Abstract
Background
Gallibacterium anatis is an opportunistic pathogen of intensively reared poultry causing oophoritis, salpingitis, peritonitis and enteritis. Gallibacterium anatis infection often remains undiagnosed. Recently multi-drug resistant isolates have been described.
Methods
A newly developed PCR restriction fragment length polymorphism assay targeting the 16S rRNA gene was used to identify and differentiate Gallibacterium isolates from chicken, turkey and partridge samples originating from 18 different geographical locations in Thuringia, Germany. Antimicrobial susceptibility to 19 compounds of different classes was assessed.
Results
Nineteen Gallibacterium isolates were investigated. In 9 birds (47.4%) Gallibacterium species were isolated exclusively while in 10 birds (52.6%) other bacterial or viral agents could be detected in addition. In one chicken a mixed infection of Gallibacterium anatis and Gallibacterium genomospecies was identified. All isolates were susceptible to apramycin, florfenicol and neomycin and resistant to clindamycin, sulfathiazole and penicillin. Resistance to sulfamethoxim, spectinomycin, tylosin and oxytetracycline was observed in 93.3%, 93.3%, 86.7% and 80.0% of the field strains, respectively.
Conclusions
The PCR-RFLP assay allows specific detection and differentiation of Gallibacterium spp. from poultry. Antimicrobial resistance of Gallibacterium spp. is highly significant in Thuringian field isolates.
Keywords: Gallibacterium, Poultry, PCR, RFLP, Antibiotic resistance
Background
Gallibacterium (G.) is a genus within the Pasteurellaceae family and is associated with a wide spectrum of avian host species based on isolations from domestic and wild birds including chickens, turkeys, geese, ducks, pheasants and partridges [1–4]. Chickens are the preferred host of Gallibacterium spp., which constitute a part of the normal flora in the upper respiratory and the lower genital tract [4–6]. G. anatis infections in chicken resulted in a variety of signs and lesions such as respiratory problems, necrosis in liver, peritonitis, salpingitis, haemorrhagic and ruptured follicles and a drop in egg production [6–9]. Mixed infections with other poultry pathogens such as Escherichia (E.) coli contributed to the major lesions with G. anatis in naturally affected chickens [9].
Genus Gallibacterium belongs to phylum Proteobacteria, class Gammaproteobacteria and family Pasteurellaceae [10, 11]. Gallibacterium was isolated for first time in 1950 from cloaca of healthy chickens and was described as haemolytic “cloaca bacterium” by Kjos-Hansen [12, 13]. Being similar to Pasteurella (P.) in several characters, it was earlier known as P. anatis. The genus name Gallibacterium was firstly given by Bisgaard in the year 1982 on the basis of certain phenotypic characters used for identification of isolates of Actinobacillus salpingitidis and avian P. haemolytica [1, 5, 14, 15]. Genus Gallibacterium was established within the family of Pasteurellaceae based on 16S rRNA gene sequences [16]. The genus includes strains belonging to G. anatis, G. genomospecies [3, 5, 10] and unnamed group V [15]. Taxon 1 designated as a third group of strains [14] and named P. anatis, which was also found closely related to A. salpingitidis and avian P. haemolytica [11].
The genus Gallibacterium comprises four species, namely G. anatis, G. melopsittaci sp. nov., G. trehalosifermentans sp. nov., and G. salpingitidis sp. nov., and three G. genomospecies [15, 17].
G. anatis can be further sub-divided into two phenotypically distinct biovars: haemolytica and the non-haemolytic biovar anatis [17]. G. anatis isolates were formerly known as strains of the avian Pasteurella haemolytica–Actinobacillus salpingitidis complex or Pasteurella anatis. Haemolytic Pasteurella-like bacteria (Gallibacterium anatis) are the causing agent of salpingitis with or without peritonitis but also of septicaemia, pericarditis, hepatitis, respiratory tract lesions and enteritis [18, 19]. G. anatis is globally distributed and has been isolated from poultry in some countries within Europe [15, 17, 20].
Various tools have been used for the identification of Gallibacterium including cultivation, fluorescence in situ hybridization (FISH), genus-specific PCR, matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry and a gtxA gene-based quantitative PCR (qPCR) assay.
Diagnosis of the Gallibacterium infection was based on isolation and identification including phenotypic characterization [17]. Phenotypic characterization involves laborious and time-consuming methods, which may also give ambiguous results due to variable outcomes. This in turn leads to difficulty in interpretation of the genus and species designations [1]. Problems during isolation are mainly related to poor growth on artificial media, subsequent overgrowth by other bacteria, and difficulties in phenotypic identification.
Although MALDI Biotyper software is able to recognize clonally related G. anatis strains isolated from different poultry flocks as well as from different organs, MALDI-TOF mass spectrometry is not very much practiced for routine diagnostics in veterinary medicine [21, 22].
The gyrB gene-based qPCR method is very useful and highly sensitive for G. anatis detection in addition to time and cost saving compared to conventional PCR and phenotypic methods [23].
Beta-subunit of DNA-dependent RNA-polymerase (rpoB) gene sequence-based classification was used within Pasteurellaceae [24] and has been recommended for this group of bacteria, particularly in cases where phenotypic identification was difficult [25]. However, phylogenetic comparison of 16S rRNA gene sequences has recently become a key character for bacterial classification [26]. They might also be used directly for identification, or result in subsequent development of polymerase chain reaction assays targeting specific regions of the aforementioned genes.
Effective means of antibiotic treatment are highly needed. The frequency of treatment failure in flocks infected with Gallibacterium seems to be a recurrent problem [27, 28]. Currently, very limited information on antimicrobial susceptibility of G. anatis is available leaving little insight into the general level of resistance for this species [27]. Emergence of antimicrobial resistance has been observed among several organisms belonging to the Pasteurellaceae family [29]. The resistance phenotypes from Gallibacterium field strains and other taxa of Pasteurellaceae demonstrated a remarkably high prevalence of multidrug resistance [27, 29–31]. Gallibacterium emerged in last few years as multi-drug resistant pathogen causing outbreaks with high mortality not only in poultry but also in other pet birds [12].
