Epithelial-to-mesenchymal transition is implicated in metastasis. Viotti et al. show that the histone methyltransferase SUV420H2 favors the mesenchymal identity in pancreatic tumor cells by silencing key drivers of the epithelial state. High levels of SUV420H2 also correlate with a loss of epithelial characteristics in invasive cancer.
Abstract
Epithelial-to-mesenchymal transition is implicated in metastasis, where carcinoma cells lose sessile epithelial traits and acquire mesenchymal migratory potential. The mesenchymal state is also associated with cancer stem cells and resistance to chemotherapy. It might therefore be therapeutically beneficial to promote epithelial identity in cancer. Because large-scale cell identity shifts are often orchestrated on an epigenetic level, we screened for candidate epigenetic factors and identified the histone methyltransferase SUV420H2 (KMT5C) as favoring the mesenchymal identity in pancreatic cancer cell lines. Through its repressive mark H4K20me3, SUV420H2 silences several key drivers of the epithelial state. Its knockdown elicited mesenchymal-to-epithelial transition on a molecular and functional level, and cells displayed decreased stemness and increased drug sensitivity. An analysis of human pancreatic cancer biopsies was concordant with these findings, because high levels of SUV420H2 correlated with a loss of epithelial characteristics in progressively invasive cancer. Together, these data indicate that SUV420H2 is an upstream epigenetic regulator of epithelial/mesenchymal state control.
Introduction
The vast majority of cancers originate in epithelial tissues, yet tumors comprise a heterogeneous mix of cell populations with varying phenotypes along the epithelial-mesenchymal continuum (Tam and Weinberg, 2013). Three distinct lines of rationale imply that, in cancer, the epithelial cell state is clinically more favorable than the mesenchymal state. First, normal epithelial cells are stationary, sharing cell–cell junctions and resting on a basement membrane, whereas mesenchymal cells are motile and more likely to migrate and invade (Thiery, 2003). Accordingly, an epithelial-to-mesenchymal transition (EMT) is often thought to accompany the progression of early cancer lesions to invasive malignancies and eventually metastasis (Yang and Weinberg, 2008). Second, the mesenchymal cell fraction in tumors possesses increased “stemness,” including superior capability for self-renewal and differentiation potency, marker expression of tissue stem cells, and elevated tumor-initiating ability (Brabletz et al., 2005; Mani et al., 2008; Rhim et al., 2012; Scheel and Weinberg, 2012). Third, mesenchymal cancer cells universally exhibit lower sensitivity to anticancer drugs than their epithelial counterparts (Yauch et al., 2005; Neve et al., 2006; Witta et al., 2006; Sayan et al., 2009), and malignant cells engage in EMT to acquire drug resistance (Singh et al., 2009; Wilson et al., 2014a,b).
Although the core signaling pathways (TGFB, NOTCH, WNT, FGF, and BMP) and transcription factors (ZEB1/2, SNAIL, SLUG, TWIST1/2, E47, and FOXC1) that regulate epithelial/mesenchymal cell states have been well characterized (Thiery et al., 2009), attempts at modulating these agents to elicit a mesenchymal-to-epithelial transition (MET) in cancers have been largely unsuccessful in cancer patients (Ginnebaugh et al., 2014). More recently, efforts have focused on manipulating the epigenetic programs that likely govern epithelial/mesenchymal cell states. Although incompletely understood, different classes of histone modifiers have been implicated in these processes in various cancers: the deacetylases HDAC1/2 (Peinado et al., 2004; von Burstin et al., 2009), the demethylases KDM1A (Lim et al., 2010; Lin et al., 2010), PHF2 (Pattabiraman et al., 2016), and LOXL2 (Peinado et al., 2005) and the methyltransferases EZH2 (Cao et al., 2008), EHMT2, and SUV39H1 (Dong et al., 2013). Histone modifiers are attractive targets for prospective therapies because they contain distinct, druggable catalytic domains with some Food and Drug Administration–approved inhibitors already in the clinic and several more in clinical trials (Dawson and Kouzarides, 2012; Jones et al., 2016).
Pancreatic cancer is one of the deadliest malignancies because it is usually detected late in the course of the disease and existing treatments are typically ineffective because of intrinsic and acquired drug resistance, as well as being poorly responsive to immunotherapy (Xiong et al., 2006; Arumugam et al., 2009; Li et al., 2013; Chen and Mellman, 2017). Priming pancreatic cancers with an epithelial-inducing agent might not only decrease invasion and metastasis and limit stemness but may also increase responses to existing cancer drugs (Singh and Settleman, 2010). Indeed, histopathological changes associated with pancreatic cancer do not appear to be strictly under genetic control (Lo et al., 2012).
We devised an arrayed screen targeting 300 epigenetic factors and identified SUV420H2 (KMT5C) as an upstream orchestrator of epithelial/mesenchymal states in pancreatic cancer cells. SUV420H2 silences several drivers of MET, and repressing SUV420H2 elicits a molecular, phenotypic, and functional cell identity shift toward the epithelial condition. Analysis of human pancreatic ductal adenocarcinoma (PDAC) samples corroborated a close link between SUV420H2 expression and epithelial/mesenchymal cell states. These findings suggest that SUV420H2 should be considered a potential target to favor MET in pancreatic cancer.
Results
Genetic screen identifies SUV420H2 as a modulator of epithelial/mesenchymal cell states in pancreatic cancer
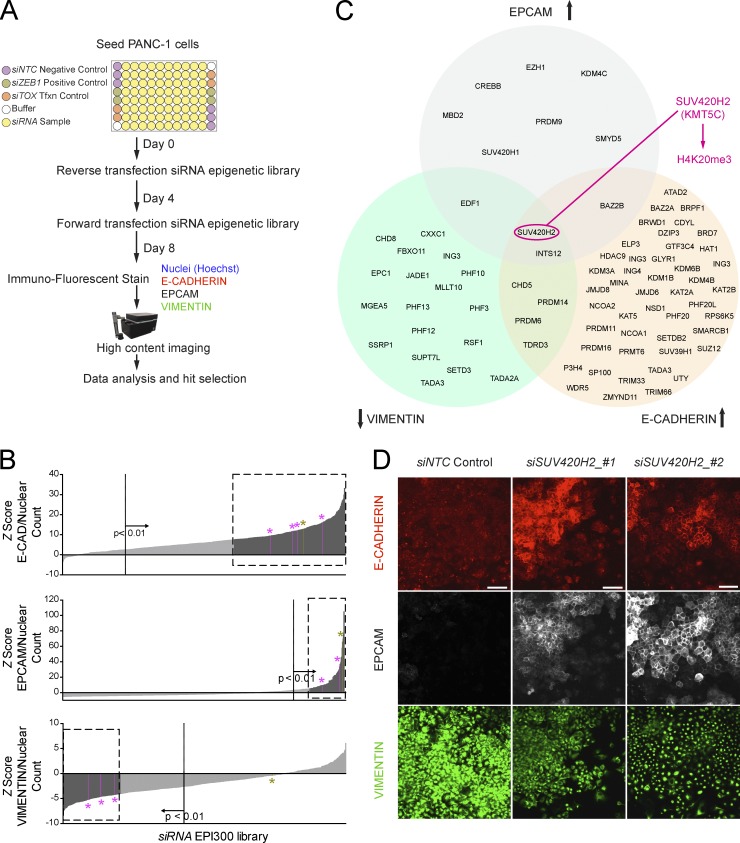
We designed an unbiased genetic screen to identify and rank epigenetic factors that modulate epithelial/mesenchymal states in pancreatic cancer (Fig. 1 A). The parental PANC-1 cell line, originally derived from the primary tumor of a patient with PDAC with invasion in the duodenal wall and peripancreatic lymph metastasis (Lieber et al., 1975), shows generally poor differentiation, high migration and invasion potential, and marker expression in line with the mesenchymal state (Deer et al., 2010; Klijn et al., 2015). Using fluorescently tagged monoclonal antibodies, we confirmed PANC-1 cells show high levels of the mesenchymal marker vimentin (VIM) and background levels of the epithelial marker E-cadherin (E-CAD) and the epithelial cell adhesion molecule (EPCAM; Fig. S1). In a course of 8 d, PANC-1 cells were subjected to two rounds of transfection by using an arrayed siRNA library targeting 300 genes involved in modulating epigenetic marks in DNA and histones (Table S1). We then subjected the cultures to immunofluorescence using antibodies raised against VIM, E-CAD, and EPCAM and used a quantitative imaging platform to seek significant changes in average fluorescent intensities per cell.
Figure 1.
Genetic screen identifies SUV420H2 as a modulator of epithelial/mesenchymal states in pancreatic cancer. (A) Diagram of arrayed siRNA screen to identify epigenetic factors modulating epithelial/mesenchymal states in pancreatic cancer. (B) Screen results for each marker depicting hit selection criteria. Only factors for which at least two of four siRNAs resulted in values in the region of interest (represented by dashed boxes) were considered for hit selection. Dotted vertical lines indicate demarcation of statistical significance (P < 0.01). Averaged results of nontargeting siRNA control (NTC) knockdown (negative control) are set as the reference (Z score = 0), olive-colored bars and asterisks indicate averaged results for ZEB1 knockdown (positive control), and pink bars and asterisks indicate values for SUV420H2 knockdown. (C) Summary of epigenetic factors that on knockdown elicit a highly significant change in marker signal, as established by selection criteria in B. (D) Original screen images for NTC and SUV420H2 knockdown depicting de novo, cell-junction localizing the E-CAD and EPCAM immunofluorescence signal, as well as the reduced VIM signal for the latter. Bars, 50 µm.
Each gene was targeted by four different siRNA sequences so we could prioritize genes whose knockdown with at least two siRNAs changed marker signal intensity in a highly significant manner (Fig. 1 B). Although knockdown of numerous genes elicited a change in a single marker and several in two markers, only two epigenetic knockdown targets (SUV420H2 and INTS12) elicited a change in all three phenotypic markers (i.e., VIM, E-CAD, and EPCAM; Fig. 1 C and Fig. S1). INTS12 is a subunit of the snRNA-processing integrator complex and recognizes posttranslational modifications of histone H3 via its PHD finger region (Hernandez, 1985) but has no known enzymatic activity. We therefore concentrated our efforts on SUV420H2 (Suppressor of variegation 4–20 homologue 2), also known as KMT5C, a well-characterized histone methyltransferase that specifically trimethylates Lys-20 of histone H4 causing transcriptional repression of associated genes (Kourmouli et al., 2004; Schotta et al., 2004, 2008). Knockdown of SUV420H2 in PANC-1 cells induced a strong de novo signal of the epithelial markers E-CAD and EPCAM, which correctly localized to cell-cell junctions and substantially diminished signal intensity of VIM (Fig. 1 D).
