Abstract
Alterations of genes regulating epigenetic processes are frequently found as cancer drivers and may cause widespread alterations of DNA methylation, histone modification patterns or chromatin structure to disrupt normal patterns of gene expression. Because of the inherent reversibility of epigenetic changes, inhibitors targeting these processes are promising anti-cancer strategies. Small molecules targeting epigenetic regulators have recently been developed and clinical trials of these agents are underway for hematologic malignancies and solid tumors. In this review we describe how the writers, readers and erasers of epigenetic marks are dysregulated in cancer and summarize the development of therapies targeting these mechanisms.
Keywords: Epigenetics, DNA methylation, Histone acetylation, Histone methylation, epigenetic readers, anti-cancer therapies
INTRODUCTION
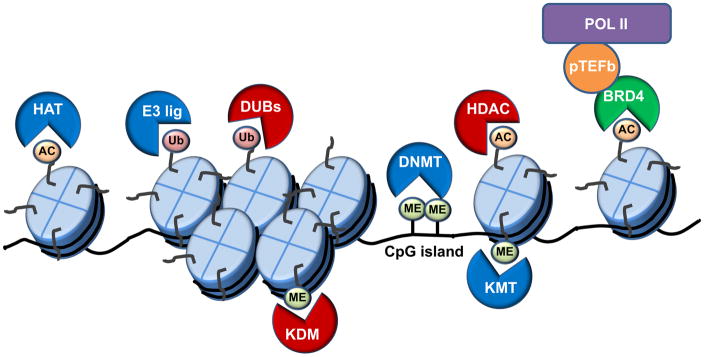
Epigenetics is the study of heritable changes in gene function that cannot be attributed to DNA sequence variations. Cellular identity and differences between cell types often rely upon systems in which there is no DNA variation. Instead, gene expression patterns are tightly controlled by the 3-dimensional architecture of chromatin and the action of multi-protein complexes especially RNA polymerase that transcribes DNA into RNA. In the nucleus, genomic DNA is wrapped around histones into nucleosome subunits that are condensed into chromatin. Highly condensed chromatin is termed heterochromatin and contains mostly inactive genes. In contrast, euchromatin has a more open structure and contains active genes. Chromatin structure is dynamically regulated by DNA methylation, nucleosome positioning and histone modifications (Figure 1).
Figure 1.
In 1974 Kornberg and Thomas discovered that DNA was packaged into the nucleosome, consisting of approximately 147 base pairs (bp) of DNA wrapped around a histone octamer core and a linker histone wrapped around another 20 bp (1). Subsequent studies revealed that histone C- and N-terminal histone tails extended beyond the nucleosome core structure and are accessible to post-translational modification (2; 3). It had been proposed for many years that histones might affect gene expression, but it wasn’t until Allis and coworkers demonstrated that histone acetylation was important for regulating mechanisms of gene expression that the significance of histone modification was realized (4). Studies have now revealed that a wide range of post-translational modifications can mark histones such as acetylation, methylation, ubiquitination or phosphorylation. Histone modifications bring about chromatin remodeling by two main mechanisms. First, marks such as acetylation or phosphorylation neutralize the positive charge of lysines, weakening interactions among nucleosomes and between DNA and histones to increase chromatin accessibility (5; 6). Second, histone modifications may serve as docking sites for additional histone modifying enzymes or specific chromatin factors that regulate chromatin architecture and/or gene expression.
DNA methylation usually occurs on cytosines that precede a guanine (CpG). The presence of endogenous palindromic CpG methylation patterns in the genome and transmission of these methylation marks through the germline has now been well established. The majority of CpG dinucleotides are concentrated within CpG-rich DNA regions termed “CpG islands” that are located near transcription start sites (TSSs) at ~70% of gene promoters (7). In addition, orphan CpG islands located far from TSSs and CpG dinucleotides in gene bodies or enhancers may be methylation targets that can regulate gene expression (8; 9). Hypermethylation of CpG islands may lead to silencing of the corresponding gene by precluding the binding of transcription factors and recruitment of methyl-CpG binding proteins that interact with repressive histone modifying enzymes (10; 11). Thus, DNA methylation plays a key role in regulating chromatin architecture and gene expression.
Epigenetic regulation is a dynamic and reversible process. Proteins that carry out these epigenetic modifications can be thought of as writers, readers and erasers. Epigenetic writers catalyze the addition of epigenetic marks onto either DNA or histones, most commonly on “tails” of histones that extend from the octamer structure. Readers recognize or are recruited to a specific epigenetic mark. Erasers remove epigenetic marks. In this review we will discuss how this network of epigenetic regulation has provided new targets for anti-cancer therapeutics.
EPIGENETIC DYSREGULATION IN CANCER
As the spectrum of mutations found in cancer has been elucidated in recent years by next generation sequencing, it is apparent that mutations, amplifications, deletions and rearrangement of genes affecting epigenetic regulation pathways frequently occur in cancer (12). Driver mutations in epigenetic regulators can lead to widespread alterations of DNA methylation or histone modification patterns that dysregulate chromatin structure and disrupt normal patterns of gene expression. DNA hypermethylation and/or repressive histone marks on a promoter can phenocopy loss of function mutations in tumor suppressor genes by silencing gene expression. Conversely, loss of DNA methylation or activating marks can significantly increase gene expression similar to what would be observed after oncogenic chromosomal translocations or gene amplification.
Unlike genetic events, epigenetic changes are reversible. Because of this inherent plasticity, deciphering how aberrant epigenetic mechanisms culminate in malignant transformation may yield novel insights into how targeting these mechanisms may be used as cancer therapy. DNA hypomethylating agents and histone deacetylase inhibitors are approved for some hematologic malignancies, including T-cell lymphoma (vorinostat 2006, romidepsin 2009), multiple myeloma (panobinostat, 2015) and myelodysplastic syndrome (MDS) (decitabine or azacitidine). Additional clinical trials of epigenetic therapies in hematologic malignancies and solid tumors as single agents and in combination with other therapies are underway.
TARGETING DNA METHYLTRANSFERASES WITH HYPOMETHYLATING AGENTS
Changes in both global and individual gene methylation patterns are often found in cancer and methylation patterns can distinguish tumor types. In general tumors show global DNA hypomethylation, due to loss of repeat region methylation and hypomethylation of specific loci (13). However, hypermethylation of specific CpG rich regions leading to silencing of tumor suppressors has been frequently reported in malignant transformation. For instance, the tumor suppressor gene Rb is silenced by hypermethylation in retinoblastoma, the cell cycle inhibitor p16 is often hypermethylated in colorectal, lung and breast carcinomas and the BRCA1 promoter was found hypermethylated in breast and ovarian cancers (14–16). Furthermore, aberrant methylation patterns outside CpG islands and hypomethylation patterns are equally important as hypermethylation in cancer. Hypomethylation of proto-oncogenes such as Hox11, c-Neu, Bcl-2, and Ras was reported in a variety of malignancies which may contribute to aberrant expression of these tumor promoting genes (17–20). The DNA methyltransferases (DNMTs), DNMT3A and DNMT3B are responsible for establishing de novo DNA methylation patterns that are maintained by DNMT1 (21; 22). Recurrent DNMT3A loss of function mutations have been reported in AML and are associated with subtle losses of DNA methylation whose functional significance remains to be ascertained (23–25).
