Large-scale knockout studies suggest that most miRNAs are phenotypically dispensable. However, Kavaler et al. show here that developmental specification toward a non-neuronal fate in the Drosophila melanogaster peripheral sensory organ lineage depends critically on mir-279/996 repression of the Notch repressor Insensible.
Abstract
Although there is abundant evidence that individual microRNA (miRNA) loci repress large cohorts of targets, large-scale knockout studies suggest that most miRNAs are phenotypically dispensable. Here, we identify a rare case of developmental cell specification that is highly dependent on miRNA control of an individual target. We observe that binary cell fate choice in the Drosophila melanogaster peripheral sensory organ lineage is controlled by the non-neuronally expressed mir-279/996 cluster, with a majority of notum sensory organs exhibiting transformation of sheath cells into ectopic neurons. The mir-279/996 defect phenocopies Notch loss of function during the sheath–neuron cell fate decision, suggesting the miRNAs facilitate Notch signaling. Consistent with this, mir-279/996 knockouts are strongly enhanced by Notch heterozygosity, and activated nuclear Notch is impaired in the miRNA mutant. Although Hairless (H) is the canonical nuclear Notch pathway inhibitor, and H heterozygotes exhibit bristle cell fate phenotypes reflecting gain-of-Notch signaling, H/+ does not rescue mir-279/996 mutants. Instead, we identify Insensible (Insb), another neural nuclear Notch pathway inhibitor, as a critical direct miR-279/996 target. Insb is posttranscriptionally restricted to neurons by these miRNAs, and its heterozygosity strongly suppresses ectopic peripheral nervous system neurons in mir-279/996 mutants. Thus, proper assembly of multicellular mechanosensory organs requires a double-negative circuit involving miRNA-mediated suppression of a Notch repressor to assign non-neuronal cell fate.
Introduction
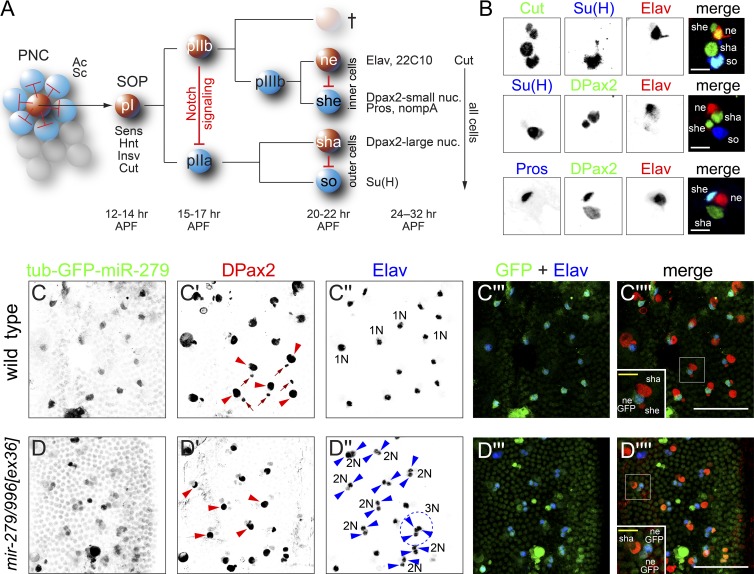
The array of mechanosensory bristle organs on the notum of Drosophila melanogaster comprises a choice model system to understand fundamental principles of developmental biology (Lai and Orgogozo, 2004). Each multicellular structure is generated via a fixed lineage initiated by a sensory organ precursor (SOP) cell (Fig. 1 A). SOPs are selected from an equivalence group known as a proneural cluster (PNC), defined by the functional activity of basic helix-loop-helix activator transcription factors. However, cell signaling mediated by the Notch receptor results in specification of individual SOPs from PNCs, with other PNC cells eventually adopting an ordinary epidermal fate. Once stably specified, the SOP executes a fixed set of asymmetric cell divisions yielding four or five distinct cell fates (Fig. 1 A). Notably, every lineage division yields a pair of different sister cell fates, such that a maximum of cell diversity is generated from a minimum of cell divisions.
Figure 1.
The non-neuronal locus mir-279/996 restricts neural fate in PNS organs. (A) Summary of mechanosensory bristle development. Spatially patterned activity of basic helix-loop-helix activators Achaete (Ac) and Scute (Sc) defines a PNC, among which Notch signaling (schematized by red repression lines) restricts neural competence to single SOP cells. The SOP undergoes a fixed set of asymmetric cell divisions to generate the four cells of the mature sensory organ (ne, neuron; sha, shaft cell; she, sheath cell; so, socket cell); a fifth glial-like cell undergoes apoptosis. Each of the cell divisions in the sensory organ lineage is made asymmetric by Notch signaling (schematized by red repression lines). Developmental times for microchaete bristle lineage stages are labeled in hours APF, and cell-specific markers used in this study are marked. (B) Examples of triple labeling of mature sensory organ cell types with distinctive markers. Bars, 10 µm. (C) The expression of a tub-GFP-miR-279 activity sensor is elevated in Elav+ neurons in the notum. In C′, examples of large DPax2+ shaft cell nuclei are labeled with red arrowheads, and small DPax2+ sheath cell nuclei are labeled with arrows. In C′′, examples of individual neuronal nuclei are labeled as 1N. (D) Expression of tub-GFP-miR-279 in mir-279/996 mutant is generally up-regulated in epidermal cells in the notum but is substantially higher in multiple sensory organ cells. This is associated with a profound cell specification defect, because most sensory organs contain two Elav+ neurons and are lacking the small DPax2+ nucleus (the sheath, which is the sister cell of the neuron). Examples of sensory organ clusters with only DPax2+ shaft cells (D′, red arrowheads,) and double neurons (2N; D′′, blue arrowheads) are indicated; a triple neuron (3N) cluster is highlighted by dotted lines. Bars: (C and D, main panels) 50 µm; (insets) 10 µm.
The mechanisms by which alternative cell fates in peripheral nervous system (PNS) bristle lineages are assigned have been studied for decades (Lai, 2004). The major regulatory strategies involve (a) cell–cell signaling via the Notch receptor, which creates fate differences via directional communication between sister cells; (b) asymmetric inheritance of cell determinants, exemplified by Numb and Neuralized, which intrinsically bias signaling capacity and thus cell fate; and (c) the expression of cell-specific transcription and chromatin factors, which direct gene expression changes that lock in differential cell fate. The aforementioned regulatory strategies all impinge on transcriptional changes that drive cell fate specification. Beyond transcriptional mechanisms, posttranscriptional regulation has impact on the peripheral sensory lineage. For instance, mutants of the RNA binding protein Musashi exhibit high-frequency socket-to-shaft transformations that resemble Notch loss of function caused by loss of regulation of tramtrack (Okabe et al., 2001).
miRNAs are also implicated in bristle organ development. For example, disruptions of 3′ UTRs in Notch target genes in the E(spl)-C and the Brd-C result in sensory organ specification defects (Klämbt et al., 1989; Leviten et al., 1997). These effects were traced to their repression by miRNA families targeting Brd, K, and/or GY boxes (Lai and Posakony, 1997; Lai et al., 1998; Lai, 2002b), and gain-of-function manipulations of several of these miRNAs also affect sensory organ fate (Lai et al., 2005). Thus far, however, only mild effects of the loss of function of these miRNAs on bristle development have been observed (e.g., with mir-7; Li et al., 2009). The effects of Brd and/or K box miRNA families might be masked by redundancy of their extensive families and/or by potential cotargeting by the distinct miRNA families regulating Notch target genes. Nevertheless, an alternative notion is that miRNA control mostly mediates robustness and is not intrinsically essential for fate specification. For example, mir-9a mutants exhibit a minor increase in some macrochaete bristle positions (Li et al., 2006; Bejarano et al., 2010), but abrogating repression of the proneural factor Senseless by miR-9a can sensitize the animal to background genomic variation (Cassidy et al., 2013).
In this study, we identify a surprisingly critical role for miRNAs in cell fate specification with the PNS. In particular, we show that mir-279/996 are required to assign the neuron sister cell fate, the sheath cell. These miRNAs share a seed region and are expressed as an operon (Sun et al., 2015), and the function of either miRNA is sufficient to support normal PNS development. Notably, we demonstrate the Notch repressor Insensible (Insb) as a critical direct target of miR-279/996 that must be suppressed to prevent ectopic neuronal commitment at the expense of its sister fate, the sheath cell. Paradoxically, insb knockouts lack apparent defects in peripheral sense organ development (Coumailleau and Schweisguth, 2014), but insb heterozygotes strongly rescue the ectopic neuron defects of the miRNA mutants. Thus, the assembly of mechanosensory organs involves an unexpected double-negative strategy, whereby miRNA-mediated repression of a nonessential gene product specifies non-neuronal cell fate.
Results
Non-neuronal activity of mir-279/996 in the mechanosensory bristle lineage
The Drosophila notum is covered with PNS bristle mechanosensory organs, including large sensilla termed macrochaetes and an array of small sensilla known as microchaetes. The microchaete field develops synchronously during early pupal stages, permitting assays of many individual sensory organs in each preparation. The patterns of key markers in the wild-type notum are summarized in Fig. 1 B. The socket and shaft are a sister pair of outer sensory organ cells and have comparably large nuclei that express Su(H) and DPax2, respectively. The sheath and neuron are a sister pair of inner sensory organ cells and have comparably small nuclei. Both the shaft and sheath express DPax2, but the sheath can be distinguished by its smaller size and its expression of Prospero (Pros). Finally, the canonical marker of the neuron is Elav.
