TBC1D7 negatively regulated cell growth as a subunit of the tuberous sclerosis complex (TSC). Ren et al. show that dTBC1D7 regulates systemic growth by modulating the biosynthesis and release of insulin-like peptide 2 independently of TSC in Drosophila.
Abstract
The insulin signaling pathway plays key roles in systemic growth. TBC1D7 has recently been identified as the third subunit of the tuberous sclerosis complex (TSC), a negative regulator of cell growth. Here, we used Drosophila as a model system to dissect the physiological function of TBC1D7 in vivo. In mutants lacking TBC1D7, cell and organ growth were promoted, and TBC1D7 limited cell growth in a cell-nonautonomous and TSC-independent manner. TBC1D7 is specifically expressed in insulin-producing cells in the fly brain and regulated biosynthesis and release of insulin-like peptide 2, leading to systemic growth. Furthermore, animals carrying the dTBC1D7 mutation were hypoglycemic, short-lived, and sensitive to oxidative stress. Our findings provide new insights into the physiological function of TBC1D7 in the systemic control of growth, as well as insights into human disorders caused by TBC1D7 mutation.
Introduction
Organisms must coordinate growth with the nutritional status of their environment to maintain tissue homeostasis. Multiple intrinsic and extrinsic signals are integrated by cells to control body size (Oldham and Hafen, 2003; Jewell and Guan, 2013). The highly conserved insulin/insulin-like growth factor (IGF) signaling pathway plays a key role in controlling systemic growth and in various aspects of organismal physiology, including longevity, stress resistance, and fecundity (in both vertebrates and invertebrates; Brogiolo et al., 2001; Oldham and Hafen, 2003; Grönke et al., 2010; Jewell and Guan, 2013; Koyama and Mirth, 2016).
In Drosophila melanogaster, eight insulin-like peptides (ILP1–8), which are homologous to vertebrate insulin/IGF, have been identified. These genes are expressed in distinct spatial and temporal patterns (Okamoto et al., 2009; Grönke et al., 2010; Colombani et al., 2012a). Studies using flies as a model have led to significant insights regarding the roles of ILPs as determinants of systemic growth (Brogiolo et al., 2001; Rulifson et al., 2002). Unlike mammalian insulin, which is secreted from the pancreas, the major fly ILPs (ILP2, ILP3, and ILP5) are specifically expressed in clusters of neurosecretory neurons and insulin-producing cells (IPCs) in the anteromedial region of the brain and are secreted into the hemolymph from IPC axon terminals (Ikeya et al., 2002; Alfa et al., 2015). The secretion of ILP from IPCs in the brain is tightly controlled by nutrient availability, thereby linking systemic growth with environmental conditions (Lee et al., 2008; Géminard et al., 2009; Rajan and Perrimon, 2012). In this way, the synthesis and secretion of ILPs by IPCs play a central role in controlling the systemic growth rate (Géminard et al., 2009; Hasygar and Hietakangas, 2014; Alfa et al., 2015). However, little is known about the mechanisms that control the biogenesis and release of ILPs by IPCs.
Circulating insulin/IGF regulates cell growth by activating the evolutionarily conserved insulin/mechanistic target of rapamycin complex 1 (mTORC1) pathway in somatic cells (Oldham and Hafen, 2003; Jewell and Guan, 2013; Kim and Neufeld, 2015). The tuberous sclerosis complex (TSC), which consists of TSC1 and TSC2, is an essential node of the insulin/mTORC1 pathway (Menon et al., 2014). mTORC1 promotes cell growth in response to both nutrients and insulin/IGF, and the TSC negatively regulates mTORC1 by functioning as a GTPase-activating protein for Rheb, an essential activator of mTORC1 (Inoki et al., 2003; Saucedo et al., 2003; Stocker et al., 2003; Zhang et al., 2003). Insulin/IGF activates mTORC1 signaling by inhibiting the TSC through the insulin receptor/phosphoinositide 3-kinase/AKT signaling cascade (Dibble and Manning, 2013). A TBC1 Domain Family Member 7 (TBC1D7) was recently identified as the third core subunit of the TSC (Dibble et al., 2012). TBC1D7 stabilizes the TSC through direct interactions with TSC1, thus TBC1D7 knockdown decreased GTPase-activating protein activity of TSC toward Rheb, which limits mTORC1 signaling and cell growth (Dibble et al., 2012; Gai et al., 2016; Qin et al., 2016). Somatic loss of heterozygosity in TSC1 or TSC2 leads to the occurrence of benign tumors in various organs (European Chromosome 16 Tuberous Sclerosis Consortium, 1993; van Slegtenhorst et al., 1997). However, loss of TBC1D7 is associated with intellectual disability and megalencephaly (Capo-Chichi et al., 2013; Alfaiz et al., 2014). These divergent phenotypes imply that TBC1D7 has important physiological functions that are distinct from its TSC complex–associated functions. Nevertheless, the physiological roles of TBC1D7 have not been well studied.
Results and discussion
With the goal of dissecting the physiological functions of TBC1D7, we identified CG6182 as the only gene with homology to TBC1D7 in the fly genome. Identical and conserved residues were 32.5% and 52.2%, respectively, when CG6182 and human TBC1D7 protein sequences were aligned (through BLASTP [http://flybase.org/blast/] analysis of human TBC1D7 and fly genomes; Fig. S1 A). Consistent with previous work, overexpressing TSC1 and TSC2 simultaneously in the compound eye of Drosophila resulted in smaller ommatidia (compared with control eyes), through down-regulation of mTORC1 (Fig. S1, B–D; Gao and Pan, 2001; Potter et al., 2001; Gao et al., 2002). However, when levels of dTBC1D7 were manipulated (either overexpressed or knocked down) in combination with TSC1 and TSC2 overexpression, the size of the ommatidia was not affected (Fig. S1, B–D). With these interesting results, we generated a null dTBC1D7 mutant (dTBC1D7KO) using CRISPR/Cas9-mediated genome editing (Fig. S1, E and F). Whereas TSC1 and TSC2 homozygous mutants are lethal, homozygous dTBC1D7 mutants were viable. These results suggest that TBC1D7 may not be a constitutive component of the TSC.
We found that dTBC1D7KO flies exhibited larger body sizes than wild-type flies at both the pupal and adult stages (Fig. 1, A and B). Mutant flies had significantly higher body weights (22%, P < 0.001) than wild-type flies regardless of sex (male flies were chosen for further investigation; Fig. 1 C). To determine whether this increase in size and weight resulted from the enlargement of particular organs or tissues, we assessed growth of the eye and wing, because growth of these tissues has been extensively characterized. Compared with wild-type flies, dTBC1D7KO flies exhibited (1) larger ommatidia, but the number of ommatidia did not differ (Fig. 1, D–F), and (2) larger average wing areas with the number of cells remaining the same (Fig. 1, G–I). Furthermore, whole-body overgrowth was rescued by the ubiquitous expression of a UAS-dTBC1D7 transgene by the daughterless-Gal4 (da-Gal4) driver (Fig. 1 J). These results indicate that dTBC1D7 is involved in regulating multiple growth processes in Drosophila.