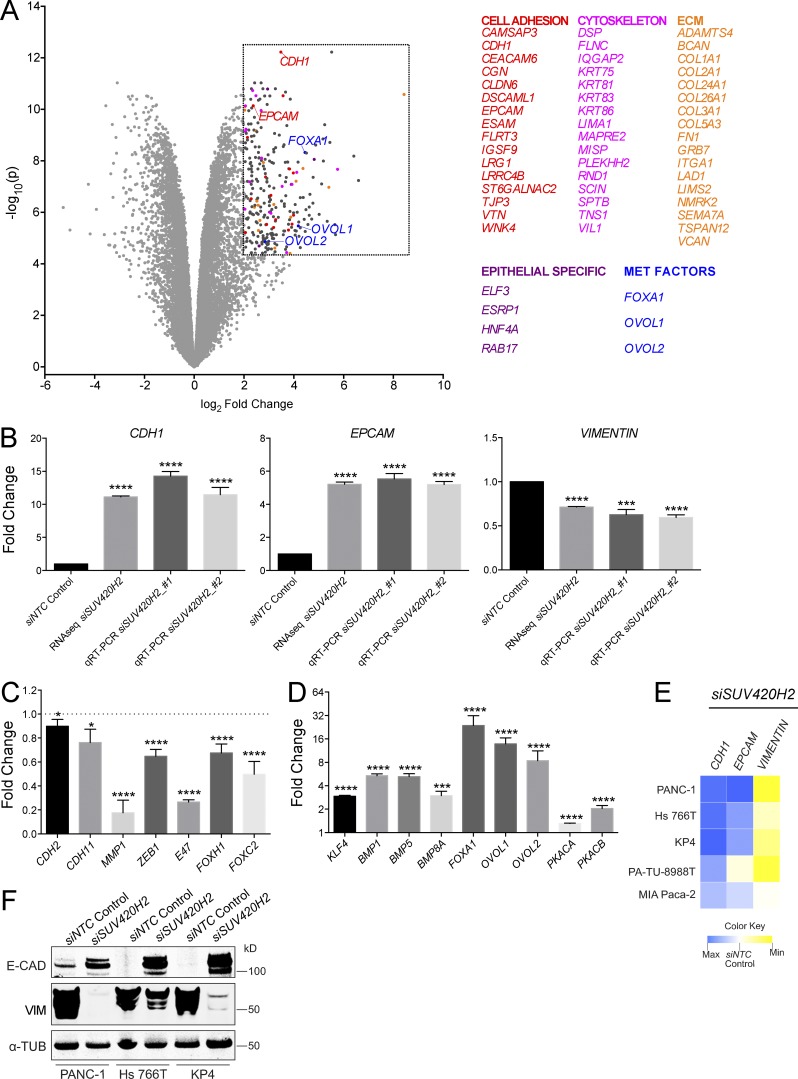
We confirmed that the SUV420H2 siRNAs used in the screen indeed diminished the expression of their intended target in PANC-1 cells on the RNA and protein level (Fig. S2, A and B). Expression profiling via RNA sequencing (RNaseq) of PANC-1 cells after SUV420H2 knockdown resulted in 266 highly and significantly up-regulated transcripts within arbitrary cutoffs: more than a fourfold change, P < 0.00005 (Table S2). Of those, a large proportion (56 of 266) encoded products that comprise or modulate cell adhesions (e.g., CDH1, EPCAM, CLDN6, and TJP3), cytoskeleton (e.g., various keratins), and extracellular matrix (e.g., various collagens, FN1, and VCAN; Fig. 2 A). Four more genes are specifically expressed in epithelial tissues (ELP3, ESRP1, HNF4A, and RAB17), and another three genes (FOXA1, OVOL1, and OVOL2) encode transcription factors previously implicated in MET (Song et al., 2010; Roca et al., 2013). Unbiased gene set enrichment analyses by Camera (Wu and Smyth, 2012) yielded “Hallmark epithelial-mesenchymal transition” as a significantly altered set among the Broad’s MsigDB Hallmark gene sets (Liberzon et al., 2015; Table S3), as well as “Reactome cell–cell junction organization” as one of the top altered signatures from MsigDB’s C2 curated signature collection (Liberzon et al., 2011; Table S4).
Figure 2.
Pancreatic cancer cells undergo molecular MET on knockdown of SUV420H2. (A) Volcano plot of RNaseq results from SUV420H2 knockdown in PANC-1 cells, compared with siNTC control. Listed genes fall within the arbitrary region of interest (dashed box), defined by a fold change >4 and P < 0.00005. (B) Expression analysis of CDH1 (gene encoding E-CAD), EPCAM, and VIM on SUV420H2 knockdown in PANC-1 cells by RNaseq and qRT-PCR. Bar graphs indicate mean ± SD. For RNaseq, n = 3 biological replicates and differences assessed by using voom+limma (see Materials and methods). For qRT-PCR, n = 3 biological replicates each averaged from three technical replicates; differences were assessed by Student’s t test compared with siNTC control. ***, P < 0.001; ****, P < 0.0001. (C) Expression analysis from RNaseq for mesenchymal markers and EMT-inducing factors in PANC-1 cells with SUV420H2 knockdown. Data are represented as normalized to the siNTC control knockdown. Bar graphs indicate mean ± SD. n = 3 biological replicates and differences assessed by using voom+limma. *, P < 0.05; ****, P < 0.0001. (D) Expression analysis from RNaseq for MET-inducing factors in PANC-1 cells with SUV420H2 knockdown. Data are represented as normalized to the siNTC control knockdown. Bar graphs indicate mean ± SD. n = 3 biological replicates and differences assessed by using voom+limma. ***, P < 0.001; ****, P < 0.0001. (E) qRT-PCR expression analysis for CHD1, EPCAM, and VIM on SUV420H2 knockdown in pancreatic cancer cell lines of mesenchymal identity. Data are represented as average fold change normalized to the siNTC control knockdown. Maxima for CDH1, EPCAM, and VIM are 16.47-, 5.50-, and 0.42-fold, respectively. Heat map data depicts means. n = 3 biological replicates each averaged from three technical replicates. (F) Western blots for E-CAD and VIM in mesenchymal pancreatic cancer cell lines with control and SUV420H2 knockdown. Loading control is α-TUBULIN (α-TUB).
RNaseq data, as well as quantitative RT-PCR (qRT-PCR), corroborated the findings of the genetic screen regarding CDH1 (E-CAD), EPCAM, and VIM on SUV420H2 knockdown; CDH1 and EPCAM were markedly up-regulated (11.1–14.3-fold and 5.2–5.5-fold, respectively), and VIM’s expression level was decreased to 0.5–0.7-fold the original values (Fig. 2 B).
Aside from VIM, RNaseq showed quantitatively variable (10–80%) but statistically significant down-regulation of other mesenchymal markers (CDH2, CDH11, and MMP1) and EMT factors (ZEB1, E47, FOXH1, and FOXC2; Fig. 2 C). SNAI1 expression was unaltered, whereas other EMT transcription factors (ZEB2, SNAI2/3, and TWIST1/2) were expressed at undetectable or negligible levels in parental PANC-1 cells and remained so after SUV420H2 knockdown. Conversely, there was statistically significant up-regulation (up to 25-fold) of various genes implicated in MET (KLF4, BMP1, BMP5, BMP8A, PKCA, and PKCB and the previously mentioned FOXA1, OVOL1, and OVOL2; Fig. 2 D).
The observed molecular transition was reversible. When PANC-1 cells were subjected to the 8-d SUV420H2 knockdown regimen followed by 8 d of recovery, expression levels of CHD1, EPCAM, and VIM largely reverted to their original levels (Fig. S2 C).
Knockdown of SUV420H2 in four other pancreatic cancer cell lines with mesenchymal identity (Klijn et al., 2015) followed by qRT-PCR for CDH1, EPCAM, and VIM indicated that aspects of MET were recapitulated in all of them (Fig. 2 E). Of those, the three cell lines with the most robust changes on the RNA level were subjected to Western blotting, showing that E-CAD and VIM were up- and down-regulated, respectively, on a protein level (Fig. 2 F). When instead SUV420H2 was knocked down in mesenchymal cancer cell lines derived from other tissues (Klijn et al., 2015), only occasional alterations of CDH1, EPCAM, and VIM expression levels were observed (Fig. S2 D).
Collectively, these results indicated that SUV420H2 is a potent epigenetic factor in controlling epithelial/mesenchymal identity status in pancreatic cancer cells, and its repression elicits a global MET from a molecular standpoint.
SUV420H2 knockdown reduces migration, invasion, stemness, and chemoresistance in pancreatic cancer cells
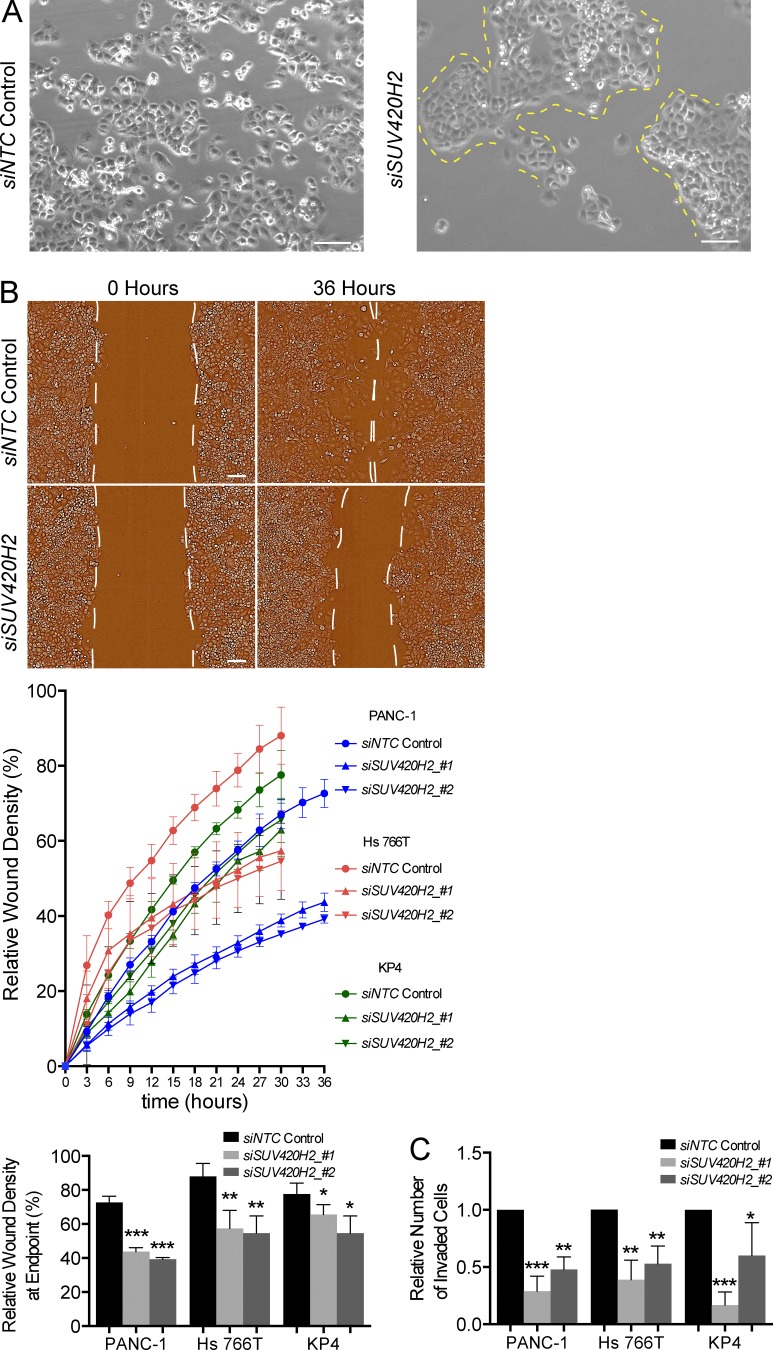
We next investigated the phenotypic and functional changes in pancreatic cancer cells after SUV420H2 knockdown. Mesenchymal cells that delaminate from cellular masses are more motile compared with their epithelial counterparts, which are typically static or move en masse (Nieto, 2013). Subconfluent parental PANC-1 cells in monolayer typically grow in loose assemblies, where the bulk of cells groups in clusters and several single cells intersperse in the spaces between those clusters (Fig. 3 A). When SUV420H2 was knocked down, PANC-1 cells grew in compact cohorts with clearly defined margins (Fig. 3 A). Scratch-wound and Matrigel-based transwell assays showed a clear reduction of migration and invasion potential of three mesenchymal pancreatic cancer cell lines on SUV420H2 knockdown (Fig. 3, B and C; and Fig. S3, A and B).
