Abstract
The open field is a classic test used to assess exploratory behavior, anxiety, and locomotor activity in rodents. Here we mapped quantitative trait loci (QTLs) underlying behaviors displayed in an open field, using a panel of 53 BXD recombinant inbred mouse strains with deep replication (10 per strain and sex). The use of these strains permits the integration and comparison of data obtained in different laboratories, and also offers the possibility to study trait covariance by exploiting powerful bioinformatics tools and resources. We quantified behavioral traits during 20 min test sessions including (1) percent time spent and distance travelled near the wall (thigmotaxis), (2) leaning against the wall, (3) rearing, (4) jumping, (5) grooming duration, (6) grooming frequency, (7) locomotion, and (8) defecation. All traits exhibit moderate heritability making them amenable to genetic analysis. We identified a significant QTL on chromosome M.m. 4 at ~104 Mb that modulates grooming duration in both males and females (LRS values of ~18, explaining 25% and 14% of the variance, respectively) and a suggestive QTL modulating locomotion that maps to the same locus. Bioinformatic analysis indicates Disabled 1 (Dab1, a key protein in the reelin signaling pathway) as a particularly strong candidate gene modulating these behaviors. We also found two highly suggestive QTLs for a sex by strain interaction for grooming duration on chromosomes 13 and 17. In addition, we identified a pairwise epistatic interaction between loci on chromosomes 12 at 36-37 Mb and 14 at 34-36 Mb that influences rearing frequency in males.
Keywords: Exploratory behavior, BXD recombinant inbred mice, quantitative trait loci, complex traits, systems genetics, Dab1, genotype x sex interaction, epistatic interaction, repetitive behavior, mouse chromosome 4
Introduction
Analyzing behaviors of rodents in a novel open field is a classic test traditionally used to assess emotionality (Hall, 1934, Hall, 1936a, Hall, 1936b). The behavior displayed is considered to be the result of two conflicting drives: curiosity and fear (Barnett & Cowan, 1976), but is now more often interpreted in terms of exploratory behavior, anxiety, and locomotor activity. Exploratory behavior is elicited by novel stimuli and consists of behavioral acts and postures that permit an animal to collect information about new aspects of the environment (Crusio & Van Abeelen, 1986). Some repetitive behaviors, such as grooming or drug-induced stereotypy, can compete with these basic exploratory behaviors. Thus, behavior in an open field has become an important aspect of the development of models of several human disorders including autism spectrum (ASD) and substance-use disorders, underscoring the importance of identifying gene variants and neural networks regulating these behaviors.
Behavior in an open field was among the first behavioral phenotypes to be analyzed by quantitative genetic methods. Classical Mendelian crosses as well as diallel crosses invariably indicated significant genetic variation among inbred strains (Broadhurst, 1969, Crusio, 2000, Crusio, 2013, Crusio & Van Abeelen, 1986, Henderson, 1986, Willis-Owen & Flint, 2006). The genetic architecture of the different behaviors consisted of additive-genetic effects as well as ambidirectional dominance, a pattern consistent with the action of multiple polymorphic genes (Crusio, 2013). Factor analyses of genetic correlations reveals the presence of three underlying processes: exploration, anxiety, and self-maintaining behavior (grooming frequency and duration; Crusio, 2000).
When molecular-genetic techniques became available that made it possible to determine the approximate locus of some of these polygenes (now termed Quantitative Trait Loci - QTLs), open-field behavior again was among the first behavioral phenotypes to be so investigated (Flint et al., 1995, Gershenfeld et al., 1997, Mathis et al., 1995, Mott & Flint, 2002, Talbot et al., 1999). Several loci have been mapped, although few genes have actually been identified (Logan et al., 2013, Williams et al., 2014, Yalcin et al., 2004). Most of these studies were carried out using different heterogeneous populations, necessitating the genotyping at many marker loci of every individual mouse studied, whereas the other studies could use only the relatively moderate numbers of recombinant inbred (RI) strains that were then available. RI strains have the advantage that all of them have already been densely genotyped (Shifman et al., 2006, Williams et al., 2001). In addition, because RI strains are fully inbred, they can be easily shared among a research community. Phenotypical data obtained by different groups can be fruitfully combined, opening the possibility to test replicability, GXE effects, and to exploit powerful bioinformatic resources to study trait covariance, pleiotropy, and genetic modulation. For example, extensive mRNA expression datasets are available for the BXD RIs for different brain regions, and these can be jointly analyzed along with behavioral, neural, or physiological phenotypes (Chesler et al., 2005, Mulligan et al., 2017). Until relatively recently, however, the number of RI strains was too small to allow mapping with sufficient power and precision. This changed about a decade ago when an extended set of RI strains became available, derived from the C57BL/6J and DBA/2J strains (BXD RIs; Peirce et al., 2004). A large number of widely-studied behaviors have been assessed in this expanded set of RI strains (Philip et al., 2010), revealing precise QTLs, though many specific aspects of behavior have yet to be examined. We therefore decided to use this resource for a large-scale systems-genetics study comprising 53 BXD strains aimed at localizing QTLs for behaviors displayed in a novel open field.
Materials and Methods
Subjects
Breeding pairs of 53 BXD strains were acquired from the University of Tennessee Health Center (Memphis, TN, USA) and the Center for Neurogenomics and Cognitive Research (Free University, Amsterdam, Netherlands). Breeding pairs of the parental strains (C57BL/6J and DBA/2J) were obtained from Charles River (L’Arbresle, France). Our aim was to test 10 males and 10 females of each strain in the open field. Because of logistical problems (for example, some strains not breeding well), we did not achieve this goal for 16l strains. Exact information about sample sizes per sex and strain is presented in supplementary worksheet 1. Briefly, at an age of 11 +/− 1 weeks, we observed the behavior of 914 mice: 451 females from 52 BXD strains and 463 males from 53 BXD strains, as well as 10 males and 10 females for each one of the parental strains. All animals used were housed and bred in the SPF mouse facility of the University of Bordeaux (Pessac) in a climate-controlled breeding room (temperature: 21+/−1 °C, humidity: 55+/−10%, 12 hour light-dark cycle with lights on at 7 am). Food (Safe, type 113, sterilized) and water (softened, sterilized) were available ad libitum. Animals were housed 2-4 in same sex/genotype groups in clear plastic cages (162×406×176 mm, Tecniplast) filled with poplar wood shavings (Souralit).
