Abstract
Spores of Bacillus cereus pose a threat to food safety due to their high resistance to the heat or acid treatments commonly used to make food microbiologically safe. Spores may survive these treatments and later resume growth either on foodstuffs or, after ingestion, upon entering the gut they are capable of producing toxins, which cause either vomiting or diarrhea. The outer layers of the spore, the spore coat and exosporium, consist primarily of proteins that may serve as potential biomarkers for detection. The major morphogenetic protein CotE is important for correct assembly and attachment of the outermost layer, the exosporium, and by extension retention of many proteins. However, characterization of the proteins affected by deletion of CotE has been limited to electrophoretic patterns. Here we report the effect of CotE deletion on the insoluble fraction of the spore proteome through liquid chromatography–Fourier transform tandem mass spectrometry (LC–FTMS/MS) analysis. A total of 560 proteins have been identified in both mutant and wild-type spore coat isolates. A further 163 proteins were identified exclusively in wild-type spore isolates indicating that they are dependent on CotE for their association with the spore. Several of these are newly confirmed as associated with the exosporium, namely BC_2569 (BclF), BC_3345, BC_2427, BC_2878, BC_0666, BC_2984, BC_3481, and BC_2570. A total of 153 proteins were only identified in ΔCotE spore isolates. This was observed for proteins that are known or likely to be interacting with or are encased by CotE. Crucial spore proteins were quantified using a QconCAT reference standard, the first time this was used in a biochemically heterogeneous system. This allowed us to determine the absolute abundance of 21 proteins, which spanned across three orders of magnitude and together covered 5.66% ± 0.51 of the total spore weight. Applying the QconCAT methodology to the ΔCotE mutant allowed us to quantify 4.13% ± 0.14 of the spore total weight and revealed a reduction in abundance for most known exosporium associated proteins upon CotE deletion. In contrast, several proteins, either known or likely to be interacting with or encased by CotE (i.e., GerQ), were more abundant. The results obtained provide deeper insight into the layered spore structure such as which proteins are exposed on the outside of the spore. This information is important for developing detection methods for targeting spores in a food safety setting. Furthermore, protein stoichiometry and determination of the abundance of germination mediating enzymes provides useful information for germination and outgrowth model development.
Keywords: Bacillus cereus, spore coat, QconCAT, CotE, quantitative proteomics
Introduction
Bacillus cereus presents a problem for the food industry due to its ability to survive common processing methods such as heat or acid treatment by forming (endo)spores. Although spores are metabolically dormant and damaged by the preservation strategies, after such treatments, when conditions become favorable for growth many can still germinate, repair incurred damage and grow out. Once back in a vegetative state they can start producing toxins either in the food product or after ingestion in the gut resulting in food poisoning causing vomiting or diarrhea, respectively. Proper detection of spores therefore determines an important benchmark for food safety.1 Spores consist of a layered structure (Figure 1) with a central core containing the DNA, surrounded by a peptidoglycan cortex layer, which in turn is surrounded by proteinaceous inner and outer coat layers. In the case of B. cereus, this compact spore structure is then surrounded by a loose layer composed of protein, lipids, and carbohydrates called the exosporium.2−4 As the outermost layers consist mainly of protein, there is a potential for these proteins to be used as spore specific biomarkers. However, to utilize spore proteins as biomarker targets for detection, it is important to know which are abundant and accessible.
Figure 1.
Schematic representation of the layered spore structure. Target proteins are indicated in their respective locations based on either experimental evidence or homology found in literature.
Our approach for analysis of the spore proteome utilizes bead-beating to disintegrate the spore’s structure and make proteins accessible to tryptic digestion. Clearly, as such this procedure eliminates any spatial information. Localization of proteins may nevertheless still be investigated by comparison of the wild type proteome to that of mutants lacking certain morphogenetic proteins. Deletion of the major morphogenetic protein CotE, which normally forms a shell around the outer coat,5 results in gross defects in the exosporium.6 Proteins normally associated with the exosporium are then likely to be affected by this perturbed assembly. When comparing the proteome of CotE knockout spores to wild type isolates, proteins that are dependent on CotE for their localization to the spore coat would either be missing or present at reduced levels in ΔCotE isolates. A similar approach has previously been used to show CotE-dependent localization of proteins in the outer coat of Bacillus subtilis(7) and the exosporium of Bacillus anthracis.5 However, the approach of these previous studies relied on solubilization of spore proteins and resolution on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), which eliminates the contribution of the insoluble fraction. Ignoring this renders an incomplete picture of the effect of CotE deletion. Furthermore, the characterization of the changes observed in these previous studies was limited to electrophoretic patterns and, with the exception of BAS2377, did not include identification of the proteins in differentially observed bands. Here, we report the first proteome wide investigation into CotE dependent localization of proteins in the spores of a B. cereus ΔCotE mutant B. cereus strain previously described by Bressuire et al.6
Most investigations into structural spore proteins are based on analyses of knockout mutants with which major morphogenetic proteins have been identified. SafA and CotE are examples of such proteins that are essential for the deposition of entire layers of the spore coat.5,6,8−11 While structural proteins such as these are likely to be abundant, less so are functional proteins such as enzymes or receptors present in the coat. Generally, spore proteins are known to be cross-linked together to form a coherent structure built on the framework created by these major morphogenetic proteins. Knockout strategies to identify protein functions might not always result in a clear phenotype as potential effects on spore structure may be masked by the remainder of the cross-linked network, determining quantitative protein abundance can give an indication of potential function of uncharacterized proteins.
Advances in mass spectrometry have allowed for the first quantitative analyses of the spore proteome;12 however, these have so far only used relative measurements that allow for comparison of different conditions but give no information on the number of proteins per spore. QconCAT approaches rely on creating a synthetic isotopically labeled protein consisting of concatenated tryptic peptides, which are quantotypic for the targeted proteins.13 This protein is then spiked into the sample and digested alongside the analyte creating a set of reference peptides for all targeted proteins in a 1:1:1 ratio at a known quantity allowing for absolute quantification of several proteins at once. Being able to select 20–25 target proteins per QconCAT construct when using two selected quantotypic peptides per target protein makes it perfectly suited for investigations of this scope as the spore proteome is estimated to consist of several hundred spore specific proteins.3,11,14 This targeted approach has an added benefit as current methods are not able to isolate only the spore specific proteins. In the case of B. cereus, this is in part due to the fact that the exosporium traps proteins from the mother cell in the so-called interspace region adding a confounding factor in the case of nontargeted methods.
QconCATs are normally implemented in a homogeneous system;13,15 however, as spore proteins are highly cross-linked, a large part of the spore proteome cannot be fully solubilized by standard methods such as SDS-extraction. By using a method first described by Abhyankhar et al.,16 it is possible to identify these proteins by using trypsin to cleave peptides exposed by prior SDS-extraction and using reducing conditions that should break cross-links such as dicysteines and dityrosines.17,18 By selecting only peptides that were frequently identified in preliminary analyses, we are able to select a set of reproducibly identifiable peptides that are likely not obstructed by cross-links. By optimizing digestion time, we can ensure completed digestion, thus enabling us to quantify protein content using a QconCAT approach in a heterogeneous system.
Beyond determining the abundance, using a QconCAT also allows for determination of protein stoichiometry. Interplay between proteins is an important factor for acquisition of spore resistance mechanics,12,17 and many spore proteins are known to interact with each other.19−21 Information about protein stoichiometry can provide insight into how proteins relate to each other in vivo and give a sense of spatial dimensions occupied. Perturbations in this stoichiometry give a measure for integrity of the spore coat, which may be connected to acquired spore characteristics such as heat resistance or germination efficiency.
To further expand upon the QconCAT application, we quantified the selected proteins in both wild type B. cereus ATCC14579 and a ΔCotE mutant B. cereus strain previously described by Bressuire et al.6 This allowed us to determine whether these proteins were partially localized in the exosporium or in fact interact with CotE and gave an indication of their accessibility.
The 21 proteins selected for quantification cover known structural proteins, enzymes, a transcription factor, and several uncharacterized proteins. These proteins are located across the various different layers of the spore though in many cases localization has not been confirmed experimentally in B. cereus (Figure 1). A short functional description of each target protein and the selected Q-peptides included in the QconCAT as well as accompanying notable bibliography are displayed in Table 1.
Table 1. Selected Proteins and Corresponding Q-Peptide Sequencesa.
