Abstract
Acinetobacter spp. occur naturally in many different habitats, including food, soil, and surface waters. In clinical settings, Acinetobacter poses an increasing health problem, causing infections with limited to no antibiotic therapeutic options left. The presence of human generated multidrug resistant strains is well documented but the extent to how widely they are distributed within the Acinetobacter population is unknown. In this study, Acinetobacter spp. were isolated from water samples at 14 sites of the whole course of the river Danube. Susceptibility testing was carried out for 14 clinically relevant antibiotics from six different antibiotic classes. Isolates showing a carbapenem resistance phenotype were screened with PCR and sequencing for the underlying resistance mechanism of carbapenem resistance. From the Danube river water, 262 Acinetobacter were isolated, the most common species was Acinetobacter baumannii with 135 isolates. Carbapenem and multiresistant isolates were rare but one isolate could be found which was only susceptible to colistin. The genetic background of carbapenem resistance was mostly based on typical Acinetobacter OXA enzymes but also on VIM-2. The population of Acinetobacter (baumannii and non-baumannii) revealed a significant proportion of human-generated antibiotic resistance and multiresistance, but the majority of the isolates stayed susceptible to most of the tested antibiotics.
Keywords: Acinetobacter, JDS3, river, water, carbapenemases
1. Introduction
The genus Acinetobacter consists of over 40 known species that can be isolated from various habitats including soil, sediment surface, and wastewater [1]. They have the ability to colonize human skin and are responsible for a growing number of nosocomial outbreaks worldwide. Although most Acinetobacter species have generally a low pathogenicity [2], according to Alsan et al. (2008), the intensive care unit (ICU) mortality rate is around 40% [3].
The most striking characteristic of Acinetobacter spp. is their natural resistance to many antibiotics and the ability to easily develop new resistances under antibiotic pressure. They overexpress efflux pumps, harbor β-lactamases, and are characterized by low membrane permeability [2]. By 2012, over 210 different β-lactamases have been identified within the genus [4]. Different Oxacillinases (OXA) enzyme families have their origin in Acinetobacter, such as OXA-21like, OXA-23like or OXA-51like [5]. These enzymes are serine hydrolases represent class D according to the Ambler classification of β-lactamases [6]. The spread of these Acinetobacter oxaxilinases into other species seems much more limited than, for example, the spread of CTX-M or NDM enzymes but is documented. This set of OXA enzyme enables Acinetobacter to adapt easily to new developed β-lactam antibiotics [4,7]. Therefore, Acinetobacter baumannii especially has become one of the problematic nosocomial pathogens. Infections with some of these strains, such as bloodstream infections and pneumonia, do not leave any further options for antibiotic treatment. Next to Pseudomonas and carbapenem-resistant Enterobacteriaceae, Acinetobacter were rated by the WHO to be the group in most urgent need of new antibiotics (http://www.who.int/medicines/publications/ global-priority-list-antibiotic-resistant-bacteria/en/) [5,7,8]. In addition to the acquisition of a seemingly infinite number of resistances, such as Pseudomonas spp., Acinetobacter is characterized by a much better ability to survive hostile conditions, e.g., survival on dry surfaces. This makes Acinetobacter an ideal candidate for survival in clinical settings and in the environment [9,10,11].
Occurrence and susceptibility of Acinetobacter spp. in clinical settings is documented quite well, whereas their distribution and proportion of resistance in the aquatic environment remains quite unclear. Nearly all studies that investigate antibiotic resistance of Acinetobacter in the environment are based on selective cultivation, masking their proportion in the population, or are based on molecular methods, with all their inherent methodological weaknesses [9,12,13,14,15]. There is some evidence that environmental transport of Acinetobacter plays a role in the spread of clinical relevant Acinetobacter strains in the environment. On the other hand, there seems to be a continuous influx of novel strains into the clinical setting with the potential of new infectious features [16,17].