Open field
Procedure
The open field was a rectangular cage (109*49*49 cm) with a clear Plexiglas front panel that was placed in a brightly-lit room (~300 lx). Thirty minutes prior to the test mice were taken to the experimental room. All tests took place between 8:30 am and 5:30 pm. Animals were placed in the center and, starting 5 sec later, observed directly and continuously for 20 min. Overall locomotor activity and time spent in the thigmotaxis zone (surface within 5 cm of the walls) were recorded automatically using the Ethovision program (version 3, over the course of the project successively upgraded to version 8; Noldus, Wageningen, The Netherlands). Rearing (standing upright on the hind legs, while forepaws are free); leaning (standing upright on the hind legs, one or two forepaws against the wall), grooming frequency (number of grooming episodes), grooming duration, and jumping, were scored manually in Ethovision by using a computer keyboard with several keys coding for the different behaviors. Grooming included all self-cleaning activities such as tail and ventrum licking, facial wiping, etc. Defecation was quantified by counting the number of fecal boli deposited during a session. The floor was wiped with water in between sessions, but not rinsed.
On any test day, animals of a given sex were tested as one batch first, followed by the batch of the other sex. The order of testing for males and females was alternated between test days.
Statistical analysis
All statistical analyses were performed with SAS 9.3 (Sas Institute Inc., 1987). Data are reported as means +/− SEM. To determine strain and sex effects, we used two-way ANOVAs with strain and sex as between-subjects factors. As a measure of the strength of strain differences, we estimated the intraclass correlation as follows. The between-strain and within-strain variance components were derived from the expected mean squares from one-way ANOVAs that were run separately for each sex using the SAS procedure GLM. The estimate of the intraclass correlation then equals ri = Vbetween strains/(Vbetween strains + Vwithin strains). The percentage of the variance between means explained by a certain QTL was calculated as the square of the correlation between the strain means and the genetic marker with the highest LRS score (note that to some extent this will be an overestimation, due to the ‘Beavis effect’; see Xu, 2003).
Bioinformatics
All of the genetic analyses were done in GeneNetwork (www.genenetwork.org; Parker et al., 2017) which is an open access bioinformatics resource for systems genetics that exists as both a repository for genetic, genomic, and phenotypic data together with a suite of statistical programs for data analysis that includes mapping and evaluating QTLs, examining phenotype/genotype correlations, and building interaction networks (Mulligan et al., 2017). Genes located within significant QTL intervals were screened with the QTLminer component of GeneNetwork (Alberts & Schughart, 2010), the Mouse Genome Informatics/Strains, SNPs and Polymorphisms database (www.informatics.jax.org/strains_SNPs.shtml), and the NCBI dbSNP database (www.ncbi.nlm.nih.gov/snp). Functional associations and gene ontology for candidate genes were further assessed using Gene (www.ncbi.nlm.nih.gov/gene), DAVID (version 6.8; david.abcc.ncifcrf.gov), WebGestalt (bioinfo.vanderbilt.edu/webgestalt/), and literature mining using PubMed (www.ncbi.nlm.nih.gov/pubmed). In order to prioritize positional candidates within the QTL intervals, the GeneWeaver database (www.geneweaver.org) was queried to find gene sets from previous studies from other labs associated with grooming and rearing. The gene sets were further analyzed using tools available at the GeneWeaver database to identify genes that are highly represented among these gene sets. The results of the GeneWeaver analysis were exported to the STRING database of known and predicted protein-protein interactions (string-db.org/) to identify the relationship of positional candidates to previously reported grooming related genes.
QTL mapping
The QTL mapping module of GeneNetwork was used to identify QTLs for the open-field data. This module enables interval mapping, composite interval mapping, and a pairwise scan option to identify epistatic effects. QTL significance was assessed using the likelihood ratio statistic (LRS) obtained after 5000 permutations and 2000 bootstrap tests. QTLs were deemed significant if Pgenome-wide<0.05 and suggestive if Pgenome-wide<0.63, which yields, on average, one false positive per genome scan. Male and female data were analyzed separately. In cases where interactions between strain and sex were present (i.e., sex differences vary over strains), we mapped the QTLs responsible for these interactions by analyzing the sex differences (measured as the difference between the male and female strain means). In all cases, outliers were winsorized.
The presence of cis-eQTLs for the genes within the QTL intervals was determined by correlating phenotype data with brain tissue-specific gene expression levels. For this, we used publicly-available data obtained from GeneNetwork, using the following datasets: amygdala [INIA Amygdala cohort Affy MoGene 1.0 ST Mar 2011 RMA], cerebellum [SJUT Cerebellum mRNA M430 (March 2005) RMA] hippocampus [Hippocampus Consortium M430v2 (June 2006) RMA], hypothalamus [INIA Hypothalamus Affy MoGene 1.0 ST (Nov10) RMA], pre-frontal cortex [VCU BXD PFC SAL M430 2.0 (Dec 2006) RMA], striatum [HBP Rosen Striatum M430V2 (April 2005) RMA], whole brain [INIA Brain mRNA 430 (June 2006) RMA, hippocampus exon [UMUTaffy Hippocampus exon (Feb 2009) RMA], striatum exon [HQF Striatum Affy Mouse Exon 1.0ST (Dec 2009) RMA]. All these datasets were based on pooled samples of males and females (except the one for the pre-frontal cortex which used males only).
Results
Raw data
All data have been deposited in the GeneNetwork database and are publicly available. GeneNetwork trait IDs (male ID followed by female ID): Locomotion: 17528, 17538; Distance covered near the wall as a percentage of total distance covered: 17529, 17539, Percentage time spent near the wall: 17530, 17540; Leaning against the wall: 17531, 17541; Rearing: 17532, 17542; Grooming, bout frequency: 17533, 17543; Grooming, duration: 17534, 17544; Jumping: 17536, 17546; and Defecation: 17537, 17547. Strain means for the different variables are presented in Figure 1 and Supplementary Figure 5.
Figure 1. BXD strain differences for grooming duration in the open field.
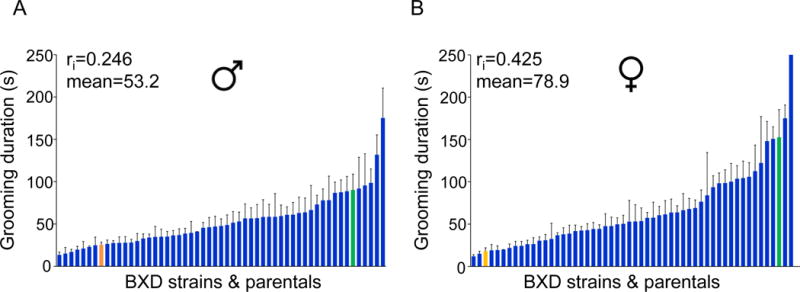
Variation in grooming duration time in the BXD mice and parental strains. C57BL/6J orange bars and DBA/2J green bars; Males (A) and females (B). Bar graphs represent the means (+/-SEM). Intra-class correlations and overall means are shown. The y-axis represents grooming duration time in seconds with BXD strains rank ordered from low to high. For enlarged versions indicating strain names, see Figures S1 A and B.