| gene ID | protein ID | protein description | peptide sequences |
|---|---|---|---|
| BC_0059 | SpoVT | Stage V sporulation protein T. Forespore specific transcriptional regulator.24,25 Identified in spore coat isolates (unpublished preliminary results). | (EHK)AVNTAASFLAK(QME) |
| (RIR)EGDPLEIFVDR(DGE) | |||
| BC_0212 | YusW | Putative uncharacterized protein. Previously identified in spore coat isolates (unpublished preliminary results). | (ITK)LSPLLQELK(FDK) |
| (DLK)LNFNEFDLK(ADY) | |||
| BC_0389 | CotB1 | Spore coat protein B1. Exosporium basal layer protein. In B. anthracis, CotB is CotE independent, found in both coat and exosporium isolates.5,26,27 | (FLK)DLIGSFVR(VNR) |
| (IVK)EEIILIAIK(HIK) | |||
| BC_0390 | CotB2 | Spore coat protein B2. Exosporium basal layer protein. In B. anthracis, CotB is CotE independent, found in both coat and exosporium isolates.5,26,27 | (ESR)VGELVSLGK(DYL) |
| (HIK)SVSQVVK(CKK) | |||
| BC_0987 | BC_0987 | Putative uncharacterized protein. Previously identified in spore coat isolates (unpublished preliminary results). | (ISR)TFVSLEPNR(TTK) |
| (RSR)NTFFPTQNELVEISR(TFV) | |||
| BC_1281 | CalY | Cell envelope-bound metalloprotease. Spore associated protease detected in exosporium isolates. Potentially involved in biofilms.14,28 | (KVK)FLWNWDK(QSE) |
| (WDK)QSEPVYETTLADLQK(VDP) | |||
| BC_1284 | InhA | Immune inhibitor A. Secreted virulence factor which regulates the secretome in Bacillus anthracis and thuringiensis.29,30 | (PGK)AADYGADAASGGHDNK(GPK) |
| (GLK)FEVVGQADDNSAGAVR(LYR) | |||
| BC_2064 | Alr1 | Alanine racemase 1. Quorum sensor for premature germination by l-alanine in exosporium.31−33 | (VIK)GDGISYNVTYR(TKT) |
| (VVK)ANAYGHDYVPVAK(TAL) | |||
| BC_2569 | BC_2569 | Collagen triple helix repeat protein. Previously identified in spore coat isolates (unpublished preliminary results). | (RVR)ATVDSLPIR(SRI) |
| (HLK)ANVQLVGTSTLLTR(LQI) | |||
| BC_2752 | YpeB | Hypothetical Membrane Spanning Protein. Cortex lysis partner to SleB.34 | (NSR)SSLSPALADVWR(LTS) |
| (QMK)IALDDGSIVGFSAK(EYL) | |||
| BC_2753 | SleB | Spore cortex-lytic enzyme. Key protein, together with CwlJ, for cortex degradation during germination.20,34 | (QEK)FGLPVDGLAGAK(TKQ) |
| (IQR)GASGEDVIELQSR(LKY) | |||
| BC_2872 | CotX1 | Spore coat protein X. Heavily cross-linked insoluble protein, important for structural integrity (density) of outer coat.35 | (ADR)VAQELFQK(SSI) |
| (SFK)NASVSEAAAQESK(TYQ) | |||
| BC_2889 | IunH | Inosine-uridine preferring nucleoside hydrolase. Involved in inosine- or adenosine-induced germination.33,36 | (IQR)IAVGFNYAAFK(EEF) |
| (VSR)DIVTENVYFLER(YYA) | |||
| BC_3607 | YaaH | Spore peptidoglycan hydrolase. Assists breakdown of cortex by cleaving products of SleB/CwlJ activity.20,34 | (QNK)FITNILQTAQK(YGM) |
| (LYR)ISQTYNVPLASLAK(VNN) | |||
| BC_3712 | BclC | Hypothetical Membrane Spanning Protein. Frequently found in spore coat protein isolates. BLAST matches to Hypothetical proteins. Contains collagen region with GXX repeats common in exosporium proteins.14,37 | (AGR)IPNTPSIPITK(AQL) |
| (GAR)ISVQSTLNEITIPATGNTNIR(LTV) | |||
| BC_3770 | CotE | Spore coat protein E. Major morphogenetic protein, anchors exosporium and many outer coat proteins.5 | (VER)EFVTEVVGETK(ICV) |
| (TER)VNYTDEVSIGYR(DKN) | |||
| BC_4419 | YhcN/CoxA | Sporulation cortex protein. Unknown function, forespore specific Sig F/G. B. subtilis mutants have hampered outgrowth.38 | (YER)TSYNDTHQYR(DNV) |
| (VDR)VSTVVYGNDVAIAVKPR(PRN) | |||
| BC_4420 | SafA | SpoVID-dependent spore coat assembly factor. Major morphogenetic protein, directs inner coat proteins.8,39 | (_MR)IHIVQK(GDT) |
| (HMK)QQAGAGSAPPK(QYV) | |||
| BC_4640 | YtfJ/GerW | Putative uncharacterized protein. Germination receptor, stimulus unknown.40 | (PIK)AADGSVILTVSK(VSF) |
| (IDK)IIELAPQAVDK(VKE) | |||
| BC_5056 | BC_5056 | Collagen adhesion protein. Unknown function, frequently found in spore coat protein isolates. Adhesin-like protein indicates it may be important as a virulence factor. | (MGK)VDINVYR(VLL) |
| (AHK)GTPTIQNAVVLLER(SEE) | |||
| BC_5391 | GerQ | Spore coat protein GerQ. Structural protein, potentially highly cross-linked. Important for CwlJ localization. Important for germination with Ca2+-DPA.41−43 | (KGK)QATVVMTYER(GSS) |
| (YER)GSSLGTQSYTGIIEAAGR(DHI) |
Bracketed sequence represents the native primary sequence context, which was not included in the QconCAT.
The abundance of 21 spore proteins is determined using a QconCAT approach in a heterogeneous system. In this manner, 5.66% ± 0.51 of the total spore weight has been quantified in wild type B. cereus and 4.13% ± 0.14 in a ΔCotE mutant strain. Using the obtained abundances, it is possible to classify proteins by function, structural, or otherwise. Furthermore, determining protein stoichiometry provides insight into the way either known or potential interactors may relate to each other. The potential of the most abundant proteins for use as biomarkers is assessed using the immunogenic epitope prediction algorithm SVMTriP.22
Materials and Methods
B. cereus Growth and Sporulation Conditions
B. cereus ATCC 14579 was grown in Tryptic Soy Broth medium at 30 °C. A ΔCotE knockout mutant strain previously described by Bressuire-Isoard et al.6 was grown in Tryptic Soy Broth supplemented with 275 μg/mL spectinomycin. Overnight cultures were harvested and washed once and transferred to 250 mL of chemically defined growth and sporulation medium (CDGS)23 at an optical density of 2 at 600 nm. Growth and sporulation were allowed to continue for 4 days until a > 95% spore crop was obtained. Spores were harvested by centrifugation, washed with 1.25% Tween-20 once, followed by four washes with cold mili-Q water after which spores were freeze-dried and stored at −80 °C before protein isolation.
Spore Coat Protein Isolation
Proteins in the spore coat insoluble fraction were isolated using a modified version of the method developed by Abhyankar et al.16 Lyophilized spores were resuspended in cold 10 mM Tris-HCl and subjected to two cycles of bead beating with Zirconia-Silica beads consisting of three rounds of 40 s beating, 60 s rest, resting on ice for 10 min between cycles. Beads were washed with 1 M NaCl five times to remove cytosolic proteins and nonspecific proteins.
The soluble fraction of the spore proteome was removed by SDS extraction (50 mM Tris-HCl (pH 7.8), 0.2% SDS, 100 mM Na-EDTA, 100 mM β-ME) for 10 min at 80 °C. This also leaves the insoluble fraction of the proteome more accessible to trypsin. After centrifugation, the insoluble pellet was washed three times with mili-Q water and freeze-dried.
Peptide Sample Preparation
Freeze-dried protein material was resuspended in 100 mM ammonium bicarbonate and reduced with 10 mM DTT at 55 °C for 60 min and alkylated with 55 mM iodoacetamide for 45 min in the dark. Samples were centrifuged, and the pellet was resuspended in 100 mM ammonium bicarbonate, 10% acetonitrile, and 5 μg of Trypsin was added per mg of material. At this point, QconCAT was added in different quantities as indicated. The digestion was allowed to proceed for 18 h at 37 °C unless otherwise indicated. After digestion, the supernatant was collected and the pellet was washed twice with 100 mM ammonium bicarbonate. Up to 1% TFA was added to acidify the buffer, which removed ammonium bicarbonate and inactivated trypsin. Samples were freeze-dried and resuspended in 50% ACN, 0.1% TFA and stored at −80 °C until use.
QconCAT Design
The QconCAT was designed based on initial nonquantitative analyses of the spore coat insoluble fraction (unpublished preliminary results). Reproducibly observed proteins were selected for quantification if two peptides were observed, which could be considered Q-peptides as described by Brownridge et al.13
To be able to include several proteins of interest, some liberties had to be taken with the strict requirements for Q-peptides proposed previously,13 for instance peptides starting with glutamic acid were allowed. The list of selected peptides and the corresponding proteins is displayed in Table 1. A standard GluFib peptide B (EGVNEEGFFSAR) was included for preliminary method development. A sacrificial N-terminal peptide MAGR was included to provide an initiator Methionine and protect the N-terminus of the first Q-peptide. A standard His-tag (LAAALEHHHHHH) was added to aid in purification. The QconCAT was assembled, expressed, and purified by PolyQuant GmbH (report in Supporting Information S1) and stored at −80 °C in 0.5% HAc-buffer until use.
Mass Spectrometry Analysis
Liquid chromatography–tandem mass spectrometry (LC–MS/MS) data were acquired with an Bruker ApexUltra Fourier Transform Ion Cyclotron Resonance Mass Spectrometer (Bruker Daltonics, Bremen, Germany) equipped with a 7 T magnet and a nanoelectrospray Apollo II DualSource coupled to an Ultimate 3000 (Dionex, Sunnyvale, CA, USA) High-Performance Liquid Chromatography system. For each digest, a similar amount of material was dried in a vacuum concentrator and resuspended in 100% TFA and sonicated for 15 min to disaggregate peptides, which improved chromatographic behavior.44 TFA was removed by blowing the sample dry with Argon gas. Peptides were resuspended and injected as a 40 μL of 0.1% trifluoroacetic acid solution and loaded onto a PepMap100 C18 (5 μm particle size, 100 Å pore size, 300 μm inner diameter × 5 mm length) precolumn at a flow rate of 50 μL/min. Following injection, the peptides were eluted via an Acclaim PepMap 100 C18 (3 μm particle size, 100 Å pore size, 75 μm inner diameter × 500 mm length at 60 °C) analytical column (Thermo Scientific, Etten-Leur, The Netherlands) to the nanoelectrospray source. LC Gradient profiles of up to 120 min were obtained using 0.1% formic acid/99.9% H2O (A) and 0.1% formic acid/80% acetonitrile/19.9% H2O (B) (0 min 97% A/3% B, 2 min 94% A/6% B, 110 min 70% A/30% B, 120 min 60% A/40% B, 125 min 100% B) at a flow rate of 300 nL/min. Data dependent Q-selected peptide ions were fragmented in the hexapole collision cell at an argon pressure of 6 × 10–6 mbar (measured at the ion gauge) and the fragment ions were detected in the ICR cell at a resolution of up to 60 000. In the MS/MS duty cycle, three different precursor peptide ions were selected from each survey MS. The MS/MS duty cycle time for one survey MS and three MS/MS acquisitions was about 2 s. Instrument mass calibration was better than 5 ppm over an m/z range of 250–1500.
