiting apoptosis via functioning as a sponge for miR-199a/b-5p reducing their inhibitory action on MLK3, thus enhancing MLK3 expression and its downstream targets in the NF-κB pathway[81]. Furthermore, SNHG15 is reported to be upregulated in HCC tissues compared to normal control and positively correlates with histological grade, vein invasion and poor prognosis hence, is a promising clinical biomarker for HCC[82]. BAIAP2-AS1 is overexpressed in HCC tissues with positive correlation to proliferation and invasion processes through the regulation of MAPKAP1, E2F3and RAF1 which are downstream targets of several miRNAs that BAIAP2-AS1 functions as a sponge for[83]. Additionally, one of the studies investigating deregulated lncRNAs in HCC concluded that LINC01419 and AK021443 are upregulated in early HCC stages hence concluding their involvement in cell cycle progression and tumor initiation[45]. Another interesting study investigated the levels of lnc-HEIH in HBV related HCC cases and reported to be upregulated and involved in cell cycle modulation through the association with EZH2 and suppression of p16, p27 and p21 which mainly regulate the cell cycle arrest. All these actions facilitate its oncogenic properties and promote cellular proliferation and invasion in HCC tissues[84]. A study was done revealing that hypoxic conditions in HCC tissues could modulate lncRNAs expression such as linc-ROR that’s overexpressed in hypoxic HCC tissues. Linc-ROR itself regulates cellular responses during hypoxic conditions by altering the expression of HIF-1α and its downstream target PDK1 in a positive manner through inhibiting the expression of miR-145[85]. On a further note, linc-ROR modulate chemotherapy induced apoptosis and cell survival through p53 dependent signaling[86]. Lnc-β-Catm associates with β-catenin and the methyltransferase EZH2, leading to activation of Wnt–β-catenin signaling, resulting in cancer severity in people with HCC[87].
UC338 enhances cell proliferation as it directly interacts with BMI1 and suppresses p21 expression in HCC[88]. LINC01225 in HCC, stimulates proliferation and invasion through binding to protein, leading to increase the level of epidermal growth factor receptor (EGFR) as a consequence, thus fine tuning the EGFR/MAPK signaling pathway[89]. PlncRNA-1 act as an oncogene in HCC progression, by induction of EMT signaling[90]. BANCR overexpression is associated with HCC carcinogenesis. Downregulation of BANCR would decrease cell proliferation, enhance cell apoptosis, and impair cell invasion and migration[91]. LncRNA CARLo-5 enhances proliferation, migration and invasion of HCC and its upregulation was associated with poor prognosis of HCC patients[92]. MVIH enhanced cell growth and inhibited cell apoptosis of HCC through inhibition of miR-199a expression; si-MVIH inhibited HCC cell viability and enhanced cell apoptosis, but this effect was reversed by miR-199a inhibitor as miR-199a had a direct binding ability to MVIH RNA[93]. Overexpression of ZEB1-AS1 reduces protein levels of epithelial markers and increases protein levels of mesenchymal markers, proving that ZEB1-AS1 promotes EMT and its role in tumor progression[94]. lncRNA Sox2ot may play a tumor oncogenic role in HCC as the up-regulation of lncRNA Sox2ot promotes the metastasis ability of HCC cells[95]. ZFAS1 acts as tumor promoter in HCC progression by binding miR-150 and abolishing its tumor-suppressive function. miR-150 repressed HCC cell invasion by inhibiting ZEB1 and the matrix metalloproteinases MMP14 and MMP16. Conversely, ZFAS1 activated ZEB1, MMP14, and MMP16 expression, inhibiting these effects of miR-150. This mean that upregulation of ZFAS1 in HCC enhances tumor metastasis in a miR-150–dependent manner[96]. The lincRNA-UFC1, a target of microRNA 34a, promotes proliferation and decreases apoptosis in HCC cells through interaction with the mRNA stabilizing protein HuR to regulate levels of β-catenin in HCC cells[97]. Also, URHC may function as an oncogene that regulates cell proliferation and apoptosis through down-regulating of ZAK expression, and inactivation of the ERK/MAPK pathway by URHC-ZAK regulation[98]. LncRNA-PE promotes HCC cell invasion by upregulating ZEB1, the master inducer of EMT via inhibiting transcription of miR-200a/b[99]. ZNF667 serves as oncogene, promotes the cell proliferation, invasion and migration via interference with the expression of Bcl-2 and BAX[100]. The upcoming section discusses individual lncRNA that were found to be extensively studied.
Highly up-regulated in liver cancer
Highly up-regulated in liver cancer (HULC), present on chromosome 6, could be identified as the most over-expressed lncRNA in HCC with various mechanisms in promoting tumorigenesis and carries a huge perspective in RNA-targeted therapy[101]. High levels of HULC in plasma of HCC patients could be exploited as a noninvasive biomarker for HCC diagnosis[102]. HULC is reported to have high plasma levels in HCC patients and its combination with Linc00152 and AFP would achieve better diagnostic accuracy[103]. Overexpression of HULC is reported with upregulation in HEIH which resulted in increased proliferation, invasion and snail protein expression in HepG2 cells[104]. HULC is overexpressed in HCC as it promotes growth in cancer cells through enhancing ubiquitin-specific peptidase 22 (USP22) which reduces ubiquitin-mediated degradation of COX-2 protein hence stabilizing and upregulating COX-2 protein[105]. A similar mechanism of HULC in HCC is reported where it promotes the deubiquitination of sirt1 protein through inducing USP22 which promotes the levels of sirt1 proteins necessary for autophagy induction in chemotherapy treatment. Another set of miRNAs miR-6825-5p, miR-6845-5p and miR-6886-3p work opposite to HULC and suppress USP22 and they were found to be inhibited by HULC expression which modulate the sensitivity of cancer cells to chemotherapy[106]. HULC promotes tumor progression through acting as a ceRNA for miR-200a-3p inhibiting its function hence upregulating ZEB1 expression and inducing EMT, tumor proliferation and metastasis[107]. HULC contributes to HCC angiogenesis through interaction and downregulation of miR-107 which upregulates E2F1 and finally activates SPHK1 which aids in tumor angiogenesis (as illustrated in Figure 2)[108].
Figure 2.
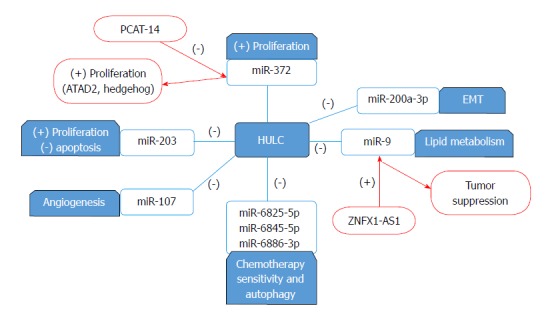
Highly up-regulated in liver cancer interactions with miRNA. Highly up-regulated in liver cancer (HULC) inhibits the actions of miR-6825-5p, miR 6845-5p and miR 6886-3p which modulates the cell sensitivity to chemotherapy and autophagy. Moreover, it inhibits the function of miR-200a-3p hence inducing EMT and metastasis. HULC downregulates miR-107 promoting angiogenesis and tumor proliferation. Overexpression of HULC diminishes miR-203 antitumor effect. While HULC contributes to lipid deregulation and tumor progression through the downregulation of miR-9, ZNFX1-AS1 upregulates miR-9 resulting in tumor suppression HULC and PCAT-14 promote proliferation through the inhibition of miR-372 however through different mechanisms. (-): Inhibits/Downregulates (+): Upregulates. EMT: Epithelia-mesenchymal transition; PCAT: Prostate cancer-associated transcript.
HULC is reported to be regulated by the Specificity protein (Sp) transcription factors that significantly modulate HCC proliferation and migration. The anti-diabetic drug, metformin downregulates Sp transcription factors and by default Sp-regulated genes such as HULC, giving metformin a strong evidence to be used in combination treatments for HCC patients[109]. One of the microRNAs that interact with HULC is miR-203 which shows a negative relation on HULC as its upregulation inhibits cell proliferation and induces cell apoptosis through downregulation of HULC and another oncogene, ADAM9. Also, the overexpression of HULC diminishes the antitumor effect of miR-203[110]. HULC contributes to the deregulation of lipid metabolism in HCC due to the upregulation of transcription factor PPARA which activates ACSL1. Moreover, an interaction with miR-9 is present as it is downregulated by HULC through promoter methylation and miR-9 itself inhibits PPARA expression as well. ACSL1 is capable of upregulating HULC via RXRA activation in hepatoma cells thus suggesting a positive feedback loop HULC/miR-9/PPARA/ACSL1/HULC is present in HCC cells[111]. HULC binds to YB-1 protein and promotes its phosphorylation through ERK and induces the translation of associated mRNAs hence increasing proliferation n HCC through promoting G1/S transition[112]. Another study suggests that HULC promotes hepatocarcinogenesis through the disruption of the circadian rhythm and upregulating circadian oscillator CLOCK in hepatoma cells[113]. Moreover, HULC is reported to be upregulated by HBx protein which results in increased cellular proliferation due to the inhibition of the tumor suppressor gene p18 on the mRNA and protein level[114]. An interesting study revealed the binding of CREB to the promoter region of HULC, activating its expression and function as an endogenous sponge to miR-372 which in turn results in further activation of HULC in an auto-regulatory loop of CREB/HULC/miR-372/PRKACB/CREB/HULC enhancing cellular proliferation (as illustrated in Figure 3)[115].
Figure 3.
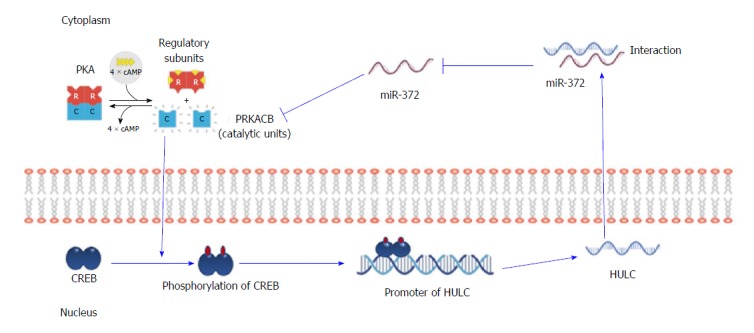
The auto-regulatory loop of highly up-regulated in liver cancer through inhibition miRNA-372 in hepatocellular carcinoma. miR-372 binding to HULC can repress the expression and activity of miR-372. Inhibition of miR-372 leads to a reduction in translational repression of its target gene PRKACB, which in turn induces phosphorylation of CREB, thereby increasing the amount of CREB that can bind to the proximal promoter of HULC to induce HULC expression. HULC: Highly up-regulated in liver cancer.
HOTAIR
HOTAIR is a lncRNA of 2158 bp length in chromosome 12 representing an antisense strand of homeobox C gene[116]. High levels of HOTAIR in HCC patients compared to normal control is significantly correlated with rapid proliferation, large tumor size and poor prognosis[117]. Another study reported that significantly high levels of HOTAIR is related with shorter recurrence-free survival period and its suppression reduced cell viability and induced apoptosis and chemotherapy sensitivity of HCC[118]. HOTAIR upregulation in HCC patients enhances tumor progression, migration and recurrence via the regulation of Wnt/β-catenin pathway[119]. Increased expression of HOTAIR in HCC patients show increased risk of recurrence and lymph node metastasis through the regulation of matrix metalloproteinase-9 (MMP-9) and vascular endothelial growth factor protein (VEGF)[120]. In a relevant study, HOTAIR overexpression in HCC patients is reported to upregulate autophagy related 3 (ATG3) and ATG7 expressions which induces cellular proliferation[121].
A study investigating the molecular mechanism of HOTAIR as an oncogene in HCC tissues revealed that it achieves its action through abolishing SETD2 expression and phosphorylation, inhibiting H3K36me3 binding to hMSH2 and SKP2 and modulating various epigenetic changes such as DNA damage repair, microsatellite instability and abnormal gene expression[122]. Interestingly, HOTAIR’s overexpression in HCC patients unveiled several molecular relations with transcription factors and miRNAs, as it is partially regulated by the transcription factor FOXC1 which binds to HOTAIR promoter regions and activates its expression. Moreover, HOTAIR is found to downregulate miR-1 and vice versa due to the presence of miR-1 binding site on HOTAIR[123]. In an extensive study, HOTAIR was found to act as a binding site to both DDX5 and Mex3b in a competitive manner where the binding of Mex3b induces the ubiquitination of SUZ12 and derepression of PRC2 targets in G2 phase of the cell cycle promoting hepatocarcinogenesis in HBV patients[124]. A study on 52 paired HCC specimens proved HOTAIR as an oncogene in HCC as its knockdown activates P16(lnk4a) and P14(ARF) downstream signaling of miR-218, enhances miR-218 expression and suppresses Bmi-1 expression hence HOTAIR suppression inhibits cell viability and tumorigenesis[125]. Additionally, it was proposed that HOTAIR promotes HCC proliferation and migration through the inhibition of RNA binding motif protein 38 (RBM38) which is crucial for biological processes and exhibits a negative relation with HOTAIR[126].
