Abstract
Aberrant activity of enhancer of zeste homolog 2 (EZH2) is associated with a wide range of human cancers. The interaction of EZH2 with embryonic ectoderm development (EED) is required for EZH2's catalytic activity. Inhibition of the EZH2-EED complex thus represents a novel strategy for interfering with the oncogenic potentials of EZH2 by targeting both its catalytic and non-catalytic functions. To date, there have been no reported high-throughput screening (HTS) assays for inhibitors acting at the EZH2-EED interface. In this study, we developed a fluorescence polarization (FP)-based HTS system for the discovery of EZH2-EED interaction inhibitors. The tracer peptide sequences, positions of fluorescein labeling, and a variety of physicochemical conditions were optimized. The high Z' factors (>0.9) at a variety of DMSO concentrations suggested that this system is robust and suitable for HTS. The minimal sequence requirement for the EZH2-EED interaction was determined by using this system. A pilot screening of an in-house compound library containing 1600 FDA-approved drugs identified four compounds (apomorphine hydrochloride, oxyphenbutazone, nifedipine and ergonovine maleate) as potential EZH2-EED interaction inhibitors.
Keywords: PRC2, EZH2, EED, protein-protein interaction, fluorescence polarization, high-throughput screening, apomorphine hydrochloride, oxyphenbutazone, nifedipine, ergonovine maleate
Introduction
Enhancer of zeste homolog 2 (EZH2) is the catalytic subunit of the polycomb repressive Complex 2 (PRC2) and implements transcriptional repression mainly by depositing tri-methylation marks at lysine 27 of histone H3 (H3K27me3)1,2,3. Accumulating evidence demonstrates that dysregulated EZH2 is intimately involved in tumorigenesis and tumor progression. Activating mutations of EZH2 result in hyper-trimethylation of H3K27, thus blocking cell differentiation and resulting in unrestrained cell growth in germinal center B-cell type diffuse large B-cell lymphoma (GCB-DLBCL) and follicular lymphoma4,5,6,7. Additionally, the over-expression of EZH2 is commonly observed in many malignant solid tumors, including those of the prostate, breast, colon, bladder, gastric, lung, and pancreas8,9,10,11,12,13,14. Moreover, the genetic depletion of EZH2 has been found to effectively suppress cancer cell growth15. These studies have raised significant interest in the discovery of small molecule inhibitors of EZH2 as novel anti-cancer agents.
Recently, several small-molecule enzyme inhibitors of EZH2 have been discovered. These inhibitors decrease cellular H3K27me3 levels and exert promising therapeutic potential against a panel of oncogenic EZH2-associated human cancers16,17,18,19. However, several studies have indicated that there is a phenotypic discrepancy between the genetic abrogation and enzyme inhibition of EZH2, and have further highlighted the dominant contributions of the non-catalytic functions in several cancer models20,21,22. Therefore, inhibitors targeting both the non-catalytic and catalytic functions of EZH2 may have broader therapeutic potentials than targeting the active site. Given that the H3K27me3 activity of EZH2 requires the assembly of a core complex with Embryonic Ectoderm Development (EED) and Suppressor of Zeste 12 (SUZ12), the blocking of these protein-protein interactions (PPIs) may serve as a novel method for inhibiting EZH2 activities23,24,25,26. The druggability of the EZH2-EED interface has been demonstrated with a stapled EZH2 peptide as well as with a small molecular inhibitor astemizole identified by structure-based virtual screening25,26. Notably, these inhibitors not only decrease H3K27me3 levels but also have a unique mechanism of action (MOA) in which the protein degradation of EZH2 is triggered after dissociation of the complex in cancer cells. A subsequent study has further revealed that the stapled peptide decreases both the H3K27me3 and EZH2 levels and impairs growth of SWI-SNF mutant cancer cells resistant to EZH2 enzyme inhibitors20. These studies emphasize the promise of targeting the EZH2-EED interaction for EZH2-dependent cancer treatment therapies. However, the poor metabolic stability and low bioavailability of the peptide inhibitor, as well as the moderate in vitro activity and cardiotoxicity of astemizole, may greatly hinder in vivo therapeutic applications27,28. Moreover, there remains an unmet need to develop novel and potent small molecular inhibitors that target the EZH2-EED interaction.
In this article, we report the development and optimization of an fluorescence polarization (FP)-based HTS assay for the discovery of small-molecule inhibitors targeting the EZH2-EED interaction. Under the optimized conditions, this assay is robust, with a high Z' factor under various DMSO concentrations, and is thus highly suitable for high-throughput drug screening.
Materials and methods
Peptide synthesis
All of the EZH2 peptides (Figure 1, Table 1) used in this article were purchased from China Peptides (Shanghai, China) and were synthesized and purified using a standard solid-phase synthesis method with a fully automatic Liberty Blue peptide synthesizer (CEM). The synthesized peptides were analyzed by reverse-phase high-performance liquid chromatography (HPLC) and assessed by mass spectrometry (MS). The purity of all of the peptides was greater than 95%. N-terminal or C-terminal fluorescein isothiocyanate (FITC)-labeled EZH2 peptides (Figure 1) were used as tracer peptides. The concentrations of tryptophan (Trp)-containing peptides were determined on the basis of the absorbance and molar extinction coefficient at 280 nm29. Notably, comparisons of the concentrations as determined by the extinction coefficient revealed differences of less than 2-fold for all of the Trp-containing peptides, when their concentrations were determined on the basis of their molecular weights. Additionally, for the peptides without tryptophan or tyrosine, the concentrations were determined on the basis of their molecular weights.
Figure 1.
