Abstract
The immune system provides defense against pathogens and functions to maintain tissue homeostasis for the life of the organism. These diverse functions of immunity are bioenergetically expensive, requiring precise control of cellular metabolic pathways. Although the initial observations in this area were made almost a century ago, studies over the last decade have elucidated the molecular basis of how extracellular signals control the uptake and catabolism of nutrients in quiescent and activated immune cells. Collectively, these studies have revealed that the metabolic pathways of oxidative metabolism, glycolysis, and glutaminolysis, preferentially fuel the cell fate decisions and effector functions of immune cells. We discuss here these findings, and provide a general framework for understanding how metabolism fuels and regulates the maturation of immune responses. A better understanding of the metabolic checkpoints that control these transitions might provide new insights for modulating immunity in infection, cancer, or inflammatory disorders.
Introduction
Metabolism fuels all biological programs, ranging from development, proliferation, differentiation, and the effector functions performed by cells and tissues in an adult organism. In these scenarios, cellular metabolism is not static but rather a dynamic process that functions to meet bioenergetic demands of any given cell at any given point-in-time. In metazoans, these metabolic adaptations are largely controlled by extracellular signals, which direct the uptake, storage, and utilization of substrates (glucose, amino acids, and fatty acids). For instance, insulin is the primary signal that mediates the uptake, storage, mobilization, and utilization of fuels in non-immune target cells, such as adipocytes, hepatocytes, and myocytes (1). In these non-immune cells, insulin regulates various programs to match the cells metabolic capacity with its functional demands. This coupling of metabolism to the performance of cellular functions is not limited to metabolic tissues. Rather, it represents a general paradigm of how metazoan cells respond to environmental signals.
For the life of the organism, the immune system continually senses and responds to environmental threats, a process that carries a considerable bioenergetic cost. For instance, the sensing of a pathogen or environmental insult results in secretion of cytokines, chemokines, and inflammatory mediators by the innate immune cells, and the clonal expansion of adaptive immune cells. Since immune cells lack significant stores of nutrients, these effector responses can only be sustained if immune cells can dramatically increase the uptake of glucose, amino acids, and fatty acids from their microenvironment. Indeed, as observed over a century ago, the elaboration of an effector innate response is critically dependent on glucose (2). Similarly, mitogen-driven proliferation of adaptive immune cells has been known for decades to be dependent on extracellular glutamine (3, 4). In both of these scenarios, this increased uptake of nutrients serves two important purposes. First, it provides the substrates for ATP synthesis to sustain the various cellular programs of activated immune cells. Second, it provides the building blocks for the synthesis of macromolecules, such as RNA, DNA, proteins, and membranes, which are necessary for the proliferation and activation of immune cells. Thus, in response to extracellular signals, a critical step in the maturation of immune cells is the reprogramming of their cellular metabolism.
In this chapter, we provide a framework for understanding the regulatory role of metabolism in the sustenance of innate and adaptive immune responses. We begin with a primer on basic cellular metabolism and how these pathways are regulated in immune cells. Subsequently, we discuss how metabolic switches and pathways control the duration and intensity of innate or adaptive immune activation, and memory cell formation. Finally, we close with the idea that cellular metabolism is not static or fixed but rather a dynamic process that provides a means for the immune system to adapt to the emerging needs of the organism. In this model, immune cells can switch back and forth between different metabolic states and preferentially utilize different metabolic substrates (glucose, amino acids, or fatty acids) to sustain their effector functions. The basic discovery that cellular metabolism, which acts through key regulatory metabolic nodes, controls immune cell function has not only introduced a novel paradigm in the field of immunology, but has also raised the prospect that inflammatory or autoimmune disorders might be amenable to metabolic therapy.
A primer on cellular metabolism
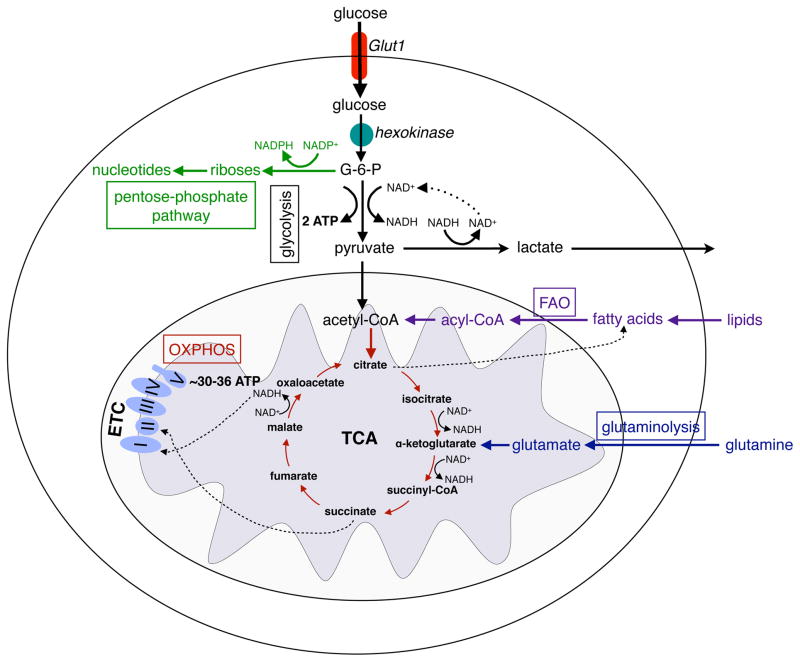
All cells, quiescent, replicating, or activated, need to produce ATP and synthesize macromolecules to maintain their basic cellular functions. These bioenergetic needs of cells are met by the interconnected pathways of glycolysis, tricarboxylic acid (TCA) cycle, and oxidative phosphorylation (OXPHOS) (Figure 1); the former occurs in the cytoplasm, whereas the later two are restricted to the mitochondria. Following the entry of glucose into cells via glucose transporters, glucose is rapidly converted to glucose 6-phosphate by hexokinases, which commits the glucose to glycolysis and prevents its leakage from the cell. Glucose 6-phosphate can then serve as an entry point for glycolysis or for the branch pentose phosphate pathway (PPP), which generates riboses necessary for the synthesis of RNA and DNA, and NADPH for fatty acid synthesis and the respiratory burst of phagocytes (Figure 1 and 2). In the sequential reactions of glycolysis, glucose is metabolized to pyruvate, liberating 2 molecules of each of pyruvate, ATP, and NADH. Although all the reactions of glycolysis are anaerobic, the fate of pyruvate is dependent on the availability of oxygen in non-malignant, resting cells. Under normoxic conditions, pyruvate is fully oxidized to CO2 and H2O via the TCA, which generates NADH that is oxidized via the OXPHOS reactions to generate ATP and NAD+. In contrast, under hypoxic conditions, pyruvate is oxidized to lactate and NAD+; while cells secrete the former, the latter sustains glycolysis. As will be discussed later, up regulation of glycolysis is a critical step in the activation of innate and adaptive immune cells, as it provides a means of increasing flux through the PPP to synthesize macromolecules and generate the antimicrobial respiratory burst.
Figure 1. Major metabolic pathways of immune cells.
Glucose enters the cells through the glucose transporter, GLUT1 and is phosphorylated to glucose-6-phosphate (G-6-P) by hexokinases. During glycolysis, G-6-P is metabolized to pyruvate, reducing NAD+ to NADH, and generating 2 ATP. In hypoxia, pyruvate is oxidized to lactate, restoring NAD+ levels in the cell. In normoxia, pyruvate is metabolized to acetyl-CoA, which is oxidized in the TCA cycle to generate NADH. In the redox reactions of OXPHOS, electrons are sequentially transferred to generate a H+ gradient across the inner mitochondrial membrane, which drives the synthesis of ATP. In contrast to glycolysis, mitochondrial OXPHOS is a highly efficient form of generating ATP, yielding ~30–36 ATP per molecule of glucose. Immune cells utilize three additional pathways, the pentose phosphate shunt (PPP), glutaminolysis, and fatty acid oxidation, to meet their metabolic and functional demands. G-6-P is the entry point for PPP, which generate riboses for nucleotide synthesis. During this process, NADP+ is reduced to NADPH, forming the critical co-factor required for ROS production via the NADPH oxidase system in neutrophils and macrophages. During glutaminolysis, glutamine is metabolized to glutamate and subsequently toα -ketoglutarate, which then enters the TCA cycle. The fate of glutamine is dependent on the activation state of the immune cell; it can either be oxidized completely to generate ATP or used to replenish the metabolic intermediates of TCA cycles, which are diverted for macromolecule biosynthesis. The β-oxidation of fatty acids yields acetyl-CoA, which enter the TCA cycle and OXPHOS pathways to generate ATP.
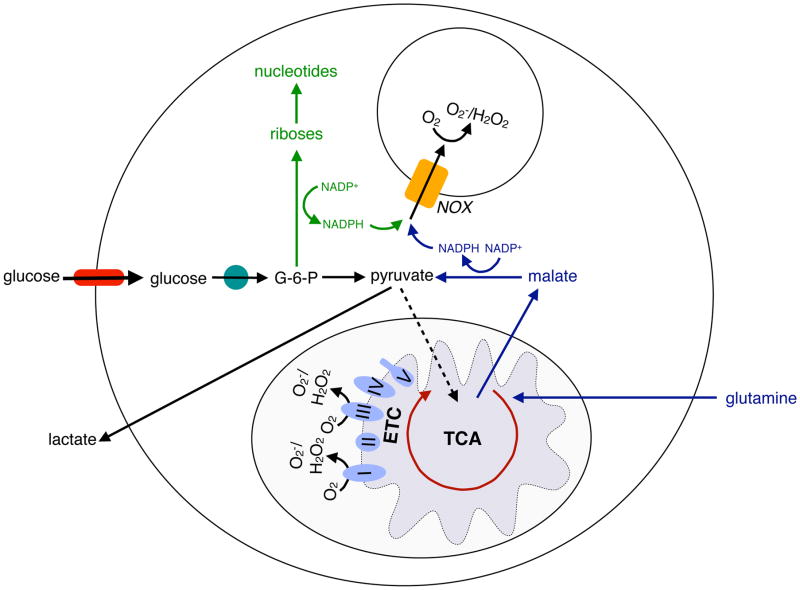
Figure 2. Metabolic pathways supporting ROS generation in macrophages and neutrophils.
Macrophages and neutrophils rely on the production of reactive oxygen species (ROS) for their bactericidal actions. NADPH, which is required for the redox reactions of NADPH oxidase (NOX) system, is generated in the PPP branch of glycolysis or from the oxidation of glutamine-derived malate to pyruvate. In this manner, the metabolic pathways of both glycolysis and glutaminolysis contribute to the microbicidal activity of macrophages and neutrophils. In addition, mitochondrial ROS, which is derived from the inefficient flow of electrons through complex I and III of the electron transport chain (ETC), is a significant source of microbicidal ROS in macrophages.
The complete oxidation of all nutrients occurs in the TCA cycle of the mitochondria, which provides the reducing equivalents as NADH for the generation of ATP via subsequent reactions of OXPHOS. Carbohydrates, amino acids, and fatty acids enter the TCA cycle after they are catabolized to acetyl coenzyme A (acetyl-CoA), which also serves as a substrate for the synthesis of isoprenoids, cholesterol, flavonoids, and fatty acids, and for the posttranslational modification (acetylation) of histones and proteins (5). In the TCA cycle, acetyl-CoA is oxidized to generate CO2 and NADH, the later fueling the electron transport chain of OXPHOS via its entry in complex I.
