Summary
Polycomb repressive complex 2 (PRC2-EZH2) methylates histone H3 at lysine 27 (H3K27) and is required to maintain gene repression during development. Misregulation of PRC2 is linked to a range of neoplastic malignancies, which is believed to involve methylation of H3K27. However, the full spectrum of non-histone substrates of PRC2 that might also contribute to PRC2 function is not known. We characterized the target recognition specificity of the PRC2 active site and used the resultant data to screen for uncharacterized potential targets. The RNA polymerase II (Pol II) transcription elongation factor, Elongin A (EloA), is methylated by PRC2 in vivo. Mutation of the methylated EloA residue decreased repression of a subset of PRC2 target genes as measured by both steady state and nascent RNA levels and perturbed embryonic stem cell differentiation. We propose that PRC2 modulates transcription of a subset of low expression target genes in part via methylation of EloA.
eTOC blurb
We show that the transcription elongation factor, Elongin A, is methylated by PRC2 in vivo. Inability to methylate EloA results in upregulation of a subset of PRC2 target genes and interferes with differentiation potential of mES cells. PRC2 modulates expression of target genes in part via methylation of EloA.

Introduction
Polycomb group (PcG) proteins form into a diverse class of multimeric, repressive, chromatin-bound complexes that play an indispensable role in differentiation and development of multicellular organisms (Margueron and Reinberg, 2011). These complexes maintain gene repression in part by modifying chromatin structure, through both physical compaction and covalent modification of histones in a tightly controlled spatial and temporal manner. Polycomb repressive complex 1 (PRC1) and PRC2 are the two major classes of PcG complexes. PRC2 possesses lysine methyltransferase (MTase) activity, the core biochemical function of which is trimethylation of lysine 27 of histone H3 (H3K27me3), a modification that leads to increased binding by PRC1 (Kuzmichev et al., 2002; Muller et al., 2002). Trimethylation of H3K27 is associated with gene repression and developmentally regulated facultative heterochromatin (Laugesen et al., 2016). In mammals, the catalytic subunits of PRC2 are EZH2, and its homolog EZH1, each of which has a SET methyltransferase domain that carries out the methylation reaction (Margueron et al., 2008; Shen et al., 2008). Methyltransferase activity of EZH2 requires at least two other subunits, SUZ12 and EED, which along with RbAp46/48 comprise the core PRC2 complex (Martin et al., 2006). The structural basis for contribution of all core subunits to the catalytic activity of the complex has recently been described at the atomic level (Ciferri et al., 2012; Justin et al., 2016).
Genetic studies have established that missense mutations that abolish the MTase activity of PRC2, without compromising the integrity of the core complex, mimic or display dominant phenotypes similar to null E(z) mutations (fly Ezh2 ortholog) ( Joshi et al., 2008). Gain and loss of EZH2 levels and activity have been reported in different types of neoplastic malignancies (Hock, 2012). Hyper-activating missense mutations in specific residues of the EZH2 SET-domain have been seen in various types of lymphomas (Morin et al., 2010), while loss or inactivating missense mutations of the Ezh2 catalytic domain have been shown to be directly involved in T-cell lymphoblastic leukemia (T-ALL) ( Simon et al., 2012). Moreover, mutation of the H3K27 substrate to methionine appears to act as a dominant mutation in pediatric glioblastoma (Justin et al., 2016). The fact that EZH2 can act both as an oncogene and tumor suppressor underscores the need to characterize context dependent roles of PRC2 that might involve distinct functions.
While methylation of H3K27 is an important function that has been shown to be necessary for developmental progression in Drosophila (Pengelly et al., 2013), it is not clear to what extent this mechanism alone is sufficient for PRC2 function during mammalian differentiation and development. The data cited above are consistent with H3K27me3 also being necessary for developmental effects in mammals, however other methylation events might contribute to regulation of certain genes and/or certain cell types. For example, other non-histone targets for methylation by PRC2 have been identified, such as RORα, GATA4, STAT3 and JARID2 ( He et al., 2012; Kim et al., 2013; Lee et al., 2012; Sanulli et al., 2015). These methylation events occur on gene-specific factors or on targeting factors, and therefore might contribute to modulating PRC2 function in specific developmental programs. Identification of uncharacterized EZH2 methyltransferase targets that are more directly involved in general regulation might provide information on alternative mechanisms that are used by PRC2 to repress genes. These might allow a more targeted and selective therapeutic approach, which could limit the undesirable consequences of complete inhibition of the PRC2-EZH2 methyltransferase machinery.
The potential for PRC2 to be involved more generally in transcriptional regulation is supported by widespread localization of the complex to the CpG rich DNA, and promoter and 5′ regions of many genes (Brookes et al., 2012; Kaneko et al., 2013; Min et al., 2011; Riising et al., 2014). PRC2 also interacts with nascent transcripts throughout the body of almost all actively transcribed genes (Beltran et al., 2016). An underexplored aspect of PRC2 function is the extent to which it might directly methylate general factors that interact with RNA polymerase II (Pol II) and modulate transcription.
One complex that interacts with Pol II contains Elongin A (EloA), and two smaller subunits, Elongin B (EloB) and Elongin C (EloC) (Aso et al., 1995). This Elongin complex interacts with the phosphorylated form of Pol II C-terminal domain (CTD) and stimulates transcription elongation (Aso et al., 1996; Kawauchi et al., 2013). The complex is also part of a ubiquitin ligase complex along with Cullin5 and Rbx2 that drives degradation of stalled Pol II (Wilson et al., 2013). Two of the subunits of the Elongin complex have recently been linked to PRC2 via their interaction with the newly identified PRC2 interacting protein EPOP (Beringer et al., 2016; Liefke et al., 2016).
Here, using a positional-scanning peptide array, we characterize the target recognition specificity of the PRC2-EZH2 complex and use that information to perform an in silico screen for potential mammalian targets of PRC2 and identified EloA as a bona fide target of PRC2 methyltransferase activity. Mouse ES (mES) cells that contain a point mutation mimicking the hypomethylated EloA state show up-regulation of a subset of lowly transcribed genes that are also upregulated in Eed−/− mES cells. These cells also show altered differentiation and growth properties. We propose, based upon mutational analysis, that methylation of EloA by PRC2 results in transcriptional down-regulation of target genes in mES cells. These findings indicate a new mechanism for PRC2 that involves methylation of EloA by PRC2 at a subset of targets whose proper level of repression is required for regulating growth and cellular differentiation.
Results
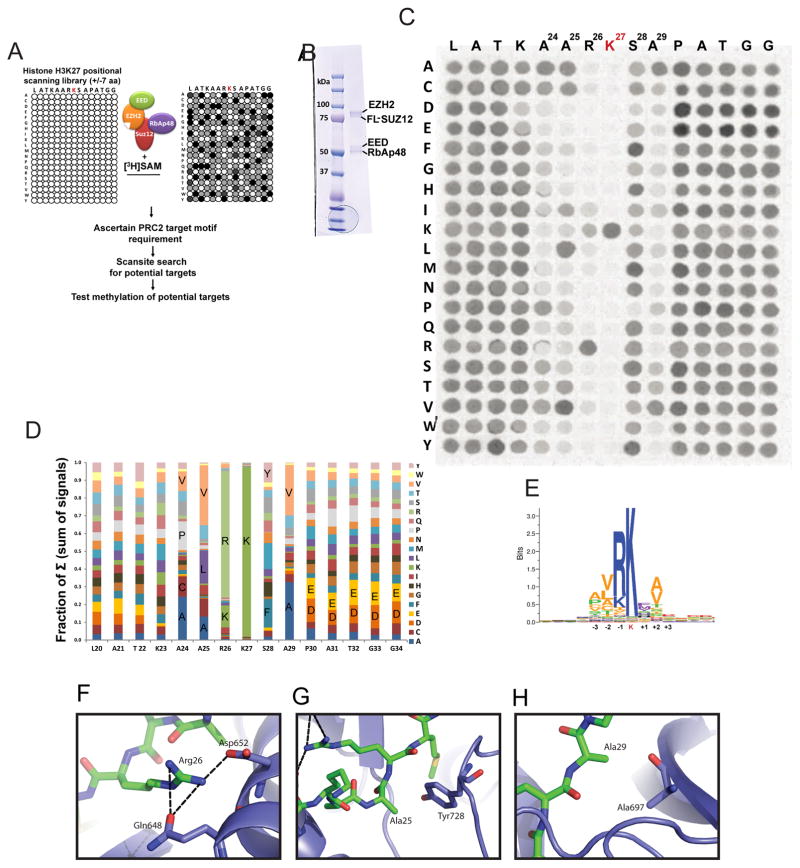
We carried out a positional scanning SPOT MTase assay to characterize the substrate preference of PRC2 active site and screen for potential targets of EZH2 in vitro (outlined in Figure 1A). Murine PRC2 complex containing four core subunits was expressed and purified from Sf9 insect cells and shown to be active (Figure 1B; S1A–C). The substrate region critical for productive interaction with the PRC2 catalytic pocket was approximated by measuring K27 methylation activity on a peptide array containing an isoleucine-scan of amino acids within 7 residues of the methylation site. Substitution of A24, A25 and R26 (positions −3, −2 and −1) and S28 and A29 (positions +1 and +2) decreased the methylation efficiency of the target peptides (Figure S1E), indicating that amino acids immediately adjacent to the methylated residue are important for PRC2 MTase activity.
Figure 1. Characterization of PRC2 Target Sequence Specificity by Positional-Scanning SPOT peptide Array.