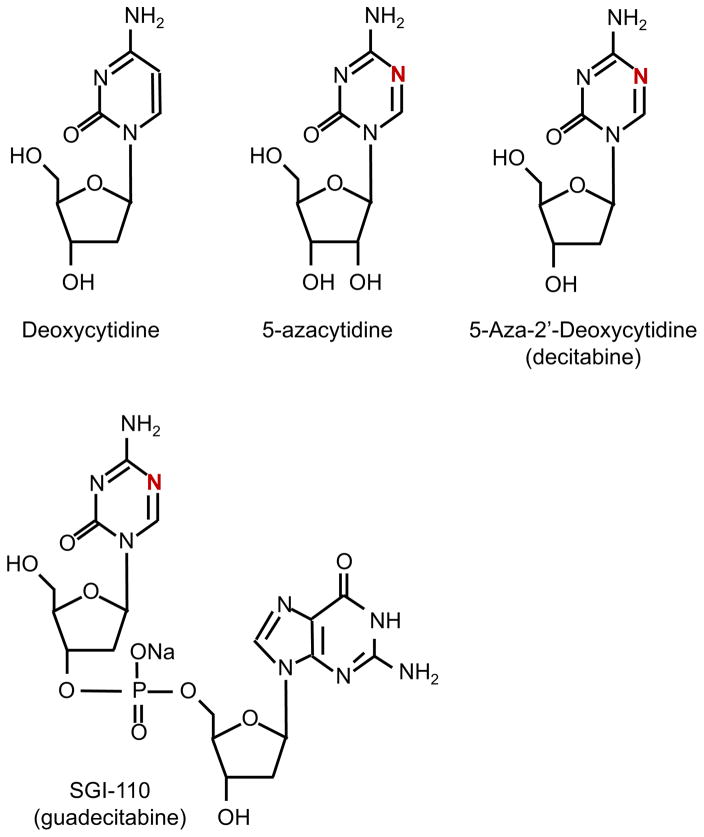
The first compounds recognized as DNMT inhibitors (DNMTi) were the cytosine analogues 5′-azacytidine (Aza, Vidaza) and 5-aza-2′-deoxycytidine (Decitabine, Dacogen). Developed originally as high-dose cytotoxic anti-leukemia agents it is now understood that at low dose these compounds inhibit DNMT activity resulting in hypomethylation. These azanucleosides substitute nitrogen for carbon at the C-5 position of the pyrimidine ring and when incorporated into DNA irreversibly bind DNMT1 resulting in DNMT1 degradation and DNA demethylation (26; 27)(Figure 2). While decitabine is mostly incorporated into DNA about 80–90% of Aza is incorporated into RNA and evidence suggests that Aza’s antineoplastic function and effect on gene expression may be due to its incorporation into both DNA and RNA (28). Studies from the 1980s showed the ability of DNMTi to reactivate silenced genes such as fetal globin genes (29; 30). Furthermore, the combination of DNMTi and HDAC inhibitors can synergistically activate genes (31). The ability of DNMTi to reactivate tumor suppressor genes was one motivation for the use of these agents in hematological and other malignancies. In 2004 and 2006 Aza and decitabine were approved for treatment of MDS respectively (Table 1) (32; 33). Clinical trials remain ongoing for both compounds as single agents and in combination therapies for hematologic malignancies and solid tumors. In addition, a second-generation analogue, SGI-110, has been developed whose active metabolite is decitabine. This compound is a dinucleotide consisting of decitabine linked by a phosphodiester bond to deoxyguanosine that protects it from drug clearance by deamination (34). SGI-110 is currently being tested in clinical trials for AML, MDS, ovarian cancer and hepatocellular carcinoma (Table 1). These agents work slowly to cause clinical response but the molecular basis by which they exert anti-cancer activity remains uncertain. While hypomethylating agents have been shown to reverse promoter methylation and reactivate silenced tumor suppressor gene expression, other mechanisms may also be important (35). For instance, treatment with decitabine causes the formation of DNA-DNMT adducts and subsequent double-stranded DNA breaks resulting in G2 arrest (36). In addition, DNMTs have been found in complexes with histone modifying enzymes and a global increase of histone H3 and H4 acetylation has been observed after treatment with Aza (37–39). Furthermore, decitabine stimulates nuclear localization of IRF7 in colon cancer cells to cause expression of toxic endogenous retroviral sequences independent of promoter methylation (40).
Figure 2.
Table 1.
DNA methylation inhibitors in cancer therapy
| Compound | Target | Clinical Status | Reference |
|---|---|---|---|
| 5-Azacytidine (Vidaza) | DNMT1 | Approved: MDS | (32) |
| 5-Aza-2′-deoxycytidine (Decitabine) | DNMT1 | Approved: MDS | (33; 124) |
| SGI-110 (Guadecitabine) | DNMT1 | Clinical trials: MDS, AML, Ovarian, HCC | NCT01261312, NCT01752933 |
| AG-120 | Mutant IDH1 | Clinical trials: AML | NCT02677922 |
| AG-221 | Mutant IDH2 | Clinical trials: MDS, AML | NCT01915498 |
Abreviations: AML, Acute myelogenous leukemia; MDS, myelodysplastic syndrome; HCC, hepatocellular carcinoma
Defects in demethylation of DNA can also lead to aberrant hypermethylation and altered expression of genes that drive neoplasia. Mutations in the enzymes IDH1 and IDH2 that normally catalyze decarboxylation of isocitrate to α-ketoglutarate occur in gliomas, AML, chondrosarcomas and cholangiocarcinoma (41–44). Mutant IDH1/2 reduces α-ketoglutarate to 2-hydroxygluterate (2-HG), a competitive inhibitor of the TET family of DNA hydroxylases (45). In addition, mutations and translocations of TET2 have been observed in numerous hematologic malignancies and associated with poor prognosis in AML (46). TET enzymes normally convert 5′-methylcytosine to 5′-hydroxymethylcytosine, an important step in cytosine demethylation and inhibition of this mechanism results in DNA hypermethylation (47). Furthermore, 2-HG has been reported to inhibit α-ketoglutarate dependent Jumonji domain lysine demethylases and activate mTOR (48; 49). Compounds specifically targeting mutant IDH have been developed (AG-120 and AG-221) and are currently in clinical trials for patients with advanced hematologic malignancies (Table 1) (50). These agents would be expected to block the production of 2-HG by the mutant enzymes and allow DNA and histone methylation patterns to normalize.
TARGETING REVERSIBLE HISTONE ACETYLATION AND HISTONE DEACETYLASE
Acetylation of lysine on histone tails is highly dynamic and important for regulation of chromatin structure, transcription and DNA repair. Two competing enzyme families, histone lysine acetyltransferases (HATs) and histone deacetylases (HDACs), regulate histone acetylation. The about 30 known HATs are classified into two groups based on their capacity to acetylate nucleosomal histones. Type A HATs are located in the nucleus and acetylate chromatin bound histones and nuclear proteins. Type B HATs acetylate newly translated, but not nucleosomal, histones H3 and H4. Based on structural and functional homology the type A HATs are further categorized into families that include: GCN5, MYST, p300/CBP, transcriptional coactivators and steroid receptor coactivators. HATs catalyze the transfer of an acetyl group from acetyl-CoA to the amino group of a histone lysine residue. Upon acetylation, the positive charge on lysine is neutralized diminishing the interaction of histones with DNA. In general this leads to a more open chromatin structure that is accessible to binding of proteins such as transcription factors. Thus, acetylation is associated with transcriptional activation while deacetylation is associated with gene repression. Numerous examples of chromosomal translocations (e.g. MLL-CBP and MOZ-CBP) or mutations (e.g. p300/CBP) involving type A HATs have been reported in hematologic malignancies and solid tumors (51–54). For instance, about 40% of diffuse large B-cell lymphomas and 41% of follicular lymphomas harbor deletions or mutations that inactivate p300/CBP (55). While type A HAT inhibitors have yet to enter clinical trials, the p300-HAT inhibitor C646 has been reported to specifically suppress growth of CBP-deficient lung and hematopoietic cancer cells (52). The only known type B HAT is a multiunit complex containing Hat-1 as its catalytic subunit that associates with the histone chaperones NASP and Asf1 in the nucleus to facilitate the deposit of histones onto DNA. In yeast, Hat-1 is required for telomeric silencing and repair of dsDNA breaks by homologous recombination (56; 57). In humans, Hat-1 is frequently amplified in cancer (cBioPortal.org) and IHC analysis of tissues from leiomyosarcoma and leiomyoma patients indicates increased Hat-1 correlates with worse survival (58). To date, no specific inhibitors of Hat-1 have been reported.
Many reports demonstrate that HDACs are overexpressed in cancers resulting in global loss of histone acetylation and silenced tumor suppressor gene expression. HDACs are divided into four classes based on their homology and structure. Classes I, II and IV are comprised of Zn-dependent HDACs while class III is made up of the NAD-dependent sirtuins. Class I HDACs (1, 2, 3 and 8) and class II HDACs (4, 5, 6, 7, 9 and 10) have been reported to play roles in tumorigenesis. Numerous synthetic or natural product HDAC inhibitors (HDACi) have been developed for cancers that display increased HDAC activity or HAT mutations. Based on their chemical structure, HDACi can be organized into groups consisting of hydroxamates, benzamides, cyclic peptides or short-chain fatty acids (Table 2). Because HDACs are Zn2+ dependent many HDACi target the Zn2+ ion in the active site of HDACs to inhibit their enzymatic activity. Nonselective broad spectrum HDACi that inhibit all zinc dependent HDACs include the hydroxamate class agents vorinostat, belinostat and panobinostat. Vorinostat induces cell cycle arrest, promotes apoptosis, sensitizes cells to other chemotherapy and has been approved to treat patients with cutaneous T-cell lymphoma (CTCL) (59). Belinostat was approved to treat peripheral T-cell lymphomas (PTCL) and panobinostat to treat multiple myeloma (MM). These drugs are also in clinical trials to treat solid tumors. Nonselective HDACi such as vorinostat can reverse aberrant epigenetic chromatin changes to reactivate tumor suppressor genes such as p21 (60). In addition, HDACs have multiple functions in the cell and also target non-histone proteins. For instance, acetylation enhances the activity of some transcription factors such as p53 and GATA-1 (61; 62). Therefore, HDACi work through many mechanisms to promote cell cycle arrest, induce differentiation and activate apoptosis pathways in cancer cells.
