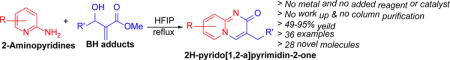
Abstract
Domino strategy has been used for the synthesis of 2H-pyrido[1,2-a]pyrimidin-2-ones. Four sequential reactions: aza-Michael addition, water elimination, intramolecular acyl substitution, and [1,3]-H shift were observed in this domino protocol. Hexafluoroisopropanol is used as a promotor and recyclable solvent in this cascade process. Availability of inexpensive 2-aminopyridines and wide variety of Michael acceptors such as commercially available acrylates and unactivated Baylis-Hillman adducts makes this methodology a huge reservoir of novel fused N-heterocycles as bioactive and potential therapeutic agents. The reaction mechanism has been proposed and rationalized by density functional theory calculation. Products are obtained up to 95% yield.
Keywords: Michael addition, domino reaction, pyridopyrimidinone, mechanism, heterocycles
Graphical Abstract
1. INTRODUCTION
Fused N-heterocycles are among the most widely used chemicals owing to its medicinal,1 agrochemical,2 and material properties.3, 4 The 2H-pyridopyrimidinone scaffold is an important class of nitrogen heterocycles. Many synthetic pyridopyrimidinones have occupied privileged position in drug development due to their unprecedented biological activities. This scaffold is an integral part of marketed drugs for the treatment of schizophrenia5 and asthma.6 Many of these molecules have been reported as anticancer,7 antimalarial,8, 9 and anticoccidial10 agents. Some of the them have also been recognized as aldose reductase inhibitors,11 estrogen-related receptors (ERRs) agonists,12 G protein signaling (RGS) protein regulators,13 HIV integrase inhibitors,14 efflux pump inhibitors,15 etc.16 Pyridopyrimidinone scaffold is also a key constituent of numerous natural products possessing wide range of biological activities including antitumor, anti-influenza, oxidative burst inhibitory, lipid droplet synthesis inhibition, and anti-obesity properties (Figure 1).17–20
Figure 1.
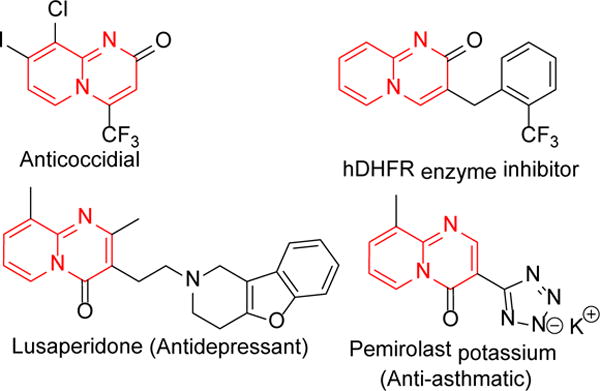
Some representative pyridopyrimidinone bearing valuable compounds
Because of the great value of these heterocycles, many groups have reported the synthesis of 4H-pyrido[1,2-a]pyrimidin-4-one scaffold but the synthesis of its regioisomer, 2H-pyrido[1,2-a]pyrimidin-2-one is scarce. Basavaiah et al. has used Baylis-Hillman (BH) derived acetates and 2-aminopyridines to synthesize these molecules in one-pot protocol.21 Microwave assisted neat reaction has also been reported to synthesize 2H-pyrido[1,2-a]pyrimidin-2-ones using the same starting materials.22 Michael addition of 2-aminopyridines on acetylene-derived electrophiles have been used to synthesize these fused pyridopyrimidinones by many groups.23, 24 Chichetti et al. have reported the solvent-free microwave synthesis of novel 6-hydroxypyrimidin-4(1H)-one derivatives using arylmalonates.25 Su et al. have reported the synthesis of these heterocycles by using Vilsmeier reagent.26 Above methods require, activated Baylis-Hillman adducts,21, 27 microwave assistance,22, 25 and added reagents.23, 24, 26 In our effort to develop domino methodologies to synthesize novel heterocycles,28, 29 herein, we present hexafluoroisopropanol (HFIP) mediated sustainable design, reagent or catalyst free, and negligible by product formation for the synthesis of 2H-pyrido[1,2-a]pyrimidin-2-ones by using 2-aminopyridines and unactivated BH-adducts.
Hexafluoroisopropanol (HFIP) is one of the fluorinated solvents with unique properties such as high hydrogen bonding donor ability, low nucleophilicity, high ionizing power, and ability to solvate water. HFIP is used to promote a wide range of reactions and this promoter and solvent helps to avoid metal catalyst and added reagents. HFIP mediated products are easily isolated in high yield and the solvent is easily recovered from the reaction medium and reused. Thus, HFIP is used as an environmentally benign solvent as it could be recovered and recycled and also most of the reactions don’t need work-up and tidy purification.30 Due to unique properties and environmentally benign nature, HFIP has been used as solvent and promoter for a wide range of reactions.31–33 We have found HFIP as the best solvent and promoter for the synthesis of 2,3-dihydro-4H-pyrido[1,2-a]pyrimidin-4-ones and thiazole derivatives.28, 29 Keep in mind the benign nature of HFIP, we have used this solvent as a promoter as well as reaction medium for this methodology.
2. Results and Discussion
Conjugate addition of 2-aminopyridines to a Michael acceptor is well known in literature.34, 35 Michael addition followed by intramolecular acyl substitution to form 4H-pyrido[1,2-a]pyrimidin-4-one has been reported by us and others.28, 36 Success of this domino methodology for the synthesis of 4H-pyrido[1,2-a]pyrimidin-4-one turned our attention to Baylis-Hillman adducts as the Michael acceptors. Baylis-Hillman adducts are the huge reservoir of Michael acceptors for numerous reactions.37 We started our study with the reaction of 2-aminopyridine with one of the simplest Baylis-Hillman adducts, methyl 2-(hydroxymethyl) acrylate. Refluxing of the reaction mixture for 12 hours showed the complete disappearance of the starting materials. Adding 5 mL acetone precipitated the product, which on filtration afforded the pure product (12) in 90% yield (Scheme 1).
Scheme 1.
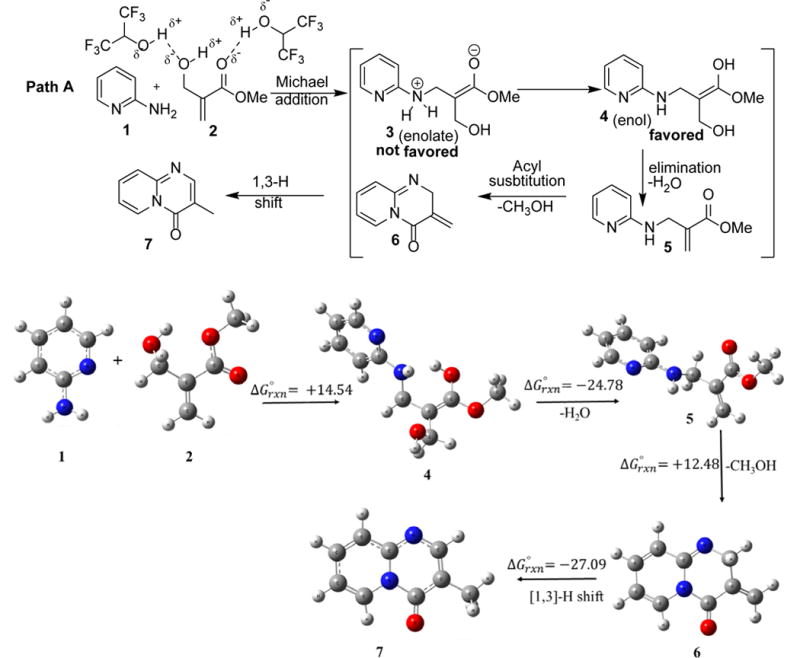
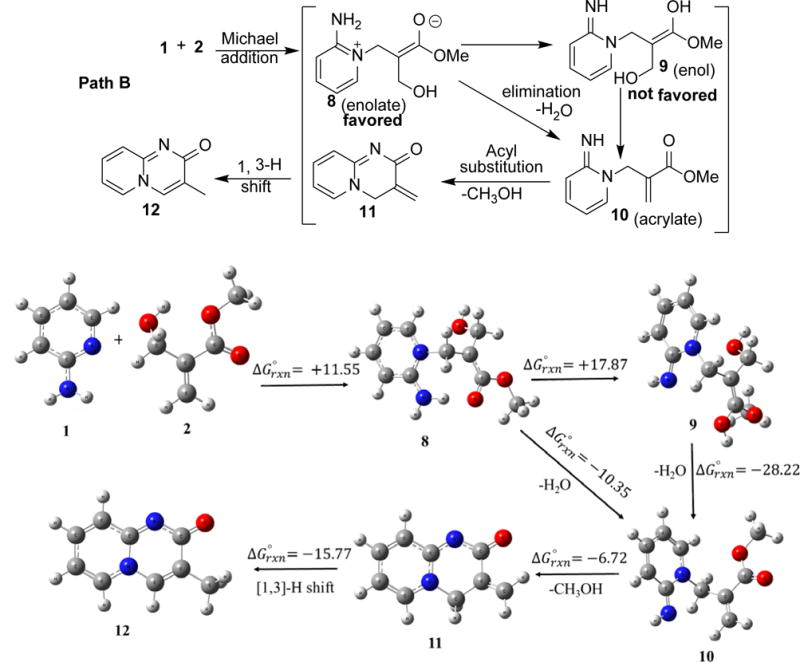
Standard free energies of reaction for Path A and Path B obtained using at M06-2X/6-31+ G(d,p) + PCM (solvent = 2,2,2-trifluoroethanol) level of theory. Note: Reaction solvent (HFIP) is not listed in the Gaussian program list. This reaction also happens in trifluoroethanol in moderate yield.