Figure 3.
Mesenchymal pancreatic cancer cells assume epithelial phenotype after SUV420H2 knockdown. (A) Bright field microscope images of PANC-1 cells grown in monolayer after NTC control or SUV420H2 knockdown. Yellow dashed lines demarcate boundaries of compact cell groups in cells treated with siSUV420H2. Bars, 50 µm. (B) Migration assay of mesenchymal pancreatic cancer cell lines with NTC control or SUV420H2 knockdown. Representative images of PANC-1 cells are shown. Cells growing in monolayer underwent two rounds of siRNA transfection at days 1 and 4 of the assay. Cells reached confluence at day 5, when a scratch wound was made, and relative wound density was recorded over the next 30 h (Hs 766T and KP4) or 36 h (PANC-1). n = 6 replicates, data depicted as mean ± SD. Differences were assessed by Student’s t test compared with siNTC control. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Bars, 50 µm. (C) Histograms depicting data from invasion assay of mesenchymal pancreatic cancer cells with NTC control or SUV420H2 knockdown. Cells growing in monolayer underwent two rounds of siRNA transfection at days 1 and 4 of the assay and were transferred at day 5 to Matrigel-coated transwell plates, and invasion was measured 30 h (Hs 766T and KP4) or 36 h (PANC-1) later. Data are normalized for total number of cells in each well. n = 6 independent experiments. Data depicted as mean ± SD. Differences were assessed by Student’s t test compared with siNTC control. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
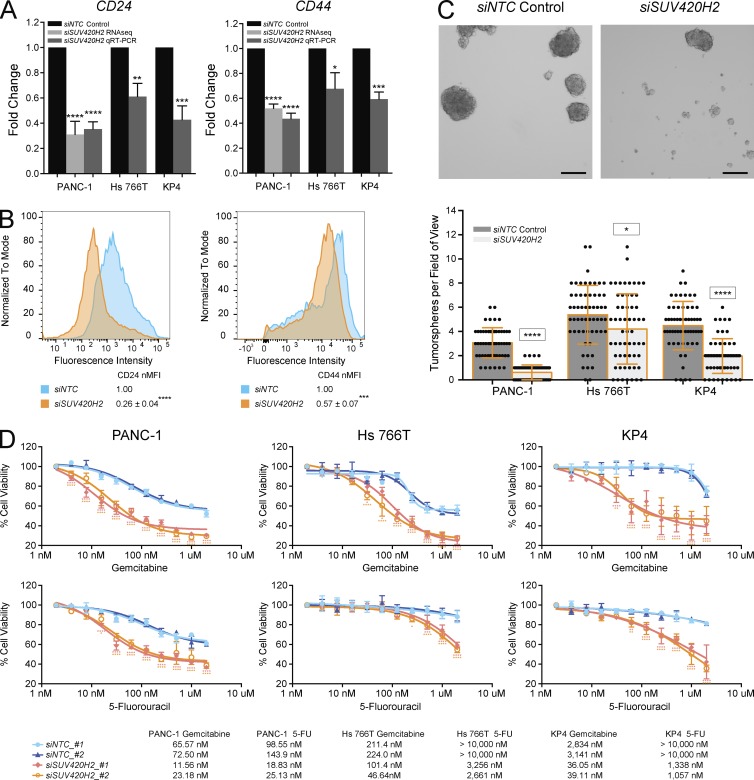
Within the bulk population of tumors, the subset of cells with mesenchymal traits has been correlated with stemness in several cancer types (Scheel and Weinberg, 2012). Pancreatic cancer cells with the profile CD24hi CD44hi are often referred to as “cancer stem cells” because of their superior ability to form tumors in xenografts and increased efficiency at forming tumorspheres, an often-used readout of stemness (Huang et al., 2008; Yin et al., 2011). On knockdown of SUV420H2, mesenchymal pancreatic cancer cells exhibited significantly decreased expression of CD24 and CD44 (Fig. 4, A and B) and had decreased potential to generate pancreatic tumorspheres (Fig. 4 C).
Figure 4.
SUV420H2 knockdown decreases stemness and anticancer drug resistance of mesenchymal pancreatic cancer cells. (A) CD24 and CD44 RNA expression levels with NTC control or SUV420H2 knockdown determined by RNaseq or qRT-PCR. For graphs, bars indicate mean ± SD. For RNaseq, n = 3 biological replicates and differences assessed by using voom+limma (see Materials and methods). For qRT-PCR, n = 3 biological replicates each averaged from three technical replicates, and differences were assessed by Student’s t test compared with siNTC control. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (B) Cell-surface protein level for CD24 and CD44 analyzed by flow cytometry in PANC-1 cells. B shows one representative panel per condition of a set of triplicates; differences were assessed by Student’s t test compared with siNTC control. ***, P < 0.001; ****, P < 0.0001. (C) Tumorsphere formation assay showing representative images from a PANC-1 experiment and quantitation in three pancreatic cancer cell lines. n = 60 fields of view considered for each cell line, composed of 20 across three biological triplicates. Bar graphs depict mean ± SD. Differences were assessed by Student’s t test compared with siNTC control in each cell line. *, P < 0.05; ****, P < 0.0001. Bars, 250 µm. (D) Cell viability assays demonstrating effects of Gemcitabine and 5-Fluorouracil (5-FU) over 72 h on mesenchymal pancreatic cancer cells with NTC control or SUV420H2 knockdown, with two siRNAs for each. n = 6 biological replicates. Significance for each drug concentration was assessed by two-way analysis of variance compared with siNTC_#1 control and indicated as pink and orange asterisks for siSUV420H2_#1 and siSUV420H2_#2, respectively. IC50 for each condition is displayed in the table. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Finally, mesenchymal cancer cells are universally more resistant to anticancer drugs than their epithelial counterparts (Singh and Settleman, 2010). SUV420H2 knockdown rendered mesenchymal pancreatic cancer cells significantly more sensitive to gemcitabine and 5-fluorouracil, two of the most commonly used chemotherapies in human PDAC (Ghadban et al., 2017; Fig. 4 D and Fig. S3 C).
Collectively, those data implied a shift from a mesenchymal to an epithelial state in pancreatic cancer cells at both the phenotypic and functional levels as a result of SUV420H2 knockdown.
SUV420H2 regulates several epithelial-promoting transcription factors
We next asked if SUV420H2 might act by selectively modulating transcription factors associated with MET. Our RNaseq analysis demonstrated that on SUV420H2 knockdown the transcription factors FOXA1, OVOL1, and OVOL2 became highly up-regulated in PANC-1 cells, and we observed increased levels of FOXA1 by Western blots in various mesenchymal pancreatic cancer cells (Fig. 2, A and D; and Fig. S3 D). Previously published observations in pancreatic cancer cells showed that expression of FOXA1 was sufficient to neutralize several E-CAD repressive mechanisms and that its knockdown induced EMT (Song et al., 2010). Other studies showed that OVOL1/2 expression facilitated MET in several cancer types by repressing ZEB1 and inducing ESRP1, which regulates RNA splicing to generate epithelial-specific isoforms (Roca et al., 2013).
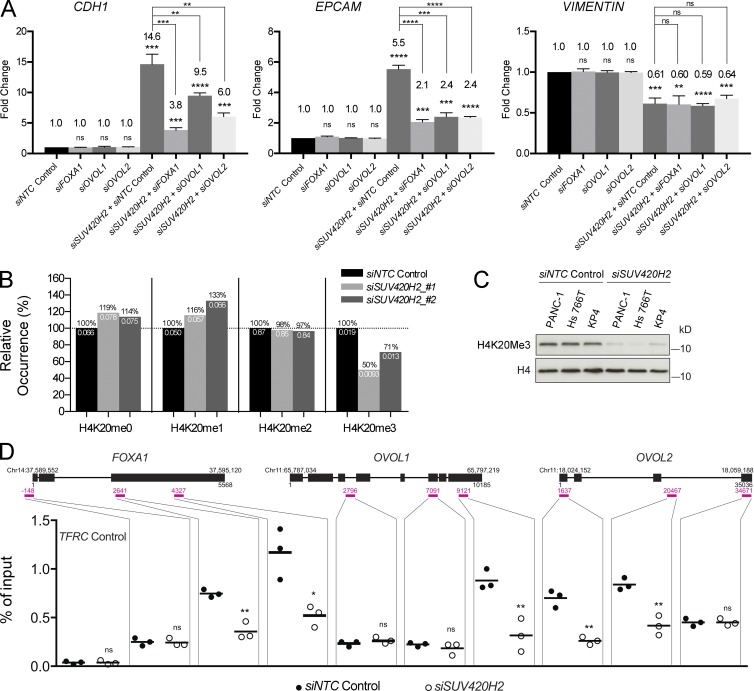
Consequently, we reasoned that if SUV420H2 was acting via one or more of these transcription factors, double-knockdown experiments should neutralize the MET-inducing effects of SUV420H2 knockdown alone. Indeed, double knockdowns in PANC-1 cells of SUV420H2 and FOXA1, OVOL1, or OVOL2 prevented key molecular features of MET (Fig. 5 A). In particular, the expression of the epithelial genes CDH1 and EPCAM was significantly reduced in all cases, even though expression of the mesenchymal gene VIM was unaffected. ZEB1 expression, which was significantly reduced in PANC-1 cells on SUV420H2 knockdown, was partially rescued when FOXA1, OVOL1, or OVOL2 was also knocked down (Fig. S3 E). Therefore, these double-knockdown “rescue” experiments argued that SUV420H2 affected CDH1, EPCAM, and ZEB1 via all three tested MET inducers.