Table 2.
Histone deacetylase inhibitors in cancer therapy
| Compound | HDAC Target | Clinical Status | Reference |
|---|---|---|---|
| Hydroxamates: | |||
| Abrexinostat | Class I, II | Clinical trials: HL, non-HL, CLL | (125) |
| Belinostat | Class I, II, IV | Approved: PTCL | (126) |
| CG200745 | Class I, II, IV | Clinical trials: Solid tumors | (127) |
| CHR-3996 | Class I | Clinical trials: Solid tumors | (128) |
| CUDC-101 | Class I, II | Clinical trials: Squamous cell carcinoma | (129) |
| CUDC-907 | Class I, II | Clinical trials: MM, lymphoma, solid tumors | (130) |
| Givinostat | Class I, II | Clinical trials: CLL, MM, HL | (131; 132) |
| MPT0E028 | HDAC1, 2, 6 | Clinical trials: Solid tumors | (133) |
| Panobinostat | Class I, II, IV | Approved: MM | (134; 135) |
| Pracinostat | Class I, II, IV | Clinical trials: AML, Prostate | (136) |
| Quisinostat | Class I, II | Clinical trials: Solid tumors, lymphoma, CTCL | (137) |
| Resminostat | Class I, II | Clinical trials: Colorectal, HCC, HL | (138) |
| Vorinostat (SAHA) | Class I, II, IV | Approved: CTLC | (139) |
| Benzamides: | |||
| Chidamide | HDAC1, 2, 3, 10 | Clinical trials: Breast, NSCLC | (140) |
| Entinostat | HDAC1, 2, 3 | Clinical trials: NSCLC, solid tumors | (141) |
| Mocetinostat | Class I, IV | Clinical trials: Hematologic and solid tumors | (142) |
| Ricolinostat | HDAC6 | Clinical trials: MM, lymphoma | (63) |
| Tacedinaline | Class I | Clinical trials: MM, lung, pancreatic | (143) |
| Cyclic Peptides: | |||
| Romidepsin | HDAC1, 2 | Approved: CTLC, PTCL | (144) |
| Fatty acids: | |||
| AR-42 | Class I, II | Clinical trials: AML, MM | (145) |
| Phenylbutyrate | Class I, II | Clinical trials: Solid, hematologic | (146) |
| Pivanex | Class I, II | Clinical trials: NSCLC, myeloma, CLL | (147) |
| Valproic acid | Class I, II | Clinical trials: Solid, hematologic | (148) |
Abbreviations: ALL, Acute lymphocytic leukemia; AML, Acute myelogenous leukemia; CLL, Chronic lymphocytic leukemia; CTLC, cutaneous T-cell lymphoma; HCC, hepatocellular carcinoma; HL, Hodgkin’s lymphoma; MDS, myelodysplastic syndrome; MM, multiple myeloma; NSCLC, Non-small cell lung cancer; PTCL, peripheral T-cell lymphoma
Selective HDACi include romidepsin that targets HDAC1 and 2 and has been approved for CTCL and PTCL patients. In addition, ricolinostat is an HDAC6 specific inhibitor in clinical trials for patients with MM or lymphoma (63). HDAC6 is a cytoplasmic deacetylase that acts on tubulin, Hsp90 and binds to ubiquitinated proteins. Ricolinostat has been especially promising for MM since these cells have high Ig production and are dependent on clearance of misfolded/aggregated proteins. Blocking HDAC6 leads to the accumulation of unfolded ubiquitinated proteins that induce cell death. Many additional HDAC inhibitors are in different phases of clinical trials either as single agents or in combination (Table 2).
TARGETING HISTONE METHYLTRANSFERASES
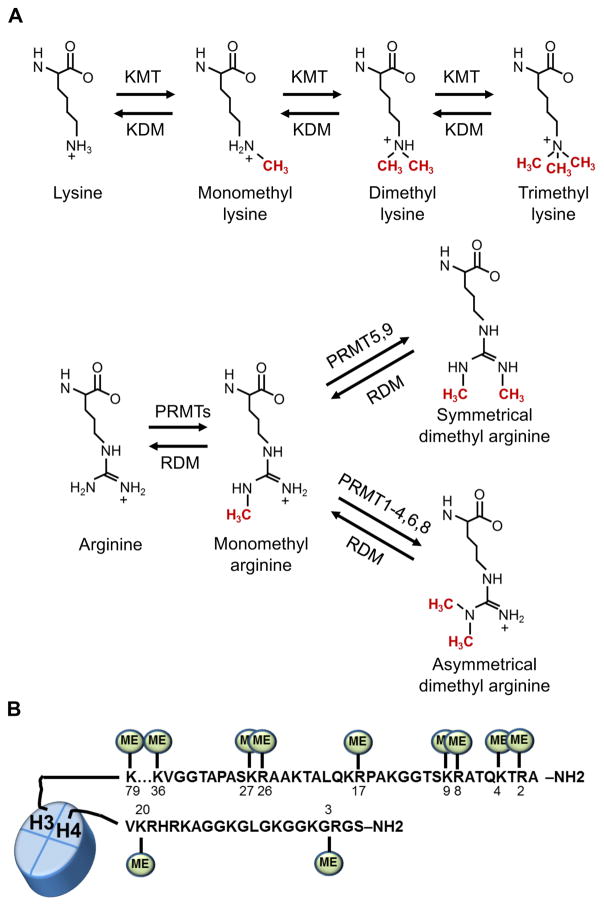
Dynamic methylation of nucleosomal histones plays an important role in regulation of gene expression. The two major families of histone methyl transferases are lysine methyl transferases (KMTs) and protein arginine methyltransferases (PRMTs). In recent years mono-, di- or tri-methylation of histone lysine methylation has been characterized to alter affinity of reader proteins to the methylated histones (Figure 3). Specific lysine methylation marks on H3K4, H3K36 and H3K79 are associated with activation of gene expression while marks associated with repression are H3K9, H3K27 and H4K20 (64). Importantly, different methylation states (e.g. me0, me1, me2, me3) of the same lysine residue occur in different contexts and may have distinct functional consequences. For instance, H3K4me3 marks are found at active or poised transcription start sites while H3K4me1 is usually found at active enhancers (65). In addition, H3K9me1 is typically found at active genes but H3K9me3 can be associated with gene repression (64).
Figure 3.
The identification of KMT genetic alterations in cancer has made small molecules that selectively inhibit oncogenic methyltransferases promising therapeutic agents. However, despite the abundance of research on the more than 50 KMTs relatively few selective inhibitors have been identified (Table 3). One promising epigenetic target is DOT1L, a H3K79 methyltransferase that has an important role in MLL (KMT2A) leukemias. Translocations of MLL are found in 5–10% of AMLs and tandem duplication of MLL is found in ~5% of de novo AML (66). MLL translocations result in fusion proteins that lack their catalytic SET domain and frequently contain elongation complex nuclear proteins such as AF9, AF10, ENL (67). This complex also contains DOT1L, and AML cells carrying these translocations display aberrant patterns of K79 methylation that activate homeobox gene expression (68). The DOT1L inhibitor Pinometostat (EPZ-5676) inhibits proliferation of cell lines with MLL rearrangements in association with decreases in H3K79me levels at Hox genes and is currently in clinical trials (69).
Table 3.