Figure 1.
dTBC1D7 mutant increased body size. (A and B) The overgrowth of the dTBC1D7KO animals was found in both pupa (A) and adult stages (B). Bars, 400 µm. (C) Quantification of body weight of 3-d-old flies. At least 200 flies were used for each genotype. (D) Scanning electron microscopy of wild-type, dTBC1D7, mutant, FRT 82B (ey-flpRh1::GFP;;FRT82B/GMR-hid CL FRT82B), and FRT82B dTBC1D7 KO/FRT82B GMR-hid (ey-flpRh1::GFP;;FRT82B, dTBC1D7KO/FRT82B, GMR-hid CL) compound eyes. Bars, 100 µm. (E) Quantification of ommatidia size. At least 20 male flies of each genotype were assayed. (F) The total number of ommatidia in rows 14–18 of compound eyes was counted. At least 20 male flies of each genotype were assayed. (G) The wings of wild-type and dTBC1D7 mutant flies. Bars, 200 µm. (H and I) Qualification of wing area (H) and cell number (I) of dTBC1D7 mutants and wild-type. Wings from at least 20 male flies of each genotype were measured. (J) Ubiquitous expression of dTBC1D7 (da>dTBC1D7: da-Gal4/UAS-dTBC1D7) rescued the increased body weight of dTBC1D7 mutants (KO: dTBC1D7KO). At least 200 male or female flies of each genotype were used. Error bars indicate the SD, and significant differences against same-sex wild-type flies were determined with Student’s t test at indicated genotypes. ***, P < 0.001; ns, not significant.
It is known that up-regulation of mTORC1 activity results in cell-autonomous overgrowth in Drosophila (Saucedo et al., 2003; Stocker et al., 2003; Vellai et al., 2003). We surmised that the overgrowth of dTBC1D7 mutants may result from the up-regulation of mTORC1 kinase activity, because TBC1D7 has been identified as the third subunit of the TSC (Dibble et al., 2012). To test this hypothesis, we first measured the phosphorylation status of a major mTORC1 substrate, the P70 ribosomal protein S6 kinase (S6K), which is thought to be indicative of mTORC1 kinase activity. Knockdown of TSC1 and/or TSC2 in S2 cells increased pS6K levels (Fig. 2 A). Conversely, pS6K levels were unchanged in both dTBC1D7 knockdown cells and dTBC1D7KO flies, suggesting that the mTORC1 kinase activity was not affected in dTBC1D7 mutant cells (Fig. 2, B–C). Collectively, these data indicate that dTBC1D7 may not be essential for TSC activity.
Figure 2.
dTBC1D7 regulated growth in a cell-nonautonomous manner. (A) Knockdown of TSC1, TSC2, or TSC1 and TSC2, but not dTBC1D7, in S2 cells by RNAi increased the phosphorylation levels of S6K (pS6K). DsRNA for GFP was used as a negative control. The efficiency of TBC1D7 knockdown in S2 cells was ∼60%, as determined by qPCR. S6K mRNA levels were not changed among the groups. (B) Western blot of pS6K in the wild-type and dTBC1D7 mutant flies. S6K mRNA levels were not changed among the groups. (C) Quantification of relative pS6K levels from three biological replicates from panel B. (D–E) Cells mutant for dTBC1D7 were the same sizes as the neighbor cells. (D) Clones of homozygous dTBC1D7KO cells marked with GFP were generated in fat body by the MARCM system, and cell boundaries were stained with phalloidin. Bar, 50 µm. (E) Quantification of the dTBC1D7KO and neighbor cells. At least 100 clones in six individuals were measured. (F–G) Overexpression of dTBC1D7 did not affect cell growth. (F) The dTBC1D7 overexpression (+dTBC1D7) clones were marked with GFP. Bar, 50 µm. (G) The cell size quantification of +dTBC1D7 clones. At least 100 clones in six individuals were measured. Bars, 50 µm. Error bars indicate SD, and significant differences were determined with Student’s t test at indicated genotypes. (H) The dTBC1D7KO fat body cells showed membrane localization of GPH. Bar, 50 µm. (I) Levels of pAKT and pS6K in fat body of wild-type, ilp21, and dTBC1D7KO flies as assessed by using Western blot analysis. ns, not significant.
We then asked whether TBC1D7 regulates growth in a cell-autonomous or nonautonomous manner. To answer this question, we first used a modified mitotic recombination approach to kill nonrecombinant retina cells by the eye-specific expression of the proapoptotic gene, hid. This generated homozygous dTBC1D7KO mutant eyes in otherwise heterozygous animals (FRT82B dTBC1D7KO/FRT82B GMR-hid; Wang et al., 2012). In contrast with whole-animal mutant dTBC1D7KO flies, which exhibit an increase in ommatidia size, compound eyes of FRT82B dTBC1D7KO/FRT82B GMR-hid flies had ommatidia of normal size (Fig. 1, D–F). Furthermore, homozygous dTBC1D7KO clones of cells were generated with the mosaic analysis with a repressible cell marker (MARCM) system (Lee et al., 1999), and clones of cells overexpressing dTBC1D7 were generated with the “flip out” method (Struhl and Basler, 1993). Thus, both dTBC1D7 loss- and gain-of-function cells were marked with GFP. When the fat body was subjected to phalloidin staining, boundaries between these clones and control cells were clearly visible. Cells lacking dTBC1D7 (Fig. 2, D and E) or overexpressing dTBC1D7 (Fig. 2, F and G) were both the same size as surrounding control cells, regardless of nutrition status. Collectively, these data indicate that dTBC1D7 may not be essential for TSC activity and that loss of dTBC1D7 induces growth in a cell-nonautonomous manner.
Growth also would be regulated by the circulating insulins. We next examined whether the increased circulating insulins in dTBC1D7 mutant flies activate the insulin signal transduction pathway by using the tGPH indicator, which fuses the PH domain of the Drosophila homologue of the general receptor for phosphoinositides-1 (GRP1) to GFP and reflects activation of cellular insulin signal transduction pathway (Britton et al., 2002; Oldham and Hafen, 2003; Saucedo et al., 2003). Similar to previous studies, under starvation conditions GPH localized to the cytoplasm and nucleus of fat body cells, indicating a limited amount of insulin secretion (Fig. 2 H). However, under the same condition, GPH instead localized to the plasma membrane of dTBC1D7 mutant cells (Fig. 2 H). To confirm that systemic insulin signaling activity is increased in dTBC1D7KO flies, we examined the activity of AKT and TORC1, which are downstream effectors of insulin signaling, in fat body. Mutation of dTBC1D7 promoted phosphorylation of Akt on Ser505 under starvation conditions, suggesting high levels of systemic insulin signaling in dTBC1D7KO animals (Fig. 2 I). However, TORC1-dependent phosphorylation of S6K Thr398 did not increase. Similarly, levels of pAKT, but not pS6K, were reduced in ilp2 mutant fat bodies, which is consistent with previous studies in which ilp2 mutants showed normal TORC1 activation in fat body (Fig. 2 I; Kim and Neufeld, 2015). This differential effect on TORC1 activation may be because mTORC1 is primarily regulated by intracellular nutrients.