Data Analysis
Raw Fourier transform (FT)-MS/MS data were processed with the MASCOT DISTILLER program, version 2.4.3.1 (64 bit), MDRO 2.4.3.0 (MATRIX science, London, UK) including the Search toolbox and the Quantification toolbox. Peak-picking for both MS and MS/MS spectra was optimized for the mass resolution of up to 60 000. Peaks were fitted to a simulated isotope distribution with a correlation threshold of 0.7, with minimum signal-to-noise ratio of 2.
The processed data were searched with the MASCOT server program 2.3.02 (MATRIX science, London, UK) against a complete B. cereus ATCC14579 predicted proteome database obtained from UniPROT (accessed January 2016) complemented with the QconCAT protein amino acid sequence. Trypsin was used as enzyme and one missed cleavage was allowed. Carbamidomethylation of cysteine, oxidation of methionine, and pyroglutamination of N-terminal glutamic acid were included as variable modification. The peptide mass tolerance was set to 25 ppm and the peptide fragment mass tolerance was set to 0.03 Da. Where applicable, the quantification method was set to the metabolic 15N labeling method to enable MASCOT to identify both the 14N target and 15N QconCAT peptides.
For identification results, identified proteins were determined using MASCOT with MudPIT peptide identification score set to a cutoff of 20. At this cutoff and based on the number of assigned decoy peptide sequences, the significance threshold was adjusted to obtain a peptide false discovery rate of 2% for all analyses. The MASCOT protein identification reports were exported as XML and then imported in a custom-made VBA software program running in Microsoft Excel. A list of all proteins identified is represented in Supplementary Table 1.
Using the quantification toolbox, the quantification of the light target peptides relative to the corresponding heavy QconCAT peptides was determined as the heavy over light isotopic ratio using Simpsons integration of the peptide MS chromatographic profiles for all detected charge states. Correlation threshold for isotopic distribution fit was set to 0.98, with a detected 15N label content of 99.6%. XIC threshold, 0.1; all charge states, on; max XIC width, 200 s. Allowed elution time shift for the heavy and light peptides was limited to 20 s.
All isotope ratios were manually validated by inspecting the MS spectral data. For target peptides, for which overall replica analyses were occasionally not MS/MS identified, the isotope ratio was manually calculated from their extracted heavy and light peptide ion chromatograms as the ratio of the areas under the curves using Bruker DataAnalysis 4.2 program (Bruker Daltonics, Bremen, Germany).
The heavy over light isotopic ratios for all target peptides over all analyses are systematically listed in Supplementary Table S2. The mass spectrometry proteomics data of the wild-type QconCAT analyses have been deposited to the ProteomeXchange Consortium45 via the PRIDE partner repository with the data set identifier PXD003569.
Method Verification
QconCAT concentration was determined by PolyQuant GmbH using a Bradford protein assay.
QconCAT approaches are usually performed in a homogeneous setting requiring complete digestion of both sample and QconCAT. The insoluble nature of a large part of the spore proteome therefore presents an issue in translating this method to spore coat analyses. Though insoluble residual material remains after performing digestion with trypsin, completed digestion of spore material was assured by optimizing digestion time until no change was observed in isotope ratios. Triplicate digests of an insoluble fraction isolate were made with digestion time varying from 16 to 22 h. Isotope ratios were plotted versus digestion time and minimal change was observed for most peptides over the selected time frame. In the ones that did change, this was mostly due to data points conceived through manual integration of the chromatogram. Overall 18 h of digestion time was selected as optimal. Data are made available in Supporting Information S2. This validates the QconCAT approach insofar that the obtained data quantifies the entirety of the proteins of interest that can be accessed by trypsin. Selective inaccessibility (i.e., cross-linking of one of the Q-peptides) is assessed by using two Q-peptides per protein and comparing the difference between them (see below). There remains a possibility that a fraction of the population of a protein of interest is completely blocked off; however, using mechanical and chemical disruption as well as enzymatic cleavage makes this possibility remote.
Statistical Approach
Acquired isotope ratios were multiplied by the amount of QconCAT used in each experiment to convert the unit to pmol/mg. Data points were lumped together for each protein across all analyses: three biological replicates with three technical replicates each, using four different QconCAT amounts for each or only one concentration of QconCAT (37.5pmol/mg of spore material) for the ΔCotE analyses. The average value and 95% confidence interval were determined using a bootstrap approach.46,47 The obtained data set was resampled using a vector length equal in size to the number of acquired data points separately for each protein and bootstrapped to a value of n = 1000 using Microsoft Excel.
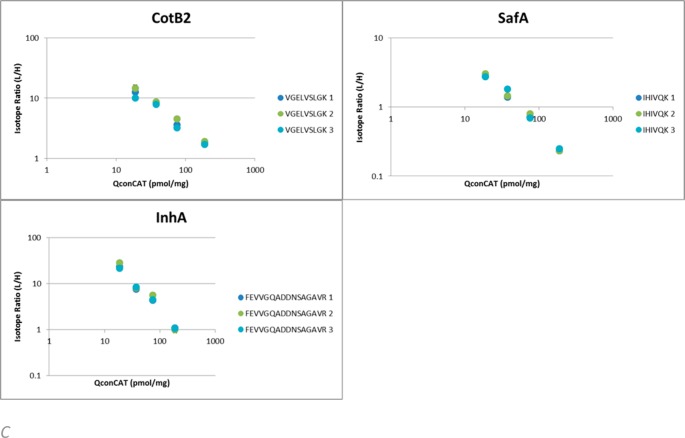
This statistical approach relies on extensive experimental sampling and allows all types of variance to be indicated under one denominator. This is enabled by quantifying across a range of QconCAT reference concentrations. To appreciate the different sources of variance, the isotope ratios for each peptide at each concentration were plotted and linearity on a double log scale was verified across the range. As the total amount of peptide material released from the sample should be the same across all technical replicates, the sum of the quantified peptides in each digestion should be the same. We can correct for any mixing/weighing error by normalizing to the average yield across each biological replicate. The resultant plots are shown in Figure 2.
Figure 2.
Per peptide quantitation results in wild type B. cereus spores. Linearity of L/H isotope ratios averaged across technical replicates for each individual Q-peptide with varying concentrations of QconCAT in wild type B.cereus spores. Error bars indicate standard deviation. (A) L/H ratios for proteins showing similar results for both Q-peptides. (B) Divergent L/H ratios between Q-peptides seen across biological replicates (1, 2, 3) indicates either selective modification through cleaving off part of the protein or more likely partial accessibility to trypsin due to structural cross-links. (C) For three proteins, only one Q-peptide was recovered and used for quantification.
Results and Discussion
Identifications
A total of 713 proteins were identified in ΔCotE isolates, having a MASCOT identification in at least two out of three replicates. Beyond these, 92 were only identified in one replicate in ΔCotE isolates and are disregarded for discussions below. A total of 560 proteins overlapped between the mutant and wild type cells. Of those proteins previously identified in wild type isolates, 163 proteins were not detected in ΔCotE isolates indicating they were CotE dependent for their association with the spore. A further 153 proteins were identified exclusively in the ΔCotE isolates, which indicates that their accessibility is higher upon removal of CotE. Proteins of interest are displayed in the tables below. All identifications across ΔCotE isolates compared to the collected identifications from wild type isolates are displayed in Supplementary Table S1.
CotE Independent Proteins
The majority of protein identifications were not dependent on CotE. Identified proteins known to be spore associated are listed in Table 2. Interestingly, these included proteins originating from all layers of the spore. Examples are SpoIVA, SafA, CotX, and InhA, which are located at the cortex boundary, inner coat, outer coat, and exosporium, respectively. This suggests that the CotE deletion does not completely remove any of the spore layers.
Table 2. Identified Proteins Known To Be Related to Spores Both in Presence and Absence of CotE.