MALAT1
LncRNA MALAT1 is located on chromosome 11q13. It has been found to participate in the carcinogenesis of several types of tumors such as lung cancer, breast cancer, nasopharyngeal carcinoma, and so on[127-129]. MALAT1 acts as an oncogene since it suppressed miR-143-3p. MALAT1 functioned as a molecular sponge for miR-143-3p, and inhibited its function. This study results suggested that MALAT1 regulated ZEB1 expression and the proliferation and invasion through miR-143-3p in HCC cells[130]. Another study showed another mechanism for how MALAT-1 acts as oncogene. MALAT1 could promote tumor growth and metastasis by activating and upregulation of LTBP3 (latent transforming growth factor β-binding protein 3), which could also be up-regulated by hepatitis B virus X protein(HBx), which is known to be involved in HCC progression. These results introduce a vital mechanism of hepatocarcinogenesis through the signaling of HBx- MALAT1/LTBP3 axis[131]. In addition, lncRNA MALAT1 act as a molecular sponge of miR-146b-5p to down-regulate its expression in HCC. MiR-146b-5p acts as tumor suppressive by inhibiting the phosphorylation of Akt mediated by TRAF6. As they found that TNF receptor associated factor 6 (TRAF6) is a direct target of miR-146b-5p in HCC. This proves that miR-146b-5p inhibits tumor growth and metastasis of HCC by targeting TRAF6 mediated Akt phosphorylation[132].
lncRNA MALAT1 plays a crucial role in tumor progression as study showed that patients with high expression level of malat-1 have high risk of tumor recurrence after liver transplant. Inhibition of MALAT1 would significantly decrease cell viability, motility, invasiveness, and increase the sensitivity to apoptosis[133]. MALAT1 is a proto-oncogene upregulated in HCC that acts by activation of Wnt pathway and promotion of the oncogenic splicing factor SRSF1. Induction of SRSF1 by MALAT1 modulates SRSF1 splicing targets, by modulating the alternative splicing of S6K1, promoting the production of antiapoptotic splicing isoforms and activating the mTOR pathway. Inhibition of SRSF1 expression or mTOR activity reduces the oncogenic properties of MALAT1, concluding that SRSF1 induction and mTOR activation may be vital for MALAT1-induced transformation[134]. Studies have indicated that the MALAT1 not only regulates tumorigenesis in HCC, but also controls cell cycle progression in hematopoietic cells. Researchers have found that MALAT1 plays different biological functions in hematopoietic cells, including the regulation of the cell cycle. MALAT1 was upregulated during liver regeneration. MALAT1 accelerated hepatocyte proliferation by enhancing cell cycle progression from the G1 to the S phase and inhibiting apoptosis in vitro. Moreover, MALAT1 was found to be regulated by p53 during liver regeneration, and hence, p53 may be a key regulator of MALAT1 activity. This study found that MALAT1 activated the Wnt/β-catenin pathway by inhibiting the expression of Axin1 and adenomatous polyposis coli (APC), causing expression promotion of cyclin D1. The results of this study suggest that MALAT1 is a crucial molecule for liver regeneration[135,136]
MEG-3
Maternally expressed gene 3 (MEG3) is the first lncRNA to be found to have tumor suppressor function, which is expressed in many human normal tissues[137]. Frequently, the expression levels of miR-26a and MEG3 were found to be downregulated in HCC tissues compared to non-malignant tissues. Upregulation of miR-26a markedly decreased the proliferation, invasion and migration of HCC cells. They thought that DNA methyltransferase 3b (DNMT3B) was a direct target gene of miR-26a. Overexpressed miR-26a suppressed the expression level of DNMT3B. Inhibited expression of DNMT3B showed similar tumor suppressive effects induced by miR-26a upregulation, and resulted in the upregulation of MEG3. Moreover, the expression levels of DNMT3B were upregulated in the HCC tissues and it was inversely correlated with miR-26a and MEG3 in HCC tissues. These findings demonstrated a new mechanism miR-26a/DNMT3B/MEG3 axis to HCC development[138].
A study confirmed that the TF, NF-κB, could bind to the MEG3 promoter region and might affect the transcription of MEG3. MEG3 overexpression would competitively “sponge” miRNA-664, relieving the inhibition of miR-664 on the transcription and translation of alcohol dehydrogenase 4 (ADH4) playing a role in inhibiting HCC cell proliferation[139]. P53 is an important tumor suppressor which is incorporated in preventing cancer through taking place as a transcription factor to regulate its target genes. In this study, MEG3 acts as a tumor suppressor in hepatoma cells through interacting with p53 protein to activate p53-mediated transcriptional activity and affect the expression of partial p53 target genes[140-143]. MEG3 regulates HCC cell proliferation and apoptosis partially through p53 accumulation.
Down-regulation of UHRF1, a new identified oncogene which is required for DNA methylation, caused MEG3 overexpression in HCC cell lines, which could be reversed by the up-regulation of UHRF1. Furthermore, overexpression of MEG3 in HCC cells partially abolished the promotion of proliferation induced by UHRF1. These data suggest that MEG3 functioning as a potential biomarker in predicting the prognosis of HCC, was regulated by UHRF1 via recruiting DNMT1 and regulated p53 expression. This means that UHRF1/DNMT1/MEG3/p53 axis signaling might be involved in HCC development[144]. Another study showed that enforced expression of MEG3 in HCC cells significantly decreased cell growth, and induced apoptosis. This study found that miR-29, which can modulate DNMT 1 and 3, could regulate MEG3 expression. Overexpression of mir-29a increased expression of MEG3. These findings show that methylation-dependent tissue-specific regulation of the lncRNA MEG3 by miR-29a may contribute to HCC growth and highlight the inter-relationship between two classes of non-coding RNA, miRNAs and lncRNAs[145]. Due to resistance of HCC to chemotherapy, this gives novel cancer treatment methods an overwhelming significance. Epigenetic therapy in cancer is useful in reversing some of the cancer defects because of reversibility of the epigenetic alterations. A study found that the “Dendrosomal curcumin” DNC dependent overexpression of miR-29a and miR-185 can decrease the expression of DNMT1, 3A and 3B and hence increases the expression of MEG3. So, DNC may enhance DNA hypomethylation and re-expression of silenced tumor suppressor genes in HCC. These findings suggest that DNC could be an effective choice for epigenetic therapy of HCC[146].
Actin filament associated protein 1-antisense RNA 1
lncRNA actin filament associated protein 1-antisense RNA 1 (AFAP1-AS1) is significantly upregulated in HCC patients with association to poor prognosis. Its knockdown inhibits cellular proliferation, invasion and metastasis and increased apoptosis in vivo[147]. Moreover, AFAP1-AS1 reported to induce cellular proliferation and invasion and inhibit apoptosis by the upregulation of RhoA/Rac2 pathway in the cell cycle, MMP-9 and PCNA protein levels and apoptotic indexes cyclinD1 along with the downregulation of Bax[148].
ANRIL (CDKN2B-AS)
A study results showed that , lncRNA ANRIL could serve as a potential therapeutic target as lncRNA ANRIL expression is significantly increased in HCC tissues and strongly correlated with advanced clinical features, and the down-regulated expression of lncRNA ANRIL could inhibit HCC cells proliferation, migration and invasion[149]. Another study showed that its overexpression may be an important factor for HCC progression, through silencing of Kruppel-like factor 2 (KLF2) by binding with PRC2[150].
ATB
lncRNA-ATB enhanced the expression of ZEB1 and ZEB2 by competitive binding to the miR-200 family and hence, induced EMT and invasion. Furthermore, lncRNA-ATB stimulated organ colonization of disseminated tumor cells by binding IL-11 mRNA, autocrine induction of IL-11, and triggering STAT3 signalling. Generally, lncRNA-ATB promotes the invasion-metastases cascade[151].
Colon cancer associated transcript 1
Colon cancer associated transcript 1 (CCAT1) is found to be overexpressed in HCC patients and could promote cellular proliferation and migration in tumor cells in vitro[152]. Additionally, CCAT1’s overexpression in HCC patients is regulated by c-myc expression that binds to the E-box of CCAT1 promoter region, increasing it expression and cellular proliferation and tumorigenesis effect[153]. In an interesting study, CCAT1 is found to exert its effect in HCC through acting as a molecular sponge for let-7, inhibiting its functions and restoring the levels of HMGA2 and c-myc hence enhancing cell proliferation and migration[154]. CCAT1 regulated CDK1 expression through functioning as a ceRNA and inhibiting miR-490-3p in HCC hence promoting cellular proliferation and invasion[155].
CCAT2
CCAT2 is referred to as an oncogene as it is overexpressed in HCC, promoting cellular proliferation, invasion and metastasis while inhibiting apoptosis[156]. The upregulation of CCAT2 present in HCC patients induces tumor growth and invasion via enhancing EMT through regulating its factors, increasing vimentin and Snail2 expressions while decreasing E-cadherin expression hence accelerating HCC progression[157].
DANCR
DANCR is upregulated in HCC tumor tissue and plasma levels of patients with better diagnostic value and accuracy than alpha fetoprotein (AFP). High levels of DANCR promotes microvascular invasion and metastasis and its knockdown inhibits cellular proliferation and metastasis through inhibiting β-catenin signaling pathway[158]. DANCR is also found to increase the stem-like characters of HCC cells aiding tumorigenesis, proliferation and invasion via the interaction with CTNNB1 which reverses the inhibitory effect miR-214, miR-320a and miR-199a exerted on CTNNB1[159].
EGFR
It was found the EGFR-AS1 was up regulated in HCC, furthermore it was determined that it promotes HCC development by improving the ability of invasion and proliferation of HCC cell, Beside the fact that it affects the cell cycle. To sum up, EGFR-AS1 may act as a prognostic factor in HCC and , on the other hand, it was observed that the inhibition of EGFR-AS1 in HCC cells significantly impeded cells proliferation and invasion in vivo which might provide a potential possibility for targeted therapy for HCC[160]. Another study showed that lnc-epidermal growth factor receptor (EGFR) up regulation in tergs correlates positively with the tumor size and expression of EGFR/Foxp 3, but negatively with INF-y expression in patients and xenografted mouse models. Lnc-EGFR stimulates treg differentiation, suppression CTL activity and promotes HCC growth in an EGFR-dependent manner. Lnc-EGFR links an immunosuppressive state to cancer by promoting treg cell differentiation, thus offering a potential therapeutic target for HCC[161].
Five prime to XIST
Lnc-FTX (five prime to XIST) represses Wnt/β-catenin signaling activity by competitively sponging miR-374a and inhibits HCC cell EMT and invasion. Furthermore, lnc-FTX binds to the DNA replication licensing factor MCM2, impeding DNA replication and inhibiting proliferation in HCC cells. These findings suggest that lnc-FTX can function as a tumor suppressor in HCC by physically binding miR-374a and MCM2[162]. A study showed that the lncRNA Ftx exerted the oncogenic function via miR-545. As RIG-I is a downstream mediator of miR-545 function in HCC. Additionally, lncRNA Ftx or miR-545 caused upregulation of cell cycle regulator Cyclin D1 and downregulation of p27 by activation of PI3K/Akt. These findings suggest that lncRNA Ftx/miR-545 axis promotes HCC progression by activation of Akt signaling pathway by targeting RIG-I [163].
GAS5
A study showed that lncRNA GAS5 was down-regulated in the HCC, The aberrant expression of GAS5 was linked to a poor prognosis[164]. Another study showed that the expression level of GAS5 is significantly downregulated in HCC tissues compared to adjacent normal controls. GAS5 overexpression decreases hepatoma cell proliferation and invasion, promoted the apoptosis of hepatoma cells, downregulated the vimentin level and upregulated E-cadherin level in hepatoma cells[165]. Overexpression of GAS5 decreases the migration and invasion of HCC cells and high expression of miR-21 eliminates GAS5-mediated suppression of HCC cell migration and invasion.This mean that GAS5 acts as a tumor suppressor in HCCs through negative regulation of miR-21[166].
H19
lncRNA H19 is paternally imprinted and maternally expressed[167], and is located on chromosome 11p15.5, which is always linked to various diseases and higher incidence of tumorigenesis. The studies have proven that LncRNA H19 acts as an oncogene or a suppressor gene. lncRNA H19 promotes bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression[168]. In contrast, H19 acts as tumor suppressor, lncRNA H19 may suppress HCC metastasis through enhancing hnRNPU/PCAF/RNAPol II, and activating miR-200 family[169]. Moreover, another study showed that inhibition of lncRNA H19 and miR-675 may promote migration and invasion of HCC cells through activating the AKT/ GSK-3β/Cdc25A signaling pathway[170].
