Gong et al. show in multiple cell types that a mitochondrial-associate peptide, humanin, activates chaperone-mediated autophagy (CMA) by stabilizing the interaction between HSP90 and CMA substrates, thereby protecting cells from stressor-induced cell death.
Abstract
Chaperone-mediated autophagy (CMA) serves as quality control during stress conditions through selective degradation of cytosolic proteins in lysosomes. Humanin (HN) is a mitochondria-associated peptide that offers cytoprotective, cardioprotective, and neuroprotective effects in vivo and in vitro. In this study, we demonstrate that HN directly activates CMA by increasing substrate binding and translocation into lysosomes. The potent HN analogue HNG protects from stressor-induced cell death in fibroblasts, cardiomyoblasts, neuronal cells, and primary cardiomyocytes. The protective effects are lost in CMA-deficient cells, suggesting that they are mediated through the activation of CMA. We identified that a fraction of endogenous HN is present at the cytosolic side of the lysosomal membrane, where it interacts with heat shock protein 90 (HSP90) and stabilizes binding of this chaperone to CMA substrates as they bind to the membrane. Inhibition of HSP90 blocks the effect of HNG on substrate translocation and abolishes the cytoprotective effects. Our study provides a novel mechanism by which HN exerts its cardioprotective and neuroprotective effects.
Introduction
Chaperone-mediated autophagy (CMA) is an autophagic pathway that allows selective degradation of soluble proteins in lysosomes (Kaushik et al., 2011), thereby contributing to the cellular quality control and maintenance of cellular energy balance. CMA starts with the recognition of substrate proteins containing a pentapeptide motif by the cytosolic heat shock cognate chaperone of 70 kD (hsc70). The substrate–chaperone complex is targeted to a lysosomal receptor protein, the lysosome-associated membrane protein type 2A (LAMP-2A), inducing the organization of single-span LAMP-2A into a multimeric translocation complex (Bandyopadhyay et al., 2008, 2010). Heat shock protein 90 (hsp90) at the cytosolic side of the lysosomal membrane enhances substrate binding, and at the luminal side, it confers stability to LAMP-2A while transitioning from a monomeric to a multimeric form (Bandyopadhyay et al., 2008, 2010). Upon formation of the translocation complex, the substrates are delivered into the lysosome with the assistance of a luminal chaperone (lys-hsc70). Lysosomal levels of LAMP-2A are rate limiting for CMA and are controlled in large extent by changes in the degradation of LAMP-2A at the lysosomal membrane (Cuervo and Dice, 2000b; Cuervo et al., 2003).
CMA is induced during conditions of stress such as nutritional deprivation, oxidative stress (Bandyopadhyay et al., 2008, 2010), hypoxia (Ferreira et al., 2013; Hubbi et al., 2013), and genotoxic (Park et al., 2015) and lipotoxic stress (Rodriguez-Navarro et al., 2012). Indeed, oxidative stress is one of the well-characterized stressors that activate CMA. CMA restores cellular homeostasis through efficient removal of oxidized proteins (Kiffin et al., 2004), whereas dysfunction of CMA causes the accumulation of damaged and misfolded proteins. Decline of CMA activity with age could contribute to the pathogenesis of age-related diseases such as neurodegeneration and metabolic disease (Zhang and Cuervo, 2008; Orenstein et al., 2013; Schneider et al., 2015). The intracellular mechanisms that contribute to the regulation of CMA activity have just started to be elucidated. Signaling through the mTOR–Akt–PHLPP axis modulates CMA directly at the lysosomal membrane (Arias et al., 2015), whereas the retinoic acid receptor α acts as an endogenous inhibitor of CMA from the nucleus (Anguiano et al., 2013). Considering the variety of stimuli that induce CMA, it is anticipated that multiple signaling pathways and intermediate molecules may contribute to CMA regulation.
Humanin (HN) is a 24-amino-acid biologically active peptide that was originally identified from surviving neurons in patients with Alzheimer’s disease (AD; Hashimoto et al., 2001). Six additional small HN-like peptides with cytoprotective and metabolic functions have been recently reported (Cobb et al., 2016). HN has been shown to be involved in multiple biological processes, including apoptosis, cell survival, lipid flux, and inflammation, playing a protective role in diseases such as AD, cardiovascular disease, stroke, myocardial infarction, diabetes, and cancer (Gong et al., 2014, 2015). HN and analogues have been shown to protect cells against a variety of stressors. HN, and one of the analogues with Ser14 amino acid conversion into glycine termed HNG, protect against cell death elicited by serum deprivation in PC12 cells (Kariya et al., 2002). HNG also protects neurons from oxygen-glucose deprivation, hypoxia-induced cell death, and cerebral infarction in vitro and in vivo (Xu et al., 2010). We showed that HNG offers cardioprotection under conditions of ischemia-reperfusion (I-R) in mice (Muzumdar et al., 2010) and mitigates oxidative stress in cardiomyoblasts in culture (Klein et al., 2013). Stressors such as I-R, mitochondria toxicity, or serum deprivation increase reactive oxygen species (ROS) and thereby induce significant oxidative damage; activation of CMA under these conditions contributes to effective removal of damaged cellular components and restores cellular homeostasis (Kiffin et al., 2004). Interestingly, HSP90 and translation elongation factor 1α (EF1α), two important regulators of CMA (Bandyopadhyay et al., 2008, 2010), have been identified as interacting proteins of HN in a yeast two-hybrid study (Maximov et al., 2006). Therefore, we designed a series of experiments to examine whether activation of CMA is involved in the protection offered by HN and analogues under situations of stress.
Results
HNG protects cells from oxidative stress–induced cell death
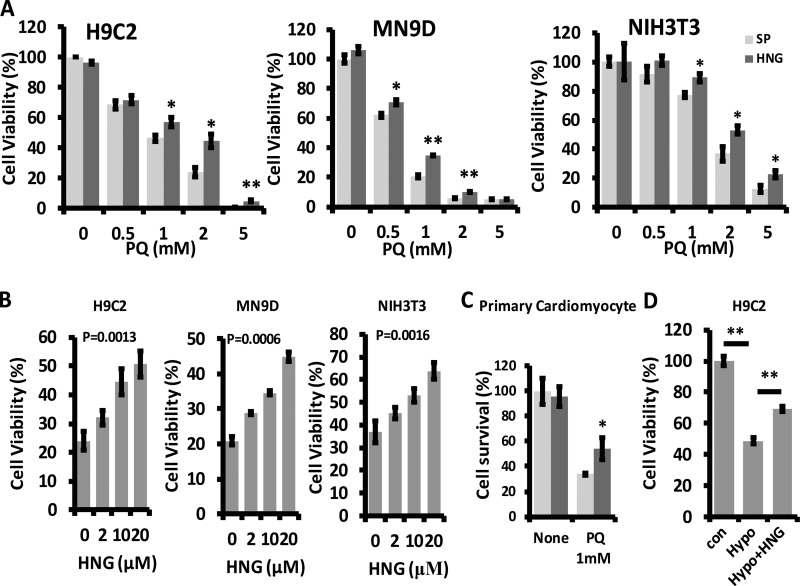
To test whether HNG treatment protects cultured cells against stressors, we preincubated H9C2 (cardiomyoblast), MN9D (dopaminergic neuronal cell), and NIH3T3 (fibroblast) cell lines with 10 µM of HNG and then challenged them with increasing doses (0.5, 1, 2, and 5 mM) of paraquat (PQ), a known oxidative stressor. We found that HNG protects against PQ-induced cell death (Fig. 1 A). At a concentration of PQ that induced cell death in 60–80% of the cells (2 mM in H9C2 and NIH3T3 and 1 mM in MN9D cells), we demonstrated that there is a significant dose-dependent response to HNG in protection against PQ-induced cell death (Fig. 1 B). Notably, our data also showed that NHG protects primary cardiomyocytes against 1 mM of PQ-induced death (Fig. 1 C). Similarly, HNG attenuated hypoxia-induced cell death in H9C2 cells (Fig. 1 D). Consistent with other research (Kariya et al., 2002), we found that HNG protects against serum deprivation–induced cell death in MN9D cells (Fig. S1 A). Furthermore, HNG protects MN9D cells against other stressor-induced cell death, including rotenone (Fig. S1 B) and H2O2 (Fig. S1 C).
Figure 1.
HNG protects cells from oxidative stress–induced cell death. (A) Effect of 10 µM HNG on 0.5, 1, 2, and 5 mM PQ-induced cell death in H9C2, MN9D, and NIH3T3 cells. (B) Dose-dependent effects of HNG on single dose of PQ-induced cell death. (C) Effect of 10 µM HNG on 1 mM PQ-induced cell death in primary cardiomyocytes. (D) Effect of 10 µM HNG on hypoxia-induced cell death in H9C2 cells. Differences with untreated samples were significant for *, P < 0.05; **, P < 0.01. Error bars show SEM.