Successful product formation led us to explore the mechanism of the reaction 2-Aminopyridine (1) can exist in its isomeric form as pyridine-2(1H)-imine (Figure 2). We carried out computational studies to find the feasibility of this transformation. Standard Gibbs free energy for this conversion is +13.0 kcal/mol in neat condition and it increases to +23.5 kcal/mol in HFIP solvent. Hence, the conversion of 2-aminopyridine (1) to pyridine-2(1H)-imine is unlikely. Therefore, 2-aminopyridine (1) is the sole reactive nucleophile in a reaction for this methodology. HFIP is a strong hydrogen bond donor30 and this highly fluorinated alcohol makes strong hydrogen bonding with the oxygens of ester and hydroxy groups of the Baylis-Hillman adduct (2). Hydrogen bonding of HFIP with the BH-adduct makes the β-carbon of the Michael acceptor (2) more electrophilic for Michael addition. Activation of the BH-adducts by this fluorinated solvent is the key factor for the facile reactions of this methodology.
Figure 2.
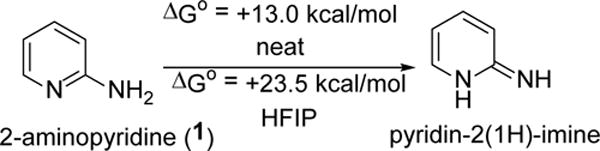
Conversion of 2-aminopyridine to pyridin-2(1H)-imine
There are two possible nucleophilic atoms, the amino nitrogen and the ring nitrogen, of 2-aminopyridine (1) for the conjugate addition to a Michael acceptor, which leads to two different paths for the reaction (Scheme 1). Path A involves the Michael addition of amino nitrogen of 2-aminopyridine, which leads to the formation of 4H-pyrido[1,2-a]pyrimidin-4-one after the intramolecular acyl substitution followed by 1,3-hydrogen shift. Path B involves the Michael addition of pyridine nitrogen of 2-aminopyridine, which would lead to the formation of 2H-pyrido[1,2-a]pyrimidin-2-one by the same sequence of reactions. To the best of our knowledge, the rationale for two different products has not been explored. We computed the feasibility of the both favored and unfavored pathways by using hybrid-density functional method (M06-2X)/6-31+G(d,p)38 + PCM as implement in Gaussian 09 suite of programs.39 The relative calculated Gibbs energy (in kcal/mol) for Path A and Path B are shown in Scheme 1.
For Path A, the calculation showed that Michael addition to the BH adduct leads to the formation of enol (4) via zwitterionic enolate intermediate (3), which undergoes water elimination to form imino acrylate derivative (5). Water elimination of enol (4) to form imino acrylate (5) is highly favorable and exergonic (ΔG = -24.78 kcal/mol). Intramolecular acyl substitution to form exomethylene pyridopyrimidinone (6) is not energetically favored and endergonic (ΔG = +12.48 kcal/mol). The final molecule (7) could form by a [1,3]-H shift. For Path B, Michael addition of 2-aminopyridine (1) on BH adduct (2) is endergonic (ΔG = +11.55 kcal/mol) but more favorable than Path A by ~3 kcal/mol. This favorable energy difference favors the product formation by this pathway. Our calculations suggests that zwitterionic enolate (8) is favored and it does not convert into enol form (9). This can be explained by taking aromatic sextet into consideration as suggested by Sola et al.40 Zwitterionic enolate (8) has the sextet and sextet is not possible in its enol form (9). To the best our knowledge, this is a rare example in which enolate is favored over enol form in an acidic condition. Intermediate (8) directly undergoes water elimination to form the acrylate derivative (10). Although, water elimination is exergonic (ΔG = -10.35 kcal/mol) process to form (10) but ΔG value is much less than the similar step (4 to 5) of Path A. Water elimination causes the loss of sextet in Path B, on the other hand, water elimination in Path A does not affect the sextet of the pyridine moiety. Intramolecular acyl substitution leads to the formation of exomethylene pyridopyrimidinone (11) and it is an exergonic (ΔG = -6.72 kcal/mol) process. Similar step (5 to 6) in Path A is highly endergonic (ΔG = +12.58 kcal/mol). The energy difference (~20 kcal/mol) of intramolecular acyl substitution between Path A and Path B is due to the losing of sextet in Path A. Conversely, the electronic structure of the pyridine ring is not affected in Path B. Energetically favorable (ΔG = -15.77 kcal/mol) [1,3]-H shift leads to the formation of final molecule (12) in a domino process.
After the successful product (12) formation of 2-aminopyridine with BH-adduct, we carried out the reaction of methyl substituted 2-aminopyridines under the same reaction condition (Scheme 2). Corresponding products formed smoothly in an average of 72% yield. All the methyl substituted 2-aminopyridines reacted with the Baylis-Hillman derived Michael acceptor to give the corresponding products (13, 14, & 15). Dimethyl substituted product (15) has been synthesized in multi-gram scale without affecting the yield and purity. We also tried the reaction of 2-amino-4-ethyl pyridine with BH-adduct and the product (16) formed in 60% yield. Success of the electron donating substituents (alkyl) prompted us to try moderately electron withdrawing groups such as halogens (F, Cl, Br, & I). Fluoro substituted 2-aminopyridines reacted with the Michael acceptor to form the products (17 & 18) in very good yield. The reaction of 2-amino-5-chloropyridine with the electrophile under the refluxing condition of HFIP afforded the corresponding pyridopyrimidinone derivative (19) in 74% yield. 2-Amino-4-bromopyridine reacted smoothly with the Michael acceptor to give the corresponding product (20). Iodo substituted product (21) was formed from the corresponding 2-aminopyridine in 85% yield. Stronger electron withdrawing groups such as trifluoro methyl (CF3) did not hamper this domino process. Both 3 and 5 trifluoro methyl 2-aminopyridines also reacted smoothly to form the corresponding products (22 & 23) in 52% and 59% yields respectively. Successful product formation of monosubstituted 2-aminopyridines led us to try the disubstituted 2-aminopyridines to get densely functionalized pyridopyrimidinones. Treatment of bromo-methyl-2-aminopyridines with the Baylis-Hillman derived Michael acceptor afforded the products formation in good yields (24, 25 & 26). Halogen substituted pyridopyrimidinones, particularly bromo and iodo derivatives (20, 21, 24, & 26), could be the excellent precursors for novel scaffolds as potential bioactive agents. Derivatization of these molecules can be done with the metal (Pd, Cu, etc.) catalyzed coupling reactions.