Homozygotes of strong mir-279/996[ex36] and null [15C] alleles (Sun et al., 2015) of the miRNA operon have largely normal exterior bristle patterning. Still, because mir-279/996 exhibits non-neuronal expression in the embryonic PNS (Sanfilippo et al., 2016), we asked whether these miRNAs were spatially modulated in mechanosensory organs. For reference, a control tub-GFP-SV40 transgene was broadly expressed at 28 h after puparium formation (APF) but exhibited higher accumulation in external sensory organ cells (Fig. S1). The pattern of tub-GFP-SV40 might reflect the polyploid nature of the large external sensory cells. In contrast, a tub-GFP-miR-279 sensor that is repressible by both miR-279 (via perfect complementarity) and miR-996 (via seed matching) was preferentially detected in a distinct array of notum cells. Cells with elevated tub-GFP-miR-279, reflecting lowest levels of miR-279/996 activity, coexpressed the neuronal marker Elav (Fig. 1 C). Therefore, these miRNAs are active in non-neuronal cells in the notum.
To verify that the spatial pattern of the sensor was induced by the cognate miRNAs, we introduced tub-GFP-miR-279 into mir-279/996[ex36C] homozygotes. This resulted in general sensor derepression in epidermal cells and elevated expression of GFP in multiple sensory lineage cells (Fig. 1 D). Therefore, these miRNAs are functionally active within non-neuronal cells of peripheral sense organs.
Deletion of mir-279/996 transforms sheath cells into neurons at high frequency
While studying the tub-GFP-miR-279 sensor pattern in mir-279/996 mutants, we were surprised to notice strong misspecification of lineage cells. Notably, we observed that a majority of sensory organs exhibited loss of small DPax2+ nuclei (Fig. 1, compare C′ with D′, arrowheads) accompanied by ectopic Elav+ nuclei (Fig. 1, compare C″ with D″, arrowheads). This implied defective asymmetry of pIIIb division (Fig. 1 A), with high-frequency conversion of sheath cells into neurons.
We studied this further using additional markers. The total numbers of sensory organ cells (as marked by Cut+ nuclei) were largely unaffected (Fig. S2), as were socket (marked by Su(H)+ nuclei, Fig. S2) and shaft (marked by large DPax2 nuclei) cell fates (Fig. 1, C and D). This implied that the major defect lay in the assignment of internal cell fates. Consistent with this, staining for Pros showed high frequency of sensilla lacking this independent sheath marker in double neuron organs (Fig. 2, A′ and B′). However, we also observed a minor population of sensory organs with three or more Elav+ nuclei (e.g., Fig. 1 D′′, circled sensory organ labeled 3N), which were variably missing either or both outer cell markers. Notably, the apparent transformation of outer cells into inner cells/neurons and the profound sheath-to-neuron transformations both resemble loss of Notch signaling (Fig. 1 A).
Figure 2.
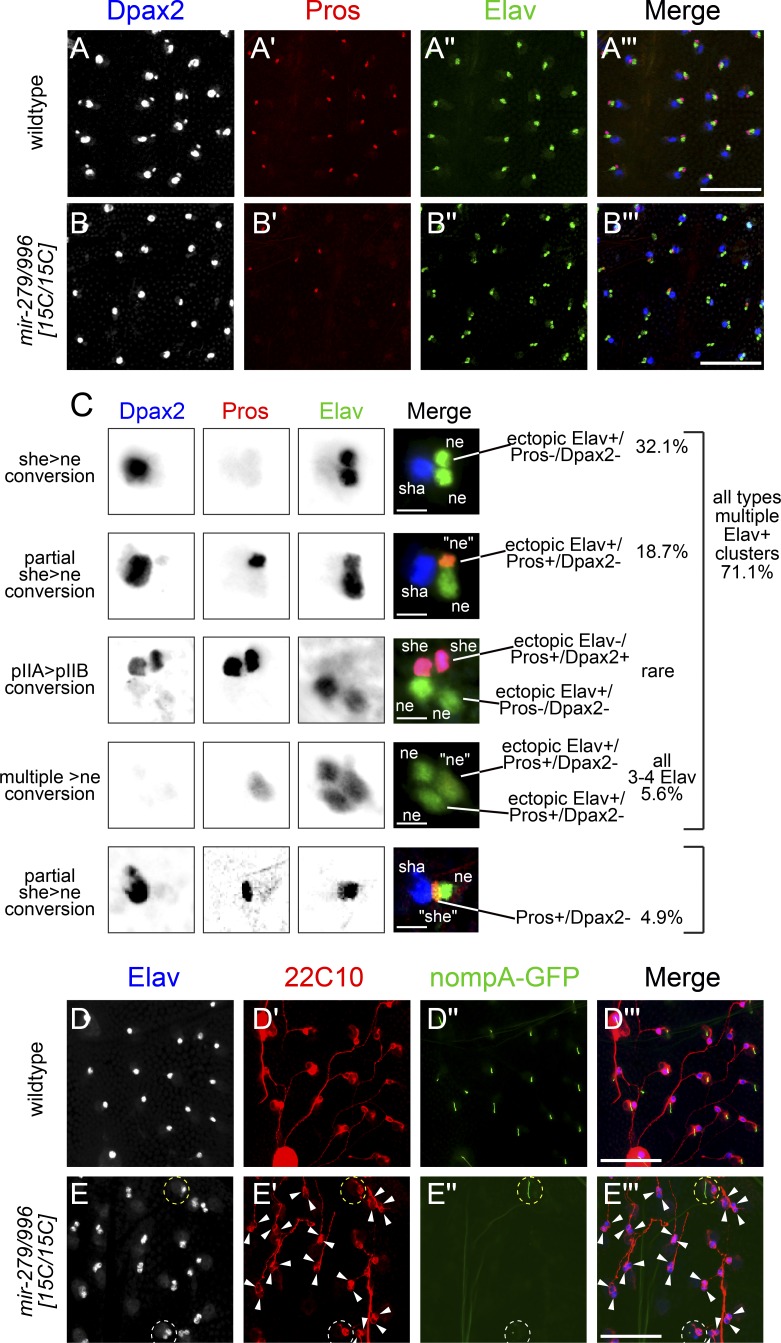
Profound sensory organ cell fate defects in mir-279/996 mutants. Shown are pupal nota at >28 h APF stained for cell-specific markers. (A and B) Wild-type (A) and mir-279/996[15C/15C] (B) nota stained for DPax2 (large shaft and small sheath nuclei), Pros (sheath nuclei), and Elav (neuronal nuclei) reveal a strong sheath-to-neuron transformation in the mutant, largely without affecting outer cell fates. Bars, 50 µM. (C) Higher magnification of mir-279/996[15C/15C] mutant sensory organs organized into characteristic classes of aberrant expression of cell-specific markers. In general, these represent conversion of non-neural cell types (especially sheath cells) into neuronal fate. Quantification of their frequency is provided at right, and their interpretation with respect to the canonical sensory lineage (see Fig. 1 A) is provided at left (wt, n = 792; mir-279/996[15C/15C], n = 593; see also Fig. 7 G for further phenotypic analysis). Bars, 10 µM. (D and E) Staining for the terminal markers of the neuron (22C10) and the sheath cell (nompA-GFP) in wild type (D) and mir-279/996[15C/15C] (E) indicates that mutant sensory organs differentiate as neurons and fail to elaborate the dendritic cap of the sheath. Dotted white circle in E′ highlights an exceptional case in which an ectopic Elav+ neuron is not surrounded by 22C10 reactivity; however, in the vast majority of preparations, the ectopic Elav+ cells express equivalent amounts of 22C10. Dotted yellow circle in E′ highlights a mutant sensory organ that elaborates a nompA-GFP sheath cell dendritic cap. Bars, 50 µM.
Systematic enumeration of aberrant fate combinations revealed a range of phenotypes, of which the dominant class of mutant organ contained no sheath/two neurons (Fig. 2 C, top). However, the diversity of mutant sensory organs was striking and included other combinations consistent with cell transformations associated with Notch loss of function at other lineage divisions (Figs. 1 A and 2 C). In particular, a curious aspect of the mir-279/996 mutant phenotype was revealed when costaining for DPax2/Elav along with Pros as another sheath cell marker. Although many previously reported PNS manipulations result in “clean” transformations of terminal lineage markers (Lai and Orgogozo, 2004), we noticed a substantial population of sensory organs contained cells expressing mixed terminal markers. In particular, DPax2 was more frequently lost from sheath cells than Pros. That is, out of >70% of sensory organs exhibiting two or more Elav+ cells, a quarter of these contained an Elav+/Pros+ nucleus, which normally does not occur (Fig. 2 C). Very occasionally, an ectopic Elav+ cell retained DPax2, or sometimes DPax2 and Pros, but these were rare. Moreover, ∼5% of sensory organs did not express high level of ectopic Elav but contained a sheath-like cell that expressed Pros, but not DPax2.
Although Elav is typically used as the de facto marker of postmitotic neurons in Drosophila, we recently observed that miR-279/996 repress spatially ubiquitous expression of Elav (Sanfilippo et al., 2016). Given the unexpected uncoupling of sheath cell markers in a population of sensory organs, we addressed whether cells that ectopically expressed Elav reflect true neurons as opposed to merely cells with derepressed Elav. To do so, we stained for 22C10 as a marker of neuronal differentiation (Hummel et al., 2000). Indeed, sensory organs with multiple Elav+ cells essentially always exhibited 22C10 reactivity around each of the ectopic neuronal nuclei (Fig. 2, D and E). Because the axons from neurons of different sensory organs fasciculate tightly, it was often not possible to distinguish multiple axonal tracts from an individual double-neuron organ. However, the 22C10 staining around the cell bodies was generally indistinguishable, suggesting a comparable extent of neuronal differentiation. Because Pros and 22C10 were marked using monoclonal antibodies, we were not able to colabel these antigens. However, the numerical evidence indicates that in mir-279/996 mutants, most Elav+/Pros+ cells will mature as neurons instead of adopting sheath cell character.