Strain and sex differences
For all variables measured, strain differences were significant. Differences between sexes were observed for percent distance in the thigmotaxis zone, rearing, grooming frequency, grooming duration, and defecation. Significant sex*strain interactions were observed for locomotion, rearing, grooming duration, and defecation (locomotion: strain: F53,804=17.60, P<0.001; sex: F1,804=0.31, ns; strain * sex: F53,804=2.13, P<0.001; percent distance in the thigmotaxis zone: strain: F53,804=11.76, P<0.001; sex: F1,804=4.92, P<0.05; strain * sex: F53,804=1.10, ns; percent time in the thigmotaxis zone: strain: F53,804=9.94, P<0.001; sex: F1,804=2.97, ns; strain * sex: F53,804=1.24, ns; leaning: strain: F53,804=8.35, P<0.001; sex: F1,804=0.04, ns; strain * sex: F53,804=0.96, ns; rearing: strain: F53,804=8.94, P<0.001; sex: F1,804=25.68, P<0.001; strain * sex: F53,804=1.46, P<0.05; grooming frequency: strain: F53,804=4.46, P<0.001; sex: F1,804=11.55, P<0.001; strain * sex: F53,804=1.22, ns; grooming duration: strain: F53,804=8.55, P<0.001; sex: F1,804=20.08, P<0.001; strain * sex: F53,804=2.88, P<0.001; jumping: strain: F53,804=3.47, P<0.001; sex: F1,804=0.59, ns; strain * sex: F53,804=0.55, ns; defecation: strain: F53,804=8.06, P<0.001; sex: F1,804=36.04, P<0.001; strain * sex: F53,804=1.72, P<0.01).
Mapping of significant QTLs
The clear strain differences indicate the presence of significant genetic variation, offering the opportunity to attempt to localize QTLs for these phenotypes. A significant QTL peak was obtained for grooming duration on Chr 4 in both males and females (LRS=17.9 and LRS=18.1, respectively; Figure 2A and B) accounting for 25% of the variance in males and 14% in females (Figure 1). Although B6 animals show smaller grooming durations than D2 animals, strains that inherited the B haplotype at this locus had increased grooming time. The QTL region, including peaks and shoulders, for males spanned 97-112 Mb and the highest LRS value occurred at 104 Mb, whereas for females the interval on Chr 4 ranged from 101-125 Mb and peaked at ~103 Mb. Composite interval mapping controlling for this QTL in males and females did not reveal any secondary QTLs.
Figure 2. Whole genome scans of trait data for grooming duration and locomotion.
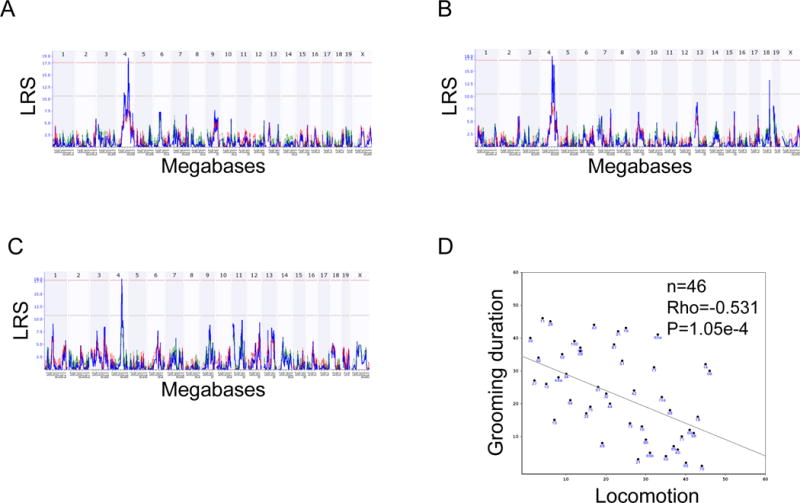
A-C: Whole genome scans for grooming duration (A: males, B: females) and locomotion (C: females). The x-axis represents chromosome number and megabase position (tick marks represent 25 Mb increments, each chromosome starting at 0) and the y-axis represents the likelihood ratio statistic (LRS) of linkage. Blue lines represent LRS across the genome. The pink and gray horizontal lines are approximate threshold values which are used to assess whether a peak is significant (P<0.05) or suggestive (P<0.63) respectively. D: Spearman rank correlation plot for grooming duration and locomotion. For enlarged versions, see Figures S2 A–D.
QTL Region Analysis
GeneNetwork was queried for the set of all traits with LRS > 10 occurring within the same Chr 4 interval. From this search, the trait that mapped most closely to the region of our QTL interval was locomotor activity (horizontal distance travelled) that had also been measured in an open field (Philip et al., 2010). The QTL interval for locomotion in that study mapped within the interval that we observed here for grooming duration (Figure 2C) and the two traits correlated inversely (Figure 2D).
The QTL interval was reviewed to define potential candidate genes based on currently known functional associations with phenotypes related to grooming and locomotor activity (Supplementary worksheet 2). Within the 97-112 Mb region of Chr 4 there are 146 genes and predicted genes (GeneNetwork database queried September 28, 2016 and MGI database queried September 29, 2016). Of the genes for which functional information is currently available, only Dab1 is associated with both locomotor and grooming phenotypes. Elavl4 and Lepr are connected with locomotor behavior. Lrp8, the reelin receptor, also occurs within the QTL interval but it is not known to be associated with either phenotype.
Dab1 is a key component of the reelin signaling pathway, involved in neuronal migration and positioning during development. In the QTL interval, Dab1 occurs around the QTL peak at ~103.839/104.166 Mb (SNP: rs32341666 single nucleotide variant G/T, with B6 having a G and D2 a T).
Genes within the QTL interval were also examined for the presence of cis-eQTLs in brain-related expression datasets specific for the BXDs (Supplementary worksheet 3). If such cis-eQTLs exist for any of the genes implicated in the previous section, this would increase the likelihood that these genes modulate the trait, since cis regulation indicates a difference in gene expression levels. For Dab1, there were no cis-eQTLs in either amygdala, cerebellum, hippocampus, hypothalamus, pre-frontal cortex, striatum, or whole brain. There were however Dab1 cis-eQTLs in two datasets of exon level gene expression in the hippocampus and striatum. There were 4 other genes within the Chr. 4 interval associated with behavior (Elavl4, Sgip1, Insl5, Lepr), 2 of which (Elavl4, Sgip1) also showed cis-eQTLs in the hippocampus and striatum exon-level datasets.