Histone methylation/demethylation inhibitors in cancer therapy
| Compound | Target | Cancer Type | Reference |
|---|---|---|---|
| HMT inhibitors: | |||
| EPZ00477 | DOT1L | MLL rearranged leukemias | (149) |
| Pinometostat (EPZ-5676) | DOT1L | Clinical trials: hematologic malignancies | (150) NCT01684150 |
| SGC0946 | DOT1L | MLL rearranged leukemias | (151) |
| DZNep | EZH2 | Breast, colon, prostate | (152) |
| GSK126 | EZH2 | DLBCL | (77) |
| GSK343 | EZH2 | Ovarian | (77) |
| EPZ005687 | EZH2 | EZH2 mutant non-HL | (78) |
| EI1 | EZH2 | DLBCL | (76) |
| Tazemetostat (EPZ-6438) | EZH2 | Clinical trials: Solid tumors, DLBCL, HL, non-HL | (79) NCT02889523 |
| UNC1999 | EZH2 | DLBCL | (153) |
| BIX-01294 | G9A | leukemia, bladder | (154) |
| BRD4770 | G9A | Pancreatic | (155) |
| UNC0638 | G9A | AML, breast | (156) |
| Chaetocin | SUV39H1 | lymphomas | (87) |
| GSK3326595 | PRMT5 | solid tumors, non-HL | NCT02783300 |
| AMI-408 | PRMT1 | AML | (93) |
| MS023 | PRMT1,3,4,6,8 | (94) | |
| HDM inhibitors: | |||
| ORY-1001 | LSD1 | Clinical trials: AML | (157) |
| GSK2879552 | LSD1 | Clinical trials: AML, Relapsed/Refractory Small Cell Lung Carcinoma | (157) NCT02034123 NCT02034123 |
| GSK354, GSK690 | LSD1 | AML | (101) |
| NCD25, NCD38 | LSD1 | MDS | (158) |
| Tranylcypromine | LSD1 | Clinical trials: AML, MDS | (157) NCT02273102 |
| 4SC-202 | HDAC-LSD1 | Clinical Trials: hematologic malignancies | (102) NCT01344707 |
| GSK-J1, GSK-J4 | JmjC domain proteins | (103) | |
| EPT-103182 | KDM5B | Hematologic and solid cancers | (106) |
Abbreviations: AML, Acute myelogenous leukemia; DLBCL, Diffuse large B-cell lymphoma; HL, Hodgkin’s lymphoma; MDS, myelodysplastic syndrome
Enhancer of Zeste 2 (EZH2) is a catalytic component of the polycomb repressive complex 2 (PRC2) where it catalyzes di- and tri-methylation of H3K27 that represses transcription. EZH2 gain of function is associated with poor prognosis in breast and prostate cancer and an activating mutation in the enzymatic SET domain of EZH2 was observed in 22% of patients with diffuse large B-cell lymphoma (DLBCL) and 7 percent of follicular lymphomas (70–72). This EZH2 mutation increases the ability of the protein to create the repressive H3K27me3 mark and is associated with a global increase in H3K27 methylation in lymphoma cells. This in turn blocks expression of genes important for cell cycle control and B cell differentiation, contributing to lymphomagenesis. By contrast, loss of function EZH2 mutations have been reported in myeloid malignancies and T-ALL, associated with widespread loss of the H3K27me3 mark suggesting that EZH2 may have either oncogenic and tumor suppressive properties depending on its cellular context and perhaps also due to the action of other co-occurring mutations (73). EZH2 is also frequently overexpressed in advanced cases of breast, prostate and other solid tumors, often in association with an epithelial to mesenchymal phenotype change (71; 72). In these cases an increase in H3K27me3 is not observed and there is evidence that in advanced prostate cancer, EZH2 contributes to gene activation by altering androgen receptor function in a manner that requires the enzymatic activity of the EZH2 protein (74).
The overexpression of EZH2 as well as its mutation motivated the development of small molecule inhibitors of EZH2 (EZH2i). EZH2i 3-Deazaneplanocin A (DZNep) is an adenosine analogue that binds to the enzyme in place of the S-adenosyl methionine (SAM) and causes degradation of the protein. DZNep induces cell cycle genes, inhibit HOXA9 expression and induce apoptosis of primary AML cells (75). In addition, DZNep in combination with panobinostat prolonged survival of mice xenografted with AML cells (75). More potent and selective SAM competitive EZH2 inhibitors EI1 and GSK126 are have been tested in cell culture and xenografts. EI1 treated cells display global loss of H3K27 methylation, and DLBCL cells carrying a gain of function EZH2 undergo apoptosis when treated with EI1 (76). Similarly, GSK126 inhibited EZH2, proliferation of mutant DLBCL cell lines and growth of these cells in xenografted mice (77). Tazemetostat (EPZ-6438) is another EZH2i that has shown promise in xenografts and is currently being evaluated in phase I/II trials as a single agent and in combination with dexamethasone (78–80). EZH2i may have uses in several situations including overexpression or mutation of the gene repressive protein EZH2, as well as tumors having loss of function mutations of gene activator proteins such as the H3K27 demethylase KDM6A (UTX) or inactivating mutations of the SWI/SNF proteins acting to restore a balance between forces mediating gene repression and activation (81; 82).
Other small molecule KMT inhibitors that have been developed as potential anticancer therapeutics include compounds targeting H3K9 KMTs (Table 3). Increased expression of the H3K9-specific KMT G9a was reported in lung cancer cell lines and treatment with inhibitor BIX-01294 reduces H3K9 methylation (83; 84). Similarly, SETDB1 is frequently amplified in melanoma and lung cancers and treatment with mithramycin down-regulates SETDB1 to inhibit proliferation (85; 86). In addition, the natural product chaetocin was identified as an inhibitor of SUV39H, a KMT that regulates erythroid and B-cell differentiation (87). These agents are currently being used for in vitro and in vivo preclinical studies.
PRMTs are frequently overexpressed in cancer and thus have also become attractive targets for anticancer strategies. The PRMT family has nine members (PRMT1-9) that methylate arginine to form monomethylarginine. Type I PRMTs (PRMT1-6 and 8) catalyze the formation of asymmetric dimethylarginine while type 2 PRMTs (PRMT5 and 9) form symmetric dimethylarginine (Figure 3). Histone arginine methylation marks can be activating (H4R3me2a, H3R2me2s, H3R17me2a, H3R26me2a) or repressive (H3R2me2a, H3R8me2a, H3R8me2s, H4R3me2s) and PRMT dysfunction has been observed in a variety of cancers. Overexpression of PRMT1 and PRMT4 (CARM1) was reported in NSCLC, and PRMT4 can drive the expression of the c-Myc pathway by deregulating SWI/SNF in breast cancer (88; 89). Increased PRMT5 expression was observed in lymphoma, leukemia, glioblastoma and prostate cancer where it is reported to activate c-Myc and other oncogenic transcription factors (90). In addition, PRMT7 has been reported to inhibit E-cadherin expression and promote EMT in breast cancer (91). The only PRMT inhibitor in clinical trials is GSK3326595 (formerly EPZ015938) that is being evaluated for patients with solid tumors and non-Hodgkin lymphoma (92). Several inhibitors have shown promise in preclinical studies such as AMI-408 a PRMT1 inhibitor that suppressed the transformation function of MLL-GAS7 or MOZ-TIF2 fusions in AML models, and the PRMT5 inhibitor EPZ015666 that displays antitumor activity in xenografts of mantle cell lymphoma (93). In addition, the compound, MS023 was recently reported to inhibit asymmetric arginine dimethylation while increasing monomethylation and symmetric dimethylation, but its anti-cancer properties have yet to be characterized (94).
TARGETING HISTONE DEMETHYLATION
Two classes of lysine demethylases (KDMs) govern demethylation of histones: the amine oxidases that include lysine-specific demethylase 1 (LSD1) and the α-ketoglutarate-dependent Jumonji domain (JmjC) containing demethylases. The LSD-family KDMs only demethylate mono- and dimethylated lysines while the JmjC demethylases remove methyl from all three states of lysine methylation (95; 96). Target specificity of KDMs is often regulated by their participation in different complexes. Mutations or dysregulation of KDMs are reported in a variety of cancers, making these enzymes an attractive target for anti-cancer therapies.
The histone demethylase LSD1 (KDM1A) is highly expressed in several cancers and is specifically required for terminal differentiation of hematopoietic cells (97). LSD1 generally demethylates H3K4me1/2 thus repressing transcription, but when LSD1 interacts with the androgen receptor its enzymatic activity switches to H3K9me1/2 thereby stimulating transcription (98). LSD1 is also a substrate for G9a KMT and methylation of LSD1 stimulates recruitment of CDH during androgen-dependent gene expression (99). LSD1 has a C-terminal amine oxidase-like domain that is structurally related to monoaminoxidases (MAO). Thus, the MAO inhibitor tranylcypromine (TCP) inhibits LSD1, but its use is limited by indiscriminate anti-MAO activity (100). Several more selective TCP derivatives have been developed and some have entered clinical trials (Table 3). ORY-1001 has been shown to reduce H3K4me2 and LSD1 target gene expression and reduce tumor growth in xenografts (101). GSK2879552 promotes differentiation and inhibits proliferation of AML cells (101). In addition, LSD1 inhibitors GSK354 and GSK690 have recently been reported to inhibit cell growth in vitro (101). Interestingly, the small molecule HDAC inhibitor 4SC-202 has a dual function to inhibit LSD1 with similar potency and is in clinical trials for patients with advanced hematologic malignancies (102).