We confirmed this notion by examining a transgenic marker for the fully differentiated sheath cell fate, nompA-GFP. NompA is required for mechanosensory signal transduction, and nompA-GFP localizes to the dendritic cap of the sheath cell (Chung et al., 2001). A minority of sensory organs expressed nompA-GFP in mir-279/996[15C/15C] nota, demonstrating a paucity of differentiated sheath cells (Fig. 2, D and E). These findings indicate that miRNA deletion can disconnect a canonical transcription factor lineage marker with its associated terminal cell fate (e.g., in differentiated neurons that express Pros).
Overall, >75% of notum sensory organs in mir-279/996[15C] animals exhibited defective lineage fates. Therefore, this ranks as one of the most severe developmental defects observed for an individual miRNA mutant. Moreover, given that the Drosophila mechanosensory organ lineage has been intensively studied for decades as a leading model system for developmental cell specification (Lai and Orgogozo, 2004), it is perhaps surprising that such a critical cell specification component escaped attention.
Clonal analysis of mir-279/996 in the notum
The phenotypes in whole-animal mutants might theoretically be caused by contributions of miRNA function outside of the peripheral sense organs. To test the autonomy of their function for mechanosensory organ development, we used clonal analysis. Because miRNAs can be stable for up to days, mitotic clones are not expected to be informative for distinguishing roles within lineage divisions separated by a few hours (Fig. 1). However, clonal analysis can resolve whether the miRNAs are required autonomously within developing sensory organ territories.
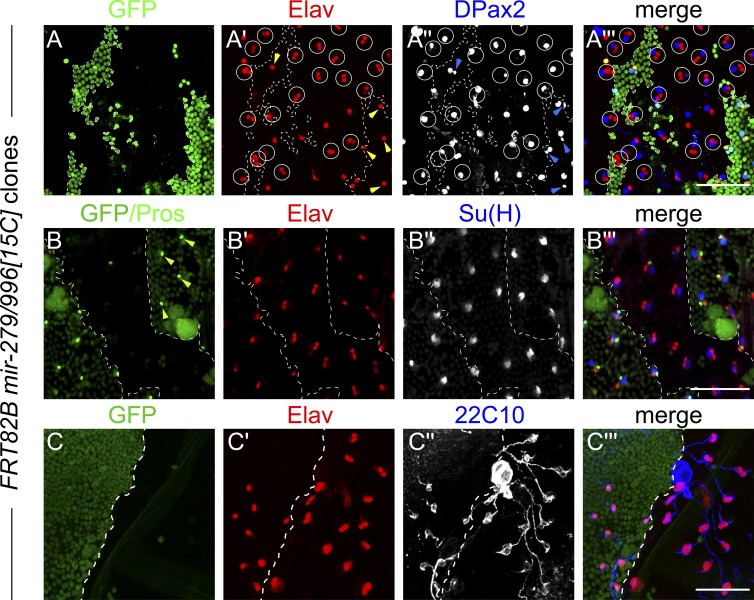
We generated mir-279/996[15C] null clones using the Minute technique and observed high frequency sensory organs bearing transformation of small DPax2 nuclei into ectopic Elav nuclei (Fig. 3 A), whereas control GFP+ territories showed normal distributions of cell markers. Triple staining with Pros/Elav/Su(H) (with Pros colabeled in the GFP clonal marker channel) confirmed specific loss of Pros within the miRNA mutant clones, concomitant with double Elav sensory organs (Fig. 3 B). Outer cell fates (large DPax2 nuclei [Fig. 3 A′′] and Su(H) nuclei [Fig. 3 B′′]) were unaffected in miRNA deletion clones. Finally, we observed that within miRNA mutant clones, ectopic Elav+ nuclei were ensheathed by 22C10 signals (Fig. 3 C). Thus, the specific sheath-to-neuron cell transformations are autonomous to mir-279/996 mutant sensory organs.
Figure 3.
Clonal analysis of the mir-279/996[15C] null allele. Shown are notum clones generated using the Minute technique, in which heterozygous tissue is labeled by GFP and mutant territories are marked by absence of GFP (dotted borders). (A) Control territories show sensory organs with single Elav nuclei (yellow arrowheads, neurons) single small DPax2 nuclei (blue arrowheads, sheath cells), whereas mir-279/996 null clones show high-frequency sheath-to-neuron conversions (circles). (B) Control territories show individual Pros nuclei (arrowheads; Pros is costained in the GFP channel), but these are mostly missing in the miRNA mutant territory. Su(H), a socket marker, is largely unaffected. (C) The sensory organs with double Elav nuclei in mir-279/996 null clones are reliably ensheathed by 22C10 reactivity. Bars, 50 µM.
Both miR-279 and miR-996 contribute to mechanosensory organ cell specification
We recently clarified that all phenotypes previously attributed to specific deletions of mir-279 (which retain the mir-996 locus) involve concomitant loss of mature miR-996 (Sun et al., 2015). To test whether mechanosensory organ development might represent a setting that distinguishes the function of these miRNAs, we further examined our allelic series of mir-279 and mir-996 mutants. We observed ∼42% sheath-to-neuron conversions in the strong allelic combination 15C/ex36, ∼29% conversion in stronger hypomorphic combination ex36/ex117, and <2% conversion in the milder hypomorph ex117/ex117 (Fig. S3). These relative defects parallel the effects of these mutants on miR-279/996 levels (Sun et al., 2015) and imply that both miRNAs contribute to sheath cell specification.
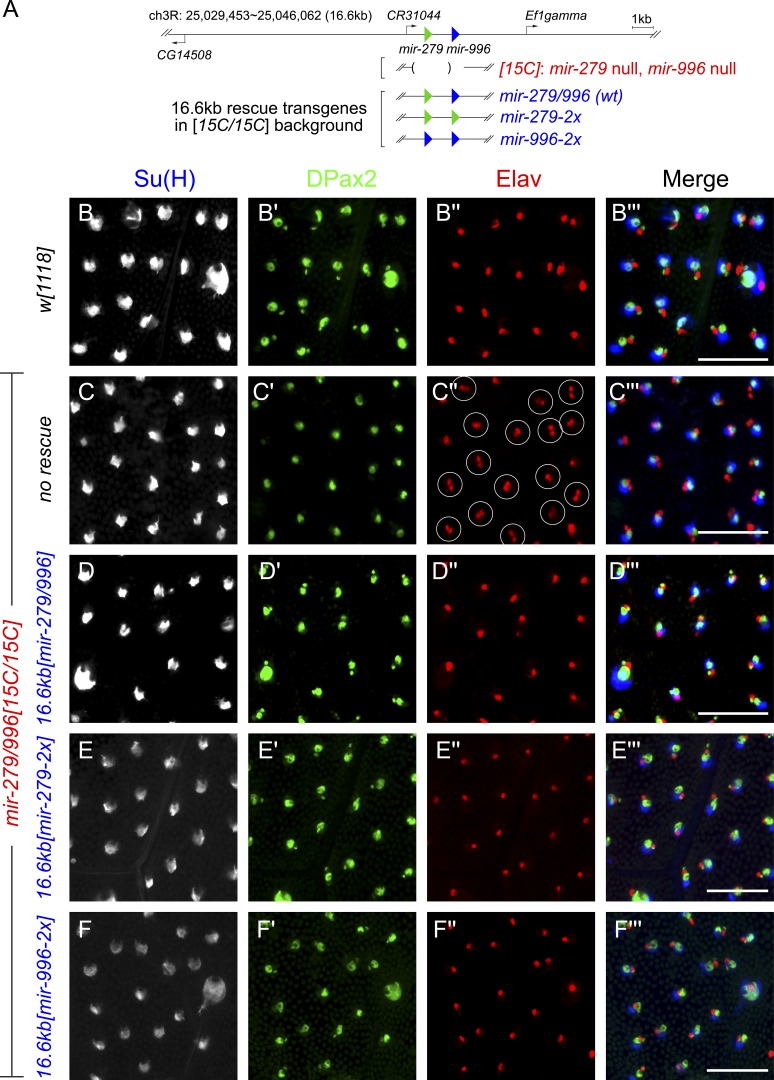
We performed reciprocal tests using genomic rescue transgenes (Fig. 4 A). We first introduced a 16.6-kb mir-279/996 transgene into the null 15C/15C background and showed this fully restores wild-type distribution of sensory organ fates (Fig. 4, B–D). We then performed similar analyses using similar genomic transgenes engineered to produce only mir-279 or mir-996 (Fig. 4 A). Both of these provide similar, complete rescue of sheath cell fate, indicating that these miRNAs are largely functionally overlapping for this critical developmental function (Fig. 4, E and F). These data help to explain why miRNA-mediated control of sensory organ development might have escaped previous notice. First, the profound neural misspecification defect in the miRNA mutant is not visible from the exterior of the animal. Second, the phenotype is not revealed unless both miR-279 and miR-996 are inactivated.
Figure 4.