(A) Outline of the PRC2 MTase positional-scanning SPOT assay.
(B) Coomassie stained gel of immunoaffinity purified core PRC2-EZH2 complex.
(C) Result of PRC2 SPOT peptide assay with H3K27 peptides as the primary target of PRC2. Position of H3K27 and adjacent residues are shown above the membrane. The amino acid that each residue is substituted to is presented on the left side of the membrane. Dots represent methylation levels of PRC2 at each position. The experiment was carried out in duplicate using independent preparations of purified PRC2-EZH2 on different membranes with similar results (Figure S3A). Reaction conditions: 10nM PRC2, 0.25μM of 3H-SAM, 30min incubation.
(D) Graph showing contribution of each amino acid to the sum (Σ) of total methylation signal at each position.
(E) Sequence Logo (SeqLogo) representation of PRC2 target sequence preference motif.
(F) Structure of PRC2 bound to target histone (PDB: 5HYN) showing formation of hydrogen bonds between Arg26 with Gln648 and Asp652.
(G) Depicts proximity of Tyr728 of EZH2 to Ala25 explaining preference for hydrophobic and mainly non-bulky amino acids at this position.
(H) Depiction of H3 Ala29 proximity to Ala697 of EZH2, showing suitability of hydrophobic or neutral amino acids at this position.
Amino acid substitutions define a target sequence for PRC2
To further define the target sequence, we measured MTase activity using a positional-scanning SPOT peptide array (Frank, 2002; Rathert et al., 2008) of histone H3K27 peptides (Figure 1C). A library of 15 amino acid peptides corresponding to histone H3K27 and the adjacent residues (+/− 7 residues flanking H3K27) was synthesized, and each native residue was substituted with each of the other 19 amino acids (i.e., 15×20=300 spotted peptides). We incubated this membrane with purified PRC2-EZH2 complex in the presence of 3H-SAM in a competitive MTase assay under steady-state enzymatic conditions (Figure 1C). We established that the reaction conditions have linear kinetics for both the incubation time and SAM concentration (Figure S1D). We quantified the extent of methylation for each unique peptide by measuring the 3H-incorporation intensity at each spot. The methylation activity of PRC2 towards these targets is presented as a fraction of sum of signal intensity at each position (Figure 1D).
As expected, substitution of H3K27 resulted in complete loss of detectable methylation by PRC2 (Figure 1C). At Position −1, arginine (R) was critical; another basic residue, lysine (K), was able to partially substitute for R (Figure 1C; 1D). The preference for arginine and lysine at position −1 is concordant with recently solved structural data characterizing the binding pocket of EZH2 at high resolution (Justin et al., 2016). The terminal nitrogen of R26 establishes three hydrogen bonds with Gln648 and Asp652 of EZH2, which stabilizes peptide binding (Figure 1F), while lysine might be able to only establish two of these bonds, explaining the lower rate of methylation with Lys at the −1 position. Shorter or polar side chains would not be able to establish these contacts. At the −3 position (A24) non-bulky, non-polar residues with hydrophobic side chains (A, C, V, P) were preferred over other amino acids (Figure 1C; 1D) and at position A25, hydrophobic and predominantly aliphatic residues were well-tolerated (A,V,L) (Figure 1C, 1D; 1E). The structure shows that A25 faces the bulky side chain of Tyr728 in the lysine access channel, explaining why bulkier residues are not well-tolerated and disrupt MTase activity (Figure 1G).
Positions 28 and 29 (+1 and +2) were also important, consistent with previous observations that phosphorylation of serine 28 by MSK1 decreases methylation and PRC2 binding (Lau and Cheung, 2011). Substitution of Ser28 at position +1 with the phosphoserine-mimic aspartic acid (D), recapitulated the decrease in methylation (Figure 1C; 1D). Proline substitution at S28 diminished methylation of H3K27, perhaps due to the inherent conformational rigidity of proline not being tolerated (Figure 1C; 1D). Surprisingly, substitution of S28 with single-ring aromatic amino acids (F, Y, H) resulted in higher or similar levels of methylation (Figure 1C, 1D; S3A). It is possible that aromatic residues at the +1 position enhance methylation through stabilization of pi-stacking non covalent interactions with one of the aromatic side chains of the lysine27 pocket (Justin et al., 2016). The +2 position was also important for methylation (Figure 1C; 1D), with the native A29 and valine being well tolerated and to a lesser degree other hydrophobic or neutral, but small, amino acids (C, T and S). These residues might be better-tolerated because the opposing Ala697 of EZH2 is positioned on an alpha helix (Figure 1H).
Almost all amino acid exchanges were well-tolerated at position P30 to G34. However, we observed that acidic residues at P30–G34 resulted in elevated methylation of H3K27, while substitution with basic residues was slightly disfavored at these positions (Figure 1C; 1D). The substrate preference of PRC2 (Figure 1E) shows that the natural H3 sequence does not have an optimal amino acid at each position.
While these data cannot be used to fully evaluate all possible target sequences for PRC2, as they are based upon a single starting sequence, they allow prediction of which sequences might serve as efficient substrates for PRC2 in other proteins. We used this information as a basis to screen for uncharacterized PRC2 targets.
In addition to EZH2, mammals also possess EZH1, a highly conserved functional paralog of this SET-domain containing subunit (Margueron et al., 2008) (Figure S2A). EZH1 can be assembled into a PRC2-EZH1 complex that methylates H3K27 (Shen et al., 2008; Son et al., 2013) and is present in all types of cells and tissues but plays a lesser role than PRC2-EZH2 in histone methylation (Margueron et al., 2008; Shen et al., 2008). We purified PRC2-EZH1, characterized the substrate preference and specificity as described above for canonical PRC2 (Figure S2; S3B), and found a strong conservation of residue preference between the two homologs (Figure S2; S3). This suggests that the non-redundant functions of these two paralogs is likely not due to differences in catalytic site substrate preference, but might result from divergent protein-protein interaction within the less conserved domains of the homologs (Figure S2A).
Identification of Non-histone targets of PRC2
We used the methylation efficiencies for each amino acid substitution to generate an efficiency matrix and performed a Scansite database search of nuclear proteins (Obenauer, 2003) and identified 339 potential nuclear targets of PRC2 (Table S1). We identified K116 of JARID2, previously identified as a bona fide target site of PRC2 methylation (Sanulli et al., 2015), validating the approach (Table S1). Using peptides as targets, we verified that more than 100 of these sequences could be methylated by PRC2 and that the calculated ‘optimal’ sequence was methylated more efficiently than the native sequence of H3 (Figure 2A; 2B, Table S1). To determine if these potential targets are methylated as full-length proteins, we expressed and purified 21 of these proteins and observed methylation of about half of them by PRC2 in vitro (Figure 2C; S3C). A diverse set of proteins were methylated, including NCOA6, ID2, TBP, ELL, Elongin A, PIAS4 and NPM1 (Figure 2C, S3C). The sequences surrounding predicted target lysine for these factors were diverse (Figure 2F). We used site-directed mutagenesis to introduce K to R or K to A point mutations in the Scansite-predicted target lysine of a subset of full length proteins and demonstrated that the predicted lysine residue was the sole or major PRC2 methylation site on each putative target protein (Figure 2D).
Figure 2. Identification and Characterization of Non-histone Methylation Substrates of PRC2.
(A) PRC2 MTase assay on peptides representing 339 potential nuclear targets of PRC2. Native histone H3K27 (H3, blue circle), H3K27A (K>A, red circle) and optimal target sequence (Opt, green circle) are highlighted. List of targets and their respective methylation intensity provided in Table S1.
(B) Methylation efficiency of native, K27A and Optimal peptides by PRC2 presented as bar graph (arbitrary unit, n=3, error bar denotes s.e.m).
(C) Methylation of a subset of potential PRC2 targets as full length proteins. Coomassie stained gel (left panel) and fluorography of dried gel (right panel). Arrows and asterisks denote automethylation of EZH2 and methylation of targets, respectively.
(D) Examining methylation of predicted target lysine residues by site-directed mutagenesis (left: coomassie stained gel, right: fluorography of dried gel.
(E) THOC1 and PSMC6 are methylated upon methylation and binding of PRC2 to H3K27me3.
(F) Position and sequence of tested Scansite predicted targets.
We tested target candidates that showed no detectable methylation by core PRC2-EZH2 in a modified protocol that included bacterially-purified core histones. The addition of core histones might increase methylation activity due to methylation of H3K27 in the reaction and subsequent EED binding to that residue, which is known to increase catalytic activity of PRC2 (Margueron et al., 2009; Scelfo et al., 2015). Under this regime, two additional potential targets, PSMC6 and THOC1, were identified as methylated in the presence of H3 but not in isolation (Figure 2E). Much like for histone methylation, EED stimulated the ability of PRC2 to methylate other substrates. Moreover, it is plausible that the allosteric regulation induced by EED binding to histone H3K27me3 may prime the complex for acquisition of new targets in a specific chromatin-bound manner.
General transcription factors as methylation targets of PRC2
Given the role for the PcG proteins in maintaining gene repression, we were particularly intrigued by the PRC2 methylation targets that were general RNA polymerase transcription and elongation factors. These might provide an avenue for direct repression or downregulation of the transcription machinery by the PcG complex and included TBP, Elongin A and ELL (Figure 2C, 3A).
Figure 3. Elongin A K754 is Methylated by PRC2 in vivo.
(A) Testing methylation of full length Elongin A, TBP and Rpb1 by PRC2. Left: coomassie stain, Right: Methylation signal.