In addition to NADH, succinate, an intermediate of the TCA cycle, contributes to the generation of mitochondrial proton gradient and ATP synthesis via its entry in complex II. In aggregate, the complete oxidation of glucose by the TCA cycle yields enough reducing equivalents to generate ~30–36 ATPs by the electron transport chain of OXPHOS. Thus, in aerobic conditions, oxidative metabolism is bioenergetically favorable way of generating ATP in metazoan cells. In addition to the catabolism of nutrients, glycolysis and TCA cycle provide the intermediates for the biosynthesis of riboses (via the PPP), fatty acids (using citrate to generate acetyl-CoA by ATP citrate lyase), and nonessential amino acids (Figure 1). For instance, glycolysis intermediates 3-phosphoglycerate and pyruvate serve as precursors for the synthesis of serine, cysteine, glycine, and alanine, whereas the TCA cycle intermediates oxaloacetate and α-ketoglutarate are used to synthesize aspartate, asparagine, proline, and arginine. Since these biosynthetic reactions require extraction of intermediates from glycolysis and the TCA cycle (termed cataplerosis), their continual replenishment is necessary to sustain these essential metabolic pathways (6, 7). For glycolysis, this need is met by increasing metabolic flux through the glycolytic pathway, as one observes in activated innate and adaptive immune cells. In contrast, two anaplerotic reactions are used to replenish the TCA cycle; the first converts pyruvate to oxaloacetate via pyruvate carboxylase, whereas second converts glutamate to α-ketoglutarate via glutamate dehydrogenase. Since activated immune cells increase their uptake of glucose (used to generate pyruvate) and glutamine (to generate glutamate), this increase in substrate availability provides a means of sustaining the necessary anaplerotic reactions. Moreover, because anaplerotic flux must be greater than cataplerotic flux to replenish the intermediates of the TCA cycle, neither glucose nor glutamine is fully oxidized to generate ATP in activated immune cells (4). Rather, even under aerobic conditions, excess flux through these pathways is maintained by the oxidation of pyruvate to lactate, which generates the necessary NAD+ to sustain both glycolysis and the TCA cycle. This preferential reliance on glycolysis and glutaminolysis, which is in excess of what is required to support cellular ATP levels, is often referred to as the Warburg effect or Warburg metabolism (8, 9).
Architectural principles for metabolic control of immune cells
The primordial functions of immune cells in host defense and tissue homeostasis provide the instructive cues that control their metabolic programs. To facilitate the dissection of these complex regulatory networks, it is useful to group these signals into distinct functional categories (Table 1). In its simplest form, immune activation can be divided into four principle components: inducers (signals that initiate the inflammatory response), sensors (proteins that detect the presence of inducers), mediators (molecules that signal to activate the effector responses), and effectors (in this case, the downstream metabolic programs that facilitate the maturation of the desired effector phenotype) (10, 11). The typical inducers of innate and adaptive immune cells include pathogen-associated molecular patterns (PAMPs), processed antigens, cytokines, and growth factors, which signal to immune cells to initiate effector responses. These signals are in turn sensed by cell surface receptors, such as the pattern recognition receptors (PRRs), antigen receptors (TCRs and BCRs), cytokine receptors, and co-stimulatory molecules (like CD28), resulting in the activation of distinct signaling pathways. In nearly all cases, these signaling pathways converge onto a small group of conserved metabolic regulators, such as the PI3K-Akt-mTOR pathway, to initiate the effector metabolic adaptations. While the acute metabolic adaptations are achieved by posttranslational modification of metabolic enzymes, long-term metabolic programming of immune cells requires activation of transcriptional regulators and co-activators to drive changes in gene expression that stabilize the appropriate substrate utilization programs. In this manner, inducers trigger metabolic switches to metabolically prime the immune cells, thereby providing the permissive conditions for enactment of their cell fate programs. A final point of consideration is that these metabolic changes are not permanent. Unlike the metabolic transitions that occur in cancer (a genetically driven switch to aerobic glycolysis) or the exercise-induced fiber-type switches in skeletal muscle, many immune cells are capable of rapidly shifting between glycolysis and OXPHOS in response to external signals.
Table 1. Architectural principles for metabolic control in immune cells.
From an immunological and metabolic perspective, the activation of innate and adaptive immune can be broadly divided into 4 principle components, including the inducers (signals that activate immune cells), the sensors (proteins that detect inducers), the mediators (proteins and metabolites that transduce the signals downstream of the sensors), and the effectors (the metabolic effector responses that support the functional state of immune cells). In a combinatorial manner, individual immune cell differentially utilize these components to achieve their desired functional outcome.
| Cell | Inducers | Sensors | Mediators | Effectors | Outcome |
|---|---|---|---|---|---|
| Neutrophil | PAMPs, Chemokines | PRRs (TLRs) | HIF-1α | Glycolysis, Glutaminolysis | ROS |
| Mast cell | PAMPs, IgE cross-linking, Cytokines, Growth factors | PRRs, FcεRI | unknown | Glycolysis | Degranulation, Cytokine production |
| Resting dendritic cell | Growth factors (GM-CSF, FLT3) | Growth factor receptors | unknown | FAO | Growth, Survival Activation, Ag |
| Activated dendritic cell | PAMPs | PRRs (TLRs) | PI3K/Akt HIF-1α |
Glycolysis | Presentation, Cytokine production |
| Classically activated macrophage (CAM) | PAMPs | PRRs (TLRs, NODs) | HIF-1α | Glycolysis, Glutaminolysis | ROS, Cytokine production |
| Alternatively activated macrophage (AAM) | IL-4, IL-13, Parasites | IL-4Rα, IL-13Rα | STAT6 PPARs PGC1β |
FAO | Differentiation |
| Naive CD4 + T cell | IL-7, Ag | IL-7R, TCR | PI3K/Akt | Mitochondrial OXPHOS, FAO | Survival |
| Activated CD4 + T cell | Ag, CD3/CD28 | TCR | PI3K/Akt/mTOR ERK/MAPK c-Myc HIF-1α |
Glycolysis, Glutaminolysis, Mitochondrial OXPHOS | Activation, Proliferation, Cytokine production |
| Memory CD8 + T cell | IL-15 | IL-15R, TRAF6 | AMPK | FAO | Survival, Quiescence |
| B cell | Ag, PAMPs | BCR, PRRs (TLRs) | PI3K/Akt | Glycolysis | Activation, Proliferation |
In discussing the metabolic states of individual immune cell subsets, a natural divide forms as we consider the differences between innate and adaptive immune cells. While both groups transition from quiescent/naïve to activated/effector states in response to inducers, adaptive immune cells are unique in their ability to rapidly proliferate and subsequently generate a memory response.
Innate cells
The innate immune system, consisting of circulating and tissue resident cells, serves as a first line defense against foreign antigens and actively participates in the maintenance of tissue homeostasis. These cells express a range of PRRs, such as the toll-like receptors (TLRs), which allows for sensing of PAMPs and initiation of complex host defense programs (12). Being professional phagocytes, innate immune cells are also responsible for the clearance of pathogenic bacteria. As will be discussed in detail below, the metabolic programs enacted by these quiescent and activated innate immune cells are not only distinct but also well suited to match the metabolic demands of their effector states. Congruent with this idea, genetic perturbations that prime or inhibit innate immune cell metabolism have a profound effect on their ability to undergo activation, and to perform their functions in host defense and tissue homeostasis.
Neutrophils
Neutrophils, which constitute about 50–60% of circulating leukocytes, are the first responders to infectious stimuli (13). As such, the primary function of these short-lived cells is to phagocytose and destroy invading pathogens, such as bacteria and fungi. Upon pathogen sensing, the latent cytosolic NADPH oxidase complex (NOX) becomes activated in these cells, allowing for generation of the microbicidal H2O2 (13) (Figure 2). Interestingly, as early as the 1950’s, it was appreciated that neutrophils primarily utilize glucose and glycolysis to fuel their respiratory burst (14, 15). Consistent with this notion, mitochondrial density in neutrophils is low and inhibitors of OXPHOS do not alter their rates of oxygen consumption or H2O2 production (16–18). In contrast, inhibition of glycolytic metabolism by 2-deoxyglucose (2-DG), a glucose molecule that is taken up by glucose transporters but cannot undergo glycolysis, severely impairs the ability of neutrophils to infiltrate tissues, and phagocytose and kill the ingested bacteria (19, 20). Similarly, the addition of glutamine to the medium has been shown to enhance the phagocytic and bacterial killing ability of cultured neutrophils (21, 22). Since both glucose and glutamine are not completely oxidized to generate ATP, it suggests that the increased uptake of these nutrients is necessary to support other cellular functions. Indeed, the engagement of Warburg metabolism is the primary means by which activated neutrophils synthesize NADPH to support their respiratory burst (Figure 2). First, the PPP branch of glycolysis allows cells to generate NADPH from catabolism of glucose. Second, neutrophils employ glutaminolysis, which utilizes steps from the TCA cycle and the malate-aspartate shuttle, to degrade glutamine to malate. Subsequently, malate is oxidized to generate pyruvate and NADPH by NADP+-dependent malate dehydrogenase (Figure 2). Thus, the biochemical pathways of glycolysis and glutaminolysis provide the means of rapidly generating the reducing equivalents of NADPH for the microbicidal NADPH oxidase system. In addition, NADPH oxidase was recently shown to be required for the process of ‘netosis’, whereby neutrophils release their mitochondrial DNA in net-like structures to facilitate bacterial trapping (23, 24).
Although the importance of glycolysis and glutaminolysis has been firmly established in the microbicidal actions of neutrophils, the downstream mediators that mediate these responses remain poorly understood. For instance, the molecular pathways that link pathogen sensing by PRRs to induction of glycolysis and glutaminolysis have not been identified. However, hypoxia-induced factor 1α (HIF-1α) has been implicated in the transcriptional responses of neutrophils to various bacteria in hypoxic and normoxic conditions. HIF-1α, which is induced by hypoxia or infection, transcriptionally activates the glucose transporter Glut1 and the glycolytic enzyme phosphoglyceratekinase (PGK), resulting in the enhancement of glycolytic metabolism in neutrophils (25). In its absence, both glycolysis and ATP production are severely compromised in these cells. In addition to regulating glycolytic metabolism, HIF-1α also controls the expression of a number of antimicrobial factors in neutrophils, including the serine proteases neutrophil elastase and cathepsin G, cathelicidin-related antimicrobial peptide, and inducible nitric oxide (Nos2) (26). Accordingly, mice lacking HIF-1α in myeloid cells are more susceptible to infection by bacteria, such as group A Streptococcus (26). This coordinated regulation of glycolytic and antimicrobial programs by HIF-1α provides an elegant example of how a single transcription factor reprograms cellular metabolism to support the effector functions of an immune cell.
