Abstract
Objectives
The detection of an STI agent in a urogenital tract (UGT) specimen from a young child is regarded as being indicative of sexual abuse. However, the probabilities of contamination events that could conceivably lead to STI positive specimens in the absence of sexual contact are unclear. The objective was to estimate the potential for fingers that have come in contact with Chlamydia trachomatis-positive urine to detectably contaminate C. trachomatis-negative urine.
Methods
The study design was based on self-experimentation. Dilutions of C. trachomatis elementary bodies (EBs) were prepared. A participant contacted an EB dilution then a urine surrogate specimen. The experiment was performed by three participants using three C. trachomatis isolates, of genotype E, F and B. Two surrogate urine contact methods were used to mimic contamination of a carer assisting with a child’s urine collection. All EB dilutions and urine surrogate specimens were subjected to C. trachomatis assay and quantification in a real-time PCR-based diagnostic system.
Results
The amplimer crossing point (Cq) for EB dilutions was 10.0±1.6 less than for corresponding finger contacted urine specimens, which corresponds to ~10 µL of EB suspension transferred. This was largely independent of participant identity, C. trachomatis strain or EB dilution. Hand decontamination led to large reductions in EBs transferred, but transfer remained consistently detectable. Recent Cq data from C. trachomatis-positive clinical urine specimens were collated, and 20% clearly contained sufficient C. trachomatis to detectably contaminate another specimen by finger-mediated transfer, as in this experiment.
Conclusions
This study directly demonstrated the potential for urine contaminated fingers to convert a C. trachomatis-negative urine specimen to C. trachomatis positive as a result of contact. Accordingly, procedures for urine specimen collection, particularly from children, need to be designed to prevent contamination.
Keywords: chlamydia trachomatis, urine, children, sexual abuse
Key messages.
Human fingers contaminated with Chlamydia trachomatis can detectably contaminate urine surrogate specimens by contact.
The relationship between level of finger contamination and urine surrogate contamination is highly consistent.
Three contaminated fingers, dried on paper towel, transfer the equivalent of approximately 10 µL C. trachomatis suspension to a urine surrogate specimen.
It is critical that urine specimen collection procedures, in particular from children, should be in accordance with procedures designed to prevent contamination from any source.
Introduction
The exquisite sensitivity of nucleic acid amplification tests for STIs can lead to the perception that positive results can arise from specimen contamination events. This is particularly significant in the Australia’s Northern Territory (NT). Approximately 30% of the NT population are Indigenous Australians, with about half living in remote communities. The notification rates for STIs are high in the NT, especially in the Indigenous population. For example, the rate of Chlamydia trachomatis notifications in Indigenous people in the NT is approximately five times higher than the notification rate in the Australian population as a whole.1
The ‘Little Children Are Sacred’ report from 20072 detailed widespread concern of child sexual abuse in NT remote Indigenous communities. This led to large-scale and controversial interventions by the Australian Commonwealth Government, which are ongoing in modified form. Long-standing tensions between the imperatives of child protection and avoidance of disruption and stereotyping of communities and families remain.3 This may manifest in difficulties formulating service provider responses to instances of detection of STI agents in the urine of young children, when there is no other evidence of sexual abuse. While recognising that an STI in a child is a strong indicator of sexual abuse, an absence of other indicators can lead some service providers to raise the possibility of a urine specimen or anatomical site contamination. Studies outside Australia have lent credence to this concept.4–6
We are investigating conceivable mechanisms that diagnostic specimens may become contaminated. We have previously reported the potential of contaminated toilet–bathroom facilities to give rise to contaminated specimens,7 and also addressed the question as to whether C. trachomatis derived from ocular infections rather than sexual contact can be reliably identified by genotyping.8 9 Here, we test another conceivable mechanism; that a carer with C. trachomatis contaminated fingers who assists a child to provide a urine specimen may convert a specimen from C. trachomatis negative to C. trachomatis positive, as a result of contact between their fingers and the specimen.
