Abstract
Long noncoding RNAs (lncRNAs) are a class of non-protein-coding transcripts with the length longer than 200 nucleotides. Growing evidence suggests that lncRNAs, which were initially thought to be merely transcriptional “noise”, participate in a wide repertoire of biological processes. It has been well established that lncRNAs not only play important roles in genomic regulation, transcription, posttranscriptional processes but are also implicated in the pathogenesis of human diseases including cardiovascular diseases, diabetes, neurodegenerative disorders, and cancer. However, the pathological role of lncRNAs in skeletal and dental diseases is just beginning to be uncovered. In the present review, we outline the current understanding of the established functions and underlying mechanisms of lncRNAs in various cellular processes. Furthermore, we discuss new findings on the role of lncRNAs in osteoblastogenesis and osteoclastogenesis as well as their involvement in skeletal and dental diseases. This review intends to provide a general framework for the actions of lncRNAs and highlight the emerging evidence for the functions of lncRNAs in skeletal and dental diseases.
Keywords: lncRNA, Osteoblastogenesis, Osteoclastogenesis, Skeletal and dental diseases
Background
The term noncoding RNA (ncRNA) refers to diverse RNA molecules that do not encode any protein. ncRNAs include infrastructural RNAs such as transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs) [1]. Increasing evidence indicates that additional regulatory ncRNAs such as lncRNAs exist and play important roles in regulating chromatin architecture/epigenetic memory, transcription, and mRNA splicing, stability, and translation [2].
lncRNAs are among the least understood RNAs despite their pervasive transcription in the genome. The most updated annotation of the human genome (Version 27, GRCh38) identifies 27,908 lncRNA transcripts from 15,778 lncRNA genes (http://www.gencodegenes.org/stats/current.html). Like mRNAs, many polyadenylated lncRNAs are transcribed by RNA polymerase II (Pol II) and are often alternatively spliced into multiple isoforms. But genes coding for lncRNAs also have characteristics distinct from those for mRNAs: lncRNA genes have fewer but longer exons, tend to be expressed at lower levels, and exhibit less-conserved sequences [3]. However, some lncRNAs that are expressed in development- and tissue-specific patterns have highly conserved promoter regions and splice sites [4].
lncRNAs do not possess any apparent protein-coding potential and are mostly expressed at low levels; they are thus characterized largely by bioinformatic approaches. Advances in high-throughput RNA sequencing technologies provide systems with which RNA transcription can be observed in an unbiased manner [5]. lncRNAs have been shown to regulate various biological processes via distinct mechanisms [6], whereas mutations or aberrant expression of lncRNAs have been implicated in the pathogenesis of a wide range of human diseases [7–9].
In this review, we briefly present the current knowledge of the functions and mechanisms of lncRNAs. We then review the role of lncRNAs in osteoblastogenesis and osteoclastogenesis based on data from multiple studies. Finally, we discuss the important roles of lncRNAs in the etiology of skeletal and dental disorders.
Functions and mechanisms of lncRNAs
The discovery of the myriad roles of lncRNAs has made it increasingly clear that lncRNAs can function via numerous paradigms and are key regulatory molecules in cells [6]. They not only participate in nuclear events such as chromatin modification and transcription [10], but also reside in the cytoplasm, where they interact with RNA-binding proteins or modulate mRNA translation. Here, we outline a concise scheme of the functions of lncRNAs for a better understanding of what roles lncRNAs play in osteoblastogenesis, osteoclastogenesis, as well as skeletal and dental diseases.
lncRNAs in chromatin modification
In the nucleus, lncRNAs target some chromatin remodeling complexes and guide them to specific genomic loci, leading to changes in gene transcription. A classic example is a long intergenic noncoding RNA (lincRNA) termed X inactive-specific transcript (XIST), which is transcribed from one of the two X chromosomes in female mammals (Fig. 1a). It recruits polycomb group complexes, such as polycomb repressive complex 2 (PRC2) [11], to the female X chromosome, leading to transcriptional silencing in cis across a majority of the chromosome. In contrast, lncRNA HOTAIR, which is transcribed from the antisense strand of homeobox C (HOXC) locus, recruits PRC2 in trans to the HOXD cluster for epigenetic repression [12].
Fig. 1.
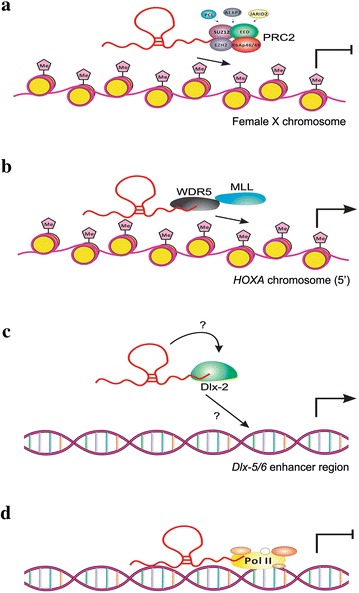
Schematic representation of how lncRNAs may function in genome regulation. a The participation of lncRNA XIST in chromosome silencing. XIST recruits PRC2 to the female X chromosome, leading to H3K27me3 formation (presented as pink pentagon with “Me”) and silencing of the chromosome. b Chromatin modification by lncRNA HOTTIP. HOTTIP interacts with adaptor protein WDR5 and targets WDR5/MLL complexes, inducing H3K4me3 (presented as a pink pentagon with “Me”) and transcription of 5′HOXA genes. c Evf-2 as an example of lncRNAs that facilitate transcriptional activation. Combination of Evf-2 with the protein Dlx-2 forms an Evf-2/Dlx-2 complex, which targets the Dlx-5/6 enhancer region and promotes transcription. Question marks indicate that the specific role of Evf-2 in the process remains to be elucidated. d Alu ncRNA can act as a potent transcriptional repressor. Alu RNA contains RNA polymerase II (Pol II, enzyme that synthesizes mRNAs in eukaryotes) binding arms and modular repression domains, allowing it to bind Pol II and block RNA synthesis
lncRNAs also associate with other chromatin regulators. An example is the lineage-specific imprinting mediated by the lncRNA Kcnq1ot1, a nuclear and moderately stable transcript from the paternal chromosome. In addition to interacting with members of the PRC2 complex, Kcnq1ot1 also recruits chromatin regulators such as G9a methyltransferase to mediate repressive histone modifications, including the trimethylation of lysine 27 on histone H3 (H3K27me3) and trimethylation of lysine 9 on histone H3 (H3K9me3) in the Kcnq1 domain [13].