The aim of this study was to use a molecular biological identification method allowing specific detection and differentiation of Gallibacterium spp. A PCR restriction fragment length polymorphism (PCR-RFLP) assay was established which allowed an easy and reliable detection and differentiation of Gallibacterium spp. Moreover, the antibiotic susceptibility profiles of Gallibacterium isolates of chickens, turkey and partridge raised in federal state of Thuringia, Germany were determined.
Methods
Isolation and characterization of bacterial strains
In this study, 120 tissue samples were collected from poultry suffering from respiratory signs and reproductive disorders as a routine diagnosis in 18 different locations in the state of Thuringia, Germany. The collected samples were investigated specifically for Gallibacterium spp. and other bacteria and viruses causing similar symptoms. Bacterial isolates were cultured from several organs of diseased and perished chickens, turkey and partridge. The age of birds, organs of isolation and pathological findings for Gallibacterium positive samples were demonstrated in Table 1. Pre-enriched non-selective medium (buffered peptone water; Oxoid, Wesel, Germany) was inoculated with the collected samples and incubated for 24 h at 37 °C under aerobic condition. A loopful of liquid medium was streaked out on 5% Columbia blood agar (Oxoid), Dextrose Starch Agar medium (Oxoid) and MacConkey agar (Oxoid), and then incubated aerobically for 24 h at 37 °C.
Table 1.
Isolated Gallibacterium strains, investigated organs and pathological findings in the hosts
| Isolate | Host | Age | Investigated organs | Pathological findings |
|---|---|---|---|---|
| Ge 002 | Chicken | Adult | Lung, ovary, spleen | Swollen kidney, ovarian degeneration, liver congestion, haemorrhagic enteritis |
| Ge 004 | Chicken | Juvenile | Proventriculus, lung | Airsacculitis, ovarian atrophy, enlargement of heart, parenchym congestion, swollen kidneys |
| Ge 006 | Chicken | Adult | Larynx, liver, lung, ovary | Laryngitis, swollen kidney, splenomegaly, polyseroritis |
| Ge 023 | Chicken | Adult | Heart, liver, lung, spleen | Ascites, pulmonary oedema, hydropericarditis, hepatomegaly with perihepatitis, haemorrhagic enteritis |
| Ge 024 | Chicken | Adult | Larynx, ovary | Airsacculitis, splenomegaly, ovarian degeneration |
| Ge 081 | Chicken | Adult | Ovary, spleen | Lung abscess, enteritis, serositis |
| Ge 082 | Chicken | Adult | Ovary, heart, kidney, liver, spleen | Enteritis, pulmonic abscess, serositis |
| Ge 095 | Chicken | Adult | Lung | Nasal sinusitis, pulmonary oedema, swollen liver |
| Ge 100 | Chicken | Juvenile | Lung | Enteritis, hepatitis, hepatomegaly and splenomegaly |
| Ge 103 | Chicken | Juvenile | Lung, spleen | Liver congestion, pulmonary oedema, haemorrhagic enteritis, swollen kidneys, yolk sac persistence |
| Ge 134 | Chicken | Juvenile | Air sac, brain, eye, heart, larynx, lung | Appendicitis, conjunctivitis, hepatitis, pulmonary oedema, rhinitis |
| Ge 156 | Chicken | Adult | Kidney, lung, spleen | Crop inflammation, enteritis, parenchymitis |
| Ge 168 | Chicken | Juvenile | Kidney, lung, trachea | Conjunctivitis, uveitis, airsacculitis and enteritis, laryngitis, pneumonia, hepatomegaly, nephritis, splenomegaly |
| Ge 173 | Chicken | Adult | Heart, lung | Liver paleness |
| Ge 186 | Chicken | Adult | Proventriculus, lung | Air sacculitis, enteritis |
| Ge 223 | Chicken | Adult | Proventriculus, lung | Hepatomegaly, nephritis, enteritis, spleen inflammation |
| Ge 297 | Chicken | Juvenile | Brain, larynx, trachea | Ovarian atrophy, enteritis, laryngitis |
| Ge 160 | Partridge | Juvenile | Brain, lung | Pneumonia, airsacculitis, enteritis, hepatitis, nephritis |
| Ge 258 | Turkey | Juvenile | Nasal cavity | Enteritis, septicaemia, serositis, swollen parenchymes |
Phenotypical characterization of isolated strains was performed as previously reported [32, 33]. Suspected bacterial colonies were 0.5–1.5 mm in diameter, bright translucent, low convex and mostly showed beta-haemolysis on blood agar. When observed obliquely with transmitted light these colonies showed concentric rings on Dextrose Starch Agar medium and pink colonies on MacConkey agar. Gallibacterium were growing on Columbia agar as fine, circular smooth-edged colonies of grey-white colour with conspicuous beta-haemolysis. After 48 h or sub-cultivation on MacConkey agar, large flat grey colonies with a diameter of 4–5 mm with an amber raised centre were observed. These microorganisms were overlooked during microbiological investigations because initially small Gallibacterium colonies were overgrown by other microorganisms. Gram as well as Giemsa staining was used for detection of morphologically characteristic appearance of examined suspected colonies. Gram staining showed Gram-negative, coccoid to pleomorphic rods. In all cases bacteria produced catalase, and the majority of the isolates formed oxidase, phosphatase, nitrate reduction and sugar fermentation with acid production.
Typical or suspected colonies were picked for further biochemical identification using API 20 NE (bioMerieux, Nürtingen, Germany).
The collected specimens were tested for Escherichia coli, Mycoplasma gallisepticum, Mycoplasma synoviae, Clostridium perfringens and adenovirus using specific recommended methods for each organism [34].