(B) Identification of EloAK754 as the target of PRC2 methylation in vitro. Top panels: methylation signal for native, K754A (two different concentrations) and K754R EloA point mutants, bottom: coomassie stain.
(C) EloA is methylated in a functional Elongin complex found in vivo. Top panel depicts time-course graph of EloA methylation (n=3,s.e.m.). Bottom panels: Fluorography and coomassie stain of a representative in vitro MTase reaction.
(D) Elongin A is methylated at K754 in vivo. Left: Detection of EloAK754me signal by immunoblotting after immunoprecipitation of EloA in WT and Ezh2−/− mES CJ7 cells. Right: Western Blot analysis of EZH2, EloA, histone H3 and H3K27me3 levels in mES cell lysates.
(E) Immunoprecipitation of transiently-transfected FLAG-tagged WT and EloAK754R constructs in 3T3 cells showing specificity of ELoAK754me1 antibody signal (Pooled EloA K754me2, 3 antibodies).
(F) Left: Inhibition of EZH2 MTase activity by GSK343 (6μM) decreases the levels of detectable Elongin A K754me1. Right: Western blot showing decrease in H3K27me3 levels upon GSK343 treatment.
(G) Target methylation site of EloA is conserved in metazoan organisms, but absent in unicellular yeast species lacking PRC2 (Kalign MSA analysis, ClustalW output format).
Elongin A was an attractive potential target for PRC2. This transcription elongation/ubiquitin ligase factor forms a complex with EloBC and enhances the catalytic rate of transcription elongation by Pol II (Aso et al., 1995, 1996). Potentially relevant to a role in Polycomb-Group function, EloA plays a role in development. EloA−/− embryos die during embryogenesis with defects in formation of cranial and spinal nerves (Yasukawa et al., 2012) and EloA mutant flies display defects in Hox gene regulation (Chopra et al., 2009), both phenotypes that are consistent with aberrant PcG function.
We identified lysine 754 of murine Elongin A (K754) as the putative target site for methylation by PRC2. Lysine 754 is located in the C-terminus of the polypeptide near a domain that has been shown to be necessary and sufficient for the transcription elongation activity of the molecule (Aso et al., 1996). Elongin A is methylated by PRC2 in vitro (Figure 2C; 3A). Substitution of the predicted target residue with A or R resulted in decreased EloA methylation, indicating that K754 is the major, but not only, site of PRC2 methylation in vitro (Figure 3B). Alternatively, the K to R substitution may prime the −1 position K for methylation. Mass spectrometry analysis of the in vitro methylated EloA confirmed methylation of K754 by PRC2 (Figure S4A). To examine if Elongin A is also methylated by PRC2 when Elongin A is in complex, we performed the MTase reaction on the heterotrimeric Elongin complex (EloA, B and C) and found that the complex is also targeted by PRC2, albeit with slower reaction kinetics (Figure 3C).
To determine whether Elongin A methylation takes place in vivo, we generated antibodies against different methylation states (mono-, di- and tri-) of Elongin K754 and validated the specificity and sensitivity of these antibodies using in vitro peptide spot assays (Figure S4B). We immunoprecipitated Elongin A from wild type (wt) and Ezh2−/− mES cells and performed immunoblotting using these methyl-specific antibodies. In contrast to the in vitro MTase reaction, where we detected all degrees of methylation by PRC2 on target peptide (Figure S4C), we found only monomethylation of Elongin A at K754 in wild type ES cells (Figure 3D). This could reflect a limit of antibody sensitivity, or instead that di- and tri-methylation do not occur either in vivo or specifically in ES cells (see discussion). The mono-methylation was lost in Ezh2−/− ES cells, indicating that PRC2-EZH2 is responsible for carrying out this modification in vivo. To support this finding we transiently expressed FLAG-tagged Elongin A in a mouse embryonic fibroblast cell line, immunoprecipitated Elongin A, and performed immunoblotting using antibodies against all degrees of Elongin A K754 methylation. As in the experiments performed in ES cells, we detected Elongin A only in the monomethylated state (Figure 3E). Additionally, we performed the same experiment using transiently expressed Elongin A K754R and saw a complete loss of methylation. This showed that this residue is necessary for methylation and validated the specificity of the methyl-specific antibodies (Figure 3E). We also examined methylation of EloA in 3T3 fibroblast cultured in the presence or absence of GSK343, a specific and potent inhibitor of EZH2 (Verma et al., 2012). In cells treated with the inhibitor, the level of EloAK754me was reduced, supporting PRC2-EZH2 as the responsible methyltransferase (Figure 3F). Lysine 754 of EloA and its adjacent residues are conserved in metazoan organisms, but absent in unicellular eukaryotic yeast species, which lack polycomb group proteins (Figure 3G) (Simon and Kingston, 2013). This degree of conservation is consistent with the hypothesis that methylation is important in regulating EloA function in multicellular organisms.
TATA-binding protein (TBP) was also an attractive potential target, as it has a role in transcription by all three types of eukaryotic polymerases (Hochheimer and Tjian, 2003) through nucleation and formation of the preinitiation complex (PIC). Previous reports had shown that TBP, and/or its associated TAFs, physically interact with PcG proteins(Breiling et al., 2001; Saurin et al., 2001). Moreover, the highly-conserved and surface-exposed TBP K236, which is targeted by PRC2 in vitro, was shown to be important for transcription in yeast (K138) (Kim and Roeder, 1994) While we were able to see methylation of TBP at this residue in vitro, we were unable to detect methylation of TBP in vivo (Figure S4F–J), consistent with findings of an earlier report (Zhao et al., 2008).
Mutation of the methylation site of EloA leads to de-repression of a subset of PRC2 targets
To examine possible physiological roles for EloAK754 methylation by PRC2, we used ES cells and a cell culture differentiation system. The CRISPR/Cas9 gene editing system was used to generate two different point mutants in murine ES cells, EloA K754R and EloA K754M, both of which abolish methylation at K754. Mutations of H3K27 to either R or to M have been shown to have a significant phenotype in Drosophila and mammals (Herz et al., 2014; Lewis and Allis, 2013) with the arginine mutant eliminating methylation but maintaining positive charge, and the methionine mutant generating a neomorphic phenotype believed to be caused by altering PRC2 function. We verified the presence of the desired mutations and homozygosity of the selected clones by restriction fragment length polymorphism (RFLP) and sequencing (Figure S5A; S5B). Global levels of histone H3K27 trimethylation are not altered in either cell line, indicating that the EloAK754M phenotype does not mimic that of H3K27M (Figure S5G). Cells containing methylation-deficient EloA remain undifferentiated as determined by alkaline phosphatase staining (S5C). They express full-length Elongin A proteins (Figure S5D; S5E), however, both methylation-deficient lines show defects in cell proliferation, and form smaller stem cell colonies (Figure S5F). Both point mutated cell lines express approximately half the amount of EloA as WT, which might reflect stability or synthesis effects (Figure S5D; S5E). We therefore compared (see below) the phenotypes of these lines to those of EloA null and EloA+/− cell lines, the latter of which also expresses approximately half the amount of EloA protein as WT (Figure S5E).
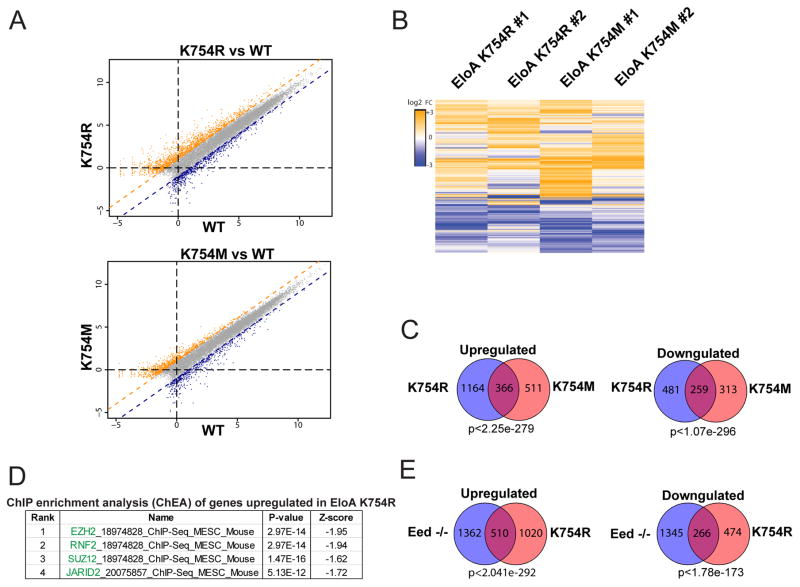
We examined gene expression patterns by performing RNA sequencing (RNA-seq) analysis in WT (control) and EloA K754 edited cells. Comparison of gene expression levels in mutant and WT cells identified genes that were both up and down regulated by more than two-fold in the mutant lines (Figure 4A). More genes were up-regulated than down regulated (Figure 4B; 4C), and the patterns showed similar trends when the K754R mutation was compared to the K754M mutation. Therefore, we focus on the analysis of the K754R mutation below. ChIP enrichment analysis (ChEA) (Kuleshov et al., 2016) for genes upregulated in EloA K754R ES cells revealed that the top-ranked and most significantly enriched ChIP gene sets are for targets of EZH2, Ring1b(RNF2), SUZ12 and JARID2 (Figure 4D, S6A; S6B). Three of these proteins are PRC2 components and one (Ring1b) is a central component of PRC1, which is targeted by PRC2 methylation (Figure 4D). We conclude that mutation of EloAK754 leads primarily to up-regulation of genes that are bound by PRC2.
