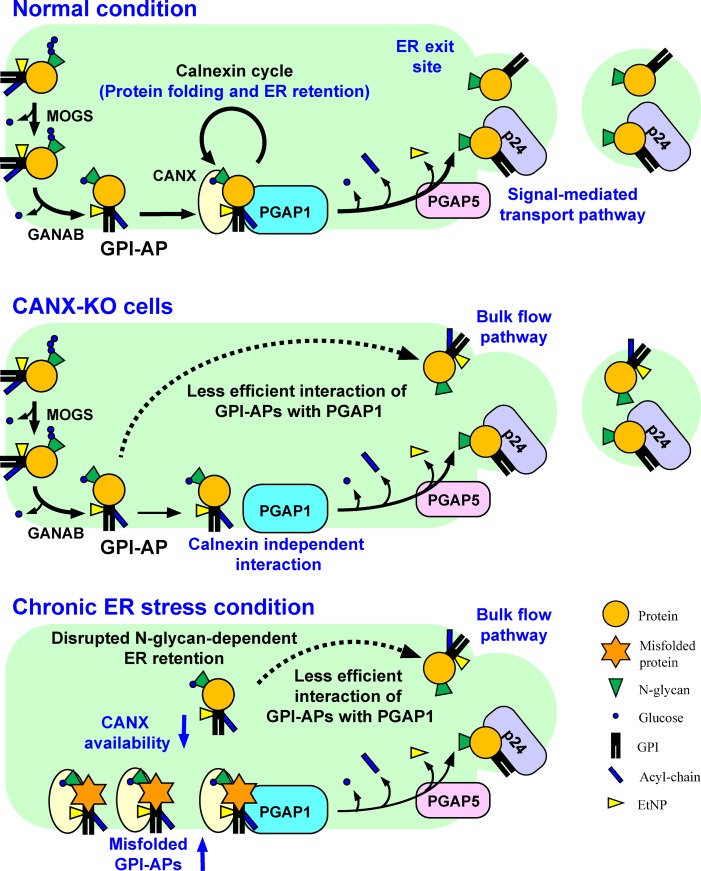
N-Glycosylation and GPI anchoring of proteins occur in the endoplasmic reticulum (ER). Liu et al. revealed N-glycans participate in quality control and temporal ER retention of GPI-anchored proteins (GPI-APs), ensuring their correct folding and GPI processing before exiting from the ER. Chronic ER stress induced exposure of unprocessed GPI-APs on the cell surface.
Abstract
Glycosylphosphatidylinositol (GPI) anchoring of proteins is a conserved posttranslational modification in the endoplasmic reticulum (ER). Soon after GPI is attached, an acyl chain on the GPI inositol is removed by post-GPI attachment to proteins 1 (PGAP1), a GPI-inositol deacylase. This is crucial for switching GPI-anchored proteins (GPI-APs) from protein folding to transport states. We performed haploid genetic screens to identify factors regulating GPI-inositol deacylation, identifying seven genes. In particular, calnexin cycle impairment caused inefficient GPI-inositol deacylation. Calnexin was specifically associated with GPI-APs, dependent on N-glycan and GPI moieties, and assisted efficient GPI-inositol deacylation by PGAP1. Under chronic ER stress caused by misfolded GPI-APs, inositol-acylated GPI-APs were exposed on the cell surface. These results indicated that N-glycans participate in quality control and temporal ER retention of GPI-APs, ensuring their correct folding and GPI processing before exiting from the ER. Once the system is disrupted by ER stress, unprocessed GPI-APs become exposed on the cell surface.