HNG stimulates autophagy
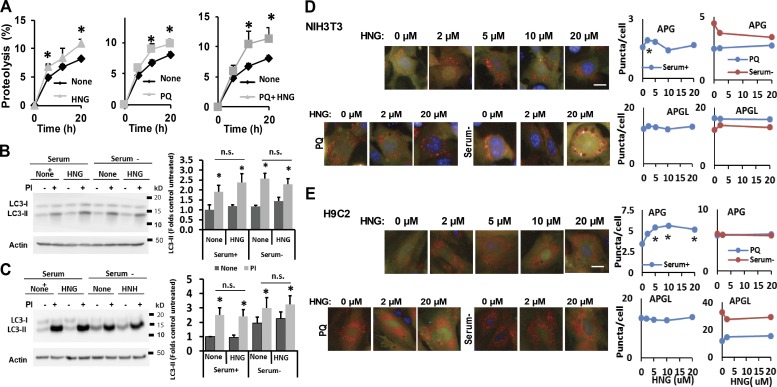
To determine whether part of the observed protective effect in response to stressors could be mediated through up-regulation of autophagy, we first investigated possible changes in total lysosomal protein degradation in presence of HNG. We found that the rate of degradation of long-lived proteins, typical autophagy substrates, was significantly increased in cells maintained under basal conditions supplemented with 20 µM HNG (Fig. 2 A). A similar increase in protein degradation in response to HNG was observed in cells subjected to mild oxidative stress by exposure to PQ (Fig. 2 A; note that the increase in protein degradation attained upon supplementation of PQ-treated cells with HN was significantly higher than the one observed with PQ only). These results support the ability of HNG to enhance both basal and inducible autophagy.
Figure 2.
HNG stimulates lysosomal protein degradation but not macroautophagy. (A) Long-lived protein degradation in NIH3T3 cells treated with HNG, PQ alone, and combination of PQ and HNG. n = 6. (B and C) Effect of HNG on LC3-II turnover in NIH3T3 (B) and H9C2 (C) cells treated or untreated with protease inhibitors (PIs). Left: Representative immunoblots. Right: Quantifications of changes in LC3-II normalized for actin. n = 3–5. (D and E) Effect of HNG on autophagic flux measured in NIH3T3 (D) and H9C2 (E) transduced with a lentivirus expressing the tandem reporter mCherry-GFP-LC3 and cultured the presence of the indicated concentrations of HNG alone (none), with PQ (10 mM), or in the absence of serum. Left: Representative images. Insets show higher-magnification images. Right: Quantification of changes in the mean number of autophagosomes (APGs; yellow puncta) or autophagolysosomes (APGLs; red puncta only) per cell section quantified with high-content microscopy in n = 1,200 cells per condition. Bars, 10 µm. Values are means + SEM. Please note that SEM in D and E are not visible because they were <0.1% of the total value. Differences with untreated cells are significant for *, P < 0.05.
Because different autophagic pathways coexist in most mammalian cells, we next investigated whether the HNG-induced increase in protein degradation was through activation of macroautophagy, the most studied form of autophagy. Immunoblotting for light chain 3 (LC3) protein, used to monitor macroautophagy (Kabeya et al., 2000), revealed no significant differences in the steady-state levels of LC3-II (the lipid-conjugate form of the protein found in autophagosomes) in mouse fibroblasts exposed or not to HNG and maintained either under basal conditions or in serum deprivation media to induce macroautophagy (Fig. 2 B). Measurement of the degradation rates of LC3-II (Tanida et al., 2005) by comparing its levels in fibroblasts treated or untreated with lysosomal protease inhibitors demonstrated no changes in autophagic flux upon HNG treatment (Fig. 2 B). Similar results in LC3 flux were also observed in cardiomyoblasts exposed to HNG (Fig. 2 C). We confirmed these data using a tandem fluorescent LC3 construct (mCherry-GFP-LC3; Kimura et al., 2007) that allowed measurement of autophagosome content (as puncta positive for both fluorophores) and autophagosome maturation into autolysosomes (as puncta positive only for mCherry as GFP fluorescence is quenched upon autophagosome–lysosome fusion because of the acid lysosomal pH environment). In agreement with the biochemical data, addition of increasing concentrations of HNG did not have a significant effect in the number of autophagosomes or flux through the autophagic system in mouse fibroblasts maintained under basal conditions (serum+) or upon induction of autophagy by nutrient deprivation (serum−) or mild oxidative stress (PQ; Fig. 2 D). In the case of cardiomyoblasts in culture, we only noticed a gradual HNG-dependent increase in the content of autophagosomes under basal conditions, but the fact that it was not associated with an increased number of autolysosomes suggests that the overall autophagic flux was not enhanced in the presence of HNG (Fig. 2 E). HNG did not significantly change the number of autophagosomes or autolysosomes in H9C2 cells exposed to serum deprivation or oxidative stress (PQ; Fig. 2 E). We next knocked down the rodent homologue of HN (rHN) in NIH3T3 cells using two different siRNAs and selected the #2 siRNA (that resulted in >70% of knockdown of rHN; Fig. S2 A) to transfect NIH3T3 cells stably expressing the mCherry-GFP–LC3-II tandem reporter. Reducing rHN levels did not affect macroautophagic activity under normal serum deprivation conditions or in response to increasing doses of PQ (Fig. S2, B–D). In addition, our results also showed that HNG-induced lysosomal proteolysis is not sensitive to 3-methyl adenine (3-MA), a well-characterized inhibitor of macroautophagy (Fig. S2, E and F). In light of these findings, we concluded that most of the increase in degradation of long-lived proteins observed in the presence of HNG could not be attributed to changes in macroautophagy activity.
HNG induces CMA
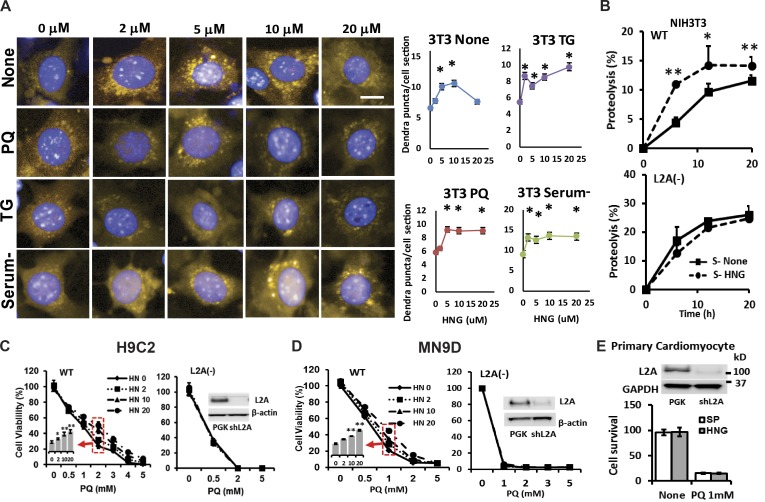
We next investigated the possible effect of HNG on CMA, the other autophagic pathway that is activated in response to cellular stressors. Activation of CMA can be monitored in cultured cells expressing a KFERQ-dendra fluorescent reporter as a change in the fluorescence pattern from diffuse to punctate when this fluorescent CMA substrate is delivered to lysosomes via CMA (Koga et al., 2011). Quantification of the number of fluorescent puncta per cell is a reliable measurement of CMA activity (Koga et al., 2011). Under basal conditions, we found that addition of HNG to fibroblasts expressing the CMA reporter resulted in a dose-dependent increase in the number of fluorescent puncta per cell at doses ≤10 µM HNG (Fig. 3 A). Higher concentrations of HNG lost part of the stimulatory effect on basal CMA. In contrast, in cells exposed to stressors such as PQ (to induce oxidative stress), thapsigargin (to induce ER stress), or serum removal, addition of HNG lead to a significant increase in CMA in fibroblasts, which was sustained even at higher HNG concentrations (Fig. 3 A). Addition of HNG to cardiomyoblasts maintained under basal or induced conditions resulted in a pattern of CMA activation similar to that observed in fibroblasts (Fig. S3, A and B).
Figure 3.
HNG induces CMA, and induction of CMA contributes to the cytoprotective effects of HNG. (A) NIH3T3 cells stably transduced with lentivirus carrying a KFERQ-dendra reporter used to monitor CMA activity were incubated with indicated concentrations of HNG in the absence or presence of PQ, thapsigargin (TG), or serum deprivation (serum−). Left: Representative images. Bar, 10 µm. Right: Quantification of changes in the mean number of puncta per cell section quantified with high-content microscopy in n = 1,500 cells per condition. Differences are significant for *, P < 0.001. (B) Long-lived protein degradation in NIH3T3 cells WT (top) and knocked down for LAMP-2A (L2A−; bottom) supplemented or not supplemented with 10 µM HNG. n = 4. (C and D) Effects of different doses of HNG on cell viability in response to PQ on WT or L2A− H9C2 (C) and MN9D cells (D). (E) Effects of HNG on cell viability in response to PQ on L2A− primary cardiomyocytes. Differences with untreated cells were significant for *, P < 0.05; **, P < 0.001. Error bars show SEM.