Figure 4. Methylation of EloA by PRC2 tunes expression of Target Genes.
(A) Scatter plot of differentially expressed genes in EloA K754R and EloA K754M vs. WT mES cells. Points represent mean RPKM (reads per kilobase per million) value of two biological replicates, upregulated and downregulated genes in orange and blue, respectively.
(B) Heat map representation of the union of differentially-expressed genes in two biological replicates of EloA K754R and K754M edited mES cells (fold change larger than 2).
(C) Venn diagrams showing the degree of overlap between upregulated and downregulated classes of differentially expressed genes (log2 of fold change) in Elongin A point mutants.
(D) ChIP enrichment analysis (ChEA) showing the top four factors enriched by ChIP-seq at genes that are upregulated in EloA K754R cells.
(E) Venn diagrams showing the degree of overlap between differentially expressed genes in Eed −/− and EloAK754-edited mES cells, p-value derived from hypergeometric test.
To further explore the interaction between EloA mutants and PRC2, we compared mRNA expression levels between Eed−/− and EloA K754R mutants in the same genetic background (CJ7 mESC). There is a significant overlap between differentially regulated genes in both cell lines (Figure 4E), with 510 genes being up-regulated in both. This observed increase in steady-state RNA level might reflect chromatin level increase in transcription from these genes. To examine this, we measured nascent transcript levels using BrU-Seq (Tani et al., 2012) by a short pulse (10 min) of transcription with 5-bromouridine (BrU), which allows measurement of nascent transcription following precipitation of these newly made transcripts and high-throughput sequencing. Bru-seq results revealed that up-regulation of a subset of genes in both the Eed−/− and the K754 edited cells is due to increased nascent transcription (Figure 5, S6C).
Figure 5. Example of PRC2 bound genes upregulated in Eed−/− and EloAK754R.
Genome browser profiles of 7 representative, expressed genes upregulated in K754R edited and Eed −/− mES cells that show PRC2 enrichment (red EZH2 ChIP-seq track). Olig2 depicts a silent gene that is only upregulated in Eed−/− mES cells. RNA-seq values represent average RPKM (bin size=50bp, n=2). Bru-seq tracks (green) represent nascent transcription signal from a 10min 5-bromouridine pulse experiment. Only the intronic RPKM signals were used for differential gene expression comparison for Bru-seq.
We show, as individual examples, an analysis of seven genes bound by PRC2 that are upregulated in Eed−/− and EloAK754R edited cells, as well as Olig2, a silent gene that is only upregulated in Eed−/− mES cells (Figure 5). Consistent with the RNA-seq data, the genes upregulated in K754R cells also showed upregulation as assessed by Bru-seq, while Olig2 again showed upregulation only in Eed−/− cells. These genes were either downregulated, or not significantly upregulated in EloA−/− mES cells as assessed by RNA-seq (Figure 5). Therefore, the increase in the level of mRNA from these genes in EloAK754R edited cells is not a result of lower EloA protein level. We note that we could not reach an unequivocal conclusion on whether EloA is present at these genes by ChIP (data not shown). This might be due to relatively low level of transcription at these genes and the inherent difficulty associated with detection of transcription elongation factors distribution along the body of low expression genes and/or a subpar ChIP-grade antibody.
EloAK754 methylation is involved in development and early differentiation of ES cells
Given that many of the upregulated genes in EloA K754R are developmentally regulated (Figure S6D), we chose to use an embryoid body (EB) formation assay as a model for determining early developmental potential of these cells. EB formation was severely compromised 5 days after LIF withdrawal in Eed−/− cells, underscoring the well-established role of PRC2 in development (Figure 6A). Both EloA K754 edited mES cells also formed fewer and smaller EBs 5 days after LIF withdrawal in comparison to WT cells (Figure 6A; 6B). Moreover, while EloA+/− and EloA K754 edited cells have similar levels of EloA protein in mESCs, EBs from the latter are significantly smaller in comparison to genetically-matched wt EBs, indicating that their smaller size is likely due to the point mutation at this residue and not expression level of EloA (Figure 6B).
Figure 6. EloA K754 mutations perturb differentiation of ES cells.
(A) Point mutant EloA K754 cells form smaller and fewer EBs. Brightfield microscopy of EBs 5 days after LIF withdrawal. ES cells from different backgrounds were genetically-matched to corresponding WT mESC cells (CJ7 or CCE cells). Scale bars, 500 μm.
(B) Mean projected area (horizontal line) of EBs relative to corresponding WT EBs (WT=1, n=24–53, error: s.d, P values (****p<0.0001, ***p<0.0002, unpaired two-tailed t test.)
(C) Heat map representation of RNA-seq results of the union of differentially-expressed genes in day 5 EBs from the average of two biological replicates (fold change larger than 2), showing different gene expression clustering patterns between EloA K754R and K754M edited mES cells and EloA null and EloA +/− cells.
D) Representative IGV signal tracks at 4 upregulated genes (Zim3, Pnrc2, Rxra and Plac1) in day 5 EloA K754R edited and Eed −/− EBs. Paternally-imprinted genes, as well as genes implicated in placenta development are significantly over-represented in Eed −/− and EloA K754-edited cells (Figure. S6E).
E) Speculative model depicting downregulation of low expression target genes through methylation of EloA by PRC2. EloA methylation may recruit a hypothetical, repressive, EloAK754me-reader (red oval), reducing expression of targeted genes. Alternatively, allosteric changes induced by methylation of EloA may evict a positive transcription factor (green oval), leading to downregulation and fine tuning of transcription at the targeted gene. Therefore, absence of PRC2 (Eed −/−) or inability to methylate EloA K754 (EloA K754R), result in upregulation of targeted genes (mid and bottom panels, respectively).
To more thoroughly examine these lines, we carried out RNA-seq on day 5 EBs (Figure 6C). The K754R mutation results in dysregulation of a large subset of genes. In Eed−/− and EloA K754 day 5 EBs there was an overlap of 158 upregulated genes (p < 8.9e–28), 98 of which were not upregulated in EloA+/− and EloA −/− cells. These genes showed a significant enrichment for placental development and paternal imprinting (Figure S6E). Representative RNA-seq tracks of this class of genes are presented in Figure 6D.
Discussion
We identified Elongin A as a bona fide substrate for PRC2 in vivo. Methylation of EloA is needed for appropriate gene regulation by PRC2 in ES cells and for proper differentiation of ES cells. As EloA interacts with the CTD of RNA Polymerase II (Kawauchi et al., 2013), we propose that regulation occurs by a methylation induced alteration in the ability of EloA to impact transcription via the general transcription machinery. This results in increased downregulation of a subset of PRC2 targets, most of which are already expressed at low levels in WT cells. Methylation of EloA might increase repression either by conferring a repressive activity on this protein or by generating a binding moiety for a repressive partner protein. Alternatively, methylation of EloA might impair the ability of EloA to activate target genes, thereby decreasing expression, again either by altering EloA itself or impacting the recruitment of a partner protein (Figure 6E). Regardless of the details of the mechanism, this methylation event is necessary for the full spectrum of repression by PRC2 in ES cells and is also necessary for normal differentiation of ES cells into embryoid bodies.
PRC2 has the highest specificity constant towards unmodified H3K27 peptide (Sneeringer et al., 2010) and higher degrees of methylation by PRC2 are thought to require more stable and longer interaction between the target and PRC2 (Laugesen et al., 2016). PRC2 and EloABC constitute functionally antagonistic complexes, perhaps resulting in considerably more limited interaction between PRC2 with EloA, as compared to H3, and therefore providing a possible explanation for why EloA K754me1 is the only form of EloA methylation that we detect in vivo (Figure 3D; 3E).
Methylation of histone H3K27 by PRC2 has been shown to be necessary for repression of developmental genes (Pengelly et al., 2013). Many genes are upregulated in PRC2 null cells but are not upregulated in EloA K754 edited cells (e.g. Olig2, Figure 5), consistent with the long proposed general repressive role for histone H3K27 methylation in PRC2-mediated repression. We propose that at a subset of PRC2 target genes that are incompletely silenced, methylation of EloA and H3K27 work together to generate full repression (Figure 6E). This might result from a combination of repressive effects generated by alterations in the chromatin template along with repressive effects generated via the general transcription machinery and EloA. It is possible that further analysis of PRC2 substrates and function will uncover other important regulatory interactions.
PcG proteins were originally identified in Drosophila as factors that repress Hox gene expression (Simon and Kingston, 2009). Mutations in these factors resulted in well characterized body-patterning defects as a result of aberrant expression of key developmental regulators. These genetic observations gave rise to a simplified functional paradigm wherein PRC2 maintains gene silencing via its ability to methylate H3K27 and recruit PRC1. However, genome-wide analyses in flies and mammalian cells have shown that PRC2 binds to many other loci, including the upstream and 5′-end region of H3K4me3 modified active bivalent genes (Ku et al., 2008; Schuettengruber et al., 2007). In ES cells, a significant proportion of these genes are active and display productive transcription elongation (Min et al., 2011). Moreover, a growing body of evidence has revealed that PRC2 binds to nascent RNA during transcription (Beltran et al., 2016; Davidovich et al., 2013; Kaneko et al., 2013), bringing PRC2 to the vicinity of transcriptionally active, Ser2 and Ser5-phosphorylated Pol II, which is also bound to Elongin A at active genes (Kawauchi et al., 2013). These studies place PRC2 in regions where it could contribute in different ways to transcriptional regulation of a wide variety of genes. The data presented here are consistent with a role for PRC2 in the repression of transcription through methylation of Elongin A at K754, as the majority of PRC2 positive genes that are differentially-regulated in Elongin A K754R are upregulated.