While four compounds are in clinical trials for LSD1, inhibitors of JmjC domain containing demethylases have been more difficult to develop. Most of the identified inhibitors are pan-specific metal chelators that are also competitive for cofactor 2-oxogluterate binding and only active in the low micromolar range. Two such compounds are GSK-J1 and its prodrug GSK-J4 that inhibit KDM6A (UTX) and KDM6B (JMJD3) but also display lesser activity against KDM5A and 5B (103). These proteins are all implicated in cancer. For instance, inactivating mutations of KDM6A have been reported in AML, multiple myeloma and bladder cancer, while KDM6B is highly expressed in TALL and metastatic prostate cancer (81; 104). In addition, a KDM5A (JARID1) Nup98 fusion protein is found in 10% of pediatric acute megakaryoblastic leukemias (105). Recently, a potent and selective inhibitor of KDM5B, EPT1013182, has been reported to have antiproliferative effects in cell lines and inhibit growth in multiple myeloma xenograft models (106).
TARGETING READERS OF EPIGENETIC MARKS
Epigenetic reader proteins recognize and bind chromatin or histone modifications to either directly induce chromatin structural changes (e.g. compaction, remodeling) recruit secondary chromatin modifiers or serve as scaffold proteins for various nuclear processes such as transcription, replication or repair. Acetylated lysines are binding sites for proteins containing bromodomains such as the Bromodomain and Extra Terminal (BET) proteins BRD2, BRD3, BRD4 and BRDT. Readers that recognize methyl-lysine include MBT domain, Tudor domain, and Chromodomain proteins (107). Chromatin reader proteins can include several types of reader domains and binding at a specific chromatin site may depend on adjacent histone modifications.
BET family proteins are important for regulation of transcription and cell proliferation. BRD4 stimulates transcriptional elongation by recruitment of the P-TEFb complex to chromatin binding sites where it phosphorylates and increases the processivity of RNA polymerase II (108). In addition, recent studies suggest that BRD4 also recruits NSD methyltransferases to increase H3K36 methylation that activates gene expression (109). Several reports indicate that BET family members disruption is associated with cancer. BRD2 is overexpressed in B-cell lymphoma and translocations of BRD3 and BRD4 drive rare midline carcinomas (110). Recently small molecules have been developed that inhibit the tandem bromodomains of BET proteins (Table 4) (111; 112). One of the first BET inhibitors developed, JQ1, induced terminal differentiation of leukemic stem cells in primary AML samples and prevented ovarian carcinoma growth in xenografts (113; 114). Furthermore, JQ1 globally reduced androgen receptor (AR) target gene transcription and promoted apoptosis in cells with activated AR (115). In addition, dBET, a phthalimide-coupled JQ1, was demonstrated to induce BRD4 protein degradation and delay leukemia progression in mice (116). Recently, new classes of improved BET inhibitors have been developed. Three of these, I-BET762, CPI-0610 and OTX015 are in clinical trials for hematologic malignancies (117). In addition, TEN-010 is in clinical trials for NUT midline carcinomas and advanced solid tumors.
Table 4.
Inhibitors of epigenetic readers used as cancer therapy
| Compound | Target | Cancer Type | Reference |
|---|---|---|---|
| JQ1 | BET proteins | Prostate, AML with MLL translocations, MM, Nut midline carcinoma | (111; 113–115) |
| I-BET762 | BET proteins | Clinical trials: hematologic malignancies, NUT midline carcinoma and solid tumors | (117) NCT01943851 NCT01587703 |
| OTX015 | BRD2, 3, 4 | Clinical trials: AML | NCT01713582 |
| CP-0610 | BET proteins | Clinical trials: AML, MDS, MM, lymphoma | NCT01949883 NCT02158858 NCT02157636 |
| TEN-010 | BET proteins | Clinical trials: Nut midline carcinomas, solid tumors | NCT01987362 |
| dBET-1 | BET proteins | Leukemias | (116) |
| I-BET151 | BET proteins | MM | (117) |
| GSK2801 | BAZ2B | ||
| UNC669 | L3MBTL1 | (118) | |
| UNC1215 | L2MBTL3 | (119) |
Abbreviations: AML, Acute myelogenous leukemia; MDS, myelodysplastic syndrome; MM, Multiple myeloma
Inhibitors of methyl-lysine readers have also been developed but have not advanced beyond preclinical studies yet. L3MBTL1 and L3MBTL3 are members of the MBT proteins and their loss has been shown to contribute to hematopoietic malignancies. UNC125 binds to the aromatic cage of L3MBTL1 that is common to lysine methyl readers and inhibits L3MBTL1 peptide binding (118). Another MBT inhibitor, UNC1679, has been reported to have a much higher affinity for L3MBTL3 than L3MBTL1 and inhibits GFP-L3MBTL3 chromatin binding in cells (119). Importantly, when take together these results suggest that targeting epigenetic readers may be a useful strategy to counteract the effect of aberrant histone acetylation/methylation profiles in cancer.
CONCLUSIONS AND FUTURE DIRECTIONS
Genome sequencing of patient tumor samples has revealed that alterations of genes regulating the cellular epigenetic state are frequently initiating events in cancer and subclones carrying these mutations are likely to persist after treatment. Thus, much recent work has been towards improving our understanding of the defective epigenetic mechanisms in cancer cell populations in order to develop more effective cancer therapies. Hypomethylating agents, HDAC inhibitors and agents that reverse cancer associated histone modifications have significantly increased our arsenal to treat cancers, particularly hematologic malignancies. In addition, emerging therapeutic strategies take advantage of crosstalk between different epigenetic mechanisms. For instance, cooperation between DNA methylation and histone deacetylation in gene expression has led to clinical trials that test combinations of HDAC inhibitors and DNA hypomethylating agents (120). These combination therapies are attractive over single agent therapies because they may synergistically reactivate silenced tumor suppressor gene expression, which allows the use of lower dosages that may reduce side effects and the likelihood of acquired resistance.
Many other mechanisms of epigenetic regulation have been described that may impact cancer. For example, in addition to lysine acetylation and methylation, histones are subject to other posttranslational modifications such as sumoylation, ubiquitination and/or phosphorylation but additional studies are necessary to determine the affect of these mechanisms on tumorigenesis. In addition, subunits of the SWI/SNF chromatin remodeler complex are frequently mutated in cancers (121). Cancer cells harboring SWI/SNF mutations become dependent on EZH2, and EZH2 inhibitors have proven effective in these cells (122). Furthermore, RNA as well as DNA is subject to covalent modification such as methylation and non-coding RNAs such as snRNAs involved in splicing, miRNAs involved in silencing and lncRNAs involved in gene expression have profound and heritable effects on gene expression. Therapeutic strategies may be developed to target these mechanisms as their significance in cancer becomes better understood.
As promising as the current epigenetic cancer therapies are, significant challenges remain. The precise role specific writers, readers and erasers play in different types of cancer at different stages of differentiation is not well understood, and no chemical inhibitors have been identified for many KDMs and KMTs that could be important targets in cancer. In addition, epigenetic writers, readers and erasers have targets in addition to those associated with chromatin. For instance, LSD1 can also demethylate non-histone proteins such as p53 and DNMT1. In addition, many proteins are regulated by acetylation and may be targets for KATs and HDACs including oncogenes and tumor suppressors such as MYC, p53 and PTEN (123). Thus, the effect of inhibiting epigenetic mechanisms may be due to cytoplasmic as well as nuclear processes. Furthermore, it remains uncertain whether the efficacy of agents targeting epigenetic mechanisms is specifically linked to any changes in gene expression. For instance it is not certain why cells harboring DNMT3A mutations that causes hypomethylation are sensitive to hypomethylating agents. Many epigenetic therapies elicit a DNA damage response and may simply be functioning as cytotoxic agents. In the future, a better comprehension of how epigenetic mechanisms are disrupted in cancer could guide a more mechanistic based rationale for use of specific inhibitors as anticancer therapies.