Induction of CMA contributes to the cytoprotective effects of HNG
Measurement of long-lived protein degradation in control cells or cells unable to perform CMA (knocked down for the lysosomal receptor for this pathway, LAMP-2A) confirmed that the stimulatory effect of HNG in protein degradation was no longer evident when HNG was added to CMA-incompetent cells (Fig. 3 B; note that the higher degradation rates in L2A− cells were previously shown to be a result of the compensatory up-regulation of macroautophagy in these cells; Massey et al., 2006). These findings support that CMA is the form of autophagy responsible for the observed HNG-dependent increase in protein degradation. To further demonstrate whether the protective effects of HNG on stress-induced cell death are mediated through activation of CMA, we treated both WT and L2A− H9C2 cells with 2, 10, and 20 µM of HNG in the presence of increasing doses of PQ. We found that the cytoprotective effects of HNG upon challenge with PQ (Fig. 3 C) and hypoxia-induced cell death (Fig. S4, A and B) are completely lost in L2A− H9C2 cells. We confirmed that the cytoprotective effects of HNG are also lost in L2A− MN9D cells (Fig. 3 D) and in primary cardiomyocytes knocked down for L2A (Fig. 3 E) in the presence of PQ.
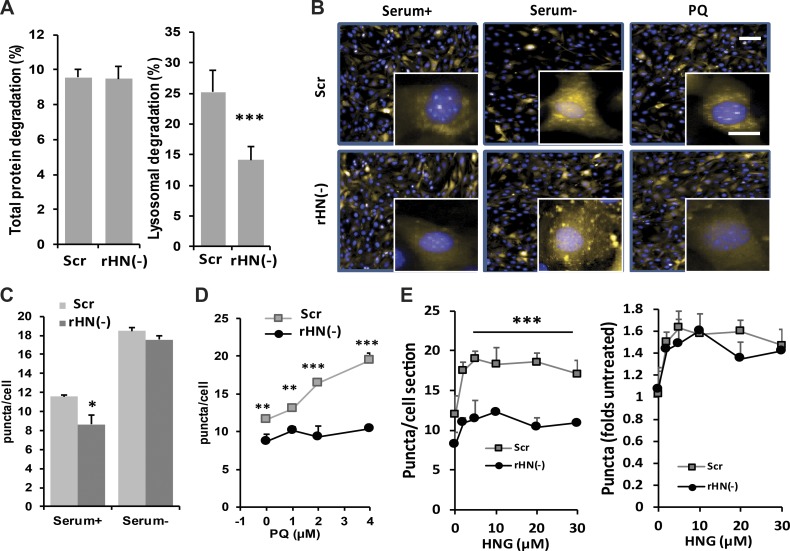
Knockdown of endogenous rHN induces defect of CMA
To investigate the role of HN in CMA, we used siRNA to knock down endogenous rHN in NIH3T3 cells (Fig. S2 A). Loss of rHN (rHN−) did not affect total protein degradation but reduced lysosome-dependent protein degradation (Fig. 4 A). Because the rHN− cells preserved normal rates of macroautophagy (Fig. S2), we speculated that CMA may be responsible for the fraction of lysosomal degradation loss in absence of rHN. CMA activity was also examined in rHN− NIH3T3 cells expressing the KFERQ-PS-dendra CMA reporter under normal conditions, serum deprivation, and mild oxidative stress (PQ). We found a significant decrease in basal and PQ-induced CMA activity in cells deficient for rHN (Fig. 4, B–E). Interestingly, rHN− cells were still able to activate CMA in response to serum deprivation (Fig. 4 C), suggesting that although HN potentiates nutrient-induced CMA, it is not necessary for this response. In contrast, HN is essential for CMA activation during oxidative stress (Fig. 4 D). In addition, we observed that rHN− cells were still capable of activating CMA in response to exogenous supplementation of HNG (Fig. 4 E; note that CMA activity in untreated control and rHN− cells has been given an arbitrary value of 1 to be able to compare the magnitude of their response to exogenous HNG [right]). These data support that endogenous and exogenously administered HNG act through similar mechanisms in their activation of CMA and that HNG supplementation is a feasible way to induce CMA in conditions with reduced endogenous HN.
Figure 4.
Effect of rHN knockdown on CMA. (A) Analysis of total protein degradation (left) and lysosomal protein degradation (right) in NIH3T3 cells control or knocked down of endogenous rHN using siRNA rHN− cells. (B–E) Analysis of CMA activity in control and rHN− NIH3T3 cells stably expressing the KFERQ-PS-dendra CMA reporter maintained in serum-supplemented (+) or serum-depleted (−) media or in the presence of 5 mM PQ. (B) Representative images. Inserts show higher magnification. Bars: (main images) 50 µm; (insets) 10 µm. Nuclei are stained with DAPI. (C) Quantification of the number of puncta per cell in basal conditions and in response to serum removal. (D) Dose-dependence effect of the same cells exposed to increasing concentrations of PQ. (E) Dose-dependent effect of adding increasing concentrations of HNG to the same cells. Number of puncta per cell (left) and increase in number of puncta normalized to untreated conditions (right). All values are presented as means + SEM and come from the quantification of nine different fields (∼3,500 cells total per condition) in triplicate wells. Differences between rHN− and control were significant for *, P < 0.05; **, P < 0.01; ***, P < 0.001.
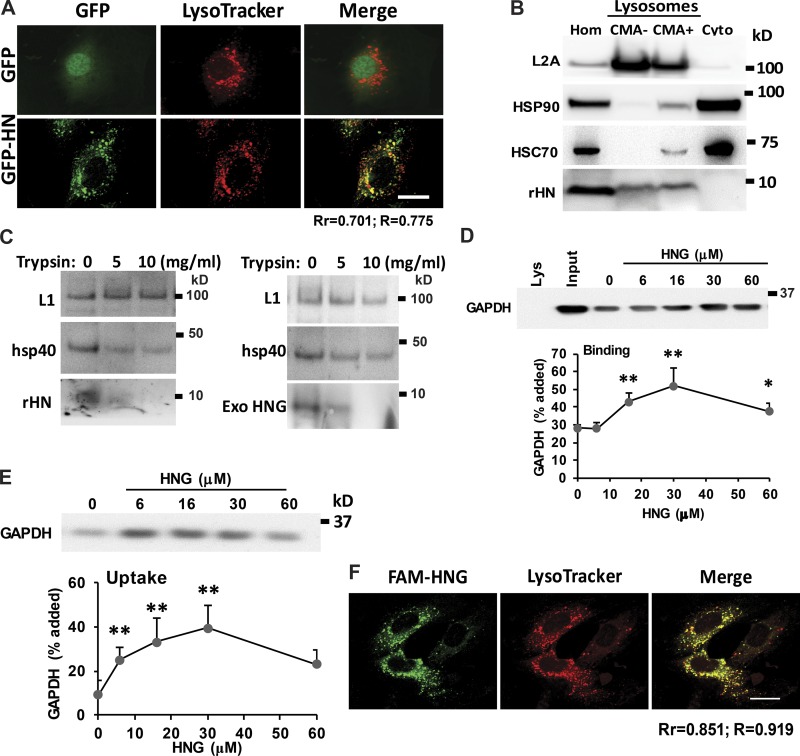
HN localizes at the lysosomal surface
To study the mechanism by which HN activates CMA, we established a stable H9C2 cell line with overexpression of GFP-HN fusion protein using lentivirus and found that overexpression of HN protects H9C2 cells from PQ-induced cell death (Fig. S4 C). We demonstrated that GFP-HN fusion protein colocalizes with acid compartments (LysoTracker labeled; Fig. 5 A), indicating possible association of HN with endosomal/lysosomal compartments. The cell fractionation studies further demonstrated that a part of cellular HN can be detected in both CMA+ (high CMA activity) and CMA− (low CMA activity) lysosomal fractions (Fig. 5 B). To further investigate the topology of the lysosome-associated HN, we treated intact lysosomes with increasing concentrations of trypsin. The susceptibility of the lysosome-associated HN to trypsin supports its location in the cytosolic side of the lysosomal membrane, where it is accessible to trypsin (Fig. 5 C). In fact, HN shows even higher sensitivity than hsp40, a well-characterized chaperone known to associate to the surface of lysosomes. We confirmed that trypsin was not permeating through the lysosomal membrane because levels of LAMP-1, detected using an antibody against the luminal part of this protein, remained unchanged even in the presence of the highest concentration of trypsin (Fig. 5 C). Both exogenously added HNG and lysosome-associated endogenous HN showed similar cytosolic topology based on its sensitivity to trypsin (Fig. 5 C).
Figure 5.
HN localizes to lysosomal surface and increases CMA substrate uptake into the lysosomes. (A) Colocalization of HN and lysosomes in H9C2 cells. (B) Immunoblot for rHN, HSP90, and L2A in homogenate (Hom), cytosol (Cyto), and lysosomes with low (CMA−) and high (CMA+) CMA activity isolated from rat liver. (C) Immunoblot for the indicated proteins of rat liver lysosomes untreated (left) or preincubated with HNG (right) and incubated with the indicated concentrations of trypsin for 10 min at 25°C. Immunoblots for rHN (left) or HNG (right). LAMP-1 (L1) and hsp40 are shown as examples of luminal and surface-associated proteins, respectively. (D and E) Intact rat liver lysosomes were preincubated (E) or not (D) with protease inhibitors or were incubated with a fixed concentration of GAPDH in the presence of increasing concentrations of HNG. At the end of the incubation, lysosomes were recovered by centrifugation and subjected to immunoblotting for GAPDH. Representative immunoblots and quantifications of the binding (D) and uptake (E) calculated from the amount of GAPDH added to the incubation media bound to lysosomes without protease inhibitors (binding) or the difference in association between lysosomes preincubated or not with protease inhibitors (uptake). (F) Colocalization of exogenous added FAM-labeled HNG with lysosomes in H9C2 cells. Bars, 10 µm. Colocalization was analyzed in both Pearson’s correlation coefficient (Rr) and Manders’s overlap coefficient (R). Values are means and SEM of three to five different experiments. Differences with untreated control samples were significant for *, P < 0.05; **, P < 0.01.