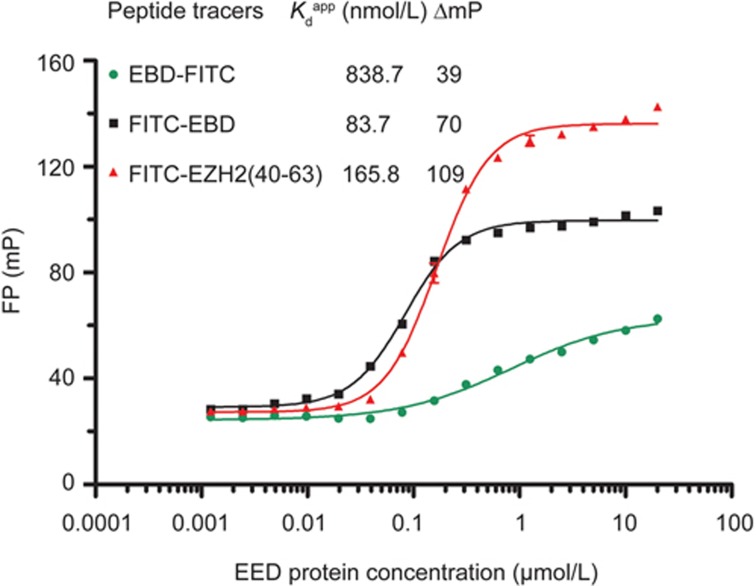
The optimization of the peptide tracer. Fluorescence polarization (FP) saturation binding curves of three different FITC-labeled peptide tracers (20 nmol/L) incubated with serial dilutions of EED (0.001 to 20 μmol/L) in HEPES buffer at room temperature. The FP values were recorded at 2 h. The experiments were repeated three times, and the data were analyzed in GraphPad Prism 5.0.
Table 1. The binding affinities of various peptide truncations of EED-binding domain (EBD) determined by FP competitive assay. Ki is the inhibition constant.
| No | Peptide sequence | Ki (μmol/L) |
|---|---|---|
| 1 | Ac-KTMFSSNRQKILERTETLNQEWKQRRIQPV-NH2 | 0.050 |
| 2 | Ac-TMFSSNRQKILERTETLNQEWKQR-NH2 | 0.079 |
| 3 | Ac-FSSNRQKILERTETLNQEWKQR-NH2 | 0.113 |
| 4 | Ac-SNRQKILERTETLNQEWKQR-NH2 | >200 |
| 5 | Ac-RQKILERTETLNQEWKQR-NH2 | >200 |
| 6 | Ac-KILERTETLNQEWKQR-NH2 | >200 |
| 7 | Ac-LERTETLNQEWKQR-NH2 | >200 |
| 8 | Ac-TMFSSNRQKILERTETLNQEWK-NH2 | 2.862 |
| 9 | Ac-TMFSSNRQKILERTETLNQE-NH2 | >200 |
| 10 | Ac-TMFSSNRQKILERTETLN-NH2 | >200 |
| 11 | Ac-TMFSSNRQKILERTET-NH2 | >200 |
| 12 | Ac-TMFSSNRQKILERT-NH2 | >200 |
| 13 | Ac-TMFSSNRQKILER-NH2 | >200 |
| 14 | Ac-FSSNRQKILERTETLNQEWK-NH2 | 15.501 |
Protein expression and purification
The DNA sequence encoding human EED amino acids 81-441, which interact with the EED-binding domain (EBD) of the human EZH2 protein, was amplified by polymerase chain reaction (PCR) from full-length DNA and cloned into plasmid pET28a-SUMO (Novagen) carrying an N-terminal 6× His-SUMO tag, as described previously26. The recombinant proteins were overexpressed in Escherichia coli BL21(DE3) (Novagen) cells, which were induced with 0.4 mmol/L isopropyl β-D-thiogalactoside (IPTG) overnight at 16 °C. The bacteria were sonicated in pre-cooled lysis buffer (25 mmol/L HEPES pH 8.0, 150 mmol/L NaCl, 1 mmol/L DTT, 20 mmol/L imidazole, 100 μg/mL PMSF) and centrifuged at 18000 r/min for 30 min at 4 °C. The samples were loaded on Ni-NTA resin (5 mL prepacked HisTrap FF column, GE Healthcare, Sweden ), and the recombinant proteins were eluted with 90 mmol/L imidazole. The EED proteins were separated by size exclusion chromatography (Superdex 75, GE Healthcare) in a buffer containing 25 mmol/L HEPES, pH 8.0, 150 mmol/L NaCl, and 1 mmol/L DTT. The proteins were concentrated to 10 mg/mL and stored at -80 °C.
FP Measurements
All FP measurements were performed on a multifunctional microplate reader (EnVision, Perkin Elmer) in black 384-well microplates (Corning, Cat No 3575) with 40 μL of the assay solution per well. 480-nm excitation and 535-nm emission filters were used for the FP measurements. The FP values were calculated according to the following equation30,31:
where S is the parallel emission intensity, P is the perpendicular emission intensity, and G is the grating factor32. The value of the G factor ranged from 0.8 to 1.2.
FP binding assay
In the FP saturation binding experiments, 20 nmol/L FITC-labeled EZH2 peptides (tracers) were mixed with increasing concentrations of EED (from 0.001 μmol/L to 20 μmol/L) in triplicate. The FP signals were recorded at 30-min intervals until 4 h after incubation at room temperature or 15 °C to 40 °C. The optimal FP buffer was 25 mmol/L HEPES, pH 8.0, and 150 mmol/L NaCl with additives (0.1 mg/mL BSA and 0.01% NP40). Four other buffers, ie, citrate (pH 5.1), PIPES (pH 6.2), Bis-tris (pH 7.0), and Bis-tris propane (pH 9.3), at the same concentration of 25 mmol/L were used for the FP assay buffer optimization. The same concentrations of NaCl, BSA, and NP40 were also included in the four buffer solutions. Different concentrations of DMSO (from 1% to 10%) were tested to evaluate the assay stabilities. All of the experiments were independently repeated at least three times. The FP values were plotted against the log of the protein concentrations, and the dissociation constant (apparent Kd) and the dynamic range (ΔmP) were obtained from the resulting sigmoidal curve as analyzed in GraphPad Prism 5.