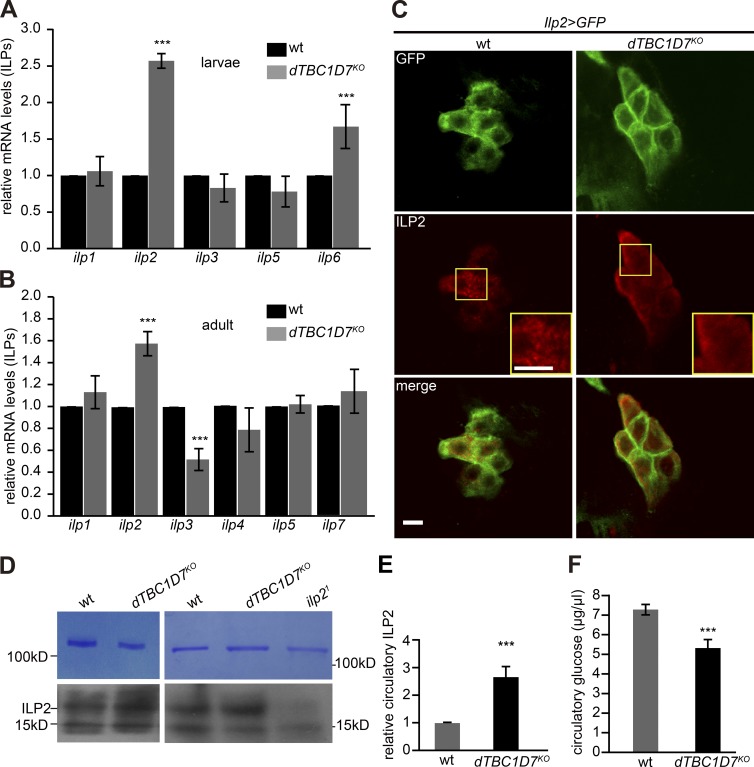
It has been established that extracellular signals, such as ILPs secreted by IPCs in Drosophila, control systemic growth in cell nonautonomous ways (Brogiolo et al., 2001; Ikeya et al., 2002; Rulifson et al., 2002; Delanoue et al., 2016; Koyama and Mirth, 2016). Previous work has revealed that the Drosophila genome encodes eight ILPs (ILP1–8) and that these ILPs are synthesized and secreted in different tissues at different developmental stages to regulate growth and development (Okamoto et al., 2009; Colombani et al., 2012). Thus, we measured levels of transcription for all ilp genes in dTBC1D7 mutants using quantitative PCR (qPCR). Interestingly, we found that ilp2 mRNA levels were significantly up-regulated in dTBC1D7 mutants compared with wild-type controls, in both larvae and adult flies (2.5- and 1.5-fold, respectively) (Fig. 3, A and B). Many studies have shown that the synthesis and secretion of ILP2 by IPC neurons is the key regulator of growth in flies (Ikeya et al., 2002; Rulifson et al., 2002; Grönke et al., 2010; Koyama and Mirth, 2016). We therefore used immunostaining to detect ILP2 in IPC neurons that were marked with GFP (Ilp2-Gal4/UAS-GFP) and found that in wild-type flies ILP2 appeared in a granule-like pattern, very similar to patterns reported for human IPCs (Park et al., 2014). In contrast, ILP2 granules were almost undetectable in dTBC1D7 mutants (Fig. 3 C). Thus, increased levels of ILP2 production and secretion by IPC neurons may underlie the increased growth observed in dTBC1D7 mutants.
Figure 3.
Secretion of ILP2 was elevated in the dTBC1D7 mutant flies. (A–B) Transcriptional levels of ilps in the wild-type and dTBC1D7 mutant animals at (A) larval stage and (B) adult stage were measured by qPCR. The expression of ilp6 was not detected in adult animals. Five independent replicates were used for quantification. (C) Brain staining of ILP2 in the wild-type and dTBC1D7KO flies with GFP expression in ILP2 cells (Ilp2>GFP: Ilp2-Gal4/UAS-GFP). Flies were dissected and immunostained for GFP (green) and ILP2 (red). High magnification pictures are shown in yellow frames. Bar, 50 µm. (D) Measurement of circulatory ILP2 in the hemolymph in the wild-type and dTBC1D7KO and ilp21 flies. The top PAGE gels were stained with Coomassie brilliant blue, and one 100-kd band of putative larval serum protein was used as loading control. The bottom PAGE gels were used for Western blot against ILP2. (E) Quantification of circulatory ILP2 from five biological replicates based on panel D. (F) Determination of circulatory glucose in hemolymph of the wild-type and dTBC1D7 mutant flies. Six biological replications were used for qualification. Error bars indicate SD. Significant differences were determined with Student’s t test at indicated genotypes. ***, P < 0.001; ns, not significant.
Furthermore, we directly measured levels of circulating ILP2 in hemolymph from wild-type, dTBC1D7KO, and ilp2 mutant flies by Western blot. The ILP2 Western blot analysis revealed that levels of circulating ILP2 were increased in dTBC1D7 mutant flies (Fig. 3, D and E). Insulins are well known to control circulating glucose levels in various organisms (Koyama and Mirth, 2016). Indeed, the concentration of glucose in the hemolymph of dTBC1D7KO animals was significantly lower than in wild-type animals (Fig. 3 F). Collectively, our data establish that circulating insulin was elevated in dTBC1D7 mutants.
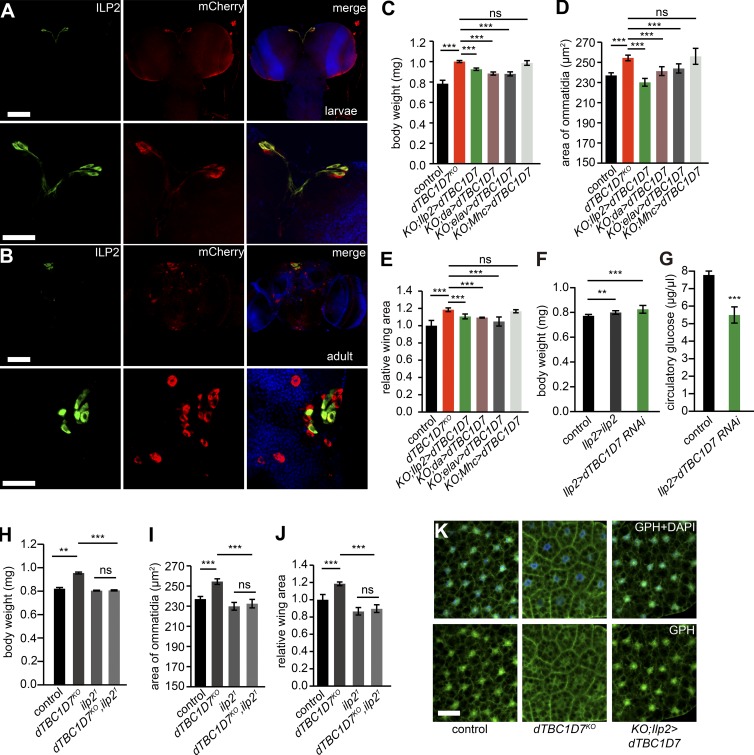
To determine how dTBC1D7 regulates the synthesis and release of ILP2, we first characterized the expression pattern of dTBC1D7 in flies. Using CRISPR/Cas9 genome editing, we generated an mCherry knock-in allele, in which mCherry expression is driven by the native dTBC1D7 promoter (dTBC1D7-mCherry; Fig. S2, A–C). mCherry Western blotting first revealed that dTBC1D7 expression was specifically enriched in the brain and ovaries (Fig. S2 D). Staining brains from larvae and adults with antibodies against mCherry and ILP2 showed that mCherry primarily localized to a subset of neurons that almost all contained ILP2 (Fig. 4, A and B). This suggests that dTBC1D7 is expressed in and may function in the IPC neurons. Encouragingly, expression of dTBC1D7 in (1) IPC neurons (Ilp2-Gal4/UAS-dTBC1D7), (2) all neurons (elav-Gal4/UAS-dTBC1D7), or (3) whole flies (da-Gal4/UAS-dTBC1D7) rescued the systemic overgrowth of dTBC1D7KO mutants. In contrast, expressing dTBC1D7 in muscle (Mhc-gla4/UAS-dTBC1D7) did not rescue the dTBC1D7KO overgrowth phenotypes (Fig. 4, C–E). Similar to dTBC1D7 mutant animals, tissue-specific overexpression of ILP2 in IPC neurons resulted in increased body weight, and expression of an dTBC1D7 RNAi transgene in IPC neurons resulted in increased body weight and lower levels of circulating glucose (Fig. 4, F and G). It is worth mentioning that direct overexpression of ILP2 had less effect on body weight than dTBC1D7 knockdown, which may be because the secretion of ILP2 is regulated (Cao et al., 2014). IPC-specific knockdown of dTBC1D7 in IPC neurons did not fully recapitulate the dTBC1D7KO phenotypes, which might imply that other dTBC1D7-expressing cells contribute to systemic growth regulation as well. Importantly, the overgrowth phenotypes of dTBC1D7 mutants (including body weight, ommatidia size, and wing area) were completely reversed by introducing the ilp2 mutation (Fig. 4, H–J). Furthermore, membrane localization of the GPH signaling after starvation treatment could be rescued by IPC-specific expression of dTBC1D7 (Fig. 4 K). The epistasis analysis between dTBC1D7 and ilp2 indicated that the induction of growth in dTBC1D7 mutants required ILP2. These results suggest that TBC1D7 functions in IPC neurons and that this IPC-neuron–specific function is sufficient to control growth.