HNG increases lysosome substrate binding and uptake in vitro
To further understand how HN increases CMA activity, we directly studied the effect of HN in substrate binding and uptake steps in a previously established cell-free system using isolated lysosomes and purified GAPDH, a well-known CMA substrate (Cuervo et al., 1997). We incubated GAPDH with intact lysosomes previously treated or untreated with protease inhibitors in the presence of increasing concentrations of HNG. At the end of the incubation, the amount of GAPDH associated to lysosomes in which protein degradation had not been blocked was used to estimate binding of the CMA substrate to the lysosomal membrane because all the GAPDH translocated into the lysosomes was rapidly degraded. Uptake was calculated by subtracting the amount of protein associated with lysosomes in the absence of protease inhibitors from the amount of protein in the presence of protease inhibitors (measures protein bound to the lysosomal membrane as well as that in the lysosomal compartment). We found that HNG supplementation resulted in a marked increase in CMA substrate binding and translocation into the lysosomes (Fig. 5, D and E; and Fig. S4 D). We observed a similar stimulatory effect of HNG using a second in vitro system that recapitulates binding, uptake, and degradation in the same assay (Kaushik and Cuervo, 2009). In this case, lysosomes were incubated in the presence of increasing concentrations of HNG with a cocktail of radiolabeled cytosolic proteins enriched in lysosomal substrates. As these proteins translocate across the lysosomal membrane, they are degraded, and CMA can be determined as the conversion of radiolabeled proteins into free radiolabeled amino acids (Fig. S4 E). To investigate whether exogenously added HNG can reach lysosomes, we added fluorescein (FAM)-labeled HNG into the media of H9C2 cells for 30 min and fixed the cells for confocal microscopy. We found that the exogenously added HNG partially colocalizes with LysoTracker (Fig. 5 F), suggesting that, in addition to the well-characterized binding of HNG to plasma membrane receptors, a fraction of the circulating HNG can be internalized and reach lysosomes. Overall, our findings support that HNG activates CMA directly in the lysosomal compartment by accelerating binding and translocation of cytosolic substrates across the lysosomal membrane.
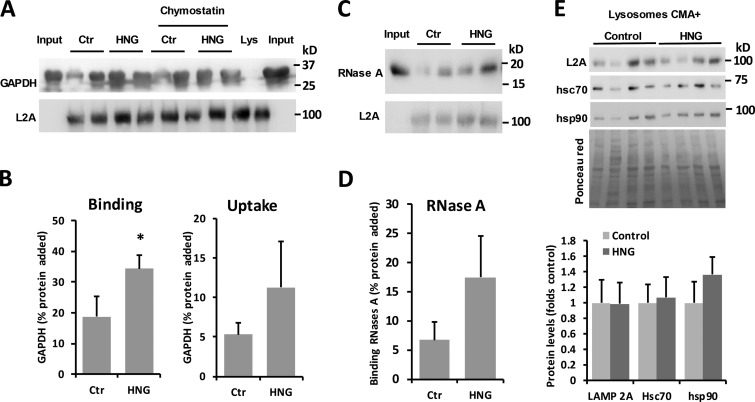
HNG increases CMA activity in vivo
To further study whether HN is an activator of CMA that also increases substrate binding and uptake in vivo, we gave rats a single dose of HNG (2 mg/ml) or scrambled peptide (SP) intraperitoneally and harvested tissues 6 h after injection. After treatment, rat liver lysosomes were isolated, and their CMA activity was measured upon incubation with the well-characterized CMA substrate GAPDH in the presence or absence of lysosomal protease inhibitors. Binding of the CMA substrate to the lysosomal membrane is estimated by the amount of substrate associated to lysosomes without protease inhibitor, and the uptake is calculated by subtracting the amount of protein associated with lysosomes in the absence of protease inhibitors from the amount of protein in the presence of protease inhibitors. Our data demonstrate that HNG treatment significantly increases the association of GAPDH with lysosomes and that this higher binding (P < 0.05) leads to a trend of increased lysosomal internalization of the substrate (P = 0.2) as expected (Fig. 6, A and B). Analysis of the lysosomal association for a second CMA substrate, RNase A, confirmed a similar trend for higher association of this protein in the lysosomes from HNG-treated rats (Fig. 6, C and D). To start investigating the possible mechanisms behind enhanced CMA upon in vivo HNG treatment, we compared the steady-state levels of several of CMA effectors at lysosomes. Levels of LAMP-2A, HSC70, and HSP90 were comparable in CMA-active (CMA+) lysosomes isolated from HNG-treated and -untreated rats (Fig. 6 E). Total hepatic levels of these three proteins (Fig. S5 A) and their abundance in homogenates and lysosome-enriched fractions from the kidney, heart, and brain (Fig. S5, B–D) also remained unchanged upon HNG treatment.
Figure 6.
Effect of in vivo treatment with HNG on hepatic CMA. (A–D) CMA activity in the liver from rats treated with 2 mg/kg of HNG or SP for 6 h. Immunoblots for LAMP-2A (A and C) and GAPDH (A) or RNase A (C) in lysosomes isolated from livers of untreated (control; Ctr) or HNG-treated rats and incubated with the two different substrates. Quantification of the binding (B and D) and uptake (B) of the substrates in n = 4 (B) or n = 2 (D) animals. (E) Immunoblots for the indicated proteins in CMA-active lysosomes isolated from the same animals as in A. Bottom shows quantification of the levels of the indicated proteins in HNG-treated animals relative to those in control untreated. n = 4. All values are means + SEM. Differences with untreated animals were significant for *, P < 0.05.
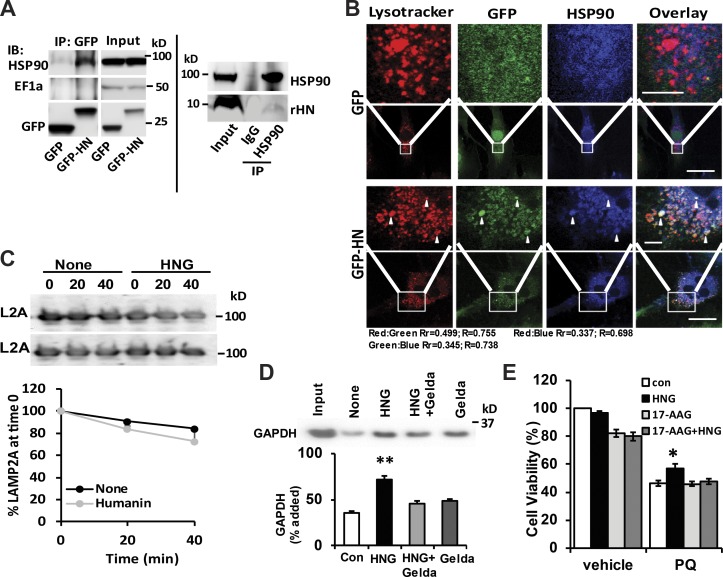
HSP90 contributes to the cytoprotective effects of HN
Previous research has shown interaction of HN with HSP90 and EF1α (Maximov et al., 2006), proteins also present at the lysosomal membrane and known to participate in CMA (Bandyopadhyay et al., 2008, 2010). Coimmunoprecipitation (coIP) experiments in cells expressing GFP-HN show that HSP90 but not EF1α, a known CMA regulator (Bandyopadhyay et al., 2010), can be precipitated with HN using the GFP antibody (Fig. 7 A, left). We confirmed that this interaction also occurs between endogenous rHN and HSP90 using similar coIP approaches in mouse liver (Fig. 7 A, right). Moreover, we show by immunofluorescence using confocal microscopy that part of the colocalization of HN with HSP90 occurs in lysosomes (Fig. 7 B). HSP90 associates both with the cytosolic and the luminal side of the lysosomal membrane (Bandyopadhyay et al., 2008). Intralysosomal HSP90 contributes to maintaining LAMP-2A stability at the lysosomal membrane when this single-span membrane protein transitions from the monomer to the multimeric form to organize into the translocation complex (Bandyopadhyay et al., 2008). Next, we evaluated whether activation of CMA by HNG results from a stabilizing effect on LAMP-2A in lysosomes. Analysis of the levels of LAMP-2A in intact lysosomes incubated in the presence or absence of HNG did not reveal a stabilizing effect for HNG on LAMP-2A (Fig. 7 C), suggesting that the functional interaction of HNG with HSP90 was likely on the cytosolic side of the lysosomal membrane, directly participating in the process of substrate binding and translocation across the lysosomal membrane (Agarraberes and Dice, 2001). Indeed, in freshly isolated lysosomes, we found that the increased substrate binding and uptake induced by HNG is completely blocked by the addition of an inhibitor of HSP90, geldanamycin (Figs. 7 D and S4 F). Furthermore, we showed that 17-AAG, another potent inhibitor of HSP90, abolishes the protective effect of HNG on PQ-induced cell death in H9C2 cells (Fig. 7 E), suggesting that at least part of the beneficial effect of HNG depends on active hsp90 activity on CMA. These findings demonstrate that activation of CMA by HN is for the most part, mediated by HSP90.