FP competition assay and quality assessment
For the competitive binding assay, a mixture containing 20 nmol/L FITC-labeled EZH2 peptides and 625 nmol/L EED was incubated with serial dilutions of unlabeled EZH2 peptides or compounds for 2 h at room temperature. The FP values were determined, and the IC50 values, ie, the concentrations required for 50% displacement of the tracer, were calculated in GraphPad Prism 5. The Ki values of competitive inhibitors were calculated on the basis of a previously reported method33.
For the high-throughput screening assay performance indicator determination, the Z' factor was calculated according to the following equation30,31:
where SDn and SDp are the standard deviations, and μn and μp represent the means of the FP values obtained from the negative and positive controls, respectively. Each 384-well plate contained 190 negative control wells (20 nmol/L tracer and 625 nmol/L protein), 190 positive control wells (20 nmol/L tracer, 625 nmol/L protein, and 10 μmol/L unlabeled EZH2 peptide competitor), and 4 free tracer wells (20 nmol/L tracer only). Different concentrations of DMSO (from 0% to 4%) were added to determine the effect of DMSO on the Z' factor. All experiments were repeated three times on three separate days.
Pilot screening of an in-house compound library
In the FP-based high-throughput screening assay, 0.5 μL of the compounds (20 mmol/L in DMSO stock for a library of 1600 compounds) were transferred into each well of 384-well assay plates with a robotic delivery system (JANUS, PerkinElmer). Mixtures of 39.5 μL containing 625 nmol/L EED and 20 nmol/L tracer were dispensed into the compound wells with a reagent dispenser (Multidrop Combi, Thermo Fisher Scientific). In each assay plate, the DMSO and unlabeled peptides were used as negative and positive controls, respectively. Free tracer wells (20 nmol/L tracer only) were also set up in each assay plate. After incubation for 2 h at room temperature, the FP values were measured. The inhibitory activities were calculated using the following equation: %Inhibition=100×(mPn–mPs)/(mPn–mPp); where mPn, mPp, and mPs represent FP values of the negative controls, positive controls, and compound samples, respectively.
Results
Optimization of the tracer peptide
Crystallographic studies of either the isolated EED protein or the whole PRC2 complex, as well as biochemical results, have suggested the critical importances of the EED binding domain (EBD) of EZH2 (residues 39–68) and the WD40 repeat domain of EED protein (residues 81–441) in the formation of the EZH2-EED complex, which is required to maintain the catalytic competent conformation of EZH224,34,35,36,37. On the basis of the sequence of EBD, we first designed two peptide tracers with FITC labeling on the N-terminus or C-terminus of the EBD peptide. The binding affinities of both tracers to the EED protein were determined with FP binding assays. As illustrated in Figure 1, the C-terminal FITC-labeled EBD peptide had an apparent Kd value of 838.7 nmol/L, which was approximately ten-fold weaker than that of the N-terminal FITC-labeled EBD peptide, which had an apparent Kd value of 83.7 nmol/L. These data indicated that the N-terminal labeling, and not the C-terminal labeling, preserved the strong binding affinity between EBD and EED, as observed in a previous study24. However, the relatively low signal dynamic range obtained from the FITC-EBD tracer peptide (ΔmP=70) may have limited the sensitivity of the assay during the HTS. Because a large molecular volume difference after binding was required to maximize the FP signals, the long EBD peptide may have resulted in a relatively higher initial FP value in its free state, and a smaller change in the FP signal for the EBD-EED complex during the lifetime of fluorescein would thus be expected. We hypothesized that a decrease in the molecular weight of the tracer peptide might improve the signal dynamic range of the FP assay. In the EBD-EED complex structure (Figure S1), we found that the first residue (K39) at the N-terminus of the EBD was missing, probably because of the lack of direct interactions with EED. The attachment of the FITC labeling at this highly flexible position may have led to a “propeller effect” that decreased the polarization of the FITC-EBD-EED complex. Additionally, the EBD interacted with EED primarily via its long N-terminal α–helix (residues 40–63) through extensive H-bond and hydrophobic interactions, whereas the last five residues at the C-terminus formed a rigid coil that adopted van der Waals contacts with EED. A previous mutational analysis has revealed that most of the “hot spot” residues are located in the N-terminal α–helix24. Thus, we predicted that the N-terminal α–helix might play a dominant role in the formation of the EBD-EED complex. On the basis of this information, we re-designed the tracer peptide by removing both the K39 and the last five residues at the C-terminal from the EBD peptide, and this truncated tracer peptide was designated FITC-EZH2 (40–63). Compared with the parental FITC-EBD peptide, the FITC-EZH2 (40–63) maintained most of the binding affinity (apparent Kd=165.8 nmol/L) and produced an approximately 55% increase in the signal dynamic range (ΔmP =109). Therefore, FITC-EZH2 (40–63) was chosen for the subsequent FP assay development.