Introduction
Monitoring of protein folding and processing is critical for protein stability and function to maintain cellular homeostasis. Most secretory and membrane proteins are synthesized in the ER, where they are folded and oligomerized. Several molecular chaperones are required for proper protein folding in the ER. Before exiting from the ER, proteins are monitored by a quality control system that ensures correct folding (Ellgaard and Helenius, 2003). Asparagine (N)-linked protein glycosylation, one of the major posttranslational modifications occurring in the ER, plays key roles in quality control of newly synthesized glycoproteins (Helenius and Aebi, 2004; Tannous et al., 2015; Lamriben et al., 2016). The Glc3Man9GlcNAc2 (Glc, glucose; Man, mannose; GlcNAc, N-acetylglucosamine) oligosaccharide moiety is transferred to nascent polypeptides by the oligosaccharyltransferase (OST) complex. After this transfer, two Glc residues are trimmed by α-glucosidase I and II, respectively. The resulting Glc1Man9GlcNAc2 structures on proteins are recognized by calnexin and calreticulin, two lectin-like chaperones, thus promoting protein folding (Helenius and Aebi, 2004; Tannous et al., 2015). Once the remaining Glc residues on protein N-glycans are further trimmed by α-glucosidase II, calnexin and calreticulin dissociate from the proteins. Once folded correctly, the proteins are transported from the ER to the Golgi apparatus. However, proteins remaining in an unfolded state are reglucosylated by UDP-Glc:glycoprotein glucosyltransferase (UGGT) and then become bound again to calnexin and calreticulin for refolding. This series of reactions is called the calnexin–calreticulin cycle and is of central importance for glycoprotein folding.
Anchoring of glycosylphosphatidylinositol (GPI) to proteins is another type of posttranslational modification conserved in eukaryotic cells (Kinoshita et al., 2008; Ferguson et al., 2009). In mammalian cells, ∼150 proteins on the cell surface are modified by GPI anchors. GPI is biosynthesized and transferred to proteins in the ER. Then, GPI-anchored proteins (GPI-APs) are transported from the ER to the plasma membrane through the Golgi apparatus. During this transport, the GPI anchors are structurally remodeled in the ER and Golgi (Fujita and Kinoshita, 2012). In particular, the first reaction, mediated by the enzyme post-GPI attachment to proteins 1 (PGAP1), affects GPI-AP transport and subsequent processing. In GPI biosynthesis, an acyl chain is added to the 2 position of inositol in GPI (Murakami et al., 2003). Soon after GPI attachment to proteins, however, the acyl chain is removed by the GPI-inositol deacylase PGAP1 (Chen et al., 1998; Tanaka et al., 2004). This reaction occurs in most mammalian cell types, with some exceptions, such as GPI-APs on erythrocytes, CD52 on the spleens, and alkaline phosphatase on some cancer cells (Walter et al., 1990; Wong and Low, 1992; Treumann et al., 1995; Schröter et al., 1999). Mutations in PGAP1 cause delayed GPI-AP transport from the ER to the Golgi (Tanaka et al., 2004). GPI-inositol deacylation by PGAP1 and subsequent GPI–glycan remodeling by PGAP5 are critical for binding of GPI-APs to their cargo receptors, the p24 family of proteins, for efficient sorting into the ER exit sites (Fujita et al., 2009, 2011). Defects in PGAP1 also impair fatty acid remodeling of GPI-APs in the Golgi (Maeda et al., 2007).
In mice, mutations in the Pgap1 gene caused severe developmental defects in forebrain formation, typical phenotypes associated with otocephaly and holoprosencephaly. This effect was probably attributable to perturbation of Cripto and/or Wnt signaling (Ueda et al., 2007; Zoltewicz et al., 2009; McKean and Niswander, 2012). It was reported that PGAP1 defects in human patients caused intellectual disability and encephalopathy (Murakami et al., 2014; Granzow et al., 2015; Kettwig et al., 2016). These results suggest that the fine structure of GPI is important during development. In lymphoblastoid cell lines from heterozygous parents of patients with PGAP1 mutations, fractions of inositol-acylated GPI-APs were increased, indicating that PGAP1 heterozygous mutation causes haploinsufficiency and that normal PGAP1 expression is limited and regulated at low levels (Murakami et al., 2014). Therefore, efficient interaction of newly synthesized GPI-APs with PGAP1 is required for correct processing of GPI moieties.