Methods
The study design basis was simulation by self-experimentation performed by three members of the investigator team (termed participants). Three C. trachomatis isolates were used. They were all derived from the ‘Mother-Child’ study, a survey of C. trachomatis in mothers and their babies, performed in the north of the NT between 1986 and 1992.10–12 Isolate E_Aus56 is serovar E, isolate F_Aus51 is serovar F and isolate B_Aus45 is serovar B. All three are urogenital tract isolates and B_Aus45 has been genome sequenced.12
Each participant contacted suspensions of live C. trachomatis with their bare fingers, and then immediately after contacted a 10 mL aliquot of a urine surrogate solution, made as described by Martino and coworkers.13 The relative quantities of C. trachomatis DNA in the suspensions and urine surrogate solutions were then determined. The experimentation was performed on three different days, with 2 weeks between each occurrence. On each day of experimentation, a single C. trachomatis isolate was tested with all three participants. In advance of experimentation, the isolate to be used was grown to high infection levels in McCoy cells (90%–95% of host cells infected) as previously described.12 Elementary bodies (EBs) were released from host cells and concentrated, as previously described,12 and a 100-fold dilution series in urine surrogate prepared yielding dilutions of 100–10−12 for E-Aus56 and F_Aus51 and 100–10−14 for B_Aus45. Between 30 and 60 min in advance of the experiment, the experimenters washed their hands in a general purpose hand and body wash (Avagard 9230-D (3M)), rinsed with tap water, then rinsed their hands with 0.5% chlorhexidine gluconate in 70% ethanol (Avagard 9250 P (3M)) and allowed this to dry.
Two different methods of contact between the experimenter’s C. trachomatis contaminated fingers and urine surrogate were used: (1) dipping the contaminated fingers into 10 mL urine surrogate in a urine pot for 10 s (method A—dipping method (performed first, using the left hand)) and (2) running 10 mL of urine surrogate over the contaminated fingers into a container (method B—pouring method (performed second, using the right hand)). These methods were first used to generate negative control (N1) specimens, where there was no prior contact between fingers and an EB suspension on that day. Then, commencing with the most dilute EB suspension, the participant contacted each EB dilution with the first, second and third fingers, dried the fingers briefly on a paper towel, then contacted a 10 mL urine surrogate specimen with the same fingers. On completion of both contact methods with all dilutions of EB suspension, the participant decontaminated their hands using the same procedure as the pre-experimentation hand decontamination. After this, on the same day, the participant generated ‘postexperiment, postdecontamination’ (N2) specimens by contacting fresh 10 mL urine surrogate with the same fingers of each hand as had been used previously. In this instance, the dipping method was used with both hands.
In the interests of practicality and safety, each experiment was performed using a class II biosafety cabinet by participant and two assistants. This ensured the participant touched nothing with their fingers in the course of the experiment apart from the EB dilutions, the urine surrogate samples and the paper towel used for drying.
The EB dilutions and the urine surrogate specimens were all subjected to analysis for C. trachomatis DNA, using the Roche Molecular Diagnostics Cobas 4800 CT/NG commercial diagnostic system (Roche Diagnostics Australia). This provides quantitation information in the form of the amplimer concentration ‘crossing point’ (Cq). Provided there is doubling of the amplimer concentration with each PCR cycle, the Cq is inversely proportional to the log2(amplimer), with a gradient of −1. In general, Cq results ≥40 were not included in subsequent analyses. We reasoned that results right at the limit of detection of the Roche instrument are potentially due to contamination from, for example, the air in the biohazard hood, and may confound rather than enhance our efforts to reliably determine the relationship between C. trachomatis loads in EB suspensions and corresponding finger-contacted urine surrogate specimens. Similarly, Cq values from positive reactions where a less diluted EB suspension yielded a negative reaction were excluded from further analysis. All results, including those from specimens that yielded positive results with Cq values ≥40, are presented in the supplementary data.