Figure 5.
SUV420H2 silences MET transcription factors via the H4K20me3 repressive mark. (A) qRT-PCR expression analysis of CDH1, EPCAM, and VIM in PANC-1 cells in double-knockdown rescue experiments. Data are normalized to siNTC control; bar graphs indicate mean ± SD. n = 3 biological replicates each averaged from three technical replicates; differences were assessed by Student’s t test compared with siNTC control, unless otherwise indicated. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (B) Mass spectrometry analysis of global K4K20 methylation patterns in PANC-1 cells with control or SUV420H2 knockdown. Black values above the bars represent the relative occurrence compared with the siNTC control for each mark; white values below the bars represent the absolute values from the mass spectrometry assay. (C) Western blot for global H4K20me3 levels in mesenchymal pancreatic cancer cells with siNTC control and SUV420H2 knockdown. H4 is the loading control. (D) ChIP-qPCR results quantifying levels of H4K20me3 on FOXA1, OVOL1, and OVOL2 genes in PANC-1 cells, with control or SUV420H2 knockdown. Gene diagrams include genomic locus information. Three regions per gene were chosen for probing, shown as pink lines and numbers, indicating the location relative to the gene sequence. Each dot represents a value for one biological replicate, averaged from three technical replicates. Black lines indicate means. Differences were assessed by Student’s t test compared with siNTC control for each probe. *, P < 0.05; **, P < 0.01; ns, not significant (P ≥ 0.05).
To confirm that SUV420H2 controls shifts in the epithelial/mesenchymal state via histone marks, we investigated global patterns of H4K20 histone methylation marks in PANC-1 cells by histone mass spectrometry. Knockdown of SUV420H2 with two different siRNAs was effective at reducing methyltransferase activity and resulted in 29–50% overall global decreases in H4K20me3, the repressive mark produced by SUV420H2 (Fig. 5 B). Conversely, there was a global increase in the unmethylated and monomethylated states of H4K20, both of which have been associated with an activated transcriptional state for a variety of genes (Karachentsev et al., 2005; Talasz et al., 2005; Li et al., 2011). Western blots in PANC-1, as well as two other mesenchymal pancreatic cancer lines (Hs 766T and KP4), showed a reduction in the H4K20me3 mark on SUV420H2 knockdown (Fig. 5 C).
We went on to analyze the status of the H4K20me3 mark specifically on FOXA1, OVOL1, and OVOL2 by chromatin immunoprecipitation coupled with quantitative PCR (ChIP-qPCR; Fig. 5 D and Fig. S3 F). Being a known repressive histone mark, trimethylation of H4K20 around their respective genes could be the mechanism through which SUV420H2 controlled expression of these three transcription factors. Histone mark profiles can vary across the sequence of a gene; consequently, we adopted a ChIP-qPCR protocol probing multiple regions per gene in our experiments (Milne et al., 2009). SUV420H2 knockdown in PANC-1 cells resulted in significant reductions in signal in at least one of three loci in each gene tested (Fig. 5 D). This suggested that SUV420H2 controlled the H4K20me3 mark on each of our three candidate MET transcription factors and therefore their capacity to be transcribed. In contrast, SUV420H knockdown had no direct effect on H4K20me3 levels on CDH1 or EPCAM, suggesting that SUV420H2 acts indirectly on these epithelial markers (Fig. S3 G). H4K20me3 levels at the ZEB1 locus were also unaltered, indicating no direct activity of SUV420H2 in controlling the expression of ZEB1 (Fig. S3 G).
Together, these results suggested that SUV420H2 controls the dynamics of H4K20me3 marks on specific genes encoding transcription factors that control MET.
SUV420H2 gain of function induces EMT in pancreatic cancer cells
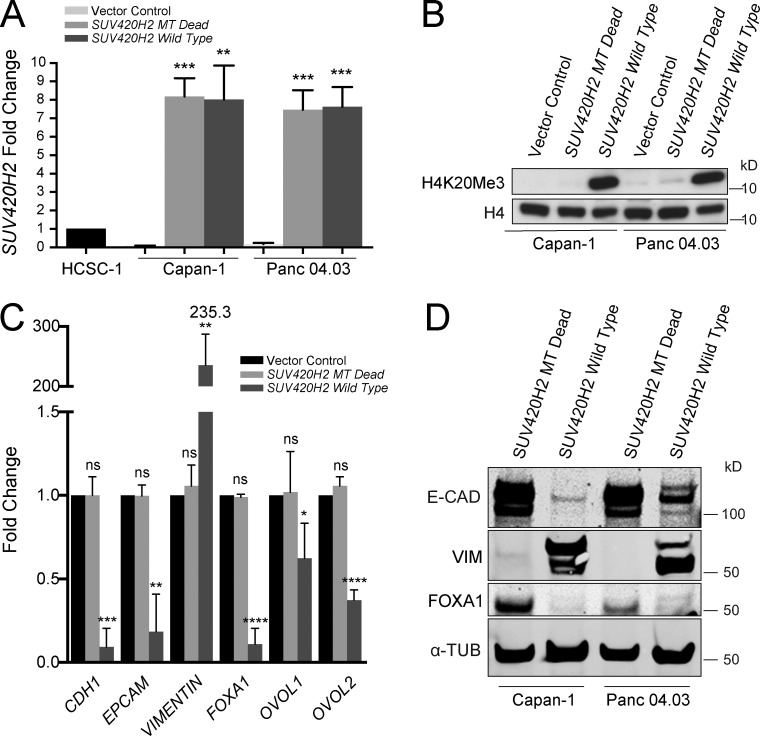
To test whether SUV420H2 gain of function could elicit the opposite effect we observed with its knockdown, we chose two epithelial pancreatic cancer cell lines (Klijn et al., 2015) and induced expression of a wild-type SUV420H2 and a methyltrasnferase dead allele of SUV420H2 using overexpression vectors. For the latter, we designed a sequence with an in-frame 42-bp deletion, resulting in a 14–amino acid excision within the catalytic domain of SUV420H2. We confirmed that the overexpression constructs could induce expression of the SUV420H2 alleles (Fig. 6 A) and that the wild-type allele, but not the kinase-dead version, resulted in increased H4K20me3 marks in transfected cells (Fig. 6 B). qRT-PCR showed that epithelial Capan-1 and Panc 04.03 cell lines strongly repressed the epithelial markers CDH1 and EPCAM and induced VIM on SUV420H2 overexpression (Fig. 6 C and Fig. S3 H). In addition, in both cell lines there was significant reduction in FOXA1, OVOL1, and OVOL2, the three MET transcription factors we had previously mechanistically implicated with SUV420H2 (Fig. 6 C and Fig. S3 H). Western blots for E-CAD, VIM, and FOXA1 confirmed these trends on a protein level in both epithelial cell lines (Fig. 6 D).
Figure 6.
Overexpression of SUV420H2 in epithelial pancreatic cancer cells. (A) qRT-PCR for SUV420H2 in two epithelial pancreatic cancer cell lines transfected with an overexpression plasmid containing an SUV420H2 sequence with a deletion in the methyltransferase region (MT Dead) or a wild-type sequence. The qRT-PCR region probed is upstream of the deletion, near the 5′ end of SUV420H2. Data are normalized to HCSC-1, a cervical cancer cell line robustly expressing SUV420H2. Bar graphs indicate mean ± SD. n = 3 biological replicates each averaged from three technical replicates; differences were assessed by Student’s t test compared with cells transfected with empty vector. **, P < 0.01; ***, P < 0.001. (B) Western blot for H4K20me3 levels in epithelial pancreatic cancer cells with vector control, MT Dead, and wild-type SUV420H2 overexpression. H4 is the loading control. (C) qRT-PCR analysis of expression of epithelial/mesenchymal factors in Capan-1 cells transfected with vector control, MT dead, and wild-type SUV420H2 overexpression plasmid. Data normalized to empty vector transfection level for each gene assayed. Bar graphs indicate mean ± SD. n = 3 biological replicates each averaged from three technical replicates; differences were assessed by Student’s t test compared with cells transfected with empty vector. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (D) Western blots for E-CAD, VIM, and FOXA1 in epithelial pancreatic cancer cell lines exposed to MT dead and wild-type SUV420H2 overexpression plasmid with SUV420H2 knockdown. The loading control is α-TUBULIN. ns, not significant (P ≥ 0.05).
The observed changes were only observed after overexpression of the wild-type allele of SUV420H2 and not with the methyltransferase dead allele, directly implicating the catalytic activity of SUV420H2 in the transitions in epithelial/mesenchymal cell identity in pancreatic cancer cells.
Together, these experiments showed that up-regulation of SUV420H2 can induce EMT in epithelial pancreatic cancer cells because of its catalytic activity and ensuing H4K20me3 marks.
Pancreatic adenocarcinoma progression correlates expression of SUV420H2 with EMT
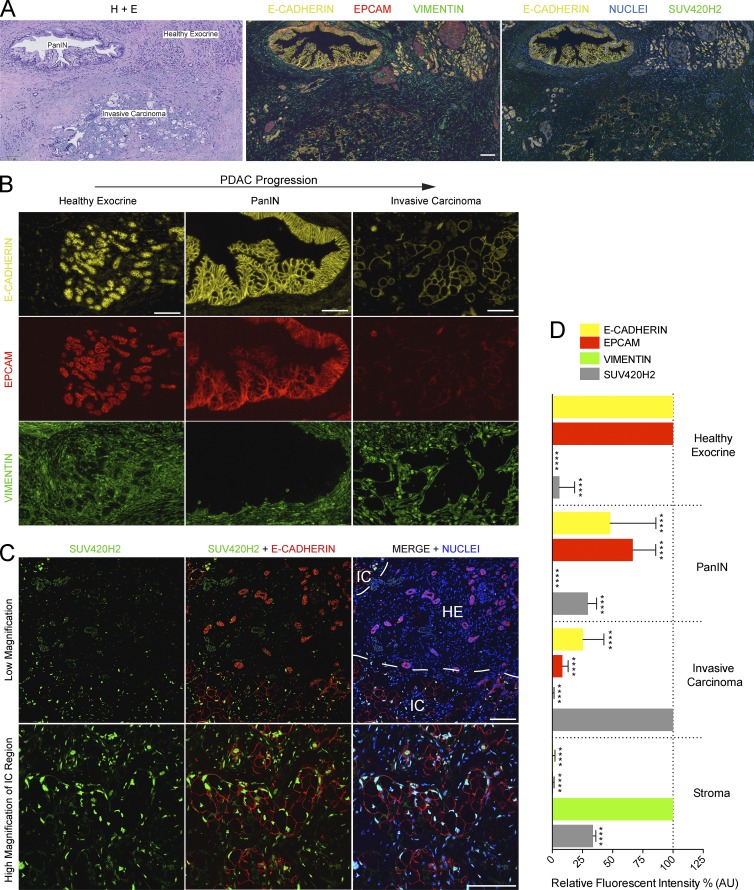
To determine if the regulatory activities of SUV420H2 in pancreatic cancer cell lines correlated with any features of clinical human PDAC, we examined the expression and localization of SUV420H2 and epithelial/mesenchymal markers in archival tumor samples (clinical details on analyzed samples outlined in Table S5). Using the same three antibodies for E-CAD, EPCAM, and VIM as we used for the original in vitro screen, we performed immunofluorescence on sections of PDAC samples, which contained regions of healthy exocrine tissue, early cancer lesions, as well as domains of advance invasive cancer (Fig. 7 A). In healthy exocrine epithelia, E-CAD and EPCAM showed strong signal at cell–cell junctions, and little or no VIM staining was observed (Fig. 7, B and D; and Fig. S4). In pancreatic intraepithelial neoplasia (PanIN) regions, levels of E-CAD and EPCAM were still relatively robust at cell–cell junctions between neighboring epithelial cells (Fig. 7, B and D; and Fig. S4). As expected, the individual epithelial cells within the PanIN lesions appeared to gradually lose their canonical epithelial morphology. PanIN epithelial cells, like healthy epithelia, were negative for VIM.