Figure 7.
HSP90 contributes to the cytoprotective effects of HN. (A) CoIP of HN and HSP90 using GFP antibody in GFP-only or GFP-HN–overexpressing H9C2 cells (left) and HSP90 antibody in mouse liver lysate (right). IP, immunoprecipitation. (B) Colocalization of LysoTracker, HSP90, and HN in H9C2 cells. Bars: (main images) 10 µm; (magnifications) 2 µm. Arrowheads show colocalization. (C) L2A stability in isolated rat liver lysosomes incubated in an isotonic buffer in the presence or absence of HNG. Top: Representative immunoblots (IBs). Bottom: Quantification. n = 2. (D) Immunoblot and quantification of substrate uptake in the lysosomes isolated from rat liver with the presence of geldanamycin (Gelda), HNG, and HNG + geldamycin. (E) H9C2 cell viability at the absence and presence of HNG and 17-AAG with or without 1 mM PQ. Colocalization was analyzed in both Pearson’s correlation coefficient (Rr) and Manders’s Overlap coefficient (R). Differences with control were significant for *, P < 0.05; **, P < 0.01. Error bars show SEM.
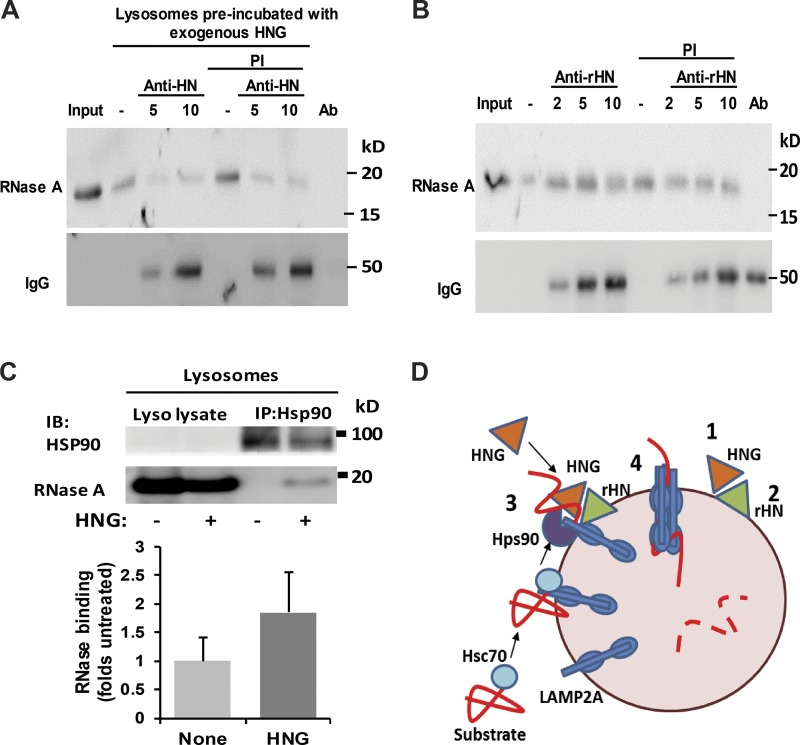
Lysosome-associated HN participates in CMA substrate binding and uptake
Based on these findings, the convergence of HNG and hsp90 in lysosomes seems required for HNG-dependent activation of CMA. To further elucidate the contribution of exogenously added HNG and endogenous rHN to CMA activation, we next used antibodies to block these proteins at the lysosomal membrane and analyzed their impact on CMA. Rat liver lysosomes were first incubated with HNG and recovered by centrifugation to eliminate unbound HNG from the media. The HNG-decorated lysosomes were next incubated with anti-HN antibody, and after washing out the unbound antibody, they were presented with the CMA substrate RNase A. As shown in Fig. 8 A, blockage of HNG and rHN with HN antibody markedly reduced binding and uptake of the CMA substrate. In contrast, when antibodies against the endogenous rHN were used, binding of RNase A to lysosomes remained unperturbed, but uptake was markedly reduced (Fig. 8 B). We confirmed that both antibodies bind to the lysosomal membrane as they can be detected in the immunoblot membranes using secondary antibodies. These results support a direct role on CMA, albeit with some differences on the steps affected, for lysosome-bound HNG and rHN.
Figure 8.
Effect of lysosome-associated HN on CMA. (A) Rat liver lysosomes were preincubated with HNG to allow for its binding to the lysosomal membrane and, after centrifugation to eliminate unbound HNG, were incubated with increasing concentrations of the antibody against HN. After the treatment, lysosomes were incubated in the presence or absence of protease inhibitor (PI) with RNase A and subjected to immunoblot against RNase A. (B) Rat liver lysosomes were preincubated with increasing concentrations of the antibody against rHN and then with RNase A in the presence or absence of protease inhibitor. Samples were processed as in A. Secondary antibody was used to confirm binding of antibodies against HN (A) or rHN (B) to the lysosomal membrane. Ab: antibody only. (C) Rat liver lysosomes incubated with RNases A in presence (+) or absence (−) of HNG were subjected to immunoprecipitation (IP) for HSP90 and immunoblot (IB) for HSP90 and RNase A. Top: Representative immunoblot. Bottom: Quantification of the amount of RNase coimmunoprecipitated with HNG. Values are expressed as folds of untreated lysosomes. Error bars show SEM. (D) Proposed model of the stimulatory effect of HN on CMA: Cytosolic HNG enhances CMA by stabilizing the interaction of HSP90 with the CMA substrates while LAMP2A transitions into the multimeric complex carrying along the substrate proteins. A fraction of the endogenous HN (rHN in our study) associated to the lysosomal membrane is required to complete substrate translocation.
HNG stabilizes HSP90 binding to CMA substrate
We next used similar in vitro assays with purified lysosomes to determine the step or steps targeted by HNG and HSP90 in their coordinated effect on CMA. Rat liver lysosomes incubated with RNase A in the presence or absence of HNG were subjected to coIP using HSP90 antibody, and the presence of RNase A in the pulled down fraction was analyzed by immunoblotting. We found that addition of HNG increases the interaction of HSP90 and the CMA substrate RNase A at the lysosome (Fig. 8 C), suggesting that HNG increases substrate binding and uptake through the enhancement of HSP90 and substrate interaction.
Discussion
Our study demonstrates that HN increases long-lived protein degradation through stimulation of both basal and inducible autophagy. The effects of HN on intracellular protein degradation are not mediated through macroautophagy, the most studied form of autophagy, as shown in our studies in both NIH3T3 and H9C2 cell lines through assessment of LC3 turnover, a marker of macroautophagy. We demonstrate instead that the effect of HN on intracellular lysosomal proteolysis is through increase in another form of autophagy, CMA, and that activation of this pathway is required, in large part, for the protective effect of HN against a variety of stressors including PQ, H2O2, rotenone, starvation, and hypoxia. We also found that (a) exogenously added HNG binds to the surface of lysosomes and is required for the HNG-stimulatory effect on CMA; (b) a fraction of endogenous rHN associates to lysosomes, and it is necessary to complete substrate uptake as part of the activation of CMA by exogenous HNG; (c) the stimulatory effect of HNG on CMA requires functional HSP90 in the lysosomes; and (d) HNG stabilizes the interaction of HSP90 with the CMA substrates at the lysosomal membrane. Collectively, our findings support a possible dual effect of HNG on substrate binding and translocation into lysosomes by stabilizing the substrate–HSP90–LAMP-2A complex during the organization of LAMP-2A into the CMA translocation complex (Fig. 8 D). Lastly, in support of its possible future therapeutic suitability, we also demonstrate that this stimulatory effect of HNG on CMA can also be attained in vivo through immunoprecipitation injection.
CMA is closely linked to oxidative stress (Kiffin et al., 2004), but the mechanisms that mediate its activation in these conditions remain unknown. HN has been shown to decrease infarct size in mouse models of stroke (Xu et al., 2008) and myocardial I-R (Muzumdar et al., 2010) in vivo, clinical scenarios with significant hypoxia and oxidative stress. In addition, HN decreases oxidative stress and preserves mitochondrial integrity in H9C2 cells by reducing ROS levels and activating antioxidants through nonreceptor-mediated tyrosine kinases Abl and Arg (Klein et al., 2013). We show in this study that HN treatment increases both basal CMA and CMA induced by different stressors including starvation and oxidative stress. The effects of HN on cytoprotection and enhancement of long-lived protein turnover are completely blocked in CMA-incompetent cells, confirming their dependence on CMA. Indeed, activation of CMA could be a potential mechanism mediating the cytoprotective effects in vivo under conditions of hypoxia and/or oxidative stress.
Increased oxidative stress and dysfunction of CMA have been implicated in the pathophysiology of Parkinson’s disease (PD; Cuervo and Wong, 2014). In our in vitro system using differentiated MN9D cells, a dopaminergic neuronal cell line, we show that HN protects cells from death induced by multiple oxidative stressors including PQ, H2O2, and serum removal as well as rotenone, a toxin closely linked to PD. Neuroprotective effects in the studies described in our research are consistent with protective effects offered by HN on neuronal cell damage induced by AD-associated insults and stroke (Kariya et al., 2002; Sponne et al., 2004; Tajima et al., 2005; Xu et al., 2006). Our study demonstrating the protective effects of HN on dopaminergic neuronal cells highlights possible therapeutic potential for HN in PD through targeting CMA.