Optimization of the assay buffer and pH conditions
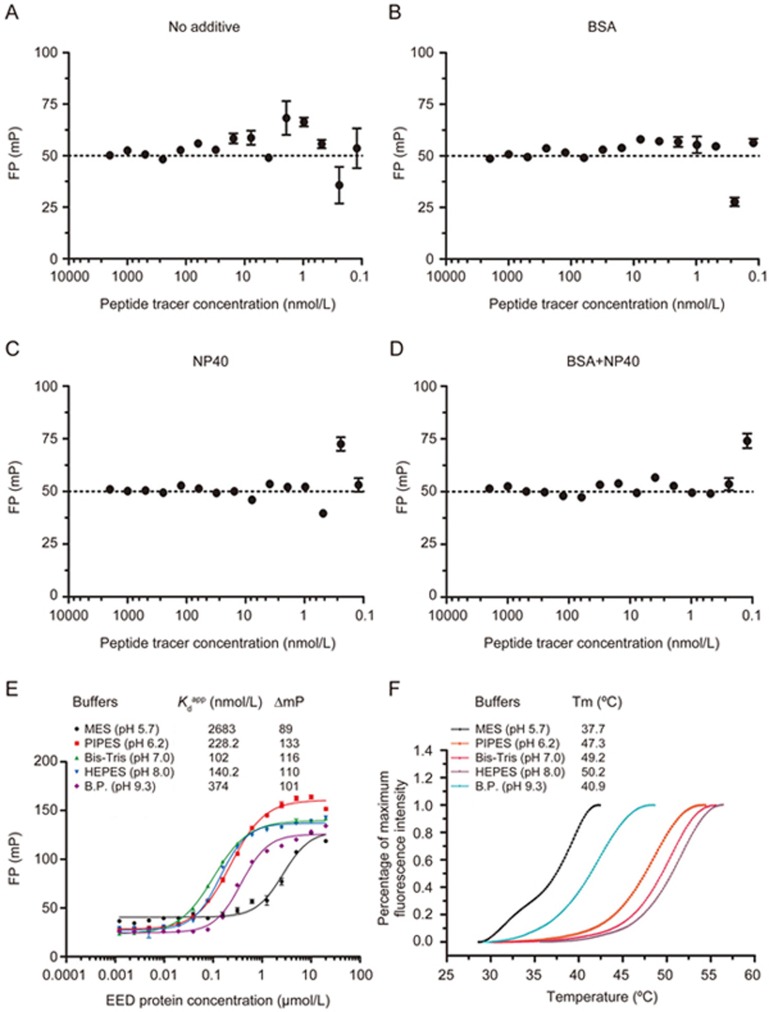
To select the optimal conditions for FITC-EZH2(40-63) by using FP binding assays, we determined the FP values of FITC-EZH2(40-63) as a function of its concentration and compared the influences of various additives. An apparent variation of the FP signal was observed when the tracer peptide concentration was diluted below 31.25 nmol/L (Figure 2A). However, when either BSA or NP40 was included in the assay buffer, the lowest concentrations that provided reliable FP signals decreased to 3.9 nmol/L and 0.98 nmol/L, respectively, and the combination of both additives further lowered this value to 0.49 nmol/L (Figure 2B–2D). These observations suggested that the additives significantly improved the detection limit of the FP signal, probably by decreasing the non-specific binding or aggregation of the tracer peptide. Previous studies have also demonstrated that the use of more than one additive with distinct chemical properties may more effectively decrease compound aggregation in the HTS38; hence, both BSA and NP40 were added into the FP buffer in our assay development. Moreover, considering the potential influences of the differences in batch purities of the tracer peptide and the technical variations in the making of the dilution, we selected 20 nmol/L of peptide tracer for use in the FP binding assay.
Figure 2.
FP buffer and pH optimization. The FP values of FITC-EZH2(40-63) are plotted as a function of the peptide tracer concentrations in HEPES buffer in the absence of additives (A) and in the presence of 0.1 mg/mL BSA (B), 0.01% NP40 (C), or both 0.1 mg/mL BSA and 0.01% NP40 (D). (E) FP saturation binding curves of FITC-EZH2(40-63) with different concentrations of EED in five different buffers. B.P. represents Bis-tris propane. The FP values were measured at 2 h. Three separate experiments were performed, and the data were analyzed in GraphPad Prism 5.0. (F) The melting curves of EED in five different buffers. In each well of 96-well assay plates, 2 μmol/L EED and 5× SYPRO Orange (Invitrogen) were mixed in 20 μL of 25 mmol/L buffers and 150 mmol/L NaCl. The assay plate was heated from 25 °C to 95 °C at a ramp rate of 0.05 °C/s. The changes in the fluorescence intensities of SYPRO Orange were monitored as a function of temperature. The melting temperature (Tm) was calculated with the Boltzmann fitting method in the Protein Thermal Shift Software v1.2. The experiments were performed in triplicate and independently repeated three times.
To determine the effects of different pH conditions in the FP binding assay, we selected 5 different buffer systems with pH values ranging from 5.7 to 9.3. The peptide-EED binding was apparently affected by the acidic (pH 5.7) and alkaline (pH 9.3) solutions, especially in the acidic conditions in which the binding was nearly abolished. Nevertheless, both the FP dynamic range and the binding affinity exhibited good tolerance within the pH range of 6.2 to 8.0 (Figure 2E). A protein thermal shift assay was used to explore whether the differential protein stabilities in the distinct pH conditions might account for the variations in the binding affinities. As demonstrated in Figure 2F and Figure S2, markedly higher Tm values (more than 10 °C) in the pH range from 6.2 to 8.0 compared with the acidic and alkaline conditions were observed, thus suggesting that the best EED protein stability was achieved within the pH range of 6.2 to 8.0. The clear correlation between the protein stability and binding affinity (Figure S2) emphasized the importance of selecting the optimal pH conditions that produced the best protein stability for maintaining high binding affinity in the FP assay. Considering both the protein stability and the binding affinity, we selected HEPES buffer (pH 8.0) for the subsequent FP assay development. Notably, we found that for compounds with limited solubility in HEPES (pH 8.0) buffer, both the PIPES (pH 6.2) and Bis-Tris buffers (pH 7.0) could also be used if an increased solubility was observed. For example, astemizole and its derivatives, ie, the small molecular inhibitors of EZH2-EED interactions identified in our previous study, were found to exhibit severe precipitation and to form aggregates in HEPES buffer. The PIPES buffer, in which these compounds have much better solubility, was subsequently used to achieve more accurate evaluation of their activities26.