DNA extraction
For isolation of chromosomal DNA, bacterial cultures from a plate were re-suspended in 200 μl of phosphate-buffered saline and the isolation procedure was performed with High Pure PCR Template Purification Kit (Roche Diagnostics, Mannheim, Germany) according to the instructions of the manufacturer.
PCR and DNA sequencing of 16S rRNA genes
16S rRNA genes were partially amplified by using primers 41f and 1066r (Table 2) [35]. After an initial denaturation at 96 °C for 60 s, 35 cycles of denaturation (96 °C for 15 s), primer annealing (55 °C for 60 s) and primer extension (72 °C for 90 s) were carried out and followed by final elongation step at 72 °C for 60 s. PCR was performed using 2 μl chromosomal DNA, 5 μl 10 x Taq buffer (Genaxxon bioscience GmbH, Biberach, Germany), 0.2 μl Taq DNA polymerase (Genaxxon bioscience GmbH), 2 μl dNTP mix (2 mM each; Carl Roth GmbH, Karlsruhe, Germany) and 1 μl of both primers (10 mM; Jena Bioscience, Jena, Germany) in a volume of 50 μl. PCR products were analysed by gel electrophoresis on 1% agarose gels, stained with ethidium bromide and visualised under UV light. The resulting ca. 1 kb fragment was excised and DNA was extracted using QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) according to manufacturer’s instruction. DNA sequencing of purified PCR products was carried out by cycle sequencing procedure with the BigDye™ Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Darmstadt, Germany). 41f, 1066r, Galli-1 and Galli-2 (Table 2) were used as sequencing primers. Nucleotide sequences were determined on an ABI Prism 310 Genetic Analyzer (Applied Biosystems).
Table 2.
Primers used in this study
| Primer | Nucleotide sequence | Aim | Amplicon length |
|---|---|---|---|
| 41f | 5´-GCT CAG ATT GAA CGC TGG CG-3′ | Amplification, sequencing | ca. 1000 bp |
| 1066r | 5´-ACA TTT CAC AAC ACG AGC TG-3′ | ||
| Galli-1 | 5´-CAA GCC GAC GAT CTC TAG-3′ | Sequencing | |
| Galli-2 | 5´-TTC GCA CAT GAG CGT CAG-3′ | ||
| Galli-3 | 5′-ATA GTA TCG AGA GAT GAA AGG GGT GG-3′ | Amplification | 684–686 |
| Galli-5 | 5′-TAT CAC GTT TGC TTC GAG AGC C-3´ |
Alignment
Multiple sequence alignments were done using AlignX of Vector NTI Suite 8.0 (Informax Inc., Oxford, UK). Based upon sequence data of investigated isolates, dendrograms were generated for both G. species in this study and other related organisms (P. multocida, P. anatis, Avibacterium paragallinarum, Avibacterium avium, G. anatis, G. genomospecies and Bisgaard taxa) with cluster tree neighbour-joining analysis using the bioinformatics tools of Geneious R9.0.5.1 analysis.
Detection and differentiation of Gallibacterium species
Primers used for a Gallibacterium spp. specific PCR assay were chosen on the basis of an alignment of 16S rRNA genes of related avian Pasteurellaceae species. PCR was carried out by using primers Galli-3 and Galli-5 (Table 2) under the following conditions: 3 μl DNA extract, 5 μl 10 x Taq buffer (Genaxxon bioscience GmbH), 0.2 μl Taq DNA polymerase (5 u; Genaxxon bioscience GmbH), 2 μl dNTP mix (2 mM each; Carl Roth GmbH) and 1 μl of each primer (10 mM), and the volume made up to 50 μl by addition of bi-distilled water. The following temperature-time programme was used for amplification: After an initial denaturation step at 96 °C for 60 s, 35 cycles of denaturation (96 °C for 15 s), primer annealing (66 °C for 60 s) and primer elongation (72 °C for 60 s) were performed. PCR was terminated by an elongation step at 72 °C for 60 s. PCR products were analysed as described above. The high annealing temperature in the PCR process avoided a cross-amplification with DNA of other avian Pasteurellaceae species. The resulting amplicons had lengths of approximately 680 bp for G. anatis and G. genomospecies.
For identification of G. anatis and G. genomospecies and conformation of affiliation to genus Gallibacterium digestion of the PCR amplicon with RsaI restriction enzyme (New England Biolabs, Frankfurt, Germany) was used. For differentiation of Gallibacterium into G. anatis and G. genomospecies, digestion of the PCR products with XbaI restriction enzyme (New England Biolabs) was performed. 8 μl of PCR products were digested with 1 μl Rsa I or Xba I using 2 μl of recommended buffers in separate 20 μl reactions. Restriction reactions were carried out at 37 °C for 2 h. Restriction products were analysed by gel electrophoresis on a 1.5% agarose gel, ethidium bromide staining and visualization under UV light.
PCR specificity and limit of detection
A total of 30 bacterial strains including 15 G. anatis (14 field strains and one reference strain), four G. genomospecies, five Pasteurella multocida, Pasteurella anatis, Avibacterium avium, Avibacterium paragallinarum, Avibacterium gallinarum, Mannheimia haemolytica and Riemerella columbina were used in this study. The sensitivity of the PCR reaction was determined by analysing serial dilutions (1:10) of genomic DNA.
Antimicrobial susceptibility testing
The antimicrobial susceptibility of 15 Gallibacterium isolates was tested by using the broth dilution method against 19 antibiotics of different classes (Table 3). Four out of 19 Gallibacterium isolates could not be re-cultivated after applying phenotypic identification, but DNA was extracted for further identification.
Table 3.