Interestingly, we have found a previously unknown association of HN with lysosomes. Although we cannot fully discard that part of the stimulatory effect of HNG on CMA could occur through signaling pathways or other integrated cellular mechanisms, the fact that addition of HNG to isolated lysosomes is sufficient to reproduce its activating effect on CMA suggests that lysosome-associated HN is primarily responsible for CMA activation in intact cells. This is also confirmed by our data showing that exogenously added HNG colocalizes with lysosomes and that blocking of HNG on the lysosomal membrane attenuates the effects of HNG on CMA substrate binding and uptake. Differences on the effect on CMA between endogenous and exogenously added HNG could be caused by differences on their respective accessibility to the CMA substrate. Blockage of exogenously added HNG diminishes both substrate binding and uptake, whereas blockage of lysosome-associated endogenous rHN only disrupts substrate translocation. We propose that, once as part of the HSP90–substrate complex, HN no longer modulates substrate binding but is required to facilitate its internalization into lysosomes. Future studies are needed to understand the basis for HNG effect on substrate internalization.
Contrary to other CMA regulators that only associate with the subpopulation of lysosomes that participate in CMA, we have found endogenous HN bound at the surface of both lysosomes active and inactive for CMA. These two groups of lysosomes have been shown to functionally interconvert, whereby CMA-inactive lysosomes can become active in conditions of maximal CMA activation. In fact, CMA-inactive lysosomes already contain LAMP-2A, an essential component for CMA, and become active by recruiting some of the missing CMA components (Cuervo et al., 1997) such as HSC70 and HSP90. The fact that, as in the case of LAMP-2A, HN is present in the membrane of both groups of lysosomes but that activation of CMA only occurs in the CMA+ group supports our idea that HSP90 and HN act in partnership in the activation of CMA.
In summary, we identify HN, a mitochondrial-derived peptide recognized to play a critical protective role in age-related diseases, to be an endogenous activator of CMA. Through interacting with HSP90, HN increases CMA via enhanced HSP90 and substrate interaction and therefore increases substrate binding and uptake into the lysosomes as well as protects cells from stressor-induced cell death. We have previously shown that the levels of HN decline with age in both humans and rodents (Muzumdar et al., 2009). It is interesting to speculate that the previously described decrease in CMA activity with age (Cuervo and Dice, 2000a) could also be in part related to the decrease in HN levels. Our study not only provides novel mechanisms for the protective effect of HN in cardiovascular disease, but it also highlights a potential protective role for HN in other age-related diseases, such as cerebrovascular diseases, as well as other neurodegenerative diseases, namely AD and PD, where oxidative stress and the accumulation of abnormal proteins contribute to the pathogenesis, and defective CMA activity has been previously reported (Cuervo and Wong, 2014).
Materials and methods
Cells and reagents
NIH3T3 mouse fibroblasts, H9C2 cardiomyoblasts, and MN9D dopaminergic neuronal cells were from ATCC. NIH3T3 clones stably RNA-interfered for LAMP-2A (L2A−) were generated as described previously (Massey et al., 2006; Singh et al., 2009). Stable knockdowns of LAMP-2A in H9C2 and MN9D cells were obtained using lentiviral vector–mediated stable RNAi directed specifically against the LAMP-2A exon as described previously (Massey et al., 2006; Singh et al., 2009). Knockdown of endogenous rHN was obtained using siRNA against rodent HN rHN: 1, 5′-CAUCAUCUCUAGCGUAAUAtt-3′; and 2, 5′-CAGUGAAAUUGACCUUUCAtt-3′ (provided by K.-W. Lee, Florida Hospital, Orlando, FL; lowercased letters were not part of the original sequences but added artificially). Geldanamycin (Tocris) and 17-AAG (Sigma-Aldrich), both HSP90 inhibitors, were added to the cells 1 h before HNG treatment in cell viability experiments and to lysosomes 10 min before HNG and the substrate. SP and HNG were synthesized by Genscript.
Cell culture
NIH3T3 mouse fibroblasts were obtained from ATCC (Massey et al., 2006). Fibroblasts were cultured in complete DMEM (Sigma-Aldrich), 10% heat-inactivated newborn calf serum (HyClone), and 1% penicillin/streptomycin/fungizone (Invitrogen) and were maintained at 37°C at 5% CO2. H9C2 cardiomyoblasts (ATCC) and MN9D dopaminergic neuronal cell lines (a gift from D. Casper, Montefiore Medical Center, Bronx, NY) were cultured in high-glucose DMEM (Thermo Fisher Scientific) in the presence of 10% FBS (Bio-West) and 1% penicillin/streptomycin (Thermo Fisher Scientific). Serum removal was performed by thoroughly washing the cells with HBSS (Invitrogen) and placing them in serum-free complete medium. H2O2 or PQ (Sigma-Aldrich) were added to the culture medium to induce oxidative stress. All the cell lines were tested for mycoplasma contamination using DNA staining protocol with Hoechst 33258 dye.
Animals
Adult (4 mo) Wistar male rats (Charles River Laboratories) were starved for 48 h before organelle isolation by removing food but maintaining water ad libitum. Rats were injected with 2 mg/ml of HNG or SP immunoprecipitation in the morning and tissues, including liver, kidney, heart, and brain, were collected 6 h later for analysis or lysosome isolation. All animal studies were approved by the Institutional Animal Care and Use Committee at the Albert Einstein College of Medicine.
Antibodies
The sources of antibodies used in this study are LAMP-2A (Invitrogen); GAPDH (Abcam); RNase A (Rockland Immunochemicals); HSP90, β-actin, LC3B, and EF-1α (Cell Signaling Technology); hsc70 (Novus Biological); rHN (generated with residues 20–35 of the rHN peptide and no cross-reaction with HN and HNG; GeneTex); and mTOR CST and GFP antibody (Thermo Fisher Scientific). Antibody against HN or HNG was generated by Genscript using residues 7–24 of the HN peptide.
Plasmids and reagents
Flag-tagged HN was synthesized and constructed into the PGK vector and then packed into lentivirus at the Gene Therapy Core Facility of the Albert Einstein College of Medicine. Sources of chemicals were as previously described (Massey et al., 2006; Singh et al., 2009).
Hypoxia
Hypoxic chamber was used on cultured cells to imitate in vivo cardiac ischemia. In brief, WT or L2A− H9C2 cells treated with SP (FRGGETRARAMPLIDLSPLCLLKV) or HNG were incubated in hypoxic chamber filled with 94.5% N2, 5% CO2, and 0.5% O2 for 2 h, and cell viability was measured by cell countingas indicated in the Cells and reagents section.
Cell viability assay
Resazurin was used to measure cell proliferation, viability, and cytotoxicity after induction of oxidative stress and treatment with HNG. In brief, one tenth of the resazurin dye was added to the growth media, and cells were incubated for additional 1–4 h in the incubator at 37°C. Absorbance was read using a spectrophotometer set to 570 nm.
Macroautophagy analysis
Macroautophagy activity was estimated in two ways.
Immunoblot for LC3-II
Cells untreated or treated with the mixture of proteinase inhibitors ammonium chloride (20 mM; American Bioanalytical) and leupeptin (100 µM; Fisher Bioreagents) were immunoblotted for LC3-II for the indicated times. The level of LC3-II (lipidated LC3) was quantified and normalized by actin levels. LC3 flux was quantified as the increase in LC3-II/actin values in response to treatment with the lysosomal proteolysis inhibitors.
Direct fluorescence
Cells transduced with lentivirus carrying the mCherry-GFP-LC3 tandem construct were analyzed by direct fluorescence. The tandem reporter consisting of recombinant mCherry and GFP fused to the LC3 protein (mCherry-GFP-LC3) was generated as described previously (Kimura et al., 2007). Cells were plated on coverslips or glass-bottomed 96-well plates, and fluorescence was read in both channels. Puncta positive for both fluorophores corresponded with autophagosomes, whereas those only positive for the red fluorophore corresponded with autolysosomes. Autophagic flux was determined as the conversion of autophagosomes (yellow) to autolysosomes (red-only puncta) as described previously (Kimura et al., 2007).
Primary cardiomyocyte isolation
Adult primary cardiomyocytes were isolated from male Sprague-Dawley rats (body weight 200–300 g) as described previously (Liu et al., 2016).
Intracellular protein degradation
Measure of intracellular degradation was performed by metabolic labeling (with [3H] leucine [2 µCi/ml]; PerkinElmer] for 48 h at 37°C) and pulse-chase experiments as described previously (Auteri et al., 1983). After labeling, cells were extensively washed and maintained in medium with an excess of unlabeled leucine. Aliquots of the medium taken at different times were precipitated in trichloroacetic acid, and proteolysis was measured as the amount of acid-perceptible radioactivity transformed in acid-soluble radioactivity at each time. Lysosomal-dependent degradation was inhibited using 20 mM NH4Cl and 100 µM leupeptin (N/L).