EloB and EloC are also components of other ubiquitin-ligase complexes that lack EloA. A recent study (Beringer et al., 2016) showed that EloB and EloC indirectly bind to PRC2 by associating with the BC-box containing, PRC2-auxillary factor EPOP (Liefke and Shi, 2015; Liefke et al., 2016). However, EloA was not found to be part of this complex. EPOP and EloA both bind to EloC through their respective BC-box domains and therefore may comprise two distinct and mutually exclusive complexes with EloBC. Together our work and these additional studies indicate that PRC2 interacts with all members of the Elongin complex in distinct manners with distinct functional outcomes.
It is intriguing that mutation of K754 impacts many genes targeted by PRC2 but does not impact all EloA targets (data not shown). It is possible that there is an EloA function that requires methylation that is not operative on all genes. Examining the in vivo elongation rate of Pol II at affected genes, and the occupancy of the EloA WT and mutant proteins, might help to provide hints as to the mechanistic role for methylation of EloA.
STAR methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Robert Kingston (Kingston@molbio.mgh.harvard.edu)
Purification of PRC2 complex and target proteins
Ezh2 (BC003772), Eed (BC012966), Suz12 (BC064461) cDNA plasmids were obtained from Open Biosystems (clone IDs: 3492110, 3991086, 6821922 respectively) and subcloned into pFastBac1 baculovirus expression plasmid, with an N-terminal FLAG-tag sequence added to Suz12. Due to sequence identity, we used the human RbAp48 pFastBac1 generated by our Lab and described elsewhere (Davidovich et al., 2013). Sf9 cells grown in Hyclone CCMIII medium at 28°C were co-transduced with media stock (P3) virus particles of all PRC2 subunits. To maintain the equal stoichiometry of the purified complex subunits, 1/6 of the FLAG-Suz12 viral stock was added relative to other subunits. After 42 hours, Sf9 cells were harvested and PRC2 complex was isolated from the nuclear extract as described elsewhere (Abmayr et al., 2006). In Brief, 500μl of equilibrated M2 anti-FLAG agarose beads (50% slurry) was added to isolated nuclei (from 1–2 Liter of Sf9 cell suspension) in BC600 (20mM HEPES pH7.9, 20% glycerol, 1.5mM MgCl2, 0.6M KCl, 0.2mM EDTA, 0.2mM PMSF, 0.5 mM DTT and Roche complete protease inhibitor tablets), after 3 hours, the bead-extract mix was passed through Econo-Pac chromatography columns (Bio-Rad) and packed beads were washed with 15–20× bead volume of Wash buffer in the following order: BC600(x2)-BC1200-BC1500-BC1200-BC600-BC300 (wash buffers also contained 0.05% NP40). PRC2 complex was eluted by incubation with 250μl of BC300 containing 400μg/ml FLAG peptide for 15min (x2=500 μl total eluate volume). Eluted complex was further concentrated using Amicon Ultra centrifuge filters. Stoichiometry and concentration of the complex was determined by running the purified complex on SDS-PAGE gel along with a serial dilution of purified BSA protein (100–1000μg, in 100μg increments), and concentration was determined by quantification of stained band intensity using ImageJ software.
Potential PRC2 target proteins were cloned in pFastBac1 as full-length proteins with an N-terminal FLAG-tag (C-terminal for ID2). The cDNAs for the following target genes were obtained from Open Biosystems (numbers denote clone ID: mTBP: 3498542, mNCOA6:30544739, NCOA1:30628581, mELL:4159557, hELL: 5170024, hADNP: 30342099, mPIAS4: 4236082, hPIAS4:5261627, mEloA/TCEB3:5387774, mEloB/TCEB2:6823854, mEloC/TCEB1: 1399237, mHMGB3:3980194, mID2:6515664, mYY1: 3156776, mTHOC1:4010605, hSpt6:EHS1001-99864830, hBCLAF1:40146802, POL2RB: 3346389). Human TBP gene plasmid was from a lab stock (human TFIID, gift from the Tjian lab). Rpb1 was sub-cloned from a FLAG-Pol-II WT plasmid from Addgene (#35175). pCAG-IPG ELoA, a kind gift from Dr. Teijiro Aso was also used for cloning mouse ELOA/TCEB3 in mammalian expression vectors. Site-directed mutagenesis of target lysine residues was performed using the Quickchange II XL site-directed mutagenesis kit (Agilent Technologies) on pFastBac1 plasmids harboring the gene of interest.
After 42 hrs of transduction with target P3 viral particles stock, cells were lysed and flash-frozen in BC500 (50mM TRIS PH 8.0, 0.5M NaCl, 20% glycerol, 0.05% NP40, 0.2mM EDTA, 0.2mM PMSF, 0.5 mM DTT and Roche complete protease inhibitor tablets), extracts were thawed, spun down and the extract was incubated with 250–300μl of equilibrated M2-agarose beads for 3–4 hrs, and target proteins were purified using Econo-Pac chromatography columns as described above and elsewhere (Grau et al., 2011). The following proteins were purchased as purified full length proteins: NPM1 (Abnova H00004869-P0), human SETDB1 (Origene Tech, TP326620), human PSMC6/Sug2 (Abcam, ab40575), human FOXO1 (GST-FKHR) (Millipore, 14-343). Elongin complex (Elongin ABC) was purified as described above for PRC2, by coinfection of Sf9 cells with limiting amounts of FLAG-EloA viral particles as well as HA-EloB and EloC viral stocks. Complexes were purified using M2 bead affinity purification as described above.
SPOT array synthesis, assay and quantification
Isoleucine-scanning, positional-scanning and 339 potential nuclear targets arrays were synthesized as 15-mer peptides at Koch Institute/MIT Biopolymer and Proteomics facility on cellulose membrane using Intavis SPOT synthesis peptide arrayer system. Each spot comprised of approximately 40 nmol (50–60 μg for an average 15-mer peptide).
Initial time course, SAM and enzyme titration reactions for determining reaction conditions and biochemical properties of the purified enzyme were carried out on individual spots in a single well of a 24-well plate in 200μl of 1× MTase buffer (25mM HEPES PH 7.0, 3mM DTT, 2mM MgCl2), with 10nM PRC2 and 0.25μM 3H SAM. Reactions were carried out at 25°C on a shaker. Upon completion, reactions were washed 4× using reaction buffer containing 50mM NH4CO3 for 30–40 min total. Spots were incubated with 500μl of AMPLIFY solution (Amersham/GE, NAMP100) for 15 min, dried on Whatman paper and exposed to a tritium storage phosphor screen (GE healthcare) for 24–48 hrs.
For quantification of spot membrane methylation levels, 3H methylation intensity volume registered on the storage phosphor screen was detected and quantified using the array spot detection setting of Typhoon phosphorimager. To determine the relative contribution of each one of the 20 amino acids at any given position of the positional-scanning peptide array, the volume intensity of each particular amino acid was divided by the sum (Σ) of all 20 amino acid methylation signal and presented as fraction of Σ. If amino acids are not favored/disfavored at a given position, the contribution of each amino acid to the total signal would be 0.05 or .
Full length protein MTase reactions
MTase reactions were carried out in 10–15 μl total volume in 1× methyltransferase buffer (10% glycerol, 25mM HEPES PH 7.9, 2mM MgCl2, 1mM MDTT). Unless stated, 20–50 nM of Enzyme complex and 500 nM of substrates (H3, nucleosome or proteins of interest) were used for each reaction. 1–2μl of 3H labeled Adenosyl-L-Methionine (SAM) (6.72 μM, Specific Activity: 55–85Ci/mmol) (PerkinElmers NET155H250UC), spiked with cold SAM (2–3μM total SAM concentration) was used as methyl donor in the reactions. Reactions were incubated at 30°C for 60 min and stopped with either high concentration of cold SAM (NEB, B9003S) or by addition of 6× SDS loading buffer. Completed reactions were loaded and separated on SDS-PAGE gel, stained with coomassie blue, destained and incubated with AMPLIFY fluorography solution (Amersham/GE, NAMP100) for 15–30 min. Gels were dried and exposed at −80°C for 1–4 days.
Scansite Database Search for PRC2 Targets
We generated matrices with various degrees of stringency based on the methylation rate of each amino acid substitution at each residue of the positional-scanning library. Tab-delimited matrices text files were uploaded as input motif in Scansite database search (Scansite.mit.edu) (Obenauer, 2003). The 339 potential murine nuclear protein targets were pooled and the predicted target lysine sequences (+/−7a.a.) were spotted as 15-mer peptides on cellulose membranes for validation.
Cell culture, Transfection
Murine wild type (WT) CJ7, Ezh2−/− and Eed−/− ES cells were the gift of the Orkin Lab (Harvard Medical School, Boston, MA) (Shen et al., 2008). EloA−/−, EloA+/− and WT CCE mES cells were kindly provided by Dr. Teijiro Aso (Kochi Medical School, Kochi, Japan)(Yasukawa et al., 2012). Cells were grown and maintained on 0.2% gelatin coated MEF-seeded 6-well plates in ES-media (1× DMEM knockOut medium, 15% hyclone FBS, 1× GlutaMAX, 1× NEAA, 1× pen/strep, 1–1.6×103 Units/ml of mLIF (Millipore), 55μM 2BME supplemented with 1μM and 3μM of PD0325901 and CHIR99021, respectively. Mouse embryonic fibroblast 3T3-L1 cells (ATCC CL-173) were grown in DMEM with 10% FCS. Transient or stable cell line transfection of mESC and 3T3-L1 cells was done using Xfect mESC transfection reagent and Xfect transfection reagent (Clontech Laboratories), respectively, according to the manufacturer’s instructions.