In this study, we aimed to understand the regulatory mechanisms of GPI-inositol deacylation. Mammalian haploid genetic screens identified seven genes required for efficient GPI-inositol deacylation by PGAP1. In particular, some encode proteins involved in N-glycan processing and protein folding. Impaired calnexin cycle and chronic ER stress conditions caused exposure of inositol-acylated GPI-APs on the cell surface. Calnexin interacts with PGAP1 and recognizes N-glycans on GPI-APs, facilitating their GPI-inositol deacylation. Once the system is impaired by chronic ER stress, unprocessed GPI-APs are expressed on the cell surface. Our findings clarified a functional linkage between N-glycan and GPI and a mechanism whereby properly folded GPI-APs are efficiently processed and transported from the ER.
Results
A haploid genetic screen identified seven factors required for efficient GPI-inositol deacylation
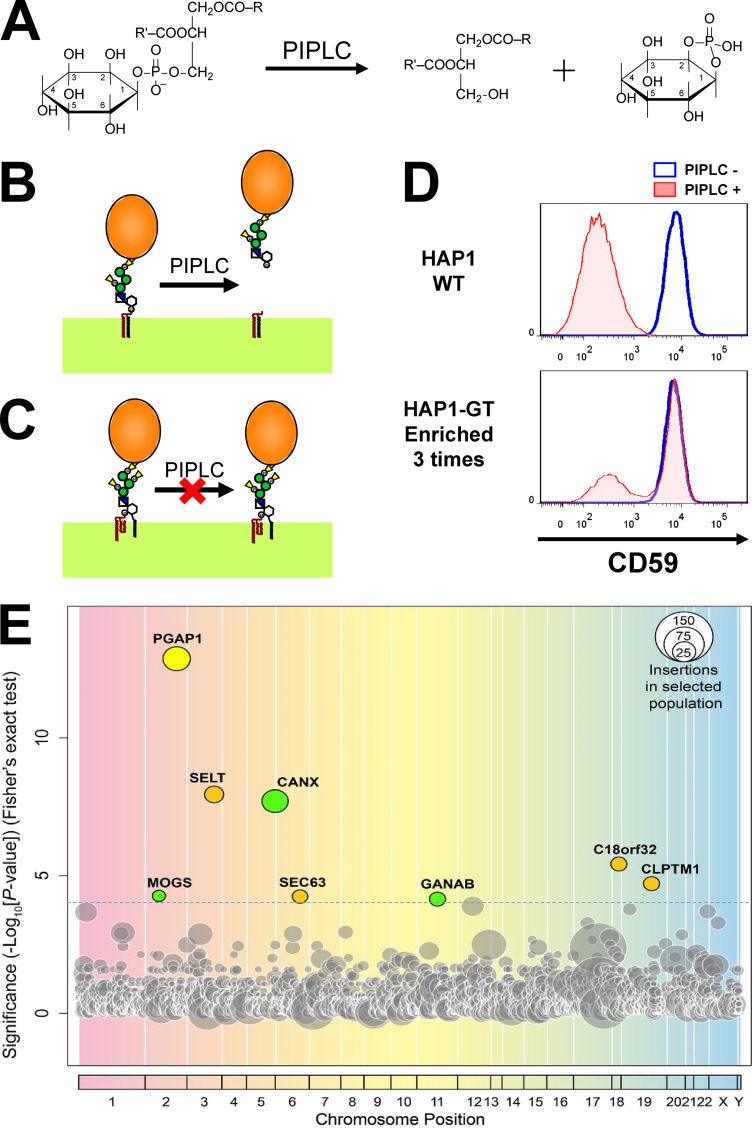
Because GPI-inositol deacylation is the first reaction after GPI attachment to proteins and is required for GPI-AP sorting into the ER exit sites, we predicted that GPI structures and their attachment to proteins would be monitored through this remodeling enzyme. We first tried to identify factors required for GPI-inositol deacylation. To detect whether the inositol portion of GPI anchors is acylated, bacterial phosphatidylinositol (PI)-specific phospholipase C (PIPLC) is generally used (Menon, 1994; Ferguson et al., 2009). To hydrolyze PI, PIPLC reacts with the hydroxyl group on the 2 position of the inositol ring instead of water, producing inositol-1,2-cyclic phosphate (Fig. 1 A; Heinz et al., 1995). If an acyl chain is attached to the 2 position of inositol, PIPLC cannot cleave the substrate. When WT cells were treated with PIPLC, GPI-APs were cleaved and released from the cell surface (Fig. 1 B). In contrast, surface expression of GPI-APs was not changed in PGAP1 mutant cells (Fig. 1 C; Tanaka et al., 2004). Using the sensitivity of GPI-APs to PIPLC, we tried to identify factors regulating GPI-inositol deacylation.