Participation in the Joint Danube Survey 2013 (JDS3) offered the possibility of isolating Acinetobacter from the total course of one of Europe’s longest rivers. This chance was taken to generate an initial picture of the resistance proportion within Acinetobacter spp. and to get an idea of how far acquired antibiotic resistances of clinical relevance have spread in the aquatic environment.
2. .Material and Methods
2.1. Sample Collection
All samples were taken during the research expedition of the Joint Danube Survey 2013 (JDS3). The survey was organized by the International Commission for the Protection of the Danube River (ICPDR), Vienna. The water samples were taken between 12 August and 26 September 2013, from 68 sampling sites along the River Danube, starting at Böfinger Halde (Germany) downstream to the delta (Romania). At each sampling site, samples were collected at three sampling points (left, middle, right), in sterile 1 L glass flasks from 30 cm below the river surface. From each flask, duplicate volumes of 45 mL of river water were filled into sterile non-toxic 50 mL plastic vials (Techno Plastic Products AG, TPP, Trasadingen, Switzerland), containing 5 mL of glycerine (final conc. 10% v/v). The vials were completely mixed by hand and immediately stored at −20 °C on board of the cruise ship until analysis in the laboratory. After transfer to the laboratory (beginning in October 2013), the samples were stored at −80 °C. Fourteen sampling sites, four of them downstream of megacities (Vienna, Budapest, Belgrade and Bucharest), two at the beginning as well as two at the delta, four rural sampling sites, and two after confluence of two biggest tributaries (Drave, Tisa) were chosen for investigation (Table 1).
Table 1.
JDS3 sampling sites chosen for isolation and their assignment to the upper-, middle-, or downstream stretches (SP = sampling point; us = upstream; ds = downstream). Country codes: Germany: DE; Austria: AT; Hungary: HU; Croatia: HR; Serbia: RS; Romania: RO; Bulgaria: BG.
| SP | Name of SP | River (km) | Country |
|---|---|---|---|
| JDS2 | Kelheim, gauging station | 2415 | DE |
| JDS3 | Geisling power plant | 2354 | DE |
| JDS8 | Oberloiben | 2008 | AT |
| JDS10 | Wildungsmauer (Vienna) | 1895 | AT |
| JDS22 | ds Budapest | 1632 | HU |
| JDS28 | us Drava | 1384 | HR/RS |
| JDS36 | ds Tisa/us Sava | 1200 | RS |
| JDS38 | us Pancevo (Belgrade) | 1159 | RS |
| JDS49 | Pristol/Novo Salo | 834 | RO/BG |
| JDS57 | ds Ruse | 488 | RO/BG |
| JDS59 | ds Arges (Bucharest) | 429 | RO/BG |
| JDS63 | Siret | 154 | RO |
| JDS67 | Sulina Arm | 26 | RO |
| JDS68 | St. Gheorge Arm | 104 | RO |
2.2. Isolation of Acinetobacter
The frozen samples were thawed, and 15 mL (left, middle, and right 5 mL each) were plated in 0.5 mL portions on selective agars. For the isolation of Acinetobacter 0.5 mL from left, middle, and right were plated on five agar-plates of CHROMagar™ (Oxoid, Germany) each. Growth conditions were 37 ± 1 °C for 18–24 h. Colonies were picked according to the manufacturer’s instructions and subcultured on Columbia blood-agar (in house production). Identification of Acinetobacter was carried out by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) as described previously [18].
2.3. Susceptibility Testing
For inoculation, colonies were picked from an overnight pure culture on Colombia blood-agar (non-selective medium) with a sterile loop and suspended in sterile saline (0.85% NaCl w/v in water) to the density of a McFarland 0.5 standard (DensiCheck, Biomerieux, Vienna, Austria). The suspension was plated on Mueller-Hinton II agar using an automatic plate rotator (Retro C80, Biomerieux, Vienna, Austria). Antibiotic test disks were stamped on the agar surface. The plates were incubated at 36 °C for 16–20 h. After incubation, inhibition zones were determined. In case of testing susceptibility with Etest®, the same procedure for preparing the plates was carried out. Interpretation of zone-diameters and Etest® was carried out according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and if no EUCAST breakpoints were available Clinical Laboratory Standards Institute (CLSI) criteria were used for interpretation (Table 2) [19,20].