Optimizations of incubation time, temperature and DMSO tolerance
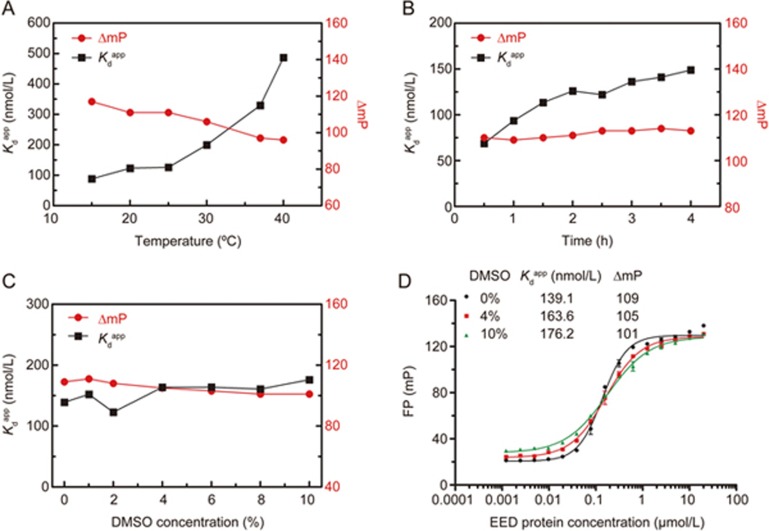
The stability of the FP saturation binding assay was evaluated at different incubation temperatures and incubation times at room temperature (25 °C). The apparent Kd values increased, and the ΔmP values significantly decreased when the incubation temperature was above 30 °C, probably through decreasing protein stability (Figure 3A). As illustrated in Figure 3B, the binding affinity and dynamic range were stable after 2–3.5 h of incubation at room temperature, although the binding affinity decreased slightly from 30 min to 2 h (the change was within two-fold). Moreover, the both FP signal and total fluorescence intensity were stable during this period (Figure S3). On the basis of the time course of the apparent Kd values, the incubation time was optimized to 2 h for both the FP binding and the screening assay. DMSO, which is commonly used for dissolving compounds in high-throughput drug discovery, was also tested in terms of stability in the FP binding assay. The FP binding curves were well maintained even in the presence of 10% DMSO. The good tolerance to DMSO may provide opportunities to achieve more accurate FP measurement when evaluating compounds with solubility issues.
Figure 3.
Stability of the FP binding assay. Three separate experiments were performed, and the data were analyzed in GraphPad Prism 5.0. (A) The effect of incubation temperature on binding affinity and dynamic range. FP values were recorded at 2 h. (B) The time courses of binding affinity and dynamic range at room temperature (25 °C). (C) DMSO tolerance of the FP binding affinity and dynamic range. (D) FP saturation binding curves of FITC-EZH2 (40–63) with increasing concentrations of EED in the absence or presence of 4% and 10% DMSO. Three separate experiments were performed, and the data were analyzed in GraphPad Prism 5.0.
Establishment of the FP competitive assay and Z` factor evaluation
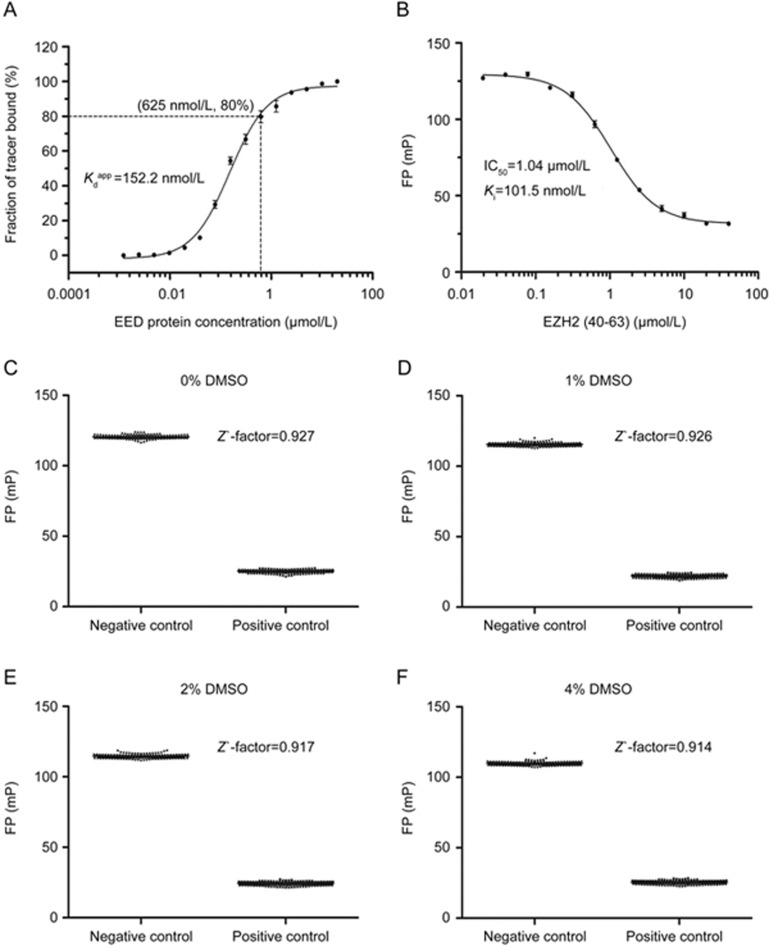
In the FP competitive assay, the protein concentration is generally above the Kd value for saturation of at least 50% of the tracer peptide in the high assay window33. To increase the sensitivity of our assay, we selected 625 nmol/L (four-fold the Kd) of EED in the FP competitive binding assay, corresponding to 80% saturation of the tracer peptide, which yielded an ∼100 mP dynamic range (Figure 4A). Under the optimized conditions described above, the unlabeled peptide corresponding to FITC-EZH2(40-63) dose-dependently displaced the tracer peptide in the FP competitive assay, with a Ki value of 101.5 nmol/L, a value similar to the apparent Kd derived from the FP binding assay. These data suggested that the FITC labeling apparently did not affect the binding affinity of the tracer peptide with EED, and the FP competitive assay could thus be used to accurately quantify the abilities of small molecular compounds to inhibit the EZH2-EED interaction.