Figure 7.
Epithelial/mesenchymal markers and SUV420H2 in human PDAC. (A) Serial sections through a human PDAC sample with a healthy exocrine epithelium, early, or invasive cancer region. One section was exposed to hematoxylin and eosin (H+E) stain and two sections with immunofluorescence for different markers. Bar, 100 µm. (B) Immunofluorescence for E-CAD, EPCAM, and VIM in different stages of PDAC progression. Bars, 100 µm. (C) Immunofluorescence for SUV420H2 and E-CAD in a section of human PDAC. Low-magnification images show a region with healthy exocrine displaying pancreatic acini (strong E-CAD signal, low SUV420H2 signal) and an invasive cancer region with dysplastic malignant cells (low E-CAD stain in cancer cells, strong SUV420H2 signal). High-magnification images show dysplastic cells with low E-CAD stain show a strong nuclear SUV420H2 signal. Bars, 100 µm. (D) Quantitation of fluorescent signal for E-CAD, EPCAM, VIM, and SUV420H2 in progressive stages of PDAC (see Fig. S4 for more sample images). n = 8 PDAC samples from separate patients, analysis of one healthy exocrine region, one PanIN, and one invasive carcinoma region in each; signal was quantified in 16 cells per region for a total of 128 measurements per marker, per stage. Measurements for 128 stromal cells in total collected evenly from all images. Data were normalized to the stage/tissue with strongest signal for each marker analyzed. Bar graphs depict mean ± SD. Differences were assessed by Student’s t test compared with normalizer. ****, P < 0.0001. IC, invasive carcinoma; HE, residual exocrine.
In invasive carcinoma regions (Fig. 7, B and D; and Fig. S4), epithelial cells displayed a decidedly dysmorphic cell shape and showed weak E-CAD signal and no EPCAM signal. We could not detect any malignant cells that unequivocally displayed de novo VIM expression. VIM levels were strong in stromal cells at all stages analyzed. These observations indicate that, concomitant with pancreatic cancer progression, cells lost epithelial marker expression and morphology and exhibited a phenotype consistent with one that is more mesenchymal in nature, although they were devoid of VIM staining.
The fact that we did not observe cancer cells clearly positive for VIM could have several explanations. First, in the samples analyzed, cells might not have made a full transition to the mesenchymal state but rather displayed a partial EMT that might be sufficient to delaminate from the normal tissue epithelium. Alternatively, a full transition does happen, but because of the sporadic and asynchronous nature of the transition, it is difficult to capture in static images. Finally, VIM might be expressed in cancer cells but at levels that are undetectable when imaged adjacent to stromal cells, which exhibit strong VIM staining. Nevertheless, we detected a gradual increase in nuclear levels of ZEB1 in progressively invasive pancreatic cancer cells, which indeed again supported the notion that an EMT accompanies PDAC progression (Fig. S5 A).
Strikingly, immunofluorescence for SUV420H2 in human PDAC resulted in background/low signal in residual healthy exocrine, but moderate signal in PanIN cells, and strong, nuclear signal in invasive carcinoma cells, showing a clear and gradual increase of SUV420H2 protein levels in parallel with tumor promotion (Fig. 7, C and D; and Fig. S4). Together with the aforementioned findings, this described a direct correlation between pancreatic cancer development, increasing amounts of SUV420H2, and a shift away from the epithelial and toward the mesenchymal cell state.
Finally, we queried public databases for SUV420H2 transcript levels in human cancers. Data compiled from the largest PDAC study in cBioPortal (Cerami et al., 2012; Gao et al., 2013) suggests that 18% of pancreatic cancers have amplifications of SUV420H2, and records derived from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) as well as internal Genentech datasets show significantly elevated levels of SUV420H2 in a vast array of cancer types, including bladder, breast, colon, kidney, lung, liver, prostate, stomach, uterus, and others (Fig. S5 B).
Collectively, the data from pancreatic cancer showed an association between EMT and elevated expression of SUV420H2 during the pathological progression of PDAC, and data from other cancers show a global correlation of SUV420H2 with malignancies in a vast array of tissues.
Discussion
Our data suggest that SUV420H2 acts as an epigenetic orchestrator of epithelial/mesenchymal cell states in pancreatic cancer. As summarized in a working model in Fig. S5 C, SUV420H2, via its repressive mark H4K20me3, silences expression of the key MET-promoting transcription factors FOXA1, OVOL1, and OVOL2 (Fig. S5 C, left). When SUV420H2 expression is attenuated, its repressive mark disappears, thereby enabling expression of FOXA1, OVOL1, and OVOL2 and activating the epithelial transcriptional program (Fig. S5 C, right). The three MET-promoting transcription factors are likely to directly induce de novo transcription of structural epithelial markers. This has been shown for FOXA1, which binds the regulatory region of CDH1 in pancreatic cancer to control its expression (Song et al., 2010). The activation of the epithelial-like MET program also includes attenuation of EMT factors such as ZEB1, whose transcription was previously shown to be repressed directly by OVOL1/2 via direct promoter binding (Roca et al., 2013). The same study found that OVOL1/2 induced ESRP1, leading to processing of epithelial-specific splice variants. Alternative splicing mediated by ESRPs is a hallmark of epithelial differentiation (Warzecha and Carstens, 2012; Bebee et al., 2015). In agreement with this interpretation, our expression-profiling experiment revealed a sharp increase in ESRP1 levels on knockdown of SUV420H2 (Fig. 2 A).
It remains unclear how SUV420H2 controls expression of the mesenchymal marker VIM. A previous study showed that OVOL1/2 represses VIM (Lee et al., 2014), but in our double knockdown experiments, VIM expression was not rescued by simultaneous siRNA knockdown of SUV420H2, FOXA1, or either of the OVOL genes. Thus, VIM expression appears to be independent of the three SUV420H2-controlled MET transcription factors in our context. It is unlikely that SUV420H2 acts directly on the VIM locus to control its expression, considering H4K20me3 is a repressive mark, and there is a positive correlation between the two factors. Instead, an as yet unidentified parallel signal is likely to play a role. It is also worth noting that, although SUV420H2 knockdown in PANC-1 cells induced robust expression of the epithelial markers, VIM levels were only partially reduced but never completely eliminated, suggesting that cells had achieved a hybrid or transitional state. Conceivably, a MET program was induced but did not go to completion, a transition referred to as “partial MET” (Jolly et al., 2015). Full MET might require other players or take longer time to achieve. Nonetheless, the acquisition of epithelial characteristics was sufficient to reduce cell invasion and migration and increase sensitivity to anticancer drugs.
Although SUV420H2 orchestrates the induction of the transcriptional program controlling MET, a key remaining question is what controls expression of SUV420H2 itself. In pancreatic cancer, the gradual increase in SUV420H2 levels as the disease progresses must reflect a response to a broader biological mechanism. Because only 18% of PDAC occurrences contain amplifications of SUV420H2 (Cerami et al., 2012; Gao et al., 2013), amplification does not seem likely to explain the elevated levels of SUV420H2 in the majority of PDAC cases. Thus, amplification-independent mechanisms leading to overexpression are likely to be more common. In general, the processes involved in the induction of MET have not been well characterized (Nieto, 2013), so the identification of the signals responsible for enhancing SUV420H2 expression will likely shed light on the conditions that control mesenchymal and epithelial balance in pancreatic cancer.
The analysis of TCGA data on SUV420H2 expression in different cancer types emphatically showed near-universal, statistically significant elevation in malignancies compared with matched healthy tissues (Fig. S5 B). Interestingly, previous studies have generally reported a decrease in H4K20 trimethylation during the tumorigenic process in many cancer types, to the extent that global loss of H4K20 trimethylation was proposed to be a common hallmark of many human tumor cells (Fraga et al., 2005). One fact that might reconcile these observations is that, even though the levels of SUV420H2 might increase in cancers, concomitant high expression of demethylases might outcompete the methyltrasnferase activity of SUV420H2, resulting in overall lower H4K20me3 levels. This does not appear to be the case in pancreatic cancer, however, and to our knowledge a decrease in H4K20me3 has never been reported for pancreatic malignancies.
It should also be noted that the data reporting a decrease in H4K20me3 in cancers have primarily focused on H4K20me3 marks on repetitive sequences of the genome, particularly in centromeric repeats. Consequently, it is possible that from a global cancer cell standpoint that levels of H4K20me3 decrease, but marks in particular loci such as the MET-transcription factors should increase. A regulator complex that guides SUV420H2 to specific genomic sites could achieve this. A precedent for such a mechanism has been reported, in which SUV420H2 associates with a long noncoding RNA that guides the methyltransferase to specific genes (Bierhoff et al., 2014).
It is also important to note that, even though SUV420H2 transcript levels appear elevated in virtually all types of malignancies, qRT-PCR analysis of epithelial/mesenchymal markers in cancer cell lines from various tissues other than the pancreas rarely resulted in significant changes upon SUV420H2 knockdown (Fig. S2 D). Published work in breast cancer cells has even shown an opposite effect of SUV420H2 regarding epithelial/mesenchymal cell identity, actually rendering cells more epithelial and less invasive (Yokoyama et al., 2014; Shinchi et al., 2015). This would argue for context-dependent activity of SUV420H2 and its mark H4K20me3. In some cancers, there appears to be mechanistic disconnect between SUV420H2 and epithelial/mesenchymal cell identity. Future work might elucidate new functions of SUV420H2 in other cancers, possibly with new targets for methyltransferase activity or scaffolding functions for protein complexes that might help explain its elevated levels across cancer types.