Grooming positional candidate evaluation
To further evaluate the genes within the QTL interval for an association with grooming, fifteen gene sets from previous experiments related to grooming behavior were retrieved from the GeneWeaver database: (GS181097: Mouse GO:2000821 regulation of grooming behavior, GS185500: Mouse GO:0007625 grooming behavior, GS197887: Human GO:2000821 regulation of grooming behavior, GS202324: Human GO:0007625 grooming behavior, GS238017: Human [MeSH] Grooming, : D006120 GS224143: Rat Anxiety related response QTL 19 [Anxrr19 Published QTL Chr 10], GS224144: Rat Anxiety related response QTL 18 [Anxrr18 Published QTL Chr 2], GS224524: Rat Anxiety related response QTL 20 [Anxrr20 Published QTL Chr 18], GS163449: Mouse MP:0001441 increased grooming behavior, GS166329: Mouse MP:0009327 abnormal maternal grooming, GS166392: Mouse MP:0001440 abnormal grooming behavior, GS168966: Mouse MP:0001442 decreased grooming behavior, GS168968: Mouse MP:0001443 poor grooming). Two gene sets were omitted from further analysis (GS136298: Maternal Performance and GS236065: [MeSH] Hair Preparations) due to lack of relevance.
In the first analysis, the union of these gene sets was obtained and intersected with the complete list of positional candidates to determine whether any of the positional candidates had already been associated with grooming. No elements were found in this intersection. Therefore we tested the putative relationship with Dab1 to grooming-related genes as follows: Among the 13 gene sets above, highly represented genes were found using the “Gene Set Graph” tool in GeneWeaver (Figure 3A). The 9 most highly connected genes were Dlg4, Hoxb8, Shank3, Nrxn1, Hprt, Ppt1, Ddo, Nmur2, Oxt, of which Dlg4, Hoxb8, Shank3, Hprt, and Ppt1 are associated with both grooming and locomotor activity. The set of 9 genes was entered into EMBL STRING along with the candidate Dab1. The network was expanded by one step and a limit of 5 interactors to search for the existence of a short path between the candidate and known (experimentally-derived) grooming genes. Dlg4, the most frequently annotated gene in GeneWeaver grooming data is one step from Dab1 through the Grin1, Grin2a, and Grin2b NMDA subunit genes (Figure 3B). Therefore it is a plausible functional candidate for grooming behavior based on molecular proximity to known genes. The relationship is through known interactions among homologues of Dab1 and NMDA subunits in other species, and co-occurrence in PubMed abstracts.
Figure 3. Relationship of Dab1 with grooming-related genes.
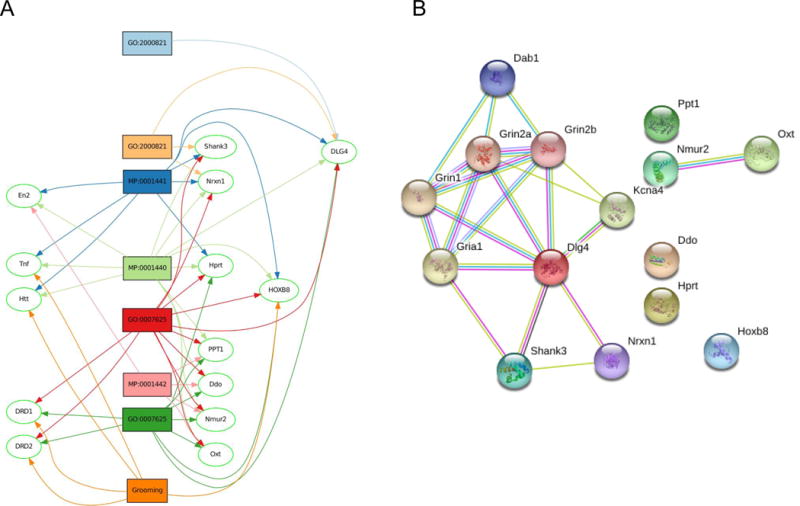
GeneSet graph intersection (A). Analysis of thirteen gene sets associated with grooming behavior revealed 9 highly represented genes (Dlg4, Hoxb8, Shank3, Nrxn1, Hprt, Ppt1, Ddo, Nmur2, Oxt). STRING molecular interaction network (B) illustrating the shortest path of the highly represented genes associated with grooming behavior and Dab1. Known interactions indicated by pink edges are experimentally confirmed, and those indicated by blue edges are from curated databases. Relationships indicated by green edges are from text mining, and black edges from gene co-expression analysis. For enlarged versions, see Figures S3 A and B.
Mapping of suggestive QTLs
For females, suggestive QTLs were found for locomotion (Chr 4 and Chr 9), percent thigmotaxis time (Chr 4 and Chr 15), percent thigmotaxis distance (Chr 3, Chr 4, Chr 5, Chr 8, Chr 11, and Chr 13), grooming frequency (Chr 4, Chr 18), grooming duration (Chr 18), leaning (Chr 5), and defecation (Chr 4) (Supplementary Figure 6). For males suggestive QTLs were found for locomotion (Chr 4), percent thigmotaxis time (Chr 4), percent thigmotaxis distance (Chr 3), grooming frequency (Chr 3 and Chr 18), leaning (Chr 4 and Chr 13), defecation (Chr 9) and rearing (Chr 15) (Supplementary Figure 6).
Suggestive QTL peaks common between males and females occurred for locomotion (Chr 4), grooming frequency (Chr 18), percent thigmotaxis time (Chr 4), and percent thigmotaxis distance (Chr 3; Supplementary Figure 6).
Mapping of QTLs for sex by strain interactions
For grooming duration, defecation, locomotion, and rearing we found significant sex by strain interactions, warranting a search for the underlying genes. QTL mapping of these data revealed highly suggestive QTLs for grooming duration on Chr 13 (LRS=15.54, 30-46 Mb) and Chr 17 (LRS=15.54, 72-78 Mb) and some barely suggestive QTLs for the other traits (defecation, locomotion, and rearing; Supplementary Figure 8). The genes within the Chr 13 and Chr 17 intervals were examined for association with sex related characteristics. The Chr 13 interval contained 102 genes. Two of these, Bmp6 (Bone morphogenetic protein 6) and Mak (male germ cell association kinase) are linked with male genitalia development and male germ cell processes, respectively. The Chr 17 interval was narrow and only contained 20 genes, one of them, Srd5a2 (steroid alpha-reductase 2) is associated with sexual differentiation and male and female genitalia development. Bmp6, Mak, and Srd5a2 each differ at 2 or 3 SNP polymorphisms between the parental strains, C57BL/6J and DBA/2J.