Figure 4.
FP competitive binding assay and determination of the Z' factors. (A) Selection of the EED concentration for the FP competitive binding assay based on the FP saturation binding curve at 80% fractions of tracer bound. (B) FP competitive binding curve. Mixtures of 20 nmol/L FITC-EZH2(40-63) and 625 nmol/L EED were incubated in the presence of different concentrations of unlabeled peptide EZH2(40-63) (20 nmol/L to 40 μmol/L). The FP values were recorded at 2 h. (C–F) Z' factors for evaluation of the FP-based high-throughput screening assay in the absence or presence of 1%, 2%, and 4% DMSO. The FP values of both negative and positive controls are shown in a scatter plot. All experiments were performed at least three times. The data were analyzed in GraphPad Prism 5.0.
To determine whether the FP competitive assay was suitable as a high-throughput screening assay, the Z' factor, a statistical parameter accessing both the assay range and the data variation in HTS assays31, was determined in a 384-well format, and the unlabeled peptide EZH2 (40–63) was used as a positive control. Generally, assays with Z' factors less than 0.5 are not recommended for HTS, whereas assays with Z' factors of 0.7–1 are considered high-quality assays. Because the concentrations of DMSO in biochemical HTS assays are usually below 5%, we evaluated the effects of DMSO on the Z' factor at concentrations of up to 4% (Figure 4C–4F). The Z' factors for the FP competitive assays were found to be more than 0.9 even in the presence of 4% DMSO. These results clearly demonstrated that the FP competitive assay is robust and suitable for HTS with DMSO concentrations up to 4%.
Determination of the minimal sequence requirement for EZH2-EED interaction
To validate the utilization of the established FP competitive assay, we first determined the minimal sequence requirement of the EBD peptide, which mediates the interaction with EED. In agreement with the observations from the FP binding experiments (Figure 1), the removal of K39 and the last five amino acids maintained most of the binding affinity of peptide 2 (Table 1) relative to the parental EBD peptide (peptide 1). Although the further truncation of two residues (T40 and M41) at the N-terminus (peptide 3) had a minimal effect on binding affinity, the removal of the F42 and S43 (peptides 4–7) completely abolished its binding ability with EED, a result in line with findings from previous mutation studies suggesting that F42 is a 'hot-spot' residue in the EZH2-EED complex24,26. Additionally, the truncation of the last two residues (Q62 and R63) at the C-terminus (peptide 8) caused an approximately 36-fold decrease in binding affinity, thus indicating the importance of the H-bond or electrostatic interactions between these two residues and EED24. Nevertheless, the binding activity was lost after the further removal of W60 and K61. Considering both the N-terminal and C-terminal truncation data, we speculate that the sequence from F42 to K61 of EZH2 is the core region that associates with EED. As expected, a peptide consisting of this core region (peptide 14) maintained its interaction with EED, albeit with a weaker binding activity (Ki =15.5 μmol/L).
Pilot screening of an in-house compound library
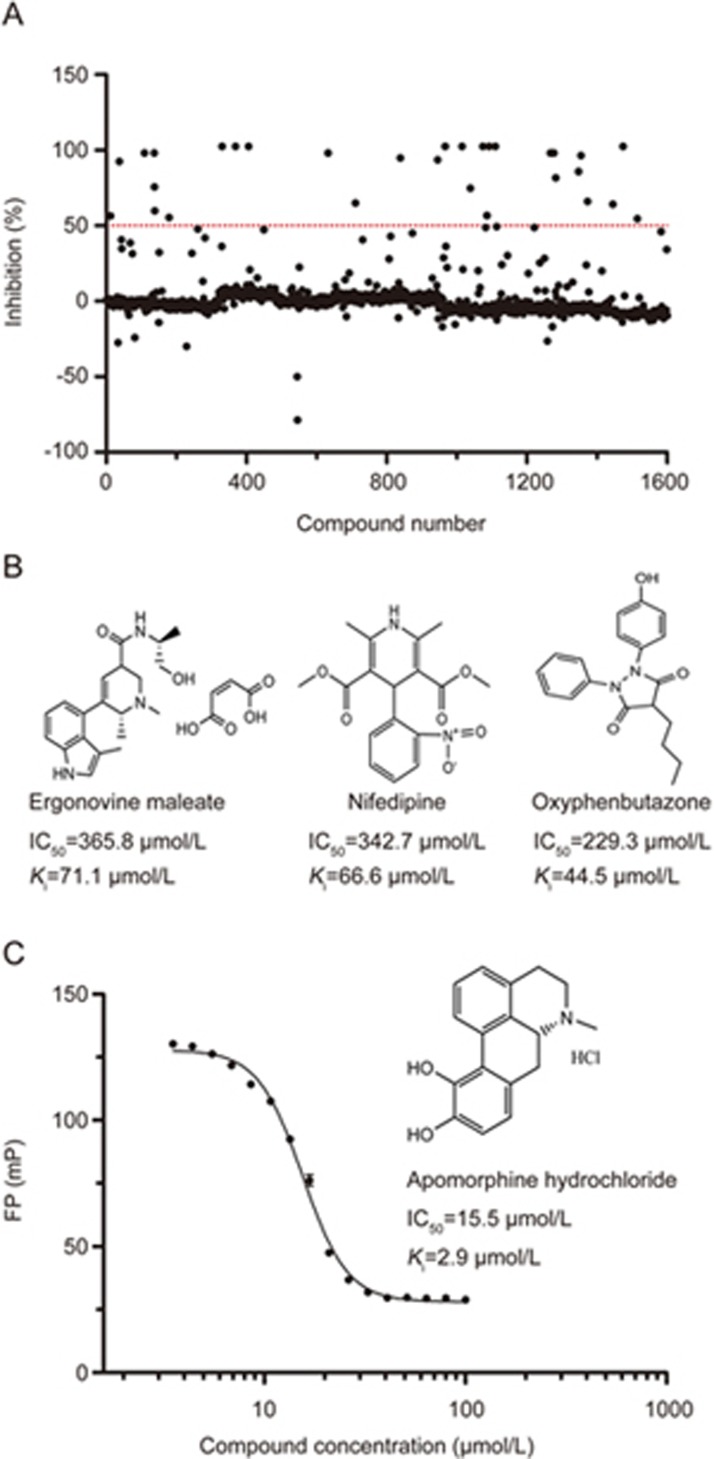
The FP-based HTS assay was then applied in a pilot screening of an in-house drug library of 1600 compounds with known biological functions (Table S1). As illustrated in Figure 5A, a primary screening of the 1600 compounds in duplicate identified 31 potential hits (% inhibition >50%). After exclusion of auto-fluorescent compounds on the basis of total fluorescence at 100 μmol/L and 10 μmol/L in the secondary screening, 4 compounds demonstrated dose-dependent inhibition of the EZH2-EED interaction. Among these compounds, apomorphine hydrochloride exhibited the greatest inhibitory activity, with an IC50 value of 15.5 μmol/L and a Ki value of 2.9 μmol/L (Figure 5B and 5C). This pilot screening further demonstrated that the FP-based competitive assay can be used as a robust HTS assay for the discovery of inhibitors of the EZH2-EED interaction.