| protein description | ID |
|---|---|
| 1-pyrroline-5-carboxylate dehydrogenasea | rocA |
| Alanine racemase 1ab | alr1 |
| Catalase | BC_1155 |
| Cell envelope-bound metalloprotease (Camelysin)ab | BC_1281 |
| Cell surface proteinab | BC_3547 |
| Cell surface proteinab | BC_2639 |
| Cell wall hydrolase cwlJa | BC_5390 |
| Cell wall-associated hydrolase | BC_2849 |
| Collagen adhesion proteina | BC_5056 |
| CotJA proteina | BC_0823 |
| CotJB proteina | BC_0822 |
| CotJC proteina | BC_0821 |
| d-alanyl-d-alanine carboxypeptidasea | BC_4075 |
| Enolasea | eno |
| Ferredoxin--nitrite reductasea | BC_1424 |
| Germination proteasea | gpr |
| Hypothetical Cytosolic Proteina | BC_3986 |
| Hypothetical Cytosolic Proteinab | BC_3195 |
| Hypothetical Exported Proteinab | BC_1334 |
| Hypothetical Membrane Spanning Proteinab | BC_3712 |
| Hypothetical Membrane Spanning Proteina | BC_2752 |
| Hypothetical Membrane Spanning Proteinab | BC_1221 |
| IG hypothetical 17193a | BC_3534 |
| IG hypothetical 18063a | BC_1029 |
| Immune inhibitor Aab | BC_1284 |
| Inosine-uridine preferring nucleoside hydrolasea | BC_3552 |
| Inosine-uridine preferring nucleoside hydrolaseab | BC_2889 |
| N-acetylmuramoyl-l-alanine amidase | BC_2823 |
| N-acetylmuramoyl-l-alanine amidase | BC_1494 |
| Nonheme chloroperoxidasea | BC_4774 |
| Oligopeptide-binding protein oppAa | BC_3586 |
| Oligopeptide-binding protein oppA | BC_3585 |
| Oligopeptide-binding protein oppA | BC_3584 |
| Oligopeptide-binding protein oppAa | BC_2026 |
| Protein translocase subunit YajCa | BC_4410 |
| Putative hydrolasea | BC_3133 |
| Putative stage IV sporulation protein | BC_4303 |
| Putative uncharacterized proteina | BC_5391 |
| Putative uncharacterized proteina | BC_4640 |
| Putative uncharacterized proteina | BC_4419 |
| Putative uncharacterized proteina | BC_4387 |
| Putative uncharacterized proteinab | BC_4047 |
| Putative uncharacterized proteinab | BC_3992 |
| Putative uncharacterized proteinab | BC_3582 |
| Putative uncharacterized proteina | BC_3090 |
| Putative uncharacterized proteina | BC_2969 |
| Putative uncharacterized proteina | BC_2745 |
| Putative uncharacterized proteinab | BC_2493 |
| Putative uncharacterized proteina | BC_2481 |
| Putative uncharacterized proteinab | BC_2426 |
| Putative uncharacterized proteinab | BC_2267 |
| Putative uncharacterized proteinab | BC_2266 |
| Putative uncharacterized proteinab | BC_2149 |
| Putative uncharacterized proteina | BC_2099 |
| Putative uncharacterized proteina | BC_2095 |
| Putative uncharacterized proteina | BC_1391 |
| Putative uncharacterized proteinb | BC_1245 |
| Putative uncharacterized proteinab | BC_0996 |
| Putative uncharacterized proteinab | BC_0987 |
| Putative uncharacterized proteinab | BC_0944 |
| Putative uncharacterized proteina | BC_0825 |
| Putative uncharacterized proteina | BC_0212 |
| Small, acid-soluble spore protein 2 | sasP-2 |
| Spore coat protein Bab | BC_0390 |
| Spore coat protein Bab | BC_0389 |
| Spore coat protein Da | BC_1560 |
| Spore coat protein Gab | BC_2030 |
| Spore coat protein Xab | BC_2874 |
| Spore coat protein Xab | BC_2872 |
| Spore coat protein Yab | BC_1222 |
| Spore coat protein Yab | BC_1218 |
| Spore coat-associated protein Na | BC_1279 |
| Spore cortex-lytic enzymea | sleB |
| Spore peptidoglycan hydrolase (N-acetylglucosaminidase)a | BC_3607 |
| Sporulation-specific N-acetylmuramoyl-l-alanine amidasea | BC_2207 |
| SpoVID-dependent spore coat assembly factor SafAa | BC_4420 |
| Stage IV sporulation protein Aa | BC_1509 |
| Stage V sporulation protein AD | BC_5148 |
| Stage V sporulation protein AD | BC_4067 |
| Stage V sporulation protein AF | BC_4064 |
| Stage V sporulation protein S | BC_3776 |
| Stage V sporulation protein T | BC_0059 |
| Superoxide dismutasea | BC_1468 |
| Superoxide dismutase [Cu–Zn] | BC_4907 |
| Thiol peroxidasea | BC_4639 |
| Trigger factora | tig |
| UPF0145 protein BC_5181a | BC_5181 |
| Zinc proteasea | BC_3787 |
| Zinc proteasea | BC_3786 |
| Zn-dependent hydrolasea | BC_3977 |
| Zn-dependent hydrolasea | BC_1613 |
Indicates proteins previously identified by Abhyankar et al.14
Indicates proteins previously identified in exosporium isolates.
CotE Dependent Localization
Comparing proteins isolated from wild type spores to those isolated from CotE knockout spores with perturbed exosporia can reveal proteins that are dependent on CotE for their association with the spore, and by extension are likely localized in the exosporium. Important to note is that removing the exosporium or damaging its integrity also eliminates the interspace region of the spore. Proteins indicated to depend on CotE for their association with the spore are therefore not necessarily spore specific and could merely be part of the mother cell cytosol trapped in the interspace. Interpretation then requires background knowledge of the function of identified proteins to determine whether they are likely spore specific. Identified proteins with a known function in sporulation or spore biogenesis, showing dependence on CotE are displayed in Table 3. The full list of identified proteins is available in Supplementary Table S1.
Table 3. Identified Proteins Known To Be Related to Spores Exclusively Identified in Wild Type Isolates.
| protein description | ID |
|---|---|
| Collagen triple helix repeat protein | BC_3481 |
| Collagen triple helix repeat protein | BC_2570 |
| Collagen triple helix repeat proteinab | BC_2569 (BclF) |
| Collagen-like triple helix repeat proteinab | BC_3345 |
| Hypothetical Glycosyltransferasea | BC_3515 |
| Immune inhibitor A | BC_0666 |
| Immune inhibitor A | BC_2984 |
| N-acetylmuramoyl-l-alanine amidase | BC_2822 |
| N-acetylmuramoyl-l-alanine amidase | BC_3677 |
| Peptidoglycan N-acetylglucosamine deacetylase | BC_1960 |
| Putative uncharacterized proteinab | BC_2427 |
| Putative uncharacterized proteinab | BC_2878 |
| Putative uncharacterized proteina | BC_0063 |
| Putative uncharacterized proteina | BC_2375 |
| Putative uncharacterized proteinabc | BC_p0002 |
| Small acid-soluble protein gamma type | sspE |
| Small acid-soluble spore protein | BC_1984 |
| Spore coat protein Ec | BC_3770 (CotE) |
| Stage IV sporulation protein B | BC_4172 |
Indicates proteins previously identified by Abhyankar et al.14
Indicates proteins previously identified in exosporium isolates.
Indicates a protein that is removed during creation of the CotE knockout strain.
An alternate strategy was previously reported by Abhyankar et al.14 who identified proteins in purified exosporium fractions of B. cereus as well as combined spore coat and exosporium isolates. By using French press to apply a shear stress, it is possible to remove parts of the exosporium; however, this method is not capable of completely removing the exosporium. Likely some fragments remain at the cap region48 where the exosporium is supposedly anchored to the coat. This was reflected in their results as the number of proteins missing from spore coat isolates where a French press step was applied was quite limited. Only three proteins, InhA, CalY, and a Putative uncharacterized protein BC_1591, were specific to exosporium containing isolates. InhA and CalY are quantified using QconCAT and discussed below. BC_1591 was identified in only one replicate analysis making biological interpretation of this particular identification difficult. The results obtained in this study complement the results obtained previously as several proteins were both detected in exosporium isolates and are missing from ΔCotE spore isolates as displayed in Table 3. This strongly suggests that BC_2569 (BclF), BC_3345, BC_2427, and BC_2878 are exosporium proteins. Beyond that, several proteins can be localized to the exosporium based on homology to known exosporium proteins. For instance, Immune inhibitor A (BC_1284) is a known exosporium component, its homologues BC_0666 and BC_2984 are thus likely to be exosporium associated. Furthermore, BC_3481 and BC_2570 both contain collagen-like triple helix repeat domains frequently found in exosporium proteins37 are therefore likely exosporium proteins. Other proteins will require further evidence to be appointed to the exosporium.
CotE Dependent Accessibility
Apart from proteins specifically being identified in the wild type, there are also a number of proteins that were exclusively identified in the ΔCotE isolates. As CotE is a major structural protein that interacts with many different proteins directly, lacking CotE might result in interacting proteins to become less tightly bound (i.e., lacking cross-links) or no longer being encased in CotE or CotE dependent proteins. Therefore, they can become more accessible to tryptic digestion and become newly identified. The resultant new identifications specific to the ΔCotE mutant isolates are displayed in Table 4. The full list of identified proteins is available in Supplementary Table S1.
Table 4. Identified Proteins Known To Be Related to Spores Exclusively Identified in ΔCotE Isolates.
| protein description | ID |
|---|---|
| Antisigma F factor | spoIIAB |
| Antisigma factor antagonist | BC_4074 |
| Oligopeptide-binding protein oppA | BC_2848 |
| Prespore specific transcriptional activator rsfA | BC_3922 |
| Putative uncharacterized proteina | BC_0263 |
| Putative uncharacterized proteina | BC_1708 |
| SpoIISA like protein | BC_2436 |
| Stage III sporulation protein D | BC_5282 |
| Stage V sporulation protein S | BC_2142 |
| Stage VI sporulation protein D | BC_4467 |
Indicates proteins previously identified by Abhyankar et al.14
SpoVID is involved in spore coat deposition; specifically it is at the interface between the cortex and coat layers where it anchors the coat layer and is known to interact with CotE directly.8,49 Identification of this protein corroborates the notion that coat integrity is compromised allowing trypsin to access this protein. Other spore related proteins specific to ΔCotE isolates are mostly regulatory proteins important early during sporulation.3 Expression of CotE or lack thereof does not affect this early regulation50 once again indicating it is increased accessibility which is causing these proteins to be identified. Altogether, this suggests these proteins are encased by CotE yet it is apparently not the (sole) reason why they are retained in the spore.
Protein Abundance
Quantitative analyses were performed by combining the spore coat insoluble fraction isolates with different concentrations of QconCAT (18.75, 37.5, 75, 187.5 pmol per milligram dry weight of insoluble coat material). Isotope ratios for each peptide at each concentration were plotted and linearity on a double log scale was verified across the range (Figure 2A), these also show the variance due to experimental errors to be small. Variance between two Q-peptides of the same protein also becomes apparent and, though small for most proteins, differed significantly for CotB1, SpoVT, IunH, BclC, YhcN, BC_5056 (Figure 2B). Ratios of both peptides were used for calculating the quantified value for the protein with the exception of CotB2, SafA, and InhA where only one peptide was reliably measured (Figure 2C). Using the higher of the two quantified peptides in these cases is worthy of consideration if the observed variance is biological in nature as discussed below. Having confirmed the linear fit to the isotope ratios, we calculated the copy number of proteins per milligram insoluble fraction by multiplying the isotope ratios with the amount of QconCAT added. To convert these obtained quantities of proteins per milligram of spore dry weight to copy numbers per spore, we need to know the dry weight of a single spore which in the case of B. cereus is estimated to be around 1 picogram.51 As spores can differ in size,52 this may have a larger variance than the data obtained here. Assuming the insoluble fraction represents the majority of the spore’s dry weight the unit (pmol/mg) of the numbers obtained above can then be converted to molecules per spore by multiplying with a conversion factor of 600 (pmol/mg = 6 × 1023 molecules/1021 spores)(Table 5).