Multivariate analyses
A factor analysis of open-field traits was performed on strain means for the BXD and parental lines (Table 1) in order to clarify the multivariate structure of the data. For both the male and female data sets two factors had an eigenvalue >1, which were subjected to an orthoblique Harris-Kaiser rotation (Table 1). Results for males and females were very similar. Factor 1 shows positive loadings of both thigmotaxis-related variables and may represent anxiety. Factor 2 had sizeable loadings for grooming (duration and frequency) and defecation, as well as for jumping, leaning, and locomotion, but with an opposite sign. This factor may represent exploration. No strong loadings were found for rearing in females, but it loaded on Factor 1 in males, with a sign opposite to thigmotactic tendencies.
Table 1.
Factor analysis of open field-related variables
| Trait |
Females factor 1 |
factor 2 |
Males factor 1 |
factor 2 |
|---|---|---|---|---|
| % time in Thigmotaxis zone | 0.96 | −0.05 | 0.97 | 0.01 |
| % distance in Thigmotaxis zone | 0.96 | 0.04 | 0.94 | −0.04 |
| Rearing | −0.24 | −0.15 | −0.39 | 0.22 |
| Grooming duration | 0.01 | 0.76 | 0.12 | −0.70 |
| Grooming frequency | −0.11 | 0.75 | −0.09 | −0.37 |
| Defecation | 0.19 | 0.46 | −0.05 | −0.34 |
| Jumping | −0.04 | −0.25 | 0.10 | 0.40 |
| Leaning | 0.29 | −0.43 | 0.29 | 0.59 |
| Locomotion | −0.15 | −0.59 | −0.09 | 0.77 |
Harris-Kaiser rotation method; factor loadings >l0.30l are in bold
QTL mapping of combined open-field traits
The factor scores were used to detect possible additional QTLs (Supplementary Figure 7). No significant QTLs were obtained for any of the male or female factors. For females, suggestive QTLs occurred on Chr 3 and Chr 4 for Factor 1 and Chr 4 for Factor 2. For males, a suggestive QTL was obtained on Chr 4 for Factor 2. The Factor 2 QTLs occurred around the same Mb location observed for the significant QTLs produced for grooming duration (and locomotion) on Chr 4.
Epistatic QTLs
Epistatic effects are non-additive interactions between multiple QTL loci where the combined effect is either greater or smaller than the summed effects of each gene alone (Mather & Jinks, 1982). A significant pairwise-epistatic interaction was found for rearing in males between loci on Chr 12 and Chr 14 (LRS 1=1.557, LRS 2=0.878, LRS interact =33.754, LRS full=35.908, p<0.05, Figure 4).
Figure 4. Two-dimensional plot of LRS scores for pairs of loci involved in epistatic interaction for rearing in males.
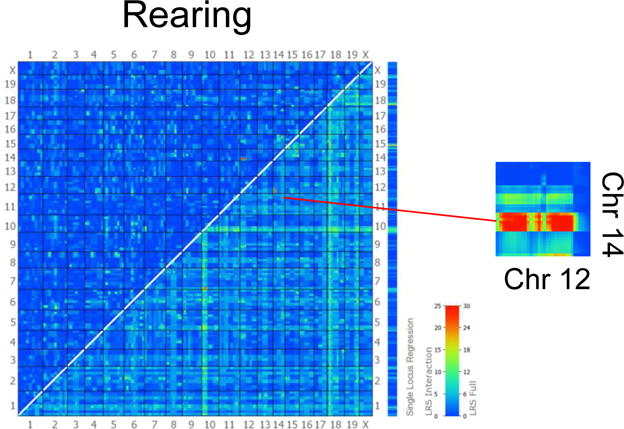
The lower right half of the plot gives a summary of LRS values of the full model, representing cumulative effects of both loci and their possible interaction. The upper left side of the plot indicates the LRS values for the presence of epistatic interactions. Both sets of x and y axes are labeled with chromosome number. Zoomed thumbnail indicates Chr12/Chr14 interaction colored in red. For an enlarged version, see Figure S4.
A QTL miner search using GeneNetwork at the 36.628917 - 37.595106 Mb location (MGI: 35.925620-36.689444 Mb) on Chr 12 rendered 9 or 10 genes within the interval: Agr3: anterior gradient 3, Agr2: anterior gradient 2, Tspan13: tetraspanin 13, Bzw2: basic leucine zipper and W2 domains 2, Ankmy2: ankyrin repeat and MYND (myeloid, Nervy, and DEAF-1)-type Zn2+ finger domain containing 2, Lrrc72: leucine rich repeat containing 72, Sostdc1: sclerostin domain containing 1 Ispd: isoprenoid synthase domain containing, and two unclassified genes, D630036H23Rik and 1700101O05Rik. Additional support for the Chr 12 locus influencing rearing was found in a query of the GeneWeaver database where a gene list and interval associated with a Chr 12 QTL for rearing overlaps with the interval identified in this study (Gene Set- GS136584/PMID:14694905).
For the Chr 14 interval, a QTL miner search using GeneNetwork at the 34.026418 - 34.609256 Mb location (MGI: 33214026-33640754) resulted in 5 or 6 genes: Arhgap22: Rho GTPase activating protein 22, Mapk8: mitogen-activated protein kinase 8, Ptpn20: protein tyrosine phosphatase, non-receptor type 20, Frmpd2: FERM and PDZ domain containing 2, as well as two unclassified genes, 6030458A19Rik and A930006J02Rik. In an attempt to find a possible functional association among genes from both chromosomal regions, a PubMed literature search for co-occurrence and ontology resulted in 2 genes with known neurological associations, Mapk8 and Bzw2. Another potential link could be Arhgap22 and Agr2 which are implicated in cell growth and cell proliferation. Only a weak epistatic interaction was observed for rearing in females (LRS1= 0.037, LRS2 =4.242, LRS interact = 25.024, LRS full = 29.557, ns).
Discussion
In this study we aimed to identify genetic components of open-field behaviors using the extended BXD RI strains. We found one significant QTL: for grooming duration on Chr 4 for both males and females. Inspection of data from other laboratories available in the GeneNetwork database showed that a significant QTL was previously found in this interval for locomotion-related traits in an open-field. Of the candidate genes in this region, only Dab1 has been associated with both grooming and locomotion. Dab1scm (scrambler) is a spontaneous mutant that produces a truncated form of the Dab1 protein due to aberrant splicing. Scrambler mice have neuroanatomical abnormalities including degeneration of the cerebellum, hippocampus, and neocortex, with concomitant behavioral problems including abnormal motor coordination, balance, and gait (Sweet et al., 1996). Similar problems are not found in conditional Dab1 KO mice, which have normal Dab1 expression during development, although Dab1 deficiency in adulthood induces hyperactivity and decreased anxiety (Imai et al., 2016). In addition, it has been observed that scrambler mice have reduced grooming durations compared to wild-type controls (Strazielle et al., 2012). The Dab1 gene undergoes complex splicing which generates many functional isoforms with different combinations of signaling domains (Gao & Godbout, 2013). The Dab1 variant 2 relevant to this study, results from an alternative splicing event that excludes exon 7 which is predicted to delete 13 amino acids containing a consensus tyrosine phosphorylation site.