Isolation of subcellular fractions
Lysosomes with high activity for CMA were isolated from rat liver by centrifugation of a light mitochondrial–lysosomal fraction in a discontinuous metrizamide density gradient by the modified method described previously (Cuervo et al., 1997). Preparations with >10% broken lysosomes, measured by β-hexosaminidase latency, were discarded. Lysosomal matrices and membranes were isolated after hypotonic shock and centrifugation. Cytosolic fractions were obtained by centrifugation for 1 h at 100,000 g of the supernatant after separation of the mitochondria–lysosome–enriched fraction.
Cell imaging
H9C2 cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin and infected with GFP only or GFP-HN lentivirus for 48 h. Cells were then plated and grown on coverslips for immunofluorescence. Lysosomes were labeled with LysoTracker (Thermo Fisher Scientific). Cells were washed with PBS, fixed with 10% formalin for 20 min at room temperature, and permeabilized with 1% FBS and 0.1% Triton X-100 in PBS for 30 min at room temperature under mild agitation. Thereafter, cells were incubated with anti-HSP90 (1:50; Cell Signaling Technology) overnight at 4°C. After extensive washing, cells were exposed to Alexa Fluor 308–conjugated goat anti–rabbit (1:500; Thermo Fisher Scientific) for 1 h at room temperature under continuous agitation and protected from light. Cells were embedded and mounted in mounting medium (Thermo Fisher Scientific) and analyzed by a confocal microscope (Olympus). Colocalization analysis was done using ImageJ software (National Institutes of Health), and Intensity Correlation Analysis was presented in both Rr (Pearson’s correlation coefficient) and R (Manders’s Overlap coefficient), with 1 indicating reliable colocalization.
For high-content microscopy, cells were plated in glass-bottomed 96-well plates (PerkinElmer), fixed with 4% paraformaldehyde for 30 min, permeabilized, and blocked with 1% BSA and 0.01% Triton X-100 in PBS. Images were captured with a high-content microscope (Operetta; PerkinElmer), and quantification was performed with the manufacturer’s software in ≥1,200 cells (approximately nine fields). In all cases, focal plane thickness was set at 0.17 µm (mean cell thickness was 1 µM), and sections with maximal nucleus diameter were selected for quantification. Values are presented as the number of puncta per cell section that in our acquisition conditions represented 10–20% of the total puncta per cell. The number of particles/puncta per cell was quantified using the manufacture’s software in nonsaturated images.
Immunoblotting
Total cell lysates were prepared in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% NaDoc, 0.1% SDS, and 50 mM Tris, pH 8) containing protease and phosphatase inhibitors. Protein concentration was measured using BSA as a standard (Lowry et al., 1951). Samples were subjected to SDS-PAGE, transferred to nitrocellulose membrane, blocked with low-fat milk, and incubated with primary antibody overnight. The proteins were visualized by using peroxidase-conjugated secondary antibodies and chemiluminescent reagent (PerkinElmer) in a LAS-3000 Imaging System (Fujifilm). Densitometric quantification was performed on unsaturated images using ImageJ.
RNAi
Lentivirus constructs carrying shRNA against LAMP-2A were generated as described previously (Massey et al., 2008) and delivered to cultured cells upon packaging into replication-deficient lentiviral particles to generate LAMP-2A–knockdown clones. The shRNA transfer vector plasmid contained the hairpin (sense–loop–antisense) against the LAMP-2A sequence 5′-CTGCAATCTGATTGATTA-3′ corresponding with bases 1,331–1,359. The hairpin (sense–loop–antisense) for these sequences was inserted in the multicloning region of the pSuper vector (Ambion). Cultured cells were transfected by the calcium phosphate method and selected for stable transfectants by resistance to geneticin. Control cells were transduced with lentiviral particles carrying only the empty vector.
CMA measurement
CMA was measured using three different procedures.
Reporter assays
Cells transduced with a lentivirus carrying the photoswitchable KFERQ-PS-dendra2 CMA reporter (a brighter modified version of the original reporters; Koga et al., 2011) were photoswitched with a 405-nm light-emitting diode (Norlux) for 4 min with the intensity of 3.5 mA (current constant). After 16 h, cells were fixed with 4% paraformaldehyde, and images were acquired with a high-content microscope (Operetta).
Degradation of radiolabeled substrates by intact lysosomes
Lysosomes were incubated with a pool of [3H]-labeled cytosolic proteins in MOPS buffer (10 mM MOPS, pH 7.3, 0.3 M sucrose, 1 mM dithiothreitol, and 5.4 µM cysteine) for 20 min at 37°C (Patel and Cuervo, 2015). After precipitation in acid, proteolysis was calculated as the amount of acid-precipitable radioactivity (protein) transformed to acid-soluble (amino acid) during the incubation.
Uptake of CMA substrates by intact lysosomes
Direct uptake of proteins by CMA was measured using a previously reported in vitro system (Patel and Cuervo, 2015) where intact lysosomes pretreated or not with protease inhibitors were incubated in 10 mM MOPS, pH 7.2, and 0.3 M sucrose buffer, with purified proteins for 20 min at 37°C with or without HSP90 inhibitor (Geldanamycin). At the end of the incubation, lysosomes were recovered by centrifugation and subjected to immunoblot. Binding was calculated as the amount of substrate protein bound to the lysosomal membrane in the absence of protease inhibitors, and uptake was calculated by subtracting the amount of protein associated with lysosomes in the absence (protein bound to the lysosomal membrane) from the presence of protease inhibitors (protein bound to the lysosomal membrane and taken up by lysosomes). For the antibody blocking experiments, uptake of substrates was analyzed after lysosomes were preincubated with antibodies against HNG or rHN.
Intracellular protein degradation
Measure of intracellular degradation was performed by metabolic labeling for 48 h at 37°C and pulse-chase experiments as described previously (Auteri et al., 1983).
Isolation of subcellular fractions
Lysosomes with high activity for CMA were isolated from rat liver by the modified method described previously (Cuervo et al., 1997).
Statistical analyses
All numerical results are reported as means ± SEM. When comparing two groups, a two-tailed post hoc test (Tukey’s) was applied to determine individual differences between the mean. One-way or two-way ANOVA was used when one or two parameters were compared between groups. Differences were considered to be statistically significant when P ≤ 0.05.
Online supplemental material
Fig. S1 shows the cytoprotective effects of HNG against other stressors. Fig. S2 demonstrates that knockdown of rHN does not affect macroautophagy. Fig. S3 verifies the activation of CMA by exogenous HNG in H9C2 cells under basal and stress conditions. Fig. S4 shows the effects of HN on stress-induced cell survival in CMA-deficient cells and HN-overexpression cells and the effect of HNG on substrate uptake with inhibition of HSP90. Fig. S5 demonstrates that treatment of HNG in vivo does not change the protein levels of major molecules involved in CMA.
Supplementary Material
Acknowledgments
This work was supported in part by grants from Children’s Hospital of Pittsburgh of the University of Pittsburgh Medical Center Health System (to Z. Gong and R. Muzumdar), the Richard King Mellon Foundation Institute for Pediatric Research (Children’s Hospital of Pittsburgh of the University of Pittsburgh Medical Center), and by a Transatlantic Network of Excellence grant by Fondation Leducq (15CVD03 to B. Kühn and RA15CVD04 to A.M. Cuervo), National Institutes of Health R-01AG035114 and R56AG044519, a CHP Foundation grant (to R. Muzumdar), National Institutes of Health grant AG021904 (to A.M. Cuervo) and Proteostasis of Aging Core grant AG038072.
The authors declare no competing financial interests.
Author contributions: Z. Gong, I. Tasset, A.M. Cuervo, and R. Muzumdar designed the experiments, performed statistical analysis, and composed the manuscript. Z. Gong, E. Tas, and L. Cui performed cytoprotective assays, interaction experiments, colocalization experiments, in vivo experiments, and other biochemical experiments. I. Tasset, A. Diaz, J. Anguiano, and R. Kuliawat performed macroautophagy and CMA analysis, long-lived protein degradation analysis, and substrate binding and uptake assays. H. Liu and B. Kühn helped with the primary cardiomyocyte isolation.