Table 5. Protein Abundance in Molecules Per Spore and Percentage of Total Weighta.
| molecules per spore |
% weight |
|||
|---|---|---|---|---|
| proteins | wild type | ΔCotE | wild type | ΔCotE |
| CotB1 | 1.9 × 105 ± 1.7 × 104 | 1.4 × 105 ± 7.6 × 103 | 0.60 ± 5.6 × 10–2 | 0.44 ± 2.4 × 10–2 |
| InhA | 1.6 × 105 ± 1.9 × 104 | 7.9 × 104 ± 1.6 × 103 | 2.3 ± 0.27 | 1.1 ± 2.3 × 10–2 |
| CotB2 | 1.5 × 105 ± 1.1 × 104 | 2.3 × 105 ± 3.3 × 103 | 0.42 ± 3.0 × 10–2 | 0.64 ± 9.2 × 10–3 |
| CotX | 1.5 × 105 ± 7.0 × 103 | 1.3 × 105 ± 3.0 × 103 | 0.40 ± 1.9 × 10–2 | 0.35 ± 8.1 × 10–3 |
| SasS | 1.0 × 105 ± 4.7 × 103 | 3.1 × 104 ± 8.8 × 102 | 0.25 ± 1.1 × 10–2 | 7.5 × 10–2 ± 2.1 × 10–3 |
| GerQ | 8.9 × 104 ± 4.5 × 103 | 1.7 × 105 ± 2.6 × 103 | 0.24 ± 1.2 × 10–2 | 0.45 ± 6.9 × 10–3 |
| CalY | 5.6 × 104 ± 8.6 × 103 | 2.6 × 104 ± 1.8 × 103 | 0.20 ± 3.1 × 10–2 | 9.4 × 10–2 ± 6.7 × 10–3 |
| CotE | 3.5 × 104 ± 1.6 × 103 | 0.12 ± 5.5 × 10–3 | ||
| BclC | 2.7 × 104 ± 2.9 × 103 | 2.5 × 104 ± 2.6 × 103 | 0.34 ± 3.6 × 10–2 | 0.32 ± 3.3 × 10–2 |
| SafA | 2.6 × 104 ± 9.0 × 102 | 1.5 × 104 ± 2.6 × 102 | 0.29 ± 9.8 × 10–3 | 0.16 ± 2.8 × 10–3 |
| YhcN | 1.9 × 104 ± 1.3 × 103 | 2.1 × 104 ± 1.9 × 103 | 7.9 × 10–2 ± 5.3 × 10–3 | 8.6 × 10–2 ± 7.8 × 10–3 |
| BC_5056 | 1.5 × 104 ± 1.5 × 103 | 9.4 × 103 ± 1.1 × 103 | 8.9 × 10–2 ± 8.7 × 10–3 | 5.4 × 10–2 ± 6.2 × 10–3 |
| YaaH | 1.1 × 104 ± 4.2 × 102 | 1.4 × 104 ± 4.5 × 102 | 9.1 × 10–2 ± 3.3 × 10–3 | 0.11 ± 3.6 × 10–3 |
| Alr1 | 1.1 × 104 ± 3.3 × 102 | 1.0 × 104 ± 1.9 × 102 | 8.1 × 10–2 ± 2.4 × 10–3 | 7.5 × 10–2 ± 1.4 × 10–3 |
| YusW | 7.2 × 103 ± 3.8 × 102 | 7.9 × 103 ± 3.6 × 102 | 2.1 × 10–2 ± 1.1 × 10–3 | 2.3 × 10–2 ± 1.1 × 10–3 |
| IunH | 5.6 × 103 ± 6.6 × 102 | 5.5 × 103 ± 5.1 × 102 | 3.4 × 10–2 ± 4.0 × 10–3 | 3.3 × 10–2 ± 3.1 × 10–3 |
| SpoVT | 5.1 × 103 ± 3.7 × 102 | 6.8 × 103 ± 2.9 × 102 | 1.7 × 10–2 ± 1.2 × 10–3 | 2.2 × 10–2 ± 9.6 × 10–4 |
| YtfJ | 4.0 × 103 ± 1.4 × 102 | 5.0 × 103 ± 1.3 × 102 | 9.4 × 10–3 ± 3.2 × 10–4 | 1.2 × 10–2 ± 2.9 × 10–4 |
| BclF | 3.6 × 103 ± 1.9 × 102 | 8.8 × 102 ± 6.6 × 101 | 3.2 × 10–2 ± 1.7 × 10–3 | 7.8 × 10–3 ± 5.9 × 10–4 |
| YpeB | 3.0 × 103 ± 2.9 × 102 | 3.2 × 103 ± 1.1 × 102 | 2.5 × 10–2 ± 2.4 × 10–3 | 2.7 × 10–2 ± 8.9 × 10–4 |
| SleB | 2.3 × 103 ± 7.4 × 101 | 3.0 × 103 ± 2.7 × 101 | 1.1 × 10–2 ± 3.5 × 10–4 | 1.4 × 10–2 ± 1.3 × 10–4 |
Margins indicate 95% confidence intervals of the bootstrapped average.
To understand how much of the spore is covered by these analyses, we made a weight comparison for each protein by multiplying copy numbers with protein masses (Table 5). This indicated 5.66% ± 0.51 of the total spore weight is covered by these proteins with InhA taking up 2.33% ± 0.27 as the largest contributor. The total amount of protein per spore which is estimated at around 70 wt %53 indicating around 8% of the entire spore protein weight is quantified using only the 21 proteins included in this QconCAT analysis. As the total number of proteins we identify from spore samples ranges in the hundreds (700 in a single LC–MS/MS analysis), this signifies a substantial part was covered using this limited set of proteins.
The rather unique properties of some spore proteins do not lend them suitable for use within a QconCAT strategy due to them containing either a large number of cysteines, repeat sequences prone to missed cleavage, regions very rich in lysine, etc. This made it impossible to include several proteins frequently identified in initial analyses such as CotX2, CotY1, CotY2, CwlJ, and CotAlpha a.o. To complete the quantitative assessment of the spore coat, other methods such as AQUA will need to be employed.54
Quantitative Dependence
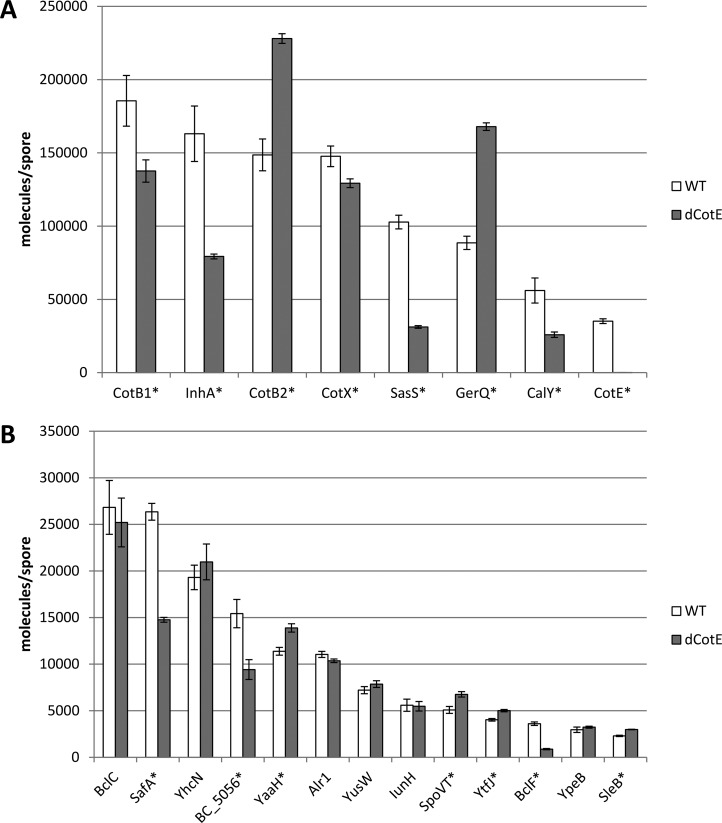
We used the Qcstel01 synthetic QconCAT protein to quantify 21 proteins from across all layers of the spore. In this manner, we can determine CotE dependence or interaction beyond the yes/no answer of identification as described above. Many proteins were significantly different in abundance compared to wild type, as displayed in Figure 3. Though CotE and BclF light peptides were not identified in the ΔCotE mutant strain, the method for quantification searches for light peptides matching the heavy peptides introduced with the QconCAT. This results in CotE being quantified at an extremely low level corresponding to noise levels and BclF at severely reduced levels.
Figure 3.
Difference in protein abundance in ΔCotE versus WT. (A) High abundant proteins. (B) Medium to low abundant proteins. Error bars indicate 95% confidence interval of the bootstrapped average. Proteins indicated with ∗ are significantly different from WT.
Fourteen out of 20 proteins quantified (excluding CotE itself) are significantly affected by the ΔCotE mutation. The proteins most severely affected include known exosporium proteins (CotB1, InhA, CotB2, CalY, BclF). This corresponds to the visual phenotype of spores having defects in their exosporium. Likewise, levels of candidate exosporium proteins SasS and BC_5056 are indeed reduced in ΔCotE isolates. This indicates that though their incorporation in the spore is not wholly dependent on CotE, they are severely affected by its absence. It is likely that the reduced amounts recovered from ΔCotE isolates are maintained in the small fragments of exosporium which still form despite the absence of CotE.6
Previously, CotB was indicated to be CotE independent in B. anthracis.5,26,27B. cereus CotB shows the same behavior qualitatively; however, quantitative differences exist. Interestingly, unlike its orthologue CotB1, CotB2 is actually quantified at a higher level than in wild type. Following the general theme of proteins detected exclusively in ΔCotE isolates being more accessible to trypsin, the same thing appears to be happening quantitatively to CotB2. What is striking is that CotB1 is actually less abundant in ΔCotE isolates indicating that although they are orthologues, their interactions appear different.