Scheme 2.
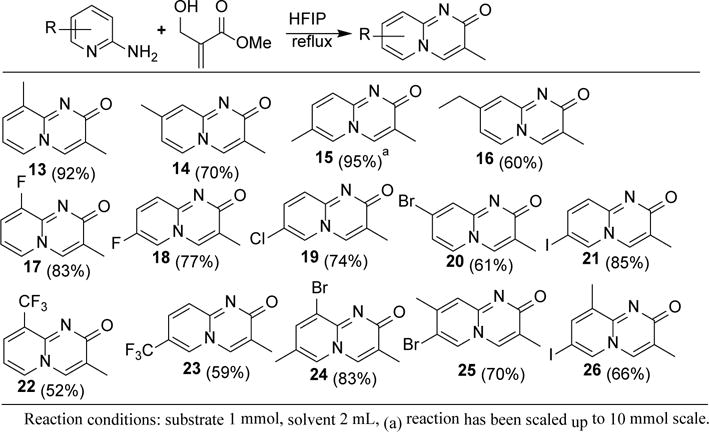
Reaction of substituted 2-aminopyridines with α-hydroxymethyl acrylate
After the successful derivatization of α-hydroxymethyl acrylate, we tried the domino reaction of benzaldehyde-derived Baylis-Hillman adduct (methyl 2-[hydroxy(phenyl)methyl] acrylate) with 2-aminopyridines to expand the scope of this methodology (Scheme 3). The reaction worked expectantly and the product (27) was obtained by filtration in 93% yield. Methyl substituted aminopyridines reacted efficiently with this electrophile to give the desired products (28 & 29). 3-Fluoro and 5-fluoro derived aminopyridines reacted with electrophile to give the corresponding products (30 & 31) in 67% and 86% yields respectively. Reaction of the 2-amino-5-chloropyridine with the benzaldehyde-derived Baylis-Hillman adduct gave the desired pyridopyrimidinone (32) in 76% yield. Bromo substituted products (33 & 34) were obtained by the reaction of 2-aminopyridines with Michael acceptors in expected average yield. 5-Iodo substituted nucleophile reacted with Baylis-Hillman adduct to give the analogous product (35) in 76% yield. Withdrawing group such as CF3 did not hamper the product (36 & 37) formation. Polysubstituted nucleophiles also gave the desired products (38 & 39) in 72% and 68% yield respectively. These small molecules have the potential to be further functionalized to generate large number of novel scaffolds.
Scheme 3.
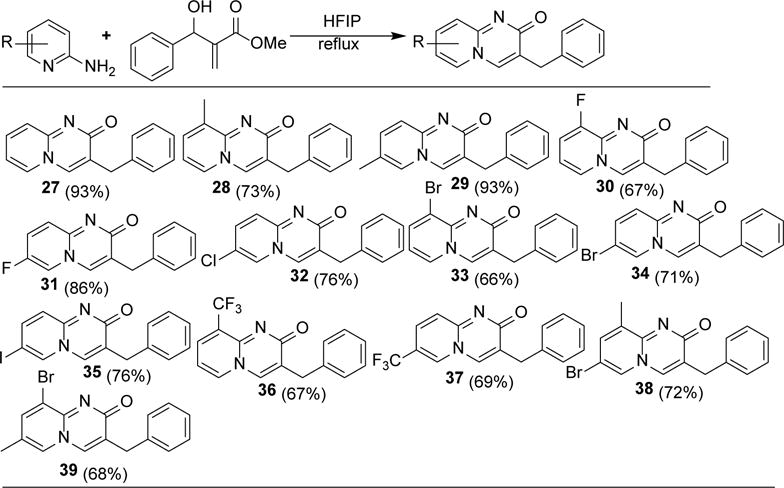
Synthesis of benzyl substituted pyridopyrimidinones
To study the robustness of this methodology, we tried 4-nitro substituted Baylis-Hillman adduct as an electrophile (Scheme 4). The methodology is equally effective for this substrate. Methyl substituted 2-aminopyridines reacted to give the corresponding products (40 & 41) in an average of 70% yield. Moderately electron withdrawing groups, halogens, gave the corresponding products (42, 43, & 44) and pure products are obtained by simple filtration.
Scheme 4.
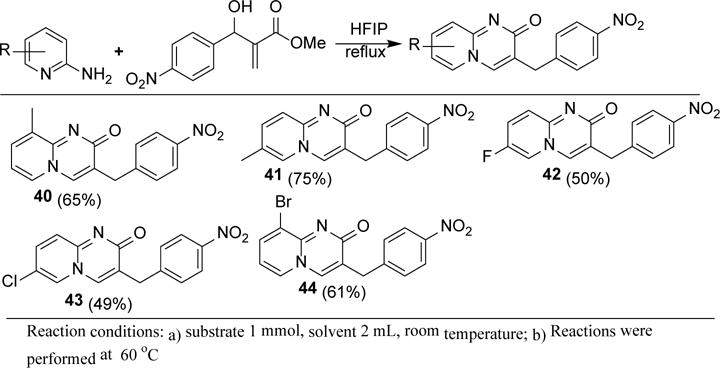
Synthesis of dihydropyrido-pyrimidinones using aza-Michael-cyclization strategy
Last but not least, we tested the scope of this methodology to synthesize multiple ring system by reacting 2-aminoquinoline with the BH-adduct. The reaction underwent smoothly to give the expected product (45) in 54% yield (Scheme 5).
Scheme 5.
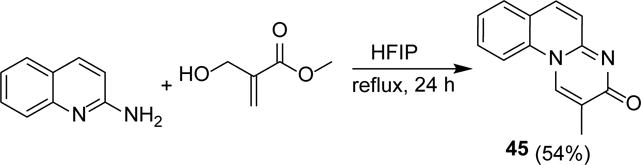
Synthesis of tricyclic pyrido-pyrimidinone derivative
Several attempts to grow crystals for X-ray single crystal diffraction failed. We confirmed the structures (13) by HMBC experiment (Figure 3).
Figure 3.
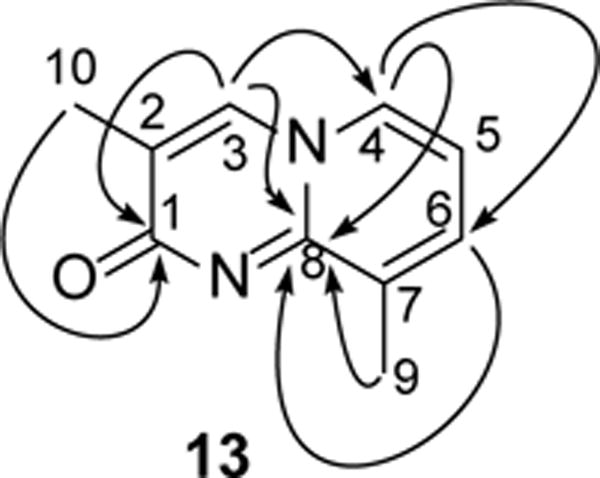
Key HMBC Correlations (H→C) of 13
The carbon signal at δ 169.5 (C-1) shows correlation with proton signals at δ 7.68 (H–3) and δ 2.20 (H-10) confirming the 1,3 relationships between these two carbon center. The carbon signals at δ 134.5 (C-3) and δ 129.9 (C-4) has correlations with proton signals at δ 7.50 (H-4) and δ (H-3) respectively, this mutual correlation confirms that these two carbons are spaced two bonds away from each other. Correlation of carbon signal at δ 151.2 (C-8) with proton signals at δ 7.68 (H-3), δ 7.50 (H-4) confirms that the C-8 carbon is the bridging carbon between the two rings. Further correlation of proton signal at δ 2.49 (H-9) with carbon signal at δ 151.2 (C-8) along with other correlation as mentioned in the table clearly confirms the structure of compound 13 (Table 1).