However, some lncRNAs function in chromatin activation rather than chromatin silencing. Enhancers are regulatory elements that increase the expression of target genes [14]. An enhancer-like lncRNA termed HOTTIP has been identified as a key intermediate that transmits information from higher order chromosomal looping into chromatin modifications. HOTTIP is transcribed from the distal 5′tip of the HOXA locus and is brought into close proximity to multiple HOXA genes by chromosomal looping of the HOXA 5′end. It directly binds the adaptor protein WDR5 and targets WDR5/MLL complexes across the HOXA locus, leading to histone H3 lysine 4 trimethylation (H3K4me3) and gene transcription [15] (Fig. 1b).
lncRNAs in transcription regulation
As shown above, lncRNAs indirectly influence transcription through chromatin modification. However, some lncRNAs regulate transcription directly. The 3.8-kb lncRNA Evf-2 is transcribed from the Dlx-5/6 ultraconserved region. Evf-2 has been found to activate the transcriptional activity of the Dlx-5/6 enhancer by cooperating with a homeodomain protein Dlx-2 (Fig. 1c). This single-stranded RNA and the ultraconserved protein form an Evf-2/Dlx-2 complex and then the complex targets the Dlx-5/6 enhancer. But whether the complex helps Dlx-2 bind to the enhancer site or only helps stabilize the protein requires further investigation [16]. Nevertheless, some lncRNAs serve as transcriptional repressors [6]. In response to heat shock, the Alu ncRNA binds Pol II and enters complexes at promoters, ultimately blocking all detectable RNA synthesis (Fig. 1d). An interesting thing is that although there are sequence discrepancies between the Pol II-binding domains of Alu RNA and B2 RNA (Alu RNA-like sequences in mouse), both of them repress transcription by Pol II. So the mechanism by which Alu RNA functions may not be sequence specific [17].
lncRNAs in pre-mRNA splicing
In the nucleus, lncRNAs are implicated in posttranscriptional regulatory steps, including pre-mRNA splicing, mRNA capping, polyadenylation, and nuclear export. Pre-mRNA splicing is a key process to increase proteome diversity in higher eukaryotes [18]. The lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) has been proposed to regulate alternative splicing (AS) by modulating the distribution of active serine/arginine-rich (SR) proteins in nuclear speckle domains. SR proteins are essential splicing factors that function in both constitutive splicing and AS [19, 20]. Previous studies indicated that MALAT1 modulated AS of endogenous pre-mRNAs by regulating SR splicing factors phosphorylation, as well as altering the distribution and ratio of phosphorylated versus non-phosphorylated pools of SR proteins (Fig. 2a). These changes may lead to alterations in the expression of specific isoforms of proteins in cells [21].
Fig. 2.
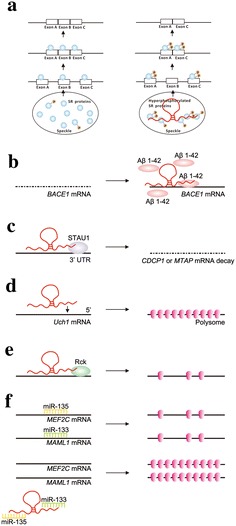
A schematic diagram illustrating the involvement of lncRNAs in posttranscriptional processes. a lncRNA MALAT1 is implicated in pre-mRNA splicing. MALAT1 changes the expression and ratio of phosphorylated (orange circle with the letter “p”) versus dephosphorylated serine/arginine-rich (SR) proteins (splicing factors can regulate splicing; blue circle), thus altering the splicing mode of pre-mRNAs. b lncRNAs also influence mRNA stability. lncRNA BACE1-AS associates with cell stressor Aβ 1–42 and stabilizes BACE1 mRNA (dashed line represents unstable mRNA). c lncRNAs may cause mRNA decay. lncRNA 1/2-sbsRNA mediates mRNA decay by binding protein STAU1, with further base-pairing with an Alu element at the 3′-untranslated regions (3′-UTRs) of CDCP1 or MTAP mRNA (dashed line represents unstable mRNA). lncRNAs can serve as activators d or repressors e in mRNA translation. Antisense Uchl1 RNA associates mRNA with active polysomes, resulting in the promotion of translation. Conversely, lincRNA-p21 enhances interaction between translational repressor Rck and mRNAs such as CTNNB1 and JUNB, giving rise to polysome size reduction and translation repression. f lncRNAs also act as miRNA sponges, leading to derepression of miRNA targets. As indicated, lncRNA linc-MD1 “sponges” miR-133 and miR-135, antagonizing the miRNA-mediated translation suppression
Another example of involvement of lncRNA in AS is sno-lncRNA, a class of nuclear-enriched intron-derived lncRNAs transcribed from a critical region of chromosome 15 (15q11-q13). This region is specifically deleted in Prader–Willi Syndrome (PWS). sno-lncRNAs are flanked by snoRNA sequences at both ends [22]. Studies have demonstrated that at least some of these sno-lncRNAs acted as molecular sinks of the splicing regulator Fox2, a member of the Fox family. sno-lncRNAs binded to Fox2 and altered the splicing patterns. Importantly, sno-lncRNA knockdown leaded to changes in Fox2-regulated splicing, while the overall gene expression levels were unaltered [23].
lncRNAs in mRNA protection and decay
As an intermediate, mRNA carries information from genes to ribosomes for protein synthesis. However, it is unstable and the concentration of mRNA depends on the balance between the rates of synthesis and degradation [24]. lncRNAs are increasingly recognized as important modulators of both mRNA synthesis and degradation.
Functional characterization of the lncRNA BACE1-AS has revealed the action of lncRNAs in maintaining mRNA stability. BACE1-AS is a conserved antisense transcript partner of β-site amyloid precursor protein cleaving enzyme 1 (BACE1), which is a crucial enzyme in the pathogenesis of Alzheimer’s disease. When exposured to amyloid-β 1–42 (Aβ 1–42), which can induce oxidative stress, elevated BACE1-AS levels increase BACE1 mRNA stability and generate additional Aβ 1–42 through a posttranscriptional feed-forward mechanism [25] (Fig. 2b).