Figure 5.
Pilot screening of an in-house compound library. (A) Overview of the primary high-throughput screening. The dashed line in the figure indicates 50% inhibition. The hits (>50% inhibition) were further validated in the second screening. (B) Three inhibitors with IC50 values of 200∼400 μmol/L and Ki values of 40-80 μmol/L. (C) Inhibition curve of apomorphine hydrochloride, which had the best inhibitory activity among the 1600-drug library . The FP values were measured at 2 h. Three separate experiments were performed, and the data were analyzed in GraphPad Prism 5.0.
Discussion
In this study, an FP-based HTS assay was developed for identifying small molecular inhibitors of the EZH2-EED interaction. The tracer peptide and various physicochemical conditions, such as pH, temperature, incubation time and DMSO resistance, were systematically optimized to maximize the sensitivity and reproducibility of the assay. The assay was miniaturized for HTS, and the high Z' factors (>0.9) with commonly used DMSO concentrations indicated that this assay is robust and well suited for HTS. By using this established assay with optimized conditions, the minimal sequence requirement for EZH2-EED interactions was first determined. A pilot screening was performed to identify several compounds that dose-dependently inhibited the EZH2-EED interaction. Apomorphine hydrochloride, the most potent inhibitor identified in the screening, exhibits agonist activities toward more than 15 human targets39,40 in addition to being a non-selective dopamine agonist used for Parkinson's disease. These findings provide the first evidence indicating that apomorphine may modulate PRC2 activity by blocking the EED-EZH2 interaction. However, more detailed validation and target engagement investigations, such as those using a red-shifted dye with a longer half-life in the excited state in the FP assay41, evaluation of the direct binding with EED via biophysical assays, and assessment of the changes in histone H3K27me3 levels, are needed to explore the role of the EZH2-EED interaction in the clinical effects of apomorphine.
Collectively, we believe that this robust FP-based HTS assay may greatly accelerate the drug discovery process for compounds targeting the EZH2-EED interaction, and these inhibitors might possess broader therapeutic potential than EZH2 inhibitors. Moreover, anti-tumor synergy effects can also be expected in the combination of EZH2 PPI and EZH2 inhibitors25,26. Additionally, the minimal peptide sequence for EZH2-EED interaction, as well as the potential hits from the pilot screening, may serve as good starting points for further development.
Author contribution
Xiang-qian KONG and Cheng LUO designed the research; Mao-rong ZHU, Dao-hai DU, and Xiang-qian KONG performed the research; Jun-chi HU, Lian-chun LI, Jing-qiu LIU, and Hong DING contributed new analytical tools and reagents; Mao-rong ZHU, Dao-hai DU and Xiang-qian KONG performed analysis and interpretation of the data; Mao-rong ZHU, Dao-hai DU and Xiang-qian KONG wrote the manuscript; Cheng LUO, Hua-liang JIANG, and Kai-xian CHEN reviewed the manuscript. All authors read and approved the manuscript.
Acknowledgments
The authors disclose the following financial support for this research and/or authorship of this article: the Ministry of Science and Technology of China (2015CB910304 to Hua-liang JIANG), the National Natural Science Foundation of China (21472208 and 81625022 to Cheng LUO, 21210003, 81230076 and 91313000 to Hua-liang JIANG), and the Fund of the State Key Laboratory of Toxicology and Medical Countermeasures, Academy of Military Medical Science (TMC201505 to Cheng LUO). We are extremely grateful to the National Centre for Protein Science Shanghai (Shanghai Science Research Center, Protein Expression and Purification System) for providing instrument support and technical assistance.
Footnotes
Supplementary information is available at the website of Acta Pharmacologica Sinica.
Supplementary Information
Supplementary Figure S1–S2.
List of compound information in the pilot screening.