HOXA transcript at the distal TIP
The upregulation of HOXA transcript at the distal TIP (HOTTIP) in HCC could be a result of loss in miR-125b, upregulation in HOXA13, downregulation of miR-192 and miR-204 or several other interactions with ncRNAs[171]. HOTTIP is discovered to be significantly upregulated in HCC patients with positive relation to levels of HOXA13. This increased loop of HOTTIP/HOXA13 enhances cell proliferation and metastasis in HCC patients[172]. Furthermore, HOTTIP is shown to be the downstream target of miR-192 and miR-204 which in turn positively modulate glutaminase (GLS1). miR-192 and miR-204 negatively regulate HOTTIP hence inhibiting GLS1 mediated glutaminolysis which interrupts HCC proliferation and cell viability[173]. The knockdown of HOTTIP caused by overexpression of miR-125b inhibited HCC proliferation, migration and tumorigenicity along with some HOXA gene expression[174].
Nuclear enriched abundant transcript 1
Nuclear enriched abundant transcript 1 (NEAT1) was concluded to be upregulated in HCC patients accelerating cellular proliferation, metastasis and vaso-invasion. Additionally, the high levels were associated with the expression of MTDH, NM23 and MALAT1[175]. The expression of NEAT1 in HCC patients was significantly increased which promoted cell viability and inhibited apoptosis. The mechanism underlying this effect could be the inhibition of miR-129-5p by NEAT1 through regulating VCP/ IκB which are the downstream targets of the miRNA[176]. The high levels of NEAT1 in HCC patients promote cellular proliferation and migration through binding to U2AF65 and regulating hnRNP A2 expression[177].
Long intergenic non-coding RNA 00152
Long intergenic non-coding RNA 00152 (linc00152) is significantly upregulated in HCC patients with fine diagnostic accuracy between HCC samples and control[103]. Linc00152 promotes cellular proliferation and tumor growth through the phosphorylation and activation of the mechanistic target of rapamycin(mTOR) signaling pathway by binding to EpCAM promoter and upregulating its expression[178].
P21 (CDK-interacting protein 1)
lincRNA-p21 is one of the lncRNAs, which contains longer than 200 nucleotides. It is initially identified as a direct transcriptional target of p53 and acts as a translational suppressor by direct binding to the target mRNA[179]. A study showed that overexpression of lincRNA-p21 led to downregulation of Notch signal-related proteins Hes-1 and NICD and the expression of EMT-related proteins E-cadherin and Claudin-1 was increased, and the expression of proteins N-cadherin and snail was reduced causing inhibition of invasion[180]. Another study showed that lincRNA-p21 negatively regulated miR-9 expression level, and miR-9 was upregulated in human HCC tissues and cells. The knock down of miR-9 suppressed HCC migration and invasion in vitro, since E-cadherin was a direct target of miR-9, the expression level of E-cadherin was found to be regulated by lincRNA-p21 and miR-9. The data suggested that lincRNA-p21 inhibits migration and invasion of HCC by regulating miR-9-mediated E-cadherin cascade signaling pathway[181].
PCAT-1
A study showed that overexpression of PCAT1 led to increase cell proliferation and migration, and inhibit apoptosis. Findings suggest that PCAT1 acts as an tumor promotor in HCC and silencing PCAT-1 may be a potential novel therapeutic strategy for HCC[182]. Another study suggests that the increased expression of PCAT-1 was correlated with advanced clinical parameters and poor overall survival of HCC patients, indicating that PCAT-1 up-regulation may serve as a novel biomarker of poor prognosis in HCC patients[183].
PVT1 (Pvt1 oncogene)
A study on serum lncRNA for diagnosis of HCC concluded that PVT1 and uc002mbe.2 in combination served as an accurate diagnostic biomarker that can be associated with tumor size and BCLC value with a predictive ability better than AFP[184]. Human PVT1 is upregulated in HCC tissues and is associated with poor prognosis, enhanced cellular proliferation and migration which is achieved through binding and stabilizing NOP2, a cell cycle gene, all under the control of TGF-β1[185]. As an oncogene, PVT1 enhances tumor growth and migration through functioning as an endogenous sponge for miR-186-5p abolishing its inhibitory action on YAP1, restoring its upregulation in HCC hence promoting tumorigenesis[186].
SNHG1
SNHG1 overexpression in HCC is correlated with large tumor size, poor differentiation and poor prognosis. SNHG1 promoted cell proliferation and inhibited cell apoptosis through the inhibitory action on p53 and p53 target genes[187]. SNHG1 is found to be significantly upregulated in HCC aiding in cellular proliferation, invasion a migration in HepG2 cells which is achieved through the possible inhibition of miR-195 proving to be one of its downstream targets[188].
SNHG6
SNHG6 is reported to be among the dysregulated genes in HCC and its expression is related to co-expressed genes on 8q involved in structural integrity of ribosome and translation[189]. Among various SNHG6 transcripts, SNHG6-003 only showed to be oncogenic enhancing cell proliferation and drug resistance. SNHG6-003 functions as a ceRNA, acting as a sponge for miR-26a and miR-26b inhibiting their functions hence, modulating the levels of transforming growth factor-β-activated kinase 1(TAK1). Additionally, SNHG6-0003 shows a significant co-expression pattern with TAK1 as both are upregulated in various kinds of cancers[190].
SNHG20
SNHG20 is significantly upregulated in HCC patients and positively correlated to tumor size, cellular proliferation, invasion and poor prognosis. Its knockdown results in remarkable inhibition of cellular proliferation and invasion in SK-Hep-1 cells[191]. The consequence of high levels of SNHG20 on HCC cells occurs due to the binding of SNHG20 to the enhancer of zeste homolog 2 (EZH2) that regulates E-cadherin expression along with SNHG20’s ability to modulate ZEB1, ZEB2, N-cadherin and vimentin in a positive relation[192].
Sprouty 4-intron transcript 1
Sprouty 4-intron transcript (SPRY4-IT) was significantly increased in HCC cells, and overexpression of SPRY4-IT1 can promote cell proliferation and increase the cell invasion ability via promoting EMT progression and interacting with Estrogen Related Receptor α (ERR-α)[193]. Moreover, it can suppress Twist1 and vimentin expression and increase E-cadherin expression[194].
The taurine upregulated gene 1
The taurine upregulated gene 1 (TUG1) many studies have found that TUG1 plays vital roles in many human cancers, such as HCC, osteosarcoma, glioma and bladder cancer[195-198]. A study showd that TUG1 was up-regulated in HCC tissues than that in corresponding non-tumor tissues. The findings suggest that TUG1 upregulation was induced by nuclear transcription factor SP1 and TUG1 can regulate KLF2 expression in the epigenetic level by binding to PRC2[195].
TCF7
LncRNA TCF7, as a T cell factor, is well known for its function in T lymphocyte development and multipotential hematopoietic cell self-renewal[199]. A study found that IL-6 transcriptionally activated the expression of lncTCF7 in HCC cells by activating STAT3, a transcription activator which binds to promoter regions of lncTCF7. Moreover, STAT3 knock down reduced lncTCF7 expression. Importantly, RNA interference-based attenuation of lncTCF7 prevented IL-6-induced EMT and cell invasion. These findings highlights the existence of an aberrant IL-6/STAT3/lncTCF7 signaling axis that leads to HCC aggressiveness through EMT induction[200]. Another study showed that lncTCF7 can promote liver CSC self-renewal and tumor progression by activation of Wnt signaling through recruiting the SWI/SNF complex to the TCF7 promoter[201].
UCA1
A study indicated that UCA1 expression may potentially promote the EMT in HCC, by upregulating the expression of Snail2. MiR-203 was a target of UCA1 and UCA1 upregulated Snail2 by negatively regulating miR-203. This indicated a novel UCA1/miR203/Snail2 signaling pathway regulatory network in HCC[202]. A study discovered that UCA1, upregulated by HBx, displayed a crucial role in G1/S transition in both hepatic and hepatoma cells. Moreover, a positive correlation between the expression of UCA1 and HBx and a negative correlation between UCA1 and p27 were observed in HCC specimens, suggesting the significance of UCA1 in HBx-mediated hepatocarcinogenesis. Also, they demonstrated that UCA1 repressed p27 expression at least partly through associating with chromatin-modifying complexes PRC2 component EZH2 in HCC cells. So, they provided a significant mechanism of hepatocarcinogenesis through the signaling of HBx-UCA1/EZH2-p27Kip1 axis[203]. UCA1 acts as an oncogene by promoting malignant progression of human HCC, UCA1 acts as an endogenous sponge to reduce miR-216b expression, resulting in derepression of FGFR1 expression and activation of FGFR1/ERK signaling pathway in HCC, providing a novel signaling pathway UCA1-miR-216b-FGFR1-ERK in HCC[204].
XIST
During previous studies, it is concluded that XIST and its activator JPX are downregulated in HCC patients and their levels serve as a biomarker of poor prognosis[205]. XIST modulates the regulation of miR-181a on PTEN as siXIST enhances cellular proliferation and migration in HCC patients via binding to miR-181a at multiple binding sites[206]. XIST functions as a tumor suppressor through interacting with miR-92b and inhibiting its miR-92b/Smad7 axis hence inhibiting its oncogenic effect on HCC cells[207]. On the contrary to previous studies, XIST was reported upregulated in HCC patients associated with poor prognosis and enhanced tumorigenesis. This is achieved through activation of AKT signaling via modulation of miR-139-5p/PDK1 axis as XIST has a reciprocal repression with miR-139-5p[208].
INTERPLAY OF lncRNAs WITH miRNAs
Emerging evidences proved an intensive interaction between lncRNAs and microRNAs as summarized in Table 1.
Table 1.
Interplay between long non-coding RNAs and miRNAs
| lncRNA | miRNA | Effect/ relationship | Pathway |
| HULC | miR-6825-5p, miR-6845-5p, miR-6886-3p | Inhibitory | Increasing chemotherapy sensitivity through USP22 and sirt1 levels controlling autophagy |
| miR-200a-3p | Inhibitory | Increasing ZEB1 expression, EMT and metastasis | |
| miR-107 | Inhibitory | Upregulates E2F1, activates SPHK1 and angiogenesis | |
| miR-203 | Inhibitory | Inhibits cellular proliferation and induces apoptosis | |
| miR-9 (ZNFX1-AS1) | Inhibitory | Positive feedback loop of HULC/miR-9/PPARA/ACSL1/HULC for lipid metabolism regulation | |
| miR-372 (PCAT-14) | Inhibitory | Auto-regulatory loop of CREB/HULC/miR-372/PRKACB/CREB/HULC enhancing cellular proliferation | |
| XIST | miR-181a | Inhibitory | Binds to several binding sites on miR-181a modulating its function |
| miR-92b | Inhibitory | Suppresses miR-92b/Smad7 oncogenic axis | |
| miR-139-5p | Inhibitory | Modulate the miR-139-5p/PDK1/AKT axis | |
| CCAT1 | Let-7 | Inhibitory | Restoring HMGA2 and c-myc levels and enhancing cell proliferation |
| miR-490-3p | Inhibitory | Regulate CDK1 and HCC proliferation | |
| AF113014 | miR-20a | Interaction | Regulate Erg2 expression |
| CRNDE | miR-384 | Inhibitory | Enhances expression of NF-κB and p-AKT |
| DANCR | miR-214, miR-320a, miR-199a | Cancels its effect | Cancels the inhibitory effect they exert on CTNNB1 hence enhancing tumorigenesis |
| FTX | miR-374a | inhibitory | represses Wnt/β-catenin signaling pathway |
| miR-545 | interaction | Tumor promotor through activation of PI3K/Akt | |
| GAS5 | miR-21 | inhibitory | downregulated the vimentin level and the upregulated E-cadherin level |
| GIHCG | miR-200a/b/429 | Inhibitory | Associates with EZH2 and silences miRNA |
| H19 | miR-200 | activation | tumor-suppressive by mediating hnRNPU/PCAF/RNAPol II |
| miR-675 | inhibitory | Activation of the AKT/ GSK-3β/Cdc25A signaling pathway | |
| HOTAIR | miR-1 | Inhibitory | |
| miR-218 | Inhibitory | Inhibits P16(lnk4a) and P14(ARF) expression, activates Bmi-1 and tumorigenesis | |
| HOTTIP | miR-192, miR-204 | Negative | Inhibit HOTTIP expression, GLS1 and HCC proliferation |
| miR-125b | Negative | Inhibits HOTTIP expression and HOXA genes | |
| Linc00052 | miR-128, miR-485-3p | Complementary base pairing | Modulating NTRK3 expression |
| Linc-ROR | miR-145 | Inhibitory | Enhances HIF-1α/PDK1 expression |
| MALAT-1 | miR-143-3p | inhibitory | regulated ZEB1 expression |
| miR-146b-5p | inhibitory | targeting TRAF6 mediated Akt phosphorylation | |
| MEG-3 | miR-29a | interaction | modulate DNMT 1 and 3 |
| miR-26a | Negative | Modulates miR-26a/DNMT3B/MEG3 | |
| NEAT1 | miR-129-5p | Inhibitory | Inhibits the miRNA through regulating VCP/IκB, its downstream pathway |
| lincRNA-p21 | miR-9 | inhibitory | Tumor suppressive |
| PCAT-14 | miR-372 (HULC) | Inhibitory | Regulates ATAD2 and hedgehog pathway |
| PTENP1 | miR-17, miR-19b, miR-20a | Inhibitory | Inhibiting PIK3T/AKT pathway and inducing autophagy |
| PVT1 | miR-186-5p | Inhibitory | Restores YAP1 expression levels |
| SNHG1 | miR-195 | Inhibitory | |
| SNHG6 | miR-26a, miR-26b | Inhibitory | Modulate TAK1 expression |
| SNHG12 | miR-199a/b-5p | Inhibitory | ceRNA, Enhancing MLK3 expression and NF-κB pathway |
| TP73-AS1 | miR-200a | Inhibitory | miR-200a dependent HMGB1/RAGE regulation |
| UCA1 | miR-203 | interaction | UCA1/miR203/Snail2 signaling pathway regulatory network |
| miR-216b | inhibitory | activation of FGFR1/ERK signaling pathway | |
| Unigene56159 | miR-140-5p | Inhibitory | ceRNA, Restoring slug expression and EMT |
| ZNFX1-AS1 | miR-9 | Positive | Tumor suppression |
HOTTIP: HOXA transcript at the distal TIP; lncRNAs: Long non-coding RNAs; PCAT: Prostate cancer-associated transcript; HULC: Highly up-regulated in liver cancer; XIST: X-inactive specific transcript; TCF7: Transcription factor 4; TUG1: Taurine upregulated gene 1; EGFR: Epidermal growth factor receptor; CCAT1: Colon cancer associated transcript 1; MALAT1: Metastasis-associated lung adenocarcinoma transcript 1; UCA1: Urothelial carcinoma-associated 1; SPRY4-IT: Sprouty 4-intron transcript; SNHG: Small nucleolar rna host gene.