Table 1.
Correlation for 1H (400 MHz), 13C (100 MHz) and HMBC correlation data of compound 13 in CDCl3.
| Position | δH (J in Hz) | δC | HMBC (C →H) |
|---|---|---|---|
| 1 | – | 169.5 | H-10, H-3 |
| 2 | – | 126.6 | – |
| 3 | 7.68 (s) | 134.5 | H-4, H-10 |
| 4 | 7.50 (d, 8) | 129.9 | H-3, H-6 |
| 5 | 6.74 (t, 8) | 112.6 | H-4 |
| 6 | 7.39 (d, 8) | 133.5 | H-4, H-9 |
| 7 | – | 133.3 | H-5 |
| 8 | – | 151.2 | H-9, H-6, H-4 |
| 9 | 2.49 (s) | 18.2 | H-6 |
| 10 | 2.20 (s) | 14.9 | H-3, H-20 |
For medicinal chemistry research, further derivatizations of these molecules are required to enhance or optimize their pharmacological properties. So we tried the reaction of some molecules (15) in multi-gram scale. The scaling up of the reaction to multi-gram scale does not require any special modification and products were isolated in pure form without any difficulty. Additionally, the reaction solvent was easily recovered, and recycled by simple distillation to achieve the best possible Environmental Factor (E).41
3. Conclusion
In this article, we designed and developed a strategy for the synthesis of pyridopyrimidinones and their derivatives. We have also proposed the mechanism and rationalized by density functional theory calculation. The use of HFIP facilitates the efficient Michael addition, cyclization, elimination, and rearrangement in a domino process. Ease of synthesis and scalability to multigram scale are key factors to utilize this methodology for further scope. Variability of both electrophiles (Baylis-Hillman adducts) and nucleophiles (2-aminopyridines) will be help to generate a library of new scaffolds required for the drug discovery and biological study. The developed reaction is clean and products could be isolated by simple filtration. Bromo and iodo substituted products will be the starting point for the metal catalyzed coupling reaction to generate a large number of new scaffolds as potential bioactive compounds. These findings will be reported in due course.
4. Experimental
4.1. General information
All commercial chemicals and solvents are reagent grade and were used without further treatment unless otherwise noted. 1H NMR spectra were obtained with a Varian Mercury-300MHz with TMS as internal standard. 13C NMR spectra were obtained with a Varian Mercury-75MHz with TMS as internal standard. The ESI-FTMS Mass spectra were recorded Bruker ApexII-FTMS system. Low resolution mass spectra were recorded in Shimadzu QQQ instrument.
4.2. General procedure for the synthesis of pyridopyrimidinones
A mixture of 2-aminopyridine derivative (1.0 mmol) and Baylis-Hillman adduct (1.1 mmol) in HFIP (2 mL) was refluxed overnight to get the solid precipitate. Filtration gave the pure product without further purification. If the solid product did not form then HFIP was evaporated and ethyl acetate or methanol was added to get the solid precipitate, which on filtration afforded the pure compounds.
3-Methyl-2H-pyrido[1,2-a]pyrimidin-2-one (12)
This molecule is known but no data have been reported.26 Yield 0.144 g (90%), white, 1H NMR (300 MHz, DMSO-d6): δ 8.31 (s, 1H), 8.13 (d, J = 6.5 Hz, 1H), 7.65 (t, J = 7.5 Hz, 1H), 7.15 (d, J = 9.0 Hz, 1H), 6.95 (t, J = 6.7 Hz, 1H), 1.96 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 168.4,151.1, 136.5, 136.0, 134.0, 124.8, 123.0, 113.0, 14.7. ESI Mass (m/z) for C9H8N2O: [M+H]calc = 161.1, [M+H]expt = 161.1.
3,9-Dimethyl-2H-pyrido[1,2-a]pyrimidin-2-one (13)
Yield 0.160g (92%), yellow, 1H NMR (300 MHz, DMSO-d6): δ 8.28 (s, 1H), 7.99 (d, J = 6.7 Hz, 1H), 7.52 (d, J = 6.8 Hz, 1H), 6.85 (t, J = 6.8 Hz, 1H), 2.26 (s, 3H), 1.95 (s, 3H), 13C NMR (75 MHz, DMSO-d6 + CDCl3): δ = 168.4, 150.7, 136.3, 134.4, 131.9, 131.0, 124.6, 112.6, 18.0, 14.6. HRMS (ESI-FTMS Mass (m/z): C9H11N2O: [M+H]calc = 175.0866, [M+H]expt = 175.0865.
3,8-Dimethyl-2H-pyrido[1,2-a]pyrimidin-2-one (14)
Yield 0.121g (70)%, brownish, 1H NMR (300 MHz, DMSO-d6): δ 8.24 (s, 1H), 8.03 (d, J = 6.9 Hz, 1H), 6.95 (s, 1H), 6.82 (d, J = 5.5 Hz, 1H), 2.32 (s, 3H), 1.94 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 168.5, 151.1, 147.8, 135.6, 133.2, 124.2, 120.8, 115.4, 21.2, 14.7. HRMS (ESI-FTMS Mass (m/z): C9H11N2O: [M+H]calc = 175.0866, [M+H]expt = 175.0863.
3,7-Dimethyl-2H-pyrido[1,2-a]pyrimidin-2-one (15)
Yield 0.165g (95%), yellowish, 1H NMR (300 MHz, DMSO-d6): δ8.20 (s, 1H), 7.96 (s, 1H), 7.55 (d, J = 9.1 Hz, 1H), 7.10 (d, J = 9.1 Hz, 1H), 2.21 (s, 3H), 2.06 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 168.4, 150.1, 139.2, 135.7, 131.1, 124.7, 122.6, 122.2, 17.3, 14.8. HRMS (ESI-FTMS Mass (m/z): C9H11N2O: [M+H]calc = 175.0866, [M+H]expt = 175.0861.
8-Ethyl-3-methyl-2H-pyrido[1,2-a]pyrimidin-2-one (16)
Yield 0.112g (60%), brownish, 1H NMR (300 MHz, CDCl3): d 7.70 (s, 1H), 7.59 (d, J = 7.0 Hz, 1H), 7.08 (s, 1H), 6.67 (dd, J = 1.8, 7.0 Hz, 1H), 2.67 (q, J = 7.5 Hz, 2H), 2.16 (s, 3H), 1.27 (t, J = 7.5 Hz, 3H), 13C NMR (75 MHz, CDCl3): δ = 169.3, 152.5, 151.4, 133.9, 131.2, 126.1, 120.8, 114.6, 28.3, 14.7, 13.2. HRMS (ESI-FTMS Mass (m/z): C11H13N2O: [M+H]calc = 189.1022, [M+H]expt = 189.1022.
9-Fluoro-3-methyl-2H-pyrido[1,2-a]pyrimidin-2-one (17)
Yield 0.147g (83%), light brown, 1H NMR (300 MHz, DMSO-d6 + TFA-D): δ 8.38 (s, 1H), 7.99 (d, J = 6.9 Hz, 1H), 7.61 (t, J = 7.7 Hz, 1H), 6.95–6.38 (m, 1H), 1.98 (s, 3H), 13C NMR (75 MHz, DMSO-d6 + TFA-D): δ = 161.7, 148.7 (d, 1J = 251.8 Hz), 141.2 (d, 2J = 25.1 Hz), 139.1, 132.4 (d, 4J = 5.0 Hz), 127.0, 125.2 (d, 2J = 15.9 Hz), 118.0 (d, 2J = 6.7 Hz), 13.3. HRMS (ESI-FTMS Mass (m/z): C9H8FN2O: [M+H]calc = 179.0615, [M+H]expt = 179.0614.