Table 2.
List of tested antibiotics, concentration on the disc (Sensi-DiscTM paper discs, BD, Vienna, Austria) or Etest® (Biomerieux) and antibiotic classes.
| Antibiotic | Concentration | Antibioti Classes |
|---|---|---|
| piperacillin/tazobactam | 100 µg/10 µg | β-lactam |
| cefotaxime | 30 µg | β-lactam |
| ceftazidime | 30 µg | β-lactam |
| cefepime | 30 µg | β-lactam |
| imipenem | 10 µg | β-lactam |
| meropenem | 10 µg | β-lactam |
| amikacin | 30 µg | aminoglycoside |
| gentamicin | 10 µg | aminoglycoside |
| trimethoprim/sulfamethoxazole | 1.25 µg/23.75 µg | folate synthesis inhibitors |
| ciprofloxacin | 5 µg | quinolone |
| levofloxacin | 5 µg | quinolone |
| tigecycline | Etest | tetracyclin |
| tetracycline | 30 µg | tetracycline |
| colistin | Etest | polypeptide antibiotic |
Etest for tigecycline was carried according to Altun et al. (2014) [21].
Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as control strains in all performed tests.
2.4. Determination of β-Lactamase Genes
Determination of resistance genes was carried out for all Acinetobacter spp. isolates that revealed a resistance to at least one tested carbapenem. PCR detection and gene identification were performed for five different β-lactamases gene families, blaCTX-M-1group, blaCTX-M-2group, blaCTX-M-9group, blaGES, blaKPC, blaOXA-23, blaOXA-24, blaOXA-51, blaOXA-48, blaOXA-58, blaNDM, blaSHV, blaTEM, and blaVIM., PCR and sequencing procedures were performed as described previously [22,23,24,25,26,27].
3. Results
In total, 262 Acinetobacter were isolated. Acinetobacter baumannii was the most common species with 135 isolates. Acinetobacter johnsonii was second most with 62 isolates; all other species were represented by less than 20 isolates; Acinetobacter haemolyticus 19 isolates, Acinetobacter junii 17 isolates, Acinetobacter lwoffii 16 isolates, Acinetobacter radioresistens four, Acinetobacter ursingii two and seven isolates where no distinct species identification was possible. Non-baumannii Acinetobacter spp. were subsumed for further analyses.
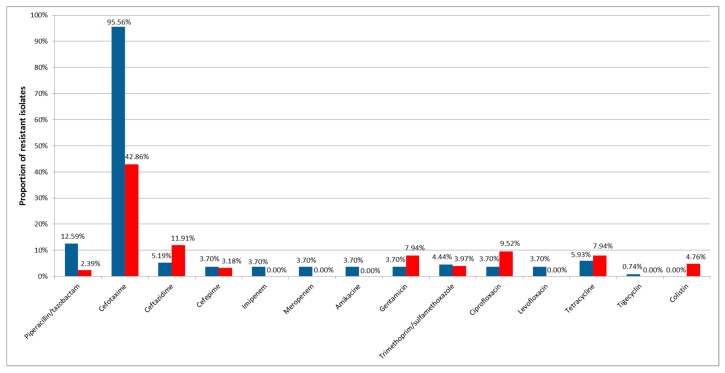
Susceptibility testing revealed that resistance to the most tested antibiotics was rare in Acinetobacter baumannii and non-baumannii Acinetobacter spp. river water isolates. The only resistance present in both groups in more than 10% of the isolates was to cefotaxime with 95.6% (129/135) of Acinetobacter baumannii and 43.3% (55/127) non-baumannii Acinetobacter spp. In addition, resistances to ceftazidime in non-baumannii (12.6%, 16/127) and to piperacillin/tazobactam in Acinetobacter baumannii (12.6%, 17/125) were present in more than 10% in one of the sample subgroups (Figure 1).