Figure 4.
dTBC1D7 expressed and functioned in IPC neurons. (A) Staining of mCherry and ILP2 in the brain of dTBC1D7 knock-in larvae (dTBC1D7-mCherry), in which mCherry expression was driven by the native promoter of dTBC1D7. (B) Staining of mCherry and ILP2 in the brain of dTBC1D7-mCherry flies at the adult stage. Bars: (top) 50 µm; (bottom) 100 µm. (C–E) Rescue of the overgrowth phenotypes of dTBC1D7 mutant by expression of dTBC1D7 in all cells (da>dTBC1D7: da-Gal4/UAS-dTBC1D7), neuronal cells (elav>dTBC1D7: elav-Gal4/UAS-dTBC1D7), and IPC neurons (Ilp2>dTBC1D7: Ilp2-Gal4/UAS-dTBC1D7); muscle expression (Mhc>dTBC1D7: Mhc-Gal4/UAS-dTBC1D7) failed to rescue the growth phenotypes. Body weight (C), ommatidia size (D), and wing area (E) were quantified. The “control” refers to the Ilp2-Gal4/+ flies. (F) Knockdown dTBC1D7 (Ilp2>dTBC1D7RNAi: Ilp2-Gal4/UAS-dTBC1D7RNAI) or overexpression of ilp2 in IPC neurons (Ilp2>ilp2: Ilp2-Gal4/UAS-ilp2) increased body sizes. At least 200 male flies of each genotype were used. The “control” refers to the Ilp2-Gal4/+ flies. Levels of dTBC1D7 transcripts in the Ilp2>dTBC1D7RNAi brain were reduced 50% compared with control brains. (G) Circulatory glucose levels decreased in hemolymph of the Ilp2>dTBC1D7RNAi flies. Five biological replications were used for quantification. (H–J) Mutation of ilp2 (ilp21) rescued growth phenotypes, included body weight (H), ommatidia size (I), and relative wing area (J) of dTBC1D7KO animals. At least 20 male flies of each genotype were assayed. Significant differences were determined with Student’s t test. **, P < 0.01; ***, P < 0.001; ns, not significant. (K) Expression of dTBC1D7 in IPC neurons reduced membrane localization of GPH in dTBC1D7KO flies. “Control” refers to Ilp2-Gal4/+ flies. Bar, 25 µm.
Next, we sought to determine whether the ability of dTBC1D7 to control growth depends on its status as a component of the TSC. The same as TBC1D7 in mammals, dTBC1D7 interacted with TSC1 in vitro (Fig. 5 A), and expression of human TBC1D7 in IPC neurons (Ilp2-Gal4/UAS-hTBC1D7) rescued the systemic overgrowth of dTBC1D7KO mutants, including body weight and ommatidia and wing sizes. This indicates that TBC1D7 mediates conserved functions (Fig. 5, B–D). Work in mammals has shown that L114Q or L114R mutations in hTBC1D7 disrupt interactions between hTBC1D7 and TSC1 (Glatter et al., 2011; Gabernet-Castello et al., 2013; Santiago Lima et al., 2014). Bioinformatics analysis suggested that I125 within dTBC1D7 is L114 in hTBC1D7. Indeed, dTBC1D7I125Q failed to interact with TSC1 in S2 cells (Fig. 5 A). Therefore, we expressed dTBC1D7I125Q via Ilp2-Gal4 in the dTBC1D7KO mutant background and found that dTBC1D7I125Q rescued the overgrowth phenotype to the same extent as wild-type dTBC1D7 (Fig. 5, E–G). This suggests that the function of dTBC1D7 in IPC neurons is independent of the TSC. Because it is very difficult to directly measure levels of mTORC1 kinase activity in IPC neurons, we genetically manipulated mTORC1 kinase activity in IPC cells. The TSC negatively regulates activity of the Rheb GTPase, and overexpression of Rheb increases intercellular mTORC1 activity in Drosophila (Saucedo et al., 2003; Stocker et al., 2003). Here, we found that, in contrast to induction of systemic overgrowth by knockdown of dTBC1D7KO in IPCs, overexpression of Rheb in IPC neurons (via Ilp2-Gal4) did not affect systemic growth (Fig. 5, H and I). These biochemical and genetic results provide strong evidence that TBC1D7 regulates systemic growth in IPC neurons independently of the TSC.
Figure 5.
dTBC1D7 controlled systemic growth independent of TSC. (A) dTBC1D7I125Q presented loss of interaction with TSC1 in S2 cells. (B–D) Expression of human TBC1D7 in IPC neurons (Ilp2-Gal4/UAS-hTBC1D7) rescued the systemic overgrowth phenotypes of dTBC1D7 mutants, including body weight (B), ommatidia size (C), and relative wing area (D). (E–G) Expression of TBC1D7I125Q in IPC neurons (Ilp2>dTBC1D7I125Q: Ilp2-Gal4/UAS-dTBC1D7I125Q) rescued the overgrowth phenotypes of the dTBC1D7KO (KO: dTBC1D7KO) animals. Body weight (E), wing area (F), and ommatidia size (G) were quantified. (H–I) Knockdown dTBC1D7 in IPC neurons increased body weight (H) and wing size (I), whereas overexpression of Rheb in the IPC neurons did not. The “control” refers to the Ilp2-Gal4/+ flies. At least 20 flies of each genotype were quantified. Significant differences were determined with Student’s t test. ***, P < 0.001; ns, not significant. Error bars indicate SD.
Numerous studies have reported that up-regulation of this highly conserved insulin signaling pathway results in decreased fitness and reduced lifespan (Broughton et al., 2005; Bai et al., 2012). These speculations, together with our observation that IPC neurons of dTBC1D7 mutants secrete more ILP2, motivated us to test whether manipulation of dTBC1D7 expression in IPC neurons affects longevity and stress resistance. We first monitored fly lifespan under normal conditions and found that loss of TBC1D7 shortened lifespan in both females and males (males were used for further analysis; Fig. S3 A). Similarly, overexpression of ILP2 and knockdown of dTBC1D7 in IPC neurons also shortened life span.
Growth and aging processes are also controlled by a variety of stresses in the environment (Holzenberger et al., 2003; Broughton et al., 2010; Bai et al., 2012; Laplante and Sabatini, 2012). Thus, we next exposed dTBC1D7 mutant flies to oxidative stress. This type of stress was induced by using 5 mM paraquat, which is toxic and causes mitochondrial dysfunction (Holzenberger et al., 2003; Grönke et al., 2010). The dTBC1D7 mutant flies exhibited a greatly reduced life span in response to paraquat treatment compared with controls (Fig. S3 B). These results suggest that dTBC1D7 is an important regulator of stress responses and life span in IPC neurons within the Drosophila central nervous system.
Aging is generally associated with a decline in mitochondrial function, and various studies have demonstrated that the aging process eventually disrupts muscle tissue homeostasis (Cai et al., 2006). Here, we found that mitochondria from 30-d-old dTBC1D7 mutants were vacuolated and had disrupted cristae when compared with wild-type flies of the same age (Fig. S3 C). Consistent with this mitochondrial degeneration, behavioral assays conducted with 30-d-old flies revealed that the climbing activities of dTBC1D7 mutants were significantly reduced compared with wild-type flies (Fig. S3 D). These data represent subcellular and behavioral evidence that the disruption of TBC1D7 function results in outcomes associated with the aging process. This suggests that TBC1D7 regulates processes that integrate growth and aging.