Cut-off values, MIC distribution and MIC50/MIC90 of 15 Gallibacterium isolates
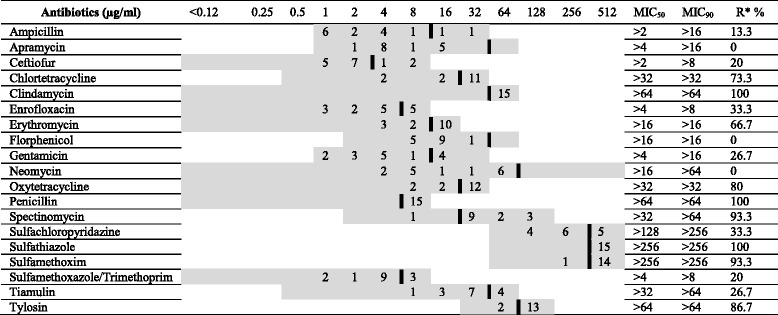
A thick black line indicates the cut-off between clinically sensitive and insusceptible strains
Grey shadow area indicated the test range (μg/ml) of each antimicrobial agent
MIC50 = (n × 0.5)
MIC90 = (n × 0.9)
*R: Resistance rate
Determination of minimum inhibitory concentration (MIC) was performed according to the CLSI standard M31-A2 [36] using a commercially prepared dehydrated panel for Enterobacteriaceae (Sensititre-TREK Diagnostic Systems, Cleveland, USA). The plates were incubated for 20–24 h at 35 °C under aerobic conditions. The MIC was defined as the lowest concentration avoiding visible growth. In categorizing the MIC results, CLSI cut-offs for resistance of P. multocida were used [36]. Specific cut-offs for respiratory disease agents were used when available. The cut-offs used are indicated in Table 3. Actinobacillus pleuropneumoniae ATCC 27090 was included as control.
Results
Isolation and phenotypic identification of Gallibacterium spp
Nineteen Gallibacterium isolates were cultivated and phenotypically identified from 19 diseased or perished birds. The panel of hosts consisted of 17 chickens (Gallus gallus f. domesticus), one partridge (Perdix perdix L.) and one turkey (Meleagris gallopavo f. domesticus) with different pathological lesions (Table 1), from 18 different farms in Thuringia, Germany. The age of the birds was between 3 weeks and one year. Nine birds were juvenile and 10 of them adult. The reported signs were respiratory symptoms (cold, dyspnea), diarrhoea, anorexia, emaciation of the affected birds and mortality. Gallibacterium strains were isolated from lung (14 out of 19), spleen (6/19), ovaries (5/9) and brain (3/19) of diseased birds. Mostly, a septicaemic course of the bacterial infection was diagnosed.
The isolation of Gallibacterium in different Galliformes species is connected with different pathological findings. The most commonly detected pathological findings were enteritis (11/19) and swollen parenchymes of liver and spleen (10/19). Sometimes, airsacculitis (5/19), ovarian atrophy (4/9) and petichael haemorrhage of different organs (5/19) were observed.
Species identification using the API 20 NE system resulted in classification as Mannheimia haemolytica with more than 90%. All isolates showed beta-haemolysis.
In 9 cases only Gallibacterium was isolated. In other cases pathogens like mycoplasmas, Clostridium perfringens, E. coli and adenoviruses were detected besides Gallibacterium (Table 4). Other bacterial colonies were collected but could not identified biochemically as Gallibacterium.
Table 4.
Results of species identification via DNA sequencing and PCR-RFLP and occurrence of other pathogens in investigated hosts
| Isolates | DNA sequencing | PCR-RFLP results | Other detected pathogensa |
|---|---|---|---|
| Ge 002 | n. d.d | G. anatis | E. coli |
| Ge 004 | + | G. anatis | – |
| Ge 006 | n. d.d | G. anatis | E. coli; adenovirus |
| Ge 023 | + | G. anatis | M. synoviae c |
| Ge 024 | + | G. anatis | – |
| Ge 081 | + | G. genomospecies | E. coli; adenovirus |
| Ge 082 | + | G. genomospecies | E. coli; adenovirus |
| Ge 095 | + | G. anatis | E. coli; C. perfringens; adenovirus |
| Ge 100 | + | G. anatis | E. coli |
| Ge 103 | + | G. anatis | – |
| Ge 134 | + | G. anatis | adenovirus |
| Ge 156 | + | G. anatis | – |
| Ge 168 | + | G. anatis | – |
| Ge 173 | + | G. anatis | – |
| Ge 186 | n. d.d | G. genomospecies/G. anatis | – |
| Ge 223 | + | G. anatis | – |
| Ge 297 | + | G. anatis | M. gallinarum b |
| Ge 160 | + | G. anatis | – |
| Ge 258 | + | G. anatis | C. perfringens |
aDetected by bacteriological, culture or cell culture methods
bDetection of antibodies against M. gallisepticum
cDetection of antibodies against M. synoviae
dn. d. not determined
Molecular biological identification of Gallibacterium
PCR assay based on 16S rRNA genes was established for genus-specific identification of Gallibacterium isolates. With primer pair Galli-3 and Galli-5 PCR products of approximately 680 bp were obtained for G. anatis and G. genomospecies. The high annealing temperature in the PCR process avoided a cross-reaction with DNA of other avian Pasteurellaceae species. The specificity of PCR amplification system was tested using DNA of other avian pathogens as PCR templates giving negative results. With this PCR assay it was possible to detect 650 fg/μl of template Gallibacterium DNA under described conditions which is equivalent to 880 cfu.
16S rRNA genes of 16 isolated Gallibacterium were partially amplified and sequenced. Species identification was carried out via BLAST search (http://www.ncbi.nlm.nih.gov/BLAST/). The multiple sequence alignments were demonstrated in phylogenetic analysis which showed the relation of isolated Gallibacterium from clinical cases and other related organisms. The relatedness of isolated Gallibacterium and other avian Pasteurellaceae species based on sequence profiles of 16S rRNA gene was demonstrated in Fig. 1.
Fig. 1.