Another type of lncRNA exerts its function by facilitating mRNA decay. Staufen 1 (STAU1) is a double-stranded RNA binding protein that binds to a subset of mRNAs and targets them for STAU1-mediated mRNA decay (SMD) [26]. Half-STAU1-binding site RNA (1/2-sbsRNA) is a polyadenylated lncRNA that induces mRNA decay by recruiting STAU1 to target mRNAs. STAU1-binding sites can be formed by imperfect base-pairing between an Alu element of an mRNA target of SMD and another Alu element in 1/2-sbsRNAs. The 1/2-sbsRNA-regulated mRNAs such as CUB-domain-containing protein 1 (CDCP1) mRNA and methylthioadenosine phosphorylase (MTAP) mRNAs can thus be degraded [27] (Fig. 2c).
lncRNAs in translation activation and repression
mRNA translation is the final step of protein synthesis. Recent studies have demonstrated that some lncRNAs control protein synthesis by either post-transcriptional activation or repression of mRNA translation in the cytoplasm.
The nuclear-enriched lncRNA, antisense Uchl1, forms sense-antisense pairs by pairing with the ubiquitin carboxy-terminal hydrolase L1 (Uchl1) gene. Under stress conditions, antisense Uchl1 lncRNA shuttles from the nucleus to the cytoplasm. It then binds the 5′end of the Uchl1 mRNA and promotes the association of this overlapping sense protein-coding mRNA with active polysomes for translation [28] (Fig. 2d).
On the contrary, lincRNA-p21 inhibits the translation of target mRNAs encoding β-catenin (CTNNB1) and JunB (JUNB) after HuR (also known as embryonic lethal abnormal vision 1, ELAVL1) is silenced. HuR is a ubiquitous RNA-binding protein that functions in cell proliferation, survival, and carcinogenesis, as well as in stress and immune responses [29, 30]. HuR exerts its functions mainly by interacting with a subset of mRNAs, and further increasing their stability and modulating their translation [31]. HuR also enhances the decay of lincRNA-p21. Therefore, in the absence of HuR, stable lincRNA-p21 inhibits the translation of CTNNB1 and JUNB mRNAs by enhancing their interaction with the translational repressor Rck, which may result in polysome size reduction and even ribosome “drop-off” [32] (Fig. 2e).
lncRNAs in miRNA biology
microRNAs (miRNAs) are endogenous 19–23-nucleotide RNAs that negatively regulate gene expression at the post-transcriptional level. They interact with partially complementary sequences in the 3′-UTR of a target mRNA, leading to translational repression, mRNA cleavage, and mRNA decay [33, 34]. Recent reports have demonstrated that lncRNAs may prevent the repressive effects of miRNAs on their targets [35–37]. lncRNAs function as competing endogenous RNAs (ceRNAs) to sequester miRNAs, thereby protecting the target mRNAs from degradation [38].
linc-MD1 has been implicated as a ceRNA that competes for shared miRNAs with mRNAs. Therefore, it can be regarded as an activator in mRNA translation. Linc-MD1 “sponges” miR-133 and miR-135 to regulate the mRNA translation of mastermind-like-1 (MAML1) and myocyte-specific enhancer factor 2C (MEF2C), respectively (Fig. 2f). With the finding that both MAML1 and MEF2C are critical genes for normal myogenic differentiation [39], linc-MD1 is postulated to be involved in the control of myoblast differentiation [40]. The well-known lncRNA H19 has been identified as a novel activator of the Wnt/β-catenin pathway by serving as a miRNA sponge. H19 antagonizes the functions of miR-141 and miR-22, both of which are negative modulators of the Wnt/β-catenin pathway and osteogenesis. The presence of H19 leads to the derepression of their shared target gene, β-catenin, and eventually promotes osteoblast differentiation [41].
Modulation of osteoblastogenesis and osteoclastogenesis by lncRNAs
Mesenchymal stem cells (MSCs) have the potential to differentiate into multiple cell types, including osteoblasts, chondrocytes, adipocytes, and neurocytes [42]. Osteoclasts are derived from hematopoietic precursor cells of the monocyte-macrophage lineage. They are large, multinucleated, terminally differentiated cells, functioning as the sole bone-resorbing cells [43]. Skeletal development and adult bone remodeling depend on the coordinated function of osteoblasts and osteoclasts, which differentiate from precursor cells in the mesenchymal osteoblastic lineage [44] and the hematopoietic osteoclastic lineage [45], respectively.
The involvement of lncRNAs in the differentiation of MSCs into osteoblasts has been unveiled over the past decade (Table 1). Analysis of lncRNA expression profiles has revealed significant differences between untreated and bone morphogenetic protein 2 (BMP-2)-treated C3H10T1/2 MSCs [46]. In the study, the authors used BMP-2 to induce early osteoblastogenesis, and compared the differential expression profiles of lncRNA by microarray and bioinformatic approaches. Over 100 differentially expressed lncRNAs were identified. A subset of 24 lncRNAs was determined to concurrently change with their nearby coding genes, which are involved in osteoblastogenesis. For example, mouselincRNA0231 and its nearby gene epidermal growth factor receptor (EGFR), which suppressed osteoblast differentiation via regulating Runx2 and Osterix, were downregulated after BMP-2 treatment. A similar correlation was observed between NR_027652 and mouselincRNA0243 with their respective nearby coding genes DLK1 and IL-5, respectively. Another study demonstrated that anti-differentiation ncRNA (ANCR) regulated Runx2 expression and osteoblastogenesis. ANCR interacted with the enhancer of zeste homolog 2 (EZH2). The recruitment of ANCR with EZH2 catalyzed H3K27me3 in Runx2 gene promoter, resulting in the inhibition of Runx2 expression and subsequent osteoblastogenesis [47].
Table 1.