Endogenous expression of either mir-279 or mir-996 fully rescues the mir-279/996 double mutant. (A) Summary of 16.6-kb genomic rescue transgenes bearing a wild-type mir-279/996 sequence or that have been modified to produce only mir-279 or only mir-996. These were assayed in the mir-279/996[15C/15C] deletion background. (B–F) Staining of ∼32 h APF nota with the socket cell marker Su(H), the shaft/sheath cell marker DPax2, and the neuronal marker Elav. (B) Wild-type pattern. (C) mir-279/996[15C/15C] shows high-frequency conversion of small DPax2 nuclei (sheath) into ectopic neurons (circled organs), whereas outer cell fates (socket and shaft) are relatively unaffected. This defect is fully rescued by wild-type mir-279/996 transgene (D), mir-279-only transgene (E), and mir-996-only transgene (F). Bars, 50 µM.
mir-279/996 regulate Notch signaling in the peripheral sensory lineage
miR-279/996 were reported to exert phenotypically substantial regulation of various neural transcription factors, of Ras signaling, and JAK/STAT signaling in various developmental contexts (Cayirlioglu et al., 2008; Luo and Sehgal, 2012; Sun et al., 2015). However, the massive sheath-to-neuron conversion we observed precisely phenocopies loss of Notch signaling during asymmetric division of the pIIIb cell (Lai, 2004; Lai and Orgogozo, 2004). Moreover, as mentioned, we sometimes observed stronger neurogenic phenotypes consistent with conversion of outer cell fates into neurons (Fig. 2 C); such phenotypes suggest Notch loss of function at multiple steps in the mechanosensory organ lineage (Fig. 1 A).
To gain evidence of the relationship between miR-279/996 and the Notch pathway, we performed genetic interaction experiments. Null mutants of mir-279/996 exhibit only mild disturbances in exterior bristle patterning, such as occasional missing macrochaete bristles, but have relatively normal microchaete numbers (Fig. 5, A and B). In addition, whereas Notch is haploinsufficient for wing development, Notch null heterozygotes support relatively normal peripheral neurogenesis (Fig. 5 C) with only modestly increased microchaete density when an allele of mir-279/996 is removed (Fig. 5 D). Strikingly, N[55E11]/+; mir-279/996[15C/15C] mutants exhibit a synergistic defect evident as fully penetrant (100% out of n = 41 animals examined), massive, adult notum balding (Fig. 5 E). This implied that miR-279/996 plays a broader endogenous role in sensory organ development and is active at other stages in the sensory organ lineage. Indeed, analysis of a transcriptional reporter consisting of nuclear GFP knocked into a 16.6-kb mir-279/996 genomic backbone revealed both epidermal expression and elevated expression in all sensory lineage cells (Fig. S4). We note that the transcriptional reporter is detected in neurons, even though the functional miRNA sensor indicates they are not active in this cell type (Fig. 1 C); we cannot distinguish whether this reflects perdurance of the transcriptional reporter or perhaps regulated miRNA processing. Nevertheless, these data provide evidence that the miRNA locus is expressed more broadly in the sensory lineage and is thus positioned to impact sensory cell fates beyond the sheath cell.
Figure 5.
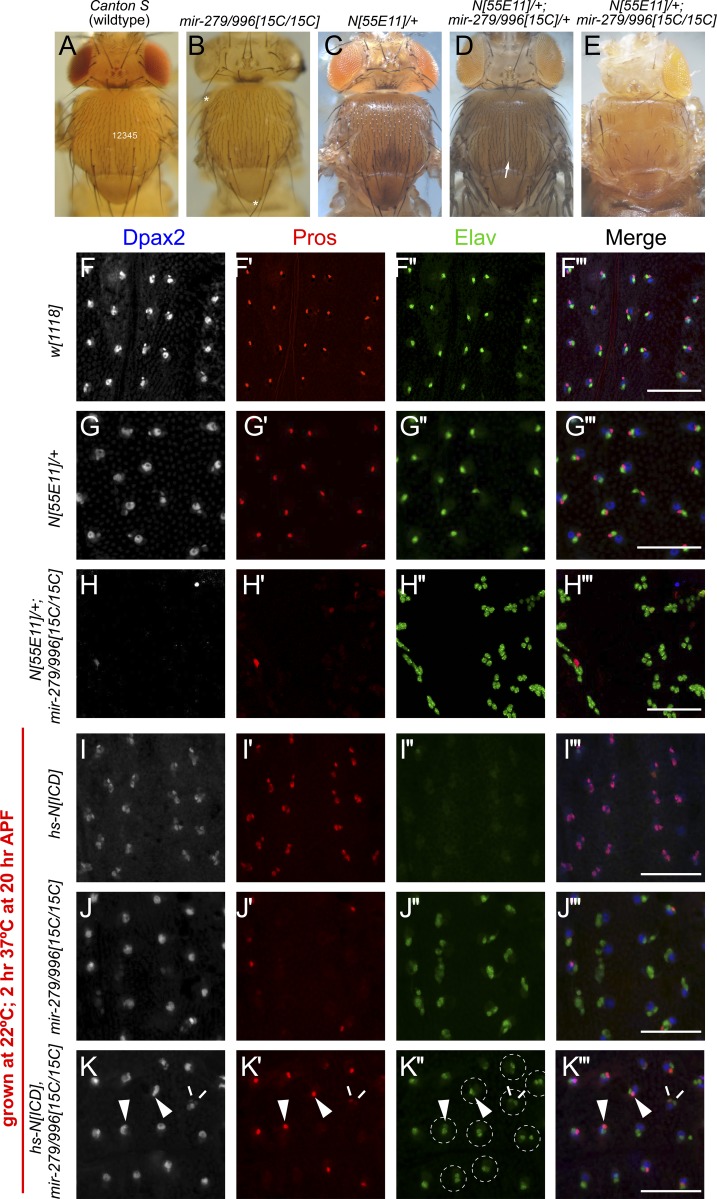
miR-279/996 facilitates the activity of nuclear Notch in the PNS lineage. (A–E) Images of adult nota. (A) Wild-type fly shows the normal pattern of external mechanosensory bristles. In each heminotum, there are five rows of small microchaete bristles from the center of the notum to the dorsocentral macrochaete (large) bristles. (B) The mir-279/996 null mutant exhibits mostly normal exterior bristle patterning, but some macrochaetes are lost (asterisks). Notch heterozygote also exhibits mostly normal bristle patterning (C), whereas double heterozygote of Notch and mir-279/996 exhibits a mild increase in microchaete density (D); arrow points at an extra partial row of bristles. (E) Notch heterozygote in the mir-279/996 null shows extreme loss of exterior bristle structures. (F–K) Shown are pupal nota at >28 h APF stained for cell-specific markers. (F and G) Normal pattern of DPax2 (large shaft and small sheath nuclei), Pros (sheath nuclei), and Elav (neuronal nuclei) in control w[1118] (F) and N[55E11]/+ (G). (H) N[55E11]+; mir-279/996[15C/15C] notum shows loss of most DPax2 and Pros staining and development of “all-Elav” sensory organ lineages. Thus, the exterior balding of the adult flies of this genotype (E) is caused by conversion of most lineage cells into neurons. (I–K) Pupal nota that were subjected to a 2 h heat shock during pIIIb division. (I) An appropriately timed pulse of hs-NICD results in relatively uniform transformation of neurons into sheath cells. (J) Heat shock treatment of mir-279/996 null animals shows the typical sheath-to-neuron phenotype of this mutant. (K) Parallel treatment of hs-NICD, mir-279/996 null animals reveals a large population of double neuron organs (dotted circles) and/or lineages that coexpress Pros with detectable Elav reactivity (arrowheads). A rare case of double Pros/double Elav organ is indicated with double lines. Epistasis of the miRNA phenotype indicates that NICD cannot function effectively without miR-279/996. Bars, 50 µM.
There are several cellular possibilities for how notum balding phenotypes might arise, some of which are oppositely acting with respect to Notch pathway activity (Fig. 1 A). For example, the SOP might fail to be specified, which could occur under Notch gain-of-function circumstances. On the other hand, conversion of outer cell fates into inner cell fates can also yield external balding. An extreme version of this would be generation of multiple neuron sensory structures, the Notch loss-of-function “neurogenic” defect. We assessed cell fate specification in N[55E11]/+ 28–32 h pupae and found it to be similar to control w[1118] (Fig. 5, F and G). In strong contrast, N[55E11]/+; mir-279/996[15C/15C] pupae exhibited strong loss of DPax2 and Pros reactivity and the presence of a majority of sensory clusters composed entirely of Elav+ neurons (Fig. 5 H). Therefore, in the absence of this miRNA locus, loss of one copy of Notch causes the failure of non-neuronal cell fate acquisition throughout the lineage of PNS mechanosensory organs, reflecting reiterative loss of Notch signaling.
We conducted reciprocal tests of the behavior of the activated Notch intracellular domain (NICD) in mir-279/996[15C] mutants. In particular, given the powerful activity of NICD, one might imagine that it could rescue the neurogenic defect of mir-279/996[15C] mutants. Because NICD can efficiently extinguish the endogenous SOP fate, its constitutive expression precludes analysis of lineage cell fates. To address whether NICD can restore sheath cells in mir-279/996[15C] mutants, we exploited temporal control afforded by a hs-NICD transgene. We were able to time a heat pulse at 37°C at ∼20hr APF in animals grown at 22°C, such that wild type exhibited normal development whereas hs-NICD/+ flies exhibited high frequency transformation of neurons into sheath cells (80%, n = 105 sensory organs; Fig. 5 I). This reflects a gain of the Notch-inhibited daughter fate of the pIIIb (Fig. 1 A). We still observed the typical ectopic neuron phenotype of mir-279/996[15C/15C] mutants after this heat-shock treatment (Fig. 5 J). However, induction of hs-NICD did not induce double sheath phenotype in mir-279/996[15C] homozygotes (<1% double sheaths, n = 246 sensory organs). Instead, the double Elav+ phenotype of the miRNA mutant was epistatic in the presence of ectopic NICD (Fig. 5 K), even though Pros could be observed in some of these Elav+ cells (Fig. 5 K, insets), as seen in the miRNA mutant alone (Fig. 2 C).