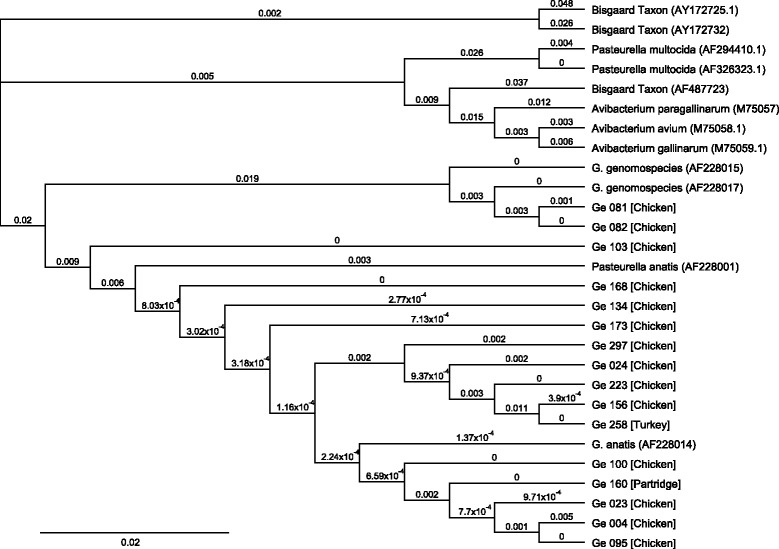
Dendrogram based on sequence profiles of 16S rRNA genes of 16 Gallibacterium isolates in this study and related avian Pasteurellaceae species and performed with cluster tree neighbour-joining analysis using the bioinformatics tools of Geneious R9.5.0.1 analysis
Gallibacterium anatis was identified in 16 birds and Gallibacterium genomospecies in 2 chickens (Ge 081 and Ge 082). Both adult hens infected by G. genomospecies were kept at the same farm. In one case a mixed culture of G. anatis and G. genomospecies was detected (Ge 186) (Table 4).
Differentiation of Gallibacterium spp
PCR–RFLP assay based on 16S rRNA genes was established for differentiation of Gallibacterium isolates. Use of Galli-3 and Galli-5 primers resulted in approximately 680 bp amplicons. Using RsaI for digestion of amplicons of both species, similar restriction patterns (210 bp and 470 bp) were obtained for G. genomospecies and G. anatis (Fig. 2). Closely related microorganisms like Bisgaard taxon 40 and 14 have no RsaI restriction site and were not cut (data not shown). Differentiation of Gallibacterium isolates into G. anatis and G. genomospecies was carried out using XbaI restriction of PCR products. G. genomospecies has an unique restriction site resulting in fragments of 216 bp and 470 bp while G. anatis amplicons were not digested (Fig. 3).
Fig. 2.
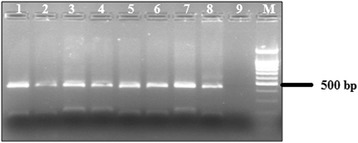
Identification of G. anatis and G. genomospecies by restriction analysis of PCR products with Rsa I; (1 – Ge 002; 2 – Ge 006; 3 – Ge 081; 4 – Ge 082; 5 – Ge 100; 6 – Ge 160; 7 – Ge 186; 8 – Ge 223; 9 – negative control; M – 100 bp DNA ladder)
Fig. 3.
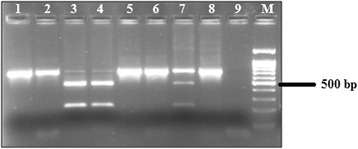
Differentiation between G. anatis and G. genomospecies by restriction analyses of PCR products with XbaI (1 – Ge 002; 2 – Ge 006; 3 – Ge 081; 4 – Ge 082; 5 – Ge 100; 6 – Ge 160; 7 – Ge 186; 8 – Ge 223; 9 – negative control; M– 100 bp DNA ladder)
Antimicrobial susceptibility test
The results of antimicrobial susceptibility testing of 15 Gallibacterium isolates against 19 antibiotics were demonstrated in Table 3. Four out of 19 Gallibacterium isolates could not be re-cultivated after applying identification. All tested isolates were susceptible to apramycin, florphenicol and neomycin, 80.0% of isolates were susceptible to ceftifur and sulfamethoxazole/trimethoprim. 73.3% of isolates were susceptible to gentamicin and tiamulin. Most of isolates showed susceptibility (66.7%) to enrofloxacin and 33.3% were susceptible to erythromycin.
All tested isolates were resistant to clindamycin, sulfathiazole and penicillin. High resistance rates were found to sulfamethoxim and spectinomycin with 93.3%, tylosin with 86.7% and oxytetracycline with 80.0%.
Discussion
To the best of our knowledge, occurrence of and diseases caused by Gallibacterium, species are not well known in Germany. In this study, a total of 19 suspected Gallibacterium isolates from different morbid and dead birds were subjected to phenotypic and molecular characterization.
Molecular biological investigation of the isolates resulted in identification of these strains as representatives of the genus Gallibacterium within the family Pasteurellaceae.
Gallibacterium can be isolated from clinically healthy chickens in farms with moderate or low levels of biosecurity [1, 6]. Furthermore, Gallibacterium was isolated from birds with various pathological lesions connected with salpingitis, oophoritis, peritonitis, pericarditis, hepatitis, enteritis, upper respiratory tract lesions, and septicaemia [3, 7, 17, 37]. In this study salpingitis, with or without peritonitis, was not detected in any of the cases, but different pathological findings could be found, including enteritis, serositis, hepatitis and nephritis. Various organs of different species were affected. Gallibacterium was found as a harmless commensal [3, 5] or in mixed infections with other poultry pathogens such as E. coli that shared the major lesions with G. anatis in naturally affected chickens [9]. In this study Gallibacterium was isolated alone in 9 cases and in 10 cases other pathogens were additionally detected: E. coli, Clostridium perfringens, Mycoplasma species and adenovirus.