Generation of Elongin A methyl-specific antibodies
Peptide synthesis, rabbit immunization and affinity purification for all antibodies was performed by New England Peptide (NEP) (Gardner, MA). In brief, rabbits were immunized with EloA K754me1 peptide (Ac-VK(KMe)IAPMMAKC-amide), EloA K754me2 peptide (Ac-VK(KMe2)IAPMMAKC-amide) or EloA K754me3 peptide (Ac-VK(KMe3)IAPMMAKC-amide). After three rounds of immunization, exsanguinated serums were subjected to multiple rounds of negative affinity purification using the unmodified EloA K754 peptide (Ac-VKKIAPMMAKC-amide), followed by positive affinity purification with corresponding methyl-specific peptides. Reactivity, specificity and sensitivity of the antibodies was examined by spotting 50 to 0.4 pmol of peptides in 5× serial dilutions on nitrocellulose membranes and performing Western Blot (Figure S4B). For generation of TBP K236 antibodies, the following peptides were synthesized and used for immunization: TBP K236me1 (Ac-CRLAAR(KMe)YARVVQK-amide), TBP K236me2 (Ac-CRLAAR(KMe2)YARVVQK-amide ) and TBP K236me3 (Ac-CRLAAR(KMe3)YARVVQK-amide) and purified as described for Elongin A K754 methyl antibodies.
CRISPR/Cas9 generation and selection of mutant mESC
Optimal CRISPR guide RNA, proximal to Elongin A K754 with lowest off-target score was identified using CRISPR Design software (crispr.mit.edu). The selected CRISPR RNA (CrRNA) and universal tracrRNA were ordered from IDT. High concentration Cas9 (5mg/ml) was obtained from PNA Bio. For homology directed repair (HDR), single stranded DNA template (ssODN) was ordered as 200nt Ultramer from IDT with homology arms of roughly equal length from the mutation site (shown in Figure S5A). For RFLP screening of selected clones, MscI restriction site was abolished (sense mutation) downstream of K754 and included into the sequence of the ordered ssODN. Five nmol of universal tracrRNA and 2nmol of crRNA were resuspended at 200μM in RNase-free duplex buffer (25 and 10 μl, respectively). Ten microliter of the tracrRNA was added to the reconstituted crRNA tube, gently vortexed, placed at 95°C and gradually cooled down to RT to allow hybridization.
For Crispr/Cas9 RNP assembly, 5μl of hybridized crRNA/tracrRNA (500pmol) was added to 50μg of Cas9 protein (312pmol), mixed gently and left at RT for 25min. Upon completion of RNP complex assembly, 2μl of 50μM ssODN/HDR template along with 3μl of 50ng/μl linear puromycin (Clontech) was added to the RNP assembly. While the RNP was being assembled, low passage CJ7 mESC were de-MEFed and 1×106 mES cell was resuspended in 100–110 μl of nucleofection reagent (Lonza).
The CRISPR/Cas9 RNP mix along with ssODN and linear puro were added to the nucleofection mix and nucleofected using A-30 program. 500μl of ES media was added to the nucleofected mix. Each nucleofection reaction was seeded in 4 or 5 separate wells of a 6-well plate containing puromycin-resistant DR4 MEF. This was done to simplify colony picking and prevent overcrowding of the plate. After 24 hours, puromycin was added to the cells at 1μg/ml and selection was done for 48hrs (media changed after 24hrs). Cells were grown for 4–6 more days in ES media without drug selection. One hundred and twenty colonies were selected under a Nikon SMZ1500 stereoscope with a 37°C thermoplate. Using a P10 pipette, each colony was detached and added to 75μl of trypsin in a 96-well plate. After 15–20 min the resuspended colonies were added to 1ml of ES media in MEF-containing 24-well plate. Cells were grown for 2–4 days with daily media change. Healthy and confluent cells in 24-well plates were trypsinized, quenched and spun down at 190 rcf for 4min and resuspended in 500–1000 μl of ES freeze media (10%DMSO, 90% hyclone FBS serum). About 1/3 of the resuspended cells was added to 1% gelatin-coated feeder-free plates containing 1ml media for expanding and RFLP screening. The plate containing the remainder of ES cells in freezing media was wrapped in parafilm, placed in a Styrofoam box and stored at −80°C.
After cells reached confluency in the MEF-free 1% gelatin plate, cells were washed once with PBS, 100μl of Quickextract DNA extraction solution (Epibio) was added to each well. Cell extract was transferred into a 96-well plate, sealed, heated at 65°C for 5min, followed by 98°C for 2 min. 0.5–1.5 μl of this extract was used as template in a 25 μl PCR reaction using Takara 2 Emerald PCR master mix (Clontech) with primers covering the HDR site (+/− 500 bp). Restriction digestion was carried out in the same tube upon completion of PCR by adding restriction digestion buffer to 1× and incubation with 0.5 μl of MscI (NEB). PCR Plates were incubated at 37°C for 90min. 5–10 μl of the RFLP reaction was ran on gel (Figure S5B) and uncut PCR products were selected, TOPO cloned and sequenced for verification of point mutation. Once it was determined, which clones have the mutation of interest, the corresponding plates were removed from −80°C, 1ml of warm media was added to the selected wells to resuspend, spun down, and expanded into 6-well plates.
Alkaline Phosphatase staining was carried out using Vector laboratories Kit (Vector Laboratories, SK-5100) for 30min according to the manufacturer’s protocol and image acquisition was done on a stereoscope. Cell proliferation was done by plating 2–5×104 MEF-depleted ES cells on a 1% gelatin coated 12-well plate and cells in each well were trypsinized and automatically counted using Biorad TC20 automated cell counter.
Embryoid Body (EB) formation Assays
EB formation assay was carried out following the hanging drop method. In brief de-MEFed cells were resuspended in LIF-free IMDM media at 7.5×103cells/ml and incubated as 25μl hanging droplets. Morphology and size of cells was examined under a brightfield microscope after the indicated time course.
RNA-seq and ChIP-seq
For RNA-seq, 1μg of total RNA was depleted of ribosomal RNA using RiboZero Gold magnetic kit (Illumina Inc) according to the manufacturer’s protocol. cDNA was prepared using TruSeq total RNA kits (Illumina) or NEBNext Ultra RNA library prep kit for Illumina (NEB) and library was prepared as described previously (Bowman et al., 2013). Single-end 50 cycles sequencing reads (SR50) was carried out on an Illumina HiSeq2500 instrument at the MGH next generation sequencing core facility.