Structural proteins SafA and GerQ are components of the inner coat layer and were significantly affected by deletion of CotE. Interestingly, where SafA was less abundant in ΔCotE isolates, GerQ was actually more abundant. As previously mentioned, the likely explanation is related to the nature of the interaction between proteins. GerQ has been indicated to be cross-linked by transglutaminase, an enzyme located across all coat layers, resulting in glutamyl-lysine isopeptide cross-links17,18 in B. subtilis. Although the specific N-terminal lysines indicated to be cross-linked55 are not present in the GerQ orthologue used by B. cereus, 19 out the first N-terminal 50 amino acids are glutamines indicating this function is likely maintained. CotE could then either be cross-linked to GerQ directly, or it is responsible for retention of proteins normally cross-linked to GerQ. In both scenarios, knocking out CotE would directly or indirectly make GerQ more accessible to trypsin. This generic effect of higher abundance in ΔCotE isolates is also seen among several other quantified proteins associated with the inner coat layers such as SleB, YtfJ, and YaaH but is most prevalent in the case of GerQ. Contrary to GerQ’s dense structure, SafA has an extended conformation in which the N-terminus spans the inner coat to interact with SpoVID at the spore cortex boundary.3 Potentially part of the SafA present is not directly attached to SpoVID and relies on outer coat integrity for its retention to the coat. Though the ΔCotE spores still maintain an outer coat layer, as mentioned above, the integrity of this layer is affected if this abundant protein is removed.
Of the proteins that show no significant difference in their abundance, IunH deserves special mention. IunH is an enzyme involved in modulating germination in response to inosine by hydrolyzing the germinant and reducing the signal triggering germination.33,36 This allows the spore to prevent premature germination in an environment where the environment is not suitable for growth. Bressuire-Isoard et al.6 report inosine triggered germination is impaired in the ΔCotE knockout strain. As IunH abundance appears not to be affected by deletion of CotE it seems that this phenotype of impaired germination is likely correlated to the germination receptors rather than the signal of the germinant itself.
Functional Classification by Protein Abundance
Protein abundance is determined across three orders of magnitude showing significant differences depending on the role of the protein (Figure 3). Structural proteins (CotB, CotX, CotE, SafA, GerQ) are generally more abundant than proteins with enzymatic functions (YaaH, SleB, Alr1, IunH) as is to be expected. Proteins located in the exosporium layer are also generally more abundant, which correlates to the size of the exosporium compared to the coat layers.
The uncharacterized protein BC_0987 was identified in many spore coat protein isolates indicating it may be spore associated and was therefore included in the QconCAT. It is determined to be as highly abundant as known structural proteins indicating, for lack of conserved (enzymatic) domains, it may be a structural spore protein. BLAST analysis reveals a homologue of this protein can be found across the B. cereus group. Strangely, there is a gene coding for a (nearly) identical protein (Accession number CKG89570.1) found in a singular isolate classified as Streptococcus pneumoniae(56) casting doubt onto whether or not BC_0987 is an actual spore protein as S.pneumoniae is incapable of producing spores. Interestingly, other verified spore proteins, such as CotX and CotB, also have nearly identical homologues (CJK03595.1 and CJA37784.1) in this particular isolate of S. pneumoniae indicating it perhaps has several residual nonfunctional spore protein homologues or this isolate is simply misclassified. Though the function of BC_0987 needs to be further confirmed via traditional knockout analysis, based on the data shown here we propose to rename this protein to structural abundant spore protein SasS.
BC_2569 contains collagen-like regions, which relates it to one of the best characterized exosporium proteins BclA. BclA is glycosylated protein, rich in GXX repeats known to be a major structural component of the exosporium in Bacillus anthracis.57 BclA is targeted to the exosporium by an N-terminal sequence, which is similar to an N-terminal sequence found in BC_2569 (LLGPTLPAIPP vs consensus LI/VGPTL/FPPIPP).58 Applying the same reasoning for classification as a structural protein to BC_2569 suggests it is in fact not a structural protein, or at least not a major one like BclA. Nonetheless, its structure can be classified as belonging to the group of Bcl proteins and Abhyankar et al.14 previously also identified it in B. cereus exosporium isolates. Here we propose to rename it BclF.
Alr1 and IunH are enzymes which modify germinant molecules, l-alanine and inosine, respectively, to prevent the initiation of germination prematurely.31,36 As such they are of crucial importance for the survival of the bacterium. Kinetics of triggering germination in response to stimulus with germinants rely on the conversion rate, which depends on the enzyme’s kinetic parameters and its abundance. The obtained abundances may therefore be useful to reinforce models for protein germination.
Stoichiometry: Interactors Have Similar Abundance
Proteins known to interact or function in a similar process appeared to be present at near equimolar ratios. For instance, paralogs CotB1 and CotB2 are roughly equally abundant (5:4) and, though structurally different, are thought to fulfill a similar role as a structural component of the exosporium.59 Their equal abundance suggests they may interact directly and have equally important roles in ensuring spore structure.
CotE and SafA are two of the major morphological proteins of the spore coat, each being required for deposition of an entire coat layer.5,8,39 In Bacillus subtilis, the two proteins have been shown to be anchored to the developing spore through the protein SpoVID though not at the same binding site.49,60 They appear to be present in a roughly 4:3 ratio CotE:SafA numerically suggesting the nonequimolar stoichiometry in interaction. This means either a fraction of the CotE present in spores does not interact with SpoVID or there is space for more copies of CotE to interact compared to SafA. Considering their respective protein masses, we can get an indication of the relative space occupied by the respective proteins. Multiplying the observed numeric ratio with respective protein masses, the proteins show a 2:5 ratio CotE/SafA. This indicates it is likely that there is indeed space for multiple copies of CotE to interact with one copy of SpoVID. To get a full picture of protein stoichiometry of the morphogenetic proteins in the spore coat layers and update the proposed structural model, quantification of SpoVID and SpoIVA is pending. Unfortunately, in our hands SpoVID and SpoIVA were not reproducibly detected when selecting proteins to include in the QconCAT and were therefore not included in the quantitative analyses. Hence, determining full stoichiometry of these morphogenetic proteins awaits complementary approaches.
YpeB and SleB are known to interact34 and appear to be present in a 5:4 ratio. YaaH is known to break down products of SleB and CwlJ19 and is more abundant. It would be interesting to see if the combined copy numbers of SleB and CwlJ equal that of YaaH or if these enzymes are kinetically balanced instead. Unfortunately, CwlJ is structurally incompatible with QconCAT analysis as possible tryptic peptides are either N-terminal, contain cysteines, or end in RR which are prone to missed cleavage and was therefore not included.
Biomarker Targets: Immunogenicity Prediction
The obtained data are of interest for practical applications for spore recognition by proposing potential protein biomarkers. As a ubiquitous food pathogen, proper detection of B. cereus is important for food safety. The golden standard for detection (ISO 7932) is still enumeration of colonies which is a time-consuming approach.1 Furthermore, spores show heterogeneity in their germination response, and so-called persistors and superdormant spores61−63 are difficult to assess with this approach. A way to circumvent these issues as well as potentially increasing sensitivity and assay speed would be to use biomarker detection instead. As the majority of the outside of the spore consists of proteins, these would be a prime target. To assess the potential biomarker targets, we used the SVMTriP algorithm22 to predict immunogenic epitopes of the most abundant proteins (CotB1, CotB2, InhA, CotX, and BC_0987), the results of which are shown in Table 6. Luckily, these proteins are all located on the outermost layers of the spore indicating they would likely be accessible to antibodies. Contrary to a top-down approach where immunization would take place using whole spores as antigens and then selecting which antibodies bind selectively, the approach used here using specific quantified biomarker protein allows for minimization of cross reactions thereby increasing specificity.
Table 6. Epitope Predictionsa.
| protein | location | epitope | score |
|---|---|---|---|
| CotB1 | 54–73 | KHLKSITKNAKECGSSDCEW | 1.000 |
| 100–119 | GPEKVEGILQDVSCDFVTLI | 0.548 | |
| CotB2 | 5–24 | LCCDQIKCLVGETVKVNLRG | 1.000 |
| 123–142 | GDEVIYVIKSHIKSVSQVVK | 0.932 | |
| 46–65 | HGELVYYQLKHVKSLVKKVK | 0.702 | |
| InhA | 436–455 | DYEKLNKGIGLATYLDQSVT | 1.000 |
| 692–711 | AIVGTLNGKPTVESSTRFQI | 0.903 | |
| 513–532 | FKSLYEIEAEYDFLEVHAVT | 0.846 | |
| 613–632 | TPQFKLDGFAVSNGTEKKSH | 0.832 | |
| 643–662 | GSDNALKFARGPVYNAGMVV | 0.766 | |
| 260–279 | DALKAAVDSGLDLSEFDQFD | 0.744 | |
| 18–37 | TQSAYAETPANKTATSPVDD | 0.708 | |
| 555–574 | TNGKWIDKSYDLSQFKGKKV | 0.697 | |
| 189–208 | LFGNEPFTLDDGSKIETFKQ | 0.671 | |
| 90–109 | LTKEASDFLKKVKDAKADTK | 0.662 | |
| CotX1 | 36–55 | QESKTYQISEESITIVDSAD | 1.000 |
| 106–125 | QINKQETVIRNSRNVTVTTT | 0.534 | |
| BC_0987 | none predicted |
Immunogenic regions with location within their respective protein and score based on the SVMTriP algorithm.22 Epitopes recommended by the algorithm are indicated in bold. These were submitted to BLAST and shown to be Bacillus cereus group specific with one exception in S. pneumoniae (see Results and Discussion).
Though most Q-peptides belonging to the same protein showed the same abundance, in some cases two Q-peptides for the same protein differed significantly, as illustrated in Figure 2. Spore proteins are known to be cross-linked together to form a coherent structure, Abyankhar et al.12 previously showed that digestion efficiency can be correlated to the degree of cross-linking of certain proteins. Though the analyses performed here do not show a transition in digestibility there is a similar trend in that accessibility to trypsin at one end of the protein appears selectively limited. This may indicate potential cross-linked regions, which were reproducible across biological replicates. This accessibility criterion can be taken into account when developing antibodies. Applicability of the proposed biomarkers now depends on the quality of the antibodies raised.