Acknowledgments
This work was funded by NIH grant CA180475, CA195732 and a Leukemia and Lymphoma Specialized Center of Research (JDL)
References
- 1.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–71. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 2.Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984;311:532–7. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 3.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 4.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–51. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 5.Luger K, Richmond TJ. The histone tails of the nucleosome. Current opinion in genetics & development. 1998;8:140–6. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 6.Steger DJ, Workman JL. Remodeling chromatin structures for transcription: what happens to the histones? BioEssays: news and reviews in molecular, cellular and developmental biology. 1996;18:875–84. doi: 10.1002/bies.950181106. [DOI] [PubMed] [Google Scholar]
- 7.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1412–7. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, et al. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS genetics. 2010;6:e1001134. doi: 10.1371/journal.pgen.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nature genetics. 2007;39:61–9. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 10.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. The New England journal of medicine. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez C, Borgel J, Court F, Cathala G, Forne T, Piette J. CTCF is a DNA methylation-sensitive positive regulator of the INK/ARF locus. Biochemical and biophysical research communications. 2010;392:129–34. doi: 10.1016/j.bbrc.2009.12.159. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nature reviews. Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. Journal of the National Cancer Institute. 2000;92:564–9. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 15.Greger V, Passarge E, Hopping W, Messmer E, Horsthemke B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Human genetics. 1989;83:155–8. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- 16.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, et al. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nature medicine. 1995;1:686–92. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 17.Hanada M, Delia D, Aiello A, Stadtmauer E, Reed JC. bcl-2 gene hypomethylation and high-level expression in B-cell chronic lymphocytic leukemia. Blood. 1993;82:1820–8. [PubMed] [Google Scholar]
- 18.Nishigaki M, Aoyagi K, Danjoh I, Fukaya M, Yanagihara K, et al. Discovery of aberrant expression of R-RAS by cancer-linked DNA hypomethylation in gastric cancer using microarrays. Cancer research. 2005;65:2115–24. doi: 10.1158/0008-5472.CAN-04-3340. [DOI] [PubMed] [Google Scholar]
- 19.Zhou H, Chen WD, Qin X, Lee K, Liu L, et al. MMTV promoter hypomethylation is linked to spontaneous and MNU associated c-neu expression and mammary carcinogenesis in MMTV c-neu transgenic mice. Oncogene. 2001;20:6009–17. doi: 10.1038/sj.onc.1204830. [DOI] [PubMed] [Google Scholar]
- 20.Watt PM, Kumar R, Kees UR. Promoter demethylation accompanies reactivation of the HOX11 proto-oncogene in leukemia. Genes, chromosomes & cancer. 2000;29:371–7. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1050>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 21.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 22.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–4. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 23.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, et al. DNMT3A mutations in acute myeloid leukemia. The New England journal of medicine. 2010;363:2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nature genetics. 2011;43:309–15. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 25.Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer cell. 2014;25:442–54. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:6993–7. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Molecular and cellular biology. 2005;25:4727–41. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Schaefer M, Hagemann S, Hanna K, Lyko F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer research. 2009;69:8127–32. doi: 10.1158/0008-5472.CAN-09-0458. [DOI] [PubMed] [Google Scholar]
- 29.Ley TJ, DeSimone J, Anagnou NP, Keller GH, Humphries RK, et al. 5-azacytidine selectively increases gamma-globin synthesis in a patient with beta+ thalassemia. The New England journal of medicine. 1982;307:1469–75. doi: 10.1056/NEJM198212093072401. [DOI] [PubMed] [Google Scholar]
- 30.Ginder GD, Whitters MJ, Pohlman JK. Activation of a chicken embryonic globin gene in adult erythroid cells by 5-azacytidine and sodium butyrate. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:3954–8. doi: 10.1073/pnas.81.13.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nature genetics. 1999;21:103–7. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 32.Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2002;20:2429–40. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 33.Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106:1794–803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 34.Yoo CB, Jeong S, Egger G, Liang G, Phiasivongsa P, et al. Delivery of 5-aza-2′-deoxycytidine to cells using oligodeoxynucleotides. Cancer research. 2007;67:6400–8. doi: 10.1158/0008-5472.CAN-07-0251. [DOI] [PubMed] [Google Scholar]
- 35.Tsai HC, Li H, Van Neste L, Cai Y, Robert C, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer cell. 2012;21:430–46. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palii SS, Van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-Aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Molecular and cellular biology. 2008;28:752–71. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer research. 2006;66:6361–9. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 38.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nature genetics. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 39.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic acids research. 2003;31:2305–12. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell. 2015;162:961–73. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tefferi A, Lasho TL, Abdel-Wahab O, Guglielmelli P, Patel J, et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia. 2010;24:1302–9. doi: 10.1038/leu.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. The Journal of pathology. 2011;224:334–43. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 43.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, et al. IDH1 and IDH2 mutations in gliomas. The New England journal of medicine. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. The oncologist. 2012;17:72–9. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer cell. 2010;17:225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, et al. Mutation in TET2 in myeloid cancers. The New England journal of medicine. 2009;360:2289–301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 47.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer cell. 2010;18:553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–8. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carbonneau M, LMG, Lalonde ME, Germain MA, Motorina A, et al. The oncometabolite 2-hydroxyglutarate activates the mTOR signalling pathway. Nature communications. 2016;7:12700. doi: 10.1038/ncomms12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stein EM, DiNardo C, Altman JK, Collins R, DeAngelo DJ, et al. Safety and Efficacy of AG-221, a Potent Inhibitor of Mutant IDH2 That Promotes Differentiation of Myeloid Cells in Patients with Advanced Hematologic Malignancies: Results of a Phase 1/2 Trial. Blood. 2015;126:323. [Google Scholar]
- 51.Wang J, Iwasaki H, Krivtsov A, Febbo PG, Thorner AR, et al. Conditional MLL-CBP targets GMP and models therapy-related myeloproliferative disease. The EMBO journal. 2005;24:368–81. doi: 10.1038/sj.emboj.7600521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogiwara H, Sasaki M, Mitachi T, Oike T, Higuchi S, et al. Targeting p300 Addiction in CBP-Deficient Cancers Causes Synthetic Lethality by Apoptotic Cell Death due to Abrogation of MYC Expression. Cancer discovery. 2016;6:430–45. doi: 10.1158/2159-8290.CD-15-0754. [DOI] [PubMed] [Google Scholar]
- 53.Sobulo OM, Borrow J, Tomek R, Reshmi S, Harden A, et al. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3) Proceedings of the National Academy of Sciences of the United States of America. 1997;94:8732–7. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Borrow J, Stanton VP, Jr, Andresen JM, Becher R, Behm FG, et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nature genetics. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 55.Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–95. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kelly TJ, Qin S, Gottschling DE, Parthun MR. Type B histone acetyltransferase Hat1p participates in telomeric silencing. Molecular and cellular biology. 2000;20:7051–8. doi: 10.1128/mcb.20.19.7051-7058.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qin S, Parthun MR. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Molecular and cellular biology. 2002;22:8353–65. doi: 10.1128/MCB.22.23.8353-8365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lai C, Lin C, Chao A. HAT1 as a target for combination therapy with pazopanib and hyperthermia. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2016;34 (supl; abstr e17120) [Google Scholar]
