In yeast, the TRAPP complexes activate Rab1 with TRAPPII also activating Rab11, but less is known about the two TRAPPs in metazoans. Riedel et al. show that in Drosophila melanogaster, TRAPPIII is an essential Rab1 activator, and TRAPPII activates Rab1 and Rab11 and becomes essential when an unrelated Rab11 activator is deleted.
Abstract
Originally identified in yeast, transport protein particle (TRAPP) complexes are Rab GTPase exchange factors that share a core set of subunits. TRAPPs were initially found to act on Ypt1, the yeast orthologue of Rab1, but recent studies have found that yeast TRAPPII can also activate the Rab11 orthologues Ypt31/32. Mammals have two TRAPP complexes, but their role is less clear, and they contain subunits that are not found in the yeast complexes but are essential for cell growth. To investigate TRAPP function in metazoans, we show that Drosophila melanogaster have two TRAPP complexes similar to those in mammals and that both activate Rab1, whereas one, TRAPPII, also activates Rab11. TRAPPII is not essential but becomes so in the absence of the gene parcas that encodes the Drosophila orthologue of the SH3BP5 family of Rab11 guanine nucleotide exchange factors (GEFs). Thus, in metazoans, Rab1 activation requires TRAPP subunits not found in yeast, and Rab11 activation is shared by TRAPPII and an unrelated GEF that is metazoan specific.
Introduction
Rab GTPases control many aspects of subcellular organization. They are typically active on only one particular organelle or vesicle class and so direct the subcellular localization of a wide range of proteins, including membrane traffic machinery, molecular motors, and regulators of phosphoinositide levels or the activity of other GTPases (Stenmark, 2009; Hutagalung and Novick, 2011; Gillingham et al., 2014). This role in spatial organization of the cell requires specific guanine nucleotide exchange factors (GEFs) to activate each Rab in only the correct location (Barr, 2013; Ishida et al., 2016). GEFs for several Rabs have been identified, and among the best studied are the transport protein particle (TRAPP) complexes (Barrowman et al., 2010; Brunet and Sacher, 2014; Kim et al., 2016). The first TRAPP subunit was identified in yeast in a screen for mutations that interact with a mutation in a SNARE protein, and the corresponding protein was found to be part of a large protein complex that was termed TRAPP (Rossi et al., 1995; Sacher et al., 1998). Subsequent work reported the existence of three different TRAPP complexes in yeast (Barrowman et al., 2010; Choi et al., 2011; Brunet and Sacher, 2014; Kim et al., 2016). All three share a heptameric core of six proteins (Bet3 being present twice), with TRAPPI having no further subunits, TRAPPII having four additional subunits called Tca17, Trs65, Trs120, and Trs130, and TRAPPIII having one additional subunit, Trs85. The shared TRAPP subunits are essential for membrane traffic through the Golgi apparatus, and consistent with this, TRAPPI was found to act as a GEF for Ypt1 (yeast Rab1), a GTPase essential for ER to Golgi and intra-Golgi traffic (Wang et al., 2000; Kim et al., 2006; Cai et al., 2008). TRAPPIII was initially reported to have a more specific role in activating Ypt1 during autophagy, but recent work suggests that TRAPPI may not exist in vivo and that TRAPPIII is responsible for the majority of Rab1 exchange activity in both secretion and autophagy (Meiling-Wesse et al., 2005; Lynch-Day et al., 2010; Thomas et al., 2017). In contrast, TRAPPII was proposed to act later in the Golgi as a GEF for the closely related GTPases Ypt31 and Ypt32, yeast orthologues of Rab11 (Jones et al., 2000; Morozova et al., 2006). This conclusion was initially questioned, but recent biochemical studies have shown both Rab1 and Rab11 GEF activity for TRAPPII from filamentous fungi and budding yeasts (Wang and Ferro-Novick, 2002; Pinar et al., 2015; Thomas and Fromme, 2016).
The shared core TRAPP subunits that are sufficient to act on Ypt1/Rab1 are very highly conserved in evolution and appear to be a universal feature of eukaryotic cells (Koumandou et al., 2007). Mammals have orthologues of all of the yeast TRAPP subunits, including those specific to TRAPPII and TRAPPIII. In addition, coprecipitation experiments have identified two further TRAPP subunits that are not present in yeast (Gavin et al., 2002; Scrivens et al., 2011). Examination of the proteins associated with each mammalian TRAPP subunit revealed that they form two complexes related to yeast TRAPPII and TRAPPIII, with there being no evidence that mammals have a complex equivalent to TRAPPI, i.e., just the core subunits (Choi et al., 2011; Bassik et al., 2013; Borner et al., 2014). Mammalian TRAPPII contains seven core subunits and orthologues of Trs120 (TRAPPC9) and Trs130 (TRAPPC10). Mammalian TRAPPIII contains the same seven core subunits and an orthologue of Trs85 (TRAPPC8) plus three further subunits: TRAPPC13 (an orthologue of yeast Trs65) and the two subunits not found in yeast (TRAPPC11 and TRAPPC12).
The precise roles of TRAPPII and TRAPPIII in mammals are not fully resolved. When assembled in vitro, the core subunits of the mammalian TRAPP complexes have exchange activity on Rab1 (Kim et al., 2006), and mammalian TRAPPII has been reported to have the same activity when immunoisolated from cells but to have no activity on Rab11 (Yamasaki et al., 2009). Moreover, there are also some striking differences to the yeast system. The most obvious is the existence of the two additional subunits in TRAPPIII, TRAPPC11 and TRAPPC12, and these seem unlikely to have minor roles as at least TRAPPC11 is essential for secretion and cell viability (Wendler et al., 2010; Scrivens et al., 2011; Kim et al., 2016). In contrast, the TRAPPII subunits Trs120 and Trs130 are both essential for growth in yeast, and yet in mammals they do not appear to be required for cell viability even though Rab11 is an essential protein (Jankovics et al., 2001; Kim et al., 2016). Indeed, loss-of-function mutations in human TRAPPC9 are not lethal but cause mental retardation (Mir et al., 2009; Mochida et al., 2009; Philippe et al., 2009).
The TRAPP complex subunits found in humans are well conserved across metazoans, and so we have used the tractable genetic system of Drosophila melanogaster to investigate TRAPP in metazoans. We found that Drosophila contain two TRAPP complexes that have the same composition as the human complexes, and we combined genetics and expression of recombinant TRAPP complexes to investigate their function in vivo and their activity in vitro.
Results
Purification of Drosophila TRAPP complexes
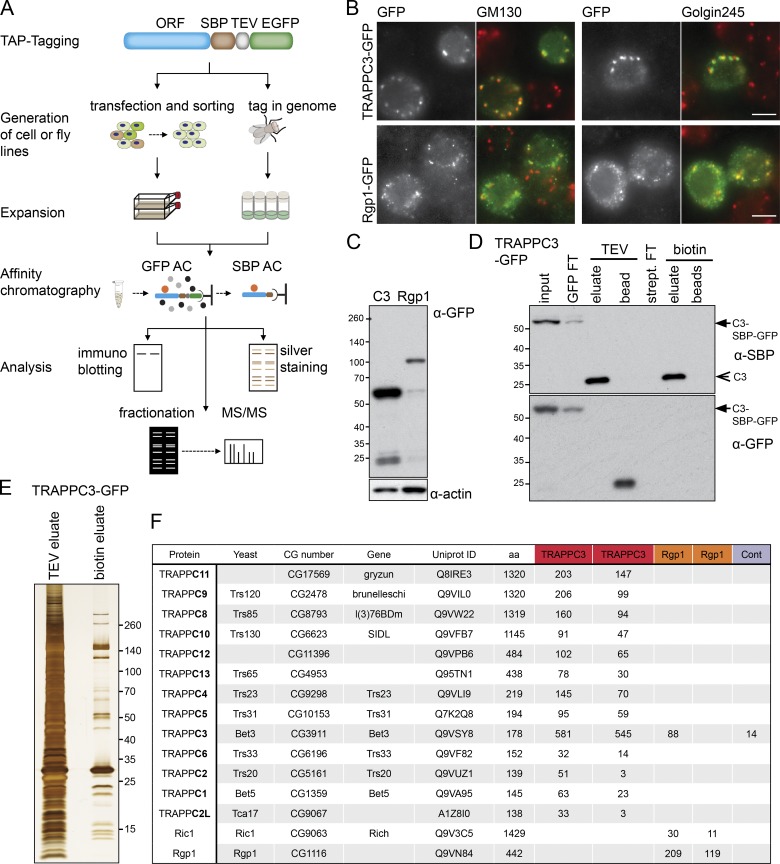
To investigate the composition of the TRAPP complexes in Drosophila, we used a tandem affinity purification (TAP) approach to isolate them from the S2 Drosophila cell line (Fig. 1 A). Stably transfected cell lines were generated expressing the TRAPPC3 subunit with a C-terminal TAP tag based on GFP and the streptavidin-binding peptide (SBP) tag. TRAPPC3 was chosen because its yeast orthologue, Bet3, is a core component of all TRAPP complexes, and C-terminal tagging does not compromise its activity (Sacher et al., 1998). In parallel and as a control, we also generated cells lines expressing TAP-tagged forms of the Drosophila orthologue of Rgp1, a subunit of another complex proposed in other species to have GEF activity on a different Golgi Rab: in yeast, Rgp1 forms a complex with Ric1 that has exchange activity on Rab6, and both proteins are conserved in flies (Siniossoglou et al., 2000; Tong et al., 2011).
Figure 1.
The Rab GEFs of the Drosophila Golgi apparatus. (A) Cartoon of the SBP-GFP TAP tag and the strategy used to make stable cell lines or fly lines by CRISPR and then analyzing the tagged proteins in cell lines or flies. AC, affinity chromatography; MS/MS, tandem mass spectrometry. (B) Widefield micrographs of cells transiently transfected with TAP-tagged Golgi GEFs and labeled with antibodies to the indicated Golgi markers. Images are representative of at least two independent experiments, with at least three micrographs obtained from each. Bars, 5 µm. (C) Immunoblot of stable cell lines expressing SBP-GFP–tagged TRAPPC3 or Rgp1. (D) TAP of TRAPPC3 from stably transfected cell lines. The purification was on anti-GFP beads with elution by TEV protease and then on streptavidin beads with elution by biotin. The starting tagged protein (arrows) and the final purified protein (arrowhead) are indicated. FT, flowthrough. (E) Silver-stained protein gel comparing single-step affinity chromatography of TRAPPC3–SBP-GFP using the GFP tag (TEV eluate) with TAP using both the GFP and the SBP tag (biotin eluate). Molecular masses are given in kilodaltons. (F) Mass spectrometry analysis of TAPs of SBP-GFP–tagged TRAPPC3 and Rgp1. For each protein that was identified by mass spectrometry, the total spectral counts are shown for the TRAPPC3 or Rgp1 purifications. Duplicates are shown. CG, computed gene; Cont., untransfected cells.
Both TAP-tagged TRAPPC3 and Rgp1 were localized to the Golgi apparatus when expressed in S2 cells (Fig. 1, B and C). Thus, both were purified from S2 cell extracts by first binding them to anti-GFP beads and eluting with protease and then by binding them to streptavidin beads and eluting with biotin. This two-step purification allowed a high degree of enrichment under conditions that are nondenaturing and hence should maintain interactions with stable binding partners (Fig. 1, D and E).
Drosophila TRAPPC3 is found in two distinct TRAPP complexes
To identify the proteins that copurified with TRAPPC3 and Rgp1, we used mass spectrometric analysis of tryptic peptides in two independent experiments (Fig. 1 F). Rgp1 was found to copurify with Rich, the Drosophila orthologue of yeast Ric1. No other proteins were found to be specifically associated with Rgp1, suggesting that in metazoans, this complex comprises the same two subunits as found in yeast. A previous study of Rich found no binding to Drosophila Rgp1, but the form of Rgp1 tested lacked the N-terminal 82 residues present in the construct used in this study (Tong et al., 2011).