Figure 1.
Haploid genetic screens identified factors required for GPI-inositol deacylation. (A) Schematic representation of the reaction catalyzed by bacterial PIPLC. PIPLC cleaves PI. For hydrolysis, PIPLC utilizes the hydroxyl group at the 2-position of the inositol ring, producing diacylglycerol and inositol-1,2-cyclic phosphate. (B) Mature GPI-APs were cleaved and released from the plasma membrane after PIPLC treatment. (C) When the 2-position of the inositol ring on GPI-AP was modified with an acyl chain, PIPLC does not react with the GPI-APs. (D) Flow cytometric analysis of a GPI-AP, CD59, in parental HAP1 cells (WT) and in the bulk population of mutagenized HAP1 cells (HAP1-GT) that were enriched three times. The red shading and blue line indicate with and without PIPLC treatment, respectively. (E) Significance of the enrichment of gene trap insertions in enriched PIPLC-resistant cells compared with in nonselected cells plotted as a bubble plot. The horizontal line shows the chromosomal position of the genes, and the vertical line the significance of enrichment of each gene (p-values). The size of the bubble shows the number of independent insertion sites in enriched cell populations. Genes significantly enriched in the PIPLC-resistant population (P < 0.0001) are colored. Yellow, PGAP1 encoding GPI-inositol deacylase; green, genes for glucose trimming and calnexin; brown, other genes.
To identify such target factors, we used genetic approaches in the HAP1 human haploid cell line (Carette et al., 2011). Using gene-trapping methods, which disrupt genes by inhibiting normal splicing, on the HAP1 cells, mutant cells were obtained and then screened for phenotypes of interest (Carette et al., 2011; Rong et al., 2015). We applied these methods to analyze the factors required for GPI-inositol deacylation. After obtaining a mutant pool from HAP1 cells mutagenized with gene-trap vectors, we enriched for the mutant cells that had GPI-APs remaining on the cell surface even after PIPLC treatment. In the WT cells, levels of one GPI-AP, CD59, were significantly decreased by PIPLC treatment. However, in enriched mutant cell populations, CD59 remained expressed on the cell surface after PIPLC treatment (Fig. 1 D). From the enriched populations, the insertion sites of gene-trapping vectors were determined by deep sequencing. Comparing the gene-trap insertion sites in the enriched mutants with those in the nonselected population, we identified genes enriched by this screening. As expected, PGAP1 was identified first, indicating that the screening method had worked well (Fig. 1 E and Table S3). In addition, seven genes (SELT, CANX, C18orf32, CLPTM1, MOGS, SEC63, and GANAB) were significantly enriched in this screening.