Based on the foregoing, we propose Dab1 as a likely gene candidate for the Chr 4 QTL. The significance of this finding is further supported by GWAS in human that have implicated DAB1 in ASD (Shen et al., 2016), as well as with plasma amyloid beta concentrations, one of the pathological hallmarks of Alzheimer disease (Chouraki et al., 2014). A possible pathway for the involvement of Dab1 in grooming is the association of Dab1 with NMDA receptors (Chen et al., 2005) through its connection with Grin subunits (cf. Figure 3b). In turn, NMDA receptors have been implicated in the regulation of grooming and other repetitive behaviors (Jaramillo et al., 2016, Kim et al., 2016).
The implication of Dab1 and the reelin pathway in this QTL for grooming duration is intriguing because this pathway is known to be associated with human autism. Grooming has been the subject of detailed genetic analyses due to its association with neuropsychiatric disorders such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, Obsessive Compulsive Disorder, Attention Deficit Disorder, and ASD (Kalueff et al., 2016, Roth et al., 2013). Self-grooming represents more than 15% of the waking activity in mice and after sleep is their main activity. Mice also groom in stressful situations such as novelty-based anxiety tests (e.g., elevated plus maze and open field), holeboard-decision making, etc. (reviewed in Kalueff et al., 2016, Roth et al., 2013). Our findings provide evidence for a construct validity of grooming as an ASD-related behavior in the mouse and further highlight the utility of the BXD RI population, systems genetic analysis, and our candidate gene prioritization and network augmentation approach as a means of studying this disorder.
Further findings from this study remain to be resolved. We also found two highly suggestive QTLs for the sex by strain interaction for grooming duration on Chr 13 and 17. Several strong candidates within these QTL intervals are Bmp6, Mak, and Srd5a2, associated with sexual differentiation, male germ cell processes, and male/female genitalia development.
There was a significant epistatic interaction between loci on Chr 12 and 14 for rearing behavior. Although these loci contain only few genes, no strong candidates could be identified. Two possible interacting gene pairs are Mapk8/Bzw2 and Arhgap22/Agr2. The Ahr gene in the Chr 12 interval has been reported as modulating both horizontal and vertical movement (Williams et al. 2014). However, the effect found in that study was a main effect of the QTL whereas the effect that we report here is only significant in interaction with the Chr 14 locus and specific for rearing. No effect of the Chr 12 locus was found on locomotor activity in our study.
Only suggestive QTLs were obtained for several of the other traits, despite the large number of strains used in this study. Most likely, this indicates a polygenic inheritance with each gene having only a very small contribution to the trait, confirming the results of previous research (Crusio, 2000, Crusio, 2013, Crusio & Van Abeelen, 1986).
In summary, the present study demonstrates the power of the BXD RIS approach for QTL mapping and analysis of behavioral traits that share both face and biomolecular construct validity with human behavioral disorders. The wealth of information available on these strains, ranging from a plethora of behavioral, neuronal, and physiological phenotypes to extensive gene-expression data from different brain regions, allows a powerful system genetics approach (Schughart & Williams, 2017) to identify the genetic and molecular processes underlying behavioral phenotypes. This strategy provides a bridge from behavioral genetic analysis into extensive biomolecular knowledge from functional genomics.
Supplementary Material
Worksheet S1. BXD strains and sample sizes.
Worksheet S2. Genes within the Chromosome 4 interval.
Worksheet S3. Cis-expression QTLs for genes within the Chromosome 4 interval.
Figure S1 BXD strain differences for grooming duration in the open field.
Enlarged version of Figure 1 A and B.
Figure S2 Whole genome scans and zoomed images of trait data for grooming duration and locomotion.
Enlarged versions of Figure 2 A–D. Panels A-C, below: significant QTL interval zoomed. Colored rectangles depict individual genes, red and green lines represent the additive genetic contribution; red lines indicate negative values (C57BL/6J alleles increasing trait values) and green lines indicate positive values (DBA/2J alleles increasing trait values). Gray lines are shown when the parental strain is unknown. The orange hash marks on the x-axis signify the SNP density (sequence difference between the two parental strains). The yellow bars represent the relative frequency of peak LRS at a given location from 2000 bootstrap resamples.
Figure S3 Relationship of Dab1 with grooming-related genes.
Enlarged version of Figure 3.
Figure S4 Two-dimensional plot of LRS scores for pairs of loci involved in epistatic interaction for rearing in males.
Enlarged version of Figure 4.
Figure S5 BXD strain differences in the open field.
Variation in the open field in the BXD strains and their parental strains (C57BL/6J and DBA/2J, indicated by orange and green bars respectively) for locomotion (A and B), thigmotaxis percent distance (C and D), thigmotaxis percent time (E and F), Leaning (G and H), jumping (I and J), defecation (K and L), rearing (M and N) grooming frequency (O and P). Bars represent mean +/- SEM. Intra-class correlations and overall mean are shown for each trait. The y-axis represents measurement and units for each of the traits and the x-axis lists the BXD strains which are rank ordered from low to high.
Figure S6 Whole genome scans of trait data for which suggestive QTLs were obtained.
Whole genome scans (top: males, bottom: females) for A and B: locomotion, C and D: thigmotaxis percent time, E and F: thigmotaxis percent distance, G and H: grooming frequency, I and J: grooming duration, K and L rearing, M and N: jumping, O and P: leaning, Q and R: defecation. The x-axis represents chromosome number and megabase position and the y-axis represents the likelihood ratio statistic (LRS) of linkage. Blue lines represent LRS across the genome. The pink and gray horizontal lines are approximate threshold values which are used to assess whether a peak is significant (P<0.05) or suggestive (P<0.63), respectively.
Figure S7. Whole genome scans for the factor scores.
Shown are scans for males (top) and females (bottom). A and B: factor 1, C and D: factor 2.
Figure S8. Whole genome scans for sex by strain interaction.
Shown are scans for the difference between male and female strain means for A: grooming duration, B: defecation, C: locomotion, and D: rearing.
Acknowledgments
This study was supported by the following grants: NIMH R01 MH072920 to WEC, NIAAA U01 AA016662 and U01 AA013499 to RWW, NIAAA R01 AA018776 to EJC, and NIAAA R01 AA021951 to LL. Drs. Sabine Spijker and August B. Smit (Free University of Amsterdam) from the Neuro-BSIK Mouse Phenomics Consortium (BSIK03053) generously provided several BXD strains (for the origin of these strains, see Loos et al., 2014). We thank Raphael Pineau and Laetitia Medan for expert animal care and Alexis Cornuez for help with behavioral testing.