Emerging studies have demonstrated the manipulation of lncRNAs expression in different models. Mice models as well as in vivo and in vitro models are summarized in Table 2.
Table 2.
Different models used in studies
| lncRNA | Study | Model | Ref. |
| HULC | Molecular mechanism of HEIH and HULC in the proliferation and invasion of hepatoma cells. | In vitro | [104] |
| lncRNA HULC promotes the growth of hepatocellular carcinoma cells via stabilizing COX-2 protein | In vitro | [105] | |
| lncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. | In vitro and in vivo (nude mice) | [106] | |
| lncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. | In vitro and in vivo (nude mice) | [107] | |
| lncRNA HULC promotes tumor angiogenesis in liver cancer by up-regulating sphingosine kinase 1 (SPHK1) | In vitro and in vivo (nude mice) | [108] | |
| Specificity protein (Sp) transcription factors and metformin regulate expression of the long non-coding RNA HULC | In vitro | [109] | |
| miR-203 suppresses the proliferation and metastasis of hepatocellular carcinoma by targeting oncogene ADAM9 and oncogenic long non-coding RNA HULC. | In vitro | [110] | |
| Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. | In vitro and in vivo (nude mice) | [111] | |
| Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. | In vitro | [112] | |
| HOTAIR | Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. | In vitro | [117] |
| Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. | In vitro | [118] | |
| Long non-coding RNA HOTAIR is a marker for hepatocellular carcinoma progression and tumor recurrence. | In vitro and in vivo (nude mice) | [119] | |
| Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression | In vitro | [120] | |
| The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma | In vitro | [121] | |
| LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2 | In vitro and in vivo (mice) | [122] | |
| HOTAIR, a long non-coding RNA driver of malignancy whose expression is activated by FOXC1, negatively regulates miRNA-1 in hepatocellular carcinoma. | In vitro and in vivo (mice) | [123] | |
| Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. | In vitro and in vivo (mice) | [125] | |
| Long non-coding RNA HOTAIR promotes cell migration and invasion via down-regulation of RNA binding motif protein 38 in hepatocellular carcinoma cells. | In vitro | [126] | |
| SNHG | SNHG3 correlates with malignant status and poor prognosis in hepatocellular carcinoma. | In vitro | [80] |
| Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. | In vitro | [81] | |
| Long noncoding RNA SNHG15, a potential prognostic biomarker for hepatocellular carcinoma | In vitro | [82] | |
| Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis | In vitro | [187] | |
| Expression of Long Non-Coding RNA (lncRNA) Small Nucleolar RNA Host Gene 1 (SNHG1) Exacerbates Hepatocellular Carcinoma Through Suppressing miR-195 | In vitro | [188] | |
| The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. | In vitro | [190] | |
| Up-regulation of LncRNA SNHG20 Predicts Poor Prognosis in Hepatocellular Carcinoma. | In vitro | [191] | |
| Long non-coding RNA SNHG20 predicts a poor prognosis for HCC and promotes cell invasion by regulating the epithelial-to-mesenchymal transition. | In vitro | [192] | |
| MALAT1 | Long Non-Coding RNA MALAT1 Regulates ZEB1 Expression by Sponging miR-143-3p and Promotes Hepatocellular Carcinoma progression. | In vitro | [130] |
| HBx-related long non-coding RNA MALAT1 promotes cell metastasis via up-regulating LTBP3 in hepatocellular carcinoma. | In vitro and in vivo (Male BALB/C nude mice) | [131] | |
| Down-regulation of miR-146b-5p by long noncoding RNA MALAT1 in hepatocellular carcinoma promotes cancer growth and metastasis. | In vitro and in vivo (nude mice) | [132] | |
| Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. | In vitro | [133] | |
| MEG3 | Armored long non-coding RNA MEG3 targeting EGFR based on recombinant MS2 bacteriophage virus-like particles against hepatocellular carcinoma. | In vitro and in vivo (BALB/c nude mice) | [137] |
| MicroRNA-26a inhibits proliferation and metastasis of human hepatocellular carcinoma by regulating DNMT3B-MEG3 axis. | In vitro | [138] | |
| Overexpression of Long Non-Coding RNA MEG3 Inhibits Proliferation of Hepatocellular Carcinoma Huh7 Cells via Negative Modulation of miRNA-664. | In vitro | [139] | |
| Long Noncoding RNA MEG3 Interacts with p53 Protein and Regulates Partial p53 Target Genes in Hepatoma Cells | In vitro | [141] | |
| The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma. | In vitro | [144] | |
| microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. | In vitro | [145] | |
| AFAP1-AS1 | Long noncoding RNA AFAP1-AS1 indicates a poor prognosis of hepatocellular carcinoma and promotes cell proliferation and invasion via upregulation of the RhoA/Rac2 signaling. | In vitro and in vivo (mice) | [148] |
| Critical role for the long non-coding RNA AFAP1-AS1 in the proliferation and metastasis of hepatocellular carcinoma. | In vitro and in vivo (mice) | [147] | |
| ANRIL | High expression of long non-coding RNA ANRIL is associated with poor prognosis in hepatocellular carcinoma. | In vitro | [149] |
| ATB | A long noncoding RNA activated by TGF-beta promotes the invasion metastasis cascade in hepatocellularcarcinoma. | In vitro and in vivo (nude mice) | [151] |
| CCAT1 | Aberrant Expression of CCAT1 Regulated by c-Myc Predicts the Prognosis of Hepatocellular Carcinoma. | In vitro | [153] |
| CCAT1 promotes hepatocellular carcinoma cell proliferation and invasion. | In vitro | [152] | |
| Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. | In vitro | [154] | |
| CCAT2 | Long non-coding RNA CCAT2 is associated with poor prognosis in hepatocellular carcinoma and promotes tumor metastasis by regulating Snail2-mediated epithelial-mesenchymal transition. | In vitro | [157] |
| Long non-coding RNA CCAT2 functions as an oncogene in hepatocellular carcinoma, regulating cellular proliferation, migration and apoptosis | In vitro | [156] | |
| DANCR | Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. | In vitro and in vivo (mice) | [159] |
| DANCR Acts as a Diagnostic Biomarker and Promotes Tumor Growth and Metastasis in Hepatocellular Carcinoma | In vitro and in vivo (mice) | [158] | |
| EGFR | The long noncoding RNA, EGFR-AS1, a target of GHR, increases the expression of EGFR in hepatocellular carcinoma. | In vitro and in vivo | [160] |
| FTX | Ftx non coding RNA-derived miR-545 promotes cell proliferation by targeting RIG-I in hepatocellular carcinoma. | In vitro and in vivo (nude mice) | [163] |
| GAS-5 | Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. | In vitro | [165] |
| Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. | In vitro | [164] | |
| H19 | Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. | In vitro and in vivo (Nude mice) | [169] |
| HOTTIP | Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. | In vivo (mice) | [174] |
| MiRNA-192 [corrected] and miRNA-204 Directly Suppress lncRNA HOTTIP and Interrupt GLS1-Mediated Glutaminolysis in Hepatocellular Carcinoma. | In vitro and in vivo (mice) | [173] | |
| Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. | In vitro | [172] | |
| Linc00152 | LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. | In vitro and in vivo (mice) | [178] |
| NEAT1 | Long non-coding RNA NEAT1 promotes hepatocellular carcinoma cell proliferation through the regulation of miR-129-5p-VCP-IκB. | In vitro and in vivo (mice) | [176] |
| Long noncoding RNA NEAT1 promotes cell proliferation and invasion by regulating hnRNP A2 expression in hepatocellular carcinoma cells. | In vitro and in vivo (mice) | [177] | |
| P21 | lincRNA-p21 inhibits invasion and metastasis of hepatocellular carcinoma through Notch signaling-induced epithelial-mesenchymal transition. | In vitro and in vivo (nude mice) | [180] |
| LincRNA-p21 inhibits invasion and metastasis of hepatocellular carcinoma through miR-9/E-cadherin cascade signaling pathway molecular mechanism. | In vitro | [181] | |
| PCAT1 | Upregulation of long non coding RNA PCAT-1 contributes to cell proliferation, migration and apoptosis in hepatocellular carcinoma. | In vitro | [182] |
| Prognostic significance of long non-coding RNA PCAT-1 expression in human hepatocellular carcinoma. | In vitro | [183] | |
| PRAL | Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma | In vitro and in vivo (mice) | [44,209] |
| PVT1 | Long non-coding RNA PVT1 serves as a competing endogenous RNA for miR-186-5p to promote the tumorigenesis and metastasis of hepatocellular carcinoma. | In vitro | [186] |
| Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. | In vitro and in vivo (mice) | [185] | |
| SPRY4-IT1 | Overexpression of the long non-coding RNA SPRY4-IT1 promotes tumor cell proliferation and invasion by activating EZH2 in hepatocellular carcinoma | In vitro | [210] |
| TCF7 | Long noncoding RNA lncTCF7, induced by IL-6/STAT3 transactivation, promotes hepatocellular carcinoma aggressiveness through epithelial-mesenchymal transition | In vitro | [200] |
| The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. | In vitro | [201] | |
| TUG1 | Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. | In vitro | [195] |
| UCA1 | HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling | In vitro | [203] |
| Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. | In vitro and in vivo (Mice) | [204] | |
| XIST | Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. | In vitro | [206] |
| MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST | In vitro and in vivo (mice) | [207] | |
| Long non-coding RNA XIST promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma. | In vitro and in vivo (mice) | [208] |
HOTTIP: HOXA transcript at the distal TIP; lncRNAs: Long non-coding RNAs; PCAT: Prostate cancer-associated transcript; HULC: Highly up-regulated in liver cancer; XIST: X-inactive specific transcript; TCF7: Transcription factor 4; TUG1: Taurine upregulated gene 1; EGFR: Epidermal growth factor receptor; CCAT1: Colon cancer associated transcript 1; MALAT1: Metastasis-associated lung adenocarcinoma transcript 1; UCA1: Urothelial carcinoma-associated 1; SPRY4-IT: Sprouty 4-intron transcript; SNHG: Small nucleolar rna host gene.
CONCLUSION
In conclusion, despite the various unanswered questions about ncRNAs, specifically lncRNAs; their role in HCC has been exhaustively researched. This research reached to a conclusion that lncRNA act as an important regulator in various biological processes rendering its dysregulation a factor in various diseases, including HCC. HCC-related lncRNAs play a crucial role in tumor initiation, progression and treatment. Their functions could be exploited for diagnosis as well as treatment giving them an exceptional potential in targeted therapy of HCC. During the years, various lncRNAs have been studied and several molecular mechanisms have been understood, however, further researches need to be done to enlighten more research calibers about lncRNAs giving it the distinction it deserves to intrigue the biopharmaceutical field. During this review, certain lncRNAs such as HULC, HOTAIR, MALAT1 and MEG3 showed extensive roles related to HCC tumorigenesis. Moreover, in this review, the focus was showing the interplay between lncRNAs and other ncRNAs contributing to HCC progression or treatment. Complex pathways have been discovered that included lncRNAs affecting miRNAs and proteins aiding or suppressing HCC. These pathways are good candidates for targeted therapy in HCC patients. Moreover, the serum and tissue levels of lncRNAs serve as an accurate diagnostic marker for HCC patients that could overcome AFP’s accuracy in the future. Finally, all the work that is being done adds to the functional benefits of lncRNAs and the beneficial roles in diagnosis, treatment and prognosis of HCC.