The difference between the Cq for an EB suspension and the Cq for a corresponding finger contacted urine surrogate specimen was defined as the ΔCq. Assuming amplimer doubling with every PCR cycle, the volume of EB suspension transferred to the urine surrogate (in µL)=10 000/2ΔCq, where 10 000 is the volume of each urine surrogate specimen in µL and 2ΔCq is the ratio of target concentration between EB suspension and derived urine surrogate specimen. A similar approach was used to calculate the reduction in C. trachomatis load on the fingers concomitant with hand decontamination after the completion of the transfer experiment. In this case, the parameter ΔCq(N2) is the difference between the Cq value for a urine surrogate specimen arising from transfer from the undiluted EB suspension (ie, the final data point in the transfer experiment) and the Cq for the urine surrogate specimen contacted by the postdecontamination fingers. The numerical reduction in load is thus 2ΔCq(N2).
The statistical methods used were primarily one-way analysis of variance (ANOVA) and Tukey ‘Honest Significant Difference’ (HSD) for comparisons of more than two categories and the t-test for comparison of two categories. In general, ΔCq values were used rather than calculated values for volumes of EB dilution transferred. This is because the statistical methods used are reliant on normal distribution of data. It was determined that the distribution of ΔCq values was consistent with normality, according to the Shapiro-Wilks test (p=0.92). In contrast, the distribution of deduced transferred volumes was inconsistent with normality, with a long tail towards high values.
Results
Initial inspection of the data revealed a consistent relationship between the Cq values derived from EB suspensions, and Cq values derived from the corresponding finger contacted urine surrogate specimens. Results from a typical experiment are shown in figure 1, and all results are shown in supplementary information.
Figure 1.
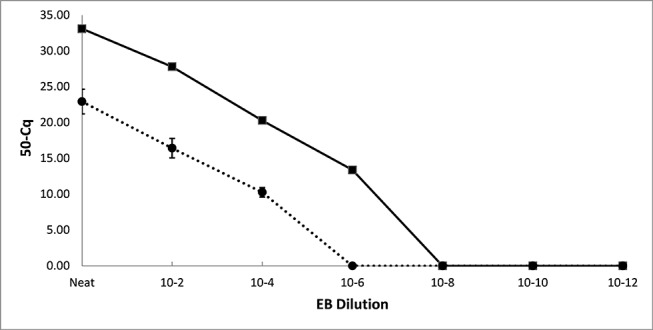
Typical result from an EB transfer experiment for one strain, with the solid line representing Cq values from dilutions of an EB suspension and the dotted line representing the mean Cq from the corresponding finger contacted urine surrogate specimen, for all three participants. EB, elementary body.
We tested the assumption that there was amplimer doubling in every PCR cycle. A linear regression of log EB dilution versus Cq for all Cq values <40 was performed. This yielded Cq=3.3 × log EB dilution +16.0 (R2=0.98). Because 3.3 = log210, it was concluded that the assumption of target doubling in each PCR cycle is justified, so allowing the volumes of EB suspensions transferred to be calculated as described in the Methods.
Collated results from all specimens apart from the N1 pre-experimentation specimens and the N2 posthand decontamination specimens are summarised in table 1. In accordance with the Methods, six Cq values from finger contacted urine surrogate specimens were excluded from analysis as they had Cq values≥40 (three specimens), yielded positive reactions where a less diluted EB specimen yielded a negative reaction (one specimen) or fulfilled both criteria (two specimens) (see online supplementary data). The mean ΔCq value across the entire experiment (not including the N1 and N2 specimens) was 10.0±1.6, which equates to ~10 µL of EB dilution being transferred to the urine surrogate specimens, with an inferred range of volumes transferred being ~1–100 µL. The ΔCq values from participant 1 were significantly lower than the ΔCq values from the other two experimenters, indicating that participant 1 transferred significantly more EBs than the other two. Similarly, the ΔCq for the pouring method was slightly but significantly less than for the dipping method. In contrast, there was no significant difference in the ΔCq values between the three different C. trachomatis strains. The 10−2 dilutions gave rise to significantly higher ΔCq values than either the undiluted suspension or 10−4 diluted suspension.