In summary, we describe for the first time the physiological function of TBC1D7 in regulating ILP biogenesis and secretion, which in turn affects growth, stress response, and life span. Although biochemical studies have demonstrated that TBC1D7 physically associates with the TSC (Santiago Lima et al., 2014; Gai et al., 2016; Qin et al., 2016), clinical manifestations of TBC1D7-related diseases [namely, intellectual disabilities and megalocephaly with unknown mechanisms (Capo-Chichi et al., 2013; Alfaiz et al., 2014)] are very different from those associated with TSC1 or TSC2 mutations, which usually result in tumorigenesis through the constitutive activation of mTORC1 (European Chromosome 16 Tuberous Sclerosis Consortium, 1993; van Slegtenhorst et al., 1997). In Drosophila, it has been reported that CG6182 is the homologue of mammalian TBC1D7 (Housden et al., 2015; Vinayagam et al., 2016). After searching for TBC1D7 homologues in the fly genome, we found that CG6182 is the only homologue of human TBC1D7. Moreover, expression of human TBC1D7 can substitute for dTBC1D7 to control systemic growth. Using the Drosophila model system, we demonstrate that dTBC1D7 plays a critical role in regulating neuropeptides such as ILP2 in the specific IPC neurons and that dTBC1D7 performs this role independent of the TSC. In mammals, TBC1D7 has been shown to contribute to TSC1/TSC2 function (Dibble et al., 2012; Santiago Lima et al., 2014). However, databases of protein expression patterns indicate that TSC1/2 and TBC1D7 are not always expressed together. TSC1 and TSC2 are expressed throughout the body, whereas TBC1D7 localizes to specific regions, such as the hippocampus in mammalian brains (Lein et al., 2007). This suggests that TBC1D7 may play a conserved TSC1/2-independent role in neuron-related physiological processes in Drosophila and mammals. In fact, mammalian studies have shown that insulin/IGF signaling is essential for neurogenesis, learning, and sensory perception (Holzenberger et al., 2003; Fernandez and Torres-Alemán, 2012). Thus, our results have important implications for human health and help explain why mutations in human TBC1D7 cause intellectual disabilities.
In mammals, multiple IGFs (including one insulin, two IGFs, three relaxins, and several ILPs) have been shown to function separately (Fernandez and Torres-Alemán, 2012), and in flies recent studies have strongly suggested that the transcription and release of individual ILPs from IPCs may be regulated independently by different triggers (Géminard et al., 2009; Broughton et al., 2010; Alfa et al., 2015). However, it has not yet been determined how ILP2, ILP3, and ILP5 are differentially regulated in IPCs. Here we demonstrated that dTBC1D7 functions in IPCs to selectively regulate the expression of ILP2. Therefore, dTBC1D7 is the first IPC cell-autonomous factor that has been shown to regulate the expression of a selective ILP. Thus, dTBC1D7 may represent a key mechanism by which nutrient signaling is relayed to ultimately affect ILP2 expression and release. Although our work reveals that dTBC1D7 regulates ILP2 biosynthesis in vivo, the molecular mechanisms remain to be discovered.
Materials and methods
Drosophila stocks
The following fly strains were used in this study: w1118, UAS-tsc1, UAS-tsc2, and UAS-Rheb flies, which have been described previously (Wang et al., 2012). The ilp21 (BL30881), Mhc-Gal4 (BL55133), elav-Gal4 (BL25750), da-Gal4 (BL55851), MARCM82B (BL44408), tGPH (BL8163), Ilp2-Gal4 (BL37516), ey-Gal4 UAS-FLP; FRT82B GMR-hid (BL5253) and hs-flp; UAS-dcr2; actin>>CD2>>Gal4 UAS-GFP (BL4780), GMR-Gal4 (BL8605) flies were obtained from the Bloomington Stock Center. The nos-Cas9 (TH00787.N) flies and dTBC1D7 (TH0439) RNAi flies were obtained from TsingHua Fly Center. The dTBC1D7KO and dTBC1D7-mCherry flies were generated by a Cas9/sgRNA system. The UAS-dTBC1D7, UAS-dTBC1D7I125Q, and UAS-ilp2 flies were generated by site-specific transgenic technique.
Generation of knockout and knock-in flies
The dTBC1D7KO mutations were generated by the Cas9/sgRNA system (Xu et al., 2015). Two recognition sequences of guiding RNA to the dTBC1D7 locus were designed (sgRNA1: 5′-TGCTAGGGTGTTACCAGCGT-3′; sgRNA2: 5′-AAGATATGGGACAAAGTCTG-3′) and cloned into the U6b-sgRNA-short vector. The plasmids were injected into the embryos of nos-Cas9 flies and screened by PCR to identify the dTBC1D7KO deletions with a pair of primers (5′-CAATCGATTCGCATACCCTAC-3′ and 5′-CGCCATAGAGGATGTGTTC-3′).
The dTBC1D7-mcherry knock-in flies were generated as shown in Fig. S2. In brief, sgDNA sequence was designed and cloned into the U6b-sgRNA-short vector. Two (∼1.5 kb) fragments of dTBC1D7 genomic region (−1,500 to +30 and +30 to +1,550; +1 is the transcription initiation site) were subcloned into a PDM19-mCherry vector so that they were separated by the 3XP3-mCherry marker. The two plasmids were co-injected into the embryos of nos-Cas9 flies, and the subsequent mCherry-positive progenies were screened and crossed with hs-Cre;CDX/TM3 flies to delete the 3XP3 promoter region. The dTBC1D7-mCherry flies were confirmed by PCR by using the following primers: 5′-GCATAATTTATCGCGGGCCTGCATG-3′ and 5′-CATGGTCTTCTTCTGCATTACG-3′.
Both dTBC1D7KO and dTBC1D7-mcherry flies were backcrossed with wild-type flies (w1118) for six generations before preforming experiments. Each time, female progeny carrying the dTBC1D7KO or dTBC1D7-mcherry chromosome were selected for the next cross.
Starvation treatment
For monitoring fly development, eggs were collected every 4 h, as described (Géminard et al., 2009). Larvae were raised at a standard density of ≤40/tube in standard cornmeal/sugar medium. We used 1% yeast in 1% nonnutritive agar without any additives as starvation medium. For starvation, third instar larvae (90 h after egg deposition) were selected, washed in water, and shifted to starvation medium for 5 h. Then, fat body tissues were dissected and homogenized in 10 mM Tris-HCl lysis buffer (pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 25 mM NaF, and 1 mM Na3VO4 with 1× proteinase inhibitor cocktail [Sigma-Aldrich]).
Generation of transgenic flies
The dTBC1D7 and ilp2 cDNA sequences were amplified from LD17019 and GH11579 cDNA clones obtained from DGRC (Drosophila Genomics Resource Center) and subcloned into the pUAST-attB vector between the EcoRI and XbaI sites. The UAS-dTBC1D7, UAS-dTBC1D7I125Q, and UAS-ilp2 constructs were injected into M (vas-int.Dm)ZH-2A;M(3xP3-RFP.attP) ZH-86Fb embryos, and transformants were identified on the basis of eye color.
Cell culture, dsRNA synthesis, and immunoprecipitation
S2 cells were grown in Schneider’s Drosophila medium (Sigma-Aldrich) with 10% fetal bovine serum (Gibco BRL), and transfected with Vigofect reagent (Vigorous Biotechnology). dsRNA was synthesized in vitro and transfected as reported (Kulkarni et al., 2006). Cells were lysed with 10 mM Tris-HCl lysis buffer (pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 25 mM NaF, and 1 mM Na3VO4 with 1× proteinase inhibitor cocktail [Sigma-Aldrich]). Immunoprecipitations were performed with HA beads (Sigma-Aldrich), Flag beads (Sigma-Aldrich), and GFP beads (Chromotek).