7-Fluoro-3-methyl-2H-pyrido[1,2-a]pyrimidin-2-one (18)
Yield 0.137g (77%), brownish, 1H NMR (300 MHz, DMSO-d6 + CDCl3): δ 8.43 (s, 1H), 8.22 (s, 1H), 7.82 (t, J = 7.5 Hz, 1H), 7.22 (dd, J = 5.5 Hz, 1H), 1.97 (s, 3H), 13C NMR (75 MHz, DMSO-d6 + CDCl3): δ = 168.2, 151.3 (d, 1J = 234.9 Hz), 149.4, 136.0, 128.9 (d, 2J = 24.6 Hz), 125.2, 124.7 (d, 3J = 7.9 Hz), 120.5 (d, 2J = 40.9 Hz), 14.7. HRMS (ESI-FTMS Mass (m/z): C9H8FN2O: [M+H]calc = 179.0615, [M+H]expt = 179.0612.
7-Chloro-3-methyl-2H-pyrido[1,2-a]pyrimidin-2-one (19)
Yield 0.144g (74%), white, 1H NMR (300 MHz, DMSO-d6): δ 8.45 (d, J = 2.2 Hz, 1H), 8.22 (s, 1H), 7.72 (dd, J = 2.3, 9.6 Hz, 1H), 7.17 (d, J = 9.6 Hz, 1H), 1.96 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 168.2, 149.8, 137.0, 135.6, 131.8, 125.0, 124.4, 118.7, 14.7. HRMS (ESI-FTMS Mass (m/z): C9H8ClN2O [M+H]calc = 195.0320, [M+H]expt = 195.0322.
8-Bromo-3-methyl-2H-pyrido[1,2-a]pyrimidin-2-one (20)
Yield 0.145g (61%), brownish, 1H NMR (300 MHz, DMSO-d6 + TFA-D): δ 8.30 (s, 1H), 8.09 (d, J = 7.3 Hz, 1H), 7.47 (s, 1H), 7.20 (d, J = 6.9 Hz, 1H), 1.95 (s, 3H), 13C NMR (75 MHz, DMSO-d6 + TFA-d): δ = 159.1, 147.1, 138.8, 138.2, 137.1, 127.4, 122.5, 118.1, 13.4. HRMS (ESI-FTMS Mass (m/z): calcd for C8H8BrN2O [M+H]calc = 240.9794, [M+H]expt = 240.9797.
7-Iodo-3-methyl-2H-pyrido[1,2-a]pyrimidin-2-one (21)
Yield 0.243g (85%), white, 1H NMR (300 MHz, DMSO-d6): δ 8.50 (d, J = 1.7 Hz, 1H), 8.19 (s, 1H), 7.81 (dd, J = 1.8, 9.4 Hz, 1H), 6.96 (d, J = 9.4 Hz, 1H), 1.73 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 168.2, 149.8, 143.4, 138.1, 135.2, 124.8, 124.3, 75.7, 14.7. HRMS (ESI-FTMS Mass (m/z): C9H8IN2O [M+H]calc = 286.9676, [M+H]expt = 286.9678.
3-Methyl-9-(trifluoromethyl)-2H-pyrido[1,2-a]pyrimidin-2-one (22)
Yield 0.118g (52%), white, 1H NMR (300 MHz, DMSO-d6): δ 8.38 (s, 1H), 8.35 (d, J = 6.8 Hz, 1H), 8.16 (d, J = 7.2 Hz, 1H), 7.04 (t, J = 7.0 Hz, 1H), 1.98 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 167.4, 147.2, 138.4, 136.6, 136.5 (q, 3J = 5.4 Hz), 125.3, 122.8 (q, 1J = 270.3 Hz), 120.0 (q, 2J = 30.6 Hz), 111.0, 14.7. HRMS (ESI-FTMS Mass (m/z): C10H8F3N2O [M+H]calc = 229.0583, [M+H]expt = 229.0585.
3-Methyl-7-(trifluoromethyl)-2H-pyrido[1,2-a]pyrimidin-2-one (23)
Yield 0.134g (59%), white, 1H NMR (300 MHz, DMSO-d6 + TFA-D): δ 9.44 (s, 1H), 8.74 (s, 1H), 7.84 (dd, J = 1.7, 9.3 Hz, 1H), 7.80 (d, J = 9.3 Hz, 1H), 2.14 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 159.1, 148.8, 138.5, 138.3, 136.1 (q, 3J = 21.0), 128.3, 122.4 (q, 1J = 270.3 Hz), 119.9 (q, 2J = 36.4 Hz), 117.2, 13.5. HRMS (ESI-FTMS Mass (m/z): C10H8F3N2O [M+H]calc = 229.0583, [M+H]expt = 229.0585.
9-Bromo-3,7-dimethyl-2H-pyrido[1,2-a]pyrimidin-2-one (24)
Yield 0.210g (83%), brownish, 1H NMR (300 MHz, DMSO-d6): δ 8.22 (s, 1H), 8.07 (s, 1H), 8.00 (s, 1H), 2.20 (s, 3H), 1.97 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 168.3, 147.2, 141.8, 136.4, 131.4, 125.1, 122.2, 116.3, 17.0, 14.5. HRMS (ESI-FTMS Mass (m/z): C10H10BrN2O [M+H]calc = 254.9951, [M+H]expt = 254.9953.
7-Bromo-3,8-dimethyl-2H-pyrido[1,2-a]pyrimidin-2-one (25)
Yield 0.177g (70%), white, 1H NMR (300 MHz, DMSO-d6): δ 8.52 (s, 1H), 8.17 (s, 1H), 7.15 (s, 1H), 2.08 (s, 3H), 1.94 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 168.3, 150.2, 147.0, 134.9, 133.6, 124.5, 122.1, 109.7, 22.2, 14.8. HRMS (ESI-FTMS Mass (m/z): C10H10BrN2O [M+H]calc = 254.9951, [M+H]expt = 254.9953.
7-Iodo-3,9-dimethyl-2H-pyrido[1,2-a]pyrimidin-2-one (26)
Yield 0.198g (66%), yellowish, 1H NMR (300 MHz, DMSO-d6): δ 8.36 (s, 1H), 8.18 (s, 1H), 7.74 (s, 1H), 2.25 (s, 3H), 1.95 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 168.1, 149.5, 141.1, 136.0, 135.5, 132.7, 124.6, 75.7, 17.6, 14.7. HRMS (ESI-FTMS Mass (m/z): C10H10IN2O [M+H]calc = 300.9832, [M+H]expt = 300.9835.
3-Benzyl-2H-pyrido[1,2-a]pyrimidin-2-one (27)
Analytical data are in very good agreement with reported data.27 Yield 0.219g (93%), beige, 1H NMR (300 MHz, DMSO-d6): δ 8.22 (s, 1H), 8.16 (d, J = 6.7 Hz, 1H), 7.65 (t, J = 7.9 Hz, 1H), 7.30–7.20 (m, 5H), 7.15 (d, J = 6.1 Hz, 1H), 6.93 (t, J = 6.6 Hz, 1H), 3.72 (s, 2H), 13C NMR (75 MHz, DMSO-d6): δ = 167.6, 151.1, 139.2, 136.9, 136.7, 134.4, 129.3, 128.7, 127.9, 126.6, 122.9, 113.1, 34.1. HRMS (ESI-FTMS Mass (m/z): C15H13N2O [M+H]calc = 237.1022, [M+H]expt = 237.1025.
3-Benzyl-9-methyl-2H-pyrido[1,2-a]pyrimidin-2-one (28)
Analytical data are in very good agreement with reported data.27 Yield 0.182g (73%), brownish, 1H NMR (300 MHz, DMSO-d6): δ 8.22 (s, 1H), 8.04 (d, J = 6.6 Hz, 1H), 7.54 (d, J = 6.9 Hz, 1H), 7.28–7.19 (m, 5H), 6.85 (t, J = 6.8 Hz, 1H), 3.72 (s, 2H), 2.27 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 167.5, 150.7, 139.3, 137.2, 134.6, 132.3, 131.0, 129.3, 128.7, 127.7, 126.6, 112.6, 34.0, 18.1. HRMS (ESI-FTMS Mass (m/z): C16H15N2O [M+H]calc = 251.1179, [M+H]expt = 251.1181.