Table 1.
Ranges of ΔCq values and inferred volumes of EB dilutions transferred
| Category (n) | Mean ΔCq (SD) | ΔCq range | Mean µL transferred (inferred from±2xΔCq SD range)(inferred from observed ΔCq range) | |
| All (49) | 10.0 (1.6) | 6.7–13.3 | 9.8 (1.1–89.7)(1.0–96.2) | |
| Participant 1 (17) | 8.8 (0.9) | 6.7–9.8 | 22.4 (6.4—78.1)(11.2–96.2) | p<0.0001* Participant 1 vs participant 2: p<0.01† Participant 1 vs participant 3: p<0.01† Participant 2 vs participant 3: not significant† |
| Participant 2 (16) | 10.5 (1.2) | 8.6–13.0 | 7.1 (1.4–32.6)(1.2–25.8) | |
| Participant 3 (16) | 10.9 (1.6) | 7.7–13.3 | 5.2 (0.6–48.1)(1.0–48.1) | |
| Dipping (23) | 10.6 (1.7) | 7.4–13.3 | 6.4 (0.6–68.0)(1.0–59.2) | p=0.015‡ |
| Pouring (26) | 9.5 (1.3) | 6.7–12.4 | 13.8 (2.3–83.7)(1.0–96.2) | |
| E_Aus56 (17) | 9.7 (1.6) | 6.7–12.2 | 12.0 (1.3–110.5)(2.1–96.2) | Not significant; p=0.41* |
| F_Aus51 (15) | 10.0 (1.6) | 7.4–13.3 | 9.8 (1.1–89.8)(1.0–59.2) | |
| B_Aus45 (17) | 10.4 (1.5) | 8.4–13.0 | 7.4 (0.9–59.2)(1.2–29.6) | |
| Undiluted (18) | 9.4 (1.7) | 6.7–12 | 14.8 (1.4–156.3)(2.4–96.2) | p=0.004* 100 vs 10−2: p<0.01† 100 vs 10−4: not significant† 102 vs 10−4: p<0.05† |
| 10−2 diluted (18) | 11.0 (1.5) | 9.0–13.3 | 4.8 (0.6–39.1)(1.0–19.5) | |
| 10−4 diluted (13) | 9.5 (1.0) | 7.7–11.1 | 13.8 (3.5–55.2)(4.6–48.1) |
The 49 data points were collated according to different criteria to reveal possible correlates. This encompasses data only from specimens that yielded a positive C. trachomatis test with a Cq <40.
*One way analysis of variance.
†Tukey HSD.
‡t-test (two tailed).
sextrans-2016-053081supp001.pdf (295.3KB, pdf)
The C. trachomatis diagnostic test yielded a positive reaction for five of the 18 N1 specimens, with Cq values ranging from 39.6 to 40.8 (see online supplementary data for complete results). All the positive reactions were from specimens obtained immediately before the pouring experiments, after the dipping experiment had been completed.
All of the N2 postdecontamination specimens yielded positive PCR reactions, with Cq values ranging from 32.5 to 40.0. After exclusion of the single Cq value of 40.0, the reductions in C. trachomatis DNA resulting from hand decontamination were quantified. The mean of the ∆Cq(N2) values was 10.3±2.1, with an absolute range of 6.4–14.8. This equates to a mean reduction of 1271-fold and an absolute range of 85.0-fold to 28526-fold.
To assess the clinical implications of this study, we compared our results to 30 successive Cq values from analysis of specimens analysed in the course of clinical service provision. The Cq values were derived from the same instrument as used to analyse the urine surrogate specimens in this study. We conservatively classed any specimen with a Cq <40–10=30 as clearly having potential to contaminate a finger sufficiently to potentially give rise to a false positive. Six of the 30 (20%) of the Cq values were ≤30, indicating that such potential exists.