Footnotes
All authors declare that they do not have any conflict of interest.
References
- Alberts R, Schughart K. QTLminer: identifying genes regulating quantitative traits. BMC Bioinform. 2010;11:516. doi: 10.1186/1471-2105-11-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett SA, Cowan PE. Activity, exploration, curiosity and fear: An ethological study. Interdisc Sci Rev. 1976;1:43–62. [Google Scholar]
- Broadhurst PL. Psychogenetics of emotionality in the rat. Ann N Y Acad Sci. 1969;159:806–824. doi: 10.1111/j.1749-6632.1969.tb12980.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Beffert U, Ertunc M, Tang TS, Kavalali ET, Bezprozvanny I, Herz J. Reelin modulates NMDA receptor activity in cortical neurons. J Neurosci. 2005;25:8209–8216. doi: 10.1523/JNEUROSCI.1951-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, Threadgill DW, Manly KF, Williams RW. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- Chouraki V, De Bruijn RF, Chapuis J, Bis JC, Reitz C, Schraen S, Ibrahim-Verbaas CA, Grenier-Boley B, Delay C, Rogers R, Demiautte F, Mounier A, Fitzpatrick AL, Berr C, Dartigues JF, Uitterlinden AG, Hofman A, Breteler M, Becker JT, Lathrop M, Schupf N, Alperovitch A, Mayeux R, van Duijn CM, Buee L, Amouyel P, Lopez OL, Ikram MA, Tzourio C, Lambert JC. A genome-wide association meta-analysis of plasma Ab peptides concentrations in the elderly. Mol Psychiatr. 2014;19:1326–1335. doi: 10.1038/mp.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crusio WE. Genetic dissection of mouse exploratory behavior. In: Schröder H, Steckler T, van der Staay FJ, Walther M, editors. Behavioural Phenotyping of Mouse Mutants. Office of the Associate Dean of Scientific Affairs, University of Cologne; Cologne: 2000. p. 31. [Google Scholar]
- Crusio WE. The genetics of exploratory behavior. In: Crusio WE, Sluyter F, Gerlai RT, Pietropaolo S, editors. Behavioral Genetics of the Mouse: Genetics of Behavioral Phenotypes. Cambridge University Press; Cambridge, United Kingdom: 2013. pp. 148–154. [Google Scholar]
- Crusio WE, van Abeelen JHF. The genetic architecture of behavioural responses to novelty in mice. Heredity. 1986;56:55–63. doi: 10.1038/hdy.1986.8. [DOI] [PubMed] [Google Scholar]
- Flint J, Corley R, DeFries JC, Fulker DW, Gray JA, Miller S, Collins AC. A simple genetic basis for a complex psychological trait in laboratory mice. Science. 1995;269:1432–1435. doi: 10.1126/science.7660127. [DOI] [PubMed] [Google Scholar]
- Gao Z, Godbout R. Reelin-Disabled-1 signaling in neuronal migration: splicing takes the stage. Cell Mol Life Sci. 2013;70:2319–2329. doi: 10.1007/s00018-012-1171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenfeld HK, Neumann PE, Mathis C, Crawley JN, Li X, Paul SM. Mapping quantitative trait loci for open-field behavior in mice. Behav Genet. 1997;27:201–210. doi: 10.1023/a:1025653812535. [DOI] [PubMed] [Google Scholar]
- Hall CS. Emotional behavior in the rat. I. Defecation and urination as measures of individual differences in emotionality. J Comp Psychol. 1934;18:385–403. [Google Scholar]
- Hall CS. Emotional behavior in the rat. II. The relationship between need and emotionality. J Comp Psychol. 1936a;22:61–68. [Google Scholar]
- Hall CS. Emotional behavior in the rat. III. The relationship between emotionality and ambulatory activity. J Comp Psychol. 1936b;22:345–352. [Google Scholar]
- Henderson ND. Predicting relationships between psychological constructs and genetic characters: an analysis of changing genetic influences on activity in mice. Behav Genet. 1986;16:201–220. doi: 10.1007/BF01065486. [DOI] [PubMed] [Google Scholar]
- Imai H, Shoji H, Ogata M, Kagawa Y, Owada Y, Miyakawa T, Sakimura K, Terashima T, Katsuyama Y. Dorsal Forebrain-Specific Deficiency of Reelin-Dab1 Signal Causes Behavioral Abnormalities Related to Psychiatric Disorders. Cerebral Cortex. 2016 doi: 10.1093/cercor/bhv334. in press. [DOI] [PubMed] [Google Scholar]
- Jaramillo TC, Speed HE, Xuan Z, Reimers JM, Liu S, Powell CM. Altered Striatal Synaptic Function and Abnormal Behaviour in Shank3 Exon4-9 Deletion Mouse Model of Autism. Autism Research. 2016;9:350–375. doi: 10.1002/aur.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalueff AV, Stewart AM, Song C, Berridge KC, Graybiel AM, Fentress JC. Neurobiology of rodent self-grooming and its value for translational neuroscience. Nature Reviews Neuroscience. 2016;17:45–59. doi: 10.1038/nrn.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Lim CS, Kaang BK. Neuronal mechanisms and circuits underlying repetitive behaviors in mouse models of autism spectrum disorder. Behav Brain Func. 2016;12:3. doi: 10.1186/s12993-016-0087-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan RW, Robledo RF, Recla JM, Philip VM, Bubier JA, Jay JJ, Harwood C, Wilcox T, Gatti DM, Bult CJ, Churchill GA, Chesler EJ. High-precision genetic mapping of behavioral traits in the diversity outbred mouse population. Genes Brain Behav. 2013;12:424–437. doi: 10.1111/gbb.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos M, Mueller T, Gouwenberg Y, Wijnands R, van der Loo RJ, Birchmeier C, Smit AB, Spijker S. Neuregulin-3 in the mouse medial prefrontal cortex regulates impulsive action. Biol Psychiatry. 2014;76:648–655. doi: 10.1016/j.biopsych.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Mather K, Jinks JL. Biometrical Genetics The Study of Continuous Variation. Chapman and Hall; London, UK: 1982. [Google Scholar]
- Mathis C, Neumann PE, Gershenfeld H, Paul SM, Crawley JN. Genetic analysis of anxiety-related behaviors and responses to benzodiazepine-related drugs in AXB and BXA recombinant inbred mouse strains. Behav Genet. 1995;25:557–568. doi: 10.1007/BF02327579. [DOI] [PubMed] [Google Scholar]