Major lncRNAs associated with osteoblastogenesis and osteoclastogenesis, as well as skeletal and dental diseases
| lncRNAs | Targets | Effects | References |
|---|---|---|---|
| MouselincRNA0231 | Runx2, Osterix | Suppresses osteoblastogenesis | [46] |
| ANCR | Runx2 | Inhibits osteoblastogenesis | [47] |
| HIF1α-AS1 | HOXD10 | Promotes osteoblastogenesis | [49] |
| DANCR | p-GSK-3β, β-catenin | Blocks odontoblast-like differentiation of hDPCs | [50] |
| DANCR | IL-6, TNF-α | Positively regulates osteoclastogenesis | [55] |
| MEG3 | SLC39A1 | Inhibits osteogenic differentiation of BMSCs | [65] |
| Hotair | PRC2, LSD1 complex | Repressor of skeletal malformation | [67] |
| SOX9nc2 | SOX9 | Promotes chondrogenesis | [71] |
| H19 | COL2A1 | Stimulates chondrocyte anabolism | [72, 73] |
| lncRNA-POIR | FoxO1 | Positive regulator of osteogenic differentiation in periodontitis | [76] |
| ANRIL | ADIPOR1, VAMP3, C110RF10 | Regulates risk variants of aggressive periodontitis | [77, 78] |
| TUSC7 | Inhibits proliferation in osteosarcoma cells | [81, 82] | |
| MALAT-1 | SFPQ, PTBP2 | Promotes proliferation, migration, or invasion in osteosarcoma cells | [83–85] |
| LINC340 | Potentially involved in ameloblastoma | [89] |
The function of the lncRNA hypoxia-inducible factor 1α-anti-sense 1 (HIF1α-AS1) in osteoblastogenesis was recently identified. HIF1α-AS1 expression was significantly repressed after overexpression of the histone deacetylase sirtuin 1 (SIRT1), an important regulator of osteoblast differentiation [48]. Lower levels of SIRT1 gave rise to the upregulation of HIF1α-AS1 in human bone marrow stem cells (BMSCs). Moreover, HIF1α-AS1 knockout inhibited the expression of HOXD10 by interfering with acetylation, suggesting the potential role of HIF1α-AS1 in the activation of osteoblastogenesis [49].
Attention has also been paid to the effects of lncRNAs on dentinogenesis (Table 1). Dentinogenesis shares many similarities with osteogenesis, and consists of multiple steps including odontoblast differentiation. Chen et al. showed that lncRNAs were involved in the odontoblast-like differentiation of human dental pulp cells (hDPCs) [50]. In their study, the expression of the differentiation-antagonizing lncRNA DANCR was considerably downregulated in a time-dependent manner in the process of hDPCs differentiation into odontoblast-like cells. Furthermore, mineralized nodule formation as well as the expression of dentin sialophosphoprotein and dentin matrix protein-1 was blocked after overexpression of DANCR in hDPCs. Upregulation of DANCR also decreased the expression levels of p-GSK-3β and β-catenin. These results reveal a role of DANCR in regulating the Wnt/β-catenin pathway and modulating dentin formation.
lncRNAs also play a regulatory role in osteoclastogenesis (Table 1). In one study, microarray analysis was performed to examine the expression profiles of lncRNAs at different stages of osteoclastogenesis. Then gene ontology analysis, pathway analysis, and lncRNA-mRNA co-expression network characterization showed the co-expression of multiple lncRNAs with tumor necrosis factor ligand superfamily member (TNFSF)12 and TNFSF13 [51], factors involved in the differentiation of monocyte/macrophage precursor cells into osteoclasts [52, 53]. Circulating monocytes are directly involved in osteoclastogenesis by acting as osteoclast precursors [54]. The role of lncRNA DANCR in blood mononuclear cells has been studied. Overexpression of DANCR increased the secretion of IL-6 and TNF-α in blood mononuclear cells [55], both of which were inflammatory cytokines and important mediators of accelerated bone loss in osteoporosis [56, 57]. This suggests that DANCR can be a potential biomarker and regulatory element in circulating monocytes for osteoclastogenesis. However, further studies are needed to determine the underlying mechanisms of lncRNAs in osteoclastogenesis.
lncRNAs in skeletal and dental diseases
lncRNAs not only play critical roles in various aspects of cellular biology but are also implicated in disease pathogenesis and progression. Several lncRNAs have been functionally associated with important pathogenic processes of cardiovascular diseases [58], diabetes [59], neurodegenerative disorders [60], immune response [61], as well as several types of cancer [62, 63]. However, the identity of lncRNAs in skeletal and dental diseases is not well known. Here, we summarize new findings in the functions of lncRNAs in these diseases (Table 1).
Osteoporosis
Emerging evidence demonstrates the correlation of lncRNAs with osteoporosis, which is a common metabolic bone disorder [55]. Osteoporosis is characterized by reduced bone mineral density and increased incidence of fractures, resulting mainly from enhanced osteoclastic bone resorption activity outpacing bone formation by osteoblasts [64]. The sequence encoding lncRNA DANCR resides on human chromosome 4, located 54.8 kb upstream of USP46 and 28.7 kb downstream from ERVMER34-1 and the ANCR locus. As mentioned above, DANCR was upregulated in circulating monocytes of postmenopausal women with low bone mineral density, and could induce the expression of IL-6 and TNF-α [55]. These results suggest the important role of DANCR in the pathogenesis of osteoporosis and possibly as a biomarker for postmenopausal osteoporosis (PMOP). Another case is the involvement of lncRNA MEG3 in the pathogenesis of PMOP [65]. In this study, MEG3 expression was increased in BMSCs derived from PMOP patients and ovariectomized mice. MEG3 directly bound to and activated miR-133a-3p, thereby inhibiting the expression of SLC39A1 (a direct target of miR-133a-3p), which was regarded as a positive regulator of osteogenic differentiation. Overexpression of MEG3 inhibited osteogenic differentiation of BMSCs, which was markedly reversed by miR-133a-3p knockdown. These data indicate that lncRNAs participate in the pathogenesis of osteoporosis, which provide novel targets for the prevention and treatment of osteoporosis.
Skeletal transformation
After its first identification in primary human fibroblasts [66], the lncRNA Hotair has also been found to be important in the embryonic patterning of the skeletal system [67]. Targeted deletion of Hotair resulted in lumbosacral homeotic transformation (6th lumbar vertebrae transform to 1th sacral vertebrae, L6 → S1) in a C57BL/6 mouse model. Moreover, malformation of the metacarpals and 4th caudal vertebrae was also observed. Hotair knockdown caused derepression of multiple HoxD cluster genes in embryos and tail tip fibroblasts. Insights into the molecular basis for the observed phenotypes revealed that Hotair acted in trans to bind both PRC2 and LSD1 complex. Hotair recruited them to hundreds of genomic sites to promote coordinated H3K27 methylation and H3K4 demethylation for gene silencing. However, in another study, the skeletal malformation indicated above was not detected after Hotair knockdown in a mixed CBAxBL/6 mouse model [68]. Whether the discrepancy in the phenotypic effects of Hotair knockdown is attributed to the different genetic background of animals needs further study.