After initiation of their microbicidal respiratory burst, neutrophils rapidly undergo apoptosis and are promptly cleared from the inflammatory sites by macrophages. The timely clearance of these dying cells is critical, because in its absence inflammation can be perpetuated, resulting in unnecessary tissue damage (27, 28). Although neutrophil mitochondrial respiration is not required for their oxidative burst, maintenance of mitochondrial potential is essential for cell survival (29). Shortly after mounting a respiratory burst, neutrophil mitochondria lose their potential, resulting in the release of cytochrome c and initiation of the proteolytic cascade of apoptotic cell death (17). Thus, from the onset of their activation to their death, host defense functions performed by neutrophils are intimately linked to their underlying metabolic state. Finally, while the literature on basophil and eosinophil metabolism is scant, the available studies do suggest that similar metabolic programs might be enacted in these granulocytes (30–34).
Mast cells
Mast cells are tissue resident cells that play important functions in wound healing, and defense against pathogens, venoms, and toxins (35). In addition, their pathogenic activation contributes to allergies and anaphylaxis (36). These diverse functions of mast cells are mediated by their release of preformed and newly synthesized mediators, such as serine proteases, histamine, serotonin, leukotrienes, and thromboxane. Upon stimulation by injury, activated complement or crosslinking of IgE receptors, mast cells rapidly degranulate, releasing their cytosolic contents into the interstitium. However, following activation, these tissue resident cells do not undergo apoptosis, but instead enter a refractory period where they restore their secretory granules. Although the energy demands of degranulation and regranulation are likely to be high, relatively little is known about how they are met metabolically. Some early investigations in rat mast cells using a glycolytic inhibitor (2-DG) indicate that histamine release and cellular ATP content of activated mast cells is dependent on glycolysis (37–39). However, in these studies, a significant portion of labeled tracer glucose is also released as CO2, which is suggestive of active glucose oxidation via the TCA cycle (40). Since the metabolic demands of mast cells during degranulation and regranulation are likely to be distinct (the later also requires new membrane synthesis), it will be important to fully understand how mast cells alter their substrate utilization programs to support their effector functions.
Macrophages
As resident cells of almost every tissue in the body, macrophages provide a first line of defense against pathogens and injury, and are necessary for the maintenance of tissue homeostasis (41). Until recently, the conventional view had been that bone marrow-derived monocytes seed tissues with macrophages. However, recent lineage tracing studies revealed that nearly all tissue resident tissue macrophages in adult mice are derived from the embryonic yolk sac progenitors, and their numbers are maintained by local proliferation through out the life of the organism (42–45). Moreover, these tissue resident macrophages are highly heterogeneous in their gene expression, likely reflecting the tissue specific functions performed by these cells. For instance, red pulp macrophages in the spleen are primarily responsible for the clearance of senescent red blood cells, whereas the highly specialized osteoclasts are involved in resorption and remodeling of bone.
In addition to adopting tissue-specific fates, resident and recruited macrophages can undergo distinct activation programs in response to pathogen-derived cues. Classical (M1) and alternative (M2) are two of the best-characterized states of macrophage activation, both in terms of their functions in host defense and the regulatory cascades that control their effector responses. While bacteria-derived PAMPs, such LPS, and the type 1 cytokine interferon γ (IFNγ) drive classical (M1) macrophage activation, the type 2 cytokines interleukin (IL)-4 and -13 polarize resident and recruited macrophages to the alternative M2 state (46). Congruent with these observations, classically activated macrophages (CAMs) confer protection against bacterial pathogens by mounting a pro-inflammatory response that is characterized by release of inflammatory cytokines (IL-1β, IL-12, IL-6 and tumor necrosis factor α (TNF)), and reactive oxygen and nitrogen species (47). In contrast, alternatively activated macrophages (AAMs) participate in host defense against parasitic infections by modulating innate and adaptive immune responses, and facilitating the formation of granulation or scar tissue (46, 48). Furthermore, the preferential expression of arginase 1, mannose receptor (CD206), CLEC10A (CD301), Chi3l3 (Ym-1), and Retnla (RELMα or Fizz1) readily identifies AAMs at sites of infection and tissue damage (46, 48). Importantly, as detailed below, these effector responses of CAMs and AAMs are intimately linked to and controlled by the signaling and transcriptional pathways that are canonical regulators of cellular metabolism.
Classically activated macrophages (M1)
Although the importance of glycolysis in inflammatory activation of macrophages was first noted almost a century ago (49), its biochemical and metabolic importance had not been appreciated till recently. These initial studies, which employed inhibitors of glycolysis and measured substrate uptake, revealed that activated macrophages had high rates of glucose and glutamine uptake, and lactic acid production (50–52). Moreover, since neither glucose nor glutamine was oxidized completely to CO2, it suggested that glycolysis and glutaminolysis were not necessary for ATP generation but rather supported some other cellular function. Indeed, as discussed earlier, glycolysis (via the PPP) and glutaminolysis (via the malate-aspartate shuttle and NADP+-dependent malate dehydrogenase) support the synthesis of NADPH, which is necessary for generation of reactive oxygen species (ROS) by the NADPH oxidase system (Figure 2). Congruent with these observations, inhibition of glycolysis or absence of glutamine severely impairs ROS production and the microbicidal activity of CAMs (53–56).
Similar to neutrophils, HIF-1α has emerged as the primary regulator of glycolytic metabolism in CAMs (Figure 3). Deletion of HIF-1α in myeloid cells dramatically reduces expression of the glucose transporter Glut1 and the glycolytic enzyme phosphoglyceratekinase (PGK) under both normoxic and hypoxic conditions (25). Consequently, in response to LPS, lactate and ATP production is reduced in HIF-1α deficient macrophages, resulting in reduced clearance of Group B streptococci. In addition, macrophages lacking HIF-1α are impaired in their migration, as tested in vitro using a modified Boyden chamber (25). Although these studies did not investigate the impact of HIF-1α deficiency on macrophage ROS production, loss of HIF-1α impaired maturation of CAMs, as evidenced by reduced production of TNF (25, 26, 57). These studies collectively demonstrate that HIF-1α transcriptionally couples glycolytic metabolism to macrophages’ inflammatory and microbicidal programs. However, it remains unknown whether glutamine uptake and glutaminolysis are under the transcriptional control of HIF-1α in CAMs.
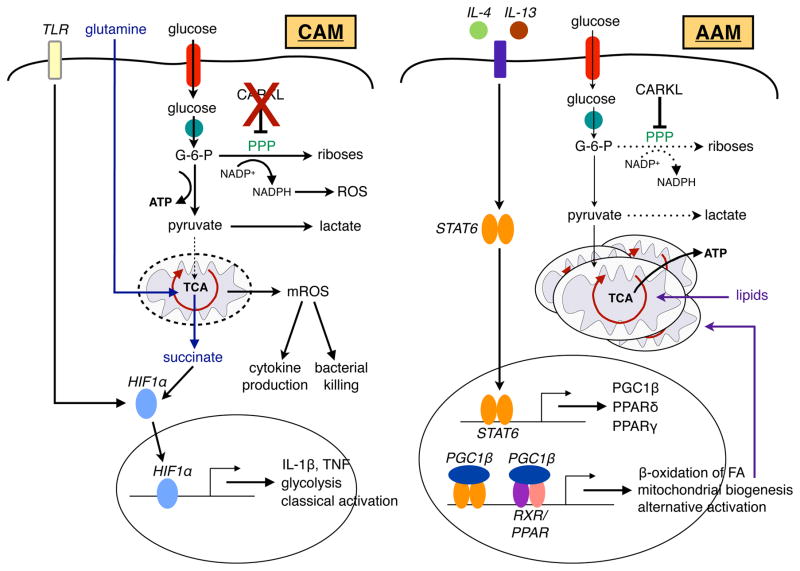
Figure 3. CAMs and AAMs use distinct metabolic programs to fuel their functions.
Stimulation of TLRs by PAMPs, such as LPS, drives the expression of glycolytic genes and inflammatory cytokines, a transcriptional program that is primarily coordinated by HIF-1α in classically activated macrophages (CAMs). This transition to glycolysis allows CAMs to generate NADPH through the PPP to support their respiratory burst. Succinate, derived from glutamine-dependent anaplerosis, furthers stabilizes HIF-1α protein, resulting in increased transcription and release of IL-1β. Moreover, downstream of TLR signaling, mitochondrial ROS (mROS) can also support the microbicidal functions of CAMs. In contrast to the glycolytic program of CAMs, alternatively activated macrophages (AAMs) preferentially rely on β-oxidation of fatty acids and mitochondrial respiration for their sustenance and functional activation. Type 2 cytokines, such as IL-4 and IL-13, signal through their cognate receptors to activate the latent STAT6 transcription factor. STAT6 promotes the metabolic transition to oxidative metabolism by inducting genes important in fatty acid oxidation (FAO) and mitochondrial biogenesis. In addition, STAT6 transcriptionally induces PGC1β, PPARγ, and PPARδ, which synergize with STAT6 to enhance expression of alternative activation markers and stabilize the metabolic switch to oxidative metabolism. Induction of CARKL in AAMs results in inhibition of PPP and glycolysis, thereby favoring the oxidative program of AAMs.
In addition to the ROS generation by NADPH oxidase system, recent studies have implicated mitochondrial ROS in the killing of intracellular bacterial pathogens (Figure 3). In the initial report, Sonoda et al. demonstrated that prolonged stimulation of macrophages with IFNγ increased expression of a large number of mitochondrial genes, whose expression was dependent on the estrogen related receptor α (ERRα) and peroxisome proliferative activated receptor-γ coactivator 1 beta (PGC-1β), two transcriptional regulators that control mitochondrial biogenesis and respiration in cells (58). Interestingly, this increase in mitochondrial respiration, which is accompanied by increased mitochondrial ROS production, was required for host defense against L. monocytogenes. Furthermore, a link between TLR signaling and the mitochondria was recently established by West et al. (59). These authors found that activation of TLR1, TLR2, and TLR4 signaling resulted in the recruitment of the mitochondria to the phagolysosome, where they contributed to the ROS production and bactericidal activity. Interestingly, the TLR signaling adaptor, tumor necrosis factor receptor-associated factor 6 (TRAF6) interacted with the ECSIT protein (evolutionarily conserved signaling intermediate in Toll pathways), resulting in the ubiquitination and enrichment of ECSIT at the periphery of the mitochondria, where it contributes to mitochondrial ROS generation (59). Consistent with these observations, depletion of TRAF6 and ECSIT in macrophages significantly reduced mitochondrial ROS production and the microbicidal activity of macrophages. Similarly, transgenic expression of mitochondrial catalase, an enzyme that reduces mitochondrial ROS production by converting H2O2 to water and oxygen, resulted in a significant increase in the replication rates of S. typhimurium in macrophages. These findings together with the observations that other mitochondrial proteins participate in antiviral immunity indicate that mitochondria and mitochondria-derived signals might be important integrators and effectors of host immunity (60).