References
- Agarraberes F.A., and Dice J.F.. 2001. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J. Cell Sci. 114:2491–2499. [DOI] [PubMed] [Google Scholar]
- Anguiano J., Garner T.P., Mahalingam M., Das B.C., Gavathiotis E., and Cuervo A.M.. 2013. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat. Chem. Biol. 9:374–382. 10.1038/nchembio.1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias E., Koga H., Diaz A., Mocholi E., Patel B., and Cuervo A.M.. 2015. Lysosomal mTORC2/PHLPP1/Akt Regulate Chaperone-Mediated Autophagy. Mol. Cell. 59:270–284. 10.1016/j.molcel.2015.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auteri J.S., Okada A., Bochaki V., and Dice J.F.. 1983. Regulation of intracellular protein degradation in IMR-90 human diploid fibroblasts. J. Cell. Physiol. 115:167–174. 10.1002/jcp.1041150210 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay U., Kaushik S., Varticovski L., and Cuervo A.M.. 2008. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol. Cell. Biol. 28:5747–5763. 10.1128/MCB.02070-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay U., Sridhar S., Kaushik S., Kiffin R., and Cuervo A.M.. 2010. Identification of regulators of chaperone-mediated autophagy. Mol. Cell. 39:535–547. 10.1016/j.molcel.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb L.J., Lee C., Xiao J., Yen K., Wong R.G., Nakamura H.K., Mehta H.H., Gao Q., Ashur C., Huffman D.M., et al. . 2016. Naturally occurring mitochondrial-derived peptides are age-dependent regulators of apoptosis, insulin sensitivity, and inflammatory markers. Aging. 8:796–809. 10.18632/aging.100943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A.M., and Dice J.F.. 2000a Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 275:31505–31513. 10.1074/jbc.M002102200 [DOI] [PubMed] [Google Scholar]
- Cuervo A.M., and Dice J.F.. 2000b Regulation of lamp2a levels in the lysosomal membrane. Traffic. 1:570–583. 10.1034/j.1600-0854.2000.010707.x [DOI] [PubMed] [Google Scholar]
- Cuervo A.M., and Wong E.. 2014. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 24:92–104. 10.1038/cr.2013.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A.M., Dice J.F., and Knecht E.. 1997. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J. Biol. Chem. 272:5606–5615. 10.1074/jbc.272.9.5606 [DOI] [PubMed] [Google Scholar]
- Cuervo A.M., Mann L., Bonten E.J., d’Azzo A., and Dice J.F.. 2003. Cathepsin A regulates chaperone-mediated autophagy through cleavage of the lysosomal receptor. EMBO J. 22:47–59. 10.1093/emboj/cdg002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira J.V., Fôfo H., Bejarano E., Bento C.F., Ramalho J.S., Girão H., and Pereira P.. 2013. STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy. 9:1349–1366. 10.4161/auto.25190 [DOI] [PubMed] [Google Scholar]
- Gong Z., Tas E., and Muzumdar R.. 2014. Humanin and age-related diseases: a new link? Front. Endocrinol. (Lausanne). 5:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Su K., Cui L., Tas E., Zhang T., Dong H.H., Yakar S., and Muzumdar R.H.. 2015. Central effects of humanin on hepatic triglyceride secretion. Am. J. Physiol. Endocrinol. Metab. 309:E283–E292. 10.1152/ajpendo.00043.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Niikura T., Tajima H., Yasukawa T., Sudo H., Ito Y., Kita Y., Kawasumi M., Kouyama K., Doyu M., et al. . 2001. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Abeta. Proc. Natl. Acad. Sci. USA. 98:6336–6341. 10.1073/pnas.101133498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbi M.E., Hu H., Kshitiz I. Ahmed A. Levchenko, and Semenza G.L.. 2013. Chaperone-mediated autophagy targets hypoxia-inducible factor-1α (HIF-1α) for lysosomal degradation. J. Biol. Chem. 288:10703–10714. 10.1074/jbc.M112.414771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., and Yoshimori T.. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728. 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariya S., Takahashi N., Ooba N., Kawahara M., Nakayama H., and Ueno S.. 2002. Humanin inhibits cell death of serum-deprived PC12h cells. Neuroreport. 13:903–907. 10.1097/00001756-200205070-00034 [DOI] [PubMed] [Google Scholar]
- Kaushik S., and Cuervo A.M.. 2009. Methods to monitor chaperone-mediated autophagy. Methods Enzymol. 452:297–324. 10.1016/S0076-6879(08)03619-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S., Bandyopadhyay U., Sridhar S., Kiffin R., Martinez-Vicente M., Kon M., Orenstein S.J., Wong E., and Cuervo A.M.. 2011. Chaperone-mediated autophagy at a glance. J. Cell Sci. 124:495–499. 10.1242/jcs.073874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiffin R., Christian C., Knecht E., and Cuervo A.M.. 2004. Activation of chaperone-mediated autophagy during oxidative stress. Mol. Biol. Cell. 15:4829–4840. 10.1091/mbc.E04-06-0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Noda T., and Yoshimori T.. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 3:452–460. 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
- Klein L.E., Cui L., Gong Z., Su K., and Muzumdar R.. 2013. A humanin analog decreases oxidative stress and preserves mitochondrial integrity in cardiac myoblasts. Biochem. Biophys. Res. Commun. 440:197–203. 10.1016/j.bbrc.2013.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H., Martinez-Vicente M., Macian F., Verkhusha V.V., and Cuervo A.M.. 2011. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat. Commun. 2:386 10.1038/ncomms1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Qin W., Wang Z., Shao Y., Wang J., Borg T.K., Gao B.Z., and Xu M.. 2016. Disassembly of myofibrils and potential imbalanced forces on Z-discs in cultured adult cardiomyocytes. Cytoskeleton (Hoboken). 73:246–257. 10.1002/cm.21298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry O.H., Rosebrough N.J., Farr A.L., and Randall R.J.. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275. [PubMed] [Google Scholar]
- Massey A.C., Kaushik S., Sovak G., Kiffin R., and Cuervo A.M.. 2006. Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA. 103:5805–5810. 10.1073/pnas.0507436103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey A.C., Follenzi A., Kiffin R., Zhang C., and Cuervo A.M.. 2008. Early cellular changes after blockage of chaperone-mediated autophagy. Autophagy. 4:442–456. 10.4161/auto.5654 [DOI] [PubMed] [Google Scholar]
- Maximov V. V., Arman I.P., and Tarantul V.Z.. 2006. Identification of the proteins interacting with neuroprotective peptide humanin in a yeast two-hybrid system. Russ. J. Genet. 42:274–277. [PubMed] [Google Scholar]
- Muzumdar R.H., Huffman D.M., Atzmon G., Buettner C., Cobb L.J., Fishman S., Budagov T., Cui L., Einstein F.H., Poduval A., et al. . 2009. Humanin: a novel central regulator of peripheral insulin action. PLoS One. 4:e6334 10.1371/journal.pone.0006334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar R.H., Huffman D.M., Calvert J.W., Jha S., Weinberg Y., Cui L., Nemkal A., Atzmon G., Klein L., Gundewar S., et al. . 2010. Acute humanin therapy attenuates myocardial ischemia and reperfusion injury in mice. Arterioscler. Thromb. Vasc. Biol. 30:1940–1948. 10.1161/ATVBAHA.110.205997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein S.J., Kuo S.H., Tasset I., Arias E., Koga H., Fernandez-Carasa I., Cortes E., Honig L.S., Dauer W., Consiglio A., et al. . 2013. Interplay of LRRK2 with chaperone-mediated autophagy. Nat. Neurosci. 16:394–406. 10.1038/nn.3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Suh Y., and Cuervo A.M.. 2015. Regulated degradation of Chk1 by chaperone-mediated autophagy in response to DNA damage. Nat. Commun. 6:6823 10.1038/ncomms7823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel B., and Cuervo A.M.. 2015. Methods to study chaperone-mediated autophagy. Methods. 75:133–140. 10.1016/j.ymeth.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro J.A., Kaushik S., Koga H., Dall’Armi C., Shui G., Wenk M.R., Di Paolo G., and Cuervo A.M.. 2012. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA. 109:E705–E714. 10.1073/pnas.1113036109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J.L., Villarroya J., Diaz-Carretero A., Patel B., Urbanska A.M., Thi M.M., Villarroya F., Santambrogio L., and Cuervo A.M.. 2015. Loss of hepatic chaperone-mediated autophagy accelerates proteostasis failure in aging. Aging Cell. 14:249–264. 10.1111/acel.12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., and Czaja M.J.. 2009. Autophagy regulates lipid metabolism. Nature. 458:1131–1135. 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sponne I., Fifre A., Koziel V., Kriem B., Oster T., and Pillot T.. 2004. Humanin rescues cortical neurons from prion-peptide-induced apoptosis. Mol. Cell. Neurosci. 25:95–102. 10.1016/j.mcn.2003.09.017 [DOI] [PubMed] [Google Scholar]
- Tajima H., Kawasumi M., Chiba T., Yamada M., Yamashita K., Nawa M., Kita Y., Kouyama K., Aiso S., Matsuoka M., et al. . 2005. A humanin derivative, S14G-HN, prevents amyloid-beta-induced memory impairment in mice. J. Neurosci. Res. 79:714–723. 10.1002/jnr.20391 [DOI] [PubMed] [Google Scholar]
- Tanida I., Minematsu-Ikeguchi N., Ueno T., and Kominami E.. 2005. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 1:84–91. 10.4161/auto.1.2.1697 [DOI] [PubMed] [Google Scholar]
- Xu X., Chua C.C., Gao J., Hamdy R.C., and Chua B.H.. 2006. Humanin is a novel neuroprotective agent against stroke. Stroke. 37:2613–2619. 10.1161/01.STR.0000242772.94277.1f [DOI] [PubMed] [Google Scholar]
- Xu X., Chua C.C., Gao J., Chua K.W., Wang H., Hamdy R.C., and Chua B.H.. 2008. Neuroprotective effect of humanin on cerebral ischemia/reperfusion injury is mediated by a PI3K/Akt pathway. Brain Res. 1227:12–18. 10.1016/j.brainres.2008.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chua K.W., Chua C.C., Liu C.F., Hamdy R.C., and Chua B.H.. 2010. Synergistic protective effects of humanin and necrostatin-1 on hypoxia and ischemia/reperfusion injury. Brain Res. 1355:189–194. 10.1016/j.brainres.2010.07.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., and Cuervo A.M.. 2008. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat. Med. 14:959–965. 10.1038/nm.1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.