Of the histone modifiers previously associated with the control of epithelial/mesenchymal cell states, their suggested mechanisms of action range from forming complexes with EMT transcription factors to directly silencing or activating expression of EMT drivers or genes encoding structural epithelial traits such as E-CAD (Peinado et al., 2005; Tam and Weinberg, 2013; Pattabiraman et al., 2016). Here, we show an alternative mechanism, namely a histone modifier silencing transcription factors that steer the MET process. From our in vitro cell work we observe that SUV420H2, being a repressor of transcription, seemingly has no direct effect on the classical EMT factors (ZEB1/2, SNAI1/2, and TWIST1/2) but instead selectively silences FOXA1 and OVOL1/2 thereby inhibiting a cell’s ability to remain in the epithelial state. In human PDAC samples, we observed a close correlation between SUV420H2 and loss of epithelial characteristics in cancer progression, which is consistent with previous work showing diminishing levels of FOXA1 with progressive pancreatic cancer (Song et al., 2010). Whether SUV420H2 also plays a broader role in development, particularly in orchestrating MET in pancreatic organogenesis and other tissues, remains to be explored. In the mouse, the SUV420H2 knockout is normal and viable, although the H4K20me3 mark is only partially abrogated in embryonic and adult tissues, suggesting the presence of compensatory methyltransferases that may be active in development but not in cancer (Schotta et al., 2008). In the context of cancer, SUV420H2 could serve as a biomarker of invasive PDAC. It is still unclear whether promoting a MET is in itself a beneficial strategy in oncology, considering that circulating tumor cells that have gone through an EMT at the primary lesion are thought to undergo MET for at the metastatic site (Tsai and Yang, 2013). Consequently, forcing a MET could conceivably facilitate metastasis outgrowth. Nonetheless, promoting the epithelial state by targeting SUV420H2 in combination with conventional chemotherapies and decreasing resistance might prove to be an effective treatment for the devastating diagnosis of pancreatic cancer.
Materials and methods
siRNA screen to identify epigenetic regulators of epithelial/mesenchymal states
PANC-1 cells (CRL-1469; ATCC) were plated at 1,000 cells per well in 96-well plates (CLS3991; Corning) prestamped in array format with the EPI300 siRNA library composed of siGENOME sequences (Dharmacon) targeting 300 genes involved in modulating epigenetic marks (see the full list of genes, siRNA sequences, and catalog numbers in Table S1). Four different siRNAs per gene were reverse transfected at 12.5 nM with DharmaFECT1 (Dharmacon) in duplicate. Each plate contained siNTC nontargeting controls (D-001810-10; Dharmacon), positive controls (siZEB1; D-006564-01; Dharmacon), and transfection controls (siControl Tox; D-001500-01; Dharmacon). 4 d later a second round of transfection was performed concomitant with a media change by using the same library and control layout. After four more days, cells were fixed with 4% paraformaldehyde, washed in PBS, and exposed to the nuclear stain Hoechst 33342 (H3570; Molecular Probes), and the following fluorescently conjugated monoclonal antibodies: rabbit Vim_488 (9854; Cell Signaling Technology), mouse E-Cadherin_555 (560064; BD), and mouse EPCAM_647 (324212; BioLegend). Images were acquired with an IN Cell 6000 (GE Healthcare) at 20× lens magnification with 0.75 NA, 9 images per well, and fluorescent signals quantified with IN Cell imaging analysis software (GE Healthcare). Replicate averaging, intra- and interplate normalization, and subsequent analysis of data and standard (Z) score calculations were performed with ScreenSifter (Kumar et al., 2013). For hit selection, we excluded siRNA sequences that substantially prevented proliferation or caused high levels of cell death (Z score of cell count less than −11.3).
RNaseq
RNaseq was performed by using PANC-1 cells with the following two knockdown conditions: siNTC (nontargeting control) and siSUV420H2 (using sequence 1; Table S1) in triplicates. Total RNA was extracted by using the Qiagen RNeasy kit per manufacturer’s protocol including the on-column DNase digestion. Quality control of samples was done to determine RNA quantity and quality before their processing by RNaseq. The concentration of total RNA samples was determined by using NanoDrop 8000 (Thermo Fisher). The integrity of RNA samples was determined by using 2100 Bioanalyzer (Agilent Technologies). Approximately 500 ng of total RNA was used as an input for library preparation by using TruSeq RNA Sample Preparation kit v2 (Illumina). The size of the libraries was confirmed by using 2200 TapeStation and High Sensitivity D1K screen tape (Agilent Technologies), and their concentration was determined by the qPCR-based method by using Library quantification kit (KAPA). The libraries were multiplexed and then sequenced on Illumina HiSeq2500 (Illumina) to generate 30M of single-end 100-bp reads.
RNaseq data processing and analysis
HTSeqGenie (Gregoire and Reeder, 2014) was used to perform filtering, alignment, and feature counting. HTSeqGenie used GSNAP (Wu and Nacu, 2010) to align reads to the human genome build hg19. Gene models were used from Genentech’s internal database based on Refseq. Gene expression was quantified by counting the number of reads mapping uniquely to any exon of a transcript from the gene. A prefiltering was done for low expression, and only genes with ≥10 read counts in at least three samples (irrespective of condition) were considered. Differential gene expression analysis was performed by using voom+limma (Law et al., 2014). The full RNaseq data discussed in this publication have been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO series accession no. GSE104542.
Gene set enrichment analysis
Camera (Wu and Smyth, 2012) was used to perform competitive gene set testing by using its parametric test. Gene set collections were used from Molecular Signatures Database (MSigDB), including the C2 collection (Liberzon et al., 2011) of canonical pathways and experimental signatures and Hallmark gene sets (Liberzon et al., 2015), which represent coherent genes for well-curated biological processes.
qRT-PCR
qRT-PCR was performed in biological triplicates, each with technical triplicates, on a QuantStudio 5 System (Thermo Fisher) after RNA isolation with the RNeasy kit (Qiagen), cDNA synthesis with SuperScript VILO (Thermo Fisher), and by using the following Taqman (Thermo Fisher) assays: CDH1 (Hs01023894_m1), EPCAM (Hs00901885_m1), VIM (Hs00185584_m1), SUV420H2 (Hs00261961_m1 or Hs00938285_g1), CD24 (Hs02379687_s1), CD44 (Hs01075864_m1), ZEB1 (Hs01566410_m1), OVOL1 (Hs00190060_m1), OVOL2 (Hs01067398_m1), FOXA1 (Hs04187555_m1), and TFRC (Hs99999911_m1), which was used as the normalizer. All cell lines used in qRT-PCR experiments were obtained from Genentech’s in-house cell line repository.
Immunoblotting
Cell lysates were extracted by boiling in 6 M Urea buffer (6 M Urea, 20 mM Tris, pH 7.5, 12.5 mM NaCl, 2.5 mM MgCl2, and 0.1% Triton X-100), supplemented with Halt Protease and Phosphatase Inhibitor cocktail. For histone and histone modifications, extraction was performed by using the EpiQuick nuclear extraction kit (Epigentek) following the manufacturer’s protocol. Protein quantification was performed by using a Pierce BCA Protein Assay kit, and equal amounts of protein lysate were loaded onto the NuPAGE Bis-Tris Gel. Immunodetection and quantification of proteins were performed by a quantitative Western blot method by using LI-COR Odyssey. Antibodies were: mouse E-CAD (Cell Signaling Technology 5296), mouse VIM (3390; Cell Signaling Technology), rabbit FOXA1 (58613; Cell Signaling Technology), rabbit α-TUBULIN (2144; Cell Signaling Technology), mouse H4K20me3 (39671; Active Motif), and rabbit histone H4 (39269; Active Motif).
Cell migration and invasion assays
Cell migration was examined by using the IncuCyte’s (Essen BioScience) ZOOM System and Cell migration kit. Cells were plated at 2,000 cells per well in 96-well ImageLock plates and exposed to a double siRNA transfection course at days 0 and 4 of plating as described for the screen. The conditions were siNTC (nontargeting control) and siSUV420H2 (using sequence 1 or sequence 2; Table S1), each in six replicates. At the fifth day, the cells were confluent, and a scratch wound was inflicted with a 96-pin woundmaker. Cells were subsequently imaged in an IncuCyte Live-Cell System, and results were analyzed with IncuCyte’s Scratch Wound Cell Migration Module, which normalizes for cell number and calculates relative wound density.
To quantify cell invasion, cells first underwent a double siRNA transfection course at days 0 and 4 of the experiment, as described for the screen. At day 5, cells were trypsinized and seeded at 50,000 cells per insert in 6-well plate invasion chambers coated with Matrigel (354481; Corning Biocoat), covered with serum-free medium, and placed in wells containing media with 10% fetal bovine serum in triplicates. After 30–36 h incubation, uninvaded and invaded cells were detached from both sides of the inserts with trypsin treatment and physical scraping, quantified by Cell Counter (Bio-Rad), and ratios of invaded versus uninvaded cells for each condition were calculated, normalizing for cell proliferation.
Flow cytometry
PANC-1 cells underwent an 8-d course of siRNA knockdown as described for the screen, and at day 8 cells were trypsinized, washed in PBS/2% serum, stained with mouse CD24-PE (555428; BD) or rat CD44-APC (17–0441-82; eBioscience) per manufacturer’s protocol and processed for routine flow cytometry in a FACSCalibur Analyzer (BD).
Tumorsphere formation
PANC-1 cells underwent siRNA transfections as described for the screen at days 1 and 4 of the assay and were subsequently detached at day 5 with trypsin, plated in ultralow attachment 6-well plates (Costar 3471) at a density of 4,000 cells per well, and cultured in stem cell media composed of DMEM supplemented with 1× B27 Supplement, 20 ng/ml human basic fibroblast growth factor, 20 ng/ml epidermal growth factor (all three from Invitrogen), and 4 µg/ml heparin calcium salt (Fisher). Spheres were imaged at day 12 of the assay by using microscopy, and only cell clusters >10 µm were considered spheres.
Cell viability
After double siRNA knockdown at day at days 0 and 4 of the experiment, the following compounds were added at day 5: gemcitabine hydrochloride (3259; Tocris Bioscience) or 5-Fluorouracil (03738; Sigma-Aldrich). Cells were exposed 72 h later to CellTiter-Glo (G7570; Promega) and assessed with a 2104 EnVision reader (PerkinElmer). Analysis and calculation of IC50 (the half maximal inhibitory concentration) was performed in Prism (GraphPad) by using nonlinear curve fitting regression algorithms.
Knockdown experiments
All knockdown experiments were performed with siRNA sequences from siGENOME (Dharmacon) with the same reagents described for the genetic screen. The siRNAs used in the screen are listed in Table S1. In other experiments, siSUV420H2 as well as siSUV420H2#1 use D-018622-24, and siSUV420H2#2 uses D-018622-25. siFOXA1, siOVOL1, and siOVOL2 use D-010319-01, D-006543-03, and D-013793-01, respectively.