Knockout (KO) of candidate genes led to partial PIPLC resistance
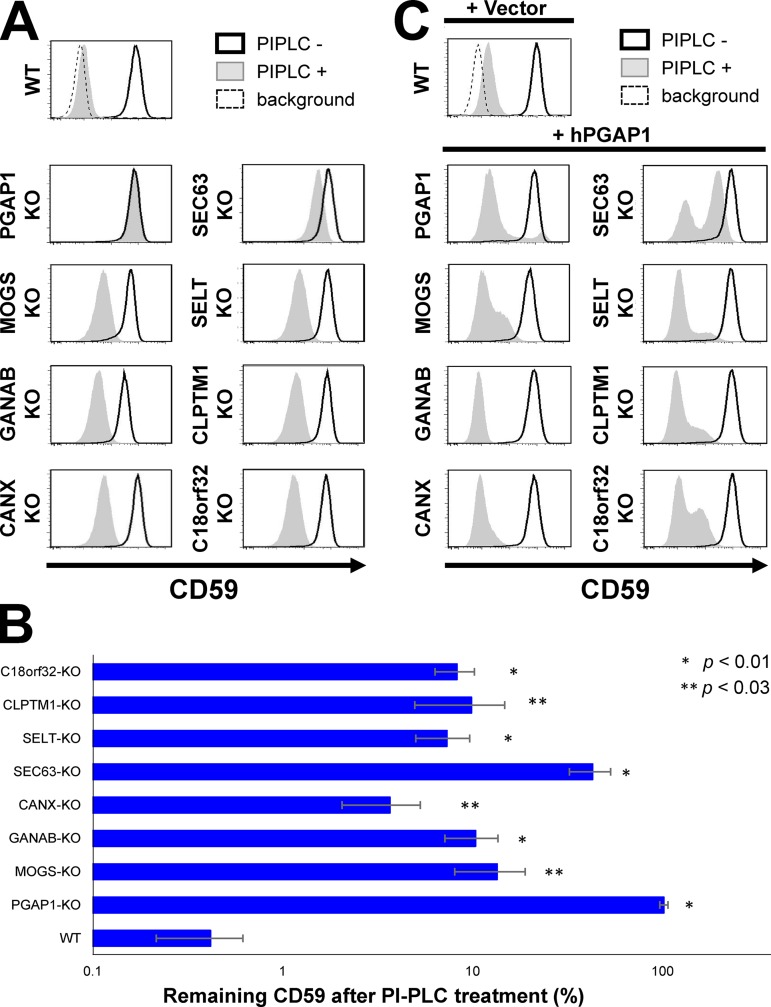
We first knocked out the candidate genes in HEK293 cells using the CRISPR-Cas9 system (Fig. S1) and tested the PIPLC sensitivity of GPI-APs (Figs. 2 A and S2 A). In the PGAP1-KO cells, the GPI-APs CD59 and decay-accelerating factor (DAF) were completely resistant to PIPLC treatment (Fig. 2, A and B; and Fig. S2 A). Cells with other genes knocked out showed partial resistance of GPI-APs against PIPLC (Fig. 2, A and B; and Fig. S2 A). The PIPLC resistance of KO cells was rescued by expression of the responsible gene (Fig. S2 B). In addition, the PIPLC resistance was rescued by PGAP1 overexpression (Fig. 2 C), suggesting that GPI-inositol deacylation was partially impaired by knocking out candidate genes. These results indicated that seven genes were required for efficient GPI-inositol deacylation of GPI-APs.
Figure 2.
KO of genes identified by screening in HEK293 cells. (A) Genes identified in a screening for GPI-APs’ resistance to PIPLC were knocked out by the CRISPR-Cas9 system. KO of the genes is confirmed in Fig. S1. The KO cells were treated with or without PIPLC, stained with anti-CD59 antibody, and analyzed by flow cytometry. Shaded areas indicate cells treated with PIPLC, solid lines indicate cells without PIPLC treatment, and dashed lines show background. (B) Percentages of CD59 remaining after PIPLC treatment of the KO cell lines are plotted. Values are means ± SD of three independent measurements, with p-values (two-tailed Student’s t test) shown on the right. (C) Overexpression of human PGAP1 in the gene KO cell lines rescued the phenotype. Human PGAP1 plasmid was transfected into the KO cells. Cells were selected with antibiotics, treated with or without PIPLC, and then stained with anti-CD59 antibody and analyzed by flow cytometry. The incomplete rescue was a result of the presence of cells showing antibiotics resistance but not expressing PGAP1.