Two mitochondria-derived signals have been implicated in the inflammatory activation of macrophages. First, Jurg Tschopp’s group demonstrated that mitochondria-derived ROS are sensed by NLRP3 inflammasome, resulting in the activation of caspase 1 and secretion of mature IL-1β (61). This release of IL-1β from macrophages was also dependent on the mitochondrial voltage-dependent anion channels, thereby providing an independent link between the mitochondria and the inflammasome. In keeping with this idea, inhibition of autophagy, which impairs the removal of ROS-generating mitochondria, resulted in activation of the NLRP3 inflammasome and cleavage of proIL-1β (61, 62). The second mitochondrial signal that has been implicated in the propagation of the effector phenotype of CAMs is succinate, an intermediate of the TCA cycle (Figure 3). In interesting studies, the O’Neill group found that stimulation of macrophages with LPS resulted in increased expression of IL-1β mRNA in a HIF-1α and glycolysis dependent manner (63). A metabolomic screen of LPS stimulated macrophages not only revealed the expected activation of the Warburg effect but also an unexpected accumulation of intermediates of the TCA cycle, in particular, succinate. Since export of succinate from the mitochondria stabilizes and activates HIF-1α, the authors utilized metabolic tracer studies to identify the precursors that contributed to the synthesis of succinate. Notably, in LPS-stimulated macrophages, glutaminolysis-derived α-ketoglutarate rather than glycolysis-derived pyruvate was found to be the primary source of labeled carbons that assimilated in succinate (63) (Figure 3). Collectively, these studies demonstrate that while glycolysis initially supports ATP, NADPH, and cytokine production in CAMs, glutaminolysis, acting through the anaplerotic reactions of the TCA cycle, is necessary for sustaining the respiratory and inflammatory programs in these cells. Since the TCA cycle is harbored in the mitochondria, these findings are consistent with the idea that mitochondrial respiration via the TCA and OXPHOS pathways supports the effector responses of CAMs.
Alternatively activated macrophages (M2)
Alternatively activated macrophages (AAMs) are induced by type 2 cytokines and participate in the host defense against parasites. Since parasitic infections are chronic and the alternative activation program is energetically demanding, both in terms of duration and intensity, Vats and colleagues postulated distinct substrates and pathways might meet the metabolic demands of AAMs (64). Microarray analysis of CAMs and AAMs revealed that genes important in fatty acid oxidation were preferentially expressed in AAMs. Metabolic studies further verified that AAMs had higher rates of fatty acid oxidation, mitochondrial mass, and mitochondrial respiration (Figure 3). Accordingly, inhibition of OXPHOS by metabolic inhibitors dramatically attenuated the expression of the alternatively activated phenotype, including the suppression of inflammatory cytokine secretion and expression of alternative activation markers (64).
Interestingly, this preferential reliance of CAMs on glycolysis and AAMs on oxidative metabolism is reminiscent of the fuel preferences of fast (type II) and slow twitch (type I) muscle fibers. For instance, type II muscle fibers rely on anaerobic glycolysis to fuel their short and rapid bursts of high intensity contractions (similar to CAMs), whereas type I muscle fibers oxidize fatty acids in their mitochondria to sustain low intensity contractions for long periods of time (similar to AAMs) (65). Interestingly, HIF-1α controls glycolytic metabolism in both CAMs and type II muscle fibers (66) whereas, as discussed below, peroxisome proliferator activated receptors (PPARs) and PGCs regulate β-oxidation of fatty acids and mitochondrial oxidative metabolism in both AAMs and type I muscle fibers (67–69). From these studies, a general principle emerges that mammalian cells utilize a conserved set of regulators for coupling metabolism to the cells effector functions, a theme that will be repeated as we consider the instructive role of metabolism in adaptive immunity.
Stimulation of macrophages with type 2 cytokines, IL-4 or IL-13, initiates a transcriptional cascade that controls and sustains their cellular program of oxidative metabolism (Table 1). Although previous studies had demonstrated that alternative macrophage activation by IL-4 and IL-13 is dependent on STAT6, the role of this transcription factor in its metabolic programming was not known. Vats et al. discovered that switch of AAMs to β-oxidation of fatty acids and oxidative metabolism was initiated by and dependent on STAT6 in two ways (Figure 3). First, STAT6 transcriptionally activated the metabolic genes important in β-oxidation of fatty acids, OXPHOS, and mitochondrial biogenesis (64). Second, STAT6 induced the expression of transcriptional regulators and co-activator proteins, such as PPARγ, PPARδ, and PGC1β, which synergized with STAT6 to sustain this metabolic switch to oxidative metabolism (64, 70–72). In addition, these canonical metabolic regulators were found to synergize with STAT6 on promoters of alternative activation genes, such as Arg1 and Clec10A, to amplify and sustain the effector phenotype of these cells.
The metabolic bias of CAMs and AAMs for glycolytic and oxidative metabolism has been subsequently verified using the metabolomics and tracer studies. Rodriguez-Padro et al. reported that the switch to glycolytic metabolism in CAMs was regulated by the phosphofructokinase 2 (PFK2) PFKFB3, which converts fructose-6-phosphate to fructose-2,6-biphosphate to stimulate glycolytic flux (73). In contrast, AAMs fail to induce the expression of PFKFB3, resulting in preservation of oxidative metabolism in these cells. Similarly, Haschemi et al. identified carbohydrate kinase-like protein (CARKL) in a kinome screen as a kinase that is rapidly down regulated in during LPS-driven classical (M1) macrophage activation (74). A sedoheptulose kinase in the pentose phosphate pathway, CARKL functions to restrict flux through the PPP, limiting the generation of NADPH and decreasing glycolytic flux (Figure 3). Since CAMs are highly dependent on glycolysis and PPP-derived NADPH for their effector functions, overexpression of CARKL metabolically reprograms macrophages towards oxidative metabolism and attenuates their inflammatory activation. Conversely, siRNA mediated knockdown of CARKL increases glycolytic metabolism, resulting in decreased expression of Mrc1, an alternative (M2) activation marker, and increased secretion of pro-inflammatory cytokines, including IL-6 and TNF (74). These studies thus provide independent verification for the importance of glycolytic and oxidative metabolism in promoting the maturation of classically (M1) and alternatively (M2) activated macrophages.
Dendritic cells
Dendritic cells (DCs), which are present in nearly all tissues of the body, continually sample their external environment, and are critical for initiating antigen-specific adaptive immune responses and for promoting tolerance (75). Similar to macrophages, tissue resident DCs exhibit great heterogeneity in gene and cell surface marker expression, which reflects their distinct capabilities for antigen processing and engagement of effector lymphocytes (76). Based on the heterogeneity in DC subsets, their bioenergetic demands are likely to be distinct and related to their tissue-specific functions. However, our knowledge of how metabolic substrates and pathways fuel DCs is currently limited, and primarily derived from studies performed using bone-marrow derived DCs. Since nearly all DC subsets capture and process external antigens in order to present them to naïve T cells, these early studies provide a useful framework for understanding how cellular metabolism can instruct DC function.
In response to TLR ligands, resting DCs undergo maturation, resulting in their migration to lymphoid tissues where processed antigens are presented to naïve T cells. Interestingly, similar to CAMs, the ligation of TLRs in DCs activates the metabolic switch for glycolytic metabolism, a program that is co-regulated by HIF-1α and the phosphatidylinositol 3’-kinase (PI3K)/Akt pathway (Table 1). Using the glycolytic inhibitor 2-DG, Jantsch et al. provided the initial evidence that glycolysis is required for the maturation of DCs (77). Notably, in the presence of 2-DG, stimulation of immature DCs with LPS failed to increase glycolysis (as quantified by lactate production), resulting in significantly lower expression of the co-stimulatory molecule CD80. This increase in glycolysis was, in part, regulated by the transcription factor HIF-1α because siRNA-mediated knockdown of HIF-1α decreased expression of Glut1, resulting in reduced proliferation of T cells in an allogenic mixed leukocyte reaction (77). In a subsequent report, Krawczyk et al. provided much stronger evidence for the importance of oxidative and glycolytic metabolism in the sustenance of immature and mature DC phenotypes, respectively (78). These authors demonstrated that LPS-induced DC maturation was associated with a shift from oxidative to glycolytic metabolism (Table 1). In fact, the suppression of β-oxidation of fatty acids and mitochondrial respiration in mature DCs was more pronounced than the increase in their glycolytic rate, the latter being mediated by the activation of PI3K-Akt pathway by the TLR ligands (78). Moreover, the switch to glycolysis was necessary for the survival of mature DCs, and their expression of co-stimulatory molecules, cytokine production, and ability to stimulate antigen-specific naïve T cells. Conversely, a shift of the metabolic state of DCs from glycolytic to oxidative via activation of adenosine monophosphate (AMP)–activated protein kinase (AMPK) was sufficient to inhibit their maturation. These findings are analogous to the metabolic dichotomy of AAMs and CAMs suggesting that switch from oxidative to glycolytic metabolism might a general mechanism by which TLR ligands activate innate immune cells.
As mentioned above, ligation of TLRs results in potent suppression of mitochondrial oxidative metabolism, suggesting that this metabolic checkpoint needs to be cleared for the complete enactment of the activated phenotype. However, how and why TLR agonists potently suppress mitochondrial OXPHOS remained unclear. In an elegant study, Everts et al. demonstrated that stimulation of immature DCs with LPS suppressed OXPHOS, resulting in metabolic reliance on glycolysis for the synthesis of ATP (79). Moreover, this shift to glycolysis was associated with increased NO production via Nos2, which potently inhibited OXPHOS and electron transport chain in these inflammatory DCs. Although this inhibition of OXPHOS by NO was required for the early shift to glycolysis in DCs, it was dispensable for the expression of their full-activated phenotype. These observations potentially suggest a dual role for glycolysis in the maturation of DCs. In the early stages of DC activation, glycolysis might function to provide the metabolic and bioenergetic intermediates necessary for DC activation. In the later stages, NO production by inflammatory DCs would result in complete inhibition of OXPHOS, necessitating ATP synthesis via glycolysis to support cell survival. Altogether, these studies suggest that the metabolic reprogramming of DCs by TLR ligands might serve multiple functions (some known and others unknown) to drive the maturation of an immune response.
Adaptive cells
Besides cytokine and chemokine production, adaptive immune cells have two additional functions that must be bioenergetically supported by cataplerotic and anaplerotic reactions of cellular metabolism. First, antigen-specific activation of adaptive immune cells results in their clonal proliferation, requiring macromolecules for the synthesis of DNA, proteins, and membranes, and for organelle biogenesis. Second, upon successful completion of the immune response, a subset of antigen-specific lymphocytes become quiescent, establishing a pool of memory cells. Since the bioenergetic demands of activated lymphocytes and memory cells are likely to be distinct, the metabolic pathways that fuel these effector functions ought to be different. Indeed, as depicted in Figure 4, lymphocytes in naïve, activated, and memory states all use distinct metabolic substrates and pathways to fuel their effector functions. Although most of the studies have focused on CD4+ T cells, parallels can be drawn to CD8+ T cells and B cells. CD4+ T cells: homeostasis Naïve CD4+ T cells are metabolically active, requiring ATP synthesis for survival and migration through the lymphatic system, where they actively sample antigens presented by DCs. The metabolic requirements of naïve CD4+ T cells are met by oxidation of glucose and fatty acids via the mitochondrial β-oxidation and OXPHOS pathways (80, 81). In the absence of antigenic stimulation, signaling via the IL-7 receptor and T cell receptor (TCR) promotes survival of naïve CD4+ T cells by inducing the expression of the glucose transporter Glut1 (Slc2a1) (82–84). This increase in Glut1 expression and glucose uptake occurs in a PI3K/Akt dependent manner, allowing cells to maintain their mitochondrial potential and ATP homeostasis (84, 85) (Figure 4). Accordingly, in the absence of adequate extrinsic signals, cell surface Glut1 expression declines, resulting in decreased glucose uptake, drop in mitochondrial membrane potential and ATP synthesis, and eventually, cell death (81, 83). Since this decline in viability occurs in the presence of sufficient glucose and O2, it suggests that growth factor signaling is necessary for maintenance of metabolic homeostasis in naïve CD4+ T cells.