Mass spectrometry
Core histones (H2A, H2B, H3, and H4) were purified from frozen cell pellets by acid extraction, ion exchange, and perchloric acid precipitation with the use of a commercial kit (Histone Purification Mini kit, 40026; Active Motif) according to the manufacturer’s instructions. The purified histones were resuspended in deionized distilled water to a final concentration of 0.5–1.0 µg/µl and stored at −80°C until use. 2-µg aliquots of endogenous histones were mixed with equal amounts of purified stable-isotope-labeled core histones purified from PC9 cells grown in media supplemented with 13C6,15N2 lysine and 13C6,15N4 arginine that serve as internal standards. Samples were prepared for mass spectrometry by propionlylation of lysines, digestion with trypsin, and derivatization of peptide N termini with phenyl isocyanate as previously described (Maile et al., 2015). Histone peptides were quantified by capillary reverse-phase liquid chromatography nanoelectrospray tandem mass spectrometry on a hybrid quadrupole-orbitrap mass spectrometer (Q-Exactive HF; Thermo Fisher) in a parallel-reaction monitoring experiment. Quantitative data on 40 distinct posttranslational modifications of histones H3 and H4 in 78 combinations were extracted via Skyline software and normalized via internal standards as previously described (Vinogradova et al., 2016). Modifications on H4K20 are reported here.
ChIP-qPCR
ChIP assays for histone modifications and data analysis were performed as previously described (Milne et al., 2009) by using EZ Chip (17–371; Millipore) and an antibody for H4K20me3 (39671; Active Motif). ChIP signal enrichment was quantified by qPCR by using the following Taqman assays: FOXA1 Hs01351917_cn, Hs00666612_cn, Hs01162620_cn; OVOL1 Hs05268126_cn, Hs02113848_cn, Hs01458465_cn; OVOL2 Hs02395224_cn, Hs07203924_cn, Hs00475512_cn; CDH1 Hs05413045_cn, Hs05411576_cn, Hs05397765_cn; EPCAM Hs04617695_cn, Hs04630814_cn, Hs02913990_cn; KLF4 Hs02900640_cn, Hs02807427_cn, Hs01469971_cn; ZEB1 Hs07440276_cn, Hs07440283_cn, Hs07438380_cn; TFRC Hs02677106_cn.
Gain-of-function experiments
Coding sequence of full-length wild-type SUV420H2 (National Center for Biotechnology Information NM_032701.3) and a methyltransferase dead allele omitting the sequence 5′-TTCATCAACCATGACTGCAAACCCAACTGCAAGTTTGTGCCT-3′ from within the SET domain-coding region were synthesized by GeneArt (Thermo Fisher) with flanking XhoI and a EcoRI restriction sites and subcloned into the overexpression vector pIRES2-ZsGreen1 (632478; Clontech). Cells were transfected with FugeneHD (Promega) and after 72-h BD fluorescence-activated cell sorted for GFP-expressing cells.
PDAC immunofluorescence imaging and analysis
Paraffin-embedded sections of human pancreatic cancer were obtained from Genentech’s tissue bank, deparaffinized, and stained with the nuclear stain and antibodies used in the screen, rabbit ZEB1 (sc-25388; Santa Cruz) with goat anti–rabbit Alexa Fluor 546 secondary antibody, and rabbit SUV420H2 (HPA052294; Sigma-Aldrich) with goat anti–rabbit Alexa Fluor 488 secondary antibody. The latter was also used on PANC-1 cells grown on glass-bottom chamber slide systems (154534PK; Thermo Fisher) after fixing in 4% paraformaldehyde. Images were captured with a confocal microscope (LSM780; Zeiss) by using Diode 405, Argon 458/488/514, HeNe543, and HeNe633 lasers, and 5× EC Plan-Neofluar dry 0.16 NA or 20× Plan Apochromat dry 0.8 NA objectives at room temperature with cells or tissues mounted in Fluoromount-G (Thermo Fisher). Images were analyzed with Zen2010 software (Zeiss), and the fluorescent signal was quantified with ImageJ.
TCGA data
TCGA RNaseq data were downloaded from the Cancer Genomics Hub at University of California, Santa Cruz, and analyzed with HTSeqGenie (Gregoire and Reeder, 2014). The results shown here are in part based on data generated by the TCGA research network. Expression values were calculated as normalized reads per kilobase gene model per million total reads (RPKMs), which represent RPKM values normalized by an adjusted total library size instead of a simple total read count. This way of normalization is akin to size-factor normalization, but the values are reported in the traditional RPKM scale. More details about this normalization can be found in the study by Srinivasan et al. (2016).
Genentech data
RNaseq reads were mapped to the GRCh 38 genome by using GSNAP with RefSeq gene models. We used voom+limma (Law et al., 2014) as implemented in the limma package for the R programming language and described in the limma user guide to calculate differential gene expression for cancer versus normal. P-values were corrected for multiple testing by using the Benjamini-Hochberg method.
Statistics
Analysis and graph preparation was done in Prism 6 (GraphPad). The statistical method used is indicated in the figure legends for each figure. For RNaseq analysis, see RNaseq data processing and analysis.
Online supplemental material
Fig. S1 shows images from primary screen described in Fig. 1 for the siRNA knockdown of various epigenetic targets. Fig. S2 depicts efficiency of SUV420H2 knockdown, the recovery experiment, and effects of SUV420H2 knockdown on different cancer cell lines of mesenchymal identity. Fig. S3 depicts representative images of migration assay, the effects of SUV420H2 knockdown on cell proliferation, FOXA1 protein levels in pancreatic cancer cells after SUV420H2 knockdown, ZEB1 expression in double-knockdown experiments, ChIP-qPCR experiments for H4K20me3 mark on various genes, and SUV420H2 gain-of-function experiments on Panc 04.03 cells. Fig. S4 depicts SUV420H2 and E-CAD immunofluorescent stains in progressive stages of PDAC. Fig. S5 depicts ZEB1 immunofluorescent stains in progressive stages of PDAC, transcript levels of SUV420H2 in various cancer types, and a working model cartoon of SUV420H2 mechanistic control of epithelial/mesenchymal cell states in pancreatic cancer. Table S1 lists siRNAs used in the study. Table S2 shows RNaseq results for highly and significantly up-regulated genes on SUV420H2 knockdown in PANC-1 cells, listed by protein coding genes (tab one) and other genes (tab two); tab three shows full results of RNAseq. Tables S3 and S4 show gene set enrichment analysis results with MsigDB Hallmark gene sets and with the MsigDB C2 curated signature collection, respectively. Table S5 lists clinical information for PDAC samples used in this study.
Supplementary Material
Acknowledgments
All authors were employed by Genentech, Inc., a member of the Roche group, during their respective contributions to the manuscript.
The authors declare no further conflict of interest.
Author contributions: M. Viotti and I. Mellman designed experiments and wrote the manuscript with input from all authors. M. Viotti, C. Wilson, D. Arnott, and J.-P. Stephan performed the experiments. Z. Modrusan, S. Jhunjhunwala, and C. Klijn provided expertise in RNAseq. M. Classon and M. McCleland contributed intellectual and technical expertise on chromatin biology. H. Koeppen provided pathology assistance. B. Haley designed and provided various molecular tools used for experiments.
References
- Arumugam T., Ramachandran V., Fournier K.F., Wang H., Marquis L., Abbruzzese J.L., Gallick G.E., Logsdon C.D., McConkey D.J., and Choi W.. 2009. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 69:5820–5828. 10.1158/0008-5472.CAN-08-2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebee T.W., Park J.W., Sheridan K.I., Warzecha C.C., Cieply B.W., Rohacek A.M., Xing Y., and Carstens R.P.. 2015. The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. Elife. 4:e08954 10.7554/eLife.08954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierhoff H., Dammert M.A., Brocks D., Dambacher S., Schotta G., and Grummt I.. 2014. Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol. Cell. 54:675–682. 10.1016/j.molcel.2014.03.032 [DOI] [PubMed] [Google Scholar]
- Brabletz T., Hlubek F., Spaderna S., Schmalhofer O., Hiendlmeyer E., Jung A., and Kirchner T.. 2005. Invasion and metastasis in colorectal cancer: epithelial-mesenchymal transition, mesenchymal-epithelial transition, stem cells and beta-catenin. Cells Tissues Organs. 179:56–65. 10.1159/000084509 [DOI] [PubMed] [Google Scholar]
- Cao Q., Yu J., Dhanasekaran S.M., Kim J.H., Mani R.S., Tomlins S.A., Mehra R., Laxman B., Cao X., Yu J., et al. 2008. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 27:7274–7284. 10.1038/onc.2008.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.S., and Mellman I.. 2017. Elements of cancer immunity and the cancer-immune set point. Nature. 541:321–330. 10.1038/nature21349 [DOI] [PubMed] [Google Scholar]
- Dawson M.A., and Kouzarides T.. 2012. Cancer epigenetics: from mechanism to therapy. Cell. 150:12–27. 10.1016/j.cell.2012.06.013 [DOI] [PubMed] [Google Scholar]
- Deer E.L., González-Hernández J., Coursen J.D., Shea J.E., Ngatia J., Scaife C.L., Firpo M.A., and Mulvihill S.J.. 2010. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 39:425–435. 10.1097/MPA.0b013e3181c15963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Wu Y., Wang Y., Wang C., Kang T., Rychahou P.G., Chi Y.I., Evers B.M., and Zhou B.P.. 2013. Interaction with Suv39H1 is critical for Snail-mediated E-cadherin repression in breast cancer. Oncogene. 32:1351–1362. 10.1038/onc.2012.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., and Lash A.E.. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga M.F., Ballestar E., Villar-Garea A., Boix-Chornet M., Espada J., Schotta G., Bonaldi T., Haydon C., Ropero S., Petrie K., et al. 2005. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 37:391–400. 10.1038/ng1531 [DOI] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadban T., Dibbern J.L., Reeh M., Miro J.T., Tsui T.Y., Wellner U., Izbicki J.R., Güngör C., and Vashist Y.K.. 2017. HSP90 is a promising target in gemcitabine and 5-fluorouracil resistant pancreatic cancer. Apoptosis. 22:369–380. 10.1007/s10495-016-1332-4 [DOI] [PubMed] [Google Scholar]
- Ginnebaugh K.R., Ahmad A., and Sarkar F.H.. 2014. The therapeutic potential of targeting the epithelial-mesenchymal transition in cancer. Expert Opin. Ther. Targets. 18:731–745. 10.1517/14728222.2014.909807 [DOI] [PubMed] [Google Scholar]
- Gregoire P., and Reeder J.. 2014. HTSeqGenie: A NGS analysis pipeline. package version 3.16.0.