- Mott R, Flint J. Simultaneous detection and fine mapping of quantitative trait loci in mice using heterogeneous stocks. Genetics. 2002;160:1609–1618. doi: 10.1093/genetics/160.4.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Mozhui K, Prins P, Williams RW. GeneNetwork: A Toolbox for Systems Genetics. Methods Molec Biol. 2017;1488:75–120. doi: 10.1007/978-1-4939-6427-7_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CC, Dickson PE, Philip VM, Thomas M, Chesler EJ. Systems Genetic Analysis in GeneNetwork.org. Current Protocols in Neuroscience. 2017;79:8, 39.31–38.39.20. doi: 10.1002/cpns.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip VM, Duvvuru S, Gomero B, Ansah TA, Blaha CD, Cook MN, Hamre KM, Lariviere WR, Matthews DB, Mittleman G, Goldowitz D, Chesler EJ. High-throughput behavioral phenotyping in the expanded panel of BXD recombinant inbred strains. Genes Brain Behav. 2010;9:129–159. doi: 10.1111/j.1601-183X.2009.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Kyzar EJ, Cachat J, Stewart AM, Green J, Gaikwad S, O’Leary TP, Tabakoff B, Brown RE, Kalueff AV. Potential translational targets revealed by linking mouse grooming behavioral phenotypes to gene expression using public databases. Progr Neuro-psychopharmacol Biol Psychiatry. 2013;40:312–325. doi: 10.1016/j.pnpbp.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute Inc SAS. SAS/STAT Guide for Personal Computers, Version. 6. Sas Institute; Cary, NC: 1987. [Google Scholar]
- Schughart K, Williams RW. Systems Genetics. Methods and Protocols. In: Walker JM, editor. Methods Molec Biol. Humana Press; New York: 2017. p. xiv.p. 609. [Google Scholar]
- Shen Y, Xun G, Guo H, He Y, Ou J, Dong H, Xia K, Zhao J. Association and gene-gene interactions study of reelin signaling pathway related genes with autism in the Han Chinese population. Autism Research. 2016;9:436–442. doi: 10.1002/aur.1540. [DOI] [PubMed] [Google Scholar]
- Shifman S, Bell JT, Copley RR, Taylor MS, Williams RW, Mott R, Flint J. A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biology. 2006;4:e395. doi: 10.1371/journal.pbio.0040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strazielle C, Lefevre A, Jacquelin C, Lalonde R. Abnormal grooming activity in Dab1(scm) (scrambler) mutant mice. Behav Brain Res. 2012;233:24–28. doi: 10.1016/j.bbr.2012.04.038. [DOI] [PubMed] [Google Scholar]
- Sweet HO, Bronson RT, Johnson KR, Cook SA, Davisson MT. Scrambler, a new neurological mutation of the mouse with abnormalities of neuronal migration. Mammalian Genome. 1996;7:798–802. doi: 10.1007/s003359900240. [DOI] [PubMed] [Google Scholar]
- Talbot CJ, Nicod A, Cherny SS, Fulker DW, Collins AC, Flint J. High-resolution mapping of quantitative trait loci in outbred mice. Nat Genet. 1999;21:305–308. doi: 10.1038/6825. [DOI] [PubMed] [Google Scholar]
- Williams EG, Mouchiroud L, Frochaux M, Pandey A, Andreux PA, Deplancke B, Auwerx J. An evolutionarily conserved role for the aryl hydrocarbon receptor in the regulation of movement. PLoS Genetics. 2014;10:e1004673. doi: 10.1371/journal.pgen.1004673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Gu J, Qi S, Lu L. The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biol. 2001;2:1–18. doi: 10.1186/gb-2001-2-11-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis-Owen SA, Flint J. The genetic basis of emotional behaviour in mice. Eur J Hum Genet. 2006;14:721–728. doi: 10.1038/sj.ejhg.5201569. [DOI] [PubMed] [Google Scholar]
- Xu S. Theoretical basis of the Beavis effect. Genetics. 2003;165:2259–2268. doi: 10.1093/genetics/165.4.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B, Willis-Owen SA, Fullerton J, Meesaq A, Deacon RM, Rawlins JN, Copley RR, Morris AP, Flint J, Mott R. Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nat Genet. 2004;36:1197–1202. doi: 10.1038/ng1450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Worksheet S1. BXD strains and sample sizes.
Worksheet S2. Genes within the Chromosome 4 interval.
Worksheet S3. Cis-expression QTLs for genes within the Chromosome 4 interval.
Figure S1 BXD strain differences for grooming duration in the open field.
Enlarged version of Figure 1 A and B.
Figure S2 Whole genome scans and zoomed images of trait data for grooming duration and locomotion.
Enlarged versions of Figure 2 A–D. Panels A-C, below: significant QTL interval zoomed. Colored rectangles depict individual genes, red and green lines represent the additive genetic contribution; red lines indicate negative values (C57BL/6J alleles increasing trait values) and green lines indicate positive values (DBA/2J alleles increasing trait values). Gray lines are shown when the parental strain is unknown. The orange hash marks on the x-axis signify the SNP density (sequence difference between the two parental strains). The yellow bars represent the relative frequency of peak LRS at a given location from 2000 bootstrap resamples.
Figure S3 Relationship of Dab1 with grooming-related genes.
Enlarged version of Figure 3.
Figure S4 Two-dimensional plot of LRS scores for pairs of loci involved in epistatic interaction for rearing in males.
Enlarged version of Figure 4.
Figure S5 BXD strain differences in the open field.
Variation in the open field in the BXD strains and their parental strains (C57BL/6J and DBA/2J, indicated by orange and green bars respectively) for locomotion (A and B), thigmotaxis percent distance (C and D), thigmotaxis percent time (E and F), Leaning (G and H), jumping (I and J), defecation (K and L), rearing (M and N) grooming frequency (O and P). Bars represent mean +/- SEM. Intra-class correlations and overall mean are shown for each trait. The y-axis represents measurement and units for each of the traits and the x-axis lists the BXD strains which are rank ordered from low to high.
Figure S6 Whole genome scans of trait data for which suggestive QTLs were obtained.
Whole genome scans (top: males, bottom: females) for A and B: locomotion, C and D: thigmotaxis percent time, E and F: thigmotaxis percent distance, G and H: grooming frequency, I and J: grooming duration, K and L rearing, M and N: jumping, O and P: leaning, Q and R: defecation. The x-axis represents chromosome number and megabase position and the y-axis represents the likelihood ratio statistic (LRS) of linkage. Blue lines represent LRS across the genome. The pink and gray horizontal lines are approximate threshold values which are used to assess whether a peak is significant (P<0.05) or suggestive (P<0.63), respectively.
Figure S7. Whole genome scans for the factor scores.
Shown are scans for males (top) and females (bottom). A and B: factor 1, C and D: factor 2.
Figure S8. Whole genome scans for sex by strain interaction.
Shown are scans for the difference between male and female strain means for A: grooming duration, B: defecation, C: locomotion, and D: rearing.