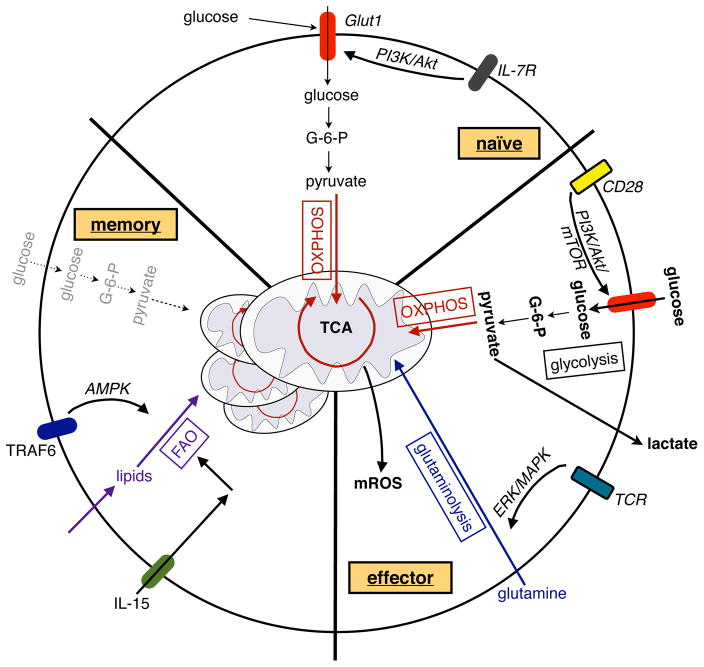
Figure 4. Metabolism of naïve, activated and memory T cells.
Signaling via the IL-7 receptor (IL-7R) maintains Glut1 expression, glucose uptake, and mitochondrial OXPHOS in naïve CD4+ T cells. Upon activation, signaling via CD28 activates the PI3K/Akt/mTOR pathway to induce glycolysis in activated T cells, whereas TCR-driven ERK/MAPK signaling initiates glutaminolysis to support T cell proliferation. In addition, glycolysis promotes the translation of cytokines in activated T cells. In metabolically restrictive environments, OXPHOS can promote the proliferation of activated T cells, whereas mROS can enhance antigen-specific T cell activation. In contrast to the activation of T cells, memory formation of CD8+ T cells is dependent on TRAF6 and IL-15 driven FAO and mitochondrial biogenesis; the former is, in part, supported by activation of the cellular energy sensor AMPK.
CD4+ T cells: activation
The importance of glycolysis and glutaminolysis in activation of T cells was initially reported 40–50 years ago. These earlier studies demonstrated that stimulation of lymphocytes by potent the mitogenic lectins, such as concanavalin A or phytohemagglutinin, dramatically increased the cellular uptake of glucose and glutamine (86–89). However, these substrates were only partially oxidized to generate ATP; glucose was largely metabolized to lactate, whereas glutamine was converted to glutamate, aspartate, and ammonia. Based on these observations, it was proposed that the increase in glycolysis and glutaminolysis might provide the necessary precursors for macromolecule synthesis, including DNA, RNA, proteins, and lipids, which are necessary for cellular proliferation (9, 90).
It was 40 years later that the seminal studies performed in the Thompson laboratory elucidated the molecular signals by which lymphocyte activation is connected to the anaplerotic reactions of glycolysis and glutaminolysis (91). These authors discovered that costimulation with anti-CD3/anti-CD28 antibodies promoted the switch to glycolytic metabolism necessary for their activation. Importantly, the engagement of CD28, but not CD3, was the critical step that allowed the cells to clear the metabolic checkpoint for glycolysis (Figure 4). Mechanistically, it was demonstrated that costimulation with anti-CD28 resulted in the activation of PI3K/Akt pathway, which induced Glut1 expression on the cell surface (91). Not only did this enhance the uptake of glucose by activated T cells, but also promoted a switch from oxidative to glycolytic metabolism. Furthermore, in the absence of CD28 ligation, constitutively active Akt was sufficient to elevate Glut1 expression and support the transition to aerobic glycolysis (91–93). Ligation of the inhibitory receptors PD-1 and CTLA-4, which inhibit T cell activation, prevented PI3K/Akt activation and the switch to glycolytic metabolism (91, 94, 95).
Although activation of glycolysis requires engagement of CD28, TCR ligation promotes glutaminolysis to support T cell proliferation (Figure 4). Consistent with previous observations, Carr et al. observed that stimulation of T cells with anti-CD3 promoted proliferation of T cells that showed a strong dependence on extracellular glutamine (96). In contrast, the expression of T cell activation markers was independent of extracellular glutamine concentration. Notably, anti-CD3 stimulation not only enhanced the uptake of glutamine by activated T cells, but also induced the expression of various enzymes necessary for its entry into the TCA cycle as α-ketoglutarate. As discussed in the preceding sections, the metabolism of glutamine to α-ketoglutarate provides a means of replenishing TCA cycle intermediates that are diverted for macromolecule synthesis (Figure 1). This commitment of T cells to glutaminolysis is mediated by activation of the ERK/MAPK pathways by anti-CD3. Thus, the proliferative response of T cells seems to be more reliant of glutamine, whereas their full activation seems to be dependent of availability of glucose.
In order to understand how T cell activation reprograms cellular metabolism, Wang et al. utilized a combination of mass-spectrum-based metabolomic and metabolic tracer studies to identify the transcriptional factor Myc as a critical regulator of glycolysis and glutaminolysis (97) (Table 1). These authors observed that stimulation of resting T cells with anti-CD3/anti-CD28 increased flux through the PPP, glycolysis, and glutaminolysis, while suppressing oxidation of pyruvate and fatty acids. In silico analyses revealed that HIF-1α and Myc might be the potential master regulators that reprogram cellular metabolism in activated T cells. Although both HIF-1α and Myc were rapidly induced upon T cell activation, only Myc was required for glycolysis, glutaminolysis, and T cell proliferation. In addition, Myc-driven glutaminolysis provided the α-ketoglutarate to fuel the biosynthesis of polyamines, which are essential for cellular proliferation. Despite the dramatic requirement of Myc in transcriptional reprogramming of activated T cells for glycolysis and glutaminolysis, there were no alterations in cytokine production in Myc-deficient T cells, suggesting that other transcriptional regulators control activation-induced functions of T cells (97).
The notion that activated T cells simply require glycolysis to support their proliferation was recently challenged by studies from the Pearce lab. These authors investigated the relative importance of glycolysis and mitochondrial respiration during T cell proliferation. As reported previously, T cell proliferation and survival was dependent on glycolysis when cells were cultured in medium containing glucose and glutamine (86, 91, 96, 98). However, if glucose was replaced with galactose, a sugar whose metabolism to requires mitochondrial OXPHOS for net ATP synthesis, then T cell proliferation became highly dependent on mitochondrial respiration (99). Despite this exquisite requirement of OXPHOS in the proliferation of blasting T cells, glycolysis was unexpectedly required for cytokine secretion, principally IFNγ and IL-2, by the activated T cells. The decrease in synthesis of IFNγ or IL-2 did not result from changes in the transcription of their mRNAs, but rather from a decrease in the translation of these messages. Since previous work had shown that the glycolytic enzyme GAPDH binds to the adenylate-uridylate (AU)-rich elements in the 3’ UTRs of IFN-γ and IL-2 mRNAs to regulate their translation (100), the authors postulated that the recruitment of GAPDH to the 3’ UTRs of cytokine genes might be dependent on glycolytic flux. Indeed, not only was the binding of GAPDH to the 3’ UTRs of IFN-γ and IL-2 mRNAs dependent on flux through glycolysis, but the translation of these messages was less efficient in states of low glycolysis, such as when T cells were maintained on galactose (99). These findings thus suggest that engagement of glycolysis in activated T cells releases a translation brake that normally restrains cytokine production in resting T cells.
Although these studies implicate mitochondrial respiration and OXPHOS rather than glycolysis in the proliferation and survival of blasting T cells, there are two important caveats that one needs to keep in mind when interpreting these data. First, the metabolism of galactose to UDP-glucose occurs in the Leloir pathway, which requires 2 ATP molecules (101). UDP-glucose then enters the glycolytic pathway to generate pyruvate and 2 ATP. Therefore, the catabolism of galactose to pyruvate does not generate ATP. In order to maintain ATP homeostasis, cells utilizing galactose must then oxidize pyruvate in the TCA cycle to generate ATP via the mitochondrial OXPHOS pathways. Therefore, it is not surprising that inhibition of ATP synthase by oligomycin impairs the survival of T cells maintained on galactose. Second, when oligomycin is used to inhibit ATP synthase, the proton gradient across the inner mitochondrial matrix cannot be dissipated. This causes flow of electrons across the OXPHOS pathways to slow down, resulting in decreased respiration. Since the mitochondrial OXPHOS and TCA cycle are interlinked, a decrease in the OXPHOS necessitates a reduction in the flux through the TCA cycle, which is required for the anaplerotic reactions of glutaminolysis. Thus, an alternative interpretation of Chang et al.’s data might be that inhibition of OXPHOS by oligomycin prevents glutaminolysis, which is known to be necessary for T cell proliferation. In this regard, it is not OXPHOS per se that is required for rapid proliferation of T cells, but rather the metabolic flexibility afforded by mitochondrial respiration that allows cells to engage in both cataplerosis and anaplerosis.
Finally, a recent report by Sena et al. provides another mechanism by which mitochondrial respiration can regulate T cell activation and proliferation. By disrupting the Uqcrfs gene, a critical component of complex III, the authors’ generated mice in which mROS was specifically reduced in CD4+ T cells (102). Interestingly, CD4+ T cells from these animals were impaired in antigen-specific activation and proliferation, because mROS was required for the activation of nuclear factor of activated T cells (NFAT) and the subsequent secretion of IL-2. However, under lymphopenic conditions, Uqcrfs−/− cells retained the ability to proliferate, suggesting that mitochondrial respiration is not per se required to support the bioenergetic demands of T cell proliferation in vivo.