For ChIP-seq, mouse ES cells were washed once with PBS, crosslinked with 1% formaldehyde in PBS for 10min at RT and quenched by adding glycine to a final concentration of 0.125M. Crosslinked cells were spun down for 3min at 3500 rpm (2850 rcf) and lysed in cell lysis buffer (50mM Tris pH 8.0, 50mM NaCl, 1mM EDTA, 5mM MgCl2, 0.25% NP-40, Roche complete EDTA-free protease inhibitor tablet (PI), 0.2mM PMSF and 0.2mM DTT (last three added fresh) at 2–2.5×107 cells/ml. Cells were left on ice for 10 min and dounced 10× using the tight pestle. Nuclei were spun down at 2850 rcf in 15ml conical tubes, and resuspended in sonication buffer (20mM Tris pH 8.0, 1% SDS, 2mM EDTA, 0.5mM EGTA, 0.5mM PMSF, PI(freshly added) at 5.0×107 nuclei/ml. Resuspended nuclei were left on ice for 10min and sonicated using the Bioruptor sonicator, at high setting 30″ on 45″ off twice, 15min each. Water and ice was changed in between each sonication. IP was performed using rabbit anti EZH2 (CST, D2C9). 2.5×106 nuclei was used per IP reaction, by adding 50 μl of sonicated chromatin to 1–1.25ml of IP buffer (20mM Tris pH 8.0, 1% Triton X-100, 150mM NaCl, 10% glycerol, 2mM EDTA) after O/N incubation with the primary antibody, protein A magnetic beads (40μl) was added to the IP reactions and the mixture was incubated for an additional 2–4hrs. Each wash was done on the rotator at RT for 5min as follows: 1× low salt wash (20mM Tris pH 7.8, 0.1% SDS, 1% Triton X-100, 2mM EDTA, 150mM NaCl), 3× high salt wash (20mM Tris pH 7.8, 0.1% SDS, 1% Triton X-100, 2mM EDTA, 500mM NaCl), 1× LiCl wash (20mM Tris pH 7.8, 1% NP-40, 1% NaDeoxycholate, 2mM EDTA, 250mM LiCl), ×2 TE wash. Elution was done on the immunoprecipitated samples twice with 250μl of elution buffer (1% SDS, 0.1M NaHCO3) on the rotator at RT. To the 500μl elution buffer, 20 μl of 5M NaCl was added and samples were incubated in a Thermomixer for 4hr to O/N at 65°C to reverse the crosslink (same was done for the input material). Next, 15μl of 1M Tris pH 7.8, 1–2 μl glycoblue (Ambion) and 2 μl of proteinase K was added to the reaction mix and incubated at 65°C for an additional 30–60min. DNA was extracted from the reversed crosslink material using phenol/chloroform extraction and EtOH precipitation, or directly after elution with elution buffer using ChIP DNA clean & concentrator (Zymo Research). IP DNA was resuspended in 100μl of H2O. For ChIP-seq, adaptor ligation to input and IP DNA and library generation was carried out as described before (Bowman et al., 2013)
Bromouridine labelling of nascent RNA (Bru-seq)
Bru-seq experiment was adapted from Tani et al.(Tani et al., 2012), with a number of modifications: Experiments were carried out on mouse ES cells gown on feeder-free, 1% gelatinized 100mm tissue culture dishes. 5-Bromouridine (BrU) was added to cells at a final concentration of 2mM (from a100mM BrU stock) and incubated for 10 min at 37°C. Media was aspirated off and cells were rapidly lysed in 4ml of Trizol, 800μl of chloroform was added to the lysed cells in 5ml centrifuge tubes, vortexed and spun down at 5200 g for 15min. Equal volume of isopropanol was added to the aqueous phase, mixed, left at RT for 5–10 min and spun down at 4°C for 20min at max speed (14000 g) in a tabletop centrifuge to precipitate RNA. Resulting RNA pellet was washed twice with 70% EtOH, air dried and resuspended in 200–400 μl Nuclease-free water. The amount of RNA was quantified using Nanodrop, and a small fraction of the total RNA was run on agarose gel to determine the quality of purified total RNA. For enrichment of Bru-RNA, Mouse αBrdU antibody (SCBT, sc-32323) was coupled to Dynabeads M-280 Sheep Anti-Mouse IgG (11201D), 1ml of magnetic beads was washed 4 × with PBS-DEPC/0.1%BSA and 500μl of Ab(200μg/ml) was incubated with 1ml of beads and yeast RNA (500ng/μl final concentration) for 2–3 hours at 4°C, washed 3× with PBS-DEPC/0.1%BSA/Tween 0.05%. The final volume was raised to 1ml. For enrichment of Bru-RNA, 25–50μg of total RNA (after preheating at 95°C and immediate cooling) was added to 50μl of antibody-bound beads in 1 ml of PBS-DEPC/0.1%BSA/Tween 0.05% and incubated for 90 min at RT in a foil-wrapped tube. Beads were washed 4× with PBS-DEPC/0.1%BSA/Tween 0.05%, 5–10min (last wash was done with Tween-free buffer). Enriched RNA was either eluted by addition of 25μl of DEPC-water and incubation at 94°C for 5min with occasional flicking/shaking, or Trizol extraction. The enriched RNA was depleted of ribosomal RNA using RiboZero Gold (Illumina) and used for generation of RNA-seq library.
in vivo Detection of EloA K754 methylation by IP
MEF-Depleted CJ7 Wt or Ezh2−/− mESC cells were grown on 1% gelatin plates in LIF-containing ES media for 2–3 days and 1.0×108 cells were used for each IP. Trypsinized cells were spun down, washed in 1× PBS and lysed (2–2.5×107cells/ml) in cell lysis buffer (50mM Tris pH 8.0, 50mM NaCl, 1mM EDTA, 5mM MgCl2, 0.1% NP-40, Roche complete EDTA-free protease inhibitor tablet, 5μM JIB 04, 0.2mM PMSF and 0.2mM DTT (last 4 added fresh) and 1/15 (v/v) of 1× RIPA buffer. Lysed cells were left on ice for 10min, flash frozen, thawed and sonicated for 10min with Bioruptor on (high setting, 30sec on, 45sec off). Benzonase was added to the sonicated cell lysate (0.15U/μl) and the extract was incubated at RT for 20min while rotating. The digested cell extract was spun down at 13,000 rpm in a refrigerated tabletop centrifuge, NaCl concentration was raised to 200mM and 30–40 μl of Goat Elongin A antibody (R-19, SCBT) and 100–120μl of equilibrated Dynabeads protein G magnetic beads were added to the extract, between 4hrs-O/N. The beads were washed 3× (15–30min total) with the Cell lysis buffer containing 200mM NaCl (w/o RIPA). Washed beads were vortexed in 1× SDS gel loading buffer and the samples were run on SDS-PAGE gels. For Western Blot, membranes were pretreated with SuperSignal Western Blot enhancer (Pierce), and primary methyl-EloA antibodies were used at 1:2,000, incubated at RT for 3 hours, washed 3× and incubated with 1:30,000–1:100,000 of secondary-HRP conjugated antibodies for 1 hr. EZH2, EloA, Histone H3 and H3K27me3 primary antibody concentrations for Western Blots were 1:3000, 1:1000, 1:3000 and 1:2000, respectively. Secondary incubation with HRP-conjugated antibody was done at 1:20,000. For histone and histone modification Immunoblots, histones were acid extracted by resuspending nuclei in 0.4N H2SO4 (1.0–2.0×107 nuclei/ml) and rotating for 3hrs in cold room. Samples were spun down in a cold table top centrifuge for 10 min at max speed. The acid extracted protein mix was neutralized by addition of half volume of 1.5M Tris pH 8.8 and equal amounts of neutralized acid-extracted histones were used for Immunoblotting.
QUANTIFICATION AND STATISTICAL ANALYSIS
RNA-Seq and ChIP-seq analysis
Sequencing was performed on an Illumina HiSeq 2500 instrument, resulting in ~30 million single-end 50 bp reads per sample on average. The splice-aware alignment program STAR (Dobin et al., 2013) was used to map sample sequencing reads (fastqs) to the mouse (mm9) reference genome. Gene expression counts were calculated using the program HTSeq (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html) based on the latest Ensembl annotation for mm9/GRCm37. The R package edgeR (Robinson et al., 2010) was used to make differential gene expression calls from these counts at a two-fold cut-off and FDR<0.05 threshold. Average gene expression values (RPKMs) for K754R and K754M EloA mutants were compared with those for wild-type mouse ES cells. The average log2 fold-change values for K754R/WT and for K754M/WT were compared against each other and against EED-null/WT. P-values for the significance of gene set overlaps were calculated using hypergeometric tests.
For ChIP-seq, sequencing was performed on an Illumina HiSeq 2500 instrument, resulting in ~30 million paired-end 50 bp reads per sample on average. Reads were aligned against the mm9 mouse genome build using BWA (Li and Durbin, 2009) All alignments were filtered for uniquely mapped reads and alignment duplicates were removed. Input-normalized coverage tracks were generated using deep Tools version 2.0.1 (Ramirez et al., 2014). Input normalized Ezh2 ChIP-seq (rabbit anti EZH2 (D2C9), Cell Signaling Technology) tracks were compared against RNA-Seq gene expression tracks for EloA mutants K754R and K754M.
Bioinformatics alignment of Elongin A for different species and human isoforms was carried out using Kalign (2.0), multiple sequence alignment software with ClustalW output format (Lassmann et al., 2009).
DATA AND SOFTWARE AVAILABILITY
All sequencing data have been deposited in GEO. GEO accession number: GSE104660
Supplementary Material
Highlights.
Characterization of PRC2 substrate preference identified potential methylation targets
The transcription elongation factor Elongin A is methylated by PRC2 in vivo
Methylation of Elongin A tunes transcription of low expression PRC2 target genes
Loss of Elongin A methylation interferes with differentiation potential in mES cells
Acknowledgments
We thank Dr. Stuart Orkin for providing Ezh2−/−, Eed−/− and CJ7 mES cells. We are also grateful to Dr. Teijiro Aso for providing EloA−/−, EloA+/−and EloA+/+ mES cells and pCAG-IPG-FLAG-mEloA plasmid construct. We thank members of the Kingston Lab for helpful discussions, in particular Chris Davis, Aaron Plys, Sara Miller and Jongmin Kim for critical reading of the manuscript, Matt Simon and Mark Borowsky for helpful discussions, the MGH NextGen Sequencing Core and Taplin mass spectrometry facility. This work was supported by NIH grants R01GM043901 and R37GM048405 to R.E.K and NIH P30 DK40561 to R.I.S.