Conclusions
We have determined 163 proteins to show a dependence on the presence of CotE for their identification in the spore coat insoluble fraction of B. cereus. In this manner, we have shown localization in the exosporium for several proteins, further expanding the list of proteins known to be exposed on the outermost layer of the spore. On the other hand, 153 proteins were only identified if CotE is knocked out suggesting they are obscured, that is, surrounded by or interacting with CotE in wild type spores.
A QconCAT approach for quantification of proteins was applied in the heterogeneous system of the spore coat insoluble fraction of wild-type B. cereus ATCC14579 and a ΔCotE knockout strain. In this manner, 21 proteins from across all layers of the spore were quantified covering 5.66% ± 0.51 of the total spore weight in wild type and 4.13% ± 0.14 in the ΔCotE mutant strain. The protein abundance spanned three orders of magnitude and suggested functional properties of proteins correlate with their abundance. The most abundant proteins are generally known structural components, which enabled us to functionally describe the previously uncharacterized protein BC_0987, now renamed SasS. The obtained information on stoichiometry of both putative and known interactors showed them to be approaching equimolar quantities, which is of importance to developing quantitative models for spore macromolecular structures.
In line with the effects observed qualitatively in the ΔCotE mutant the 20 proteins (excluding CotE) quantified using the QconCAT approach, known exosporium proteins were generally less abundant in the CotE knockout whereas inner coat associated proteins were generally more abundant.
The five most abundant proteins are considered as biomarkers for detection of B. cereus group spores and immunogenic epitopes have been predicted to be present in CotB1, CotB2, CotX, and InhA. These proteins are indicated to be present in the outermost layers of the spore making them ideal candidate biomarkers.
These results further contribute to our understanding of the spore structure, in particular of the outer layers of the spore. This may be applied in a food safety scenario where the makeup of the outside of the spore is of crucial importance for development of detection techniques.
Acknowledgments
This project is supported by NanoNextNL, a micro and nanotechnology consortium of the Government of The Netherlands and 130 partners from academia and industry. The authors would like to thank Christelle Bressuire-Isoard, Véronique Broussole, and Frederic Carlin for kindly providing us with the CotE knockout strain.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.7b00732.
Author Contributions
⊥ These authors contributed equally.
The authors declare no competing financial interest.
Notes
The mass spectrometry proteomics data of the wild-type QconCAT analyses have been deposited to the ProteomeXchange Consortium45 via the PRIDE partner repository with the data set identifier PXD003569.
Supplementary Material
References
- Opinion of the Scientific Panel on Biological Hazards on Bacillus cereus and other Bacillus spp in foodstuffs. EFSA J. 2005, 175, 1–48. [Google Scholar]
- Aronson A. I.; Fitz-James P. Structure and morphogenesis of the bacterial spore coat. Bacteriol. Rev. 1976, 40 (2), 360–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques A. O.; Moran C. P. Jr Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 2007, 61, 555–588. 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- Henriques A. O.; Moran C. P.; et al. Structure and assembly of the bacterial endospore coat. Methods 2000, 20 (1), 95–110. 10.1006/meth.1999.0909. [DOI] [PubMed] [Google Scholar]
- Giorno R.; Bozue J.; Cote C.; Wenzel T.; Moody K.-S.; Mallozzi M.; Ryan M.; Wang R.; Zielke R.; Maddock J. R.; et al. Morphogenesis of the Bacillus anthracis Spore. J. Bacteriol. 2007, 189 (3), 691–705. 10.1128/JB.00921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressuire-Isoard C.; Bornard I.; Henriques A. O.; Carlin F.; Broussolle V. Sporulation temperature reveals a requirement for CotE in the assembly of both the coat and exosporium layers of Bacillus cereus spores. Appl. Environ. Microbiol. 2016, 82, 232. 10.1128/AEM.02626-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L. B.; Donovan W. P.; Fitz-James P. C.; Losick R. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 1988, 2 (8), 1047–1054. 10.1101/gad.2.8.1047. [DOI] [PubMed] [Google Scholar]
- Ozin A. J.; Henriques A. O.; Yi H.; Moran C. P. Morphogenetic proteins SpoVID and SafA form a complex during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 2000, 182 (7), 1828–1833. 10.1128/JB.182.7.1828-1833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura D.; Kuwana R.; Takamatsu H.; Watabe K. Proteins Involved in Formation of the Outermost Layer of Bacillus subtilis Spores. J. Bacteriol. 2011, 193 (16), 4075–4080. 10.1128/JB.05310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney P. T.; Driks A.; Eichenberger P. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 2012, 11 (1), 33–44. 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. C. The Exosporium Layer of Bacterial Spores: a Connection to the Environment and the Infected Host. Microbiol. Mol. Biol. Rev. 2015, 79 (4), 437–457. 10.1128/MMBR.00050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abhyankar W.; Pandey R.; Ter Beek A.; Brul S.; de Koning L. J.; de Koster C. G. Reinforcement of Bacillus subtilis spores by cross-linking of outer coat proteins during maturation. Food Microbiol. 2015, 45, 54–62. 10.1016/j.fm.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Brownridge P.; Holman S. W.; Gaskell S. J.; Grant C. M.; Harman V. M.; Hubbard S. J.; Lanthaler K.; Lawless C.; O’cualain R.; Sims P.; et al. Global absolute quantification of a proteome: Challenges in the deployment of a QconCAT strategy. Proteomics 2011, 11 (15), 2957–2970. 10.1002/pmic.201100039. [DOI] [PubMed] [Google Scholar]
- Abhyankar W.; Hossain A. H.; Djajasaputra A.; Permpoonpattana P.; Ter Beek A.; Dekker H. L.; Cutting S. M.; Brul S.; de Koning L. J.; de Koster C. G. In Pursuit of Protein Targets: Proteomic Characterization of Bacterial Spore Outer Layers. J. Proteome Res. 2013, 12 (10), 4507–4521. 10.1021/pr4005629. [DOI] [PubMed] [Google Scholar]
- Brownridge P.; Beynon R. J. The importance of the digest: Proteolysis and absolute quantification in proteomics. Methods 2011, 54 (4), 351–360. 10.1016/j.ymeth.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Abhyankar W.; Beek A. T.; Dekker H.; Kort R.; Brul S.; de Koster C. G. Gel-free proteomic identification of the Bacillus subtilis insoluble spore coat protein fraction. Proteomics 2011, 11 (23), 4541–4550. 10.1002/pmic.201100003. [DOI] [PubMed] [Google Scholar]
- Kuwana R.; Okuda N.; Takamatsu H.; Watabe K. Modification of GerQ reveals a functional relationship between Tgl and YabG in the coat of Bacillus subtilis spores. J. Biochem. 2006, 139 (5), 887–901. 10.1093/jb/mvj096. [DOI] [PubMed] [Google Scholar]
- Fernandes C. G.; Plácido D.; Lousa D.; Brito J. A.; Isidro A.; Soares C. M.; Pohl J.; Carrondo M. A.; Archer M.; Henriques A. O. Structural and Functional Characterization of an Ancient Bacterial Transglutaminase Sheds Light on the Minimal Requirements for Protein Cross-Linking. Biochemistry 2015, 54 (37), 5723–5734. 10.1021/acs.biochem.5b00661. [DOI] [PubMed] [Google Scholar]
- Üstok F. I.; Chirgadze D. Y.; Christie G. Structural and functional analysis of SleL, a peptidoglycan lysin involved in germination of Bacillus spores: Crystal structures for the Bacillus spore lytic enzyme SleL. Proteins: Struct., Funct., Genet. 2015, 83 (10), 1787–1799. 10.1002/prot.24861. [DOI] [PubMed] [Google Scholar]
- Atrih A.; Foster S. J.; Moir A.; Chirakkal H.; O'Rourke M. Analysis of spore cortex lytic enzymes and related proteins in Bacillus subtilis endospore germination. Microbiology 2002, 148 (8), 2383–2392. 10.1099/00221287-148-8-2383. [DOI] [PubMed] [Google Scholar]
- Isticato R.; Pelosi A.; De Felice M.; Ricca E. CotE Binds to CotC and CotU and Mediates Their Interaction during Spore Coat Formation in Bacillus subtilis. J. Bacteriol. 2010, 192 (4), 949–954. 10.1128/JB.01408-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B.; Zhang L.; Liang S.; Zhang C. SVMTriP: a method to predict antigenic epitopes using support vector machine to integrate tri-peptide similarity and propensity. PLoS One 2012, 7 (9), e45152. 10.1371/journal.pone.0045152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries Y. P.; Hornstra L. M.; de Vos W. M.; Abee T. Growth and Sporulation of Bacillus cereus ATCC 14579 under Defined Conditions: Temporal Expression of Genes for Key Sigma Factors. Appl. Environ. Microbiol. 2004, 70 (4), 2514–2519. 10.1128/AEM.70.4.2514-2519.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijlander R. T.; Holsappel S.; de Jong A.; Ghosh A.; Christie G.; Kuipers O. P. SpoVT: From Fine-Tuning Regulator in Bacillus subtilis to Essential Sporulation Protein in Bacillus cereus. Front. Microbiol. 2016, 7, 1607. 10.3389/fmicb.2016.01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagyan I.; Hobot J.; Cutting S. A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J. Bacteriol. 1996, 178 (15), 4500–4507. 10.1128/jb.178.15.4500-4507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E. M.; Phadke N. D.; Kachman M. T.; Giorno R.; Vazquez S.; Vazquez J. A.; Maddock J. R.; Driks A. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J. Bacteriol. 2003, 185 (4), 1443. 10.1128/JB.185.4.1443-1454.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond C. Identification of proteins in the exosporium of Bacillus anthracis. Microbiology 2004, 150 (2), 355–363. 10.1099/mic.0.26681-0. [DOI] [PubMed] [Google Scholar]