Measurement of body weight, cell number, and cell size
All flies’ growth and crosses were performed according to standard procedures at 25°C under a 12-h light/dark cycle. Each vial contained <50 progenies to avoid overcrowding. After eclosion, males and females were separated, and 3-d-old flies were weighed with an electronic balance (AL104; Mettler Toledo). Wing size and cell number were measured as reported (Brogiolo et al., 2001). In brief, wings were dissected and mounted in Turpentine oil (Sigma-Aldrich). Pictures were acquired by using an Eclipse Ni-E Microscope (Nikon) at room temperature with a Plan Fluor 4× NA = 0.13 (for measurement of wing size) and Plan Fluor Plan Apo 20× NA = 0.75 (for measurement of cell number) objective lens and a DS-U3 camera (Nikon). The number of wing hairs on the dorsal wing surface in 10,000-µm2 was measured by ImageJ and defined as cell density (d). Each wing acquired at least three areas for calculation of the mean density (dm), and the whole wing area (S) excluding the alula and the costal cells was measured. The approximate number of cells in the whole wing area was calculated as total cell number = dm × S. We counted the number of ommatidia in the middle rows (rows 14–18) of compound eyes from scanning electron microscopy pictures (200×) as an estimation of the total number of ommatidia.
Oxidative stress resistance and life span analysis
For oxidative stress assays, 3-d-old male flies were transferred to food containing 5 mM paraquat and 5% sucrose solution (25–30 flies/vial). The flies were transferred to new food every 24 h, and fly survival was monitored.
Analysis of life span was performed according to methods previously described (Broughton et al., 2010; Grönke et al., 2010; Bai et al., 2012). More than 200 male flies were separated and raised in a 25°C, 65% humidity-controlled incubator (25–30 flies/vial) under a 12-h light/dark cycle. Dead flies were counted and removed every 2 d, and the remaining flies were flipped into new vials.
Immunofluorescence staining
Immunofluorescence assays were performed as previously described (Delanoue et al., 2016; Koyama and Mirth, 2016). In brief, tissues were dissected and fixed in PBS with 4% paraformaldehyde (Sigma-Aldrich) for 2 h at 4°C and then incubated with primary antibodies including rabbit anti–ILP2 antibody (1:200; a gift from H. Stocker, ETH Zurich, Zurich, Switzerland), rabbit anti–GFP antibody (1:200; Invitrogen), mouse anti-GFP (DSHB), or mouse anti–RFP antibody (1:200; Allele) overnight at 4°C. Finally, samples were stained with Alexa Fluor 488–conjugated, Alexa Fluor 568–conjugated, Alexa Fluor 647–conjugated secondary antibodies (1:500; Invitrogen), 20 nM phalloidin (Invitrogen), and DAPI (Invitrogen) for 2 h at room temperature. Fluorescence images were acquired at room temperature with an A1 confocal laser scanning microscope (Nikon) by using a 40× air objective (Plan Fluor NA 1.30; Nikon) and an A1+ camera (Nikon).
Hemolymph extraction and glucose determination
We obtained the hemolymph by bleeding 100 larvae 110 h after egg deposition as described previously (Delanoue et al., 2016; Koyama and Mirth, 2016). After centrifugation at 500 g for 30 min at 4°C, supernatants were transferred to a new tube, followed by additional centrifugation at 16,000 g for 20 min at 4°C. The supernatant was collected as hemolymph. Glucose levels were determined by a glucose assay kit according to its user manual (GAGO-20; Sigma-Aldrich).
Western blot analysis
Western blot analysis was performed as described previously (Huang et al., 2016). The rabbit anti-pS6K (Thr398), rabbit anti-pAKT (S505), rabbit anti–AKT antibody (1:1,000; Cell Signaling), mouse anti-actin (1:10,000; Santa Cruz Biotechnology), and rat anti–ILP2 antibody (1:8,000; a gift from P. Léopold, Institute of Biology Valrose, Parc Valrose, France) were used as primary antibodies. The blots were subsequently probed with IRDye 800 IgG, IRDye 680 IgG (LI-COR Biosciences), or peroxidase conjugate IgG (Invitrogen). Signals were detected with either an Odyssey infrared imaging system (LI-COR Biosciences) or ECL reagents (Thermo).
qPCR
Total RNA was prepared with Trizol reagent (Invitrogen) from larval fat body, larval brain, larval gut, adult head, or adult thorax. Total cDNA was synthesized with RT master mix (RR036A-1; Takara). An iQ SYBR green supermix was used for real-time PCR (Bio-Rad). The following primers were used for this study: ilp1: forward, 5′-CCCCGGAAACCACAAACTCT-3′; reverse, 5′-TAAAGCCATGGGGACACACC-3′; ilp2: forward, 5′-GGTGTGTCCCCATGGCTTTA-3′; reverse, 5′-TGCTGCTATCATCCTGCACC-3′; ilp3: forward, 5′-GTGTATGGCTTCAACGCAATG-3′; reverse, 5′-CAGCAGGGAACGGTCTTCG-3′; ilp4: forward, 5′-TGGATTTACACGCCGTGTCA-3′; reverse, 5′-GGTCTCGCACTCTAGCATCC-3′; ilp5: forward, 5′-TGCCTGTCCCAATGGATTCAA-3′; reverse, 5′-GCCAAGTGGTCCTCATAATCG-3′; ilp6: forward, 5′-GTCCAAAGTCCTGCTAGTCCT-3′; reverse, 5′-TCTGTTCGTATTCCGTGGGTG-3′; ilp7: forward, 5′-CCTGGCTGCACGTGAACTAT-3′; reverse, 5′-TGGATGGACAATACTCGGCG-3′; rpl32: forward, 5′-GCCGCTTCAAGGGACAGTATCTG-3′; reverse, 5′-AAACGCGGTTCTGCATGAG-3′; dTBC1D7: forward, 5′-AACTACACAGTGCCGAACAAC-3′; reverse, 5′-CGTACATTTCGCGTCTTTGGTC-3′; s6k: forward, 5′-ACTGGGCGCTCTCATGTTTG-3′; reverse, 5′-TTGGCTTTCAGAATGGTCT-3′.
Climbing assay
Climbing assays were performed as described (Wu et al., 2014). In brief, groups of twenty 30-d-old flies were assayed. Flies were transferred into a 100-ml dose tank containing a wet filter paper for 1 h. Then flies were shaken to the bottom of tanks, and the time taken for a median number of flies to reach a height of 15 cm was recorded. For each genotype, 100 flies were tested, and the average climbing rate (R = 15/time) was calculated.
Transmission electron microscopy (TEM) and scanning electron microscopy
TEM and scanning electron microscopy were performed with standard methods as described previously (Wang et al., 2012). For TEM, dissected muscles were prefixed with 4% paraformaldehyde and 2.5% at 4°C followed by incubation in 1% osmium tetroxide for 1 h at 4°C. Then samples were dehydrated in a series of ethanol (10, 25, 50, 75, and 100% ethanol) and embedded in LR White resin (Polysciences, Inc.). Thin sections were prepared and examined by using a JEM-1400 transmission electron microscope (JEOL) at room temperature. The images were acquired by using a Gatan camera (model 794; Gatan, Inc.).
For scanning electron microscopy, fly heads were dehydrated with a series of ethanol and immersed in hexamethyldisilazane at room temperature. Samples were mounted and coated with gold/palladium after solvent was evaporated off. The samples were examined with a JSM-5800 microscope (JEOL). The area of 10 ommatidia in high-magnification (1,000×) scanning electron microscopy pictures was measured with ImageJ, and the mean ommatidium size was calculated.
Image processing and statistical analysis
All the original fluorescent images were exported in tiff format by NIS-Elements viewer 4.0. TEM and scanning electron microscopy images were acquired and saved in tiff format embedded with a scale bar by Digital Micrograph software (Gatan, Inc.). Quantifying the area of ommatidia, wings and mosaic clones was achieved with ImageJ software.