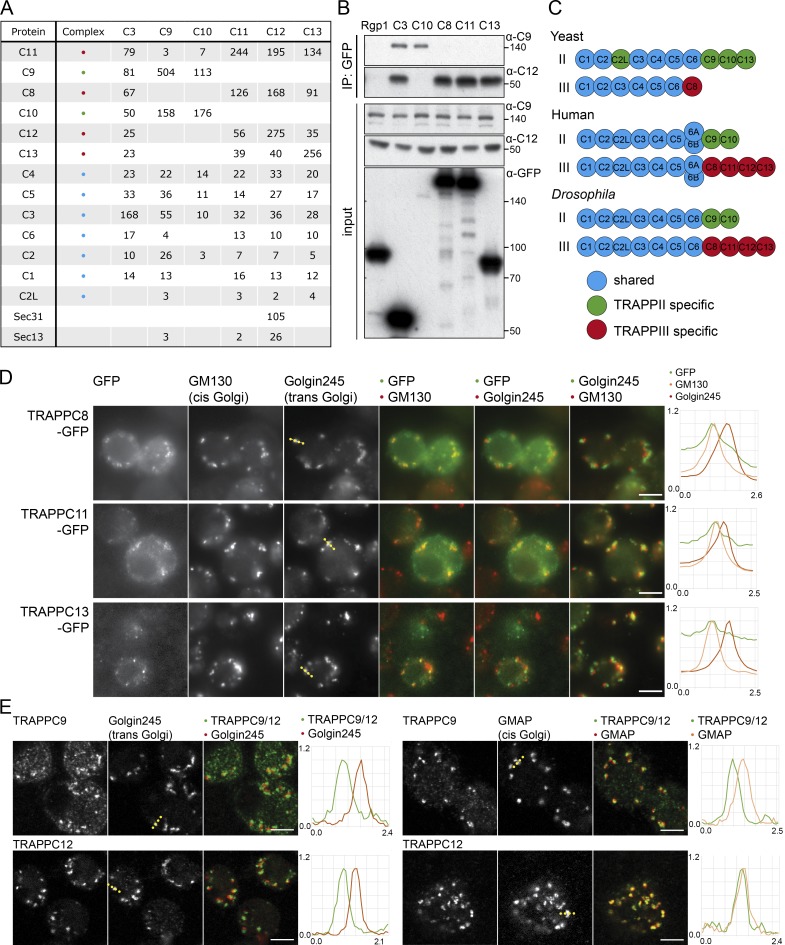
When we precipitated TRAPPC3, it was found to associate with all 12 of the Drosophila orthologues of the other known mammalian TRAPP subunits (Fig. 1 F). In other species, the TRAPP complexes consist of a shared core of small subunits (TRAPPC1–6) along with larger subunits that are complex specific (TRAPPC8–13; Brunet and Sacher, 2014; Kim et al., 2016). We thus generated stable cell lines expressing TAP-tagged forms of the larger TRAPP subunits and used two-step affinity purification and mass spectrometry to determine the subunit composition of the associated complexes (Fig. S1 A). In two independent experiments, precipitation of the large subunits revealed the same two patterns of interaction. TRAPPC9 and TRAPPC10 coprecipitated each other but not the other large subunits, whereas TRAPPC11, TRAPPC12, and TRAPPC13 consistently coprecipitated each other along with TRAPPC8 (Fig. 2 A with replicates in Fig. S1 B). All of the seven small subunits TRAPPC1–6 were detected with all of the larger subunits in most experiments, with the occasional apparent absence of a particular small subunit being likely to be stochastic because of the small number of peptides as it was not seen reproducibly. We next generated antibodies against two of the complex-specific subunits, TRAPPC9 and TRAPPC12, and could confirm the specificity of the interactions found by mass spectrometry (Fig. 2 B). This apparent division into two distinct complexes is entirely consistent with a previous analysis in human cells (Fig. 2 C), where the C9–C10 complex was termed TRAPPII and the C8–C11–C12–C13 complex termed TRAPPIII (Bassik et al., 2013). This indicates that Drosophila provide a good model system to study the role of the metazoan TRAPP complexes and their subunits, and for consistency, we will use the same complex and subunit nomenclature as that used in humans.
Figure 2.
The TRAPP complexes of Drosophila. (A) Mass spectrometry analysis of TRAPPC3, TRAPPC9, TRAPPC10, TRAPPC11, TRAPPC12, and TRAPPC13 tandem affinity purified from stably transfected cell lines. Total spectral counts for each protein are shown, with replicates shown in Fig. S1 B. Precipitations also contained lower levels of abundant chaperones and cytosolic enzymes. With the shared TRAPP subunit TRAPPC3, only three proteins apart from known TRAPP subunits were consistently present across all TAP purifications and several single-step purifications (catalase, ribosomal protein S26, and Gdi), and these were only present at trace levels in the TAP samples and so were not pursued further. (B) Immunoblot using the TRAPPC9 and TRAPPC12 antibodies of proteins associated with Rgp1, TRAPPC3, TRAPPC8, TRAPPC10, TRAPPC11, and TRAPPC13 after anti-GFP affinity purification from lysates of transiently transfected S2 cells. Molecular masses are given in kilodaltons. IP, immunoprecipitation. (C) Subunits present in the different the TRAPP complexes of yeast, flies, and humans. (D) Widefield micrographs of S2 cells transiently expressing GFP-tagged TRAPPC8, TRAPPC11, and TRAPPC13 and stained with antibodies against Golgin245 and GM130. Line scans of fluorescence intensity relative to the maximum are from representative Golgi stacks (dotted line on Golgin245 panels). The TRAPP subunits are enriched at the cis end of the stack. (E) Confocal micrographs of S2 cells labeled with antisera to endogenous TRAPPC9 (TRAPPII) or TRAPPC12 (TRAPPIII) and the Golgi proteins Golgin245 or GMAP. Line scans are from representative Golgi stacks (dotted line on Golgi marker panels). Bars, 5 µm. Images in D and E are representative of at least two independent experiments, with at least three micrographs obtained from each.
Localization of the TRAPP complexes in Drosophila cultured cells
To determine the subcellular location of the TRAPP complexes, we initially examined the TAP-tagged subunits in S2 cells. All of the four subunits that are TRAPPIII-specific were Golgi localized and showed extensive colocalization with the cis-Golgi marker GM130 but a displacement from the trans-Golgi protein Golgin245, indicating a location toward the cis end of the stack (Figs. 2 D and S1 C). Consistent with this, the antibody raised against endogenous TRAPPC12 also showed colocalization with a cis-Golgi marker (Fig. 2 E). The TRAPPII subunits proved harder to localize because when expressed at readily detectable levels, they had a diffuse distribution. We interpret this as being caused by the complex being present at lower levels, and therefore the overexpressed individual subunits were mostly mislocalized as they were mostly not incorporated into the rest of the complex. In mammalian cultured cells, TRAPPII-specific subunits were present at ∼10-fold lower levels than TRAPPIII subunits (Borner et al., 2014). However, antisera to TRAPPC9 again showed Golgi labeling close to the cis-Golgi marker (Fig. 2 E). Collectively, these results indicate that in S2-cultured cells, both TRAPP complexes are associated with the cis-Golgi rather than the trans-Golgi.
Composition and localization of TRAPP complexes in Drosophila tissues
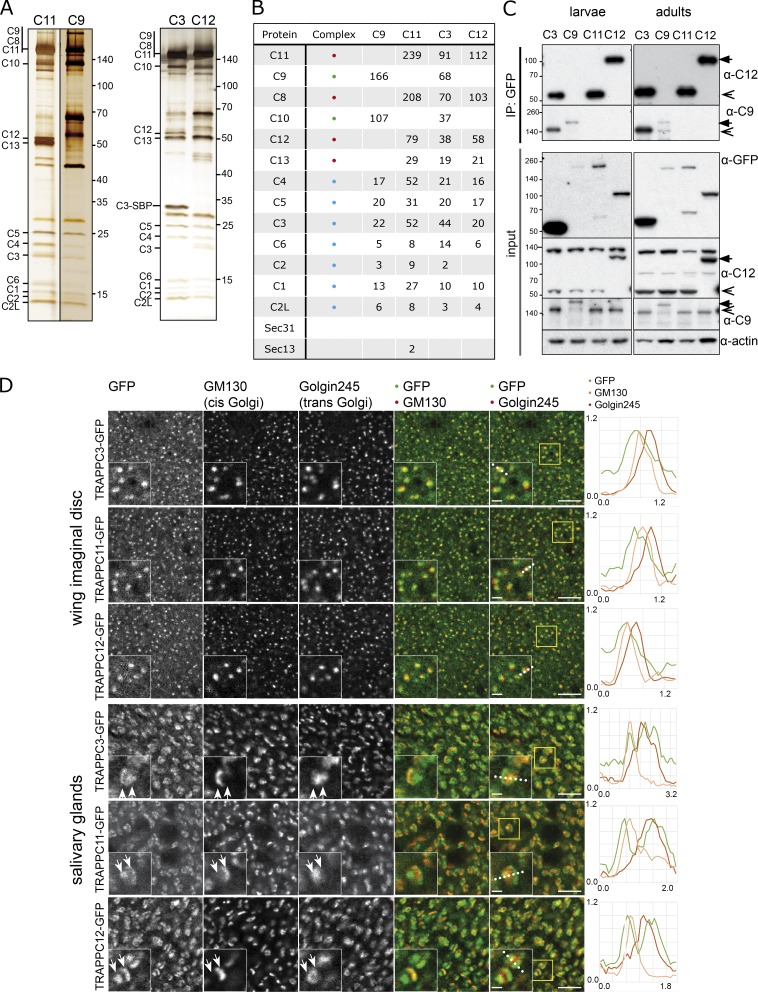
These experiments rely on cultured cells and expressing the tagged TRAPP subunits in the context of the endogenous untagged protein. To instead investigate the behavior of tagged subunits in their native situation, we used CRISPR/Cas9-mediated homologous recombination to attach the SBP-GFP TAP tag to the C terminus of TRAPP subunits genes in the Drosophila genome. For this, we selected TRAPPC3 as being in both complexes and TRAPPC9, TRAPPC11, and TRAPPC12 as complex-specific subunits. All four tagged alleles produced viable flies when homozygous, and all four resulted in the expression of TAP-tagged proteins, which could be isolated from embryos and analyzed by mass spectrometry (Fig. 3, A and B). This revealed that as in the S2 cells, TRAPPC3 associated with all TRAPP subunits, whereas the specific subunits showed the same pattern of interaction seen in S2 cells, indicating that TRAPPII and TRAPPIII exist in the native context (Fig. 3 B). This dichotomy into two complexes was also found in larvae and adults when the complexes were isolated and examined by immunoblotting (IB; Fig. 3 C).
Figure 3.
TRAPPII and TRAPPIII are present in embryos, larvae, and adults. (A) Silver-stained protein gels of TAPs of SBP-GFP–tagged TRAPPC11, TRAPPC9, TRAPPC3, and TRAPPC12 from embryos. (B) Mass spectrometry analysis of proteins associated with SBP-GFP–tagged TRAPPC11, TRAPPC9, TRAPPC3, and TRAPPC12 after TAP from embryos. Total spectral counts for each protein are shown. (C) Immunoblot analysis using the TRAPPC9 and TRAPPC12 antibodies of proteins associated with TRAPPC3, TRAPPC9, TRAPPC11, and TRAPPC12 after anti-GFP immunoprecipitation (IP) from lysates prepared from adults (left) or wandering third instar larvae (right) carrying SBP-GFP–tagged alleles. Arrows indicate GFP-tagged TRAPP subunits; arrowheads indicate native TRAPP subunits. Molecular masses are given in kilodaltons. (D) Confocal micrographs of SBP-GFP–tagged TRAPPC3, TRAPPC11, and TRAPPC12 in wing imaginal discs and salivary glands dissected from wandering third instar larvae and stained for GFP, GM130, and Golgin245. In each case, the tag was inserted at the genomic locus, and therefore, the tagged protein was expressed from its native promoter. Line scans are from representative Golgi stacks (dotted lines in GFP/Golgin245 panels). In the salivary gland, all three proteins were discretely located to both the cis and trans ends of the stack (arrows in insets), with a similar bimodal distribution also observed in follicle cells (not depicted). Bars: (main images) 5 µm; (insets) 1 µm. Images are representative of at least two independent experiments, with at least three micrographs obtained from each.
Examination of the subcellular distribution of the tagged subunits in tissues revealed that it was possible to detect TRAPPC3 and the TRAPPIII-specific subunits but not TRAPPII-specific TRAPPC9 in both wing discs and the salivary gland (Fig. 3 D). Interestingly, in salivary glands, TRAPPIII showed a striking bimodal distribution in the Golgi stack with a clear concentration at both the cis and trans sides of the stack. The trans peak colocalized better with the clathrin adapter AP-1 than Golgin245 and thus appeared to correspond with the trans-Golgi network (Figs. 3 D and S1 D). This bimodal distribution was seen with both TRAPPC11 and TRAPPC12 as well as with TRAPPC3 and clearly indicates that TRAPPIII is present on both ends of the Golgi stack in at least some tissues.
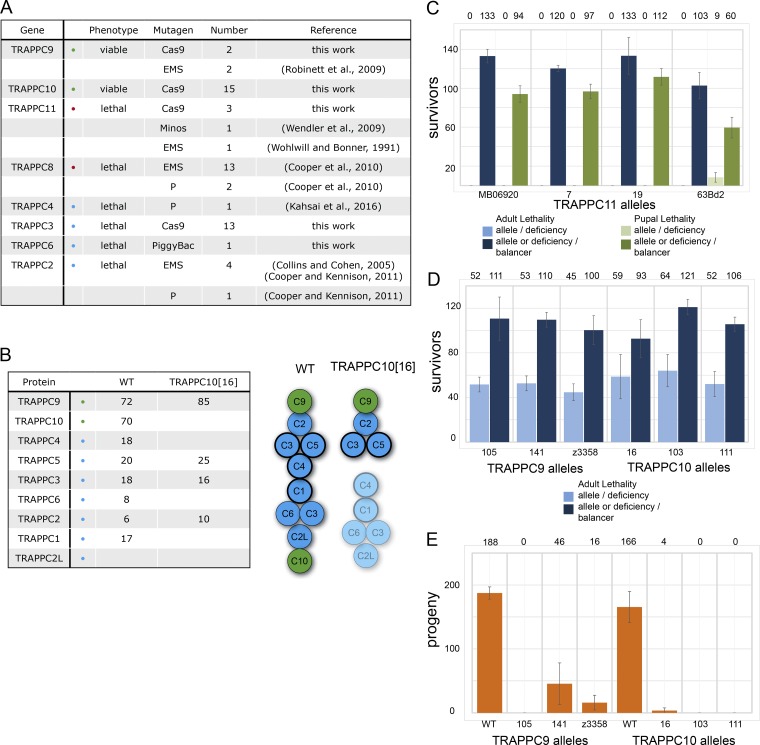
Generation of null alleles in TRAPPII- and TRAPPIII-specific subunits
Several chemical or transposon-based mutations in genes encoding TRAPP subunits have been isolated in screens for lethal genetic lesions or for defects in fertility (Fig. 4 A). This has shown that at least three of the core subunits are essential, along with TRAPPC8 and TRAPPC11 from TRAPPIII. In some cases, the genetic lesion was not reported, and by sequencing the previously reported alleles, we were able to show that the lethal ethyl methane sulfonate (EMS) mutations in TRAPPC2, TRAPPC8, and TRAPPC11 were all caused by nonsense mutations (Fig. S2 B; see also the Characterization of potential TRAPP complex alleles section of Materials and methods). In contrast, a nonsense mutation that truncates TRAPPC9 at residue 514 was reported to be viable but showed a temperature-sensitive defect in male fertility (Robinett et al., 2009). To investigate the role of the specific TRAPP complexes, we used the CRISPR/Cas9 system to generate mutations in the shared core subunit TRAPPC3 and in the complex-specific subunits TRAPPC9, TRAPPC10, and TRAPPC11 (Fig. S2, A–D). We were able to confirm the loss of TRAPPC10 protein in the relevant mutant by using TRAPPC9-GFP to precipitate the TRAPPII complex from the TRAPPC10 mutant flies (Fig. 4 B). Interestingly, in the absence of TRAPPC10, TRAPPC9 was associated with only the three core subunits, TRAPPC2, TRAPPC3, and TRAPPC5, that in mammals and yeast are present at one end of the core (Fig. 4 B; Kim et al., 2006; Cai et al., 2008). This is consistent with the finding in yeast that this part of the TRAPP core can bind to either Trs85 (TRAPPC8) or Trs120 (TRAPPC9; Tan et al., 2013; Taussig et al., 2014), therefore suggesting that TRAPPC9 has the same location in TRAPPII as TRAPPC8 occupies in TRAPPIII and raising the possibility that TRAPPC10 either enables or stabilizes the assembly of the core complex in TRAPPII.