Because the powerful NICD molecule is blunted in the absence of miR-279/996, these miRNAs are required for effective adoption of the Notch-inhibited cell fate. Overall, these data suggest intimate links between miR-279/996 and the facilitation of nuclear Notch function.
Expression of the nuclear Notch antagonist Insb is restricted to the neuron by miR-279/996
mir-279/996 mutants phenocopy Notch loss of function within the mechanosensory lineage, which implies that the miRNAs repress a negative component of the pathway in this setting. Although scores of genes are specifically required for Notch pathway function, relatively few genes mutate to Notch gain-of-function phenotypes (Lai, 2002a; Mummery-Widmer et al., 2009; Le Bras et al., 2012). Although the core positive Notch pathway component neuralized bears a conserved miR-279/996 site, none of the classically well-characterized Notch inhibitors bear conserved miR-279/996 sites.
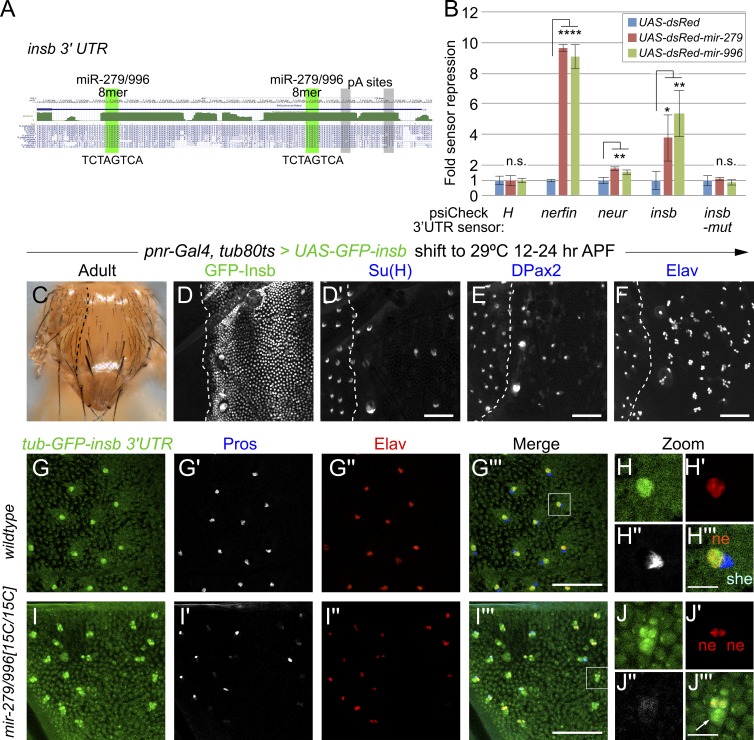
However, the anonymous gene CG6520 was recently recognized as a neural-specific Notch inhibitor and christened insb (Coumailleau and Schweisguth, 2014). Intriguingly, the insb 3′ UTR bears two optimal 8mer sites (TCTAGTCA) for miR-279/996 that are conserved across the sequenced Drosophilids (Fig. 6 A). In fact, the only other Drosophila gene predicted with two conserved 8mers for miR-279/996 is nerfin-1, a transcription factor that is critical for the generation of ectopic CO2 neurons in mir-279/996 mutants (http://www.targetscan.org); nerfin-1 bears additional conserved sites for these miRNAs.
Figure 6.
Insb is directly repressed by miR-279/996 in non-neuronal mechanosensory cells. (A) The 3′ UTR of the Notch antagonist insb contains two highly conserved, optimal seed matches for miR-279/996. (B) Luciferase-3′ UTR sensor assays in S2 cells demonstrate direct targeting of insb by miR-279 and miR-996 via their cognate shared seed matches (compare insb and insb-mut sensors). We use the 3′ UTR of the Notch antagonist H as a negative control and nerfin as a positive control. We also validate the positive Notch pathway factor neuralized (neur) as a miR-279/996 target. These data represent quadruplicate wells from a representative transfection experiment; the experiment was done three times with similar results. *, P < 0.02; **, P < 0.002; ****, P < E-07, Student’s t test; standard deviations are shown. (C–F) Spatiotemporal misexpression of GFP-insb in the central domain of the notum using pnr-Gal4 and tub-Gal80[ts]; flies were shifted to the restrictive temperature (29°C) at 12–24 h APF and left to develop further at 25°C. These flies exhibit a strong balding defect within the pnr-Gal4 domain (compare to lateral wild-type domain, to the side of the dotted line). (D–F) Staining at ∼30 h APF. The GFP-Insb domain is marked with the dotted line, but only one panel of GFP staining is shown for reference (in D). Misexpression of GFP-Insb induces strong loss of Su(H) (D′) and DPax2 (E) and conversion into multiple Elav sensory organ lineages (F). (G–I) Expression of a tub-GFP-insb-3′ UTR sensor in wild-type (G and H) and mir-279/996[15C/15C] nota (I and J); individual sensory organs are magnified in H and J. The insb sensor is posttranscriptionally repressed to yield neuronal expression. Bars: (D–G and I) 50 µM; (H and J) 10 µM.
We used sensor assays in S2 cells to assess capacity of insb to be directly repressed by miR-279/996. A Renilla luciferase reported linked to the full insb 3′ UTR was specifically repressed by cotransfection of either miR-279 or miR-996, and responses were abrogated upon mutation of both seed matches (Fig. 6 B). The degree of repression of the insb sensor by these miRNAs was lower than that of nerfin-1, which was previously shown to be an exceptionally robust target of miR-279/996, but was higher than that of neur, which also validated in this assay (Fig. 6 B).
Of relevance to the present study is that misexpression of Insb induces a similar cell transformation as deletion of mir-279/996, phenocopying a loss of Notch signaling in the bristle lineage. We recapitulated previous results showing that ectopic expression of Insb using pnr-Gal4 induces a profound balding defect (Coumailleau and Schweisguth, 2014). We performed these experiments using both constitutive Gal4 activity (not depicted) and Gal80[ts] to restrict Gal4 activity to sensory organ lineage divisions (during 12–24 h APF; Fig. 6 C). The advantage of the latter manipulation was that it simplified cellular lineage analysis by bypassing any initial defect during lateral inhibition. These analyses confirmed that external balding was caused by conversion of non-neuronal cells into neurons (Fig. 6, D–F).
Because Insb antibodies do not exist and a previously reported GFP-tagged Insb allele is no longer extant (Coumailleau and Schweisguth, 2014), we sought an alternate approach to visualize miR-279/996-mediated posttranscriptional repression of Insb in vivo. To this end, we generated a tub-GFP-insb 3′ UTR sensor transgene. Strikingly, this ubiquitously transcribed transgene yields substantially neural-restricted GFP in the notum, as indicated by its costaining with Elav and exclusion from neighboring Pros+ cells (Fig. 6, G and H). This endogenous 3′ UTR sensor exhibits a similar pattern to the artificial sensor bearing multiple perfect matches to miR-279 (Fig. 1), demonstrating the potency of this 3′ UTR to respond to miR-279/996. We verified that its pattern was induced by mir-279/996, because deletion of the miRNAs led to elevation of GFP sensor activity in multiple sensory organ cells (Fig. 6, I and J). We note that GFP was high in both small inner sensory organ nuclei, regardless of whether they were transformed into a double neuron cluster or maintained a Pros+ nucleus. Elevated GFP was also observed in large outer sensory organ nuclei. Collectively, these data demonstrate the nuclear Notch antagonist Insb is posttranscriptionally repressed by miR-279/996 into the neuron.
Insb is responsible for neurogenic lineage defects in mir-279/996 mutants
Because misexpression of Insb can convert non-neuronal cells of the sensory lineage into neurons, it has the genetic properties of a miR-279/996 target that is well positioned to account for the phenotypes of mir-279/996 mutant notum sensory organs. We sought to test whether physiological expression of Insb is relevant to mir-279/996 deletion phenotypes.
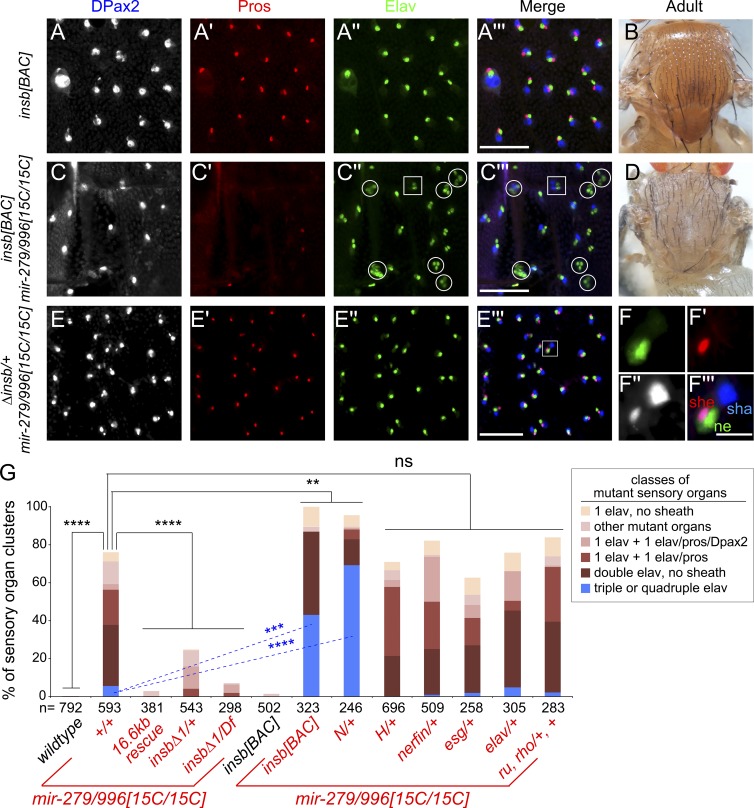
We first examined the consequences of mildly increasing level insb using a bacterial artificial chromosome (BAC) transgene. In contrast to misexpression using the Gal4-UAS system, adding a chromosomal dose of insb has no effect on normal PNS patterning (Fig. 7, A and B). However, placing this transgene into mir-279/996[15C] has a strong enhancing effect, mirroring that obtained with the Notch/+ interaction. This genetic background yields substantial pIIA>pIIB cell fate transformations, as indicated by increased frequency of three or more neuron sensory organs and/or two sheath and two neuron sensory organs (Fig. 7 C) in the central notum region, accompanied by adult balding (Fig. 7 D). These data are consistent with the fact that expression of Insb initiates in the sensory precursory cell (Coumailleau and Schweisguth, 2014) and that miR-279/996 is required in multiple Notch-dependent sensory cell divisions (Fig. 5).