Footnotes
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Egypt
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
Conflict-of-interest statement: The authors have no conflict of interest to report.
Peer-review started: November 13, 2017
First decision: December 13, 2017
Article in press: January 23, 2018
P- Reviewer: Giorgio A, Grassi G, Namisaki T, Ohkoshi S S- Editor: Gong ZM L- Editor: A E- Editor: Huang Y
Contributor Information
Aya El Khodiry, Genetic Pharmacology Research Group, Clinical Pharmacy Unit, Department of Pharmacology and Toxicology, Faculty of Pharmacy and Biotechnology, German University in Cairo, Cairo 11835, Egypt.
Menna Afify, Genetic Pharmacology Research Group, Clinical Pharmacy Unit, Department of Pharmacology and Toxicology, Faculty of Pharmacy and Biotechnology, German University in Cairo, Cairo 11835, Egypt.
Hend M El Tayebi, Genetic Pharmacology Research Group, Clinical Pharmacy Unit, Department of Pharmacology and Toxicology, Faculty of Pharmacy and Biotechnology, German University in Cairo, Cairo 11835, Egypt. hend.saber@guc.edu.eg.
References
- 1.Ledford H. Language: Disputed definitions. Nature. 2008;455:1023–1028. doi: 10.1038/4551023a. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 4.Weinhold B. Epigenetics: the science of change. Environ Health Perspect. 2006;114:A160–A167. doi: 10.1289/ehp.114-a160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen T, Li E. Structure and function of eukaryotic DNA methyltransferases. Curr Top Dev Biol. 2004;60:55–89. doi: 10.1016/S0070-2153(04)60003-2. [DOI] [PubMed] [Google Scholar]
- 6.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 7.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 8.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 9.Fatemi M, Hermann A, Gowher H, Jeltsch A. Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur J Biochem. 2002;269:4981–4984. doi: 10.1046/j.1432-1033.2002.03198.x. [DOI] [PubMed] [Google Scholar]
- 10.Ma L, Chua MS, Andrisani O, So S. Epigenetics in hepatocellular carcinoma: an update and future therapy perspectives. World J Gastroenterol. 2014;20:333–345. doi: 10.3748/wjg.v20.i2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 14.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 15.Gioia J. Epigenetics. Peanuts a Biotechnical Newsletter. 2009;9:3–5. [Google Scholar]
- 16.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15 Spec No 1:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 17.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 18.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 21.Pasmant E, Laurendeau I, Héron D, Vidaud M, Vidaud D, Bièche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 22.Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G, Zhang H, Wan X, Yang X, Zhu C, Wang A, He L, Miao R, Chen S, Zhao H. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed Res Int. 2014;2014:780521. doi: 10.1155/2014/780521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parasramka MA, Maji S, Matsuda A, Yan IK, Patel T. Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol Ther. 2016;161:67–78. doi: 10.1016/j.pharmthera.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartonicek N, Maag JL, Dinger ME. Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hauptman N, Glavač D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655–4669. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Wu Z, Fu X, Han W. Long Noncoding RNAs: Insights from Biological Features and Functions to Diseases. Med Res Rev. 2013;33:517–553. doi: 10.1002/med.21254. [DOI] [PubMed] [Google Scholar]
- 28.Gong Z, Zhang S, Zhang W, Huang H, Li Q, Deng H, Ma J, Zhou M, Xiang J, Wu M, et al. Long non-coding RNAs in cancer. Sci China Life Sci. 2012;55:1120–1124. doi: 10.1007/s11427-012-4413-9. [DOI] [PubMed] [Google Scholar]
- 29.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang X, Xiao R, Pan S, Yang X, Yuan W, Tu Z, Xu M, Zhu Y, Yin Q, Wu Y, et al. Uncovering the roles of long non-coding RNAs in cancer stem cells. J Hematol Oncol. 2017;10:62. doi: 10.1186/s13045-017-0428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48:R45–R53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu J, Song C, Duan B, Zhang X, Li D, Zhu L, Gao H. LncRNA-SVUGP2 suppresses progression of hepatocellular carcinoma. Oncotarget. 2017;8:97835–97850. doi: 10.18632/oncotarget.18279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng T, Wang D, Chen J, Tian Y, Cai X, Peng H, Zhu L, Huang A, Tang H. LncRNA-AF113014 promotes the expression of Egr2 by interaction with miR-20a to inhibit proliferation of hepatocellular carcinoma cells. PLoS One. 2017;12:e0177843. doi: 10.1371/journal.pone.0177843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni W, Zhang Y, Zhan Z, Ye F, Liang Y, Huang J, Chen K, Chen L, Ding Y. A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J Hematol Oncol. 2017;10:91. doi: 10.1186/s13045-017-0449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ge Z, Cheng Z, Yang X, Huo X, Wang N, Wang H, Wang C, Gu D, Zhao F, Yao M, et al. Long noncoding RNA SchLAH suppresses metastasis of hepatocellular carcinoma through interacting with fused in sarcoma. Cancer Sci. 2017;108:653–662. doi: 10.1111/cas.13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo P, Jing W, Zhu M, Li ND, Zhou H, Yu MX, Liang CZ, Tu JC. Decreased expression of LncRNA SRA1 in hepatocellular carcinoma and its clinical significance. Cancer Biomark. 2017;18:285–290. doi: 10.3233/CBM-160305. [DOI] [PubMed] [Google Scholar]
- 37.Huang R, Wang X, Zhang W, Zhangyuan G, Jin K, Yu W, Xie Y, Xu X, Wang H, Sun B. Down-Regulation of LncRNA DGCR5 Correlates with Poor Prognosis in Hepatocellular Carcinoma. Cell Physiol Biochem. 2016;40:707–715. doi: 10.1159/000452582. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L, He T, Yan Y, Zhang Y, Zhou X, Huang P, Kong Y, Xie M, Zhang L, Sun Q, et al. Expression and Clinical Significance of the Novel Long Noncoding RNA ZNF674-AS1 in Human Hepatocellular Carcinoma. Biomed Res Int. 2016;2016:3608914. doi: 10.1155/2016/3608914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Wang F, Hu Y. STARD13 promotes hepatocellular carcinoma apoptosis by acting as a ceRNA for Fas. Biotechnol Lett. 2017;39:207–217. doi: 10.1007/s10529-016-2253-6. [DOI] [PubMed] [Google Scholar]
- 40.Gao Y, Wang G, Zhang C, Lin M, Liu X, Zeng Y, Liu J. Long non-coding RNA linc-cdh4-2 inhibits the migration and invasion of HCC cells by targeting R-cadherin pathway. Biochem Biophys Res Commun. 2016;480:348–354. doi: 10.1016/j.bbrc.2016.10.048. [DOI] [PubMed] [Google Scholar]
- 41.Xiao C, Wang C, Cheng S, Lai C, Zhang P, Wang Z, Zhang T, Zhang S, Liu R. The significance of low levels of LINC RP1130-1 expression in human hepatocellular carcinoma. Biosci Trends. 2016;10:378–385. doi: 10.5582/bst.2016.01123. [DOI] [PubMed] [Google Scholar]
- 42.Wang T, Ma S, Qi X, Tang X, Cui D, Wang Z, Chi J, Li P, Zhai B. Long noncoding RNA ZNFX1-AS1 suppresses growth of hepatocellular carcinoma cells by regulating the methylation of miR-9. Onco Targets Ther. 2016;9:5005–5014. doi: 10.2147/OTT.S103329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong D, Sheng Y, Ding S, Chen J, Tan X, Zeng T, Qin D, Zhu L, Huang A, Tang H. LINC00052 regulates the expression of NTRK3 by miR-128 and miR-485-3p to strengthen HCC cells invasion and migration. Oncotarget. 2016;7:47593–47608. doi: 10.18632/oncotarget.10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feo F, Simile MM, Pascale RM. Focal loss of long non-coding RNA-PRAL, as determinant of cell function and phenotype of hepatocellular carcinoma. Ann Transl Med. 2016;4:183. doi: 10.21037/atm.2016.03.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H, Zhu C, Zhao Y, Li M, Wu L, Yang X, Wan X, Wang A, Zhang MQ, Sang X, et al. Long non-coding RNA expression profiles of hepatitis C virus-related dysplasia and hepatocellular carcinoma. Oncotarget. 2015;6:43770–43778. doi: 10.18632/oncotarget.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang Y, Tang GN, Zhou WP, Sun SH. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. 2013;57:1882–1892. doi: 10.1002/hep.26195. [DOI] [PubMed] [Google Scholar]
- 47.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 48.Liu L, Liao JZ, He XX, Li PY. The role of autophagy in hepatocellular carcinoma: friend or foe. Oncotarget. 2017;8:57707–57722. doi: 10.18632/oncotarget.17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang TH, Yu CC, Lin YS, Chen TC, Yeh CT, Liang KH, Shieh TM, Chen CY, Hsueh C. Long noncoding RNA CPS1-IT1 suppresses the metastasis of hepatocellular carcinoma by regulating HIF-1α activity and inhibiting epithelial-mesenchymal transition. Oncotarget. 2016;7:43588–43603. doi: 10.18632/oncotarget.9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Liu Z, Yao B, Dou C, Xu M, Xue Y, Ding L, Jia Y, Zhang H, Li Q, et al. Long non-coding RNA TUSC7 acts a molecular sponge for miR-10a and suppresses EMT in hepatocellular carcinoma. Tumour Biol. 2016;37:11429–11441. doi: 10.1007/s13277-016-4892-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lv L, Chen G, Zhou J, Li J, Gong J. WT1-AS promotes cell apoptosis in hepatocellular carcinoma through down-regulating of WT1. J Exp Clin Cancer Res. 2015;34:119. doi: 10.1186/s13046-015-0233-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang TH, Lin YS, Chen Y, Yeh CT, Huang YL, Hsieh TH, Shieh TM, Hsueh C, Chen TC. Long non-coding RNA AOC4P suppresses hepatocellular carcinoma metastasis by enhancing vimentin degradation and inhibiting epithelial-mesenchymal transition. Oncotarget. 2015;6:23342–23357. doi: 10.18632/oncotarget.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng H, Yang S, Yang Y, Yuan SX, Wu FQ, Wang LL, Yan HL, Sun SH, Zhou WP. Epigenetically silenced long noncoding-SRHC promotes proliferation of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2015;141:1195–1203. doi: 10.1007/s00432-014-1871-4. [DOI] [PubMed] [Google Scholar]
- 54.Gan Y, Han N, He X, Yu J, Zhang M, Zhou Y, Liang H, Deng J, Zheng Y, Ge W, et al. Long non-coding RNA CASC2 regulates cell biological behaviour through the MAPK signalling pathway in hepatocellular carcinoma. Tumour Biol. 2017;39:1010428317706229. doi: 10.1177/1010428317706229. [DOI] [PubMed] [Google Scholar]
- 55.He X, Zheng Y, Zhang Y, Gan Y, Zhou Y, Liang H, Wu D, Ge W, Deng J, Xu X. Long non-coding RNA AK058003, as a precursor of miR-15a, interacts with HuR to inhibit the expression of γ-synuclein in hepatocellular carcinoma cells. Oncotarget. 2017;8:9451–9465. doi: 10.18632/oncotarget.14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X, Ren Y, Yang X, Xiong X, Han S, Ge Y, Pan W, Zhou L, Yuan Q, Yang M. miR-190a inhibits epithelial-mesenchymal transition of hepatoma cells via targeting the long non-coding RNA treRNA. FEBS Lett. 2015;589:4079–4087. doi: 10.1016/j.febslet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 57.Xu Y, Zheng Y, Liu H, Li T. Modulation of IGF2BP1 by long non-coding RNA HCG11 suppresses apoptosis of hepatocellular carcinoma cells via MAPK signaling transduction. Int J Oncol. 2017;51:791–800. doi: 10.3892/ijo.2017.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu JH, Chang WH, Fu HW, Shu WQ, Yuan T, Chen P. Upregulated long non-coding RNA LOC90784 promotes cell proliferation and invasion and is associated with poor clinical features in HCC. Biochem Biophys Res Commun. 2017;490:920–926. doi: 10.1016/j.bbrc.2017.06.141. [DOI] [PubMed] [Google Scholar]