Figure 4.
TRAPPII is required for male fertility, and TRAPPIII is essential for viability. (A) Summary of mutations generated in TRAPP subunits in this and previous studies. The nature of the lesions in the EMS alleles of TRAPPC2, TRAPPC8, and TRAPPC11 were determined by sequencing the locus from the relevant mutants (Fig. S2 B). (B) Mass spectrometry analysis of TRAPPC9–SBP-GFP tandem affinity–purified from a WT or TRAPPC10[16] mutant background. Total spectral counts for each protein are shown. Subunits with a thick line in the cartoon are essential for Rab1 nucleotide exchange activity in yeast. (C) TRAPPC11 is essential for adult (blue bars) and pupal (green bars) viability. The bars show mean numbers of adult or pupal progenies calculated from two parallel crosses (pupal lethality of TRAPPC11[MB06920]/deficiency heterozygotes and TRAPPC11[63BD2]/deficiency heterozygotes) or three parallel crosses (all other heterozygotes). The mean value is shown above each bar. Note that TRAPPC11[63BD2]/deficiency heterozygotes occasionally develop into pupae, suggesting the allele is a strong hypomorph. (D) TRAPPC9 and TRAPPC10 are not essential for adult viability. The bars show the mean number of adult progeny obtained in three independent crosses. (E) TRAPP9 and TRAPPC10 are required for male fertility. The bars show the mean number of pupal progeny from three independent crosses using males with the indicated allele over the relevant deficiency. Error bars show SD.
Examination of the phenotypes of the various mutations revealed that frameshift mutations in TRAPPC3 and TRAPPC11 were homozygous lethal, confirming that TRAPPIII is essential for viability (Fig. 4 C). In contrast, a deletion of the entire TRAPPC9 coding region and a frameshift mutation in TRAPPC10 were both homozygous viable (Fig. 4 D). For both TRAPPC9 and TRAPPC10, the loss of the protein resulted in male sterility, indicating that both proteins contribute to the same activity of TRAPPII (Fig. 4 E). This is consistent with a previous study that found that a male sterile mutation corresponded to a 514-stop lesion in TRAPPC9 and indicates that this truncated form of the protein has little residual activity (Robinett et al., 2009). Collectively, these findings indicate that the TRAPPIII complex is essential for viability, but the TRAPPII complex appears only essential for normal male fertility, indicating that the two complexes have distinct roles in vivo.
In vitro assay of the exchange activity TRAPPII and TRAPPIII
To compare the Rab exchange activity of TRAPPII and TRAPPIII, we established a method to prepare the two complexes in recombinant form. For each complex, the genes encoding all of the relevant subunits were coexpressed in insect cells using a multipromotor baculovirus vector. In both cases, cleavable tags were attached to one of the subunits, and by affinity purification, we were able to produce complete TRAPPII and TRAPPIII complexes (Fig. 5 A). The fact that each complex contains all of the subunits found in the specific TAP purifications suggests that these two complexes do indeed form in vivo and that no further subunits are required to hold them together.
Figure 5.
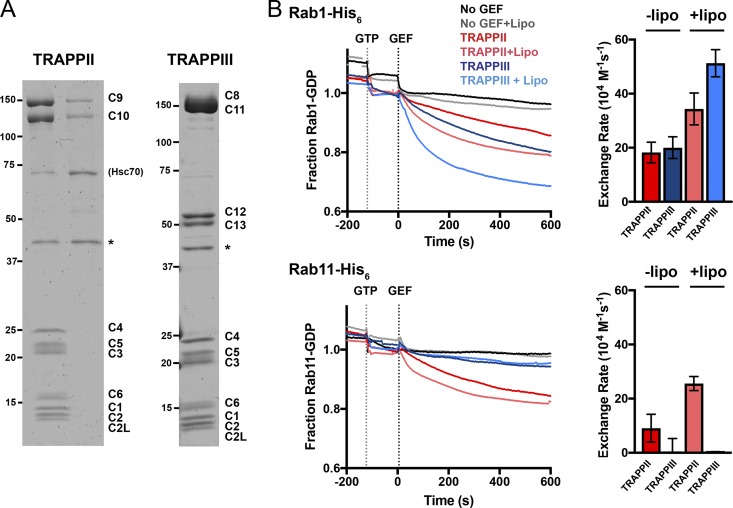
TRAPPII complex activates the Rab GTPases Rab1 and Rab11, whereas TRAPPIII only shows GEF activity toward Rab1. (A) Coomassie blue–stained protein gels of recombinant Drosophila TRAPP complexes purified from Sf9 cells coexpressing the subunits of each complex. FLAG tags on C10 or C11 allowed isolation of TRAPPII or TRAPPIII, respectively. PreScission protease (GST-HRV-3C protease) was used to cleave the tags (asterisks) and was subsequently removed using glutathione Sepharose beads. C10 also copurified with C9 in the absence of the shared subunits (TRAPPII lane). The Hsc70 chaperone (CG4264) is a contaminant of the TRAPPII purification protocol. Molecular masses are given in kilodaltons. (B) Release of mant-GDP from 250 nM of Rab-His6 by 50 nM of TRAPPII or TRAPPIII in the presence or absence of synthetic fly Golgi mix liposomes. Traces are the mean of at least three experiments. Error bars show SEM.
When the purified TRAPPII and TRAPPIII complexes were tested with Drosophila Rab1, both were able to stimulate the exchange of bound nucleotides (Fig. 5 B). A previous study of the yeast TRAPP complexes found that their exchange activity is enhanced if the relevant GTPase is attached to liposomes (Thomas and Fromme, 2016), and we also found increased nucleotide exchange with Rab1 when it was attached to the surface of liposomes. In contrast with these findings with Rab1, when we examined Rab11, we found that only TRAPPII had readily detectable exchange activity, with TRAPPIII having only minimal activity (Fig. 5 B). These activities appear specific, as neither TRAPP complex stimulated exchange on the Golgi Rabs Rab2 and Rab18 or on the endosomal Rab, Rab5 (Fig. S3, A–C). Moreover, a similar Rab11 GEF activity was found for TRAPPII when the assay was performed using a different purification procedure and a different assay for GTP exchange (Fig. S3, D and E).
Thus, it seems that as in plants and fungi, the presence of TRAPPC9 and TRAPPC10 allows the metazoan TRAPPII complex to act on Rab11. Interestingly, these two subunits form a stable complex when expressed without the shared subunits, adding further support to the notion that the two proteins act together to allow the TRAPP core complex to recognize Rab11 (Fig. 5 A).
TRAPPIII stimulates Rab1 recruitment to membranes in vivo
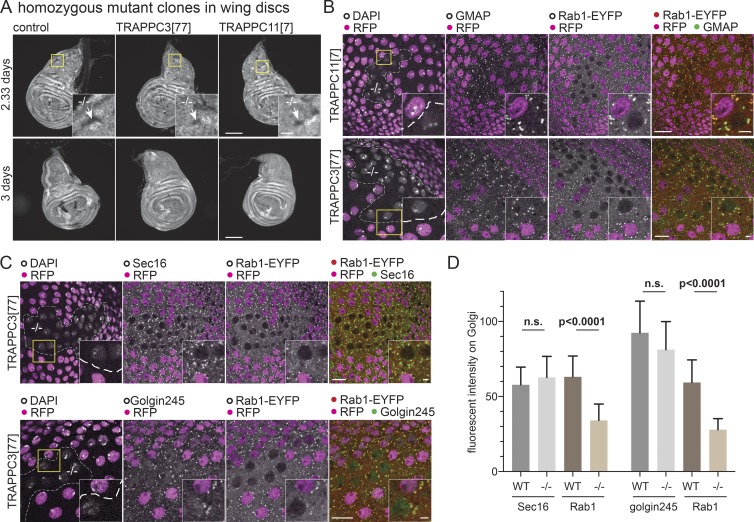
As described in the Generation of null alleles… section above, mutations in the TRAPPIII-specific subunit TRAPPC11 are lethal. However, we were able to use the FRT system in wing discs to generate clones of cells that lacked both copies of either TRAPPC3 or TRAPPC11. After 2.33 d, these clones were several cells in size, but after 3 d, clones were no longer detectable, suggesting that the cells eventually became defective and were removed (Fig. 6 A). To examine Rab1 in these clones, we generated them in a background where both alleles of Rab1 were tagged with YFP (Dunst et al., 2015). In clones of cells lacking TRAPPC3 or TRAPPC11, the levels of YFP-Rab1 on the Golgi were significantly reduced, whereas other Golgi markers were unaffected (Fig. 6, B–D). Thus, the levels of Rab1 on the Golgi are reduced in the absence of TRAPPIII, indicating that TRAPPIII is a major activator of Rab1 in vivo.
Figure 6.
TRAPPIII is required for cell growth and Rab1 localization. (A) Widefield micrographs of wing imaginal discs in which mitotic clones had been induced using the Flp-FRT system 2.33 or 3 d before fixation. This method generates homozygous clones of cells in a heterozygous background. The control clones or clones mutant for TRAPPC3[77] or TRAPPC11[7] are marked by the absence of RFP-tagged histone that is expressed from the chromosome that carries the mutation (arrows in inset zoom). Clones lacking TRAPPC3 or TRAPPC11 are only detectable at the earlier time point. Bars: (main images) 100 µm; (insets) 20 µm. (B and C) Confocal micrographs of YFP-Rab1 in clones of cells mutant for TRAPPC11[7] or TRAPPC3[77] in the peripodial cells of the wing imaginal discs costained for YFP and the Golgi markers GMAP or Golgin245 or the ER exit site marker Sec16. Mutant clones are marked by loss of RFP-tagged histone, with the nuclei stained with DAPI. Loss of TRAPPC11 or TRAPPC3 reduces YFP-Rab1 on the Golgi without affecting the other markers. Bars: (main images) 10 µm; (insets) 2 µm. Images are representative of at least two independent experiments, with at least three micrographs obtained from each. (D) Comparison of the levels on the Golgi of YFP-Rab1 and of Golgi/ER exit site markers in the homozygous mutant clones for TRAPPC3−/− versus the surrounding WT tissue. Fluorescent intensities were extracted from micrographs such as those shown in C, and in each case, n = 20 Golgi from two wing discs. Mean values are shown. Error bars show SD. The level of YFP-Rab1 showed a statistically significant reduction in the mutant clones, whereas the other markers were not significantly affected (two-tailed nonparametric Mann-Whitney test).
TRAPPII is functionally redundant with Parcas, the Drosophila orthologue of the mammalian Rab11 GEF SH3BP5
These results indicate that TRAPPII can act as an exchange factor on Rab11, but it is known that Rab11 is essential in Drosophila (Jankovics et al., 2001), whereas TRAPPC9 and TRAPPC10, the specific subunits that define TRAPPII, are clearly not essential. This implies that TRAPPII is not the only Rab11 GEF in flies, and indeed, when we examined endogenous Rab11 in the TRAPPC9-null mutant, we could not detect any obvious change in its distribution in salivary glands or wing discs (not depicted). Recently, two closely related proteins in Caenorhabditis elegans, Rei-1 and Rei-2, were identified as putative Rab11 GEFs by their ability to bind to the GDP form of Rab11, and both Rei-1 and its mammalian orthologue SH3BP5 were confirmed to have Rab11 GEF activity in vitro (Sakaguchi et al., 2015). These proteins have no detectable similarity with TRAPP subunits or any other protein family. Drosophila have a single orthologue of Rei-1 and SH3BP5, which was initially isolated as a mutant with a defect in the formation of embryonic germ cells and named poirot (Sinka et al., 2002). This gene was subsequently shown to have other roles in development, including muscle formation, and was renamed parcas (pcs; Artero et al., 2003). The original pcs allele, pcsgs, is a null mutant, but nonetheless it is adult viable, albeit with reduced egg laying (Sinka et al., 2002). We thus tested the possibility that Parcas has Rab11 GEF activity that can explain why flies are viable in the absence of TRAPPII.