The application of MALDI-TOF mass spectrometry and molecular biological methods, especially DNA–DNA hybridization and 16S rRNA sequence analysis, has contributed to an improved Gallibacterium classification [17, 21, 22]. A Gallibacterium specific PCR assay was used in this study to simplify and shorten accurate identification of suspicious Gallibacterium isolates. Molecular biological identification was carried out by sequencing 16S rRNA genes of isolates, and comparison of obtained sequence data with those of GenBank database. Using the described primer system, amplicons were produced which are specific for strains of the genus Gallibacterium. No cross-reactivity with closely related bacterial species like Bisgaard taxon 14 and 40 and other members of the Pasteurellaceae was recognized. The high annealing temperature in the PCR process avoided a cross-amplification with DNA of other avian Pasteurellaceae species. The enzymatic digestion of the amplicons with RsaI was used for confirmation of identified Gallibacterium species. Only Gallibacterium isolates possess a restriction site for RsaI in the amplified region within members of the Pasteurellaceae. This method makes identification of Gallibacterium isolates easy. Additionally, an enzymatic digestion using restriction enzyme XbaI was suited for species differentiation between species G. anatis and G. genomospecies.
Antimicrobial susceptibility reports for Gallibacterium were relatively rare. In this study, MIC and cut-off from CLSI guidelines and previous reports were used [27, 28]. The number of different techniques and antimicrobial agents made it difficult to compare previous results. Testing of 15 Gallibacterium isolates against 19 antibiotics showed that all isolates were susceptible to apramycin, florfenicol and neomycin. 80.0% of isolates were susceptible to ceftiofur and sulfamethoxazole/trimethoprim. 73.3% of isolates were susceptible to gentamicin and tiamulin and most of isolates (66.7%) were sensitive to enrofloxacin. Similar findings were observed in G. anatis isolated from both broiler flocks and broiler breeders. They were sensitive to ceftiofur (90.0%), enrofloxacin (93.0%), gentamicin (93.0%), sulfamethoxazole/trimethoprim (83.0%), florfenicol (86.0%), and sulfathiazole (82.0%) [28].
In a previous study, chloramphenicol susceptibility was observed among European strains in Denmark and very high fraction of susceptibilities towards quinolone were detected among 35.0% of Danish G. anatis isolates [27]. Moreover, high neomycin susceptibility (64.0%) was observed for Mexican strains isolated from chickens [28]. In contrast to our findings, a higher resistance to neomycin was reported in Taiwan [38]. Only 33.3% of tested isolates were susceptible to erythromycin. In previous studies, it was reported that all of chicken isolates were resistant to erythromycin [38, 39].
Results were in agreement with a study reporting M. haemolytica isolated from chicken had high sensitivities to ceftiofur (100%), enrofloxacin (96–100%), florfenicol (92–100%), and sulfamethoxazole/trimethoprim (93–100%) [39]. Similarly, strong sensitivity in G. anatis isolates from chickens to ceftiofur and enrofloxacin was reported [38].
Danish and Mexican Gallibacterium field strains have high MIC values to a broad range of antimicrobial agents including agents currently used for treatment of G. anatis infections. Most prominently, resistance to tetracycline and sulfamethoxazole was observed in 92.0% and 97.0% of field strains, respectively [27]. However, contrasting reports demonstrated moderate to high sensitivity to sulfonamides in isolates originating from broiler flocks and broiler breeders [28].
The resistance rates of isolates to tylosin and oxytetracycline were 86.7% and 80.0%, respectively, which was in agreement with previous study [28] demonstrating that G. anatis isolates from poultry revealed resistance to tylosin (100%), clindamycin (97.0%), tetracyclines (80.0–90.0%), and penicillin (70.0%). Similarly, high resistance rates in chickens have been reported for penicillin (60.0–100%) and tetracyclines (72.0–100%) [39]. In contrast moderate sensitivity to tetracycline was reported, too [38].
Most of isolates (86.7%) were resistant to spectinomycin, which was in agreement with a previous study with 0 to 50% spectinomycin sensitive chicken isolates [39]. In another study high sensitivity to spectinomycin was detected in more than 89.0% of the isolates [28].
Conclusion
The phenotypic identification methods were unable to differentiate the Gallibacterium species correctly. The presented study allowed identification and differentiation of Gallibacterium isolated from birds by integrated use of phenotypic characterization and subsequent 16S rRNA gene sequence analysis. The description of a PCR-RFLP assay improves the diagnosis and epidemiological understanding of these organisms and allowed correct identification of G. anatis and G. genomospecies. Future workings will be needed to study the prevalence of Gallibacterium species in poultry flocks. This study demonstrates that there is a need for continued monitoring of the antimicrobial susceptibilities of Gallibacterium isolates as pathogens in poultry.
Acknowledgements
The authors thank Byrgit Hofmann and Karola Zmuda for their excellent technical assistance. The authors thank Dr. Werner Herold for his great effort in sample collection. We thank Keri Clack for proof reading the manuscript.
Funding
Not applicable
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors’ contributions
HE, HH and HB participated in the conception and design of the study. HB and HH performed the farm and laboratory work. HE, HH, HB, HN and HMH analyzed the data, wrote the manuscript and contributed to manuscript discussion. All authors read and approved the final manuscript.
Ethics approval
This study was carried out in strict accordance with the recommendations of the German “Tierschutzgesetz” TierSchG from 25.05.1998 (BGBI IS.1105) and complies with the German laws and regulation regarding ethical considerations, transport, housing and experimental use of animals in research. The care and well-being of all animals was ensured by carrying out all research in compliance with all EU and member state legislation and directives.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hosny El-Adawy, Email: hosny.eladawy@fli.de.
Herbert Bocklisch, Email: herbert.bocklisch@gmx.de.
Heinrich Neubauer, Email: heinrich.neubauer@fli.de.
Hafez Mohamed Hafez, Email: Hafez.Mohamed@fu-berlin.de.
Helmut Hotzel, Email: helmut.hotzel@fli.de.