Osteoarthritis
Osteoarthritis (OA) is the clinical and pathological outcome of a range of disorders that results in structural and functional failure of synovial joints. While many risk factors (e.g., IL-1, IL-6, TNF-α, PGE2, MMPs) contribute to the onset of OA [69], the mechanism responsible for OA has not been fully elucidated. Recent studies have investigated the effects of lncRNAs on OA. Xing et al. reported that over 100 lncRNAs were up- or down-regulated in OA cartilage compared with normal cartilage based on microarray analysis. The increased expression of six lncRNAs (HOTAIR, GAS5, PMS2L2, RP11-445H22.4, H19 and CTD-2574D22.4) in the microarray data was validated by real-time PCR [70], suggesting the regulatory potential of lncRNAs in OA. SOX9nc2 is a cartilage-specific lncRNA which lies upstream of SOX9 in the genome. Depletion of the SOX9nc2 transcript by RNA interference prevented chondrogenesis and the expression of the transcription factor SOX9 [71]. Moreover, a significant correlation has been observed among the expression of lncRNA H19, miR-675, and COL2A1 in OA cartilage. Co-upregulation of H19, COL2A1, and miRNA-675 was observed in chondrocytes under hypoxic conditions, which were known to stimulate chondrocyte anabolism. When chondrocytes were treated with inflammatory factors IL-1β and TNF-α to induce chondrocyte catabolism, the expression of H19, COL2A1, and miRNA-675 was significantly decreased [72]. In addition, Dudek et al. showed that inhibition of H19 downregulated COL2A1, while overexpression of miR-675 rescued COL2A1 expression in H19-depleted human articular chondrocytes [73]. More work is needed to investigate the function and mechanisms of lncRNAs as key regulators of OA.
Periodontitis
Evidence for the relationship between periodontitis and lncRNAs is emerging. Periodontitis is a common chronic inflammatory disease initiated by a group of bacterial pathogens in dental plaque. The inflammation extends deep into tissues, damages the connective tissue and alveolar bone around teeth, and eventually leads to tooth loss [74]. Microarray analysis of lncRNA expression profile revealed a total of 8925 differently expressed lncRNAs in chronic periodontitis tissues compared with adjacent normal tissues. Further subgroup analysis showed there were 589 enhancer-like lncRNAs, 238 HOX cluster lncRNAs, as well as 1218 lincRNAs. Based on the information, the function and mechanisms of lncRNAs associated with periodontitis needs further investigation [75]. The role of a crucial lncRNA related to periodontitis, lncRNA-POIR, has recently been investigated. lncRNA-POIR expression was significantly lower in periodontal mesenchymal stem cells (PDLSCs) from periodontitis patients (pPDLSCs) than that in human periodontal MSCs (hPDLSCs). Overexpression of lncRNA-POIR promoted osteogenic differentiation of pPDLSCs. Further study revealed that lncRNA-POIR acted as a ceRNA for miR-182, thus positively regulating expression of FoxO1. The inflammatory environment, which usually occurred in periodontitis, increased miR-182 expression through NF-κB pathway, finally resulted in an imbalance in the lncRNA-POIR-miR-182 regulatory network [76]. The association between periodontitis and another lncRNA ANRIL has also been reported. ANRIL is the first shared genetic risk factor of coronary artery disease and aggressive periodontitis [77]. Bochenek et al. demonstrated that ANRIL knockdown resulted in repression of three genes ADIPOR1, VAMP3, and C11ORF10. Exploration of the identified genes highlighted a region upstream of VAMP3 within CAMTA1 (rs10864294) to be associated with increased risk of coronary artery disease and aggressive periodontitis [78]. These studies indicate the potential of lncRNAs as diagnostic biomarkers and targets for the treatment of periodontitis.
Osteosarcoma
Differences in the expression of lncRNAs in different types of tumors have been well documented [79, 80]. This finding promoted interest in addressing the potential of lncRNAs in skeletal tumors. Osteosarcoma is the most common primary malignant tumor of bone with cytogenetic complexity. The lncRNA TUSC7 (tumor suppressor candidate 7), previously named as LOC285194, was significantly downregulated in osteosarcomas. The decreased expression of TUSC7 was due to copy number loss of the genomic region on chr3q13.31. Depletion of TUSC7 promoted proliferation and inhibited apoptosis in osteosarcoma cells. TUSC7 suppression also increased osteosarcoma growth in a mouse model, and was correlated with poor survival of osteosarcoma patients [81, 82]. In addition, recent studies revealed that the lncRNA MALAT1 was dysregulated in multiple malignant tumors, including osteosarcoma. Knockdown of MALAT1 decreased proliferation, migration, and induced apoptosis in osteosarcoma. MALAT1 knockdown significantly inhibited PI3K/AKT and RhoA/ROCK signaling pathway. High expression of MALAT1 was closely correlated with pulmonary metastasis in patients with osteosarcoma [83, 84]. Interestingly, Fang et al. demonstrated that downregulation of MALAT1 induced by high dose of 17β-Estradiol promoted the binding of SFPQ to oncogene PTBP2, therefore affecting proliferation, migration or invasion in osteosarcoma cells [85]. Further discussion of lncRNAs in osteosarcoma can be found elsewhere [86, 87].
Ameloblastoma
Ameloblastoma is a benign but locally invasive odontogenic tumor of the jaws [88]. It often results in facial deformity and significant morbidity because of its high rate of recurrence and requirement for radical surgery. Considerable efforts have been made to clarify the underlying molecular mechanisms and actions of lncRNAs in ameloblastoma. The ncRNA expression profile of ameloblastoma was characterized in a well-defined ameloblastoma cohort. In this study, whole transcriptome profiling by microarray followed by real-time PCR assays validated five highly associated ncRNAs, including the lncRNA LINC340 (also known as CASC15). However, whether LINC340 is a prognostic and therapeutic marker that can improve the treatment of ameloblastoma requires further investigation [89].