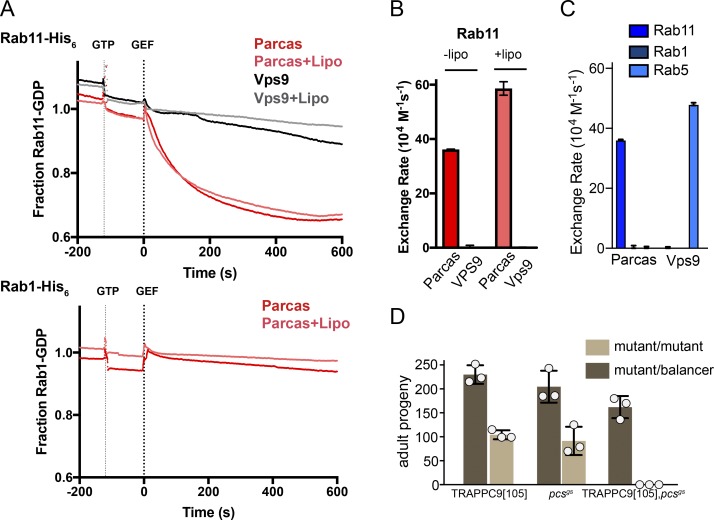
The Parcas protein is 477 residues long and it could be expressed and purified from Escherichia coli. This recombinant Parcas had GEF activity on Drosophila Rab11 but not Rab1 or Rab5, indicating that it behaves like its orthologues in C. elegans and humans (Fig. 7, A–C). The pcs gene is on the same chromosome as that encoding TRAPPC9, and so we combined the two mutations by recombination. Matings between flies carrying the single or double mutations confirmed that progeny homozygous for either of single mutations are viable; however, the double mutant stock produced no adult progeny homozygous for the chromosome carrying both mutations, indicating that loss of both genes is lethal (Fig. 7 D). These double mutant progeny completed embryogenesis but died in early larval stages. This striking genetic interaction is consistent with both TRAPPII and Pcs having Rab11 GEF activity and provides a simple explanation for why the individual loss of function mutations have a milder phenotype than loss of Rab11.
Figure 7.
Parcas and TRAPPII are both Rab11 GEFs and together serve an essential role in Drosophila. (A) Release of mant-GDP from 250 nM of Rab-His6 by 50 nM of Parcas in the presence or absence of synthetic fly Golgi mix liposomes (Lipo). The Rabex domain of the Rab5 GEF Vps9 (50 nM) was used as a negative control. Traces are means. (B) Rab11 nucleotide exchange rates calculated from the plots in A. Values are means ± SEM (Parcas, n = 3; Vps9, n = 2). (C) Nucleotide exchange rates of Parcas and the Rabex domain of Vps9 on the indicated Rabs. Parcas acts on Rab11 but not Rab1 or Rab5. (D) The mean number of adult progeny with homozygous or heterozygous genotypes obtained from three independent crosses, each using five females and three males carrying the indicated mutations. The independent values are shown along with the mean. Error bars show SD. Flies were viable if they lacked Parcas or the TRAPPII subunit TRAPPC9, but not both.
Discussion
The TRAPP complexes have emerged as fundamental components of membrane traffic machinery in eukaryotes. Since their discovery almost 20 years ago, most of the characterization of the complexes has been performed in yeast, and although recent work has shown that humans and other metazoans have two TRAPP complexes (Bassik et al., 2013), these are not identical to their yeast counterparts. In this study, we have provided functional evidence from Drosophila to show what is shared between metazoans and yeast and also to resolve some of the apparent paradoxes that have emerged from comparing the yeast and mammalian systems. We find that as in yeast, both of the Drosophila TRAPP complexes have GEF activity on Rab1, whereas TRAPPII also acts as a GEF on Rab11. In the only previous study in which mammalian TRAPP complexes were tested on Rab11, TRAPPCII was immunoisolated from cells using antiserum to TRAPPC9, and activity was found on Rab1 but not Rab11 (Yamasaki et al., 2009). However, the subunit composition of the isolated complex was not determined, and it is also possible that the antibody or attached beads inhibited access to the Rab11 substrate. A recent study has suggested that mammalian TRAPPII can act on both Rab1 and Rab18 (Rab11 was not tested), although for reasons that are not clear, we were unable to detect this Rab18 activity within the Drosophila complex (Li et al., 2017).
Beyond these shared properties, there are some clear differences between the yeast and metazoan TRAPP complexes. First, TRAPPIII contains two additional subunits, TRAPPC11 and TRAPPC12, that are absent from yeast. These two subunits are unlikely to have a metazoan-specific role as they are very widely conserved throughout eukaryotic phyla, including even filamentous fungi, and instead appear to have been lost in the budding yeast lineage (Fig. S4). TRAPPC11 is distantly related to TRAPPC10, and so it seems likely that in early eukaryotic evolution, there was an ur-TRAPP complex that had two additional subunits, and then both duplicated to make on the one hand TRAPPC8 and TRAPPC9 and on the other TRAPPC10 and TRAPPC11, and hence two TRAPP complexes. TRAPPC11 is essential for the growth of both Drosophila and mammalian cultured cells, and therefore yeast must have found a means to bypass this requirement. Given that the core of shared subunits is sufficient for Rab1 GEF activity, the role of TRAPPC11 seems most likely to be to direct the TRAPPIII Rab1 GEF activity to the correct location. It is possible that yeast have evolved an alternative mechanism to direct TRAPPIII to Golgi membranes via the core subunits (Sacher et al., 2001; Bassik et al., 2013). However, it is also possible that TRAPPC11 allows the complex to have an additional activity that budding yeast no longer requires. Our finding that in some tissues, TRAPPIII is found on the trans-Golgi as well as the cis-Golgi provides some support for this possibility as Rab1 is widely believed to function only on the cis-Golgi (Satoh et al., 2005, 2016), and consistent with this, we could only detect YFP-Rab1 on the cis-Golgi in these tissues (Fig. S5). In contrast, Drosophila Rab11 is known to be present on the TGN as well as on recycling endosomes (Emery et al., 2005; Satoh et al., 2005).
Our work has focused on TRAPPC11 as the TRAPPIII subunit that is essential but absent from yeast. The TRAPPIII subunit that is shared with yeast, TRAPPC8, has been reported to be essential in Drosophila and in mammalian cultured cells, although its yeast orthologue Trs85 is not essential, perhaps for the same reasons suggested in the previous pargaraph as to how yeast survive without TRAPPC11 (Cooper et al., 2010; Kim et al., 2016). Metazoans have two further TRAPPIII subunits: TRAPPC13, a distant orthologue of the yeast protein Trs65 that has been suggested to contribute to TRAPP complex structure (Choi et al., 2011), and TRAPPC12, a protein that is absent from budding yeasts but present in a diverse range of eukaryotic phyla, suggesting it had been lost during budding yeast evolution (Fig. S4). The role of TRAPPC12 is unclear, and it is unrelated to other TRAPP subunits and has even been suggested to have additional roles outside of the TRAPP complex (Milev et al., 2015). TRAPPC12 has also been reported to bind to the Sec13/31 component of the COPII coat (Bassik et al., 2013; Zhao et al., 2017). However, although we found this interaction when we overexpressed tagged TRAPPC12, we did not recover Sec13/31 when we precipitated TRAPPIII with other subunits (Fig. 2 A). This suggests that if this interaction is physiological, then it may represent a distinct function of TRAPPC12 or possibly an intermediate in the assembly of TRAPPIII.
A second striking difference between the yeast and metazoan TRAPP complexes is that although TRAPPII can act on Rab11 in both classes of organism, in metazoans, it shares the role of activating Rab11 with the unrelated SH3BP5 protein family. There is clearly considerable redundancy between the two, as mutations in the SH3BP5 orthologues in Drosophila and C. elegans are viable as are mutations in TRAPPII in Drosophila and mammalian cultured cells (Sinka et al., 2002; Sakaguchi et al., 2015; Kim et al., 2016). Loss of one or the other Rab11 GEF does have consequences, with loss of TRAPPII from flies causing male infertility and loss of Parcas causing defects in several developmental processes, some of which have been linked to signaling by nonreceptor tyrosine kinases (Beckett and Baylies, 2006; Robinett et al., 2009; Hamada-Kawaguchi et al., 2015). In C. elegans, loss of the two SH3BP5 orthologues causes defects in embryonic cytokinesis (Sakaguchi et al., 2015). Thus, it seems likely that each of the two types of Rab11 GEF can provide much of the necessary activation of Rab11, but each also has unique specialized roles in particular cell types. Revealing this overlap in function between the two proteins should greatly assist dissecting the function of each class of GEF. It also seems likely that TRAPPII can contribute to the activation of Rab1 in vivo as both the yeast and Drosophila complexes have activity on Rab1, at least in vitro (Thomas and Fromme, 2016). This could provide a possible explanation for our observation that at least in S2 cells, the major pool of TRAPPII is found on the cis-Golgi. Indeed, a study of mammalian TRAPPC10 found an epitope-tagged version of the protein to be present on the early Golgi (Yamasaki et al., 2009). However, it is clear that the ability of TRAPPII to activate both Rab1 and Rab11 raises a conundrum because there is a growing consensus that Rab GEFs define the location of active Rabs within the cell (Barr, 2013; Blümer et al., 2013). However, in this case, the two Rabs are widely believed to act on different compartments and recruit very different effectors (Stenmark, 2009; Hutagalung and Novick, 2011). It may be that in this case, there are additional mechanisms to restrict the recruitment of the two Rabs to a particular location before activation by TRAPP, such as a GDI displacement factor (Pfeffer, 2013). Thus, it seems likely that further investigation of the action of the TRAPP complexes will reveal new fundamental principles of how membrane traffic is organized in cells, and our study will hopefully facilitate the pursuit of such studies in metazoan systems.
Materials and methods
Flybase gene identifiers
Flybase genes used were: GM130 (FBgn0034697), Golgi microtubule–associated protein (GMAP; FBgn0027287), Golgin245 (FBgn0034854), l(3)63Bb (FBgn0002191), l(3)63Bc (FBgn0002192), l(3)63Bd (FBgn0002193), l(3)63Be (FBgn0002194), l(3)63Bh (FBgn0004564), l(3)67BDa (FBgn0002203), l(3)67BDc (FBgn0002205), l(3)67BDd (FBgn0002206), l(3)67BDe (FBgn0002207), l(3)67BDf (FBgn0002208), l(3)67BDg (FBgn0002209), l(3)67BDh (FBgn0002210), l(3)67BDi (FBgn0002211), l(3)67BDj (FBgn0002212), l(3)67BDl (FBgn0002214), l(3)67BDm (FBgn0002215), l(3)67BDn (FBgn0002216), Rab1 (FBgn0016700), Rgp1 (FBgn0037297), Ric1 (FBgn0028500), Sec13 (FBgn0024509), Sec16 (FBgn0052654), Sec31 (FBgn0033339), TRAPPC1 (FBgn0260860), TRAPPC10 (FBgn0038303), TRAPPC11 (FBgn0035416), TRAPPC12 (FBgn0037022), TRAPPC13 (FBgn0032204), TRAPPC2 (FBgn0266724), TRAPPC2L (FBgn0033605), TRAPPC3 (FBgn0260859), TRAPPC4 (FBgn0260861), TRAPPC5 (FBgn0266723), TRAPPC6 (FBgn0266722), TRAPPC8 (FBgn0260655), TRAPPC9 (FBgn0261787), pcs (FBgn0033988), and Rab11 (FBgn0015790).
Flybase allele and aberration identifiers
Flybase allele and aberration identifiers used were: TRAPPC2[1159] (FBal0183949), TRAPPC2[1] (FBal0009970), TRAPPC2[421] (FBal0183948), TRAPPC9[Z3358] (FBal0180654), TRAPPC8[12] (FBal0219692), TRAPPC11[MB06920] (FBal0217699), TRAPPC11[63Bd2] (FBal0009692), l(3)63Be[1] (FBal0009693), l(3)63Bb[6] (FBal0031842), l(3)63Bc[5] (FBal0009690), l(3)63Bh[1] (FBal0009698), w[1118] (FBal0018186), l(3)67BDn[1] (FBal0009777), l(3)67BDj[1] (FBal0009772), l(3)67BDi[1] (FBal0009770), l(3)67BDa[1] (FBal0009717), l(3)67BDl[1] (FBal0009774), l(3)67BDg[2] (FBal0009763), l(3)67BDd[1] (FBal0009750), l(3)67BDh[1] (FBal0009766), l(3)67BDf[2] (FBal0009760), l(3)67BDm[1] (FBal0009776), l(3)67BDc[2] (FBal0009746), l(3)67BDe[2] (FBal0009756), Rab1[EYFP] (FBal0314192), TRAPPC6[f00985] (FBal0161182), and pcs[gs] (FBal0140984). Aberrations were Df(3R)Exel6174 (FBab0038229) Df(2L)Exel7078 (FBab0037939), Df(3L)BSC672 (FBab0045738), Df(3R)BSC750 (FBab0045816), and Df(3L)AC1 (FBab0002292).