References
- 1.Bisgaard M. Ecology and significance of pasteurellaceae in animals. Zentralbl Bakteriol. 1993;279(1):7–26. doi: 10.1016/S0934-8840(11)80487-1. [DOI] [PubMed] [Google Scholar]
- 2.Gregersen R, Neubauer C, Christensen H, Korczak B, Bojesen A, Hess M, Bisgaard M. Characterization of Pasteurellaceae-like bacteria isolated from clinically affected psittacine birds. J Appl Microbiol. 2010;108(4):1235–1243. doi: 10.1111/j.1365-2672.2009.04518.x. [DOI] [PubMed] [Google Scholar]
- 3.Mushin R, Weisman Y, Singer N. Pasteurella haemolytica found in the respiratory tract of fowl. Avian Dis. 1980;24(1):162–168. doi: 10.2307/1589775. [DOI] [Google Scholar]
- 4.Bojesen A, Nielsen S, Bisgaard M. Prevalence and transmission of haemolytic Gallibacterium species in chicken production systems with different biosecurity levels. Avian Pathol. 2003;32(5):503–510. doi: 10.1080/0307945031000154107. [DOI] [PubMed] [Google Scholar]
- 5.Bisgaard M. Incidence of Pasteurella haemolytica in the respiratory tract of apparently healthy chickens and chickens with infectious bronchitis. Characterisation of 213 strains. Avian Pathol. 1977;6(4):285–292. doi: 10.1080/03079457708418238. [DOI] [PubMed] [Google Scholar]
- 6.Mirle C, Schoengarth M, Meinhart H, Olm U. Studies into incidence of Pasteurella haemolytica infections and their relevance to hens, with particular reference to diseases of the egg-laying apparatus. Monatsh Veterinarmed. 1991;46:545–549. [Google Scholar]
- 7.Addo P, Mohan K. Atypical Pasteurella haemolytica type a from poultry. Avian Dis. 1985;29(1):214–217. doi: 10.2307/1590710. [DOI] [PubMed] [Google Scholar]
- 8.Jordan F, Williams N, Wattret A, Jones T. Observations on salpingitis, peritonitis and salpingoperitonitis in a layer breeder flock. Vet Rec. 2005;157(19):573–577. doi: 10.1136/vr.157.19.573. [DOI] [PubMed] [Google Scholar]
- 9.Neubauer C, De Souza-Pilz M, Bojesen A, Bisgaard M, Hess M. Tissue distribution of haemolytic Gallibacterium anatis isolates in laying birds with reproductive disorders. Avian Pathol. 2009;38(1):1–7. doi: 10.1080/03079450802577848. [DOI] [PubMed] [Google Scholar]
- 10.Christensen H, Bisgaard M, Bojesen A, Mutters R, Olsen J. Genetic relationships among avian isolates classified as Pasteurella haemolytica, ‘Actinobacillus salpingitidis’ or Pasteurella anatis with proposal of Gallibacterium anatis gen. Nov., comb. nov. and description of additional genomospecies within Gallibacterium gen. Nov. Int J Syst Evol Microbiol. 2003;53(1):275–287. doi: 10.1099/ijs.0.02330-0. [DOI] [PubMed] [Google Scholar]
- 11.Mutters R, Ihm P, Pohl S, Frederiksen W, Mannheim W. Reclassification of the genus Pasteurella Trevisan 1887 on the basis of deoxyribonucleic acid homology, with proposals for the new species Pasteurella dagmatis, Pasteurella canis, Pasteurella stomatis, Pasteurella anatis, and Pasteurella langaa. Int J Syst Evol Microbiol. 1985;35(3):309–322. [Google Scholar]
- 12.Singh S, Singh B, Sinha D, Kumar V, Vadhana P, Bhardwaj M, Dubey S. Gallibacterium anatis: an emerging pathogen of poultry birds and domiciled birds. J Veterinar Sci Techno. 2016;7(3):1-7.
- 13.Kjos-Hansen B. Egglederperitonitt forårsaket av patogen "kloakkbakterie" hos høns. Nord Vet Med. 1950;2:523–531. [Google Scholar]
- 14.Bisgaard M. Isolation and characterization of some previously unreported taxa from poultry with phenotypical characters related to Actinobacillus-an Pasteurella species. Acta Pathol Microbiol Immunol Scand B. 1982;90(1):59–67. doi: 10.1111/j.1699-0463.1982.tb00081.x. [DOI] [PubMed] [Google Scholar]
- 15.Bisgaard M, Korczak B, Busse H, Kuhnert P, Bojesen A, Christensen H. Classification of the taxon 2 and taxon 3 complex of Bisgaard within Gallibacterium and description of Gallibacterium melopsittaci sp. nov., Gallibacterium trehalosifermentans sp. nov. and Gallibacterium salpingitidis sp. nov. Int J Syst Evol Microbiol. 2009;59(4):735–744. doi: 10.1099/ijs.0.005694-0. [DOI] [PubMed] [Google Scholar]
- 16.Christensen H, Foster C, Christensen J, Pennycott T, Olsen J, Bisgaard M. Phylogenetic analysis by 16S rDNA gene sequence comparison of avian taxa of Bisgaard and characterization and description of two new taxa of Pasteurellaceae. J Appl Microbiol. 2003;95(2):354–363. doi: 10.1046/j.1365-2672.2003.01986.x. [DOI] [PubMed] [Google Scholar]
- 17.Christensen H, Bisgaard M, Bojesen A, Mutters R, Olsen J. Genetic relationships among avian isolates classified as Pasteurella haemolytica, ‘Actinobacillus salpingitidis’ or Pasteurella anatis with proposal of Gallibacterium anatis gen. Nov., comb. nov. and description of additional genomospecies within Gallibacterium gen. Nov. Int J Sys Evol Microbiol. 2003;53(1):275–287. doi: 10.1099/ijs.0.02330-0. [DOI] [PubMed] [Google Scholar]