- Caro-Astorga J.; Pérez-García A.; de Vicente A.; Romero D. A genomic region involved in the formation of adhesin fibers in Bacillus cereus biofilms. Front. Microbiol. 2015, 5, 745. 10.3389/fmicb.2014.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lövgren A.; Zhang M.; Engström A.; Dalhammar G.; Landén R. Molecular characterization of immune inhibitor A, a secreted virulence protease from Bacillus thuringiensis. Mol. Microbiol. 1990, 4 (12), 2137–2146. 10.1111/j.1365-2958.1990.tb00575.x. [DOI] [PubMed] [Google Scholar]
- Pflughoeft K. J.; Swick M. C.; Engler D. A.; Yeo H.-J.; Koehler T. M. Modulation of the Bacillus anthracis Secretome by the Immune Inhibitor A1 Protease. J. Bacteriol. 2014, 196 (2), 424–435. 10.1128/JB.00690-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokova O. N.; McPherson S. A.; Steichen C. T.; Turnbough C. L. The Spore-Specific Alanine Racemase of Bacillus anthracis and Its Role in Suppressing Germination during Spore Development. J. Bacteriol. 2009, 191 (4), 1303–1310. 10.1128/JB.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorno R.; Mallozzi M.; Bozue J.; Moody K.-S.; Slack A.; Qiu D.; Wang R.; Friedlander A.; Welkos S.; Driks A. Localization and assembly of proteins comprising the outer structures of the Bacillus anthracis spore. Microbiology 2009, 155 (4), 1133–1145. 10.1099/mic.0.023333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd S. J.; Moir A. J. G.; Johnson M. J.; Moir A. Genes of Bacillus cereus and Bacillus anthracis Encoding Proteins of the Exosporium. J. Bacteriol. 2003, 185 (11), 3373–3378. 10.1128/JB.185.11.3373-3378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Butzin X. Y.; Davis A.; Setlow B.; Korza G.; Üstok F. I.; Christie G.; Setlow P.; Hao B. Activity and regulation of various forms of CwlJ, SleB, and YpeB proteins in degrading cortex peptidoglycan of spores of Bacillus species in vitro and during spore germination. J. Bacteriol. 2013, 195 (11), 2530–2540. 10.1128/JB.00259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Fitz-James P. C.; Aronson A. I. Cloning and characterization of a cluster of genes encoding polypeptides present in the insoluble fraction of the spore coat of Bacillus subtilis. J. Bacteriol. 1993, 175 (12), 3757–3766. 10.1128/jb.175.12.3757-3766.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlass P. J.; Houston C. W.; Clements M. O.; Moir A. Germination of Bacillus cereus spores in response to L-alanine and to inosine: the roles of gerL and gerQ operons. Microbiology 2002, 148 (7), 2089–2095. 10.1099/00221287-148-7-2089. [DOI] [PubMed] [Google Scholar]
- Leski T. A.; Caswell C. C.; Pawlowski M.; Klinke D. J.; Bujnicki J. M.; Hart S. J.; Lukomski S. Identification and Classification of bcl Genes and Proteins of Bacillus cereus Group Organisms and Their Application in Bacillus anthracis Detection and Fingerprinting. Appl. Environ. Microbiol. 2009, 75 (22), 7163–7172. 10.1128/AEM.01069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagyan I.; Noback M.; Bron S.; Paidhungat M.; Setlow P. Characterization of yhcN, a new forespore-specific gene of Bacillus subtilis. Gene 1998, 212 (2), 179–188. 10.1016/S0378-1119(98)00172-3. [DOI] [PubMed] [Google Scholar]
- Takamatsu H.; Kodama T.; Nakayama T.; Watabe K. Characterization of the yrbA Gene of Bacillus subtilis, Involved in Resistance and Germination of Spores. J. Bacteriol. 1999, 181 (16), 4986–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Mora J.; Pérez-Valdespino A.; Gupta S.; Withange N.; Kuwana R.; Takamatsu H.; Christie G.; Setlow P. The GerW Protein Is Not Involved in the Germination of Spores of Bacillus Species. PLoS One 2015, 10 (3), e0119125. 10.1371/journal.pone.0119125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragkousi K.; Setlow P. Transglutaminase-Mediated Cross-Linking of GerQ in the Coats of Bacillus subtilis Spores. J. Bacteriol. 2004, 186 (17), 5567–5575. 10.1128/JB.186.17.5567-5575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragkousi K.; Eichenberger P.; van Ooij C.; Setlow P. Identification of a new gene essential for germination of Bacillus subtilis spores with Ca2+-dipicolinate. J. Bacteriol. 2003, 185 (7), 2315–2329. 10.1128/JB.185.7.2315-2329.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry C.; Shepherd A.; Radford D. S.; Moir A.; Bullough P. A. YwdL in Bacillus cereus: Its Role in Germination and Exosporium Structure. PLoS One 2011, 6 (8), e23801. 10.1371/journal.pone.0023801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao S.-C.; Ma K.; Talafous J.; Orlando R.; Zagorski M. G. Trifluoroacetic acid pretreatment reproducibly disaggregates the amyloid β-peptide. Amyloid 1997, 4 (4), 240–252. 10.3109/13506129709003835. [DOI] [Google Scholar]
- Vizcaíno J. A.; Deutsch E. W.; Wang R.; Csordas A.; Reisinger F.; Ríos D.; Dianes J. A.; Sun Z.; Farrah T.; Bandeira N.; et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014, 32 (3), 223–226. 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B. Bootstrap Methods: Another Look at the Jackknife. Ann. Stat. 1979, 7 (1), 1–26. 10.1214/aos/1176344552. [DOI] [Google Scholar]
- Davison A. C.; Hinkley D. V.. Bootstrap Methods and Their Application (Cambridge Series in Statistical and Probabilistic Mathematics); Cambridge University Press: Cambridge, 1997. [Google Scholar]
- Steichen C. T.; Kearney J. F.; Turnbough C. L. Non-uniform assembly of the Bacillus anthracis exosporium and a bottle cap model for spore germination and outgrowth. Mol. Microbiol. 2007, 64 (2), 359–367. 10.1111/j.1365-2958.2007.05658.x. [DOI] [PubMed] [Google Scholar]
- de Francesco M.; Jacobs J. Z.; Nunes F.; Serrano M.; McKenney P. T.; Chua M.-H.; Henriques A. O.; Eichenberger P. Physical Interaction between Coat Morphogenetic Proteins SpoVID and CotE Is Necessary for Spore Encasement in Bacillus subtilis. J. Bacteriol. 2012, 194 (18), 4941–4950. 10.1128/JB.00914-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa T.; Serrano M.; Steil L.; Volker U.; Moran C. P.; Henriques A. O. The Timing of cotE Expression Affects Bacillus subtilis Spore Coat Morphology but Not Lysozyme Resistance. J. Bacteriol. 2007, 189 (6), 2401–2410. 10.1128/JB.01353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera M.; Zandomeni R. O.; Sagripanti J.-L. Wet and dry density of Bacillus anthracis and other Bacillus species. J. Appl. Microbiol. 2008, 105 (1), 68–77. 10.1111/j.1365-2672.2008.03758.x. [DOI] [PubMed] [Google Scholar]
- Carrera M.; Zandomeni R. O.; Fitzgibbon J.; Sagripanti J.-L. Difference between the spore sizes of Bacillus anthracis and other Bacillus species. J. Appl. Microbiol. 2007, 102 (2), 303–312. 10.1111/j.1365-2672.2006.03111.x. [DOI] [PubMed] [Google Scholar]
- Gould G. W.; Hurst A.. The Bacterial Spore; Academic Press, 1969. [Google Scholar]
- Gerber S. A.; Rush J.; Stemman O.; Kirschner M. W.; Gygi S. P. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (12), 6940–6945. 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe A.; Setlow P. Localization of the Transglutaminase Cross-Linking Sites in the Bacillus subtilis Spore Coat Protein GerQ. J. Bacteriol. 2006, 188 (21), 7609–7616. 10.1128/JB.01116-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chewapreecha C.; Harris S. R.; Croucher N. J.; Turner C.; Marttinen P.; Cheng L.; Pessia A.; Aanensen D. M.; Mather A. E.; Page A. J.; et al. Dense genomic sampling identifies highways of pneumococcal recombination. Nat. Genet. 2014, 46 (3), 305–309. 10.1038/ng.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre P.; Couture-Tosi E.; Mock M. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 2002, 45 (1), 169–178. 10.1046/j.1365-2958.2000.03000.x. [DOI] [PubMed] [Google Scholar]
- Thompson B. M.; Stewart G. C. Targeting of the BclA and BclB proteins to the Bacillus anthracis spore surface. Mol. Microbiol. 2008, 70 (2), 421–434. 10.1111/j.1365-2958.2008.06420.x. [DOI] [PubMed] [Google Scholar]
- Steichen C. T.; Kearney J. F.; Turnbough C. L. Characterization of the Exosporium Basal Layer Protein BxpB of Bacillus anthracis. J. Bacteriol. 2005, 187 (17), 5868–5876. 10.1128/JB.187.17.5868-5876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullerova D.; Krajcikova D.; Barak I. Interactions between Bacillus subtilis early spore coat morphogenetic proteins. FEMS Microbiol. Lett. 2009, 299 (1), 74–85. 10.1111/j.1574-6968.2009.01737.x. [DOI] [PubMed] [Google Scholar]
- Gould G. W. Symposium on bacterial spores: IV. Germination and the problem of dormancy. J. Appl. Bacteriol. 1970, 33 (1), 34–49. 10.1111/j.1365-2672.1970.tb05232.x. [DOI] [PubMed] [Google Scholar]
- Ghosh S.; Setlow P. Isolation and Characterization of Superdormant Spores of Bacillus Species. J. Bacteriol. 2009, 191 (6), 1787–1797. 10.1128/JB.01668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R.; Ter Beek A.; Vischer N. O. E.; Smelt J. P. P. M.; Brul S.; Manders E. M. M. Live Cell Imaging of Germination and Outgrowth of Individual Bacillus subtilis Spores; the Effect of Heat Stress Quantitatively Analyzed with SporeTracker. PLoS One 2013, 8 (3), e58972. 10.1371/journal.pone.0058972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.