Discussion
The principal finding was remarkably consistent ΔCq values of ~10 cycles, with respect to the EB suspensions and corresponding finger-contacted urine surrogate specimens. This was largely independent of suspension density, C. trachomatis isolate, participant identity or EB transfer procedure. Comparison with Cq values from a sample of C. trachomatis-positive urogenital tract (UGT) clinical specimens indicated that 20% of such specimens can contaminate a human finger sufficiently to be able to convert a C. trachomatis-negative specimen to positive as a result of contact. While ΔCq values may provide an indication of the likelihood of detectable contamination of clinical specimens, to make the results more generalisable, we inferred the volumes of EB suspensions transferred from human skin to the 10 mL urine surrogate specimens. The self-experimentation procedure transferred into each surrogate urine specimen C. trachomatis or DNA equivalent corresponding to a mean of 10 µL and a range of ~1–100 µL of EB suspension. It was shown that hand decontamination reduced the amount of C. trachomatis DNA transferred from contaminated fingers by ~1000-fold, thus reducing potential for specimen contamination. The principal strength of the study is that it was a very direct test of the potential for C. trachomatis-contaminated fingers to contaminate diagnostic specimens, with the remarkable consistency of ΔCq values, indicating integrity in the experimental design.
We considered whether the positive diagnostic tests for N1 specimens (5/18) indicated that the experiment had been confounded, such as by pre-existing finger contamination. All five positive reactions were from specimens taken after the dipping method experiments and prior to pouring method experiments. This, in combination with extremely high Cq values, suggests the likely basis for the positive N1 reactions was aerosolised C. trachomatis inside the biosafety hood arising from the dipping method experiments. While we cannot rule out that such contamination has impacted the experimental results, this can only be the case for specimens that gave Cq values close to 40 and will not have had a significant effect on the study outcomes.
The experimental design did not encompass a systematic determination of the persistence of C. trachomatis on the skin. This was primarily because it is unreasonable to expect a participant to contaminate their fingers with live C. trachomatis and then refrain from hand decontamination for a long period of time. However, the results from the N1 specimens from the experiments carried out on the second and third days of experimentation provide some insight. The three experimentation days were 2 weeks apart. No C. trachomatis DNA was detected in the N1 specimens that were obtained at the start of the second and third days of experimentation, indicating that no detectable finger contamination had persisted from the experimentation 2 weeks, or 2 weeks and 4 weeks prior. This is despite the N2 specimens obtained at the end of the experimentation in experimentation days 1 and 2 being universally positive. Thus, between experiments on different days, the amount of C. trachomatis DNA on the fingers of the participants dropped below the level in which they could contaminate specimens. Given that hand decontamination reduced the amount of C. trachomatis/C. trachomatis DNA transferred by an average of ~1000, this is unsurprising.
The assumption implied by the data analysis method was that the quantitative PCR device accurately measured C. trachomatis DNA. However, this may not be justified. It was unexpected that the distribution of ΔCq values conforms to normality more closely than the distribution of inferred volumes transferred to the urine surrogate. The volume transferred is the physical phenomenon expected to be normally distributed, while the ΔCq is the product of a log transformation. This suggests that variation in the ∆Cq values may encompass instrumental artefacts as well as variation in the volumes of amounts of EB suspensions transferred.
We observed that participant identity and EB transfer method are correlated with EBs transferred. We do not regard speculation regarding mechanisms as useful, as the effects are small and do not change translational implications. Also, we regard the higher ΔCq observed with the 10−2 dilutions of EB as not mechanistically meaningful. Phenomena such as a limited number of high affinity EB binding sites on the skin could reduce EB transfer below what would be expected when the EB density is low. However, there was no significant difference between the ΔCq values arising from undiluted suspension and most dilute (10−4diluted) suspensions, so no evidence for loss of EBs at low EB densities. The similar results from three C. trachomatis isolates suggest that our results are generalisable to the species as a whole.