3-Benzyl-7-methyl-2H-pyrido[1,2-a]pyrimidin-2-one (29)
Yield 0.232g (93%), white, 1H NMR (300 MHz, DMSO-d6): δ 8.08 (s, 1H), 8.00 (s, 1H), 7.55 (d, J = 9.1 Hz, 1H), 7.30–7.28 (m, 4H), 7.29–7.19 (m, 1H), 7.10 (d, J = 9.2 Hz, 1H), 3.71 (s, 2H), 2.19 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 167.7, 150.1, 139.4, 139.1, 136.4, 131.6, 129.3, 128.7, 127.9, 126.6, 122.5, 34.3, 17.5. HRMS (ESI-FTMS Mass (m/z): C16H15N2O [M+H]calc = 251.1179, [M+H]expt = 251.1183.
3-Benzyl-9-fluoro-2H-pyrido[1,2-a]pyrimidin-2-one (30)
Yield 0.170g (67%), white, 1H NMR (300 MHz, DMSO-d6): δ 8.30 (s, 1H), 8.04 (d, J = 6.9 Hz, 1H), 7.61 (t, J = 8.7 Hz, 1H), 7.30–7.21 (m, 5H), 6.94–6.88 (m, 1H), 3.75 (s, 2H), 13C NMR (75 MHz, DMSO-d6): δ = 167.0, 150.9 (d, 1J = 248.4 Hz), 144.7 (d, 2J = 21.7 Hz), 138.9, 137.1, 130.7 (d, 4J = 5.4 Hz), 129.3, 128.7, 128.6, 126.7, 118.3 (d, 2J = 17.4 Hz), 111.2 (d, 3J = 7.0 Hz), 34.1. HRMS (ESI-FTMS Mass (m/z): C15H12FN2O [M+H]calc = 255.0928, [M+H]expt = 255.0932.
3-Benzyl-7-fluoro-2H-pyrido[1,2-a]pyrimidin-2-one (31)
Yield 0.218g (86%), white, 1H NMR (300 MHz, DMSO-d6 + TFA-D): δ 9.13 (s, 1H), 8.55 (s, 1H), 8.45–8.40 (m, 1H), 7.71 (dd, J = 4.8, 9.8 Hz, 1H), 7.32–7.30 (m, 3H), 7.27–7.23 (m, 2H), 3.89 (s, 2H), 13C NMR (75 MHz, DMSO-d6 + TFA-D): δ = 158.3, 154.1 (1J = 242.5 Hz), 145.5, 139.0, 136.8, 134.6 (2J = 23.5 Hz), 131.3, 129.3, 128.8, 127.2, 124.5 (3J = 7.9 Hz), 32.9. HRMS (ESI-FTMS Mass (m/z): C15H12FN2O [M+H]calc = 255.0928, [M+H]expt = 255.0932.
3-Benzyl-7-chloro-2H-pyrido[1,2-a]pyrimidin-2-one (32)
Analytical data are in very good agreement with reported data.27 Yield 0.205g (76%), light brown, 1H NMR (300 MHz, DMSO-d6): δ 8.51 (d, J = 2.1 Hz, 1H), 8.11 (s, 1H), 7.73 (dd, J = 2.2, 9.6 Hz, 1H), 7.30–7.16 (m, 6H), 3.72 (s, 2H), 13C NMR (75 MHz, DMSO-d6): δ = 167.5, 149.8, 138.8, 137.2, 136.5, 132.2, 129.4, 128.8, 128.1, 126.7, 124.3, 118.8, 34.1. HRMS (ESI-FTMS Mass (m/z): C15H12ClN2O [M+H]calc = 271.0633, [M+H]expt = 271.0635.
3-Benzyl-9-bromo-2H-pyrido[1,2-a]pyrimidin-2-one (33)
Yield 0.208g (66%), brownish, 1H NMR (300 MHz, DMSO-d6 + CDCl3): δ 8.26 (s, 1H), 8.22 (d, J = 6.8 Hz, 1H), 8.11 (d, J = 7.4 Hz, 1H), 7.29–7.27 (m, 4H), 7.24–7.19 (m, 1H), 6.84 (t, J = 7.4 Hz, 1H), 3.75 (s, 2H), 13C NMR (75 MHz, DMSO-d6 + CDCl3): δ = 167.5, 148.2, 139.4, 138.9, 137.6, 134.5, 129.3, 128.7, 128.2, 126.7, 116.7, 112.8, 33.8. HRMS (ESI-FTMS Mass (m/z): C15H12BrN2O [M+H]calc = 317.0108, [M+H]expt = 317.0109.
3-Benzyl-7-bromo-2H-pyrido[1,2-a]pyrimidin-2-one (34)
Analytical data are in very good agreement with reported data.27 Yield 0.223g (71%), white, 1H NMR (300 MHz, DMSO-d6 + CDCl3): δ 8.56 (s, 1H), 8.10 (s, 1H), 7.78 (d, J = 8.1 Hz, 1H), 7.30–7.24 (m, 5H), 7.11 (d, J = 9.6 Hz, 1H), 3.72 (s, 2H), 13C NMR (75 MHz, DMSO-d6 + CDCl3): δ = 167.4, 149.8, 139.3, 138.8, 136.4, 134.2, 129.4, 128.8, 128.1, 126.7, 124.4, 105.6, 34.1. HRMS (ESI-FTMS Mass (m/z): C15H12BrN2O [M+H]calc = 317.0108, [M+H]expt = 317.0109.
3-Benzyl-7-iodo-2H-pyrido[1,2-a]pyrimidin-2-one (35)
Yield 0.275g (76%), yellowish, 1H NMR (300 MHz, DMSO-d6): δ 8.55 (s, 1H), 8.08 (s, 1H), 7.81 (d, J = 9.4 Hz, 1H), 7.29–7.28 (m, 4H), 7.28–7.21 (m, 1H), 6.96 (d, J = 9.3 Hz, 1H), 3.70 (s, 2H) 13C NMR (75 MHz, DMSO-d6): δ = 167.4, 149.8, 143.6, 138.8, 138.6, 136.1, 129.4, 128.8, 127.9, 126.7, 124.2, 79.4, 34.1. HRMS (ESI-FTMS Mass (m/z): C15H12IN2O [M+H]calc = 362.9989, [M+H]expt = 362.9992.
3-(Benzyl)-9-(trifluoromethyl)-2H-pyrido[1,2-a]pyrimidin-2-one (36)
Yield 0.203g (67%), white, 1H NMR (300 MHz, DMSO-d6): δ 8.39 (d, J = 6.7 Hz, 1H), 8.31 (s, 1H), 8.16 (d, J = 7.2 Hz, 1H), 7.31–7.21 (m, 5H), 7.03 (t, J = 7.2 Hz, 1H), 3.74 (s, 2H), 13C NMR (75 MHz, DMSO-d6): δ= 166.6, 147.3, 138.9, 138.8, 137.5, 136.8 (d, 3J = 5.2 Hz), 129.3, 128.7, 128.2, 126.7, 122.8 (d, 1J = 270.2 Hz), 120.0 (d, 2J = 30.6 Hz), 111.1, 34.0. HRMS (ESI-FTMS Mass (m/z): C16H12F3N2O [M+H]calc = 305.0896, [M+H]expt = 305.0899.