Conclusions
Over the past decade, extensive research has established that lncRNAs play important roles in diverse cellular processes. Moreover, the molecular mechanisms by which lncRNAs exert their functions have been largely elucidated. These discoveries have promoted investigators in the skeletal and dental fields to address the potential role of lncRNAs in regulating the differentiation and function of bone cells as well as in the pathogenesis of skeletal and dental diseases. However, whereas a few studies have revealed the functional role of some lncRNAs, most of the results have merely demonstrated an association of lncRNAs with either bone cell biology or the development of some skeletal and dental diseases. Hence, future investigations should focus on further establishing the functional links between lncRNAs and hard tissue diseases and elucidating the underlying molecular mechanisms. A better understanding of the regulatory roles and molecular mechanisms of lncRNAs in skeletal and dental diseases may identify new biomarkers for diagnosis and novel therapeutic targets for these disorders.
Authors’ contributions
YL wrote the manuscript. JZ, JP and PD created the figures. XY, YX and YW made the table and revised the manuscript. XF and SZ revised and approved this manuscript prior to its submission. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Not applicable.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China [Grant Number: 81470777].
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ANCR
anti-differentiation noncoding RNA
- AS
alternative splicing
- Aβ 1-42
amyloid-β 1-42
- BACE1
β-secretase-1
- BMSC
bone marrow stem cell
- BMP-2
bone morphogenetic protein 2
- CDCP1
CUB-domain-containing protein 1
- ceRNA
competing endogenous RNA
- DANCR
differentiation-antagonizing long noncoding RNA
- EGFR
epidermal growth factor receptor
- EZH2
enhancer of zeste homolog 2
- hDPC
human dental pulp cell
- HIF1α-AS1
hypoxia-inducible factor 1α-anti-sense 1
- HOX
homeobox
- hPDLSC
human periodontal mesenchymal stem cell
- H3K4me3
trimethylation of lysine 4 on histone H3
- H3K9me3
trimethylation of lysine 9 on histone H3
- H3K27me3
trimethylation of lysine 27 on histone H3
- lincRNA
long intergenic noncoding RNA
- lncRNA
long noncoding RNA
- LSD1
lysine-specific demethylase 1
- MALAT1
metastasis-associated lung adenocarcinoma transcript 1
- MAML1
mastermind-like-1
- MEF2C
myocyte-specific enhancer factor 2C
- miRNA
microRNA
- MSC
mesenchymal stem cell
- MTAP
methylthioadenosine phosphorylase
- ncRNA
non-protein-coding RNA
- OA
osteoarthritis
- PDLSC
periodontal mesenchymal stem cell
- PMOP
postmenopausal osteoporosis
- Pol II
polymerase II
- pPDLSC
patient periodontal mesenchymal stem cell
- PRC2
polycomb repressive complex 2
- PWS
Prader–Willi Syndrome
- rRNA
ribosomal RNA
- RT-PCR
real-time polymerase chain reaction
- SIRT1
sirtuin 1
- SMD
STAU1-mediated mRNA decay
- snoRNA
small nucleolar RNA
- snRNA
small nuclear RNA
- SR protein
serine/arginine-rich protein
- STAU1
Staufen 1
- TNFSF
necrosis factor ligand superfamily
- tRNA
transfer RNA
- Uchl1
ubiquitin carboxy-terminal hydrolase L1
- XIST
X inactive-specific transcript
- 1/2-sbsRNA
half-STAU-1-binding site RNA
- 3′-UTR
3′-untranslated region
Contributor Information
Yuyu Li, Email: lyysunnysmell@163.com.
Jiawei Zhang, Email: zhangjiawei_0510@126.com.
Jie Pan, Email: 1045425516@qq.com.
Xu Feng, Email: xfeng@uabmc.edu.
Peipei Duan, Email: psdarling@163.com.
Xing Yin, Email: yinxing1217@gmail.com.
Yang Xu, Email: youngf15@163.com.
Xin Wang, Email: wangxin94forever@163.com.
Shujuan Zou, Phone: +86 13808014978, Phone: +86 02885501474, Email: drzsj@scu.edu.cn.
References
- 1.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 2.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Lee C, Kikyo N. Strategies to identify long noncoding RNAs involved in gene regulation. Cell Biosci. 2012;2:37. doi: 10.1186/2045-3701-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: noncoding RNAs. BBA-Rev Cancer. 2005;1756:65–75. doi: 10.1016/j.bbcan.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 9.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 10.Chen L-L, Carmichael GG. Decoding the function of nuclear long non-coding RNAs. Curr Opin Cell Biol. 2010;22:357–364. doi: 10.1016/j.ceb.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rinn JL. lncRNAs: linking RNA to chromatin. Cold Spring Harb Perspect Biol. 2014;6:a019331. doi: 10.1101/cshperspect.a018614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-DiNardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 14.Lam MTY, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Hallegger M, Llorian M, Smith CWJ. Alternative splicing: global insights. FEBS J. 2010;277:856–866. doi: 10.1111/j.1742-4658.2009.07521.x. [DOI] [PubMed] [Google Scholar]
- 19.Fu X-D, Ares M., Jr Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy ASN. Plant serine/arginine-rich proteins and their role in pre-mRNA splicing. Trends Plant Sci. 2004;9:541–547. doi: 10.1016/j.tplants.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rearick D, Prakash A, McSweeny A, Shepard SS, Fedorova L, Fedorov A. Critical association of ncRNA with introns. Nucleic Acids Res. 2011;39:2357–2366. doi: 10.1093/nar/gkq1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin Q-F, Yang L, Zhang Y, Xiang J-F, Wu Y-W, Carmichael Gordon G, Chen L-L. Long noncoding RNAs with snoRNA ends. Mol Cell. 2012;48:219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 24.Pérez-Ortín JE, Alepuz P, Chávez S, Choder M. Eukaryotic mRNA decay: methodologies, pathways, and links to other stages of gene expression. J Mol Biol. 2013;425:3750–3775. doi: 10.1016/j.jmb.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 25.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St. Laurent Iii G, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of [beta]-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3[prime] UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 29.Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. WIREs RNA. 2010;1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukherjee N, Corcoran David L, Nusbaum Jeffrey D, Reid David W, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene Jack D. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples Pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon J-H, Abdelmohsen K, Srikantan S, Yang X, Martindale Jennifer L, De S, Huarte M, Zhan M, Becker Kevin G, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng L, Yang S-B, Xu F-F, Zhang J-H. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Canc Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xing C, Hu X, Xie F, Yu Z, Li H, Bin Z, Wu J, Tang L, Gao S. Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett. 2015;589:1981–1987. doi: 10.1016/j.febslet.2015.04.061. [DOI] [PubMed] [Google Scholar]