- Hernandez N. 1985. Formation of the 3′ end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 4:1827–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Wang C.Y., Gou S.M., Wu H.S., Liu T., and Xiong J.X.. 2008. Isolation and biological analysis of tumor stem cells from pancreatic adenocarcinoma. World J. Gastroenterol. 14:3903–3907. 10.3748/wjg.14.3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly M.K., Boareto M., Huang B., Jia D., Lu M., Ben-Jacob E., Onuchic J.N., and Levine H.. 2015. Implications of the Hybrid Epithelial/Mesenchymal Phenotype in Metastasis. Front. Oncol. 5:155 10.3389/fonc.2015.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.A., Issa J.P., and Baylin S.. 2016. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 17:630–641. 10.1038/nrg.2016.93 [DOI] [PubMed] [Google Scholar]
- Karachentsev D., Sarma K., Reinberg D., and Steward R.. 2005. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 19:431–435. 10.1101/gad.1263005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z., Liu H., Degenhardt J., Mayba O., Gnad F., Liu J., et al. 2015. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 33:306–312. 10.1038/nbt.3080 [DOI] [PubMed] [Google Scholar]
- Kourmouli N., Jeppesen P., Mahadevhaiah S., Burgoyne P., Wu R., Gilbert D.M., Bongiorni S., Prantera G., Fanti L., Pimpinelli S., et al. 2004. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J. Cell Sci. 117:2491–2501. 10.1242/jcs.01238 [DOI] [PubMed] [Google Scholar]
- Kumar P., Goh G., Wongphayak S., Moreau D., and Bard F.. 2013. ScreenSifter: analysis and visualization of RNAi screening data. BMC Bioinformatics. 14:290 10.1186/1471-2105-14-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C.W., Chen Y., Shi W., and Smyth G.K.. 2014. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15:R29 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Villarreal-Ponce A., Fallahi M., Ovadia J., Sun P., Yu Q.C., Ito S., Sinha S., Nie Q., and Dai X.. 2014. Transcriptional mechanisms link epithelial plasticity to adhesion and differentiation of epidermal progenitor cells. Dev. Cell. 29:47–58. 10.1016/j.devcel.2014.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Kong D., Ahmad A., Bao B., and Sarkar F.H.. 2013. Pancreatic cancer stem cells: emerging target for designing novel therapy. Cancer Lett. 338:94–100. 10.1016/j.canlet.2012.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Nie F., Wang S., and Li L.. 2011. Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc. Natl. Acad. Sci. USA. 108:3116–3123. 10.1073/pnas.1009353108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., and Mesirov J.P.. 2011. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 27:1739–1740. 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., and Tamayo P.. 2015. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1:417–425. 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M., Mazzetta J., Nelson-Rees W., Kaplan M., and Todaro G.. 1975. Establishment of a continuous tumor-cell line (panc-1) from a human carcinoma of the exocrine pancreas. Int. J. Cancer. 15:741–747. 10.1002/ijc.2910150505 [DOI] [PubMed] [Google Scholar]
- Lim S., Janzer A., Becker A., Zimmer A., Schüle R., Buettner R., and Kirfel J.. 2010. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 31:512–520. 10.1093/carcin/bgp324 [DOI] [PubMed] [Google Scholar]
- Lin Y., Wu Y., Li J., Dong C., Ye X., Chi Y.I., Evers B.M., and Zhou B.P.. 2010. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 29:1803–1816. 10.1038/emboj.2010.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo B., Strasser G., Sagolla M., Austin C.D., Junttila M., and Mellman I.. 2012. Lkb1 regulates organogenesis and early oncogenesis along AMPK-dependent and -independent pathways. J. Cell Biol. 199:1117–1130. 10.1083/jcb.201208080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maile T.M., Izrael-Tomasevic A., Cheung T., Guler G.D., Tindell C., Masselot A., Liang J., Zhao F., Trojer P., Classon M., and Arnott D.. 2015. Mass spectrometric quantification of histone post-translational modifications by a hybrid chemical labeling method. Mol. Cell. Proteomics. 14:1148–1158. 10.1074/mcp.O114.046573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., et al. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 133:704–715. 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne T.A., Zhao K., and Hess J.L.. 2009. Chromatin immunoprecipitation (ChIP) for analysis of histone modifications and chromatin-associated proteins. Methods Mol. Biol. 538:409–423. 10.1007/978-1-59745-418-6_21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.P., Tong F., et al. 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 10:515–527. 10.1016/j.ccr.2006.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M.A. 2013. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 342:1234850 10.1126/science.1234850 [DOI] [PubMed] [Google Scholar]
- Pattabiraman D.R., Bierie B., Kober K.I., Thiru P., Krall J.A., Zill C., Reinhardt F., Tam W.L., and Weinberg R.A.. 2016. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science. 351:aad3680 10.1126/science.aad3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H., Ballestar E., Esteller M., and Cano A.. 2004. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell. Biol. 24:306–319. 10.1128/MCB.24.1.306-319.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H., Del Carmen Iglesias-de la Cruz M., Olmeda D., Csiszar K., Fong K.S., Vega S., Nieto M.A., Cano A., and Portillo F.. 2005. A molecular role for lysyl oxidase-like 2 enzyme in snail regulation and tumor progression. EMBO J. 24:3446–3458. 10.1038/sj.emboj.7600781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim A.D., Mirek E.T., Aiello N.M., Maitra A., Bailey J.M., McAllister F., Reichert M., Beatty G.L., Rustgi A.K., Vonderheide R.H., et al. 2012. EMT and dissemination precede pancreatic tumor formation. Cell. 148:349–361. 10.1016/j.cell.2011.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca H., Hernandez J., Weidner S., McEachin R.C., Fuller D., Sud S., Schumann T., Wilkinson J.E., Zaslavsky A., Li H., et al. 2013. Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS One. 8:e76773 10.1371/journal.pone.0076773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayan A.E., Griffiths T.R., Pal R., Browne G.J., Ruddick A., Yagci T., Edwards R., Mayer N.J., Qazi H., Goyal S., et al. 2009. SIP1 protein protects cells from DNA damage-induced apoptosis and has independent prognostic value in bladder cancer. Proc. Natl. Acad. Sci. USA. 106:14884–14889. 10.1073/pnas.0902042106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel C., and Weinberg R.A.. 2012. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin. Cancer Biol. 22:396–403. 10.1016/j.semcancer.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., Reinberg D., and Jenuwein T.. 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18:1251–1262. 10.1101/gad.300704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G., Sengupta R., Kubicek S., Malin S., Kauer M., Callén E., Celeste A., Pagani M., Opravil S., De La Rosa-Velazquez I.A., et al. 2008. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 22:2048–2061. 10.1101/gad.476008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinchi Y., Hieda M., Nishioka Y., Matsumoto A., Yokoyama Y., Kimura H., Matsuura S., and Matsuura N.. 2015. SUV420H2 suppresses breast cancer cell invasion through down regulation of the SH2 domain-containing focal adhesion protein tensin-3. Exp. Cell Res. 334:90–99. 10.1016/j.yexcr.2015.03.010 [DOI] [PubMed] [Google Scholar]
- Singh A., and Settleman J.. 2010. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 29:4741–4751. 10.1038/onc.2010.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Greninger P., Rhodes D., Koopman L., Violette S., Bardeesy N., and Settleman J.. 2009. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 15:489–500. 10.1016/j.ccr.2009.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Washington M.K., and Crawford H.C.. 2010. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res. 70:2115–2125. 10.1158/0008-5472.CAN-09-2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K., Friedman B.A., Larson J.L., Lauffer B.E., Goldstein L.D., Appling L.L., Borneo J., Poon C., Ho T., Cai F., et al. 2016. Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat. Commun. 7:11295 10.1038/ncomms11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talasz H., Lindner H.H., Sarg B., and Helliger W.. 2005. Histone H4-lysine 20 monomethylation is increased in promoter and coding regions of active genes and correlates with hyperacetylation. J. Biol. Chem. 280:38814–38822. 10.1074/jbc.M505563200 [DOI] [PubMed] [Google Scholar]
- Tam W.L., and Weinberg R.A.. 2013. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat. Med. 19:1438–1449. 10.1038/nm.3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J.P. 2003. Epithelial-mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 15:740–746. 10.1016/j.ceb.2003.10.006 [DOI] [PubMed] [Google Scholar]
- Thiery J.P., Acloque H., Huang R.Y., and Nieto M.A.. 2009. Epithelial-mesenchymal transitions in development and disease. Cell. 139:871–890. 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Tsai J.H., and Yang J.. 2013. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 27:2192–2206. 10.1101/gad.225334.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova M., Gehling V.S., Gustafson A., Arora S., Tindell C.A., Wilson C., Williamson K.E., Guler G.D., Gangurde P., Manieri W., et al. 2016. An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat. Chem. Biol. 12:531–538. 10.1038/nchembio.2085 [DOI] [PubMed] [Google Scholar]
- von Burstin J., Eser S., Paul M.C., Seidler B., Brandl M., Messer M., von Werder A., Schmidt A., Mages J., Pagel P., Schnieke A., Schmid R.M., Schneider G., and Saur D.. 2009. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology. 137:361–371. [DOI] [PubMed] [Google Scholar]
- Warzecha C.C., and Carstens R.P.. 2012. Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT). Semin. Cancer Biol. 22:417–427. 10.1016/j.semcancer.2012.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C., Nicholes K., Bustos D., Lin E., Song Q., Stephan J.P., Kirkpatrick D.S., and Settleman J.. 2014a Overcoming EMT-associated resistance to anti-cancer drugs via Src/FAK pathway inhibition. Oncotarget. 5:7328–7341. 10.18632/oncotarget.2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C., Ye X., Pham T., Lin E., Chan S., McNamara E., Neve R.M., Belmont L., Koeppen H., Yauch R.L., et al. 2014b AXL inhibition sensitizes mesenchymal cancer cells to antimitotic drugs. Cancer Res. 74:5878–5890. 10.1158/0008-5472.CAN-14-1009 [DOI] [PubMed] [Google Scholar]
- Witta S.E., Gemmill R.M., Hirsch F.R., Coldren C.D., Hedman K., Ravdel L., Helfrich B., Dziadziuszko R., Chan D.C., Sugita M., et al. 2006. Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res. 66:944–950. 10.1158/0008-5472.CAN-05-1988 [DOI] [PubMed] [Google Scholar]
- Wu D., and Smyth G.K.. 2012. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 40:e133 10.1093/nar/gks461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.D., and Nacu S.. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 26:873–881. 10.1093/bioinformatics/btq057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H.Q., Carr K., and Abbruzzese J.L.. 2006. Cytotoxic chemotherapy for pancreatic cancer: Advances to date and future directions. Drugs. 66:1059–1072. 10.2165/00003495-200666080-00003 [DOI] [PubMed] [Google Scholar]
- Yang J., and Weinberg R.A.. 2008. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 14:818–829. 10.1016/j.devcel.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Yauch R.L., Januario T., Eberhard D.A., Cavet G., Zhu W., Fu L., Pham T.Q., Soriano R., Stinson J., Seshagiri S., et al. 2005. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin. Cancer Res. 11:8686–8698. 10.1158/1078-0432.CCR-05-1492 [DOI] [PubMed] [Google Scholar]
- Yin T., Wei H., Gou S., Shi P., Yang Z., Zhao G., and Wang C.. 2011. Cancer stem-like cells enriched in Panc-1 spheres possess increased migration ability and resistance to gemcitabine. Int. J. Mol. Sci. 12:1595–1604. 10.3390/ijms12031595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama Y., Matsumoto A., Hieda M., Shinchi Y., Ogihara E., Hamada M., Nishioka Y., Kimura H., Yoshidome K., Tsujimoto M., and Matsuura N.. 2014. Loss of histone H4K20 trimethylation predicts poor prognosis in breast cancer and is associated with invasive activity. Breast Cancer Res. 16:R66 10.1186/bcr3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.