3-Benzyl-7-(trifluoromethyl)-2H-pyrido[1,2-a]pyrimidin-2-one (37)
Yield 0.209g (69%), brownish, 1H NMR (300 MHz, DMSO-d6 + TFA-D): δ 9.49 (s, 1H), 8.62 (s, 1H), 8.58 (d, J = 9.3 Hz, 1H), 7.78 (d, J = 9.3 Hz, 1H), 7.59–7.33 (m, 3H), 7.30–7.27 (m, 2H), 3.90 (s, 2H), 13C NMR (75 MHz, DMSO-d6 + TFA-D): δ = 159.3, 149.0, 139.2, 138.2, 136.8, 136.6 (q, 3J = 5.3 Hz), 130.9, 129.5, 129.0, 127.3, 122.5 (q, 1J = 270 Hz), 119.6 (q, 2J = 35.3 Hz), 118.1, 33.1. HRMS (ESI-FTMS Mass (m/z): C16H12F3N2O [M+H]calc = 305.0896, [M+H]expt = 305.0899.
3-Benzyl-7-bromo-9-methyl-2H-pyrido[1,2-a]pyrimidin-2-one (38)
Yield 0.237g (72%), white, 1H NMR (300 MHz, DMSO-d6): δ 8.43 (s, 1H), 8.11 (s, 1H), 7.72 (s, 1H), 7.28–7.26 (m, 4H), 7.22–7.16 (m, 1H), 3.71 (s, 2H), 1.98 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 167.3, 149.5, 138.9, 136.9, 136.7, 133.0, 131.9, 129.3, 128.7, 127.9, 126.7, 105.4, 34.0, 17.8. HRMS (ESI-FTMS Mass (m/z): C16H14BrN2O [M+H]calc = 331.0264, [M+H]expt = 331.0264.
3-Benzyl-9-bromo-7-methyl-2H-pyrido[1,2-a]pyrimidin-2-one (39)
Yield 0.223g (68%), brownish, 1H NMR (300 MHz, DMSO-d6): δ 8.10 (s, 1H), 8.07 (s, 1H), 7.29–7.28 (m, 5H), 7.23–7.20 (m, 1H), 3.73 (s, 2H), 1.98 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 167.5, 147.2, 142.0, 138.9, 137.2, 131.8, 129.3, 128.7, 128.2, 126.7, 122.2, 116.3, 33.8, 17.0. HRMS (ESI-FTMS Mass (m/z): C16H14BrN2O [M+H]calc = 331.0264, [M+H]expt = 331.0264.
9-Methyl-3-[(4-nitrophenyl)methyl]-2H-pyrido[1,2-a]pyrimidin-2-one (40)
Yield 0.191g (65%), light brown, 1H NMR (300 MHz, DMSO-d6): δ 8.34 (s, 1H), 8.15 (d, J = 8.5 Hz, 2H), 8.04 (d, J = 6.6 Hz, 1H), 7.58 (d, J = 8.6 Hz, 2H), 6.89 (t, J = 6.9 Hz, 1H), 3.88 (s, 2H), 2.28 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 167.3, 150.9, 147.7, 146.5, 137.9, 134.8, 132.4, 131.1, 130.5, 126.4, 123.8, 112.8, 34.0, 18.1. HRMS (ESI-FTMS Mass (m/z): C16H14N3O3 [M+H]calc = 296.1030, [M+H]expt = 296.1032.
7-Methyl-3-[(4-nitrophenyl)methyl]-2H-pyrido[1,2-a]pyrimidin-2-one (41)
Yield 0.221g (75%), brownish, 1H NMR (300 MHz, DMSO-d6 + CDCl3): δ 8.30 (s, 1H), 8.18 (d, J = 11.7 Hz, 2H), 8.14 (s, 1H), 7.59–7.55 (m, 3H), 7.11 (d, J = 9.2 Hz, 1H), 3.87 (s, 2H), 2.21 (s, 3H), 13C NMR (75 MHz, DMSO-d6 + CDCl3): δ = 167.9, 150.3, 146.8, 146.5, 139.1, 136.7, 131.0, 130.2, 126.9, 123.6, 122.9, 122.8, 34.2, 17.4. HRMS (ESI-FTMS Mass (m/z): C16H14N3O3 [M+H]calc = 296.1030, [M+H]expt = 296.1032.
7-Fluoro-3-(4-nitrobenzyl)-2H-pyrido[1,2-a]pyrimidin-2-one (42)
Yield 0.157g (50%), light brown, 1H NMR (300 MHz, DMSO-d6 + TFA-D): δ 8.46 (t, J = 3.6 Hz, 1H), 8.19 (s, 1H), 8.17 (d, J = 8.4 Hz, 2H), 7.85 (dt, J = 2.5, 7.1 Hz, 1H), 7.57 (d, J = 8.4 Hz, 2H), 7.24 (dd, J = 5.5 Hz, 1H), 3.88 (s, 2H), 13C NMR (75 MHz, DMSO-d6 + TFA-d): δ = 154.1 (d, 1J = 242.3 Hz), 147.0, 145.5, 145.2, 139.7, 134.8 (d, 2J = 23.5 Hz), 130.7, 129.9, 124.8, 124.3, 124.0, 117.9 (d, 3J = 7.9 Hz), 32.8. HRMS (ESI-FTMS Mass (m/z): C15H11FN3O3 [M+H]calc = 300.0779, [M+H]expt = 300.0780.
7-Chloro-3-(4-nitrobenzyl)-2H-pyrido[1,2-a]pyrimidin-2-one (43)
Yield 0.154g (49%), brownish, 1H NMR (300 MHz, DMSO-d6): δ 8.48 (s, 1H), 8.18–8.15 (m, 3H), 7.75 (d, J = 9.2 Hz, 1H), 7.57 (d, J = 8.2 Hz, 2H), 7.19 (d, J = 9.4 Hz, 1H), 3.87 (s, 2H), 13C NMR (75 MHz, DMSO-d6): δ = 167.3, 149.9, 147.3, 146.6, 137.4, 137.1, 132.2, 130.6, 126.9, 124.4, 123.9, 118.9, 34.1. HRMS (ESI-FTMS Mass (m/z): C15H10ClN3O3 [M+H]calc = 316.0483, [M+H]expt = 316.0485.
9-Bromo-3-[(4-nitrophenyl)methyl]-4H-pyrido[1,2-a]pyrimidin-4-one (44)
Yield 0.219g (61%), brownish, 1H NMR (300 MHz, DMSO-d6): δ 8.37 (s, 1H), 8.20 (d, J = 6.7 Hz, 1H), 8.16 (d, J = 8.4 Hz, 2H), 7.59 (d, J = 8.4 Hz, 2H), 6.87 (t, J = 7.2 Hz, 1H), 3.90 (s, 2H), 13C NMR (75 MHz, DMSO-d6): δ = 167.3, 148.4, 147.4, 146.6, 139.7, 138.4, 134.6, 130.5, 126.8, 123.8, 116.8, 112.9, 33.8. HRMS (ESI-FTMS Mass (m/z): C15H10BrN3O3 [M+H]calc = 361.9958[M+H]expt = 361.9960.
2-Methyl-3H-pyrimido[1,2-a]quinolin-3-one (45)
Yield 0.121g (54%), brown, 1H NMR (300 MHz, DMSO-d6): δ 9.14 (s, 1H), 8.41 (d, J = 8.6 Hz, 1H), 8.06 (d, J = 9.4 Hz, 1Hz, 1H), 7.95 (d, J = 7.7 Hz, 1H), 7.80 (t, J = 7.3 Hz, 1H), 7.58 (t, J = 7.4 Hz, 1H), 7.13 (d, J = 9.4 Hz, 1H), 2.07 (s, 3H), 13C NMR (75 MHz, DMSO-d6): δ = 169.0, 150.6, 136.9, 134.6, 132.0, 131.6, 130.0, 126.4, 123.3, 123.1, 123.1, 115.5, 14.7. HRMS (ESI-FTMS Mass (m/z): C13H10BrN2O [M+H]calc = 211.0866, [M+H]expt = 211.0868.