- 37.Cai H, Xue Y, Wang P, Wang Z, Li Z, Hu Y, Li Z, Shang X, Liu Y. The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget. 2015;6:19759–19779. doi: 10.18632/oncotarget.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon J-H, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen H, McElhinny AS, Cao Y, Gao P, Liu J, Bronson R, Griffin JD, Wu L. The notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 2006;20:675–688. doi: 10.1101/gad.1383706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang W, Fu W, Wang Y, Sun Y, Xu L, Wong C, Chan K, Li G, Waye MM, Zhang J. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci Rep. 2016;6:20121. doi: 10.1038/srep20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem. 2010;285:25221–25231. doi: 10.1074/jbc.M110.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asagiri M, Takayanagi H. The molecular understanding of osteoclast differentiation. Bone. 2007;40:251–264. doi: 10.1016/j.bone.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 44.Marie PJ. Osteoblast dysfunctions in bone diseases: from cellular and molecular mechanisms to therapeutic strategies. Cell Mol Life Sci. 2015;72:1347–1361. doi: 10.1007/s00018-014-1801-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walsh MC, Kim N, Kadono Y, Rho J, Lee SY, Lorenzo J, Choi Y. OSTEOIMMUNOLOGY: interplay between the immune system and bone metabolism. Annu Rev Immunol. 2006;24:33–63. doi: 10.1146/annurev.immunol.24.021605.090646. [DOI] [PubMed] [Google Scholar]
- 46.Zuo C, Wang Z, Lu H, Dai Z, Liu X, Cui L. Expression profiling of lncRNAs in C3H10T1/2 mesenchymal stem cells undergoing early osteoblast differentiation. Mol Med Rep. 2013;8:463–467. doi: 10.3892/mmr.2013.1540. [DOI] [PubMed] [Google Scholar]
- 47.Zhu L, Xu P. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Bioph Res Co. 2013;432:612–617. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 48.Bäckesjö C-M, Li Y, Lindgren U, Haldosén L-A. Activation of Sirt1 decreases adipocyte formation during osteoblast differentiation of mesenchymal stem cells. J Bone Miner Res. 2006;21:993–1002. doi: 10.1359/jbmr.060415. [DOI] [PubMed] [Google Scholar]
- 49.Xu YAO, Wang S, Tang C, Chen W. Upregulation of long non-coding RNA HIF 1α-anti-sense 1 induced by transforming growth factor-β-mediated targeting of sirtuin 1 promotes osteoblastic differentiation of human bone marrow stromal cells. Mol Med Rep. 2015;12:7233–7238. doi: 10.3892/mmr.2015.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen L, Song Z, Huang S, Wang R, Qin W, Guo J, Lin Z. lncRNA DANCR suppresses odontoblast-like differentiation of human dental pulp cells by inhibiting wnt/β-catenin pathway. Cell Tissue Res. 2016;364:309–318. doi: 10.1007/s00441-015-2333-2. [DOI] [PubMed] [Google Scholar]
- 51.Dou C, Cao Z, Yang B, Ding N, Hou T, Luo F, Kang F, Li J, Yang X, Jiang H, et al. Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci Rep. 2016;6:21499. doi: 10.1038/srep21499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park J, Kwok S, Lim M, Oh H, Kim E, Jhun J, Ju JH, Park K, Park Y, Park S, et al. TWEAK promotes osteoclastogenesis in rheumatoid arthritis. Am J Pathol. 2013;183:857–867. doi: 10.1016/j.ajpath.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 53.Hemingway F, Taylor R, Knowles HJ, Athanasou NA. RANKL-independent human osteoclast formation with APRIL, BAFF, NGF, IGF I and IGF II. Bone. 2011;48:938–944. doi: 10.1016/j.bone.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 54.Fujikawa Y, Quinn JM, Sabokbar A, McGee JO, Athanasou NA. The human osteoclast precursor circulates in the monocyte fraction. Endocrinology. 1996;137:4058–4060. doi: 10.1210/endo.137.9.8756585. [DOI] [PubMed] [Google Scholar]
- 55.Tong X, Gu P, Xu S, Lin X. Long non-coding RNA-DANCR in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. Biosci Biotech Bioch. 2015;79:732–737. doi: 10.1080/09168451.2014.998617. [DOI] [PubMed] [Google Scholar]
- 56.Yu B, Chang J, Liu Y, Li J, Kevork K, Al-Hezaimi K, Graves DT, Park N, Wang C. Wnt4 signaling prevents skeletal aging and inflammation by inhibiting nuclear factor-[kappa]B. Nat Med. 2014;20:1009–1017. doi: 10.1038/nm.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pacifici R, Brown C, Puscheck E, Friedrich E, Slatopolsky E, Maggio D, McCracken R, Avioli LV. Effect of surgical menopause and estrogen replacement on cytokine release from human blood mononuclear cells. Proc Natl Acad Sci. 1991;88:5134–5138. doi: 10.1073/pnas.88.12.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Annilo T, Kepp K, Laan M. Natural antisense transcript of natriuretic peptide precursor A (NPPA): structural organization and modulation of NPPA expression. BMC Mol Biol. 2009;10:81. doi: 10.1186/1471-2199-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morán I, Akerman İ, van de Bunt M, Xie R, Benazra M, Nammo T, Arnes L, Nakić N, García-Hurtado J, Rodríguez-Seguí S, et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16:435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salta E, De Strooper B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol. 2012;11:189–200. doi: 10.1016/S1474-4422(11)70286-1. [DOI] [PubMed] [Google Scholar]