Fly stocks
Fly stocks were obtained from the Bloomington Drosophila Stock Center at Indiana University. Stocks used were: 34498 (w[1118]; P{w[+mC]=His2Av-mRFP1}III.1 P{w[+mW.hs]=FRT(w[hs])}2A), 62539 (w[1118]; TI{TI}Rab1[EYFP]; Dunst et al., 2015), 997 (Df(3L)AC1, rn[roe-1] p[p]/TM3, Sb[1]), 26848 (w[1118]; Df(3R)BSC750/TM6C, Sb[1] cu[1]), 26524 (w[1118]; Df(3L)BSC672, P+PBac{w[+mC]=XP3.WH3}BSC672/TM6C, Sb[1] cu[1]), 7581 (w[1118]; Df(2L)Exel7078/CyO), 1997 (w[*]; P{w[+mW.hs]=FRT(w[hs])}2A), 25338 (w[1118]; Mi{ET1}gry[MB06920]/TM6C, Sb[1]; Wendler et al., 2010), 5058 (Trs20[1]/TM2), 23931 (w[1118]; Trs20[1159]/TM6B, Tb[1]), 23932 (w[1118]; Trs20[421] E(smoDN)B-right[421]/TM6B, Tb[1]), 26176 (l(3)76BDm[12] red[1] e[4]/TM6B, Sb[1] Tb[1] ca[1]), 2434 (l(3)63Bd[2] e[1]/TM6B, Tb[1]), 2436 (l(3)63Be[1] e[1]/TM6B, Tb[1]), 2423 (l(3)63Bb[6]/TM6B, Tb[+] e[+]), 2429 (l(3)63Bc[5]/TM3, Sb[1]), 2443 (l(3)63Bh[1]/TM6B, Tb[1]), 2542 (l(3)67BDn[1] kni[ri-1] p[p]/TM3, Sb[1]), 2516 (l(3)67BDj[1] kni[ri-1] p[p]/TM3, Sb[1]), 2514 (l(3)67BDi[1] kni[ri-1] p[p]/TM3, Sb[1]), 2478 (l(3)67BDa[1] kni[ri-1] p[p]/TM3, Sb[1]), 2520 (l(3)67BDl[1] kni[ri-1] p[p]/TM3, Sb[1]), 2504 (l(3)67BDg[2] kni[ri-1] p[p]/TM3, Sb[1]), 2489 (l(3)67BDd[1] kni[ri-1] p[p]/TM3, Sb[1]), 2509 (l(3)67BDh[1] kni[ri-1] p[p]/TM3, Sb[1]), 2502 (l(3)67BDf[2] kni[ri-1] p[p]/TM3, Sb[1]), 2521 (l(3)67BDm[1] kni[ri-1] p[p]/TM3, Sb[1]), 2488 (l(3)67BDc[2] kni[ri-1] p[p]/TM3, Sb[1]), 2493 (l(3)67BDe[2] kni[ri-1] p[p]/TM3, Sb[1]), 7653 (w[1118]; and Df(3R)Exel6174, P{w[+mC]=XP-U}Exel6174/TM6B, Tb[1]), 18399 (w[1118]; PBac{w[+mC]=WH}Trs33[f00985]/TM6B, Tb[1]).
Construction of expression plasmids with SBP-GFP tag
The SBP-GFP tag consists of an SBP tag (Keefe et al., 2001), two tandem TEV protease recognition sites, and enhanced GFP (GenBank accession number AAB02574) with the A207K mutation to prevent dimerization. The sequence was codon optimized for expression in Drosophila, and two versions were synthesized (Integrated DNA Technologies) to tag proteins at either the N or C terminus and were then inserted into a pMT vector backbone for copper-inducible expression.
cDNA clones
cDNA clones from the Berkeley Drosophila Genome Project were obtained from Drosophila Genomics Resource Center. These clones were: Rgp1 (clone RH52122; GenBank accession number BT010211), TRAPPC3 (clone RE68712; GenBank accession number BT015980), TRAPPC8 (clone FI18195; GenBank accession number BT133116), TRAPPC9 (clone RE66325; GenBank accession number BT023878), TRAPPC10 (clone LD45339; GenBank accession number AY118591), TRAPPC11 (AT04056; GenBank accession number BT015311), TRAPPC12 (RE19263; GenBank accession number AY089596), TRAPPC13 (LD37668; GenBank accession number AY058665), and Sec31 (AT25839; GenBank accession number BT016143). RH52122 was corrected by inserting the sequence GA between nucleotides 282 and 283 of GenBank accession number BT010211. The coding sequence of this corrected clone encodes a longer version of Rgp1 corresponding with GenBank accession number NM_141259. The cDNA clone RE18773 (Berkeley Drosophila Genome Project) was corrected so that its coding sequence corresponded with GenBank accession number NM_139363. Open reading frames were amplified and inserted into corresponding expression vectors.
Antibodies
Primary antibodies used were mouse anti-SBP (1:1,000; MAB10764; EMD Millipore), rabbit antiactin (1:3,000; 8227; Abcam), mouse anti-GFP (IB, 1:1,000; immunofluorescence [IF], 1:2,000; 11814460001; Roche) mouse anti-V5 (1:2,000; R960-25; Invitrogen), rabbit anti-GM130 (1:500; ab30637; Abcam), goat anti–GMAP CRB16 and goat anti–Golgin245 CRB14 (1:2,000; Riedel et al., 2016), rabbit anti-Sec16 (1:2,000; Ivan et al., 2008), rabbit anti-TRAPPC9 AC1, rabbit anti-TRAPPC12 AC1 (IB, 1:1,000), rabbit anti-TRAPPC12 AC2 (IF, 1:100), and rabbit anti–Drosophila AP-1–γ (Hirst et al., 2009). Secondary antibodies were species-specific donkey anti-IgG, goat anti-IgG1, and goat anti-IgG2a antibodies coupled with Alexa Fluor 488, Alexa Fluor 555, or Alexa Fluor 647 (1:100; Thermo Fisher Scientific) and anti-IgG antibodies coupled with horseradish peroxidase (Santa Cruz Biotechnology, Inc.). GFP was also detected with the GFP booster Atto488 (1:400; gba488-100; ChromoTek).
Expression of tagged proteins in S2 cells for IF
Drosophila S2 cells (D.Mel-2; Invitrogen) were grown at 25°C in serum-free medium containing penicillin, streptomycin and l-glutamine (Express Five; 10486-025; Thermo Fisher Scientific). For transient expression cells were split in fresh medium and plated in 0.5-ml aliquots per well in a 24-well plate. Cells were allowed to settle for at least 60 min and then transfected with 0.4 µg of each expression plasmid using Fugene HD (Promega). Cells were incubated at 25°C for 24 h. The medium was replaced by fresh medium supplemented with copper sulfate to induce the expression of the tagged proteins. Cells were incubated for another 48 h, removed from the wells, and transferred to polytetrafluoroethylene multiwell glass slides coated with poly-L-lysine (P4707; Sigma-Aldrich). Cells were allowed to settle for 60 min, washed three times with PBS, fixed for 10 min in 4% PFA in PBS, permeabilized for 10 min in PBS with 0.5% Triton X-100, blocked for 30 min in 20% fetal calf serum in PBS, and incubated for 60 min with primary antibodies diluted in 20% fetal calf serum in PBS. For the GFP-tagged proteins, the signal was typically boosted using an anti-GFP monoclonal. The cells were then washed three times for 20 min in PBS with 0.5% Tween-20, incubated for 60 min in species-specific secondary antibodies diluted in 20% fetal calf serum in PBS, washed three times for 20 min in PBS with 0.5% (vol/vol) Tween-20, and washed once for 20 min in PBS. Cells were mounted in Vectashield (H-1000; Vector Laboratories).
To generate stably transfected S2 cells lines, cells were split in fresh medium and plated in T25 flasks. Cells were allowed to settle for 60 min and then transfected with 2.5 µg expression plasmid and 0.25 µg pCoBlast using Fugene HD. Cells were incubated for 2 d, subjected to selection for 2 wk in medium with 25 µg/ml blasticidin, and maintained in medium with 5 µg/ml blasticidin. Cell sorting was then used to generated pooled populations of cells expressing the relevant tagged protein. Transfected S2 cells were split into fresh medium without blasticidin in T75 flasks. 2 d before cell sorting, the medium was replaced with fresh medium containing 500 µM copper sulfate. Cells were removed from the flask, washed twice in fresh medium, filtered (04-004-2327; Celltrics; Partec) and sorted into groups of cells with low, intermediate, and high GFP intensity using a cell sorter. Cells were collected in medium with 5% fetal calf serum. Once cells started dividing, the medium was replaced with fresh medium (without fetal calf serum) with 5 µg/ml blasticidin.
IF microscopy
Salivary glands and wing discs were dissected from wandering third instar larvae in PBS, fixed for 30 min in 4% PFA in PBS, permeabilized three times for 20 min in PBS with 0.5% Triton X-100, blocked for 30 min in 20% fetal calf serum in PBS, incubated for 90 min with primary antibodies diluted in 20% fetal calf serum in PBS, washed three times for 20 min in PBS with 0.5% Tween-20, incubated for 60 min in species-specific fluorescently labeled secondary antibodies diluted in 20% fetal calf serum in PBS, washed three times for 20 min in PBS with 0.5% Tween-20, and washed once for 20 min in PBS. Tissues were mounted in ProLong Gold Antifade Mountant with or without DAPI (P36941 and P36934; Thermo Fisher Scientific). For comparing the distribution of different markers, each primary antibody was raised in a different species and then detected with species-specific antibodies labeled with different fluorochromes. To localize GFP-tagged proteins in tissues, the signal was boosted using Atto-488–labeled GFP booster (ChromoTek). Clones of TRAPPC3 and TRAPPC11 were generated in wing discs by heat-shocking yw hs-flp; TRAPPC3[77] FRT2A YFP-Rab1/His-RFP FRT2A YFP-Rab1 larvae and yw hs-flp; TRAPPC11[7] FRT2A YFP-Rab1/His-RFP FRT2A YFP-Rab1 larvae, respectively, at 37°C for 60 min, and wandering third instar larvae were dissected 56 or 72 h after the heat shock.
Widefield epifluorescence micrographs were obtained with an Axioplan microscope (ZEISS) equipped with a CoolSNAP HQ2 camera (Photometrics) using Micro-Manager software or a DM6 microscope equipped with a DFC365 FX camera using LAS X software (Leica Microsystems). Confocal images were taken with an SP8 microscope (Leica Microsystems) using LAS X software, and FIJI (ImageJ; National Institutes of Health) was used for subsequent analysis, including line scans of intensities across the Golgi stack (Schindelin et al., 2012). All images were acquired with a 63× 1.4 NA Plan Apochromat oil immersion lens except for whole discs (Fig. 6 A), which were with a 10× 0.4 NA Plan Apochromat dry lens. In all cases, the images shown are representative of at least two independent experiments with at least three micrographs obtained from each experiment.
Western blot from lysates of adult flies
10 adult male flies were collected on ice and homogenized with a motorized pestle in 100 µl lysis buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 1% Triton X-100, 5 mM β-mercaptoethanol, cOmplete EDTA-free protease inhibitors [18970600; Roche], and 1 mM PMSF). Lysates were cleared by centrifugation for 10 min at 20,000 g at 4°C. The protein concentration of the supernatants was determined with Bradford reagent (500-0006; Bio-Rad Laboratories). Lysates of parallel samples were adjusted to the same concentration and mixed with SDS sample buffer.
Affinity purification of GFP-tagged proteins from transiently transfected cells and analysis by Western blotting
Cells were plated in 2 ml per well in six-well plates and allowed to settle for at least 60 min before transfection with 2 µg of expression plasmid using Fugene HD (E2311). Cells were incubated at 25°C for 24 h, and the medium was then replaced with fresh medium supplemented with 100 µM copper sulfate to induce protein expression. Cells were incubated for another 48 h, collected in 1.5-ml tubes, washed in PBS, and lysed in 1 ml lysis buffer. Lysates were cleared by centrifugation for 10 min at 20,000 g at 4°C. Lysates were mixed with 5 µl equilibrated GFP-trap agarose slurry (gta-100; ChromoTek) and incubated for 60 min at 4°C with rotation. Beads were washed four times in 500 µl lysis buffer without PMSF. Proteins were eluted by incubation for 10 min at 90°C in 50 µl SDS sample buffer and analyzed by Western blotting.
Affinity purification of GFP-tagged proteins from larvae and adult flies
50 wandering third instar larva or fifty adult males were collected on ice and homogenized in 500 µl lysis buffer with a motorized pestle homogenizer. Lysates were cleared by centrifugation for 10 min at 20,000 g and 4°C. The protein concentration of the supernatants was determined with Bradford reagent. Lysates of parallel samples were adjusted to the same concentration and mixed with 20 µl equilibrated GFP-trap agarose slurry and incubated for 60 min at 4°C with rotation. Beads were washed four times in 500 µl lysis buffer without PMSF. Proteins were eluted by incubation for 10 min at 90°C in 50 µl SDS sample buffer and analyzed by Western blotting.