Footnotes
Author Contribution
M.B.A, R.E.K conceived the project. M.B.A designed and performed the experiments. A.A. and R.I.S. carried out the bioinformatics analysis. A.A. generated figures for bioinformatics analysis. J.C.C. carried out structural modeling and analysis. S.K. prepared ChIP-seq libraries. M.B.A, R.E.K and J.C.C. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abmayr SM, Carrozza MJ, Workman JL. Preparation of nuclear and cytoplasmic extracts from mammalian cells. Curr Protoc Pharmacol. 2006 doi: 10.1002/0471141755.ph1203s35. [DOI] [PubMed] [Google Scholar]
- Aso T, Lane WS, Conaway JW, Conaway RC. Elongin (SIII): a multisubunit regulator of elongation by RNA polymerase II. Science. 1995;269:1439–1443. doi: 10.1126/science.7660129. [DOI] [PubMed] [Google Scholar]
- Aso T, Haque D, Barstead RJ, Conaway RC, Conaway JW. The inducible elongin A elongation activation domain: structure, function and interaction with the elongin BC complex. EMBO J. 1996;15:5557. [PMC free article] [PubMed] [Google Scholar]
- Beltran M, Yates CM, Skalska L, Dawson M, Reis FP, Viiri K, Fisher CL, Sibley CR, Foster BM, Bartke T, et al. The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res. 2016;26:896–907. doi: 10.1101/gr.197632.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer M, Pisano P, Di Carlo V, Blanco E, Chammas P, Vizán P, Gutiérrez A, Aranda S, Payer B, Wierer M, et al. EPOP Functionally Links Elongin and Polycomb in Pluripotent Stem Cells. Mol Cell. 2016;64:645–658. doi: 10.1016/j.molcel.2016.10.018. [DOI] [PubMed] [Google Scholar]
- Bowman SK, Simon MD, Deaton AM, Tolstorukov M, Borowsky ML, Kingston RE. Multiplexed Illumina sequencing libraries from picogram quantities of DNA. BMC Genomics. 2013;14:1. doi: 10.1186/1471-2164-14-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiling A, Turner BM, Bianchi ME, Orlando V. General transcription factors bind promoters repressed by Polycomb group proteins. Nature. 2001;412:651–655. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, et al. Polycomb Associates Genome-wide with a Specific RNA Polymerase II Variant, and Regulates Metabolic Genes in ESCs. Cell Stem Cell. 2012;10:157–170. doi: 10.1016/j.stem.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra VS, Hong JW, Levine M. Regulation of Hox Gene Activity by Transcriptional Elongation in Drosophila. Curr Biol. 2009;19:688–693. doi: 10.1016/j.cub.2009.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, Lander GC, Maiolica A, Herzog F, Aebersold R, Nogales E. Molecular architecture of human polycomb repressive complex 2. Elife. 2012;1:e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinforma Oxf Engl. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank R. The SPOT-synthesis technique: synthetic peptide arrays on membrane supports—principles and applications. J Immunol Methods. 2002;267:13–26. doi: 10.1016/s0022-1759(02)00137-0. [DOI] [PubMed] [Google Scholar]
- Grau DJ, Chapman BA, Garlick JD, Borowsky M, Francis NJ, Kingston RE. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev. 2011;25:2210–2221. doi: 10.1101/gad.17288211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Shen X, Ma Q, Cao J, von Gise A, Zhou P, Wang G, Marquez VE, Orkin SH, Pu WT. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26:37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heringa J. Two strategies for sequence comparison: profile-preprocessed and secondary structure-induced multiple alignment. Comput Chem. 1999;23:341–364. doi: 10.1016/s0097-8485(99)00012-1. [DOI] [PubMed] [Google Scholar]
- Herz HM, Morgan M, Gao X, Jackson J, Rickels R, Swanson SK, Florens L, Washburn MP, Eissenberg JC, Shilatifard A. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science. 2014;345:1065–1070. doi: 10.1126/science.1255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A, Tjian R. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 2003;17:1309–1320. doi: 10.1101/gad.1099903. [DOI] [PubMed] [Google Scholar]
- Hock H. A complex Polycomb issue: the two faces of EZH2 in cancer. Genes Dev. 2012;26:751–755. doi: 10.1101/gad.191163.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao L, Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015;350:aac4383. doi: 10.1126/science.aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi P, Carrington EA, Wang L, Ketel CS, Miller EL, Jones RS, Simon JA. Dominant Alleles Identify SET Domain Residues Required for Histone Methyltransferase of Polycomb Repressive Complex 2. J Biol Chem. 2008;283:27757–27766. doi: 10.1074/jbc.M804442200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justin N, Zhang Y, Tarricone C, Martin SR, Chen S, Underwood E, De Marco V, Haire LF, Walker PA, Reinberg D, et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun. 2016;7:11316. doi: 10.1038/ncomms11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi J, Inoue M, Fukuda M, Uchida Y, Yasukawa T, Conaway RC, Conaway JW, Aso T, Kitajima S. Transcriptional Properties of Mammalian Elongin A and Its Role in Stress Response. J Biol Chem. 2013;288:24302–24315. doi: 10.1074/jbc.M113.496703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Roeder RG. Involvement of the basic repeat domain of TATA-binding protein (TBP) in transcription by RNA polymerases I, II, and III. J Biol Chem. 1994;269:4891–4894. [PubMed] [Google Scholar]
- Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, et al. Phosphorylation of EZH2 Activates STAT3 Signaling via STAT3 Methylation and Promotes Tumorigenicity of Glioblastoma Stem-like Cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide Analysis of PRC1 and PRC2 Occupancy Identifies Two Classes of Bivalent Domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T, Frings O, Sonnhammer ELL. Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 2009;37:858–865. doi: 10.1093/nar/gkn1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau PNI, Cheung P. Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc Natl Acad Sci. 2011;108:2801–2806. doi: 10.1073/pnas.1012798108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen A, Højfeldt JW, Helin K. Role of the Polycomb Repressive Complex 2 (PRC2) in Transcriptional Regulation and Cancer. Cold Spring Harb Perspect Med. 2016;6:a026575. doi: 10.1101/cshperspect.a026575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Lee JS, Kim H, Kim K, Park H, Kim JY, Lee SH, Kim IS, Kim J, Lee M, et al. EZH2 Generates a Methyl Degron that Is Recognized by the DCAF1/DDB1/CUL4 E3 Ubiquitin Ligase Complex. Mol Cell. 2012;48:572–586. doi: 10.1016/j.molcel.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Allis CD. Poisoning the “histone code” in pediatric gliomagenesis. Cell Cycle. 2013;12:3241–3242. doi: 10.4161/cc.26356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefke R, Shi Y. The PRC2-associated factor C17orf96 is a novel CpG island regulator in mouse ES cells. Cell Discov. 2015;1:15008. doi: 10.1038/celldisc.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefke R, Karwacki-Neisius V, Shi Y. EPOP Interacts with Elongin BC and USP7 to Modulate the Chromatin Landscape. Mol Cell. 2016;64:659–672. doi: 10.1016/j.molcel.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 Maintain Repressive Chromatin through Different Mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, III, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Cao R, Zhang Y. Substrate Preferences of the EZH2 Histone Methyltransferase Complex. J Biol Chem. 2006;281:8365–8370. doi: 10.1074/jbc.M513425200. [DOI] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Obenauer JC. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara MM, Simon JA. Inner workings and regulatory inputs that control Polycomb repressive complex 2. Chromosoma. 2012;121:221–234. doi: 10.1007/s00412-012-0361-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengelly AR, Copur Ö, Jäckle H, Herzig A, Müller J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science. 2013;339:698–699. doi: 10.1126/science.1231382. [DOI] [PubMed] [Google Scholar]
- Perez-Burgos L, Peters AH, Opravil S, Kauer M, Mechtler K, Jenuwein T. Generation and characterization of methyl-lysine histone antibodies. Methods Enzymol. 2003;376:234–254. doi: 10.1016/S0076-6879(03)76016-9. [DOI] [PubMed] [Google Scholar]
- Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, Helin K. Gene Silencing Triggers Polycomb Repressive Complex 2 Recruitment to CpG Islands Genome Wide. Mol Cell. 2014;55:347–360. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma Oxf Engl. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanulli S, Justin N, Teissandier A, Ancelin K, Portoso M, Caron M, Michaud A, Lombard B, da Rocha ST, Offer J, et al. Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation. Mol Cell. 2015;57:769–783. doi: 10.1016/j.molcel.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- Scelfo A, Piunti A, Pasini D. The controversial role of the Polycomb group proteins in transcription and cancer: how much do we not understand Polycomb proteins? FEBS J. 2015;282:1703–1722. doi: 10.1111/febs.13112. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome Regulation by Polycomb and Trithorax Proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 Mediates Methylation on Histone H3 Lysine 27 and Complements EZH2 in Maintaining Stem Cell Identity and Executing Pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Occupying Chromatin: Polycomb Mechanisms for Getting to Genomic Targets, Stopping Transcriptional Traffic, and Staying Put. Mol Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res Mol Mech Mutagen. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Simon C, Chagraoui J, Krosl J, Gendron P, Wilhelm B, Lemieux S, Boucher G, Chagnon P, Drouin S, Lambert R. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012;26:651–656. doi: 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, Copeland RA. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Shen SS, Margueron R, Reinberg D. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 2013;27:2663–2677. doi: 10.1101/gad.225888.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, Isogai T, Suzuki Y, Akimitsu N. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22:947–956. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen MCF, Nielsen M. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 2012;40:W281–W287. doi: 10.1093/nar/gks469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma SK, Tian X, LaFrance LV, Duquenne C, Suarez DP, Newlander KA, Romeril SP, Burgess JL, Grant SW, Brackley JA, et al. Identification of Potent, Selective, Cell-Active Inhibitors of the Histone Lysine Methyltransferase EZH2. ACS Med Chem Lett. 2012;3:1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MD, Harreman M, Svejstrup JQ. Ubiquitylation and degradation of elongating RNA polymerase II: The last resort. Biochim Biophys Acta BBA - Gene Regul Mech. 2013;1829:151–157. doi: 10.1016/j.bbagrm.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Yasukawa T, Bhatt S, Takeuchi T, Kawauchi J, Takahashi H, Tsutsui A, Muraoka T, Inoue M, Tsuda M, Kitajima S, et al. Transcriptional Elongation Factor Elongin A Regulates Retinoic Acid-Induced Gene Expression during Neuronal Differentiation. Cell Rep. 2012;2:1129–1136. doi: 10.1016/j.celrep.2012.09.031. [DOI] [PubMed] [Google Scholar]
- Zhao X, Jankovic V, Gural A, Huang G, Pardanani A, Menendez S, Zhang J, Dunne R, Xiao A, Erdjument-Bromage H, et al. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008;22:640–653. doi: 10.1101/gad.1632608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.