- 59.Cao SW, Huang JL, Chen J, Hu YW, Hu XM, Ren TY, Zheng SH, Lin JD, Tang J, Zheng L, et al. Long non-coding RNA UBE2CP3 promotes tumor metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma. Oncotarget. 2017;8:65370–65385. doi: 10.18632/oncotarget.18524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gong J, Qi X, Zhang Y, Yu Y, Lin X, Li H, Hu Y. Long noncoding RNA linc00462 promotes hepatocellular carcinoma progression. Biomed Pharmacother. 2017;93:40–47. doi: 10.1016/j.biopha.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 61.Tian F, Xu J, Xue F, Guan E, Xu X. TINCR expression is associated with unfavorable prognosis in patients with hepatocellular carcinoma. Biosci Rep. 2017;37:pii: BSR20170301. doi: 10.1042/BSR20170301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu F, Li J, Du X, Zhang W, Lei P, Zhang Q. Long noncoding RNA AB019562 promotes cell proliferation and metastasis in human hepatocellular carcinoma. Mol Med Rep. 2017;16:69–74. doi: 10.3892/mmr.2017.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Ye Y, Feng B, Qi Y. Long Noncoding RNA lncARSR Promotes Doxorubicin Resistance in Hepatocellular Carcinoma via Modulating PTEN-PI3K/Akt Pathway. J Cell Biochem. 2017;118:4498–4507. doi: 10.1002/jcb.26107. [DOI] [PubMed] [Google Scholar]
- 64.Zhao X, Liu Y, Yu S. Long noncoding RNA AWPPH promotes hepatocellular carcinoma progression through YBX1 and serves as a prognostic biomarker. Biochim Biophys Acta. 2017;1863:1805–1816. doi: 10.1016/j.bbadis.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y, Hu Y, Wu G, Yang Y, Tang Y, Zhang W, Wang K, Liu Y, Wang X, Li T. Long noncoding RNA PCAT-14 induces proliferation and invasion by hepatocellular carcinoma cells by inducing methylation of miR-372. Oncotarget. 2017;8:34429–34441. doi: 10.18632/oncotarget.16260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li S, Huang Y, Huang Y, Fu Y, Tang D, Kang R, Zhou R, Fan XG. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res. 2017;36:51. doi: 10.1186/s13046-017-0519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang J, Xie Y, Xu X, Yin Y, Jiang R, Deng L, Tan Z, Gangarapu V, Tang J, Sun B. Bidirectional transcription of Linc00441 and RB1 via H3K27 modification-dependent way promotes hepatocellular carcinoma. Cell Death Dis. 2017;8:e2675. doi: 10.1038/cddis.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu RT, Cao JL, Yan CQ, Wang Y, An CJ, Lv HT. Effects of LncRNA-HOST2 on cell proliferation, migration, invasion and apoptosis of human hepatocellular carcinoma cell line SMMC-7721. Biosci Rep. 2017;37:pii: BSR20160532. doi: 10.1042/BSR20160532. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Pan Y, Qin T, Yin S, Zhang X, Gao X, Mu L. Long non-coding RNA UC001kfo promotes hepatocellular carcinoma proliferation and metastasis by targeting α-SMA. Biomed Pharmacother. 2017;87:669–677. doi: 10.1016/j.biopha.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 70.Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B, Du Y, Gao G, Tian Y, He L, et al. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun. 2016;7:13608. doi: 10.1038/ncomms13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang F, Yang H, Deng Z, Su Y, Fang Q, Yin Z. HOX Antisense lincRNA HOXA-AS2 Promotes Tumorigenesis of Hepatocellular Carcinoma. Cell Physiol Biochem. 2016;40:287–296. doi: 10.1159/000452545. [DOI] [PubMed] [Google Scholar]
- 72.Chen Z, Yu C, Zhan L, Pan Y, Chen L, Sun C. LncRNA CRNDE promotes hepatic carcinoma cell proliferation, migration and invasion by suppressing miR-384. Am J Cancer Res. 2016;6:2299–2309. [PMC free article] [PubMed] [Google Scholar]
- 73.Lan T, Chang L, Wu L, Yuan Y. Downregulation of ZEB2-AS1 decreased tumor growth and metastasis in hepatocellular carcinoma. Mol Med Rep. 2016;14:4606–4612. doi: 10.3892/mmr.2016.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lv J, Fan HX, Zhao XP, Lv P, Fan JY, Zhang Y, Liu M, Tang H. Long non-coding RNA Unigene56159 promotes epithelial-mesenchymal transition by acting as a ceRNA of miR-140-5p in hepatocellular carcinoma cells. Cancer Lett. 2016;382:166–175. doi: 10.1016/j.canlet.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 75.Peng W, Fan H. Long noncoding RNA CCHE1 indicates a poor prognosis of hepatocellular carcinoma and promotes carcinogenesis via activation of the ERK/MAPK pathway. Biomed Pharmacother. 2016;83:450–455. doi: 10.1016/j.biopha.2016.06.056. [DOI] [PubMed] [Google Scholar]
- 76.Zhu XT, Yuan JH, Zhu TT, Li YY, Cheng XY. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. FEBS J. 2016;283:3739–3754. doi: 10.1111/febs.13839. [DOI] [PubMed] [Google Scholar]
- 77.Sui CJ, Zhou YM, Shen WF, Dai BH, Lu JJ, Zhang MF, Yang JM. Long noncoding RNA GIHCG promotes hepatocellular carcinoma progression through epigenetically regulating miR-200b/a/429. J Mol Med (Berl) 2016;94:1281–1296. doi: 10.1007/s00109-016-1442-z. [DOI] [PubMed] [Google Scholar]
- 78.Ding LJ, Li Y, Wang SD, Wang XS, Fang F, Wang WY, Lv P, Zhao DH, Wei F, Qi L. Long Noncoding RNA lncCAMTA1 Promotes Proliferation and Cancer Stem Cell-Like Properties of Liver Cancer by Inhibiting CAMTA1. Int J Mol Sci. 2016;17:pii: E1617. doi: 10.3390/ijms17101617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen ZZ, Huang L, Wu YH, Zhai WJ, Zhu PP, Gao YF. LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat Commun. 2016;7:12598. doi: 10.1038/ncomms12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang T, Cao C, Wu D, Liu L. SNHG3 correlates with malignant status and poor prognosis in hepatocellular carcinoma. Tumour Biol. 2016;37:2379–2385. doi: 10.1007/s13277-015-4052-4. [DOI] [PubMed] [Google Scholar]
- 81.Lan T, Ma W, Hong Z, Wu L, Chen X, Yuan Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. J Exp Clin Cancer Res. 2017;36:11. doi: 10.1186/s13046-016-0486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang JH, Wei HW, Yang HG. Long noncoding RNA SNHG15, a potential prognostic biomarker for hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2016;20:1720–1724. [PubMed] [Google Scholar]
- 83.Gong X, Wei W, Chen L, Xia Z, Yu C. Comprehensive analysis of long non-coding RNA expression profiles in hepatitis B virus-related hepatocellular carcinoma. Oncotarget. 2016;7:42422–42430. doi: 10.18632/oncotarget.9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 85.Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J Cell Sci. 2014;127:1585–1594. doi: 10.1242/jcs.141069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu P, Wang Y, Huang G, Ye B, Liu B, Wu J, Du Y, He L, Fan Z. lnc-β-Catm elicits EZH2-dependent β-catenin stabilization and sustains liver CSC self-renewal. Nat Struct Mol Biol. 2016;23:631–639. doi: 10.1038/nsmb.3235. [DOI] [PubMed] [Google Scholar]
- 88.Bo C, Li N, Li X, Liang X, An Y. Long noncoding RNA uc.338 promotes cell proliferation through association with BMI1 in hepatocellular carcinoma. Hum Cell. 2016;29:141–147. doi: 10.1007/s13577-016-0140-z. [DOI] [PubMed] [Google Scholar]
- 89.Wang X, Zhang W, Tang J, Huang R, Li J, Xu D, Xie Y, Jiang R, Deng L, Zhang X, et al. LINC01225 promotes occurrence and metastasis of hepatocellular carcinoma in an epidermal growth factor receptor-dependent pathway. Cell Death Dis. 2016;7:e2130. doi: 10.1038/cddis.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dong L, Ni J, Hu W, Yu C, Li H. Upregulation of Long Non-Coding RNA PlncRNA-1 Promotes Metastasis and Induces Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma. Cell Physiol Biochem. 2016;38:836–846. doi: 10.1159/000443038. [DOI] [PubMed] [Google Scholar]
- 91.Zhou T, Gao Y. Increased expression of LncRNA BANCR and its prognostic significance in human hepatocellular carcinoma. World J Surg Oncol. 2016;14:8. doi: 10.1186/s12957-015-0757-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang F, Xie C, Zhao W, Deng Z, Yang H, Fang Q. Long non-coding RNA CARLo-5 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Clin Exp Med. 2017;17:33–43. doi: 10.1007/s10238-015-0395-9. [DOI] [PubMed] [Google Scholar]
- 93.Shi Y, Song Q, Yu S, Hu D, Zhuang X. Microvascular invasion in hepatocellular carcinoma overexpression promotes cell proliferation and inhibits cell apoptosis of hepatocellular carcinoma via inhibiting miR-199a expression. Onco Targets Ther. 2015;8:2303–2310. doi: 10.2147/OTT.S86807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575–1584. doi: 10.1038/onc.2015.223. [DOI] [PubMed] [Google Scholar]
- 95.Shi XM, Teng F. Up-regulation of long non-coding RNA Sox2ot promotes hepatocellular carcinoma cell metastasis and correlates with poor prognosis. Int J Clin Exp Pathol. 2015;8:4008–4014. [PMC free article] [PubMed] [Google Scholar]
- 96.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, et al. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res. 2015;75:3181–3191. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 97.Cao C, Sun J, Zhang D, Guo X, Xie L, Li X, Wu D, Liu L. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of β-catenin in HCC cells. Gastroenterology. 2015;148:415–26.e18. doi: 10.1053/j.gastro.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 98.Xu WH, Zhang JB, Dang Z, Li X, Zhou T, Liu J, Wang DS, Song WJ, Dou KF. Long non-coding RNA URHC regulates cell proliferation and apoptosis via ZAK through the ERK/MAPK signaling pathway in hepatocellular carcinoma. Int J Biol Sci. 2014;10:664–676. doi: 10.7150/ijbs.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shen Y, Liu S, Yuan H, Ying X, Fu H, Zheng X. A long non-coding RNA lncRNA-PE promotes invasion and epithelial-mesenchymal transition in hepatocellular carcinoma through the miR-200a/b-ZEB1 pathway. Tumour Biol. 2017;39:1010428317705756. doi: 10.1177/1010428317705756. [DOI] [PubMed] [Google Scholar]
- 100.Cheng K, Chen Z, Liu L, Zhao Y, Zhang S, Wang Q, Deng Z, Tan S, Ye Q. ZNF667 Serves as a Putative Oncogene in Human Hepatocellular Carcinoma. Cell Physiol Biochem. 2017;41:2523–2533. doi: 10.1159/000475971. [DOI] [PubMed] [Google Scholar]
- 101.Yu X, Zheng H, Chan MT, Wu WK. HULC: an oncogenic long non-coding RNA in human cancer. J Cell Mol Med. 2017;21:410–417. doi: 10.1111/jcmm.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013:136106. doi: 10.1155/2013/136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li J, Wang X, Tang J, Jiang R, Zhang W, Ji J, Sun B. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell Physiol Biochem. 2015;37:687–696. doi: 10.1159/000430387. [DOI] [PubMed] [Google Scholar]
- 104.Zhang Y, Li Z, Zhang Y, Zhong Q, Chen Q, Zhang L. Molecular mechanism of HEIH and HULC in the proliferation and invasion of hepatoma cells. Int J Clin Exp Med. 2015;8:12956–12962. [PMC free article] [PubMed] [Google Scholar]
- 105.Xiong H, Li B, He J, Zeng Y, Zhang Y, He F. lncRNA HULC promotes the growth of hepatocellular carcinoma cells via stabilizing COX-2 protein. Biochem Biophys Res Commun. 2017;490:693–699. doi: 10.1016/j.bbrc.2017.06.103. [DOI] [PubMed] [Google Scholar]
- 106.Xiong H, Ni Z, He J, Jiang S, Li X, He J, Gong W, Zheng L, Chen S, Li B, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36:3528–3540. doi: 10.1038/onc.2016.521. [DOI] [PubMed] [Google Scholar]