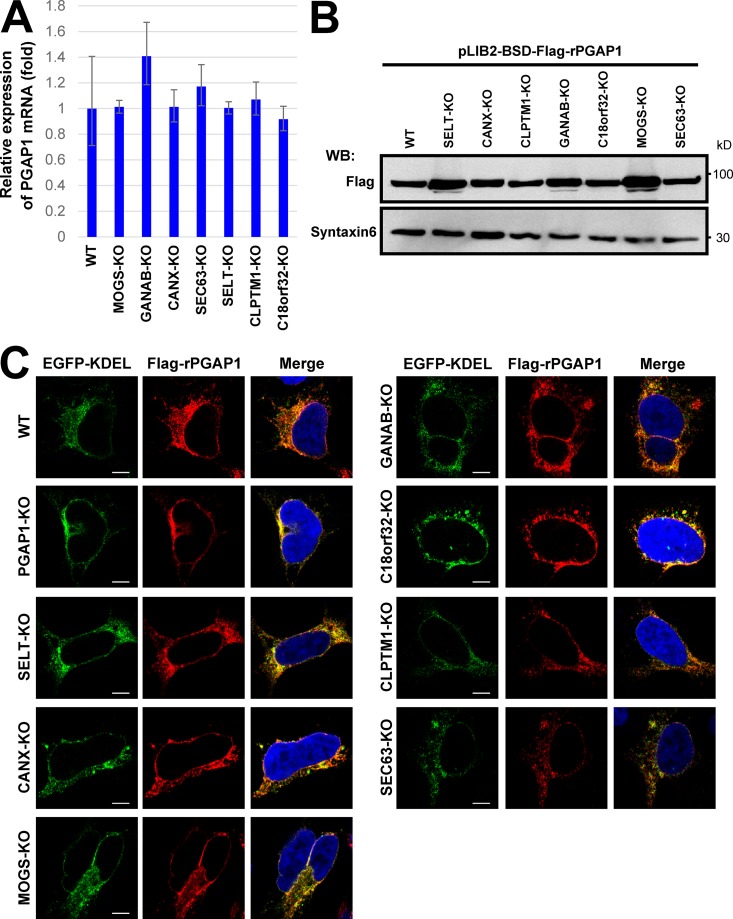
To understand the reason for partial resistance of GPI-APs against PIPLC, we investigated whether knocking out candidate genes affected PGAP1 expression, localization, or protein stability. PGAP1 expression was not changed among all KO cell lines (Fig. 3 A). Because antibodies that detect endogenous PGAP1 were not available, Flag-tagged rat PGAP1 (Flag-rPGAP1) was stably expressed and analyzed for its localization and stability. In the gene KO cell lines, levels of Flag-rPGAP1 were similar to those in WT cells at steady-state (Fig. 3 B). Flag-rPGAP1 was localized to the ER in parental HEK293 cells, and its localization did not change in all KO cell lines (Fig. 3 C). These results suggested that PIPLC resistance observed in candidate gene KO cells was not caused by PGAP1 down-regulation or by its instability.
Figure 3.
Expression, protein stability, and localization of PGAP1 were not changed in gene KO cell lines. (A) Quantitative PCR analysis of PGAP1 mRNA levels in WT HEK293FF6, MOGS-KO, GANAB-KO, CANX-KO, SEC63-KO, SELT-KO, CLPTM1-KO, and C18orf32-KO cells. GAPDH values were used to normalize the data. The bars represent RQ (relative quantification) values ± RQmax and RQmin (error bars) of triplicate samples. (B) Cells stably expressing Flag-tagged rat PGAP1 (Flag-rPGAP1) were lysed and analyzed by Western blotting (WB). Proteins were detected with anti-Flag antibodies. Syntaxin 6 was used as the loading control. (C) Cells stably expressing Flag-tagged rat PGAP1 were transfected with GFP-KDEL and immunostained with an anti-Flag antibody. Images were collected using confocal microscopy. DAPI staining was shown as blue in merged images. Bars, 5 µm.