Single-step purification and TAP from embryos and stable cell lines
Embryos were collected for 24 h on 90-mm apple juice agar plates, with plates being kept for ≤2 d in the fridge after collecting. For a single purification, embryos from 15 plates were dounce homogenized in 10 ml lysis buffer (Jencons). For affinity chromatography from stably transfected cell lines, cells were grown in two T150 flasks (Sigma-Aldrich), induced with copper sulfate at least 2 d before harvest, and grown to a density of 106 to 107 cells per ml. Cells were pelleted at 200 g for 5 min, washed in PBS, and lysed in 2 ml lysis buffer. Lysates were cleared by centrifugation for 10 min at 20,000 g and 4°C. The protein concentration of the supernatants was determined with Bradford reagent. Lysates of parallel samples were adjusted to the same concentration and mixed with 20 µl equilibrated GFP-trap agarose and then incubated for 60 min at 4°C with rotation. Beads were transferred to Mobicol columns (M1003; Mobitec) and then washed four times with 500 µl lysis buffer without PMSF. Proteins were eluted from the beads for 30 min at 4°C in 150 µl TEV protease elution buffer (lysis buffer without PMSF; 1 mM DTT and 0.1 U/µl AcTEV; 12575-15; Invitrogen). The eluate was incubated with 20 µl slurry of high-capacity streptavidin agarose beads (20357; Thermo Fisher Scientific) for 1 h at 4°C with rotation. Beads were transferred to Mobicol columns and washed four times with 500 µl lysis buffer without PMSF. Proteins were eluted from the beads by rotation for 10 min at 4°C in 150 µl in lysis buffer without PMSF and with saturating concentrations of biotin. Proteins were precipitated with chloroform/methanol. The precipitate was dried in a concentrator and resuspended in 25 µl SDS buffer.
For protein identification by silver staining, proteins were fractionated by SDS-PAGE and visualized with the Pierce Silver Stain kit (24612; Thermo Fisher Scientific). For protein identification by mass spectrometry, proteins were first fractionated by SDS-PAGE, and the gel was stained with Sypro Ruby protein gel stain (50562; Lonza). Gel lanes were cut into 1-mm slices and then into small cubes, transferred to a 96-well plate, and digested with trypsin. Tryptic peptides were identified by liquid chromatography coupled with tandem mass spectrometry by the Medical Research Council Laboratory of Molecular Biology biological mass spectrometry facility and then combined in Scaffold (Proteome Software). Identified proteins were filtered to match the criteria of having at least two exclusive unique peptides at an overall protein false discovery rate of <1%. The mass spectrometry data together with the complete Drosophila proteome from UniProt, fly genes from Flybase, and human genes from Ensembl as well as orthologous relationships from Ensembl were combined in a single custom-made relational database using Filemaker (Filemaker Inc.) in order to better distinguish background proteins from specific interactors.
Viability and fertility tests
To test viability, crosses were set up in triplicate by mating virgins heterozygous for the deficiency and a balancer with males heterozygous for the mutation and a balancer. Adults were removed after 5 d. To determine adult viability, each vial was cleared twice a day from the start of eclosion for 5 d. Adult progeny carrying either a balancer (heterozygous for the deficiency or the mutation and the balancer) or no balancer (heterozygous for the deficiency and the mutation) were scored. To determine pupal viability, pupae carrying a balancer and pupae carrying no balancer were scored on a single day just before the start of eclosion. For TRAPPC11, we used the deficiency Df(3L)BSC672, for TRAPPC10, we used the deficiency Df(3R)BSC750, and for TRAPPC9, we used the deficiency Df(2L)Exel7078. To test male fertility, crosses were set up in triplicate by mating w[1118] virgins or w[*]; betaTub60D[2] Kr[If-1]/CyO virgins with males heterozygous for the mutation and the deficiency. Adults were removed after 5 d. Pupal progenies were counted on a single day before eclosion.
Mutagenesis of TRAPP subunit–encoding genes by CRISPR/Cas9
For TRAPPC3, TRAPPC10, and TRAPPC11, we selected Cas9 target sites immediately downstream of the start codon to introduce indels. For TRAPPC9, we decided to remove the coding sequence from the genome by designing two pairs of Cas9 target sites. One target of each pair was upstream of the start codon, and the other target was downstream of the stop codon. Constructs expressing single guide RNAs were cloned into pCFD3, and constructs expressing two guide RNAs were cloned into pCFD4 (Port et al., 2014). Two guide RNA constructs were cloned for TRAPPC3, TRAPPC9, and TRAPPC11, and three guide RNA constructs were cloned for TRAPPC10.
Guide RNA–expressing constructs were injected into a stock expressing Cas9 in the germ line (Port et al., 2014). Stocks were generated from progeny of injected flies by crossing individual males heterozygous for a mutagenized chromosome to balancer females. For TRAPPC9, a total of 169 stocks were established from 44 injected flies; for TRAPPC3, a total of 90 stocks were established from 48 injected flies; for TRAPPC11, a total of 138 lines were established from 29 injected flies; and for TRAPPC10, a total of 200 stocks were established from 82 injected flies. Genomic DNA was prepared from single males taken from each of the established stocks, and the genomic DNA surrounding the Cas9 target site was amplified by PCR. The primers used were 5′-TCCAAACGTAAACAGCCGGA-3′ and 5′-GGTGGCGCTGAAATGGTTTT-3′ for TRAPPC3, 5′-TGCCAGGGGTGCATTAACAT-3′ and 5′-ACTGGACCTGCTTTTCCGTC-3′ for TRAPPC11, and 5′-GAAAGCACACCTGTTGAGCG-3′ and 5′-GTGCTCCAAGATGCTCCAGT-3′ for TRAPPC10. The resulting amplicon was sequenced with one of the two primers used for PCR. Mixed trace files indicative of the presence of an indel in heterozygous flies were first analyzed by Poly Peak Parser (Hill et al., 2014). The mutant sequences were then aligned individually to the Drosophila genome using NCBI Blast (National Institutes of Health) to determine the mutation in each allele. To screen fly lines established from the TRAPPC9 mutagenesis experiment, we used the primers 5′-ACCCATGTGAAGGAGTGCAG-3′ and 5′-ACGGATTCACCATGGACGAG-3′ that flank the coding sequence of TRAPPC9. This resulted in a PCR amplicon only for flies with a deletion of the TRAPPC9 coding sequence. This amplicon was sequenced with one of the primers used for PCR and aligned against the Drosophila genome using the UCSC genome browser.
From the TRAPPC3 lines, a total of 24 lines showed an indel, representing 11 deletion alleles, two insertion alleles, and one allele with substitutions. All of the homozygous viable lines carried indels that did not shift the reading frame. Seven of the homozygous lethal alleles, lines 17, 19, 23, 44, 76, 77, and 68 (six deletions and one insertion), carried frameshift mutations. Six of these tested negative in a complementation test with Df(3L)AC1. Line 44 has not been included in this complementation test. For TRAPPC9, four fly lines representing two alleles were found to carry a deletion of the whole coding sequence. None of these lines was lethal over the deficiency Df(2L)Exel7078, and lines 105 and 141 were kept for further studies. For TRAPPC10, a total of 29 lines with indels representing 15 alleles were isolated. None of these lines was lethal over Df(3R)BSC750. Of the 138 lines that were established for TRAPPC11, four failed to complement TRAPPC11[MB06920]. Three of the four lines represent one insertion allele and one deletion allele. For the fourth line, we were unable to determine the type of the mutation.
Genomic tagging of TRAPP subunits
TRAPPC3, TRAPPC9, TRAPPC12, and TRAPPC11 were tagged at the C terminus as for expression in S2 cells. Cas9 target sequences were selected close to the stop codon, which were 5′-GTTGCTTCAGAGGTGGCTGTGGG-3′ for TRAPPC11, 5′-TGGAGGAGAACATGACCGATTGG-3′ for TRAPPC9, 5′-ACTGGAATCAAACAACTCCAAGG-3′ for TRAPPC12, and 5′-GGAGGTCATACCAGCTGGCGAGG-3′ for TRAPPC3, and the corresponding guide RNAs were cloned into pCFD3. Donor DNA constructs were cloned with a central region encoding the tag, which was flanked by homology arms, each corresponding with 800–1,000 bp of genomic DNA. The protospacer-adjacent motif in each of these constructs was mutated to prevent Cas9 from cutting the donor DNA. Both the donor DNA and the guide RNA were injected into a stock expressing Cas9 in the germ line.
For tagging of TRAPPC3, 90 stocks from 17 injected flies; for TRAPPC9, 148 stocks from 14 injected flies; for TRAPPC11, 130 stocks from 18 injected flies; and for TRAPPC12, 92 stocks from 29 injected flies were set up. These lines were screened by PCR with two primer pairs. One primer of each pair bound to the DNA encoding the tag and the other bound in the flanking genomic DNA of the donor plasmid. Positive lines were subjected to a second PCR, with primers flanking the donor DNA to confirm that the entire donor DNA integrated into the correct place. The following primers were used: 5′-GCAATTTTGTGCGGCTTGTG-3′ and 5′-ACGCGCCGGAATCTAAATCT-3′ for TRAPPC3, 5′-TCACCGCATCATCGTCGAAA-3′ and 5′-ACTTGCCGCAAAATGCTTGT-3′ for TRAPPC9, 5′-CGGCGAGCAACTGGAAAG-3′ and 5′-GAGCTGGGCAACATACGC-3′ for TRAPPC11, and 5′-AACACTAATCCACGCCGGTT-3′ and 5′-TCGACACCCACTCAAACGAG-3′ for TRAPPC12. The amplicon of at least two lines per gene was sequenced. All lines for TRAPPC9, TRAPPC11, and TRAPPC12 were homozygous viable and were kept as stable stocks without a balancer. All lines for TRAPPC3 were homozygous viable but sterile.
Characterization of potential TRAPP complex alleles
The EMS alleles l(3)63Bd[2], l(3)63Be[1], l(3)63Bb[6], l(3)63Bc[5], and l(3)63Bh[1] were found with Flybase Cytosearch. They were previously mapped to the cytogenetic region, which also contains TRAPPC11 (Wohlwill and Bonner, 1991). Of these five mutations, l(3)63BD[2] failed to complement TRAPPC11[MB06920] and TRAPPC11[7]. We therefore changed the name of this allele to TRAPPC11[63BD2]. Flies heterozygous for TRAPPC11[63BD2] and the deficiency Df(3L)BSC672 do not develop into adulthood, but in rare cases, they pupariate. Furthermore, flies heterozygous for the CRISPR alleles TRAPPC11[19] and TRAPPC11[63BD2] in rare cases develop into adults.
The alleles l(3)67BDn[1], l(3)67BDj[1], l(3)67BDi[1], l(3)67BDa[1], l(3)67BDl[1], l(3)67BDg[2], l(3)67BDd[1], l(3)67BDh[1], l(3)67BDf[2], l(3)67BDm[1], l(3)67BDc[2], and l(3)67BDe[2] were found with Flybase Cytosearch. They were previously mapped to the same cytogenetic region as TRAPPC3 (Leicht and Bonner, 1988), but all of these alleles complemented the TRAPPC3 alleles 76, 77, and 84, indicating that none is an allele of TRAPPC3. EMS alleles of TRAPPC11[63Bd2], TRAPPC8[12], TRAPPC2[1159], TRAPPC2[1], and TRAPPC2[421] were sequenced from heterozygous flies. Natural variations to the reference genome of Drosophila were identified from the Drosophila Genetics Reference Panel via the UCSC genome browser and the Drosophila Population Genome Project via Ensembl. All above-mentioned EMS alleles had a nonsense mutation in the corresponding gene. We also obtained a fly line with a piggyBac insertion in the coding sequence of TRAPPC6. Mutants heterozygous for this allele, called TRAPPC6[f00985], and the deficiency Df(3R)6174 are lethal, suggesting that TRAPPC6 in flies is essential in contrast with yeast Trs33 and mammalian TRAPPC6A and TRAPPC6B (Kim et al., 2016).
Generation of antibodies to TRAPPC9 and TRAPPC12
Full-length TRAPPC12 corresponding with GenBank accession number AAL90334 was fused at its N terminus to GST by cloning the coding sequence of cDNA clone RE19263 into the expression vector pGEX–6P-1. The N terminus of TRAPPC9 corresponding with amino acids 20–417 of GenBank accession number ABA81812 was fused at its N terminus to GST by cloning the corresponding region of the coding sequence of cDNA clone RE66325 into the expression vector pGEX–6P-1. The expression of the GST-fusion proteins was induced overnight at 16°C in E. coli BL21 (DE3; Agilent Technologies) with 0.2 µM IPTG. Cells were harvested and resuspended in lysis buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 150 mM NaCl, 10 mM DTT, and cOmplete protease inhibitors). The lysate was sonicated and cleared by centrifugation at 50,000 g for 15 min at 4°C. GST proteins were purified from the supernatant by affinity chromatography using glutathione Sepharose 4B (GE Healthcare). Antibodies were raised in rabbits by Cambridge Research Biochemicals.
To purify TRAPPC12 antisera, GST and TRAPPC12-GST were coupled to CNBr-activated Sepharose 4B beads (GE Healthcare). GST antibodies were first removed using a GST column, and TRAPPC12 antibodies were purified with a TRAPPC12-GST column. Antibodies were eluted with 0.1 M glycine, pH 2.5, and fractions were neutralized with 1 M Tris, pH 8.8. Protein concentration was determined with Bradford reagent. Peak fractions were pooled, dialyzed in PBS at 4°C overnight, and stored at −80°C. Two sera were purified separately (TRAPPC12 AC1 and TRAPPC12 AC2). To purify TRAPPC9 antisera, the N terminus of TRAPPC9 corresponding with amino acids 23–514 of GenBank accession number ABA81812 was fused at its N terminus to His-MBP by cloning the corresponding region of the coding sequence of cDNA clone RE66325 into the expression vector pETM-41. The expression of this protein was induced overnight at 16°C in E. coli BL21 (DE3) with 0.2 mM IPTG and 0.2% glucose. Cells were harvested and resuspended in column buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 0.1 mM EDTA, 10 mM DTT, and cOmplete protease inhibitors). Cells were lysed by sonication. The lysate was cleared at 50,000 g at 4°C for 15 min. The TRAPPC9 (aa23–514)-6×His-MBP fusion protein was purified from the cleared lysate with amylose resin (E8021S; New England Biolabs, Inc.), and coupled with CNBr-activated Sepharose 4B beads. TRAPPC9 antibodies were bound to the beads and then eluted with 0.1 M glycine, pH 2.5, and fractions were neutralized with 1 M Tris, pH 8.8. Protein concentration was determined with Bradford reagent. Peak fractions were pooled, dialyzed in PBS at 4°C overnight, and stored at −80°C.