CD8+ T cells and the generation of memory
A characteristic hallmark of a successful immune response is the generation of memory T cells. Memory T cells, which are antigen-specific, can be rapidly activated when the appropriate antigen or pathogen is re-encountered, resulting in a faster and stronger immune response. From the viewpoint of metabolism, ‘memory’ is metabolically unique because memory T cells are quiescent but positioned to respond rapidly. Pearce et al. investigated the metabolic basis for CD8+ memory T cells. Using mice lacking TNF receptor-associated factor 6 (TRAF6) in T cells, the authors demonstrated that TRAF6 was specifically required for formation of antigen-specific CD8+ memory T cells but not the primary immune response against L. monocytogenes (103). Interestingly, deficiency of TRAF6 in CD8+ memory T cells was associated with a decrease in activation of AMPK, resulting in impaired fatty acid oxidation. Metformin, an indirect activator of AMPK, restored CD8+ memory T cell formation, conferring protective immunity against infection and tumors (103). Moreover, in a follow up study, van der Windt et al. demonstrated that IL-15 stimulates mitochondrial biogenesis and increases mitochondrial respiratory capacity in CD8+ memory T cells, thereby enhancing ability of CD8+ memory T cells to survive in metabolically stressful conditions (104). Together, these findings suggest that formation of memory T cells is exquisitely dependent on fatty acid oxidation and mitochondrial oxidative metabolism, whereas the effector T cell responses require glycolysis and glutaminolysis (Figure 4 and Table 1).
In most cell types, the metabolic actions of AMPK and the mammalian target of rapamycin (mTOR) are known to be antagonistic. AMPK, which is activated during starvation by an increase in cellular AMP, functions to restore homeostasis by increasing fatty acid oxidation and inhibiting pathways of protein synthesis. In contrast, mTOR (specifically mTORC1) is responsive to nutrients and growth factors, and stimulates cellular growth by promoting protein synthesis and glycolytic metabolism. Although the metabolic importance of mTORC1 in CD8+ memory T cells remains to be explored, its inhibition by rapamycin has been shown to increase their numbers and enhance their function in both mice and non-human primates (105, 106). Finally, since memory T cells play a critical role in protective vaccination, pharmacological manipulation of their metabolic state might be a useful means of improving the clinically efficacy of vaccines.
B cells
B cells, the sole producers of antibodies, serve a critical role in establishing and maintaining humoral immunity. With regards to their activation and proliferation, there are many similarities between B and T cells. As such, their metabolism in states of quiescence, activation, and memory are hypothesized to be similar to T cells, a postulate that is, in part, supported by the few publications on B cell metabolism (107, 108). For instance, it has been demonstrated that the stimulation of B cells via the B cell receptor activates the PI3K/Akt/mTOR pathway to promote glycolytic metabolism (109), a metabolic transition that is necessary for their proliferation (Table 1). However, there are number of unique features of B cells, including antibody class switching, affinity maturation, and plasma cell formation, whose metabolic basis remain poorly understood. Thus, this particular area of B cell biology warrants further investigation, as it has the potential of providing new insights into the regulation of humoral responses and for the development of vaccines.
Concluding remarks
In this chapter, we have discussed the general importance of metabolic substrates and pathways in the regulation of immune responses, ranging from the microbicidal actions of neutrophils to the distinct cell fates adopted by macrophages and T cells. In aggregate, these studies have demonstrated that activation of immune cells by environmental signals results in dramatic reprogramming of their cellular metabolism. The major goal of this metabolic rewiring is to provide the immune cell with sufficient energy (ATP) and metabolic intermediates to perform its effector functions in host defense and tissue homeostasis. In most instances, activated immune cells achieve this goal by simultaneously engaging in cataplerosis and anaplerosis; the former removes TCA intermediates to synthesize macromolecules, whereas the latter provides a means of replenishing the TCA metabolic intermediates through glycolysis and glutaminolysis. Furthermore, this dramatic reprogramming represents a metabolic checkpoint that needs to be cleared prior to the adoption of new cell fates or performance of effector functions. As such, it suggests that interference with or augmentation of certain metabolic programs might be clinically useful in dampening pathogenic autoimmunity or chronic inflammation in a diverse group of metabolic and degenerative diseases.
Despite the rapid progress over the last decade, our understanding of immune cell metabolism is still in its infancy and merits further investigation. First, our catalogue of metabolic effector responses across the entire repertoire of immune cells is incomplete. In most cases, investigations have focused on the most abundant cell populations found in blood or bone marrow rather than on tissue-resident or recruited cells that actively participate in host defense and tissue homeostasis. Second, although many of the inducers and sensors have been identified for activation of immune cells, our understanding of how these signals are integrated into a cohesive metabolic program, which supports the cells effector functions, is lacking. Third, our knowledge of metabolic control of immunity is primarily derived from studying cultured cells after challenging them with potent stimuli, such as anti-CD3/anti-CD28 antibodies, rather than stimulating them with physiologic ligands that might activate the cells in vivo. Fourth, our present data do not take into account the observed diversity in immune cell lineages or their tissue specific functions. For instance, although LPS activated macrophages and dendritic cells switch from mitochondria-based oxidative metabolism to glycolysis, it is unclear whether similar metabolic reprogramming occurs in vivo in tissue resident macrophages and dendritic cells, and whether it is linked to their known phagocytic and processing functions. Fifth, most models of immune activation are largely based on a binary switch between mitochondria-based oxidative metabolism and glycolysis/glutaminolysis. However, it is likely that the immune cells optimize flux through various metabolic pathways in a dynamic manner to fuel their emerging bioenergetic needs. Thus, it will be important to develop novel tools and experimental paradigms to explore metabolic underpinnings of immunity across the entire spectrum of innate and adaptive immune responses.
When one considers the broader importance of metabolism in fueling cellular processes, it is not surprising that the transitions of immune cells from quiescence to activation and back to memory formation (at least for the adaptive immune cells) are highly dependent on and regulated by nutrients, metabolic intermediates, and the canonical regulators of cellular metabolism. Indeed, this paradigm of coupling a cell’s metabolism to its functions has previously been explored in skeletal muscle, heart, and tissue-specific stem cells, where metabolic switching between oxidative metabolism and glycolysis controls skeletal muscle fiber types, heart function, and hematopoietic stem cell activation (68, 69, 110, 111). The conservation of this paradigm across various tissue types and cell states suggests that a small number of metabolic nodes or switches might be the critical triggers of these metabolic adaptations. As outlined in Table 1, this is indeed the case because cell-specific inducers and sensors converge on the PI3K-Akt-mTOR pathway to meditate the switch to glycolytic metabolism in all innate and adaptive immune cells undergoing inflammatory activation. While HIF-1α is the dominant transcriptional regulator of glycolytic metabolism in innate immune cells (25), Myc transcriptionally coordinates glycolysis and glutaminolysis in activated T cells (97). Conversely, the maintenance of immune cell quiescence, anti-inflammatory alternative macrophage activation, and CD8 memory formation is highly dependent on β-oxidation of fatty acids and mitochondrial oxidative metabolism, programs that are coordinated by the energy sensor AMPK and its downstream transcriptional regulators the PPARs and PGCs (64, 70, 103). However, this transcriptional coupling of metabolism to the cells effector responses is not unique to immune cells, as previous studies have identified a similarly critical role for metabolic regulators, including HIF-1α, PPARδ, AMPK, and PGC-1α, in skeletal muscle fiber type switching, exercise tolerance, and cardiac function (66–69). These findings thus suggest that the metabolic control of immune responses is representative of a much broader paradigm in biology, in which a cell’s identity, function, and destiny are all dictated by its underlying metabolic state.
Acknowledgments
The Larry L. Hillblom Foundation Postdoctoral Fellowship in part supported KG. A.C. was supported by grants from: NIH (HL076746, DK094641, and DK081405), AHA Innovative Award (12PILT11840038) and an NIH Director’s Pioneer Award (DP1AR064158). We regret that we are unable to cite all relevant citations from our colleagues due to space limitations.
Footnotes
Authorship
The authors declare that they have no competing financial interests.
References
- 1.Taniguchi C, Emanuelli B, Kahn C. Critical nodes in signalling pathways: insights into insulin action. Nature reviews. Molecular cell biology. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 2.Levene P, Meyer G. The action of leucocytes on glucose. Journal of Biological Chemistry 1912 [Google Scholar]
- 3.Ardawi M, Newsholme E. Maximum activities of some enzymes of glycolysis, the tricarboxylic acid cycle and ketone-body and glutamine utilization pathways in lymphocytes of the rat. The Biochemical journal. 1982;208:743–8. doi: 10.1042/bj2080743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newsholme E, Crabtree B, Ardawi M. Glutamine metabolism in lymphocytes: its biochemical, physiological and clinical importance. Quarterly journal of experimental physiology (Cambridge, England) 1985;70:473–89. doi: 10.1113/expphysiol.1985.sp002935. [DOI] [PubMed] [Google Scholar]
- 5.Wellen K, Hatzivassiliou G, Sachdeva U, Bui T, Cross J, Thompson C. ATP-citrate lyase links cellular metabolism to histone acetylation. Science (New York, NY ) 2009;324:1076–80. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox C, Hammerman P, Thompson C. Fuel feeds function: energy metabolism and the T-cell response. Nature reviews. Immunology. 2005;5:844–52. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- 7.DeBerardinis R, Lum J, Hatzivassiliou G, Thompson C. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Warburg O. Origin of cancer cells. Oncologia. 1956;9:75–83. [PubMed] [Google Scholar]
- 9.Vander Heiden M, Cantley L, Thompson C. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science (New York, NY ) 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–35. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 11.Odegaard J, Chawla A. The immune system as a sensor of the metabolic state. Immunity. 2013;38:644–54. doi: 10.1016/j.immuni.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janeway C, Medzhitov R. Innate immune recognition. Annual review of immunology. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 13.Segal A. How neutrophils kill microbes. Annual review of immunology. 2005;23:197–223. doi: 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valentine W, Beck W. Biochemical studies on leucocytes. I. Phosphatase activity in health, leucocytosis, and myelocytic leucemia. The Journal of laboratory and clinical medicine. 1951;38:39–55. [PubMed] [Google Scholar]