- 59.Xue K, Gu JJ, Zhang Q, Mavis C, Hernandez-Ilizaliturri FJ, et al. Vorinostat, a histone deacetylase (HDAC) inhibitor, promotes cell cycle arrest and re-sensitizes rituximab- and chemo-resistant lymphoma cells to chemotherapy agents. Journal of cancer research and clinical oncology. 2016;142:379–87. doi: 10.1007/s00432-015-2026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10014–9. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terui T, Murakami K, Takimoto R, Takahashi M, Takada K, et al. Induction of PIG3 and NOXA through acetylation of p53 at 320 and 373 lysine residues as a mechanism for apoptotic cell death by histone deacetylase inhibitors. Cancer research. 2003;63:8948–54. [PubMed] [Google Scholar]
- 62.Lamonica JM, Vakoc CR, Blobel GA. Acetylation of GATA-1 is required for chromatin occupancy. Blood. 2006;108:3736–8. doi: 10.1182/blood-2006-07-032847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Santo L, Hideshima T, Kung AL, Tseng JC, Tamang D, et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood. 2012;119:2579–89. doi: 10.1182/blood-2011-10-387365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 65.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nature reviews. Cancer. 2007;7:823–33. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 67.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Human molecular genetics. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 68.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waters NJ, Daigle SR, Rehlaender BN, Basavapathruni A, Campbell CT, et al. Exploring drug delivery for the DOT1L inhibitor pinometostat (EPZ-5676): Subcutaneous administration as an alternative to continuous IV infusion, in the pursuit of an epigenetic target. Journal of controlled release: official journal of the Controlled Release Society. 2015;220:758–65. doi: 10.1016/j.jconrel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 70.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nature genetics. 2010;42:181–5. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 72.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11606–11. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature genetics. 2010;42:722–6. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 74.Xu K, Wu ZJ, Groner AC, He HH, Cai C, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–9. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fiskus W, Wang Y, Sreekumar A, Buckley KM, Shi H, et al. Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood. 2009;114:2733–43. doi: 10.1182/blood-2009-03-213496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qi W, Chan H, Teng L, Li L, Chuai S, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21360–5. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 78.Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nature chemical biology. 2012;8:890–6. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 79.Kurmasheva RT, Sammons M, Favours E, Wu J, Kurmashev D, et al. Initial testing (stage 1) of tazemetostat (EPZ-6438), a novel EZH2 inhibitor, by the Pediatric Preclinical Testing Program. Pediatric blood & cancer. 2017:64. doi: 10.1002/pbc.26218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Knutson SK, Kawano S, Minoshima Y, Warholic NM, Huang KC, et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Molecular cancer therapeutics. 2014;13:842–54. doi: 10.1158/1535-7163.MCT-13-0773. [DOI] [PubMed] [Google Scholar]
- 81.van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature genetics. 2009;41:521–3. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim KH, Kim W, Howard TP, Vazquez F, Tsherniak A, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nature medicine. 2015;21:1491–6. doi: 10.1038/nm.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Watanabe H, Soejima K, Yasuda H, Kawada I, Nakachi I, et al. Deregulation of histone lysine methyltransferases contributes to oncogenic transformation of human bronchoepithelial cells. Cancer cell international. 2008;8:15. doi: 10.1186/1475-2867-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen P, Yao JF, Huang RF, Zheng FF, Jiang XH, et al. Effect of BIX-01294 on H3K9me2 levels and the imprinted gene Snrpn in mouse embryonic fibroblast cells. Bioscience reports. 2015:35. doi: 10.1042/BSR20150064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–7. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rodriguez-Paredes M, Martinez de Paz A, Simo-Riudalbas L, Sayols S, Moutinho C, et al. Gene amplification of the histone methyltransferase SETDB1 contributes to human lung tumorigenesis. Oncogene. 2014;33:2807–13. doi: 10.1038/onc.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nature chemical biology. 2005;1:143–5. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- 88.Wang L, Zhao Z, Meyer MB, Saha S, Yu M, et al. CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer cell. 2014;25:21–36. doi: 10.1016/j.ccr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Elakoum R, Gauchotte G, Oussalah A, Wissler MP, Clement-Duchene C, et al. CARM1 and PRMT1 are dysregulated in lung cancer without hierarchical features. Biochimie. 2014;97:210–8. doi: 10.1016/j.biochi.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 90.Tarighat SS, Santhanam R, Frankhouser D, Radomska HS, Lai H, et al. The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia. 2016;30:789–99. doi: 10.1038/leu.2015.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yao R, Jiang H, Ma Y, Wang L, Wang L, et al. PRMT7 induces epithelial-to-mesenchymal transition and promotes metastasis in breast cancer. Cancer research. 2014;74:5656–67. doi: 10.1158/0008-5472.CAN-14-0800. [DOI] [PubMed] [Google Scholar]
- 92.Chan-Penebre E, Kuplast KG, Majer CR, Boriack-Sjodin PA, Wigle TJ, et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nature chemical biology. 2015;11:432–7. doi: 10.1038/nchembio.1810. [DOI] [PubMed] [Google Scholar]
- 93.Cheung N, Fung TK, Zeisig BB, Holmes K, Rane JK, et al. Targeting Aberrant Epigenetic Networks Mediated by PRMT1 and KDM4C in Acute Myeloid Leukemia. Cancer cell. 2016;29:32–48. doi: 10.1016/j.ccell.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eram MS, Shen Y, Szewczyk MM, Wu H, Senisterra G, et al. A Potent, Selective, and Cell-Active Inhibitor of Human Type I Protein Arginine Methyltransferases. ACS chemical biology. 2016;11:772–81. doi: 10.1021/acschembio.5b00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 96.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 97.Sprussel A, Schulte JH, Weber S, Necke M, Handschke K, et al. Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia. 2012;26:2039–51. doi: 10.1038/leu.2012.157. [DOI] [PubMed] [Google Scholar]
- 98.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 99.Metzger E, Willmann D, McMillan J, Forne I, Metzger P, et al. Assembly of methylated KDM1A and CHD1 drives androgen receptor-dependent transcription and translocation. Nature structural & molecular biology. 2016;23:132–9. doi: 10.1038/nsmb.3153. [DOI] [PubMed] [Google Scholar]
- 100.Schmidt DM, McCafferty DG. trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry. 2007;46:4408–16. doi: 10.1021/bi0618621. [DOI] [PubMed] [Google Scholar]
- 101.Maes T, Carceller E, Salas J, Ortega A, Buesa C. Advances in the development of histone lysine demethylase inhibitors. Current opinion in pharmacology. 2015;23:52–60. doi: 10.1016/j.coph.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 102.Tresckow Bv, Gundermann S, Eichenauer DA, Aulitzky WE, Göbeler M, et al. First-in-human study of 4SC-202, a novel oral HDAC inhibitor in advanced hematologic malignancies (TOPAS study) Journal of Clinical Oncology. 2014;32:8559. [Google Scholar]
- 103.Heinemann B, Nielsen JM, Hudlebusch HR, Lees MJ, Larsen DV, et al. Inhibition of demethylases by GSK-J1/J4. Nature. 2014;514:E1–2. doi: 10.1038/nature13688. [DOI] [PubMed] [Google Scholar]
- 104.Ler LD, Ghosh S, Chai X, Thike AA, Heng HL, et al. Loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. Science translational medicine. 2017:9. doi: 10.1126/scitranslmed.aai8312. [DOI] [PubMed] [Google Scholar]
- 105.de Rooij JD, Hollink IH, Arentsen-Peters ST, van Galen JF, Berna Beverloo H, et al. NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia. 2013;27:2280–8. doi: 10.1038/leu.2013.87. [DOI] [PubMed] [Google Scholar]
- 106.Hancock RL, Dunne K, Walport LJ, Flashman E, Kawamura A. Epigenetic regulation by histone demethylases in hypoxia. Epigenomics. 2015;7:791–811. doi: 10.2217/epi.15.24. [DOI] [PubMed] [Google Scholar]
- 107.Kim J, Daniel J, Espejo A, Lake A, Krishna M, et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO reports. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Molecular and cellular biology. 2008;28:967–76. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, et al. NSD3-Short Is an Adaptor Protein that Couples BRD4 to the CHD8 Chromatin Remodeler. Molecular cell. 2015;60:847–59. doi: 10.1016/j.molcel.2015.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yan J, Diaz J, Jiao J, Wang R, You J. Perturbation of BRD4 protein function by BRD4-NUT protein abrogates cellular differentiation in NUT midline carcinoma. The Journal of biological chemistry. 2011;286:27663–75. doi: 10.1074/jbc.M111.246975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baratta MG, Schinzel AC, Zwang Y, Bandopadhayay P, Bowman-Colin C, et al. An in-tumor genetic screen reveals that the BET bromodomain protein, BRD4, is a potential therapeutic target in ovarian carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:232–7. doi: 10.1073/pnas.1422165112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–82. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–81. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chaidos A, Caputo V, Gouvedenou K, Liu B, Marigo I, et al. Potent antimyeloma activity of the novel bromodomain inhibitors I-BET151 and I-BET762. Blood. 2014;123:697–705. doi: 10.1182/blood-2013-01-478420. [DOI] [PubMed] [Google Scholar]