Figure 7.
Insb is a key miR-279/996 target during sensory organ development. (A–F) Reciprocal dose-sensitive genetic interactions of insb and mir-279/996 in PNS development. (A and B) insb[BAC] transgenic animals show wild-type mechanosensory development (A) and normal adult bristle patterning (B). (C) insb[BAC]; mir-279/996[15C/15C] mutant animals exhibit strongly enhanced conversion of sensory organ fates into neurons. Circles in C′′ and C′′′ highlight triple or higher-order Elav+ sensory organ clusters, which are normally very rare in the miRNA mutant; square denotes a double sheath/double neuron cluster. (D) Adult insb[BAC]; mir-279/996[15C/15C] exhibits noticeable balding consistent with multiple neuron sensory organs. (E) Heterozygosity for insb strongly suppresses mir-279/996 null PNS cell specification defects. (F) Close-up of an individual rescued mutant sensory organ cluster. Bars: (A, C, and E) 50 µM; (F) 10 µM. (G) Quantification of sensory organ marker expression across a panel of mutant genotypes. The numbers (n) of sensory organs analyzed for each genotype are indicated. Standard deviations are indicated, and statistical significance was determined by one-way ANOVA. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
We then conducted reciprocal loss-of-function assays to interrogate whether Insb, or other characterized targets, are responsible for neural cell fate conversion in mir-279/996 mutant PNS. We introduced heterozygous mutations of miR-279/996 targets and assayed for phenotypic suppression. If the miRNAs mediate their effect by fine-tuning a large cohort of targets, then no epistatic interactions are expected to exist (Dai et al., 2012). However, the existence of dose-sensitive miR-279/996 targets would support that derepression of a particular target is causal to miRNA mutant phenotypes.
Strikingly, heterozygosity for insbΔ1 strongly suppressed the specification of ectopic notum neurons and restored DPax2+/Pros+ sheath cells (Fig. 7, E and F). Moreover, the insb null condition (insbΔ1/Df) nearly completely restored normal mechanosensory organ development in mir-279/996[15C/15C] null mutants (Fig. S5). We quantify the rescue of abnormal sensory lineage complements in different insb backgrounds in Fig. 7 G. In particular, we highlight the robust rescue of the profound mir-279/996 multiple neuron defects simply by insb heterozygosity, nearly complete rescue by insb hemizygosity, and strong enhancement of triple/quadruple neuron frequency by adding an insb dose or removing a Notch allele (Fig. 7 G). These exquisite reciprocal genetic interactions pinpoint Insb as a central player that compromises Notch signaling to generate neuronal cell transformation in the miRNA mutant.
Surprisingly, genetic interaction of mir-279/996 does not extend to another critical Notch repressor. In particular, Hairless (H) is a haploinsufficient corepressor of the Notch transcription factor Su(H), and loss of one H allele is sufficient to induce Notch gain-of-function phenotypes in the shaft-socket cell division and in the wing (Bang and Posakony, 1992). However, H/+ did not rescue mir-279/996 mutants in the notum (Figs. 7 G and S5), highlighting the specificity of the dose-sensitive suppression achieved with insb mutants. The validated Notch pathway target neur (Fig. 7 G) also did not modify mir-279/996[15C/15C] (Fig. S5).
Finally, we find that many other validated miR-279/996 targets do not seem to be relevant in mechanosensory organs. In the olfactory system, deletion of mir-279/996 causes ectopic CO2-sensing neurons, a phenotype that can be largely suppressed by heterozygosity of the direct targets nerfin-1 (Cayirlioglu et al., 2008) or esg (Hartl et al., 2011). We have also reported that miR-279/996 is an in vivo repressor of ubiquitous Elav (Sanfilippo et al., 2016) and documented that miR-279/996 can repress the Ras pathway targets roughoid and rhomboid in vitro (Sun et al., 2015). However, neither nerfin-1, esg, or elav heterozygotes nor roughoid, rhomboid double heterozygotes modify the mir-279/996 notum PNS phenotype (Fig. 7 G). Thus, insb represents a highly specific target modifier in the miR-279/996 output network.
Overall, we demonstrate that a previously unappreciated double-negative circuit controls non-neural fate within the seemingly well-understood Drosophila PNS lineage.
Discussion
Phenotypic sensitivity of Notch signaling to miRNA regulation
We find that mir-279/996 plays critical roles in assigning Notch-mediated, non-neuronal cell fate in Drosophila peripheral sense organs. The severity of mir-279/996 mutant phenotypes place it among a very small number of miRNAs that function as developmental cell fate switches and contrast with the typical notion of miRNAs as fine-tuners. In retrospect, the phenotypic sensitivity of Notch signaling to miRNA-mediated regulation is consistent with the exquisite dose sensitivity of this pathway (Lai, 2004). Aside from ribosomal genes, only a handful of Drosophila genes are morphologically haploinsufficient. Remarkably, three of these are core Notch pathway members: Delta, Notch, and H. Reciprocally, clonal analyses that juxtapose cells with an extra copy of Delta or Notch against wild-type cells reveal a profound directional effect on the epidermal–neural cell fate choice (Heitzler and Simpson, 1991). Thus, mild alterations of Notch pathway activity can have a strong effect on developmental decisions.
On the other hand, there are several surprising aspects to the regulatory circuit that we define in the mechanosensory lineage. First, our finding that an essential logic of cell fate commitment in the sensory organ lineage involves miRNA-mediated repression is unexpected, given the decades of study on the genetics of sensory bristle patterning. Their development has been systematically analyzed using virtually every conceivable strategy, such as forward genetics (Jafar-Nejad et al., 2005), reverse genetics (Reeves and Posakony, 2005), genome-wide RNAi (Mummery-Widmer et al., 2009), systematic gain-of-function screening (Abdelilah-Seyfried et al., 2000), and quantitative trait loci analysis (Norga et al., 2003), to name a few. We emphasize that miR-279/996 is not a robustness factor for mechanosensory organ development, as with the well-studied mir-9a locus (Li et al., 2006; Bejarano et al., 2010; Cassidy et al., 2013), but that cell specification in this miRNA mutant fails in the majority of sensory organs during normal conditions.
On the target side, we identify Insb as a critical output of miR-279/996, whose derepression mediates ectopic neural specification. We note that most other well-documented miRNA targets are overtly critical genes whose mutation causes strong consequences. An unexpected finding here is that a profound miRNA mutant phenotype involves regulation of a nonessential factor. In particular, deletion of Insb has almost no consequence on PNS development (Coumailleau and Schweisguth, 2014), but its failure to be regulated by miR-279/996 causes massively defective sensory organ cell specification. Curiously, this genetic situation parallels one of the first studies on miRNA targeting, involving repression of the Notch regulator Bearded by Brd box seed elements. Like Insb, Bearded is a nonessential Drosophila factor whose deletion has little effect (Leviten et al., 1997), but whose misexpression inhibits Notch signaling and causes profound defects in sensory organ development (Lai and Posakony, 1997; Lai et al., 1998; Lai, 2002b). Thus, multiple critical genetic circuits in PNS cell specification involve posttranscriptional repression of nonessential genes, an apparently uneconomical strategy that likely reflects the ad hoc nature of evolutionary twists and turns that led to present day regulatory networks, as opposed to efficient design.
Conservation of miRNA function via divergent targets: antineural activity of mir-279/996
The vast majority of miRNA knockouts in diverse animals reveal mostly subtle and/or impenetrant defects, especially during the course of normal development (Miska et al., 2007; Chen et al., 2014). Therefore, even though virtually every important developmental gene bears conserved binding sites for one or more miRNAs (Friedman et al., 2009), the relative necessity for miRNA-mediated regulation during development remains mostly unclear. Very recent studies come to divergent conclusions, by either continuing to expand the breadth of the miRNA regulatory network via noncanonical sites (Kim et al., 2016), or by suggesting that most predicted targets, even apparent conserved sites, may be false positives (Pinzón et al., 2017).