- 15.Sbarra A, Karnovsky M. The biochemical basis of phagocytosis. I. Metabolic changes during the ingestion of particles by polymorphonuclear leukocytes. The Journal of biological chemistry. 1959;234:1355–62. [PubMed] [Google Scholar]
- 16.van Raam B, Verhoeven A, Kuijpers T. Mitochondria in neutrophil apoptosis. International journal of hematology. 2006;84:199–204. doi: 10.1532/IJH97.06131. [DOI] [PubMed] [Google Scholar]
- 17.Fossati G, Moulding D, Spiller D, Moots R, White M, Edwards S. The mitochondrial network of human neutrophils: role in chemotaxis, phagocytosis, respiratory burst activation, and commitment to apoptosis. Journal of immunology (Baltimore, Md : 1950) 2003;170:1964–72. doi: 10.4049/jimmunol.170.4.1964. [DOI] [PubMed] [Google Scholar]
- 18.Borregaard N, Herlin T. Energy metabolism of human neutrophils during phagocytosis. The Journal of clinical investigation. 1982;70:550–7. doi: 10.1172/JCI110647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weisdorf D, Craddock P, Jacob H. Granulocytes utilize different energy sources for movement and phagocytosis. Inflammation. 1982;6:245–56. doi: 10.1007/BF00916406. [DOI] [PubMed] [Google Scholar]
- 20.Boxer L, Baehner R, Davis J. The effect of 2-deoxyglucose on guinea pig polymorphonuclear leukocyte phagocytosis. Journal of cellular physiology. 1977;91:89–102. doi: 10.1002/jcp.1040910110. [DOI] [PubMed] [Google Scholar]
- 21.Furukawa S, Saito H, Inoue T, Matsuda T, Fukatsu K, Han I, Ikeda S, Hidemura A. Supplemental glutamine augments phagocytosis and reactive oxygen intermediate production by neutrophils and monocytes from postoperative patients in vitro. Nutrition (Burbank, Los Angeles County, Calif ) 2000;16:323–9. doi: 10.1016/s0899-9007(00)00228-8. [DOI] [PubMed] [Google Scholar]
- 22.Ogle C, Ogle J, Mao J, Simon J, Noel J, Li B, Alexander J. Effect of glutamine on phagocytosis and bacterial killing by normal and pediatric burn patient neutrophils. Journal of Parenteral and Enteral Nutrition. 1994:18. doi: 10.1177/0148607194018002128. [DOI] [PubMed] [Google Scholar]
- 23.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss D, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science (New York, NY ) 2004;303:1532–5. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 24.Kirchner T, Moller S, Klinger M, Solbach W, Laskay T, Behnen M. The impact of various reactive oxygen species on the formation of neutrophil extracellular traps. Mediators of inflammation. 2012;2012:849136. doi: 10.1155/2012/849136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cramer T, Yamanishi Y, Clausen B, Forster I, Pawlinski R, Mackman N, Haase V, Jaenisch R, Corr M, Nizet V, Firestein G, Gerber H, Ferrara N, Johnson R. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–57. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peyssonnaux C, Datta V, Cramer T, Doedens A, Theodorakis E, Gallo R, Hurtado-Ziola N, Nizet V, Johnson R. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. The Journal of clinical investigation. 2005;115:1806–15. doi: 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowe S, Allen L, Ridger V, Hellewell P, Whyte M. Caspase-1-deficient mice have delayed neutrophil apoptosis and a prolonged inflammatory response to lipopolysaccharide-induced acute lung injury. Journal of immunology (Baltimore, Md : 1950) 2002;169:6401–7. doi: 10.4049/jimmunol.169.11.6401. [DOI] [PubMed] [Google Scholar]
- 28.McGrath E, Marriott H, Lawrie A, Francis S, Sabroe I, Renshaw S, Dockrell D, Whyte M. TNF-related apoptosis-inducing ligand (TRAIL) regulates inflammatory neutrophil apoptosis and enhances resolution of inflammation. Journal of leukocyte biology. 2011;90:855–65. doi: 10.1189/jlb.0211062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maianski N, Geissler J, Srinivasula S, Alnemri E, Roos D, Kuijpers T. Functional characterization of mitochondria in neutrophils: a role restricted to apoptosis. Cell death and differentiation. 2004;11:143–53. doi: 10.1038/sj.cdd.4401320. [DOI] [PubMed] [Google Scholar]
- 30.Kominsky D, Campbell E, Colgan S. Metabolic shifts in immunity and inflammation. Journal of immunology (Baltimore, Md : 1950) 2010;184:4062–8. doi: 10.4049/jimmunol.0903002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sher R, Wadee A, Joffe M. The enhancement of eosinophil function by lymphocyte supernatants. Clinical and experimental immunology. 1983;51:525–34. [PMC free article] [PubMed] [Google Scholar]
- 32.Sumbayev V, Nicholas S, Streatfield C, Gibbs B. Involvement of hypoxia-inducible factor-1 HiF(1alpha) in IgE-mediated primary human basophil responses. European journal of immunology. 2009;39:3511–9. doi: 10.1002/eji.200939370. [DOI] [PubMed] [Google Scholar]
- 33.Venge P, Moberg L, Bjornsson E, Bergstrom M, Langstrom B, Hakansson L. Mechanisms of basal and cytokine-induced uptake of glucose in normal human eosinophils: relation to apoptosis. Respiratory medicine. 2003;97:1109–19. doi: 10.1016/s0954-6111(03)00143-4. [DOI] [PubMed] [Google Scholar]
- 34.Peachman K, Lyles D, Bass D. Mitochondria in eosinophils: functional role in apoptosis but not respiration. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1717–22. doi: 10.1073/pnas.98.4.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abraham SN, St John AL. Mast cell-orchestrated immunity to pathogens. Nat Rev Immunol. 2010;10:440–52. doi: 10.1038/nri2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galli SJ, Tsai M, Piliponsky AM. The development of allergic inflammation. Nature. 2008;454:445–54. doi: 10.1038/nature07204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansen T. Adenosine triphosphate levels during anaphylactic histamine release in rat mast cells in vitro. Effects of glycolytic and respiratory inhibitors. European journal of pharmacology. 1979;58:107–15. doi: 10.1016/0014-2999(79)90001-3. [DOI] [PubMed] [Google Scholar]
- 38.Johansen T. Utilization of Adenosine Triphosphate in Rat Mast Cells during and after Secretion of Histamine in Response to Compound 48/80. Acta Pharmacologica et Toxicologica. 1983:53. doi: 10.1111/j.1600-0773.1983.tb01132.x. [DOI] [PubMed] [Google Scholar]
- 39.Chakravarty N. Inhibition of Anaphylactic Histamine Release by 2-Deoxyglucose. Nature. 1962:194. doi: 10.1038/1941182a0. [DOI] [PubMed] [Google Scholar]
- 40.Chakravarty N, Sorensen H. Stimulation of glucose metabolism in rat mast cells by antigen, dextran and compound 48–80, used as histamine releasing agents. Acta physiologica Scandinavica. 1974;91:339–53. doi: 10.1111/j.1748-1716.1974.tb05689.x. [DOI] [PubMed] [Google Scholar]
- 41.Wynn T, Chawla A, Pollard J. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–55. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yona S, Kim K-W, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, Hume D, Perlman H, Malissen B, Zelzer E, Jung S. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hashimoto D, Chow A, Noizat C, Teo P, Beasley M, Leboeuf M, Becker C, See P, Price J, Lucas D, Greter M, Mortha A, Boyer S, Forsberg E, Tanaka M, van Rooijen N, García-Sastre A, Stanley E, Ginhoux F, Frenette P, Merad M. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jenkins S, Ruckerl D, Cook P, Jones L, Finkelman F, van Rooijen N, MacDonald A, Allen J. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science (New York, NY ) 2011;332:1284–8. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geissmann F, Manz M, Jung S, Sieweke M, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science (New York, NY ) 2010;327:656–61. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gordon S. Alternative activation of macrophages. Nature reviews. Immunology. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 47.Murray P, Wynn T. Protective and pathogenic functions of macrophage subsets. Nature reviews. Immunology. 2011;11:723–37. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Odegaard J, Chawla A. Alternative macrophage activation and metabolism. Annual review of pathology. 2011;6:275–97. doi: 10.1146/annurev-pathol-011110-130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kempner W. THE NATURE OF LEUKEMIC BLOOD CELLS AS DETERMINED BY THEIR METABOLISM. The Journal of clinical investigation. 1939;18:291–300. doi: 10.1172/JCI101045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newsholme P, Curi R, Gordon S, Newsholme E. Metabolism of glucose, glutamine, long-chain fatty acids and ketone bodies by murine macrophages. The Biochemical journal. 1986;239:121–5. doi: 10.1042/bj2390121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newsholme P, Newsholme E. Rates of utilization of glucose, glutamine and oleate and formation of end-products by mouse peritoneal macrophages in culture. The Biochemical journal. 1989;261:211–8. doi: 10.1042/bj2610211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oren R, Farnham A, Saito K, Milofsky E, Karnovsky M. Metabolic patterns in three types of phagocytizing cells. The Journal of cell biology. 1963;17:487–501. doi: 10.1083/jcb.17.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newsholme P, Gordon S, Newsholme EA. Rates of utilization and fates of glucose, glutamine, pyruvate, fatty acids and ketones bodies by mouse macrophages. Biochemical Journal. 1987;242:631–6. doi: 10.1042/bj2420631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barghouthi S, Everett K, Speert D. Nonopsonic phagocytosis of Pseudomonas aeruginosa requires facilitated transport of D-glucose by macrophages. Journal of immunology (Baltimore, Md : 1950) 1995;154:3420–8. [PubMed] [Google Scholar]
- 55.Wallace C, Keast D. Glutamine and macrophage function. Metabolism: clinical and experimental. 1992;41:1016–20. doi: 10.1016/0026-0495(92)90130-3. [DOI] [PubMed] [Google Scholar]
- 56.Michl J, Ohlbaum D, Silverstein S. 2-Deoxyglucose selectively inhibits Fc and complement receptor-mediated phagocytosis in mouse peritoneal macrophages II. Dissociation of the inhibitory effects of 2-deoxyglucose on phagocytosis and ATP generation. The Journal of experimental medicine. 1976;144:1484–93. doi: 10.1084/jem.144.6.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peyssonnaux C, Cejudo-Martin P, Doedens A, Zinkernagel A, Johnson R, Nizet V. Cutting edge: Essential role of hypoxia inducible factor-1alpha in development of lipopolysaccharide-induced sepsis. Journal of immunology (Baltimore, Md : 1950) 2007;178:7516–9. doi: 10.4049/jimmunol.178.12.7516. [DOI] [PubMed] [Google Scholar]
- 58.Sonoda J, Laganiere J, Mehl I, Barish G, Chong L-W, Li X, Scheffler I, Mock D, Bataille A, Robert F, Lee C-H, Giguere V, Evans R. Nuclear receptor ERR alpha and coactivator PGC-1 beta are effectors of IFN-gamma-induced host defense. Genes & development. 2007;21:1909–20. doi: 10.1101/gad.1553007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.West A, Brodsky I, Rahner C, Woo D, Erdjument-Bromage H, Tempst P, Walsh M, Choi Y, Shadel G, Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–80. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arnoult D, Carneiro L, Tattoli I, Girardin S. The role of mitochondria in cellular defense against microbial infection. Seminars in immunology. 2009;21:223–32. doi: 10.1016/j.smim.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 61.Zhou R, Yazdi A, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–5. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 62.Nakahira K, Haspel J, Rathinam V, Lee S-J, Dolinay T, Lam H, Englert J, Rabinovitch M, Cernadas M, Kim H, Fitzgerald K, Ryter S, Choi A. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nature immunology. 2011;12:222–30. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tannahill G, Curtis A, Adamik J, Palsson-McDermott E, McGettrick A, Goel G, Frezza C, Bernard N, Kelly B, Foley N, Zheng L, Gardet A, Tong Z, Jany S, Corr S, Haneklaus M, Caffrey B, Pierce K, Walmsley S, Beasley F, Cummins E, Nizet V, Whyte M, Taylor C, Lin H, Masters S, Gottlieb E, Kelly V, Clish C, Auron P, Xavier R, O'Neill L. Succinate is an inflammatory signal that induces IL-1b through HIF-1a. Nature. 2013;496:238–42. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, Wagner RA, Greaves DR, Murray PJ, Chawla A. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006;4:13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Picard M, Hepple RT, Burelle Y. Mitochondrial functional specialization in glycolytic and oxidative muscle fibers: tailoring the organelle for optimal function. AJP: Cell Physiology. 2012:302. doi: 10.1152/ajpcell.00368.2011. [DOI] [PubMed] [Google Scholar]