- 118.Herold JM, Wigle TJ, Norris JL, Lam R, Korboukh VK, et al. Small-molecule ligands of methyl-lysine binding proteins. Journal of medicinal chemistry. 2011;54:2504–11. doi: 10.1021/jm200045v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.James LI, Barsyte-Lovejoy D, Zhong N, Krichevsky L, Korboukh VK, et al. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nature chemical biology. 2013;9:184–91. doi: 10.1038/nchembio.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer discovery. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature genetics. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7922–7. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 124.Kantarjian H, Oki Y, Garcia-Manero G, Huang X, O’Brien S, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109:52–7. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- 125.Evens AM, Balasubramanian S, Vose JM, Harb W, Gordon LI, et al. A Phase I/II Multicenter, Open-Label Study of the Oral Histone Deacetylase Inhibitor Abexinostat in Relapsed/Refractory Lymphoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016;22:1059–66. doi: 10.1158/1078-0432.CCR-15-0624. [DOI] [PubMed] [Google Scholar]
- 126.Lee HZ, Kwitkowski VE, Del Valle PL, Ricci MS, Saber H, et al. FDA Approval: Belinostat for the Treatment of Patients with Relapsed or Refractory Peripheral T-cell Lymphoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015;21:2666–70. doi: 10.1158/1078-0432.CCR-14-3119. [DOI] [PubMed] [Google Scholar]
- 127.Kim KP, Park SJ, Kim JE, Hong YS, Lee JL, et al. First-in-human study of the toxicity, pharmacokinetics, and pharmacodynamics of CG200745, a pan-HDAC inhibitor, in patients with refractory solid malignancies. Investigational new drugs. 2015;33:1048–57. doi: 10.1007/s10637-015-0262-2. [DOI] [PubMed] [Google Scholar]
- 128.Banerji U, van Doorn L, Papadatos-Pastos D, Kristeleit R, Debnam P, et al. A phase I pharmacokinetic and pharmacodynamic study of CHR-3996, an oral class I selective histone deacetylase inhibitor in refractory solid tumors. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:2687–94. doi: 10.1158/1078-0432.CCR-11-3165. [DOI] [PubMed] [Google Scholar]
- 129.Galloway TJ, Wirth LJ, Colevas AD, Gilbert J, Bauman JE, et al. A Phase I Study of CUDC-101, a Multitarget Inhibitor of HDACs, EGFR, and HER2, in Combination with Chemoradiation in Patients with Head and Neck Squamous Cell Carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015;21:1566–73. doi: 10.1158/1078-0432.CCR-14-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Qian C, Lai CJ, Bao R, Wang DG, Wang J, et al. Cancer network disruption by a single molecule inhibitor targeting both histone deacetylase activity and phosphatidylinositol 3-kinase signaling. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:4104–13. doi: 10.1158/1078-0432.CCR-12-0055. [DOI] [PubMed] [Google Scholar]
- 131.Galli M, Salmoiraghi S, Golay J, Gozzini A, Crippa C, et al. A phase II multiple dose clinical trial of histone deacetylase inhibitor ITF2357 in patients with relapsed or progressive multiple myeloma. Annals of hematology. 2010;89:185–90. doi: 10.1007/s00277-009-0793-8. [DOI] [PubMed] [Google Scholar]
- 132.Furlan A, Monzani V, Reznikov LL, Leoni F, Fossati G, et al. Pharmacokinetics, safety and inducible cytokine responses during a phase 1 trial of the oral histone deacetylase inhibitor ITF2357 (givinostat) Molecular medicine. 2011;17:353–62. doi: 10.2119/molmed.2011.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zwergel C, Valente S, Jacob C, Mai A. Emerging approaches for histone deacetylase inhibitor drug discovery. Expert opinion on drug discovery. 2015;10:599–613. doi: 10.1517/17460441.2015.1038236. [DOI] [PubMed] [Google Scholar]
- 134.Popat R, Brown SR, Flanagan L, Hall A, Gregory W, et al. Bortezomib, thalidomide, dexamethasone, and panobinostat for patients with relapsed multiple myeloma (MUK-six): a multicentre, open-label, phase 1/2 trial. The Lancet. Haematology. 2016;3:e572–e80. doi: 10.1016/S2352-3026(16)30165-X. [DOI] [PubMed] [Google Scholar]
- 135.Assouline SE, Nielsen TH, Yu S, Alcaide M, Chong L, et al. Phase 2 study of panobinostat with or without rituximab in relapsed diffuse large B-cell lymphoma. Blood. 2016;128:185–94. doi: 10.1182/blood-2016-02-699520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Eigl BJ, North S, Winquist E, Finch D, Wood L, et al. A phase II study of the HDAC inhibitor SB939 in patients with castration resistant prostate cancer: NCIC clinical trials group study IND195. Investigational new drugs. 2015;33:969–76. doi: 10.1007/s10637-015-0252-4. [DOI] [PubMed] [Google Scholar]
- 137.Venugopal B, Baird R, Kristeleit RS, Plummer R, Cowan R, et al. A phase I study of quisinostat (JNJ-26481585), an oral hydroxamate histone deacetylase inhibitor with evidence of target modulation and antitumor activity, in patients with advanced solid tumors. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:4262–72. doi: 10.1158/1078-0432.CCR-13-0312. [DOI] [PubMed] [Google Scholar]
- 138.Brunetto AT, Ang JE, Lal R, Olmos D, Molife LR, et al. First-in-human, pharmacokinetic and pharmacodynamic phase I study of Resminostat, an oral histone deacetylase inhibitor, in patients with advanced solid tumors. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:5494–504. doi: 10.1158/1078-0432.CCR-13-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Olsen EA, Kim YH, Kuzel TM, Pacheco TR, Foss FM, et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25:3109–15. doi: 10.1200/JCO.2006.10.2434. [DOI] [PubMed] [Google Scholar]
- 140.Shi Y, Dong M, Hong X, Zhang W, Feng J, et al. Results from a multicenter, open-label, pivotal phase II study of chidamide in relapsed or refractory peripheral T-cell lymphoma. Annals of oncology: official journal of the European Society for Medical Oncology. 2015;26:1766–71. doi: 10.1093/annonc/mdv237. [DOI] [PubMed] [Google Scholar]
- 141.Knipstein J, Gore L. Entinostat for treatment of solid tumors and hematologic malignancies. Expert opinion on investigational drugs. 2011;20:1455–67. doi: 10.1517/13543784.2011.613822. [DOI] [PubMed] [Google Scholar]
- 142.Younes A, Oki Y, Bociek RG, Kuruvilla J, Fanale M, et al. Mocetinostat for relapsed classical Hodgkin’s lymphoma: an open-label, single-arm, phase 2 trial. The Lancet. Oncology. 2011;12:1222–8. doi: 10.1016/S1470-2045(11)70265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pauer LR, Olivares J, Cunningham C, Williams A, Grove W, et al. Phase I study of oral CI-994 in combination with carboplatin and paclitaxel in the treatment of patients with advanced solid tumors. Cancer investigation. 2004;22:886–96. doi: 10.1081/cnv-200039852. [DOI] [PubMed] [Google Scholar]
- 144.Piekarz RL, Frye R, Prince HM, Kirschbaum MH, Zain J, et al. Phase 2 trial of romidepsin in patients with peripheral T-cell lymphoma. Blood. 2011;117:5827–34. doi: 10.1182/blood-2010-10-312603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Guzman ML, Yang N, Sharma KK, Balys M, Corbett CA, et al. Selective activity of the histone deacetylase inhibitor AR-42 against leukemia stem cells: a novel potential strategy in acute myelogenous leukemia. Molecular cancer therapeutics. 2014;13:1979–90. doi: 10.1158/1535-7163.MCT-13-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Iannitti T, Palmieri B. Clinical and experimental applications of sodium phenylbutyrate. Drugs in R&D. 2011;11:227–49. doi: 10.2165/11591280-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Reid T, Valone F, Lipera W, Irwin D, Paroly W, et al. Phase II trial of the histone deacetylase inhibitor pivaloyloxymethyl butyrate (Pivanex, AN-9) in advanced non-small cell lung cancer. Lung Cancer. 2004;45:381–6. doi: 10.1016/j.lungcan.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 148.Bilen MA, Fu S, Falchook GS, Ng CS, Wheler JJ, et al. Phase I trial of valproic acid and lenalidomide in patients with advanced cancer. Cancer chemotherapy and pharmacology. 2015;75:869–74. doi: 10.1007/s00280-015-2695-x. [DOI] [PubMed] [Google Scholar]
- 149.Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117:4759–68. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122:1017–25. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yu W, Chory EJ, Wernimont AK, Tempel W, Scopton A, et al. Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors. Nature communications. 2012;3:1288. doi: 10.1038/ncomms2304. [DOI] [PubMed] [Google Scholar]
- 152.Tan J, Yang X, Zhuang L, Jiang X, Chen W, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes & development. 2007;21:1050–63. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Konze KD, Ma A, Li F, Barsyte-Lovejoy D, Parton T, et al. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS chemical biology. 2013;8:1324–34. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chang Y, Zhang X, Horton JR, Upadhyay AK, Spannhoff A, et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nature structural & molecular biology. 2009;16:312–7. doi: 10.1038/nsmb.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yuan Y, Wang Q, Paulk J, Kubicek S, Kemp MM, et al. A small-molecule probe of the histone methyltransferase G9a induces cellular senescence in pancreatic adenocarcinoma. ACS chemical biology. 2012;7:1152–7. doi: 10.1021/cb300139y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nature chemical biology. 2011;7:566–74. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zheng YC, Yu B, Jiang GZ, Feng XJ, He PX, et al. Irreversible LSD1 Inhibitors: Application of Tranylcypromine and Its Derivatives in Cancer Treatment. Current topics in medicinal chemistry. 2016;16:2179–88. doi: 10.2174/1568026616666160216154042. [DOI] [PubMed] [Google Scholar]
- 158.Sugino N, Kawahara M, Tatsumi G, Kanai A, Matsui H, et al. A novel LSD1 inhibitor NCD38 ameliorates MDS-related leukemia with complex karyotype by attenuating leukemia programs via activating super-enhancers. Leukemia. 2017 doi: 10.1038/leu.2017.59. [DOI] [PubMed] [Google Scholar]