References
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 2009; 10: 697–708. [DOI] [PubMed] [Google Scholar]
- Tan JZ, Yan Y, Wang XX, Jiang Y, Xu HE. EZH2: biology, disease, and structure-based drug discovery. Acta Pharmacol Sin 2014; 35: 161–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang MW. Histone lysine methyltransferases as anti-cancer targets for drug discovery. Acta Pharmacol Sin 2016; 37: 1273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc Natl Acad Sci U S A 2012; 109: 2989–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 2010; 42: 181–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan RJ, Nitta M, Borger D, Zukerberg LR, Ferry JA, Harris NL, et al. EZH2 codon 641 mutations are common in BCL2-rearranged germinal center B cell lymphomas. PLoS One 2011; 6: e28585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A 2010; 107: 20980–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens C, Solis LM, Lin H, Yuan P, Tang X, Kadara H, et al. EZH2 protein expression associates with the early pathogenesis, tumor progression, and prognosis of non-small cell lung carcinoma. Clin Cancer Res 2013; 19: 6556–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Xie D, Yin Li W, Man Cheung C, Yao H, Chan CY, et al. RNAi targeting EZH2 inhibits tumor growth and liver metastasis of pancreatic cancer in vivo. Cancer lett 2010; 297: 109–16. [DOI] [PubMed] [Google Scholar]
- Fussbroich B, Wagener N, Macher-Goeppinger S, Benner A, Falth M, Sultmann H, et al. EZH2 depletion blocks the proliferation of colon cancer cells. PLoS One 2011; 6: e21651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He LJ, Cai MY, Xu GL, Li JJ, Weng ZJ, Xu DZ, et al. Prognostic significance of overexpression of EZH2 and H3k27me3 proteins in gastric cancer. Asian Pac J Cancer Prev 2012; 13: 3173–8. [DOI] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A 2003; 100: 11606–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002; 419: 624–9. [DOI] [PubMed] [Google Scholar]
- Weikert S, Christoph F, Kollermann J, Muller M, Schrader M, Miller K, et al. Expression levels of the EZH2 polycomb transcriptional repressor correlate with aggressiveness and invasive potential of bladder carcinomas. Int J Mol Med 2005; 16: 349–53. [PubMed] [Google Scholar]
- Béguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer cell 2013; 23: 677–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garapaty-Rao S, Nasveschuk C, Gagnon A, Chan EY, Sandy P, Busby J, et al. Identification of EZH2 and EZH1 small molecule inhibitors with selective impact on diffuse large B cell lymphoma cell growth. Chem Biol 2013; 20: 1329–39. [DOI] [PubMed] [Google Scholar]
- Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol 2012; 8: 890–6. [DOI] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 2012; 492: 108–12. [DOI] [PubMed] [Google Scholar]
- Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A 2012; 109: 21360–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Kim W, Howard TP, Vazquez F, Tsherniak A, Wu JN, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med 2015; 21: 1491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Mol cell 2011; 43: 798–810. [DOI] [PubMed] [Google Scholar]
- Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 2012; 338: 1465–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Gao S, Li J, Liu D, Sheng C, Yao C, et al. Wedelolactone disrupts the interaction of EZH2-EED complex and inhibits PRC2-dependent cancer. Oncotarget 2015; 6: 13049–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Xing X, Hu M, Zhang Y, Liu P, Chai J. Structural basis of EZH2 recognition by EED. Structure 2007; 15: 1306–15. [DOI] [PubMed] [Google Scholar]
- Kim W, Bird GH, Neff T, Guo G, Kerenyi MA, Walensky LD, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol 2013; 9: 643–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Chen L, Jiao L, Jiang X, Lian F, Lu J, et al. Astemizole arrests the proliferation of cancer cells by disrupting the EZH2-EED interaction of polycomb repressive complex 2. J Med Chem 2014; 57: 9512–21. [DOI] [PubMed] [Google Scholar]
- Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov 2004; 3: 301–17. [DOI] [PubMed] [Google Scholar]
- Zinzalla G, Thurston DE. Targeting protein-protein interactions for therapeutic intervention: a challenge for the future. Future Med Chem 2009; 1: 65–93. [DOI] [PubMed] [Google Scholar]
- Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem 1989; 182: 319–26. [DOI] [PubMed] [Google Scholar]
- Zhang TT, Huang ZT, Dai Y, Chen XP, Zhu P, Du GH. High-throughput fluorescence polarization method for identifying ligands of LOX-1. Acta Pharmacol Sin 2006; 27: 447–52. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 1999; 4: 67–73. [DOI] [PubMed] [Google Scholar]
- Li D, Han Q, Yang XY, Rong DM, Lin ZJ. Effects of kappa-selenocarrageenan on membrane fluidity and ghost reseal ability of rat erythrocyte. Acta Pharmacol Sin 1994; 15: 211–4. [PubMed] [Google Scholar]
- Nikolovska-Coleska Z, Wang R, Fang X, Pan H, Tomita Y, Li P, et al. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem 2004; 332: 261–73. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. SUZ12 is required for both the histone methyl-transferase activity and the silencing function of the EED-EZH2 complex. Mol Cell 2004; 15: 57–67. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 2002; 111: 185–96. [DOI] [PubMed] [Google Scholar]
- Jiao L, Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 2015; 350: aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooun A, Gajiwala KS, Deng YL, Liu W, Bolanos B, Bingham P, et al. Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nat commun 2016; 7: 11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin MR, Glicksman MA, Fu H, Havel JJ, Du Y. Inhibition of protein-protein interactions: non-cellular assay formats. In: Sittampalam GS, Coussens NP, Nelson H, Arkin M, Auld D, Austin C, Bejcek B, et al. editors. Assay Guidance Manual. Bethesda (MD); 2004.
- Chen X, Ji ZL, Chen YZ. TTD: Therapeutic target database. Nucleic Acids Res 2002; 30: 412–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvernmo T, Houben J, Sylte I. Receptor-binding and pharmacokinetic properties of dopaminergic agonists. Curr Top Med Chem 2008; 8: 1049–67. [DOI] [PubMed] [Google Scholar]
- Banks P, Gosselin M, Prystay L. Impact of a red-shifted dye label for high throughput fluorescence polarization assays of G protein-coupled receptors. J Biomol Screen 2000; 5: 329–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1–S2.
List of compound information in the pilot screening.