Recombinant Rabs and TRAPP complexes
Rabs were expressed in the E. coli strain BL21-DE3 (Agilent Technologies). The cDNA of Drosophila Rab1 (NP_732610), Rab2 (NP_477090), Rab11 (NP_599137), Rab18 (NP_524744), and Rab5 (NP_722795) were amplified and cloned into pET28a+ in frame with a C-terminal His6 tag. The Rab C-terminal cysteine residues were replaced by His6 tags to allow membrane anchoring. Cell growth and protein expression were as described by Galindo et al. (2016). Cell pellets were resuspended in buffer A (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM MgCl2, and 1 mM DTT) with 20 mM imizadole and cOmplete protease inhibitors. After sonication, the lysate was cleared by centrifugation at 32,000 g for 30 min at 4°C. Cleared lysate were mixed with Ni-NTA beads and incubated for 1 h at 4°C. Resin was washed with 10 bed volumes of buffer A plus 40 mM imizadole and a final wash with 10 bed volumes of buffer A plus 200 mM imizadole. Eluted fractions were analyzed by SDS-PAGE, and imizadole was removed using a PD10 desalting column. Proteins were concentrated to ∼1 mg/ml, snap frozen in liquid N2, and stored at −80°C. Purified Rabs were loaded with mant-GDP (Invitrogen) as previously described by Thomas and Fromme (2016). Parcas (CG7761; NP_611010; Poirot) cDNA was synthesized (Epoch Life Science) and cloned into pGEX-6P. N-terminally GST-tagged protein was expressed in BL21-DE3 growing at 37°C and purified as described previously for the C. elegans protein (Sakaguchi et al., 2015) but eluted by incubating the glutathione Sepharose slurry overnight with PreScission protease (∼10 U/ml) at 4°C.
TRAPP complexes were expressed in Sf9 cells using the Multibac system (Nie et al., 2014). The TRAPP subunit genes were synthesized (Epoch Life Science). C1 (NP_651778), C2 (NP_648841), C3 (NP_648312), C4 (NP_609247), C5 (NP_610986), C6 (NP_650450), and C2L (NP_610662) were inserted sequentially into pACEBac1, with each expression cassette separated by 30-bp linkers to generate the pACEBac1-C1-C6 vector. The TRAPP2-specific genes C9 (NP_610044) and C10 (NP_650431) were cloned into pIDS, and the TRAPP3-specific subunits C8 (NP_649135) and C11 (NP_647785) were cloned into pIDS; meanwhile, the other two TRAPP3 subunits C12 (NP_649255) and C13 (NP_609365) were inserted into pIDC. Cre recombinase (New England Biolabs, Inc.) was used to fuse the appropriate pID vectors with pACEBac1-C1-C6 to generate a plasmids for the coexpression of all TRAPPII and TRAPPIII subunits. In these plasmids, the C3 subunits were tagged with Strep-TagII, and either C11 (TRAPPIII) or C10 (TRAPPII) was FLAG tagged. The linker sequence between each subunit and the tag included a site for PreScission protease (GST–human rhinovirus [HRV]-3C protease). Bacmids using the EMBacY baculovirus genome and virus production were made as described previously by Nie et al. (2014).
500-ml cell suspensions (2 × 106 cells/ml) were infected with 5 ml of P2 baculovirus and incubated at 27°C and 124 rpm. Cells coexpressing TRAPP core subunits, all TRAPPII subunits, or all TRAPPIII subunits were harvested after 66 h (75–80% viability) by centrifugation at 2,250 g for 10 min at 4°C. Pellets were washed once with PBS, centrifuged again, and stored at −80°C. Frozen pellets were vortexed in buffer B (50 mM Hepes-KOH, pH 7.44, 110 mM NaCl, 1 mM DTT, and 0.1% Igepal CA-630) with inhibitors (1 mM PMSF, cOmplete protease inhibitors, 0.4 µM pepstatin, 0.24 µM leupeptin, and 5 µM MG132) using 30 ml per 500 ml of initial culture, and incubated at 4°C for 10 min before being homogenizing using 15–20 strokes of a tight-fitting dounce. The lysates were clarified by centrifugation at 32,000 g for 30 min at 4°C and mixed with the relevant equilibrated slurry (either StrepTactin Superflow plus [∼400 µl per 500 ml of initial culture; QIAGEN] or anti-Flag M2 affinity gel [∼100 µl per 500 ml culture; Sigma-Aldrich]) and incubated on a rotation wheel for 1 h at 4°C. Beads were washed three times with 10 bead volumes of buffer B plus 0.05% Igepal CA-630. Bound material was eluted by washing the beads with five volumes of buffer B with 100 µg/ml of FLAG peptide or 2.5 mM desthiobiotin as appropriate. Eluted fractions were analyzed by SDS-PAGE, concentrated, and buffer exchanged. Alternatively, the bound complexes were eluted by incubating the slurry with PreScission protease (∼10 U/ml) overnight at 4°C, and the eluates then were mixed with glutathione Sepharose to remove the PreScission protease. The TRAPP core complex was purified further by gel filtration using a Superose 16/30 column (GE Healthcare) equilibrated in buffer B plus 0.05% Igepal CA-630.
GEF activity assays
GEF assays were performed in HKM buffer (20 mM Hepes-KOH, pH 7.4, 150 mM KOAc, 2 mM MgCl2, and 1 mM DTT). All reagents, Rabs, and TRAPP complexes were buffer exchanged into HKM. The activity of His-tagged Rabs was confirmed by the exchange of nonfluorescent GTP for mant-GDP using a PHERASTAR plate reader. Reactions containing 250 nM mant-GDP–labeled Rab alone, Rab and 200 µM GTP, or adding 10 mM EDTA to the former mix were set up in 96-well black-bottomed plates (Corning), and fluorescence decay was measured at 30°C. Drosophila Rab5 and the Vps9 domain of Rabex5 were used as a control. More rapid exchange rates were measured using an LS55 fluorimeter (PerkinElmer) under multiple turnover conditions. The mant-GDP–to-GTP exchange was performed by adding 250 nM mant-GDP Rab to the solution, warming the solution to 30°C for 150 s, adding 200 µM, and waiting 120 s before adding 50 nM of the corresponding TRAPP complex. Fluorescence measurements (360 nm excitation and 448 nm emission) were recorded for 20 min. Assays in the presence of liposomes were performed by preincubating 300 µM liposomes with the mant-GDP Rab before adding the GTP. All GEF assays were performed with n ≥ 3. To determine exchange rates, curves of fluorescence versus time were fitted to a single exponential curve with an additional linear drift term using the Prism7 software (GraphPad Software) as described previously by Richardson and Fromme (2015). The resulting rate constants were divided by the concentration of GEF to give the exchange rate.
Synthetic liposomes were prepared using an “insect Golgi” mixture based on previous lipidomics studies (Drin et al., 2008; Guan et al., 2013) with Ni2+-DOGS included when needed to anchor His-tagged Rabs for GEF assays. Thus, phosphatidylcholine (14 mol%), phosphatidylethanolamine (62%), phosphatidylserine (3%), phosphatidic acid (1%), phosphatidylinositol (10%), phosphatidylinositol-4-phosphate (1%), diacylglycerol (1%), ceramide (5%), cholesterol (3%), and Ni2+-DOGS (5%) were extruded through 400- and 100-nm filters (Whatman) as previously described (Christis and Munro, 2012).
Online supplemental material
Fig. S1 shows SBP-GFP–tagged TRAPP complex subunits in Drosophila S2 cells. Fig. S2 shows sequence details of EMS alleles in various TRAPP complex subunits. Fig. S3 shows Rab GEF assays with recombinant TRAPP complexes. Fig. S4 shows the evolutionary conservation of TRAPP complex subunits and Parcas/SH3BP5. Fig. S5 shows Localization of YFP-Rab1 in WT and TRAPPC9 mutant larval tissues.
Supplementary Material
Acknowledgments
We are indebted to Farida Begum, Sarah Maslen, and Mark Skehel for mass spectrometry; Fan Zhang and Maria Daly for cell sorting; Miklós Erdélyi, Jenny Hirst, Catherine Rabouille, Carmen Robinett, Katja Röper, Akiko Sato, Rita Sinka, and Daniel St. Johnston for stocks and reagents; Inmaculada Perez Dorado for artwork; Tim Stevens for phylogenetic profiles; Nick Barry, Matthias Pasche, Jon Howe, Fillip Port, and John Kilmartin for technical advice; and Alison Gillingham for comments on the manuscript.
This work was supported by the Medical Research Council (file reference number MC_U105178783) and the Wellcome Trust (095927/B/11/A). F. Riedel was also supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (reference RI 2182/1-1).
The authors declare no competing financial interests.
Author contributions: A. Galindo expressed and purified Drosophila Rabs and TRAPPs and also performed GEF assays. N. Muschalik helped with analysis of parcas and localization of TRAPP in Drosophila. F. Riedel performed all the other experimental work. A. Galindo, F. Riedel, and S. Munro conceived and planned the experiments. S. Munro wrote the manuscript.