In the face of this ongoing debate, it is notable that a small subset of miRNAs are both phenotypically pleiotropic and have multiple critical target genes in different contexts (Smibert and Lai, 2010). mir-279/996 is an exemplar of this situation, because it exhibits profound developmental and/or functional defects in multiple contexts, including various aspects of the nervous system and ovary (Cayirlioglu et al., 2008; Luo and Sehgal, 2012; Sun et al., 2015). Interestingly, our current study revealing strong, ectopic neuronal commitment in the notum mechanosensory organs is broadly analogous to the ectopic CO2 neuron defect this miRNA mutant exhibits in the olfactory system (Cayirlioglu et al., 2008). However, the mechanistic basis of antineural miR-279/996 activity is different in these settings. In particular, we show here that these miRNAs guide non-neuronal cell fate, with the sheath cell being most sensitive, by repressing the nuclear Notch inhibitor Insb. In the olfactory system, repression of the transcription factors Nerfin-1 and Escargot by miR-279/996 suppresses the formation of ectopic CO2-sensing neurons (Cayirlioglu et al., 2008; Hartl et al., 2011). Surprisingly, we present evidence that many of these developmental settings rely on different “critical targets,” as seen by the fact that strong in vivo phenotypes can be rescued by heterozygosity of individual specific targets, but cross-rescue was not observed. Thus, although one interpretation of an extensive target network is that the miRNA simultaneously but mildly suppresses all of them in a given cell, another interpretation is that a diversity of targets are functionally relevant across several different cell types. Such an in vivo situation, relevant to phenotypes and biology, might be difficult to appreciate using cell culture models.
Clearly, miRNAs are able to incorporate diverse target cohorts. A third member of the miR-279 seed family (miR-286) exhibits very distinct accumulation from miR-279/996 in that it is restricted to the early embryo (Sun et al., 2015), and mir-286 is part of a zygotically expressed cluster that facilitates clearing of maternal transcripts (Bushati et al., 2008). Nevertheless, it is instructive to consider that miR-279/996 in Drosophila has been reiteratively used as an “antineural” factor in multiple settings of sensory organ development. In contrast to typical recycling of developmental subroutines, miR-279/996 has been rewired for different critical target outputs in multiple settings to achieve thematically similar functions in suppressing neural fate. Perhaps this relates to roles even outside of the nervous system, because we have shown that a lower level of spatially broad miR-279/996 has a role in repressing ubiquitous Elav (Sanfilippo et al., 2016), which is well known as the canonical marker of postmitotic neurons in Drosophila. Members of the miR-279 seed family are present throughout nonchordate animal species (Mohammed et al., 2014), and it will be interesting to know if an antineural function of this family represents an ancient function that has been preserved through evolving target cohorts.
Materials and methods
Drosophila stocks
We used previously reported deletion alleles of the mir-279/996 locus (ex117, ex36, and 15C) and 16.6-kb rescue transgenes of the mir-279/996 locus (wt, 2x-mir-279, and 2x-mir-996; Sun et al., 2015). Other alleles and transgenes used to construct stocks analyzed in this study were tub-GFP-SV40 3′UTR (Brennecke et al., 2003; gift of S. Cohen, University of Copenhagen, Copenhagen, Denmark), hs-NICD (Struhl et al., 1993; gift of G. Struhl, Columbia University, New York, NY), H[E31] (Bang and Posakony, 1992; gift of J. Posakony, Univeristy of California, San Diego, La Jolla, CA), N[55E11] (BL#28813), neur[IF65] (Lai and Rubin, 2001), elav[5] (Yao et al., 1993; gift of M. Soller, University of Birmingham, Birmingham, England, UK), esg[k00606] (BL#10359), pnr-Gal4 (BL#3039), nerfin-1[Δ54] (Kuzin et al., 2005; gift of W. Odenwald, National Institutes of Health, Bethesda, MD), su(ve)[1] ru[1] rho[ve-1] h[1] th[1] (BL#617), UAS-GFP-insb and insbΔ1 (Coumailleau and Schweisguth, 2014; gifts of F. Schweisguth, Institut Pasteur, Paris, France), and nompA-GFP (Chung et al., 2001; gift of M. Kernan, Stony Brook University, Stony Brook, NY). Because the original insb[BAC] transgene is no longer available (Coumailleau and Schweisguth, 2014), we reinjected the insb[BAC] to generate a new transgene (BestGene). We generated the miR-279 sensor by cloning two antisense copies of the mature miR-279 sequence into tub-GFP-SV40 3′UTR vector (Brennecke et al., 2003). We generated the insb sensor by cloning its 3′ UTR and ∼250-nt downstream genomic sequence into tub-nGFP vector.
In general, flies were raised at 25°C, synchronized as white prepupae, and aged as appropriate until dissection. Clonal analysis was performed by heat-shocking 24- to 48-h larvae of the genotypes hs-FLP/+ or Y; FRT82B ubi-GFP M(3)/FRT82B mir-279/996[15C] for 30 min at 37°C. For hs-N[ICD] experiments, stocks were raised at 22°C, white prepupae were collected and aged at 25°C, then heat-shocked at 37°C from 20–22 h APF, and then returned to 25°C before dissection. For experiments involving temporal induction of transgenes, we crossed pnr-Gal4, tub-Gal80[ts] to UAS-insb-GFP, raised them at 18°C, and then shifted them to 29°C at 12–24 h APF before dissection.
Immunohistochemistry and imaging
We dissected tissues and fixed them for 15–30 min in 4% paraformaldehyde in PBS, washed extensively in PBS/0.1% Triton X-100 (PBT). Subsequent incubations with primary or secondary antibodies diluted in blocking solution (2% BSA in PBT) were conducted at room temperature for 3 h or at 4°C overnight. Tissues were washed six times with PBT in between primary and secondary antibody incubations, and finally mounted in Vectashield (Vector Labs) with DAPI (1:10,000; Roche). Primary antibodies were rat anti-Elav (1:200; DSHB), mouse anti-Cut (1:200; DSHB), mouse anti-Pros (1:200; DSHB), mouse anti-22C10 (1:200; DSHB), rabbit anti-DPax2 (1:10,000; J. Kavaler), goat anti-Su(H) (1:200; Santa Cruz Biotechnology), rabbit anti-GFP (1:1,250; Invitrogen), and chicken anti-GFP (1:1,000; Abcam). Secondary antibodies were conjugated to Alexa Fluor 488, 568, and 647 (1:500; Molecular Probes).
Images were taken with a Zeiss Axioimager.A2 fluorescence microscope using EC Plan neofluar 20× and EC Plan neofluar 40× oil lenses and Axiocam MRm camera or a Leica SP5 spectral confocal microscope using HCX PL APO 20×/0.70 and HCX PL APO 40×/1.25-0.75 lenses. Images were processed using Axiovision 4.7, ImageJ, or Adobe Photoshop CS6, including gamma adjustments, and figures were assembled in Adope Illustrator CS6.
Luciferase sensor assays
S2 cells were plated in 96-well plates and transfected with 12.5 ng/well of empty UAS-DsRed or UAS-DsRed-miRNA constructs, 12.5 ng/well psiCHECK plasmid, or 6.25 ng/well Ub-Gal4. UAS-DsRed-mir-279, UAS-DsRed-mir-996, psiCHECK-H, psiCHECK-nerfin-1, and psiCHECK-neur plasmids were described previously (Sun et al., 2015); insb luciferase sensors were made by cloning its 3′ UTR and ∼250-nt downstream genomic sequence into psiCHECK and then mutagenizing the miR-279/996 sites to create psiCHECK-insb-mut.
Transfections were performed using Effectene Transfection Reagent (Qiagen) according to the manufacturer’s instructions. Luciferase values were measured 3 d after transfection. We normalized transfection efficiencies using control firefly luciferase carried within psiCHECK, and fold repression was normalized against empty UAS-DsRed and psiCHECK empty plasmid. We present representative data from quadruplicate sensor assays, for which each set was performed at least three times and found to yield qualitatively similar results.
Online supplemental material
Fig. S1 depicts the expression of a control tub-GFP-SV40 sensor in the pupal notum. Fig. S2 provides evidence that mir-279/996 null mutants have specific defects in internal cell fates. Fig. S3 depicts a phenotypic series of mir-279/996 alleles that reveal a graded response in the severity of the ectopic neuron defects. Fig. S4 shows the expression pattern of a 16.6 kb mir-279/996-GFP transcriptional reporter in the notum. Fig. S5 summarizes a series of genetic interaction analyses that show that mir-279/996 neural-specification phenotypes are only modified by select Notch pathway mutants.
Supplementary Material
Acknowledgments
We are very grateful to Fuqu Hu for performing luciferase assays. We thank Francois Schweisguth, James Posakony, Matthias Soller, Stephen Cohen, Gary Struhl, Maurice Kernan, Ward Odenwald, the Bloomington Stock Center, and the Developmental Studies Hybridoma Bank for fly stocks and antibodies.
Work in E.C. Lai’s group was supported by the Burroughs Wellcome Fund, the National Institutes of Health (grants R01-NS074037, R01-NS083833, and R01-GM083300), and Memorial Sloan Kettering (core grant P30-CA008748).
The authors declare no competing financial interests.
Author contributions: J. Kavaler performed the bulk of genetic and cellular analyses shown in this study. H. Duan first identified the notum mechanosensory defect in mir-279/996 mutants and initiated this work. R. Aradhya and L. de Navas helped with notum staining and analysis. B. Joseph generated insb and mir-279 transgenes. B. Shklyar helped quantify phenotypes. J. Kavaler, H. Duan, L. de Navas, R. Aradhya, B. Shklyar, and E. C. Lai evaluated data. E. C. Lai designed the study and wrote the manuscript with input from the co-authors.