This study has directly shown that contact between contaminated fingers and urine specimens has potential to contaminate the specimen and underpin a positive C. trachomatis test. This provides justification for standardised procedures for collection of urine specimens from children, encompassing controlled environments, with clear guidance, and if possible, close supervision by clinical staff. In the absence of other measures such as wearing gloves, hand decontamination has been shown to greatly reduce the potential for specimen contamination. There must be clear instruction on hand washing and avoidance of contact with the urine specimen and interior of the container to all involved in collecting a child’s urine sample for STI testing. Regardless, a positive STI test in a child remains a strong indicator of child sexual assault necessitating further investigation by police, statutory child protection agencies and relevant forensic medical teams.
More broadly, this study adds to accumulating knowledge regarding mechanisms that could contaminate specimens, and so give rise to spurious STI positive results. Previous studies have focused on the potential for STI nucleic acid on clinic or laboratory surfaces to find its way into specimens.4–7 The emerging picture is that such surface contamination is commonly found but rarely leads to STI positive specimens. The difficulty of proving a negative, plus the demonstration by Andersson and coworkers7 of a non-zero frequency of specimen contamination, justifies measures to control the environment and procedure for STI specimen collection and emphasises the benefits of specimen collection under controlled conditions, particularly from young children. This current study adds to the imperative for this, by showing that potential for contact between specimen and uncovered skin should be avoided. In general, guidelines for urine specimens for STI testing simply specify ‘first catch urine’ and do not specify specific precautions for avoidance of urine contamination. In the NT, poster material available to medical practitioners specifies that patients ‘wash your hands with soap’.14 This is not children specific, and the extent that this procedure is followed is unknown. The absence of such precautions against contamination of specimens from young children could potentially be significant in the judicial system. Interestingly, guidelines for clean catch urine sampling, which is not used in STI testing, do specify avoidance of contact between specimens and fingers for example.15 Determining the most effective and culturally appropriate means of obtaining UGT specimens from young children while avoiding contamination risks could be a useful area for future research.
Footnotes
Contributors: PMG: Contributed to study conception, contributed to study design and formulation of detailed experimental procedure, carried out practical work, analysed data, wrote manuscript. RAL: Contributed to study design, contributed to study design and formulation of detailed experimental procedure, carried out practical work, analysed data. JW: Contributed to formulating detailed experimental procedure, carried out practical work, analysed data, commented on manuscript. GM: Carried out practical work, analysed data, commented on manuscript. SP: Carried out practical work, analysed data, commented on manuscript. SNT: Contributed to study conception, contributed to study design, provided clinical data. SMG: Contributed to study conception, contributed to study design, commented on manuscript. LM: Identified problem that this study addressed, contributed to study design, commented on manuscript. GS: Contributed to study design, commented on manuscript. SYCT: Contributed to study conception, contributed to study design, commented on manuscript. DCH: Contributed to study design and formulation of detailed experimental procedure, carried out practical work, analysed data, commented on manuscript. PA: Contributed to study conception, contributed to study design and formulation of detailed experimental procedure, carried out practical work, analysed data, commented on manuscript.
Funding: This study was funded by Australian National Health and Medical Research Council project grant 1060768. SYCT was supported by an Australian National Health and Medical Research Council Career Development Fellowship 1065736.
Competing interests: None declared.
Patient consent: Obtained.
Ethics approval: This study was approved by the Human Research Ethics Committee of the Northern Territory Department of Health and the Menzies School of Health Research, under ethical clearance number 2011-1673.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: All data are included in the publication itself.
Correction notice: This paper has been amended since it was published Online First. Owing to a scripting error, some of the publisher names in the references were replaced with ’BMJ Publishing Group'. This only affected the full text version, not the PDF. We have since corrected these errors and the correct publishers have been inserted into the references.