- 107.Li SP, Xu HX, Yu Y, He JD, Wang Z, Xu YJ, Wang CY, Zhang HM, Zhang RX, Zhang JJ, et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7:42431–42446. doi: 10.18632/oncotarget.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lu Z, Xiao Z, Liu F, Cui M, Li W, Yang Z, Li J, Ye L, Zhang X. Long non-coding RNA HULC promotes tumor angiogenesis in liver cancer by up-regulating sphingosine kinase 1 (SPHK1) Oncotarget. 2016;7:241–254. doi: 10.18632/oncotarget.6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gandhy SU, Imanirad P, Jin UH, Nair V, Hedrick E, Cheng Y, Corton JC, Kim K, Safe S. Specificity protein (Sp) transcription factors and metformin regulate expression of the long non-coding RNA HULC. Oncotarget. 2015;6:26359–26372. doi: 10.18632/oncotarget.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wan D, Shen S, Fu S, Preston B, Brandon C, He S, Shen C, Wu J, Wang S, Xie W, et al. miR-203 suppresses the proliferation and metastasis of hepatocellular carcinoma by targeting oncogene ADAM9 and oncogenic long non-coding RNA HULC. Anticancer Agents Med Chem. 2016;16:414–423. doi: 10.2174/1871520615666150716105955. [DOI] [PubMed] [Google Scholar]
- 111.Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, Sun B, Ye L, Zhang X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75:846–857. doi: 10.1158/0008-5472.CAN-14-1192. [DOI] [PubMed] [Google Scholar]
- 112.Li D, Liu X, Zhou J, Hu J, Zhang D, Liu J, Qiao Y, Zhan Q. Long noncoding RNA HULC modulates the phosphorylation of YB-1 through serving as a scaffold of extracellular signal-regulated kinase and YB-1 to enhance hepatocarcinogenesis. Hepatology. 2017;65:1612–1627. doi: 10.1002/hep.29010. [DOI] [PubMed] [Google Scholar]
- 113.Cui M, Zheng M, Sun B, Wang Y, Ye L, Zhang X. A long noncoding RNA perturbs the circadian rhythm of hepatoma cells to facilitate hepatocarcinogenesis. Neoplasia. 2015;17:79–88. doi: 10.1016/j.neo.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, Ye L, Zhang X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang Z, Lu Y, Xu Q, Tang B, Park CK, Chen X. HULC and H19 Played Different Roles in Overall and Disease-Free Survival from Hepatocellular Carcinoma after Curative Hepatectomy: A Preliminary Analysis from Gene Expression Omnibus. Dis Markers. 2015;2015:191029. doi: 10.1155/2015/191029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun J, Bie B, Zhang S, Yang J, Li Z. Long non-coding RNAs: critical players in hepatocellular carcinoma. Int J Mol Sci. 2014;15:20434–20448. doi: 10.3390/ijms151120434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.117 Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29:946–950. doi: 10.3892/or.2012.2219. [DOI] [PubMed] [Google Scholar]
- 118.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 119.Gao JZ, Li J, DU JL, Li XL. Long non-coding RNA HOTAIR is a marker for hepatocellular carcinoma progression and tumor recurrence. Oncol Lett. 2016;11:1791–1798. doi: 10.3892/ol.2016.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39:2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 121.Yang L, Zhang X, Li H, Liu J. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol Biosyst. 2016;12:2605–2612. doi: 10.1039/c6mb00114a. [DOI] [PubMed] [Google Scholar]
- 122.Li H, An J, Wu M, Zheng Q, Gui X, Li T, Pu H, Lu D. LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget. 2015;6:27847–27864. doi: 10.18632/oncotarget.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Su DN, Wu SP, Chen HT, He JH. HOTAIR, a long non-coding RNA driver of malignancy whose expression is activated by FOXC1, negatively regulates miRNA-1 in hepatocellular carcinoma. Oncol Lett. 2016;12:4061–4067. doi: 10.3892/ol.2016.5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang H, Xing Z, Mani SK, Bancel B, Durantel D, Zoulim F, Tran EJ, Merle P, Andrisani O. RNA helicase DEAD box protein 5 regulates Polycomb repressive complex 2/Hox transcript antisense intergenic RNA function in hepatitis B virus infection and hepatocarcinogenesis. Hepatology. 2016;64:1033–1048. doi: 10.1002/hep.28698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang H, Liang WC, Wang SS, Ko CH, Waye MM, et al. Hotair mediates hepatocarcinogenesis through suppressing miRNA-218 expression and activating P14 and P16 signaling. J Hepatol. 2015;63:886–895. doi: 10.1016/j.jhep.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 126.Ding C, Cheng S, Yang Z, Lv Z, Xiao H, Du C, Peng C, Xie H, Zhou L, Wu J, et al. Long non-coding RNA HOTAIR promotes cell migration and invasion via down-regulation of RNA binding motif protein 38 in hepatocellular carcinoma cells. Int J Mol Sci. 2014;15:4060–4076. doi: 10.3390/ijms15034060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chou J, Wang B, Zheng T, Li X, Zheng L, Hu J, Zhang Y, Xing Y, Xi T. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem Biophys Res Commun. 2016;472:262–269. doi: 10.1016/j.bbrc.2016.02.102. [DOI] [PubMed] [Google Scholar]
- 129.Jin C, Yan B, Lu Q, Lin Y, Ma L. The role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal carcinoma. Tumour Biol. 2016;37:4025–4033. doi: 10.1007/s13277-015-4227-z. [DOI] [PubMed] [Google Scholar]
- 130.Chen L, Yao H, Wang K, Liu X. Long Non-Coding RNA MALAT1 Regulates ZEB1 Expression by Sponging miR-143-3p and Promotes Hepatocellular Carcinoma Progression. J Cell Biochem. 2017;118:4836–4843. doi: 10.1002/jcb.26158. [DOI] [PubMed] [Google Scholar]
- 131.Hou Z, Xu X, Fu X, Tao S, Zhou J, Liu S, Tan D. HBx-related long non-coding RNA MALAT1 promotes cell metastasis via up-regulating LTBP3 in hepatocellular carcinoma. Am J Cancer Res. 2017;7:845–856. [PMC free article] [PubMed] [Google Scholar]
- 132.Li C, Miao R, Liu S, Wan Y, Zhang S, Deng Y, Bi J, Qu K, Zhang J, Liu C. Down-regulation of miR-146b-5p by long noncoding RNA MALAT1 in hepatocellular carcinoma promotes cancer growth and metastasis. Oncotarget. 2017;8:28683–28695. doi: 10.18632/oncotarget.15640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 134.Malakar P, Shilo A, Mogilevsky A, Stein I, Pikarsky E, Nevo Y, Benyamini H, Elgavish S, Zong X, Prasanth KV, et al. Long Noncoding RNA MALAT1 Promotes Hepatocellular Carcinoma Development by SRSF1 Upregulation and mTOR Activation. Cancer Res. 2017;77:1155–1167. doi: 10.1158/0008-5472.CAN-16-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li C, Chang L, Chen Z, Liu Z, Wang Y, Ye Q. The role of lncRNA MALAT1 in the regulation of hepatocyte proliferation during liver regeneration. Int J Mol Med. 2017;39:347–356. doi: 10.3892/ijmm.2017.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chang L, Wang G, Jia T, Zhang L, Li Y, Han Y, Zhang K, Lin G, Zhang R, Li J, et al. Armored long non-coding RNA MEG3 targeting EGFR based on recombinant MS2 bacteriophage virus-like particles against hepatocellular carcinoma. Oncotarget. 2016;7:23988–24004. doi: 10.18632/oncotarget.8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li Y, Ren M, Zhao Y, Lu X, Wang M, Hu J, Lu G, He S. MicroRNA-26a inhibits proliferation and metastasis of human hepatocellular carcinoma by regulating DNMT3B-MEG3 axis. Oncol Rep. 2017;37:3527–3535. doi: 10.3892/or.2017.5579. [DOI] [PubMed] [Google Scholar]
- 139.He JH, Han ZP, Liu JM, Zhou JB, Zou MX, Lv YB, Li YG, Cao MR. Overexpression of Long Non-Coding RNA MEG3 Inhibits Proliferation of Hepatocellular Carcinoma Huh7 Cells via Negative Modulation of miRNA-664. J Cell Biochem. 2017;118:3713–3721. doi: 10.1002/jcb.26018. [DOI] [PubMed] [Google Scholar]
- 140.el-Deiry WS. Regulation of p53 downstream genes. Semin Cancer Biol. 1998;8:345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 141.Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J, Wei L, Jin Y, Fu H, Wu Y, et al. Long Noncoding RNA MEG3 Interacts with p53 Protein and Regulates Partial p53 Target Genes in Hepatoma Cells. PLoS One. 2015;10:e0139790. doi: 10.1371/journal.pone.0139790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 143.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 144.Zhuo H, Tang J, Lin Z, Jiang R, Zhang X, Ji J, Wang P, Sun B. The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma. Mol Carcinog. 2016;55:209–219. doi: 10.1002/mc.22270. [DOI] [PubMed] [Google Scholar]
- 145.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM, Patel T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zamani M, Sadeghizadeh M, Behmanesh M, Najafi F. Dendrosomal curcumin increases expression of the long non-coding RNA gene MEG3 via up-regulation of epi-miRs in hepatocellular cancer. Phytomedicine. 2015;22:961–967. doi: 10.1016/j.phymed.2015.05.071. [DOI] [PubMed] [Google Scholar]
- 147.Lu X, Zhou C, Li R, Liang Z, Zhai W, Zhao L, Zhang S. Critical role for the long non-coding RNA AFAP1-AS1 in the proliferation and metastasis of hepatocellular carcinoma. Tumour Biol. 2016;37:9699–9707. doi: 10.1007/s13277-016-4858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang JY, Weng MZ, Song FB, Xu YG, Liu Q, Wu JY, Qin J, Jin T, Xu JM. Long noncoding RNA AFAP1-AS1 indicates a poor prognosis of hepatocellular carcinoma and promotes cell proliferation and invasion via upregulation of the RhoA/Rac2 signaling. Int J Oncol. 2016;48:1590–1598. doi: 10.3892/ijo.2016.3385. [DOI] [PubMed] [Google Scholar]
- 149.Hua L, Wang CY, Yao KH, Chen JT, Zhang JJ, Ma WL. High expression of long non-coding RNA ANRIL is associated with poor prognosis in hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:3076–3082. [PMC free article] [PubMed] [Google Scholar]
- 150.Huang MD, Chen WM, Qi FZ, Xia R, Sun M, Xu TP, Yin L, Zhang EB, De W, Shu YQ. Long non-coding RNA ANRIL is upregulated in hepatocellular carcinoma and regulates cell apoptosis by epigenetic silencing of KLF2. J Hematol Oncol. 2015;8:50. doi: 10.1186/s13045-015-0146-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 152.Zhu H, Zhou X, Chang H, Li H, Liu F, Ma C, Lu J. CCAT1 promotes hepatocellular carcinoma cell proliferation and invasion. Int J Clin Exp Pathol. 2015;8:5427–5434. [PMC free article] [PubMed] [Google Scholar]
- 153.Zhu HQ, Zhou X, Chang H, Li HG, Liu FF, Ma CQ, Lu J. Aberrant Expression of CCAT1 Regulated by c-Myc Predicts the Prognosis of Hepatocellular Carcinoma. Asian Pac J Cancer Prev. 2015;16:5181–5185. doi: 10.7314/apjcp.2015.16.13.5181. [DOI] [PubMed] [Google Scholar]
- 154.Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dou C, Sun L, Jin X, Han M, Zhang B, Li T. Long non-coding RNA colon cancer-associated transcript 1 functions as a competing endogenous RNA to regulate cyclin-dependent kinase 1 expression by sponging miR-490-3p in hepatocellular carcinoma progression. Tumour Biol. 2017;39:1010428317697572. doi: 10.1177/1010428317697572. [DOI] [PubMed] [Google Scholar]
- 156.Zhou N, Si Z, Li T, Chen G, Zhang Z, Qi H. Long non-coding RNA CCAT2 functions as an oncogene in hepatocellular carcinoma, regulating cellular proliferation, migration and apoptosis. Oncol Lett. 2016;12:132–138. doi: 10.3892/ol.2016.4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Xu Y, Wang B, Zhang F, Wang A, Du X, Hu P, Zhu Y, Fang Z. Long non-coding RNA CCAT2 is associated with poor prognosis in hepatocellular carcinoma and promotes tumor metastasis by regulating Snail2-mediated epithelial-mesenchymal transition. Onco Targets Ther. 2017;10:1191–1198. doi: 10.2147/OTT.S127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ma X, Wang X, Yang C, Wang Z, Han B, Wu L, Zhuang L. DANCR Acts as a Diagnostic Biomarker and Promotes Tumor Growth and Metastasis in Hepatocellular Carcinoma. Anticancer Res. 2016;36:6389–6398. doi: 10.21873/anticanres.11236. [DOI] [PubMed] [Google Scholar]
- 159.Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL, Yang Y, Liu H, Wang ZG, Xu QG, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63:499–511. doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- 160.Qi HL, Li CS, Qian CW, Xiao YS, Yuan YF, Liu QY, Liu ZS. The long noncoding RNA, EGFR-AS1, a target of GHR, increases the expression of EGFR in hepatocellular carcinoma. Tumour Biol. 2016;37:1079–1089. doi: 10.1007/s13277-015-3887-z. [DOI] [PubMed] [Google Scholar]