- 18.Johnson T, Fernandez-Alarcon C, Bojesen A, Nolan L, Trampel D, Seemann T. Complete genome sequence of Gallibacterium anatis strain UMN179, isolated from a laying hen with peritonitis. J Bacteriol. 2011;193(14):3676–3677. doi: 10.1128/JB.05177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Persson G, Bojesen A. Bacterial determinants of importance in the virulence of Gallibacterium anatis in poultry. Vet Res. 2015;46(1):57. doi: 10.1186/s13567-015-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mráz O, Vladík P, Bohácek J. Actinobacilli in domestic fowl. Zentralbl Bakteriol Orig A. 1976;236(2–3):294–307. [PubMed] [Google Scholar]
- 21.Alispahic M, Christensen H, Hess C, Razzazi-Fazeli E, Bisgaard M, Hess M. Identification of Gallibacterium species by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry evaluated by multilocus sequence analysis. Int J Med Microbiol. 2011;301(6):513–522. doi: 10.1016/j.ijmm.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 22.Alispahic M, Christensen H, Hess C, Razzazi-Fazeli E, Bisgaard M, Hess M. MALDI-TOF mass spectrometry confirms clonal lineages of Gallibacterium anatis between chicken flocks. Vet Microbiol. 2012;160(1–2):269–273. doi: 10.1016/j.vetmic.2012.05.032. [DOI] [PubMed] [Google Scholar]
- 23.Wang C, Robles F, Ramirez S, Riber A, Bojesen A. Culture-independent identification and quantification of Gallibacterium anatis (G. anatis) by real-time quantitative PCR. Avian Pathol. 2016;45(5):538–544. doi: 10.1080/03079457.2016.1184743. [DOI] [PubMed] [Google Scholar]
- 24.Korczak B, Christensen H, Emler S, Frey J, Kuhnert P. Phylogeny of the family Pasteurellaceae based on rpoB sequences. Int J Syst Evol Microbiol. 2004;54(4):1393–1399. doi: 10.1099/ijs.0.03043-0. [DOI] [PubMed] [Google Scholar]
- 25.Christensen H, Kuhnert P, Busse H, Frederiksen W, Bisgaard M. Proposed minimal standards for the description of genera, species and subspecies of the Pasteurellaceae. Int J Syst Evol Microbiol. 2007;57(1):166–178. doi: 10.1099/ijs.0.64838-0. [DOI] [PubMed] [Google Scholar]
- 26.Ludwig W, Klenk H. Overview: a phylogenetic backbone and taxonomic framework for procaryotic systematics. In: Boone D, Castenholz R, Garrity G, editors. Bergeys’ manual of systematic bacteriology. Volume 1. New York: Springer; 2001. pp. 49–65. [Google Scholar]
- 27.Bojesen A, Vazquez M, Bager R, Ifrah D, Gonzalez C, Aarestrup F. Antimicrobial susceptibility and tetracycline resistance determinant genotyping of Gallibacterium anatis. Vet Microbiol. 2011;148(1):105–10. [DOI] [PubMed]
- 28.Jones K, Thornton J, Zhang Y, Mauel M. A 5-year retrospective report of Gallibacterium anatis and Pasteurella multocida isolates from chickens in Mississippi. Poult Sci. 2013;92(12):3166–71. [DOI] [PubMed]
- 29.Aarestrup F, Seyfarth A, Angen Ø. Antimicrobial susceptibility of Haemophilus parasuis and Histophilus somni from pigs and cattle in Denmark. Vet Microbiolol. 2004;101(2):143–6. [DOI] [PubMed]
- 30.Schwarz S, Kehrenberg C, Salmon S, Watts J. In vitro activities of spectinomycin and comparator agents against Pasteurella multocida and Mannheimia haemolytica from respiratory tract infections of cattle. J Antimicrob Chemoth. 2004;53(2):379–82. [DOI] [PubMed]
- 31.Kehrenberg C, Walker R, Wu C, Schwarz S. Antimicrobial resistance in members of the family Pasteurellaceae. In: Aarestrup F, editor. Antimicrobl resist bacteria of animal origin. Washington, DC: ASM Press; 2006. p. 167–83.
- 32.Bisgaard M, Houghton S, Mutters R, Stenzel A. Reclassification of German, British and Dutch isolates of so-called Pasteurella multocida obtained from pneumonic calf lungs. Vet Microbiol. 1991;26(1):115–24. [DOI] [PubMed]
- 33.Dufour-Zavala L, Glisson J, Jackwood M, Pearson J, Reed W, Swayne D, Woolcock P. Isolation, identification and characterization of avian pathogens. In: Am assoc avian Pathol. Volume 5 ed; 2008. p. 12–8.
- 34.Swayne D, Glisson J, McDougald L, Nolan L, Suarez D, Nair V. Diseases of poultry. 13th ed. Ames: Wiley-Blackwell; 2013.
- 35.Anonymous. Nachweis einer gentechnischen Veränderung von Lactobacillus curvatus in Rohwurst durch Amplifikation der veränderten DNA-Sequenz mit Hilfe der PCR (Polymerase Chain Reaction) und Hybridisierung des PCR-Produktes mit einer DNA-Sonde. Amtliche Sammlung von Untersuchungsverfahren, Deutschland. LMBG. 1997, 35(L 08.00 44).
- 36.CLSI. Clinical Laboratory Standards Institute (National Committee for Clinical Laboratory Standards) Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; 2nd edition: Approved standard. Wayne; 2002;vol. 22 (M31-A2):55-58.
- 37.Bisgaard M, Dam A. Salpingitis in poultry. II. Prevalence, bacteriology, and possible pathogenesis in egg-laying chickens. Nord Vet Med. 1981;33(2):81–9. [PubMed]
- 38.Lin M, Lin K, Lan Y, Liaw M, Tung M. Pathogenicity and drug susceptibility of the Pasteurella anatis isolated in chickens in Taiwan. Avian Dis. 2001;45(3):655–8. [PubMed]
- 39.Malik Y, Chander Y, Gupta S, Goyal S. A retrospective study on antimicrobial resistance in Mannheimia (Pasteurella) haemolytica, Escherichia coli, Salmonella species, and Bordetella avium from chickens in Minnesota. J Appl Poult Res. 2005;14(3):506–11.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