The SD was used as y axis error bars for quantification of the data. Datasets were analyzed by Student’s unpaired t test, and data were considered statistically different at P < 0.05. Life span and stress assays were analyzed with SPSS software, and the log-rank method was used to calculate p-values.
Online supplemental material
Fig. S1 shows that dTBC1D7 is fly homologue of TBC1D7 and how the dTBC1D7 knockout flies were generated. Fig. S2 illustrates the schemes for generation the dTBC1D7-mCherry knock-in flies. Fig. S3 shows that dTBC1D7 regulates longevity and protects against oxidative stress.
Supplementary Material
Acknowledgments
We thank Drs. H. Stocker, E. Hafen, P. Léopold, and T. Neufield, as well as the Bloomington Stock Center and the Developmental Studies Hybridoma Bank, for providing the stocks and reagents. We thank Ping Wu from Imaging Facility of the National Center for Protein Science, Beijing, for assistance with microscopy imaging. We thank Dr. D. O’Keefe for comments on the manuscript. We thank Drs. L. Yang and Y. Tian from the Institute of Genetics and Developmental Biology for their assistance with electron microscopy. We also thank Y. Wang and X. Liu for their assistance with the fly injections.
This work was supported by a “973” grant from the Chinese Ministry of Science and Technology (2014CB849700 to T. Wang), grants from the National Natural Science Foundation of China (81670891 to T. Wang and 11674389, 11404390, and 11174089 to Y. Jiang), and a grant from the Instrument Developing Project of the Chinese Academy of Sciences (YZ201263 to Y. Jiang).
The authors declare no competing financial interests.
Author contributions: S. Ren, Z. Huang, and T. Wang designed the experiments. S. Ren and Z. Huang performed the experiments. S. Ren, Z. Huang, and T. Wang analyzed and interpreted the data. T. Wang, Y. Jiang, S. Ren, and Z. Huang wrote the manuscript.
References
- Alfa R.W., Park S., Skelly K.R., Poffenberger G., Jain N., Gu X., Kockel L., Wang J., Liu Y., Powers A.C., and Kim S.K.. 2015. Suppression of insulin production and secretion by a decretin hormone. Cell Metab. 21:323–333. 10.1016/j.cmet.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaiz A.A., Micale L., Mandriani B., Augello B., Pellico M.T., Chrast J., Xenarios I., Zelante L., Merla G., and Reymond A.. 2014. TBC1D7 mutations are associated with intellectual disability, macrocrania, patellar dislocation, and celiac disease. Hum. Mutat. 35:447–451. 10.1002/humu.22529 [DOI] [PubMed] [Google Scholar]
- Bai H., Kang P., and Tatar M.. 2012. Drosophila insulin-like peptide-6 (dilp6) expression from fat body extends lifespan and represses secretion of Drosophila insulin-like peptide-2 from the brain. Aging Cell. 11:978–985. 10.1111/acel.12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton J.S., Lockwood W.K., Li L., Cohen S.M., and Edgar B.A.. 2002. Drosophila’s insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell. 2:239–249. 10.1016/S1534-5807(02)00117-X [DOI] [PubMed] [Google Scholar]
- Brogiolo W., Stocker H., Ikeya T., Rintelen F., Fernandez R., and Hafen E.. 2001. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11:213–221. 10.1016/S0960-9822(01)00068-9 [DOI] [PubMed] [Google Scholar]
- Broughton S.J., Piper M.D., Ikeya T., Bass T.M., Jacobson J., Driege Y., Martinez P., Hafen E., Withers D.J., Leevers S.J., and Partridge L.. 2005. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. USA. 102:3105–3110. 10.1073/pnas.0405775102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton S.J., Slack C., Alic N., Metaxakis A., Bass T.M., Driege Y., and Partridge L.. 2010. DILP-producing median neurosecretory cells in the Drosophila brain mediate the response of lifespan to nutrition. Aging Cell. 9:336–346. 10.1111/j.1474-9726.2010.00558.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S.L., Tee A.R., Short J.D., Bergeron J.M., Kim J., Shen J., Guo R., Johnson C.L., Kiguchi K., and Walker C.L.. 2006. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J. Cell Biol. 173:279–289. 10.1083/jcb.200507119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Ni J., Ma W., Shiu V., Milla L.A., Park S., Spletter M.L., Tang S., Zhang J., Wei X., et al. 2014. Insight into insulin secretion from transcriptome and genetic analysis of insulin-producing cells of Drosophila. Genetics. 197:175–192. 10.1534/genetics.113.160663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo-Chichi J.M., Tcherkezian J., Hamdan F.F., Décarie J.C., Dobrzeniecka S., Patry L., Nadon M.A., Mucha B.E., Major P., Shevell M., et al. 2013. Disruption of TBC1D7, a subunit of the TSC1-TSC2 protein complex, in intellectual disability and megalencephaly. J. Med. Genet. 50:740–744. 10.1136/jmedgenet-2013-101680 [DOI] [PubMed] [Google Scholar]
- Colombani J., Andersen D.S., and Léopold P.. 2012a Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science. 336:582–585. 10.1126/science.1216689 [DOI] [PubMed] [Google Scholar]
- Delanoue R., Meschi E., Agrawal N., Mauri A., Tsatskis Y., McNeill H., and Léopold P.. 2016. Drosophila insulin release is triggered by adipose Stunted ligand to brain Methuselah receptor. Science. 353:1553–1556. 10.1126/science.aaf8430 [DOI] [PubMed] [Google Scholar]
- Dibble C.C., and Manning B.D.. 2013. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol. 15:555–564. 10.1038/ncb2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble C.C., Elis W., Menon S., Qin W., Klekota J., Asara J.M., Finan P.M., Kwiatkowski D.J., Murphy L.O., and Manning B.D.. 2012. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol. Cell. 47:535–546. 10.1016/j.molcel.2012.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Chromosome 16 Tuberous Sclerosis Consortium 1993. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 75:1305–1315. [DOI] [PubMed] [Google Scholar]
- Fernandez A.M., and Torres-Alemán I.. 2012. The many faces of insulin-like peptide signalling in the brain. Nat. Rev. Neurosci. 13:225–239. 10.1038/nrn3209 [DOI] [PubMed] [Google Scholar]
- Gabernet-Castello C., O’Reilly A.J., Dacks J.B., and Field M.C.. 2013. Evolution of Tre-2/Bub2/Cdc16 (TBC) Rab GTPase-activating proteins. Mol. Biol. Cell. 24:1574–1583. 10.1091/mbc.E12-07-0557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai Z., Chu W., Deng W., Li W., Li H., He A., Nellist M., and Wu G.. 2016. Structure of the TBC1D7-TSC1 complex reveals that TBC1D7 stabilizes dimerization of the TSC1 C-terminal coiled coil region. J. Mol. Cell Biol.. 10.1093/jmcb/mjw001 [DOI] [PubMed] [Google Scholar]
- Gao X., and Pan D.. 2001. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 15:1383–1392. 10.1101/gad.901101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Zhang Y., Arrazola P., Hino O., Kobayashi T., Yeung R.S., Ru B., and Pan D.. 2002. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat. Cell Biol. 4:699–704. 10.1038/ncb847 [DOI] [PubMed] [Google Scholar]
- Géminard C., Rulifson E.J., and Léopold P.. 2009. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 10:199–207. 10.1016/j.cmet.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Glatter T., Schittenhelm R.B., Rinner O., Roguska K., Wepf A., Jünger M.A., Köhler K., Jevtov I., Choi H., Schmidt A., et al. 2011. Modularity and hormone sensitivity of the Drosophila melanogaster insulin receptor/target of rapamycin interaction proteome. Mol. Syst. Biol. 7:547 10.1038/msb.2011.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke S., Clarke D.F., Broughton S., Andrews T.D., and Partridge L.. 2010. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 6:e1000857 10.1371/journal.pgen.1000857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasygar K., and Hietakangas V.. 2014. p53- and ERK7-dependent ribosome surveillance response regulates Drosophila insulin-like peptide secretion. PLoS Genet. 10:e1004764 10.1371/journal.pgen.1004764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzenberger M., Dupont J., Ducos B., Leneuve P., Géloën A., Even P.C., Cervera P., and Le Bouc Y.. 2003. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 421:182–187. 10.1038/nature01298 [DOI] [PubMed] [Google Scholar]