- 161.Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie Y, Wang K, Jia W, Chu WM, Sun B. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun. 2017;8:15129. doi: 10.1038/ncomms15129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu F, Yuan JH, Huang JF, Yang F, Wang TT, Ma JZ, Zhang L, Zhou CC, Wang F, Yu J, et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene. 2016;35:5422–5434. doi: 10.1038/onc.2016.80. [DOI] [PubMed] [Google Scholar]
- 163.Liu Z, Dou C, Yao B, Xu M, Ding L, Wang Y, Jia Y, Li Q, Zhang H, Tu K, et al. Ftx non coding RNA-derived miR-545 promotes cell proliferation by targeting RIG-I in hepatocellular carcinoma. Oncotarget. 2016;7:25350–25365. doi: 10.18632/oncotarget.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tu ZQ, Li RJ, Mei JZ, Li XH. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:4303–4309. [PMC free article] [PubMed] [Google Scholar]
- 165.Chang L, Li C, Lan T, Wu L, Yuan Y, Liu Q, Liu Z. Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Mol Med Rep. 2016;13:1541–1550. doi: 10.3892/mmr.2015.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hu L, Ye H, Huang G, Luo F, Liu Y, Liu Y, Yang X, Shen J, Liu Q, Zhang J. Long noncoding RNA GAS5 suppresses the migration and invasion of hepatocellular carcinoma cells via miR-21. Tumour Biol. 2016;37:2691–2702. doi: 10.1007/s13277-015-4111-x. [DOI] [PubMed] [Google Scholar]
- 167.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 168.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 169.Zhang L, Yang F, Yuan JH, Yuan SX, Zhou WP, Huo XS, Xu D, Bi HS, Wang F, Sun SH. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34:577–586. doi: 10.1093/carcin/bgs381. [DOI] [PubMed] [Google Scholar]
- 170.Lv J, Ma L, Chen XL, Huang XH, Wang Q. Downregulation of LncRNAH19 and MiR-675 promotes migration and invasion of human hepatocellular carcinoma cells through AKT/GSK-3β/Cdc25A signaling pathway. J Huazhong Univ Sci Technolog Med Sci. 2014;34:363–369. doi: 10.1007/s11596-014-1284-2. [DOI] [PubMed] [Google Scholar]
- 171.Lian Y, Cai Z, Gong H, Xue S, Wu D, Wang K. HOTTIP: a critical oncogenic long non-coding RNA in human cancers. Mol Biosyst. 2016;12:3247–3253. doi: 10.1039/c6mb00475j. [DOI] [PubMed] [Google Scholar]
- 172.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ge Y, Yan X, Jin Y, Yang X, Yu X, Zhou L, Han S, Yuan Q, Yang M. MiRNA-192 [corrected] and miRNA-204 Directly Suppress lncRNA HOTTIP and Interrupt GLS1-Mediated Glutaminolysis in Hepatocellular Carcinoma. PLoS Genet. 2015;11:e1005726. doi: 10.1371/journal.pgen.1005726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tsang FH, Au SL, Wei L, Fan DN, Lee JM, Wong CC, Ng IO, Wong CM. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver Int. 2015;35:1597–1606. doi: 10.1111/liv.12746. [DOI] [PubMed] [Google Scholar]
- 175.Guo S, Chen W, Luo Y, Ren F, Zhong T, Rong M, Dang Y, Feng Z, Chen G. Clinical implication of long non-coding RNA NEAT1 expression in hepatocellular carcinoma patients. Int J Clin Exp Pathol. 2015;8:5395–5402. [PMC free article] [PubMed] [Google Scholar]
- 176.Fang L, Sun J, Pan Z, Song Y, Zhong L, Zhang Y, Liu Y, Zheng X, Huang P. Long non-coding RNA NEAT1 promotes hepatocellular carcinoma cell proliferation through the regulation of miR-129-5p-VCP-IκB. Am J Physiol Gastrointest Liver Physiol. 2017;313:G150–G156. doi: 10.1152/ajpgi.00426.2016. [DOI] [PubMed] [Google Scholar]
- 177.Mang Y, Li L, Ran J, Zhang S, Liu J, Li L, Chen Y, Liu J, Gao Y, Ren G. Long noncoding RNA NEAT1 promotes cell proliferation and invasion by regulating hnRNP A2 expression in hepatocellular carcinoma cells. Onco Targets Ther. 2017;10:1003–1016. doi: 10.2147/OTT.S116319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ji J, Tang J, Deng L, Xie Y, Jiang R, Li G, Sun B. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6:42813–42824. doi: 10.18632/oncotarget.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Jia M, Jiang L, Wang YD, Huang JZ, Yu M, Xue HZ. lincRNA-p21 inhibits invasion and metastasis of hepatocellular carcinoma through Notch signaling-induced epithelial-mesenchymal transition. Hepatol Res. 2016;46:1137–1144. doi: 10.1111/hepr.12659. [DOI] [PubMed] [Google Scholar]
- 181.Ding G, Peng Z, Shang J, Kang Y, Ning H, Mao C. LincRNA-p21 inhibits invasion and metastasis of hepatocellular carcinoma through miR-9/E-cadherin cascade signaling pathway molecular mechanism. Onco Targets Ther. 2017;10:3241–3247. doi: 10.2147/OTT.S134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wen J, Xu J, Sun Q, Xing C, Yin W. Upregulation of long non coding RNA PCAT-1 contributes to cell proliferation, migration and apoptosis in hepatocellular carcinoma. Mol Med Rep. 2016;13:4481–4486. doi: 10.3892/mmr.2016.5075. [DOI] [PubMed] [Google Scholar]
- 183.Yan TH, Yang H, Jiang JH, Lu SW, Peng CX, Que HX, Lu WL, Mao JF. Prognostic significance of long non-coding RNA PCAT-1 expression in human hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:4126–4131. [PMC free article] [PubMed] [Google Scholar]
- 184.Yu J, Han J, Zhang J, Li G, Liu H, Cui X, Xu Y, Li T, Liu J, Wang C. The long noncoding RNAs PVT1 and uc002mbe.2 in sera provide a new supplementary method for hepatocellular carcinoma diagnosis. Medicine (Baltimore) 2016;95:e4436. doi: 10.1097/MD.0000000000004436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang F, Yuan JH, Wang SB, Yang F, Yuan SX, Ye C, Yang N, Zhou WP, Li WL, Li W, et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278–1290. doi: 10.1002/hep.27239. [DOI] [PubMed] [Google Scholar]
- 186.Lan T, Yan X, Li Z, Xu X, Mao Q, Ma W, Hong Z, Chen X, Yuan Y. Long non-coding RNA PVT1 serves as a competing endogenous RNA for miR-186-5p to promote the tumorigenesis and metastasis of hepatocellular carcinoma. Tumour Biol. 2017;39:1010428317705338. doi: 10.1177/1010428317705338. [DOI] [PubMed] [Google Scholar]
- 187.Zhang M, Wang W, Li T, Yu X, Zhu Y, Ding F, Li D, Yang T. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother. 2016;80:73–79. doi: 10.1016/j.biopha.2016.02.036. [DOI] [PubMed] [Google Scholar]
- 188.Zhang H, Zhou D, Ying M, Chen M, Chen P, Chen Z, Zhang F. Expression of Long Non-Coding RNA (lncRNA) Small Nucleolar RNA Host Gene 1 (SNHG1) Exacerbates Hepatocellular Carcinoma Through Suppressing miR-195. Med Sci Monit. 2016;22:4820–4829. doi: 10.12659/MSM.898574. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 189.Birgani MT, Hajjari M, Shahrisa A, Khoshnevisan A, Shoja Z, Motahari P, Farhangi B. Long Non-Coding RNA SNHG6 as a Potential Biomarker for Hepatocellular Carcinoma. Pathol Oncol Res. 2017 doi: 10.1007/s12253-017-0241-3. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 190.Cao C, Zhang T, Zhang D, Xie L, Zou X, Lei L, Wu D, Liu L. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene. 2017;36:1112–1122. doi: 10.1038/onc.2016.278. [DOI] [PubMed] [Google Scholar]
- 191.Zhang D, Cao C, Liu L, Wu D. Up-regulation of LncRNA SNHG20 Predicts Poor Prognosis in Hepatocellular Carcinoma. J Cancer. 2016;7:608–617. doi: 10.7150/jca.13822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Liu J, Lu C, Xiao M, Jiang F, Qu L, Ni R. Long non-coding RNA SNHG20 predicts a poor prognosis for HCC and promotes cell invasion by regulating the epithelial-to-mesenchymal transition. Biomed Pharmacother. 2017;89:857–863. doi: 10.1016/j.biopha.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 193.Yu G, Lin J, Liu C, Hou K, Liang M, Shi B. Long non-coding RNA SPRY4-IT1 promotes development of hepatic cellular carcinoma by interacting with ERRα and predicts poor prognosis. Sci Rep. 2017;7:17176. doi: 10.1038/s41598-017-16781-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Li J, Chen Y, Chen Z, He A, Xie H, Zhang Q, Cai Z, Liu Y, Huang W. SPRY4-IT1: A novel oncogenic long non-coding RNA in human cancers. Tumour Biol. 2017;39:1010428317711406. doi: 10.1177/1010428317711406. [DOI] [PubMed] [Google Scholar]
- 195.Huang MD, Chen WM, Qi FZ, Sun M, Xu TP, Ma P, Shu YQ. Long non-coding RNA TUG1 is up-regulated in hepatocellular carcinoma and promotes cell growth and apoptosis by epigenetically silencing of KLF2. Mol Cancer. 2015;14:165. doi: 10.1186/s12943-015-0431-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Han Y, Liu Y, Gui Y, Cai Z. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J Surg Oncol. 2013;107:555–559. doi: 10.1002/jso.23264. [DOI] [PubMed] [Google Scholar]
- 197.Ma B, Li M, Zhang L, Huang M, Lei JB, Fu GH, Liu CX, Lai QW, Chen QQ, Wang YL. Upregulation of long non-coding RNA TUG1 correlates with poor prognosis and disease status in osteosarcoma. Tumour Biol. 2016;37:4445–4455. doi: 10.1007/s13277-015-4301-6. [DOI] [PubMed] [Google Scholar]
- 198.Liu Q, Sun S, Yu W, Jiang J, Zhuo F, Qiu G, Xu S, Jiang X. Altered expression of long non-coding RNAs during genotoxic stress-induced cell death in human glioma cells. J Neurooncol. 2015;122:283–292. doi: 10.1007/s11060-015-1718-0. [DOI] [PubMed] [Google Scholar]
- 199.Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, Bhandoola A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wu J, Zhang J, Shen B, Yin K, Xu J, Gao W, Zhang L. Long noncoding RNA lncTCF7, induced by IL-6/STAT3 transactivation, promotes hepatocellular carcinoma aggressiveness through epithelial-mesenchymal transition. J Exp Clin Cancer Res. 2015;34:116. doi: 10.1186/s13046-015-0229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, Yan X, Ye B, Li C, Xia P, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 202.Xiao JN, Yan TH, Yu RM, Gao Y, Zeng WL, Lu SW, Que HX, Liu ZP, Jiang JH. Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. J Cancer Res Clin Oncol. 2017;143:981–990. doi: 10.1007/s00432-017-2370-1. [DOI] [PubMed] [Google Scholar]
- 203.Hu JJ, Song W, Zhang SD, Shen XH, Qiu XM, Wu HZ, Gong PH, Lu S, Zhao ZJ, He ML, et al. HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling. Sci Rep. 2016;6:23521. doi: 10.1038/srep23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, Chen J, Liu X, Wang SK. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899–7917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ma W, Wang H, Jing W, Zhou F, Chang L, Hong Z, Liu H, Liu Z, Yuan Y. Downregulation of long non-coding RNAs JPX and XIST is associated with the prognosis of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2017;41:163–170. doi: 10.1016/j.clinre.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 206.Chang S, Chen B, Wang X, Wu K, Sun Y. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer. 2017;17:248. doi: 10.1186/s12885-017-3216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhuang LK, Yang YT, Ma X, Han B, Wang ZS, Zhao QY, Wu LQ, Qu ZQ. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016;7:e2203. doi: 10.1038/cddis.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Mo Y, Lu Y, Wang P, Huang S, He L, Li D, Li F, Huang J, Lin X, Li X, et al. Long non-coding RNA XIST promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma. Tumour Biol. 2017;39:1010428317690999. doi: 10.1177/1010428317690999. [DOI] [PubMed] [Google Scholar]
- 209.Zhou CC, Yang F, Yuan SX, Ma JZ, Liu F, Yuan JH, Bi FR, Lin KY, Yin JH, Cao GW, et al. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology. 2016;63:850–863. doi: 10.1002/hep.28393. [DOI] [PubMed] [Google Scholar]
- 210.Zhou M, Zhang XY, Yu X. Overexpression of the long non-coding RNA SPRY4-IT1 promotes tumor cell proliferation and invasion by activating EZH2 in hepatocellular carcinoma. Biomed Pharmacother. 2017;85:348–354. doi: 10.1016/j.biopha.2016.11.035. [DOI] [PubMed] [Google Scholar]