Figure 1.
Percentage of resistance to tested antibiotics of isolated Acinetobacter baumannii (blue) and non-baumannii Acinetobacter spp. (red).
Resistance to fluoroquinolones showed one notable detail: all five Acinetobacter baumannii were resistant to ciprofloxacin and levofloxacin, whereas non-baumannii Acinetobacter spp. remained susceptible to the second tested fluoroquinolone. Less than 10 Acinetobacter baumannii isolates revealed resistance to all other tested antibiotics, all isolates were susceptible to colistin. In contrast to this, colistin resistance could be detected in six (4.7%) non-baumannii Acinetobacter spp., and no resistance was found to levofloxacin, imipenem, meropenem, amikacine, and tigecycline (Figure 1).
Only six (4.4%) Acinetobacter baumannii revealed susceptibility to all tested antibiotics, but only 16 (11.9%) were resistant to one or more tested antibiotics additionally to cefotaxime. Six (4.4%) isolates could be classified as multiresistant, with resistance to antibiotics from at least three different antibiotic classes, including one isolate only susceptible to colistin and four to colistin and tigecyline (Table 3).
Table 3.
Detected resistance genes in carbapenem resistant Acinetobacter baumannii; us = upstream; ds = downstream.
| Isolate | Site of Isolation | Susceptible Antibiotics | Detected β-Lactamases |
|---|---|---|---|
| JDS38AC017 | us Pancevo (Belgrade) | colistin, tigecycline | OXA-23, OXA-51, VIM-2 |
| JDS38AC018 | us Pancevo (Belgrade) | colistin | OXA-23, OXA-51, VIM-2, TEM-1 |
| JDS38AC020 | us Pancevo (Belgrade) | colistin, tigecycline | OXA-24, OXA-51, |
| JDS59AC001 | ds Arges (Bucharest) | colistin, tigecycline | OXA-23, OXA-51 |
| JDS59AC007 | ds Arges (Bucharest) | colistin, tigecycline | OXA-23, OXA-51 |
Susceptibility to all antibiotics was 10 times higher (55 isolates, 43.3%) in the non-baumannii Acinetobacter spp. group compared to the Acinetobacter baumannii group. Only 10 isolates revealed multiresistance. Two isolates with different resistance profile showed resistance to four antibiotics (JDS10AC012 to CTX, piperacillin/tazobactam, FEP, and ceftazidime; JDS38AC048 to CTX, TZP SXT and CAZ).
Five Acinetobacter baumannii were resistant to carbapenems (meropenem and imipenem). These isolates were analyzed for the presence of several β-lactamases genes, resulting in four different gene patterns. Classic intrinsic OXA carbapenemases were present in all isolates, but no gene was present in all isolates. JDS59AC007 and JDS59AC001 were positive for OXA-23 and OXA-51, and JDS38AC020 was positive for OXA-24 and OXA-51. In addition to OXA-23 and OXA-51, two isolates revealed β-lactamases from another Ambler class: JDS38AC018 and JDS38AC017 harbored both the gene for carbapenemase VIM-2 and JDS38AC017 harbored additionally the gene for the broad spectrum β-lactamases TEM-1 (Table 3).
4. Discussion
The presence of Acinetobacter with human-induced multidrug resistance phenotypes in surface water has been reported from all over the world. Their origin seems to be influenced by (treated and untreated) hospital waste water. The impact (proportion and persistence) of these strains on the Acinetobacter water population is not well documented [9,14,15,28]. This study shows for the first time the susceptibility phenotypes of Acinetobacter of a total European river system. Furthermore, our study provides a first glimpse of the anthropogenic impact on the Acinetobacter river population. In the Acinetobacter population of the River Danube, even resistance to last line antibiotics (e.g., colistin and tigecycline) is detectable, and this without using selective media by screening only a relatively small volume of water. This screening led to the detection of multidrug-resistant Acinetobacter baumannii isolates, whose multidrug resistance would normally be found and related only to intensive care units. The isolation of multiresistant Acinetobacter was limited to the area of influence of megacities, but even there the great majority of isolates remained not or only slightly influenced on their susceptibility pattern.