References
- Artero R., Furlong E.E., Beckett K., Scott M.P., and Baylies M.. 2003. Notch and Ras signaling pathway effector genes expressed in fusion competent and founder cells during Drosophila myogenesis. Development. 130:6257–6272. 10.1242/dev.00843 [DOI] [PubMed] [Google Scholar]
- Barr F.A. 2013. Rab GTPases and membrane identity: causal or inconsequential? J. Cell Biol. 202:191–199. 10.1083/jcb.201306010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrowman J., Bhandari D., Reinisch K., and Ferro-Novick S.. 2010. TRAPP complexes in membrane traffic: convergence through a common Rab. Nat. Rev. Mol. Cell Biol. 11:759–763. 10.1038/nrm2999 [DOI] [PubMed] [Google Scholar]
- Bassik M.C., Kampmann M., Lebbink R.J., Wang S., Hein M.Y., Poser I., Weibezahn J., Horlbeck M.A., Chen S., Mann M., et al. 2013. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 152:909–922. 10.1016/j.cell.2013.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett K., and Baylies M.K.. 2006. Parcas, a regulator of non-receptor tyrosine kinase signaling, acts during anterior-posterior patterning and somatic muscle development in Drosophila melanogaster. Dev. Biol. 299:176–192. 10.1016/j.ydbio.2006.07.049 [DOI] [PubMed] [Google Scholar]
- Blümer J., Rey J., Dehmelt L., Mazel T., Wu Y.-W., Bastiaens P., Goody R.S., and Itzen A.. 2013. RabGEFs are a major determinant for specific Rab membrane targeting. J. Cell Biol. 200:287–300. 10.1083/jcb.201209113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner G.H.H., Hein M.Y., Hirst J., Edgar J.R., Mann M., and Robinson M.S.. 2014. Fractionation profiling: a fast and versatile approach for mapping vesicle proteomes and protein-protein interactions. Mol. Biol. Cell. 25:3178–3194. 10.1091/mbc.E14-07-1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet S., and Sacher M.. 2014. In sickness and in health: the role of TRAPP and associated proteins in disease. Traffic. 15:803–818. 10.1111/tra.12183 [DOI] [PubMed] [Google Scholar]
- Cai Y., Chin H.F., Lazarova D., Menon S., Fu C., Cai H., Sclafani A., Rodgers D.W., De La Cruz E.M., Ferro-Novick S., and Reinisch K.M.. 2008. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane-tethering complexes. Cell. 133:1202–1213. 10.1016/j.cell.2008.04.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C., Davey M., Schluter C., Pandher P., Fang Y., Foster L.J., and Conibear E.. 2011. Organization and assembly of the TRAPPII complex. Traffic. 12:715–725. 10.1111/j.1600-0854.2011.01181.x [DOI] [PubMed] [Google Scholar]
- Christis C., and Munro S.. 2012. The small G protein Arl1 directs the trans-Golgi-specific targeting of the Arf1 exchange factors BIG1 and BIG2. J. Cell Biol. 196:327–335. 10.1083/jcb.201107115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M.T., Conant A.W., and Kennison J.A.. 2010. Molecular genetic analysis of Chd3 and polytene chromosome region 76B-D in Drosophila melanogaster. Genetics. 185:811–822. 10.1534/genetics.110.115121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G., Morello V., Casella J.-F., Gounon P., and Antonny B.. 2008. Asymmetric tethering of flat and curved lipid membranes by a golgin. Science. 320:670–673. 10.1126/science.1155821 [DOI] [PubMed] [Google Scholar]
- Dunst S., Kazimiers T., von Zadow F., Jambor H., Sagner A., Brankatschk B., Mahmoud A., Spannl S., Tomancak P., Eaton S., and Brankatschk M.. 2015. Endogenously tagged rab proteins: a resource to study membrane trafficking in Drosophila. Dev. Cell. 33:351–365. 10.1016/j.devcel.2015.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery G., Hutterer A., Berdnik D., Mayer B., Wirtz-Peitz F., Gaitan M.G., and Knoblich J.A.. 2005. Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell. 122:763–773. 10.1016/j.cell.2005.08.017 [DOI] [PubMed] [Google Scholar]
- Galindo A., Soler N., McLaughlin S.H., Yu M., Williams R.L., and Munro S.. 2016. Structural insights into Arl1-mediated targeting of the Arf-GEF BIG1 to the trans-Golgi. Cell Reports. 16:839–850. 10.1016/j.celrep.2016.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.-C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.-M., Cruciat C.-M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 415:141–147. 10.1038/415141a [DOI] [PubMed] [Google Scholar]
- Gillingham A.K., Sinka R., Torres I.L., Lilley K.S., and Munro S.. 2014. Toward a comprehensive map of the effectors of rab GTPases. Dev. Cell. 31:358–373. 10.1016/j.devcel.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X.L., Cestra G., Shui G., Kuhrs A., Schittenhelm R.B., Hafen E., van der Goot F.G., Robinett C.C., Gatti M., González-Gaitán M., and Wenk M.R.. 2013. Biochemical membrane lipidomics during Drosophila development. Dev. Cell. 24:98–111. 10.1016/j.devcel.2012.11.012 [DOI] [PubMed] [Google Scholar]
- Hamada-Kawaguchi N., Nishida Y., and Yamamoto D.. 2015. Btk29A-mediated tyrosine phosphorylation of armadillo/β-catenin promotes ring canal growth in Drosophila oogenesis. PLoS One. 10:e0121484 10.1371/journal.pone.0121484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J.T., Demarest B.L., Bisgrove B.W., Su Y.-C., Smith M., and Yost H.J.. 2014. Poly peak parser: Method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products. Dev. Dyn. 243:1632–1636. 10.1002/dvdy.24183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., Sahlender D.A., Choma M., Sinka R., Harbour M.E., Parkinson M., and Robinson M.S.. 2009. Spatial and functional relationship of GGAs and AP-1 in Drosophila and HeLa cells. Traffic. 10:1696–1710. 10.1111/j.1600-0854.2009.00983.x [DOI] [PubMed] [Google Scholar]
- Hutagalung A.H., and Novick P.J.. 2011. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91:119–149. 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida M., E Oguchi M., and Fukuda M.. 2016. Multiple types of guanine nucleotide exchange factors (GEFs) for Rab small GTPases. Cell Struct. Funct. 41:61–79. 10.1247/csf.16008 [DOI] [PubMed] [Google Scholar]
- Ivan V., de Voer G., Xanthakis D., Spoorendonk K.M., Kondylis V., and Rabouille C.. 2008. Drosophila Sec16 mediates the biogenesis of tER sites upstream of Sar1 through an arginine-rich motif. Mol. Biol. Cell. 19:4352–4365. 10.1091/mbc.E08-03-0246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovics F., Sinka R., and Erdélyi M.. 2001. An interaction type of genetic screen reveals a role of the Rab11 gene in oskar mRNA localization in the developing Drosophila melanogaster oocyte. Genetics. 158:1177–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S., Newman C., Liu F., and Segev N.. 2000. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol. Biol. Cell. 11:4403–4411. 10.1091/mbc.11.12.4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe A.D., Wilson D.S., Seelig B., and Szostak J.W.. 2001. One-step purification of recombinant proteins using a nanomolar-affinity streptavidin-binding peptide, the SBP-Tag. Protein Expr. Purif. 23:440–446. 10.1006/prep.2001.1515 [DOI] [PubMed] [Google Scholar]
- Kim J.J., Lipatova Z., and Segev N.. 2016. TRAPP complexes in secretion and autophagy. Front. Cell Dev. Biol. 4:20 10.3389/fcell.2016.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-G., Raunser S., Munger C., Wagner J., Song Y.-L., Cygler M., Walz T., Oh B.-H., and Sacher M.. 2006. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell. 127:817–830. 10.1016/j.cell.2006.09.029 [DOI] [PubMed] [Google Scholar]
- Koumandou V.L., Dacks J.B., Coulson R.M.R., and Field M.C.. 2007. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol. Biol. 7:29 10.1186/1471-2148-7-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leicht B.G., and Bonner J.J.. 1988. Genetic analysis of chromosomal region 67A-D of Drosophila melanogaster. Genetics. 119:579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Luo X., Zhao S., Siu G.K., Liang Y., Chan H.C., Satoh A., and Yu S.S.. 2017. COPI-TRAPPII activates Rab18 and regulates its lipid droplet association. EMBO J. 36:441–457. 10.15252/embj.201694866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch-Day M.A., Bhandari D., Menon S., Huang J., Cai H., Bartholomew C.R., Brumell J.H., Ferro-Novick S., and Klionsky D.J.. 2010. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA. 107:7811–7816. 10.1073/pnas.1000063107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiling-Wesse K., Epple U.D., Krick R., Barth H., Appelles A., Voss C., Eskelinen E.-L., and Thumm M.. 2005. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J. Biol. Chem. 280:33669–33678. 10.1074/jbc.M501701200 [DOI] [PubMed] [Google Scholar]
- Milev M.P., Hasaj B., Saint-Dic D., Snounou S., Zhao Q., and Sacher M.. 2015. TRAMM/TrappC12 plays a role in chromosome congression, kinetochore stability, and CENP-E recruitment. J. Cell Biol. 209:221–234. 10.1083/jcb.201501090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir A., Kaufman L., Noor A., Motazacker M.M., Jamil T., Azam M., Kahrizi K., Rafiq M.A., Weksberg R., Nasr T., et al. 2009. Identification of mutations in TRAPPC9, which encodes the NIK- and IKK-beta-binding protein, in nonsyndromic autosomal-recessive mental retardation. Am. J. Hum. Genet. 85:909–915. 10.1016/j.ajhg.2009.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida G.H., Mahajnah M., Hill A.D., Basel-Vanagaite L., Gleason D., Hill R.S., Bodell A., Crosier M., Straussberg R., and Walsh C.A.. 2009. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am. J. Hum. Genet. 85:897–902. 10.1016/j.ajhg.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova N., Liang Y., Tokarev A.A., Chen S.H., Cox R., Andrejic J., Lipatova Z., Sciorra V.A., Emr S.D., and Segev N.. 2006. TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nat. Cell Biol. 8:1263–1269. 10.1038/ncb1489 [DOI] [PubMed] [Google Scholar]
- Nie Y., Bellon-Echeverria I., Trowitzsch S., Bieniossek C., and Berger I.. 2014. Multiprotein complex production in insect cells by using polyproteins. Methods Mol. Biol. 1091:131–141. 10.1007/978-1-62703-691-7_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S.R. 2013. Rab GTPase regulation of membrane identity. Curr. Opin. Cell Biol. 25:414–419. 10.1016/j.ceb.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe O., Rio M., Carioux A., Plaza J.-M., Guigue P., Molinari F., Boddaert N., Bole-Feysot C., Nitschke P., Smahi A., et al. 2009. Combination of linkage mapping and microarray-expression analysis identifies NF-κB signaling defect as a cause of autosomal-recessive mental retardation. Am. J. Hum. Genet. 85:903–908. 10.1016/j.ajhg.2009.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinar M., Arst H.N. Jr., Pantazopoulou A., Tagua V.G., de los Ríos V., Rodríguez-Salarichs J., Díaz J.F., and Peñalva M.A.. 2015. TRAPPII regulates exocytic Golgi exit by mediating nucleotide exchange on the Ypt31 ortholog RabERAB11. Proc. Natl. Acad. Sci. USA. 112:4346–4351. 10.1073/pnas.1419168112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Chen H.-M., Lee T., and Bullock S.L.. 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA. 111:E2967–E2976. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson B.C., and Fromme J.C.. 2015. Biochemical methods for studying kinetic regulation of Arf1 activation by Sec7. Methods Cell Biol. 130:101–126. 10.1016/bs.mcb.2015.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel F., Gillingham A.K., Rosa-Ferreira C., Galindo A., and Munro S.. 2016. An antibody toolkit for the study of membrane traffic in Drosophila melanogaster. Biol. Open. 5:987–992. 10.1242/bio.018937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C.C., Giansanti M.G., Gatti M., and Fuller M.T.. 2009. TRAPPII is required for cleavage furrow ingression and localization of Rab11 in dividing male meiotic cells of Drosophila. J. Cell Sci. 122:4526–4534. 10.1242/jcs.054536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G., Kolstad K., Stone S., Palluault F., and Ferro-Novick S.. 1995. BET3 encodes a novel hydrophilic protein that acts in conjunction with yeast SNAREs. Mol. Biol. Cell. 6:1769–1780. 10.1091/mbc.6.12.1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M., Jiang Y., Barrowman J., Scarpa A., Burston J., Zhang L., Schieltz D., Yates J.R. III, Abeliovich H., and Ferro-Novick S.. 1998. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 17:2494–2503. 10.1093/emboj/17.9.2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M., Barrowman J., Wang W., Horecka J., Zhang Y., Pypaert M., and Ferro-Novick S.. 2001. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol. Cell. 7:433–442. 10.1016/S1097-2765(01)00190-3 [DOI] [PubMed] [Google Scholar]
- Sakaguchi A., Sato M., Sato K., Gengyo-Ando K., Yorimitsu T., Nakai J., Hara T., Sato K., and Sato K.. 2015. REI-1 Is a guanine nucleotide exchange factor regulating RAB-11 localization and function in C. elegans embryos. Dev. Cell. 35:211–221. 10.1016/j.devcel.2015.09.013 [DOI] [PubMed] [Google Scholar]
- Satoh A.K., O’Tousa J.E., Ozaki K., and Ready D.F.. 2005. Rab11 mediates post-Golgi trafficking of rhodopsin to the photosensitive apical membrane of Drosophila photoreceptors. Development. 132:1487–1497. 10.1242/dev.01704 [DOI] [PubMed] [Google Scholar]
- Satoh T., Nakamura Y., and Satoh A.K.. 2016. The roles of Syx5 in Golgi morphology and Rhodopsin transport in Drosophila photoreceptors. Biol. Open. 5:1420–1430. 10.1242/bio.020958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrivens P.J., Noueihed B., Shahrzad N., Hul S., Brunet S., and Sacher M.. 2011. C4orf41 and TTC-15 are mammalian TRAPP components with a role at an early stage in ER-to-Golgi trafficking. Mol. Biol. Cell. 22:2083–2093. 10.1091/mbc.E10-11-0873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Peak-Chew S.Y., and Pelham H.R.. 2000. Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J. 19:4885–4894. 10.1093/emboj/19.18.4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinka R., Jankovics F., Somogyi K., Szlanka T., Lukácsovich T., and Erdélyi M.. 2002. poirot, a new regulatory gene of Drosophila oskar acts at the level of the short Oskar protein isoform. Development. 129:3469–3478. [DOI] [PubMed] [Google Scholar]
- Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525. 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Tan D., Cai Y., Wang J., Zhang J., Menon S., Chou H.-T., Ferro-Novick S., Reinisch K.M., and Walz T.. 2013. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc. Natl. Acad. Sci. USA. 110:19432–19437. 10.1073/pnas.1316356110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taussig D., Lipatova Z., and Segev N.. 2014. Trs20 is required for TRAPP III complex assembly at the PAS and its function in autophagy. Traffic. 15:327–337. 10.1111/tra.12145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L.L., and Fromme J.C.. 2016. GTPase cross talk regulates TRAPPII activation of Rab11 homologues during vesicle biogenesis. J. Cell Biol. 215:499–513. 10.1083/jcb.201608123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L.L., Joiner A.M.N., and Fromme J.C.. 2017. The TRAPPIII complex activates the GTPase Ypt1 (Rab1) in the secretory pathway. J. Cell Biol. 10.1083/jcb.201705214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong C., Ohyama T., Tien A.-C., Rajan A., Haueter C.M., and Bellen H.J.. 2011. Rich regulates target specificity of photoreceptor cells and N-cadherin trafficking in the Drosophila visual system via Rab6. Neuron. 71:447–459. 10.1016/j.neuron.2011.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., and Ferro-Novick S.. 2002. A Ypt32p exchange factor is a putative effector of Ypt1p. Mol. Biol. Cell. 13:3336–3343. 10.1091/mbc.01-12-0577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Sacher M., and Ferro-Novick S.. 2000. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J. Cell Biol. 151:289–296. 10.1083/jcb.151.2.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler F., Gillingham A.K., Sinka R., Rosa-Ferreira C., Gordon D.E., Franch-Marro X., Peden A.A., Vincent J.-P., and Munro S.. 2010. A genome-wide RNA interference screen identifies two novel components of the metazoan secretory pathway. EMBO J. 29:304–314. 10.1038/emboj.2009.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlwill A.D., and Bonner J.J.. 1991. Genetic analysis of chromosome region 63 of Drosophila melanogaster. Genetics. 128:763–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki A., Menon S., Yu S., Barrowman J., Meerloo T., Oorschot V., Klumperman J., Satoh A., and Ferro-Novick S.. 2009. mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI-coated vesicles. Mol. Biol. Cell. 20:4205–4215. 10.1091/mbc.E09-05-0387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Li C.M., Luo X.M., Siu G.K.Y., Gan W.J., Zhang L., Wu W.K.K., Chan H.C., and Yu S.. 2017. Mammalian TRAPPIII Complex positively modulates the recruitment of Sec13/31 onto COPII vesicles. Sci. Rep. 7:43207 10.1038/srep43207 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.