- Housden B.E., Valvezan A.J., Kelley C., Sopko R., Hu Y., Roesel C., Lin S., Buckner M., Tao R., Yilmazel B., et al. 2015. Identification of potential drug targets for tuberous sclerosis complex by synthetic screens combining CRISPR-based knockouts with RNAi. Sci. Signal. 8:rs9 10.1126/scisignal.aab3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Ren S., Jiang Y., and Wang T.. 2016. PINK1 and Parkin cooperatively protect neurons against constitutively active TRP channel-induced retinal degeneration in Drosophila. Cell Death Dis. 7:e2179 10.1038/cddis.2016.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeya T., Galic M., Belawat P., Nairz K., and Hafen E.. 2002. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 12:1293–1300. 10.1016/S0960-9822(02)01043-6 [DOI] [PubMed] [Google Scholar]
- Inoki K., Li Y., Xu T., and Guan K.L.. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17:1829–1834. 10.1101/gad.1110003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell J.L., and Guan K.L.. 2013. Nutrient signaling to mTOR and cell growth. Trends Biochem. Sci. 38:233–242. 10.1016/j.tibs.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., and Neufeld T.P.. 2015. Dietary sugar promotes systemic TOR activation in Drosophila through AKH-dependent selective secretion of Dilp3. Nat. Commun. 6:6846 10.1038/ncomms7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., and Mirth C.K.. 2016. Growth-Blocking Peptides As Nutrition-Sensitive Signals for Insulin Secretion and Body Size Regulation. PLoS Biol. 14:e1002392 10.1371/journal.pbio.1002392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni M.M., Booker M., Silver S.J., Friedman A., Hong P., Perrimon N., and Mathey-Prevot B.. 2006. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat. Methods. 3:833–838. [DOI] [PubMed] [Google Scholar]
- Laplante M., and Sabatini D.M.. 2012. mTOR signaling in growth control and disease. Cell. 149:274–293. 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.O., Yang H., Georgescu M.M., Di Cristofano A., Maehama T., Shi Y., Dixon J.E., Pandolfi P., and Pavletich N.P.. 1999. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 99:323–334. 10.1016/S0092-8674(00)81663-3 [DOI] [PubMed] [Google Scholar]
- Lee K.S., Kwon O.Y., Lee J.H., Kwon K., Min K.J., Jung S.A., Kim A.K., You K.H., Tatar M., and Yu K.. 2008. Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat. Cell Biol. 10:468–475. 10.1038/ncb1710 [DOI] [PubMed] [Google Scholar]
- Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J., et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 445:168–176. 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Menon S., Dibble C.C., Talbott G., Hoxhaj G., Valvezan A.J., Takahashi H., Cantley L.C., and Manning B.D.. 2014. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 156:771–785. 10.1016/j.cell.2013.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N., Yamanaka N., Yagi Y., Nishida Y., Kataoka H., O’Connor M.B., and Mizoguchi A.. 2009. A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev. Cell. 17:885–891. 10.1016/j.devcel.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S., and Hafen E.. 2003. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 13:79–85. 10.1016/S0962-8924(02)00042-9 [DOI] [PubMed] [Google Scholar]
- Park S., Alfa R.W., Topper S.M., Kim G.E., Kockel L., and Kim S.K.. 2014. A genetic strategy to measure circulating Drosophila insulin reveals genes regulating insulin production and secretion. PLoS Genet. 10:e1004555 10.1371/journal.pgen.1004555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter C.J., Huang H., and Xu T.. 2001. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell. 105:357–368. 10.1016/S0092-8674(01)00333-6 [DOI] [PubMed] [Google Scholar]
- Qin J., Wang Z., Hoogeveen-Westerveld M., Shen G., Gong W., Nellist M., and Xu W.. 2016. Structural Basis of the Interaction between Tuberous Sclerosis Complex 1 (TSC1) and Tre2-Bub2-Cdc16 Domain Family Member 7 (TBC1D7). J. Biol. Chem. 291:8591–8601. 10.1074/jbc.M115.701870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A., and Perrimon N.. 2012. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell. 151:123–137. 10.1016/j.cell.2012.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson E.J., Kim S.K., and Nusse R.. 2002. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 296:1118–1120. 10.1126/science.1070058 [DOI] [PubMed] [Google Scholar]
- Santiago Lima A.J., Hoogeveen-Westerveld M., Nakashima A., Maat-Kievit A., van den Ouweland A., Halley D., Kikkawa U., and Nellist M.. 2014. Identification of regions critical for the integrity of the TSC1-TSC2-TBC1D7 complex. PLoS One. 9:e93940 10.1371/journal.pone.0093940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo L.J., Gao X., Chiarelli D.A., Li L., Pan D., and Edgar B.A.. 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5:566–571. 10.1038/ncb996 [DOI] [PubMed] [Google Scholar]
- Stocker H., Radimerski T., Schindelholz B., Wittwer F., Belawat P., Daram P., Breuer S., Thomas G., and Hafen E.. 2003. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 5:559–565. 10.1038/ncb995 [DOI] [PubMed] [Google Scholar]
- Struhl G., and Basler K.. 1993. Organizing activity of wingless protein in Drosophila. Cell. 72:527–540. 10.1016/0092-8674(93)90072-X [DOI] [PubMed] [Google Scholar]
- van Slegtenhorst M., de Hoogt R., Hermans C., Nellist M., Janssen B., Verhoef S., Lindhout D., van den Ouweland A., Halley D., Young J., et al. 1997. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 277:805–808. 10.1126/science.277.5327.805 [DOI] [PubMed] [Google Scholar]
- Vellai T., Takacs-Vellai K., Zhang Y., Kovacs A.L., Orosz L., and Müller F.. 2003. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 426:620 10.1038/426620a [DOI] [PubMed] [Google Scholar]
- Vinayagam A., Kulkarni M.M., Sopko R., Sun X., Hu Y., Nand A., Villalta C., Moghimi A., Yang X., Mohr S.E., et al. 2016. An integrative analysis of the InR/PI3K/Akt network identifies the dynamic response to insulin signaling. Cell Reports. 16:3062–3074. 10.1016/j.celrep.2016.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Blumhagen R., Lao U., Kuo Y., and Edgar B.A.. 2012. LST8 regulates cell growth via target-of-rapamycin complex 2 (TORC2). Mol. Cell. Biol. 32:2203–2213. 10.1128/MCB.06474-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Liu J., Zhuang N., and Wang T.. 2014. UCP4A protects against mitochondrial dysfunction and degeneration in pink1/parkin models of Parkinson’s disease. FASEB J. 28:5111–5121. 10.1096/fj.14-255802 [DOI] [PubMed] [Google Scholar]
- Xu Y., An F., Borycz J.A., Borycz J., Meinertzhagen I.A., and Wang T.. 2015. Histamine Recycling Is Mediated by CarT, a Carcinine Transporter in Drosophila Photoreceptors. PLoS Genet. 11:e1005764 10.1371/journal.pgen.1005764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gao X., Saucedo L.J., Ru B., Edgar B.A., and Pan D.. 2003. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5:578–581. 10.1038/ncb999 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.