Looking at the resistance data for invasive Acinetobacter isolates for the Danube neighboring countries reveals a rather gloomy picture: In clinical isolates, the ratio for carbapenem resistance spans from 5.5% in Germany to over 80% in Romania. In our study of the river water, however, only five isolates (less than 2% of all isolates) showed resistance to carbapenems, a very low proportion of resistant Acinetobacter spp. compared with the clinical settings [29]. This ratio corresponds with our findings in the Pseudomonas population in the river Danube, where we also found a ratio of around 2% [30,31].
In a study of the Jadro River, carried out by Maravic et al., only selected multi-drug-resistant Acinetobacter were isolated (using selective media with supplements). Comparing these isolates with the Danube isolates, there is a remediable difference as regards aminoglycoside resistance. Maravic et al. did not detect a single isolate that was resistant to the tested aminoglycoside in contrast to 5.7% (15 isolates) from the River Danube. Furthermore, only two multiresistant Danube isolates revealed no resistance to one of the tested aminoglycoside. Carbapenem resistance in Jadro River isolates was restricted to meropenem, but the number of isolates was too low to be noteworthy. Interestingly enough, the proportion of cefotaxime resistance is nearly identical with 68% in Jadro River and 66% in the river Danube [9].
5. Conclusions
Multiresistant strains can be found in our environment, in any habitat and at any time, making chances for contact high and permanent. Further investigation will show if the spread of multiresistant Acinetobacter has reached its peak and if susceptible environmental Acinetobacter will still outnumber the clinical strains or if we have to further deal with a constant increase of non-susceptible Acinetobacter in the future.
Acknowledgments
Furthermore, we would like to thank Georg Reischer, Stefan Jakwerth, and Stoimir Kolarevic for their help in sampling.
Author Contributions
C.K. and G.E.Z. conceived and designed the experiments; M.L., R.B., and G.E.Z. performed the experiments; G.E.Z. and C.K. analyzed the data; K.A. and A.H.F. collected samples and data; G.E.Z., C.K., and F.M. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Funding
The Joint Danube Survey was organized by the International Commission for the Protection of the Danube River (ICPDR). The study was supported by the Austrian Science Fund (FWF), project number P25817-B22.
References
- 1.Towner K.J. Acinetobacter: An Old Friend, but a New Enemy. J. Hosp. Infect. 2009;73:355–363. doi: 10.1016/j.jhin.2009.03.032. [DOI] [PubMed] [Google Scholar]
- 2.Peleg A.Y., Seifert H., Paterson D.L. Acinetobacter baumannii: Emergence of a Successful Pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alsan M., Klompas M. Acinetobacter baumannii: An Emerging and Important Pathogen. J. Clin. Outcomes Manag. 2010;17:363–369. [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao W.H., Hu Z.Q. Acinetobacter: A Potential Reservoir and Dispenser for Beta-Lactamases. Crit. Rev. Microbiol. 2012;38:30–51. doi: 10.3109/1040841X.2011.621064. [DOI] [PubMed] [Google Scholar]