References
- Abdelilah-Seyfried S., Chan Y.M., Zeng C., Justice N.J., Younger-Shepherd S., Sharp L.E., Barbel S., Meadows S.A., Jan L.Y., and Jan Y.N.. 2000. A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics. 155:733–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang A.G., and Posakony J.W.. 1992. The Drosophila gene Hairless encodes a novel basic protein that controls alternative cell fates in adult sensory organ development. Genes Dev. 6:1752–1769. 10.1101/gad.6.9.1752 [DOI] [PubMed] [Google Scholar]
- Bejarano F., Smibert P., and Lai E.C.. 2010. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev. Biol. 338:63–73. 10.1016/j.ydbio.2009.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D.R., Stark A., Russell R.B., and Cohen S.M.. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 113:25–36. 10.1016/S0092-8674(03)00231-9 [DOI] [PubMed] [Google Scholar]
- Bushati N., Stark A., Brennecke J., and Cohen S.M.. 2008. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 18:501–506. 10.1016/j.cub.2008.02.081 [DOI] [PubMed] [Google Scholar]
- Cassidy J.J., Jha A.R., Posadas D.M., Giri R., Venken K.J., Ji J., Jiang H., Bellen H.J., White K.P., and Carthew R.W.. 2013. miR-9a minimizes the phenotypic impact of genomic diversity by buffering a transcription factor. Cell. 155:1556–1567. 10.1016/j.cell.2013.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayirlioglu P., Kadow I.G., Zhan X., Okamura K., Suh G.S., Gunning D., Lai E.C., and Zipursky S.L.. 2008. Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science. 319:1256–1260. 10.1126/science.1149483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.W., Song S., Weng R., Verma P., Kugler J.M., Buescher M., Rouam S., and Cohen S.M.. 2014. Systematic study of Drosophila microRNA functions using a collection of targeted knockout mutations. Dev. Cell. 31:784–800. 10.1016/j.devcel.2014.11.029 [DOI] [PubMed] [Google Scholar]
- Chung Y.D., Zhu J., Han Y., and Kernan M.J.. 2001. nompA encodes a PNS-specific, ZP domain protein required to connect mechanosensory dendrites to sensory structures. Neuron. 29:415–428. 10.1016/S0896-6273(01)00215-X [DOI] [PubMed] [Google Scholar]
- Coumailleau F., and Schweisguth F.. 2014. Insensible is a novel nuclear inhibitor of Notch activity in Drosophila. PLoS One. 9:e98213 10.1371/journal.pone.0098213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q., Smibert P., and Lai E.C.. 2012. Exploiting Drosophila genetics to understand microRNA function and regulation. Curr. Top. Dev. Biol. 99:201–235. 10.1016/B978-0-12-387038-4.00008-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R.C., Farh K.K., Burge C.B., and Bartel D.P.. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19:92–105. 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl M., Loschek L.F., Stephan D., Siju K.P., Knappmeyer C., and Kadow I.C.. 2011. A new Prospero and microRNA-279 pathway restricts CO2 receptor neuron formation. J. Neurosci. 31:15660–15673. 10.1523/JNEUROSCI.2592-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzler P., and Simpson P.. 1991. The choice of cell fate in the epidermis of Drosophila. Cell. 64:1083–1092. 10.1016/0092-8674(91)90263-X [DOI] [PubMed] [Google Scholar]
- Hummel T., Krukkert K., Roos J., Davis G., and Klämbt C.. 2000. Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron. 26:357–370. 10.1016/S0896-6273(00)81169-1 [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad H., Andrews H.K., Acar M., Bayat V., Wirtz-Peitz F., Mehta S.Q., Knoblich J.A., and Bellen H.J.. 2005. Sec15, a component of the exocyst, promotes notch signaling during the asymmetric division of Drosophila sensory organ precursors. Dev. Cell. 9:351–363. 10.1016/j.devcel.2005.06.010 [DOI] [PubMed] [Google Scholar]
- Kim D., Sung Y.M., Park J., Kim S., Kim J., Park J., Ha H., Bae J.Y., Kim S., and Baek D.. 2016. General rules for functional microRNA targeting. Nat. Genet. 48:1517–1526. 10.1038/ng.3694 [DOI] [PubMed] [Google Scholar]
- Klämbt C., Knust E., Tietze K., and Campos-Ortega J.A.. 1989. Closely related transcripts encoded by the neurogenic gene complex enhancer of split of Drosophila melanogaster. EMBO J. 8:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzin A., Brody T., Moore A.W., and Odenwald W.F.. 2005. Nerfin-1 is required for early axon guidance decisions in the developing Drosophila CNS. Dev. Biol. 277:347–365. 10.1016/j.ydbio.2004.09.027 [DOI] [PubMed] [Google Scholar]
- Lai E.C. 2002a Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 3:840–845. 10.1093/embo-reports/kvf170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E.C. 2002b Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 30:363–364. 10.1038/ng865 [DOI] [PubMed] [Google Scholar]
- Lai E.C. 2004. Notch signaling: control of cell communication and cell fate. Development. 131:965–973. 10.1242/dev.01074 [DOI] [PubMed] [Google Scholar]
- Lai E.C., and Orgogozo V.. 2004. A hidden program in Drosophila peripheral neurogenesis revealed: fundamental principles underlying sensory organ diversity. Dev. Biol. 269:1–17. 10.1016/j.ydbio.2004.01.032 [DOI] [PubMed] [Google Scholar]
- Lai E.C., and Posakony J.W.. 1997. The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development. 124:4847–4856. [DOI] [PubMed] [Google Scholar]
- Lai E.C., and Rubin G.M.. 2001. neuralized functions cell-autonomously to regulate a subset of notch-dependent processes during adult Drosophila development. Dev. Biol. 231:217–233. 10.1006/dbio.2000.0124 [DOI] [PubMed] [Google Scholar]
- Lai E.C., Burks C., and Posakony J.W.. 1998. The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of enhancer of split complex transcripts. Development. 125:4077–4088. [DOI] [PubMed] [Google Scholar]
- Lai E.C., Tam B., and Rubin G.M.. 2005. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 19:1067–1080. 10.1101/gad.1291905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bras S., Rondanino C., Kriegel-Taki G., Dussert A., and Le Borgne R.. 2012. Genetic identification of intracellular trafficking regulators involved in Notch-dependent binary cell fate acquisition following asymmetric cell division. J. Cell Sci. 125:4886–4901. 10.1242/jcs.110171 [DOI] [PubMed] [Google Scholar]
- Leviten M.W., Lai E.C., and Posakony J.W.. 1997. The Drosophila gene Bearded encodes a novel small protein and shares 3′ UTR sequence motifs with multiple Enhancer of split complex genes. Development. 124:4039–4051. [DOI] [PubMed] [Google Scholar]
- Li X., Cassidy J.J., Reinke C.A., Fischboeck S., and Carthew R.W.. 2009. A microRNA imparts robustness against environmental fluctuation during development. Cell. 137:273–282. 10.1016/j.cell.2009.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang F., Lee J.A., and Gao F.B.. 2006. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 20:2793–2805. 10.1101/gad.1466306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., and Sehgal A.. 2012. Regulation of circadian behavioral output via a MicroRNA-JAK/STAT circuit. Cell. 148:765–779. 10.1016/j.cell.2011.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska E.A., Alvarez-Saavedra E., Abbott A.L., Lau N.C., Hellman A.B., McGonagle S.M., Bartel D.P., Ambros V.R., and Horvitz H.R.. 2007. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 3:e215 10.1371/journal.pgen.0030215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed J., Siepel A., and Lai E.C.. 2014. Diverse modes of evolutionary emergence and flux of conserved microRNA clusters. RNA. 20:1850–1863. 10.1261/rna.046805.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummery-Widmer J.L., Yamazaki M., Stoeger T., Novatchkova M., Bhalerao S., Chen D., Dietzl G., Dickson B.J., and Knoblich J.A.. 2009. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 458:987–992. 10.1038/nature07936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norga K.K., Gurganus M.C., Dilda C.L., Yamamoto A., Lyman R.F., Patel P.H., Rubin G.M., Hoskins R.A., Mackay T.F., and Bellen H.J.. 2003. Quantitative analysis of bristle number in Drosophila mutants identifies genes involved in neural development. Curr. Biol. 13:1388–1396. 10.1016/S0960-9822(03)00546-3 [DOI] [PubMed] [Google Scholar]
- Okabe M., Imai T., Kurusu M., Hiromi Y., and Okano H.. 2001. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 411:94–98. 10.1038/35075094 [DOI] [PubMed] [Google Scholar]
- Pinzón N., Li B., Martinez L., Sergeeva A., Presumey J., Apparailly F., and Seitz H.. 2017. microRNA target prediction programs predict many false positives. Genome Res. 27:234–245. 10.1101/gr.205146.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves N., and Posakony J.W.. 2005. Genetic programs activated by proneural proteins in the developing Drosophila PNS. Dev. Cell. 8:413–425. 10.1016/j.devcel.2005.01.020 [DOI] [PubMed] [Google Scholar]
- Sanfilippo P., Smibert P., Duan H., and Lai E.C.. 2016. Neural specificity of the RNA-binding protein Elav is achieved by post-transcriptional repression in non-neural tissues. Development. 143:4474–4485. 10.1242/dev.141978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P., and Lai E.C.. 2010. A view from Drosophila: multiple biological functions for individual microRNAs. Semin. Cell Dev. Biol. 21:745–753. 10.1016/j.semcdb.2010.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G., Fitzgerald K., and Greenwald I.. 1993. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 74:331–345. 10.1016/0092-8674(93)90424-O [DOI] [PubMed] [Google Scholar]
- Sun K., Jee D., de Navas L.F., Duan H., and Lai E.C.. 2015. Multiple in vivo biological processes are mediated by functionally redundant activities of Drosophila mir-279 and mir-996. PLoS Genet. 11:e1005245 10.1371/journal.pgen.1005245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K.M., Samson M.L., Reeves R., and White K.. 1993. Gene elav of Drosophila melanogaster: a prototype for neuronal-specific RNA binding protein gene family that is conserved in flies and humans. J. Neurobiol. 24:723–739. 10.1002/neu.480240604 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.