Supplementary Material
Acknowledgments
This work was supported by College of Science and Mathematics, Arkansas State University, Jonesboro. Arkansas Statewide MS facility, Grant Number P30 GM103450, from the National Institute of General Medical Sciences of the National Institutes of Health (NIH) for recording mass spectrometry. This publication was made possible by the Arkansas INBRE program, supported by grant funding from the National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS) (P20 GM103429). MAA thanks to provost office to complete this work by giving FRAC award and ABI-Astate for (Grant number: Start-up 200126) start-up funding. Hessa and Zakeyah are thankful to the Saudi Arabian Cultural Mission (SACM) for sponsoring their scholarship. M A Ali thanks Sejong University, Seoul and KISTI supercomputer for the computational resources.
Footnotes
Supporting information
Characterization data of new compounds, including 1H and 13C NMR spectra, HMBC and HSQC spectra of compounds 13 and HRMS
References
- 1.Li JJ. Heterocyclic chemistry in drug discovery. John Wiley & Sons; Hoboken, N.J: 2013. [Google Scholar]
- 2.Jeanmart S, Edmunds AJ, Lamberth C, Pouliot M. Bioorg Med Chem. 2016;24:317–341. doi: 10.1016/j.bmc.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 3.Chen DC, Su SJ, Cao Y. J Mater Chem C. 2014;2:9565–9578. [Google Scholar]
- 4.Liu M, Su SJ, Jung MC, Qi YB, Zhao WM, Kido J. Chem Mater. 2012;24:3817–3827. [Google Scholar]
- 5.Corena-McLeod M. Drugs R D. 2015;15:163–174. doi: 10.1007/s40268-015-0092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gohda T, Ra C, Hamada C, Tsuge T, Kawachi H, Tomino Y. Arzneimittelforschung. 2008;58:18–23. doi: 10.1055/s-0031-1296461. [DOI] [PubMed] [Google Scholar]
- 7.Ni J, Liu QS, Xie SZ, Carlson C, Von T, Vogel K, Riddle S, Benes C, Eck M, Roberts T, Gray N, Zhao J. Cancer Discov. 2012;2:425–433. doi: 10.1158/2159-8290.CD-12-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mane UR, Mohanakrishnan D, Sahal D, Murumkar PR, Giridhar R, Yadav MR. Eur J Med Chem. 2014;79:422–435. doi: 10.1016/j.ejmech.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 9.Mane UR, Li H, Huang J, Gupta RC, Nadkarni SS, Giridhar R, Naik PP, Yadav MR. Bioorg Med Chem. 2012;20:6296–6304. doi: 10.1016/j.bmc.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 10.Silpa L, Niepceron A, Laurent F, Brossier F, Penichon M, Enguehard-Gueiffier C, Abarbri M, Silvestre A, Petrignet J. Bioorg Med Chem Lett. 2016;26:114–120. doi: 10.1016/j.bmcl.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 11.La Motta C, Sartini S, Mugnaini L, Simorini F, Taliani S, Salerno S, Marini AM, Da Settimo F, Lavecchia A, Novellino E, Cantore M, Failli P, Ciuffi M. J Med Chem. 2007;50:4917–4927. doi: 10.1021/jm070398a. [DOI] [PubMed] [Google Scholar]
- 12.Peng L, Gao X, Duan L, Ren X, Wu D, Ding K. J Med Chem. 2011;54:7729–7733. doi: 10.1021/jm200976s. [DOI] [PubMed] [Google Scholar]
- 13.Blazer LL, Roman DL, Chung A, Larsen MJ, Greedy BM, Husbands SM, Neubig RR. Mol Pharmacol. 2010;78:524–533. doi: 10.1124/mol.110.065128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le G, Vandegraaff N, Rhodes DI, Jones ED, Coates JA, Lu L, Li X, Yu C, Feng X, Deadman JJ. Bioorg Med Chem Lett. 2010;20:5013–5018. doi: 10.1016/j.bmcl.2010.07.041. [DOI] [PubMed] [Google Scholar]
- 15.Askoura M, Mottawea W, Abujamel T, Taher I. Libyan J Med. 2011;6 doi: 10.3402/ljm.v6i0.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rapolu S, Alla M, Ganji RJ, Saddanapu V, Kishor C, Bommena VR, Addlagatta A. Med Chem Comm. 2013;4:817–821. [Google Scholar]
- 17.Yu G, Zhou G, Zhu M, Wang W, Zhu T, Gu Q, Li D. Org Lett. 2016;18:244–247. doi: 10.1021/acs.orglett.5b02964. [DOI] [PubMed] [Google Scholar]
- 18.Rebhun JF, Roloff SJ, Velliquette RA, Missler SR. Fitoterapia. 2015;101:57–63. doi: 10.1016/j.fitote.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Huang G, Roos D, Stadtmüller P, Decker M. Tetrahedron Lett. 2014;55:3607–3609. [Google Scholar]
- 20.Schramm A, Hamburger M. Fitoterapia. 2014;94:127–133. doi: 10.1016/j.fitote.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Basavaiah D, Satyanarayana T. Tetrahedron Lett. 2002;43:4301–4303. [Google Scholar]
- 22.Satyanarayana S, Praveen Kumar K, Lakshmi Reddy P, Narender R, Narasimhulu G, Subba Reddy BV. Tetrahedron Lett. 2013;54:4892–4895. [Google Scholar]
- 23.Katritzky AR, Rogers JW, Witek RM, Nair SK. Arkivoc. 2004:52–60. [Google Scholar]
- 24.Alanine TA, Galloway WR, Bartlett S, Ciardiello JJ, McGuire TM, Spring DR. Org Biomol Chem. 2016;14:1031–1038. doi: 10.1039/c5ob01784j. [DOI] [PubMed] [Google Scholar]
- 25.Chichetti SM, Ahearn SP, Adams B, Rivkin A. Tetrahedron Lett. 2007;48:8250–8252. [Google Scholar]
- 26.Weng YY, Ying LM, Chen QX, Su WK. Chinese Chem Lett. 2012;23:911–914. [Google Scholar]
- 27.Satyanarayana S, Praveen Kumar K, Lakshmi Reddy P, Narender R, Narasimhulu G, Subba Reddy BV. Tetrahedron Lett. 2013;54:4892–4895. [Google Scholar]
- 28.Alam MA, Alsharif Z, Alkhattabi H, Jones D, Delancey E, Gottsponer A, Yang T. Sci Rep. 2016;6:36316. doi: 10.1038/srep36316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alsharif ZA, Alam MA. RSC Advances. 2017;7:32647–32651. doi: 10.1039/c7ra05993k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonnet-Delpon D, Bégué JP, Crousse B. Synlett. 2004:18–29. [Google Scholar]
- 31.Kelley BT, Walters JC, Wengryniuk SE. Org Lett. 2016;18:1896–1899. doi: 10.1021/acs.orglett.6b00672. [DOI] [PubMed] [Google Scholar]
- 32.Shen C, Wang L, Wen M, Shen H, Jin J, Zhang P. Ind Eng Chem Res. 2016;55:3177–3181. [Google Scholar]
- 33.Khaksar S, Talesh SM. J Fluor Chem. 2012;135:87–90. [Google Scholar]
- 34.Li ML, Ren YJ, Dong MH, Ren WX. Eur J Med Chem. 2015;96:122–138. doi: 10.1016/j.ejmech.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 35.Ren W, Ren Y, Wang S. Eur J Med Chem. 2016;120:148–159. doi: 10.1016/j.ejmech.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 36.Dai L, Zhang Y, Dou Q, Wang X, Chen Y. Tetrahedron. 2013;69:1712–1716. [Google Scholar]
- 37.Basavaiah D, Veeraraghavaiah G. Chem Soc Rev. 2012;41:68–78. doi: 10.1039/c1cs15174f. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y, Truhlar DG. Theor Chem Acc. 2008;120:215–241. [Google Scholar]
- 39.T MJ, Frisch GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, et al. Gaussian 09, Revision D.01. Gaussian, Inc.; Wallingford CT: 2013. [Google Scholar]
- 40.Solà M. Frontiers in Chemistry. 2013;1:22. doi: 10.3389/fchem.2013.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delidovich I, Palkovits R. Greem Chem. 2016;18:590–593. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