- 5.Evans B.A., Amyes S.G. OXA Beta-Lactamases. Clin. Microbiol. Rev. 2014;27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ambler R.P., Coulson A.F.W., Frere J.M., Ghuysen J.M., Joris B., Forsman M., Levesque R.C., Tiraby G., Waley S.G. A Standard Numbering Scheme for the Class-a Beta-Lactamases. Biochem. J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L., Nordmann P. Carbapenem Resistance in Acinetobacter baumannii: Mechanisms and Epidemiology. Clin. Microbiol. Infect. 2006;12:826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 8.Santajit S., Indrawattana N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. Biomed. Res. Int. 2016;2016:2475067. doi: 10.1155/2016/2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maravic A., Skocibusic M., Fredotovic Z., Samanic I., Cvjetan S., Knezovic M., Puizina J. Urban Riverine Environment is a Source of Multidrug-Resistant and ESBL-Producing Clinically Important Acinetobacter spp. Environ. Sci. Pollut. Res. Int. 2016;23:3525–3535. doi: 10.1007/s11356-015-5586-0. [DOI] [PubMed] [Google Scholar]
- 10.Jawad A., Seifert H., Snelling A.M., Heritage J., Hawkey P.M. Survival of Acinetobacter baumannii on Dry Surfaces: Comparison of Outbreak and Sporadic Isolates. J. Clin. Microbiol. 1998;36:1938–1941. doi: 10.1128/jcm.36.7.1938-1941.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fournier P.E., Vallenet D., Barbe V., Audic S., Ogata H., Poirel L., Richet H., Robert C., Mangenot S., Abergel C., et al. Comparative Genomics of Multidrug Resistance in Acinetobacter Baumannii. PLoS Genet. 2006;2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin M.F., Lan C.Y. Antimicrobial Resistance in Acinetobacter baumannii: From Bench to Bedside. World J. Clin. Cases. 2014;2:787–814. doi: 10.12998/wjcc.v2.i12.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng M., Zhu M.H., Li J.J., Bi S., Sheng Z.K., Hu F.S., Zhang J.J., Chen W., Xue X.W., Sheng J.F., et al. Molecular Epidemiology and Mechanisms of Tigecycline Resistance in Clinical Isolates of Acinetobacter baumannii from a Chinese University Hospital. Antimicrob. Agents Chemother. 2014;58:297–303. doi: 10.1128/AAC.01727-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seruga Music M., Hrenovic J., Goic-Barisic I., Hunjak B., Skoric D., Ivankovic T. Emission of Extensively-Drug-Resistant Acinetobacter baumannii from Hospital Settings to the Natural Environment. J. Hosp. Infect. 2017;96:323–327. doi: 10.1016/j.jhin.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Osinska A., Harnisz M., Korzeniewska E. Prevalence of Plasmid-Mediated Multidrug Resistance Determinants in Fluoroquinolone-Resistant Bacteria Isolated from Sewage and Surface Water. Environ. Sci. Pollut. Res. Int. 2016;23:10818–10831. doi: 10.1007/s11356-016-6221-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilharm G., Skiebe E., Higgins P.G., Poppel M.T., Blaschke U., Leser S., Heider C., Heindorf M., Brauner P., Jackel U., et al. Relatedness of Wildlife and Livestock Avian Isolates of the Nosocomial Pathogen Acinetobacter baumannii to Lineages Spread in Hospitals Worldwide. Environ. Microbiol. 2017;19:4349–4364. doi: 10.1111/1462-2920.13931. [DOI] [PubMed] [Google Scholar]
- 17.Rafei R., Hamze M., Pailhories H., Eveillard M., Marsollier L., Joly-Guillou M.L., Dabboussi F., Kempf M. Extrahuman Epidemiology of Acinetobacter baumannii in Lebanon. Appl. Environ. Microbiol. 2015;81:2359–2367. doi: 10.1128/AEM.03824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jamal W., Albert M.J., Rotimi V.O. Real-Time Comparative Evaluation of bioMerieux VITEK MS Versus Bruker Microflex MS, Two Matrix-Assisted Laser Desorption-Ionization Time-of-Flight Mass Spectrometry Systems, for Identification of Clinically Significant Bacteria. BMC Microbiol. 2014;14:289. doi: 10.1186/s12866-014-0289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The European Committee on Antimicrobial Susceptibility Testing (EUCAST) Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 3.1, EU. [(accessed on 29 December 2017)];2013 Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_3.1.xls.