References
- 1. The Kirby Institute. HIV, viral hepatitis and sexually transmissible infections in Australia. Annual Surveillance Report 2015. Australia, Sydney NSW: The Kirby Institute UNSW, 2052. [Google Scholar]
- 2. Wild R, Anderson P. Ampe Akelyernemane Meke Mekarle, ’Little Children Are Sacred' Report of the board of enquiry into the protection of Aboriginal children from sexual abuse. 2007. http://www.inquirysaac.nt.gov.au/pdf/bipacsa_final_report.pdf
- 3. Brown A, Brown NJ. The Northern Territory intervention: voices from the centre of the fringe. Med J Aust 2007;187:621–3. [DOI] [PubMed] [Google Scholar]
- 4. Chan SY, Jose S, King R, et al. How likely is environmental or patient cross-contamination of Chlamydia trachomatis DNA to lead to false positive results in patients attending our clinic? Sex Transm Infect 2013;89:105–7. 10.1136/sextrans-2012-050667 [DOI] [PubMed] [Google Scholar]
- 5. Lewis N, Dube G, Carter C, et al. Chlamydia and gonorrhoea contamination of clinic surfaces. Sex Transm Infect 2012;88:418–21. 10.1136/sextrans-2012-050543 [DOI] [PubMed] [Google Scholar]
- 6. Meader E, Waters J, Sillis M. Chlamydia trachomatis RNA in the environment: is there potential for false-positive nucleic acid amplification test results? Sex Transm Infect 2008;84:107–10. 10.1136/sti.2007.027862 [DOI] [PubMed] [Google Scholar]
- 7. Andersson P, Tong SY, Lilliebridge RA, et al. Multisite direct determination of the potential for environmental contamination of urine samples used for diagnosis of sexually transmitted infections. J Pediatric Infect Dis Soc 2014;3:189–96. 10.1093/jpids/pit085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giffard PM, Brenner NC, Tabrizi SN, et al. Chlamydia trachomatis genotypes in a cross-sectional study of urogenital samples from remote Northern and Central Australia. BMJ Open 2016;6:e009624 10.1136/bmjopen-2015-009624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Giffard PM, Singh G, Garland SM. What does Chlamydia trachomatis detection in a urogenital specimen from a young child mean? Sex Transm Infect 2016:sextrans-2015-052473 10.1136/sextrans-2015-052473 [DOI] [PubMed] [Google Scholar]
- 10. Bowie WR, Caldwell HD, Jones RP, et al. Proceedings of the Seventh International Symposium on Human Chlamydial Infections. International Symposium on Human Chlamydial Infections; 1990; Harrison Hot Springs. Press Syndicate of the University of Cambridge. [Google Scholar]
- 11. Oriel D, Ridgway GL, Schachter J, et al. Proceedings of the Sixth International Symposium on Human Chlamydial Infections. International Symposium on Human Chlamydial Infections; 1986; Sanderstead, Surrey. Press Syndicate of the University of Cambridge. [Google Scholar]
- 12. Andersson P, Harris SR, Seth Smith HM, et al. Chlamydia trachomatis from Australian aboriginal people with Trachoma are polyphyletic composed of multiple distinctive lineages. Nat Commun 2016;7:10688 10.1038/ncomms10688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martino PD, Fursy R, Bret L, et al. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can J Microbiol 2003;49:443–9. 10.1139/w03-056 [DOI] [PubMed] [Google Scholar]
- 14. Anonymous. Specimen collection posters. 2011. http://digitallibrary.health.nt.gov.au/prodjspui/handle/10137/531
- 15. Kirkwood J. Clean catch urine sample and culture. 2016. http://www.healthline.com/health/urine-culture-clean-catch#overview1
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
sextrans-2016-053081supp001.pdf (295.3KB, pdf)