- 66.Mason S, Howlett R, Kim M, Olfert I, Hogan M, McNulty W, Hickey R, Wagner P, Kahn C, Giordano F, Johnson R. Loss of skeletal muscle HIF-1alpha results in altered exercise endurance. PLoS biology. 2004:2. doi: 10.1371/journal.pbio.0020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Narkar V, Downes M, Yu R, Embler E, Wang Y-X, Banayo E, Mihaylova M, Nelson M, Zou Y, Juguilon H, Kang H, Shaw R, Evans R. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–15. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin J, Wu H, Tarr P, Zhang C-Y, Wu Z, Boss O, Michael L, Puigserver P, Isotani E, Olson E, Lowell B, Bassel-Duby R, Spiegelman B. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y-X, Zhang C-L, Yu R, Cho H, Nelson M, Bayuga-Ocampo C, Ham J, Kang H, Evans R. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS biology. 2004:2. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, Chawla A. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–20. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Odegaard JI, Ricardo-Gonzalez RR, Red Eagle A, Vats D, Morel CR, Goforth MH, Subramanian V, Mukundan L, Ferrante AW, Chawla A. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab. 2008;7:496–507. doi: 10.1016/j.cmet.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kang K, Reilly SM, Karabacak V, Gangl MR, Fitzgerald K, Hatano B, Lee CH. Adipocyte-derived Th2 cytokines and myeloid PPARdelta regulate macrophage polarization and insulin sensitivity. Cell Metab. 2008;7:485–95. doi: 10.1016/j.cmet.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodriguez-Prados J-C, Traves P, Cuenca J, Rico D, Aragones J, Martin-Sanz P, Cascante M, Bosca L. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. Journal of immunology (Baltimore, Md : 1950) 2010;185:605–14. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- 74.Haschemi A, Kosma P, Gille L, Evans C, Burant C, Starkl P, Knapp B, Haas R, Schmid J, Jandl C, Amir S, Lubec G, Park J, Esterbauer H, Bilban M, Brizuela L, Pospisilik J, Otterbein L, Wagner O. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell metabolism. 2012;15:813–26. doi: 10.1016/j.cmet.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee H, Iwasaki A. Innate control of adaptive immunity: dendritic cells and beyond. Seminars in immunology. 2007;19:48–55. doi: 10.1016/j.smim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 76.Miller J, Brown B, Shay T, Gautier E, Jojic V, Cohain A, Pandey G, Leboeuf M, Elpek K, Helft J, Hashimoto D, Chow A, Price J, Greter M, Bogunovic M, Bellemare-Pelletier A, Frenette P, Randolph G, Turley S, Merad M Immunological Genome C. Deciphering the transcriptional network of the dendritic cell lineage. Nature immunology. 2012;13:888–99. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jantsch J, Chakravortty D, Turza N, Prechtel A, Buchholz B, Gerlach R, Volke M, Glasner J, Warnecke C, Wiesener M, Eckardt K-U, Steinkasserer A, Hensel M, Willam C. Hypoxia and hypoxia-inducible factor-1 alpha modulate lipopolysaccharide-induced dendritic cell activation and function. Journal of immunology (Baltimore, Md : 1950) 2008;180:4697–705. doi: 10.4049/jimmunol.180.7.4697. [DOI] [PubMed] [Google Scholar]
- 78.Krawczyk C, Holowka T, Sun J, Blagih J, Amiel E, DeBerardinis R, Cross J, Jung E, Thompson C, Jones R, Pearce E. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood. 2011;115:4742–9. doi: 10.1182/blood-2009-10-249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Everts B, Amiel E, van der Windt G, Freitas T, Chott R, Yarasheski K, Pearce E, Pearce E. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood. 2012;120:1422–31. doi: 10.1182/blood-2012-03-419747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.MacIver N, Michalek R, Rathmell J. Metabolic regulation of T lymphocytes. Annual review of immunology. 2013;31:259–83. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rathmell J, Vander Heiden M, Harris M, Frauwirth K, Thompson C. In the absence of extrinsic signals, nutrient utilization by lymphocytes is insufficient to maintain either cell size or viability. Molecular cell. 2000;6:683–92. doi: 10.1016/s1097-2765(00)00066-6. [DOI] [PubMed] [Google Scholar]
- 82.Rathmell J, Farkash E, Gao W, Thompson C. IL-7 enhances the survival and maintains the size of naive T cells. Journal of immunology (Baltimore, Md : 1950) 2001;167:6869–76. doi: 10.4049/jimmunol.167.12.6869. [DOI] [PubMed] [Google Scholar]
- 83.Jacobs S, Michalek R, Rathmell J. IL-7 is essential for homeostatic control of T cell metabolism in vivo. Journal of immunology (Baltimore, Md : 1950) 2010;184:3461–9. doi: 10.4049/jimmunol.0902593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wofford J, Wieman H, Jacobs S, Zhao Y, Rathmell J. IL-7 promotes Glut1 trafficking and glucose uptake via STAT5-mediated activation of Akt to support T-cell survival. Blood. 2008;111:2101–11. doi: 10.1182/blood-2007-06-096297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barata J, Silva A, Brandao J, Nadler L, Cardoso A, Boussiotis V. Activation of PI3K is indispensable for interleukin 7-mediated viability, proliferation, glucose use, and growth of T cell acute lymphoblastic leukemia cells. The Journal of experimental medicine. 2004;200:659–69. doi: 10.1084/jem.20040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Parenti F, Franceschini P, Forti G, Cepellini R. The effect of phytohaemagglutinin on the metabolism and gamma-globulin synthesis of human lymphocytes. Biochimica et biophysica acta. 1966;123:181–7. doi: 10.1016/0005-2787(66)90171-7. [DOI] [PubMed] [Google Scholar]
- 87.Polgar P, Foster J, Cooperband S. Glycolysis as an energy source for stimulation of lymphocytes by phytohemagglutinin. Experimental cell research. 1968;49:231–7. doi: 10.1016/0014-4827(68)90174-2. [DOI] [PubMed] [Google Scholar]
- 88.Roos D, Loos J. Changes in the carbohydrate metabolism of mitogenically stimulated human peripheral lymphocytes. II. Relative importance of glycolysis and oxidative phosphorylation on phytohaemagglutinin stimulation. Experimental cell research. 1973;77:127–35. doi: 10.1016/0014-4827(73)90561-2. [DOI] [PubMed] [Google Scholar]
- 89.Roos D, Loos J. Changes in the carbohydrate metabolism of mitogenically stimulated human peripheral lymphocytes. I. Stimulation by phytohaemagglutinin. Biochimica et biophysica acta. 1970;222:565–82. doi: 10.1016/0304-4165(70)90182-0. [DOI] [PubMed] [Google Scholar]
- 90.Hume D, Radik J, Ferber E, Weidemann M. Aerobic glycolysis and lymphocyte transformation. The Biochemical journal. 1978;174:703–9. doi: 10.1042/bj1740703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Frauwirth K, Riley J, Harris M, Parry R, Rathmell J, Plas D, Elstrom R, June C, Thompson C. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–77. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 92.Rathmell J, Elstrom R, Cinalli R, Thompson C. Activated Akt promotes increased resting T cell size, CD28-independent T cell growth, and development of autoimmunity and lymphoma. European journal of immunology. 2003;33:2223–32. doi: 10.1002/eji.200324048. [DOI] [PubMed] [Google Scholar]
- 93.Wieman H, Wofford J, Rathmell J. Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Molecular biology of the cell. 2007;18:1437–46. doi: 10.1091/mbc.E06-07-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Parry R, Chemnitz J, Frauwirth K, Lanfranco A, Braunstein I, Kobayashi S, Linsley P, Thompson C, Riley J. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Molecular and cellular biology. 2005;25:9543–53. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Francisco L, Salinas V, Brown K, Vanguri V, Freeman G, Kuchroo V, Sharpe A. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. The Journal of experimental medicine. 2009;206:3015–29. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Carr E, Kelman A, Wu G, Gopaul R, Senkevitch E, Aghvanyan A, Turay A, Frauwirth K. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. Journal of immunology (Baltimore, Md : 1950) 2010;185:1037–44. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang R, Dillon C, Shi L, Milasta S, Carter R, Finkelstein D, McCormick L, Fitzgerald P, Chi H, Munger J, Green D. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–82. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ardawi M. Glutamine and glucose metabolism in human peripheral lymphocytes. Metabolism: clinical and experimental. 1988;37:99–103. doi: 10.1016/0026-0495(88)90036-4. [DOI] [PubMed] [Google Scholar]
- 99.Chang C-H, Curtis J, Maggi L, Faubert B, Villarino A, O'Sullivan D, Huang S, van der Windt G, Blagih J, Qiu J, Weber J, Pearce E, Jones R, Pearce E. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–51. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nagy E, Rigby W. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold) The Journal of biological chemistry. 1995;270:2755–63. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- 101.Holden H, Rayment I, Thoden J. Structure and function of enzymes of the Leloir pathway for galactose metabolism. The Journal of biological chemistry. 2003;278:43885–8. doi: 10.1074/jbc.R300025200. [DOI] [PubMed] [Google Scholar]
- 102.Sena L, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman D, Wang C-R, Schumacker P, Licht J, Perlman H, Bryce P, Chandel N. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38:225–36. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pearce E, Walsh M, Cejas P, Harms G, Shen H, Wang L-S, Jones R, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–7. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.van der Windt G, Everts B, Chang C-H, Curtis J, Freitas T, Amiel E, Pearce E, Pearce E. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Araki K, Turner A, Shaffer V, Gangappa S, Keller S, Bachmann M, Larsen C, Ahmed R. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–12. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rao R, Li Q, Odunsi K, Shrikant P. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Doughty C, Bleiman B, Wagner D, Dufort F, Mataraza J, Roberts M, Chiles T. Antigen receptor-mediated changes in glucose metabolism in B lymphocytes: role of phosphatidylinositol 3-kinase signaling in the glycolytic control of growth. Blood. 2006;107:4458–65. doi: 10.1182/blood-2005-12-4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dufort F, Bleiman B, Gumina M, Blair D, Wagner D, Roberts M, Abu-Amer Y, Chiles T. Cutting edge: IL-4-mediated protection of primary B lymphocytes from apoptosis via Stat6-dependent regulation of glycolytic metabolism. Journal of immunology (Baltimore, Md : 1950) 2007;179:4953–7. doi: 10.4049/jimmunol.179.8.4953. [DOI] [PubMed] [Google Scholar]
- 109.Limon J, Fruman D. Akt and mTOR in B Cell Activation and Differentiation. Frontiers in immunology. 2012;3:228. doi: 10.3389/fimmu.2012.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cheng L, Ding G, Qin Q, Huang Y, Lewis W, He N, Evans R, Schneider M, Brako F, Xiao Y, Chen Y, Yang Q. Cardiomyocyte-restricted peroxisome proliferator-activated receptor-delta deletion perturbs myocardial fatty acid oxidation and leads to cardiomyopathy. Nature medicine. 2004;10:1245–50. doi: 10.1038/nm1116. [DOI] [PubMed] [Google Scholar]
- 111.Gan B, Hu J, Jiang S, Liu Y, Sahin E, Zhuang L, Fletcher-Sananikone E, Colla S, Wang Y, Chin L, Depinho R. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–4. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]