- 20.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing: 18th Informational Supplement. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2008. CLSI Document M100-S18. [Google Scholar]
- 21.Altun H.U., Yagci S., Bulut C., Sahin H., Kinikli S., Adiloglu A.K., Demiroz A.P. Antimicrobial Susceptibilities of Clinical Acinetobacter baumannii Isolates with Different Genotypes. Jundishapur J. Microbiol. 2014;7:e13347. doi: 10.5812/jjm.13347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biglari S., Alfizah H., Ramliza R., Rahman M.M. Molecular Characterization of Carbapenemase and Cephalosporinase Genes among Clinical Isolates of Acinetobacter baumannii in a Tertiary Medical Centre in Malaysia. J. Med. Microbiol. 2015;64:53–58. doi: 10.1099/jmm.0.082263-0. [DOI] [PubMed] [Google Scholar]
- 23.Dallenne C., Da Costa A., Decre D., Favier C., Arlet G. Development of a Set of Multiplex PCR Assays for the Detection of Genes Encoding Important Beta-Lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010;65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 24.Zarfel G., Lipp M., Gurtl E., Folli B., Baumert R., Kittinger C. Troubled Water Under the Bridge: Screening of River Mur Water Reveals Dominance of CTX-M Harboring Escherichia coli and for the First Time an Environmental VIM-1 Producer in Austria. Sci. Total Environ. 2017;593–594:399–405. doi: 10.1016/j.scitotenv.2017.03.138. [DOI] [PubMed] [Google Scholar]
- 25.Hornsey M., Phee L., Wareham D.W. A Novel Variant, NDM-5, of the New Delhi Metallo-Beta-Lactamase in a Multidrug-Resistant Escherichia coli ST648 Isolate Recovered from a Patient in the United Kingdom. Antimicrob. Agents Chemother. 2011;55:5952–5954. doi: 10.1128/AAC.05108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potron A., Nordmann P., Lafeuille E., Al Maskari Z., Al Rashdi F., Poirel L. Characterization of OXA-181, a Carbapenem-Hydrolyzing Class D Beta-Lactamase from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2011;55:4896–4899. doi: 10.1128/AAC.00481-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradford P.A., Bratu S., Urban C., Visalli M., Mariano N., Landman D., Rahal J.J., Brooks S., Cebular S., Quale J. Emergence of Carbapenem-Resistant Klebsiella Species Possessing the Class A Carbapenem-Hydrolyzing KPC-2 and Inhibitor-Resistant TEM-30 Beta-Lactamases in New York City. Clin. Infect. Dis. 2004;39:55–60. doi: 10.1086/421495. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y., Marrs C.F., Simon C., Xi C. Wastewater Treatment Contributes to Selective Increase of Antibiotic Resistance among Acinetobacter spp. Sci. Total Environ. 2009;407:3702–3706. doi: 10.1016/j.scitotenv.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Girlich D., Bonnin R.A., Bogaerts P., De Laveleye M., Huang D.T., Dortet L., Glaser P., Glupczynski Y., Naas T. Chromosomal Amplification of the blaOXA-58 Carbapenemase Gene in a Proteus Mirabilis Clinical Isolate. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.01697-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kittinger C., Lipp M., Baumert R., Folli B., Koraimann G., Toplitsch D., Liebmann A., Grisold A.J., Farnleitner A.H., Kirschner A., et al. Antibiotic Resistance Patterns of Pseudomonas Spp. Isolated from the River Danube. Front. Microbiol. 2016;7:586. doi: 10.3389/fmicb.2016.00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.European Centre for Disease Prevention and Control . Antimicrobial Resistance Surveillance in Europe 2015. Annual Report of the European Antimicrobial Resistance Surveilance Network (EARS-Net) ECDC; Solna Municipality, Sweden: 2016. [Google Scholar]