- 61.Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35:408–419. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. BBA-Gene Regul Mech. 2014;1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 63.Kitagawa M, Kitagawa K, Kotake Y, Niida H, Ohhata T. Cell cycle regulation by long non-coding RNAs. Cell Mol Life Sci. 2013;70:4785–4794. doi: 10.1007/s00018-013-1423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manolagas SC. Birth and death of bone cells: basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr Rev. 2000;21:115–137. doi: 10.1210/edrv.21.2.0395. [DOI] [PubMed] [Google Scholar]
- 65.Wang Q, Li Y, Zhang Y, Ma L, Lin L, Meng J, Jiang L, Wang L, Zhou P, Zhang Y. LncRNA MEG3 inhibited osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis by targeting miR-133a-3p. Biomed Pharmacother. 2017;89:1178–1186. doi: 10.1016/j.biopha.2017.02.090. [DOI] [PubMed] [Google Scholar]
- 66.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li L, Liu B, Wapinski Orly L, Tsai M, Qu K, Zhang J, Carlson Jeff C, Lin M, Fang F, Gupta Rajnish A, et al. Targeted disruption of hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amândio AR, Necsulea A, Joye E, Mascrez B, Duboule D. Hotair is dispensible for mouse development. PLoS Genet. 2016;12(12):e1006232. doi: 10.1371/journal.pgen.1006232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hunter DJ, Felson DT. Osteoarthritis. BMJ-Brit Med J. 2006;332:639–642. doi: 10.1136/bmj.332.7542.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xing D, Liang J, Li Y, Lu J, Jia H, Xu L, Ma X. Identification of long noncoding RNA associated with osteoarthritis in humans. Orthop Surg. 2014;6:288–293. doi: 10.1111/os.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meulenbelt IM, Bhutani N, den Hollander W, Gay S, Oppermann U, Reynard LN, Skelton AJ, Young DA, Beier F, Loughlin J. The first international workshop on the epigenetics of osteoarthritis. Connect Tissue Res. 2016;58:37–48. doi: 10.3109/03008207.2016.1168409. [DOI] [PubMed] [Google Scholar]
- 72.Steck E, Boeuf S, Gabler J, Werth N, Schnatzer P, Diederichs S, Richter W. Regulation of H19 and its encoded microRNA-675 in osteoarthritis and under anabolic and catabolic in vitro conditions. J Mol Med. 2012;90:1185–1195. doi: 10.1007/s00109-012-0895-y. [DOI] [PubMed] [Google Scholar]
- 73.Katarzyna AD, Jérôme EL, Aida MS, Christopher LM. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J Biol Chem. 2010;285:24381. doi: 10.1074/jbc.M110.111328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 75.Zou Y, Li C, Shu F, Tian Z, Xu W, Xu H, Tian H, Shi R, Mao X. lncRNA expression signatures in periodontitis revealed by microarray: the potential role of lncRNAs in periodontitis pathogenesis. J Cell Biochem. 2015;116:640–647. doi: 10.1002/jcb.25015. [DOI] [PubMed] [Google Scholar]
- 76.Wang L, Wu F, Song Y, Li X, Wu Q, Duan Y, Jin Z. Long noncoding RNA related to periodontitis interacts with miR-182 to upregulate osteogenic differentiation in periodontal mesenchymal stem cells of periodontitis patients. Cell Death Dis. 2016;7:e2327. doi: 10.1038/cddis.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schaefer AS, Richter GM, Groessner-Schreiber B, Noack B, Nothnagel M, et al. Identification of a shared genetic susceptibility locus for coronary heart disease and periodontitis. PLoS Genet. 2009;5(2):e1000378. doi: 10.1371/journal.pgen.1000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bochenek G, Häsler R, El Mokhtari N, König IR, Loos BG, Jepsen S, Rosenstiel P, Schreiber S, Schaefer AS. The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum Mol Genet. 2013;22:4516–4527. doi: 10.1093/hmg/ddt299. [DOI] [PubMed] [Google Scholar]
- 79.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38–38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19:R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pasic I, Shlien A, Durbin AD, Stavropoulos DJ, Baskin B, Ray PN, Novokmet A, Malkin D. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. 2010;70:160–171. doi: 10.1158/0008-5472.CAN-09-1902. [DOI] [PubMed] [Google Scholar]
- 82.Cong M, Li J, Jing R, Li Z. Long non-coding RNA tumor suppressor candidate 7 functions as a tumor suppressor and inhibits proliferation in osteosarcoma. Tumor Biol. 2016;37:9441. doi: 10.1007/s13277-015-4414-y. [DOI] [PubMed] [Google Scholar]
- 83.Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3 K/Akt pathway. Tumor Biol. 2015;36:1477. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]
- 84.Cai X, Liu Y, Yang W, Xia Y, Yang C, Yang S, Liu X. Long noncoding RNA MALAT1 as a potential therapeutic target in osteosarcoma. J Orthop Res. 2016;34:932–941. doi: 10.1002/jor.23105. [DOI] [PubMed] [Google Scholar]
- 85.Fang D, Yang H, Lin J, Teng Y, Jiang Y, Chen J, Li Y. 17β-estradiol regulates cell proliferation, colony formation, migration, invasion and promotes apoptosis by upregulating miR-9 and thus degrades MALAT-1 in osteosarcoma cell MG-63 in an estrogen receptor-independent manner. Biochem Biophys Res Commun. 2015;457:500–506. doi: 10.1016/j.bbrc.2014.12.114. [DOI] [PubMed] [Google Scholar]
- 86.Huynh NPT, Anderson BA, Guilak F, McAlinden A. Emerging roles for long noncoding RNAs in skeletal biology and disease. Connect Tissue Res. 2017;58:116–141. doi: 10.1080/03008207.2016.1194406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Z, Yu X, Shen J. Long non-coding RNAs: emerging players in osteosarcoma. Tumor Biol. 2016;37:2811. doi: 10.1007/s13277-015-4749-4. [DOI] [PubMed] [Google Scholar]
- 88.Mendenhall WM, Werning JW, Fernandes R, Malyapa RS, Mendenhall NP. Ameloblastoma. Am J Clin Oncol. 2007;30:645–648. doi: 10.1097/COC.0b013e3181573e59. [DOI] [PubMed] [Google Scholar]
- 89.Davanian H, Balasiddaiah A, Heymann R, Sundstrom M, Redenstrom P, Silfverberg M, Brodin D, Sallberg M, Lindskog S, Kruger Weiner C, et al. Ameloblastoma RNA profiling uncovers a distinct non-coding RNA signature. Oncotarget. 2017;8:4530–4542. doi: 10.18632/oncotarget.13889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