The calnexin cycle was required for efficient GPI-inositol deacylation
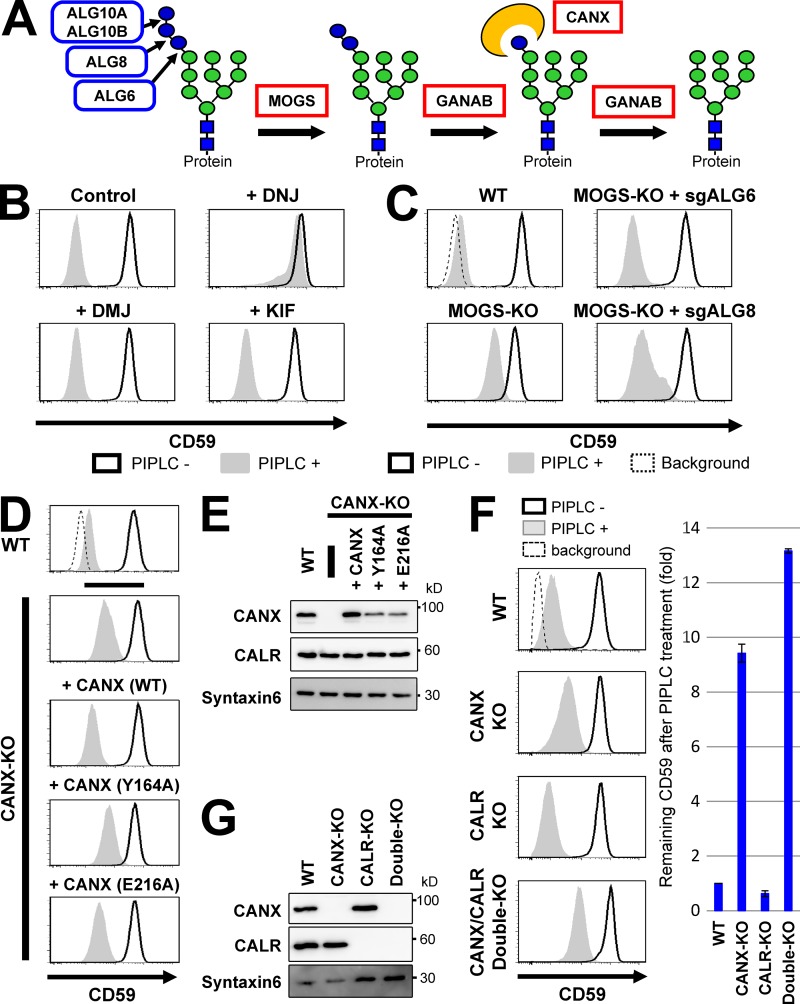
Among candidate genes, MOGS and GANAB encode α-glucosidases I and II, respectively, and both are required in the ER for trimming glucose residues from the N-glycan moieties on newly synthesized proteins (Fig. 4 A; Tannous et al., 2015). CANX encodes calnexin, a molecular chaperone involved in folding and quality control of glycoproteins in the ER (Helenius and Aebi, 2004). Calnexin recognizes glucose residues on N-glycans after two other glucose moieties are removed by α-glucosidases I and II. Therefore, MOGS, GANAB, and CANX participate in the same pathway calnexin cycle for monitoring the folding status of glycoproteins (Helenius and Aebi, 2004; Tannous et al., 2015; Lamriben et al., 2016). When these three genes are mutated, newly synthesized proteins cannot enter the calnexin cycle. Therefore, we next analyzed the relationship between this pathway and GPI-inositol deacylation.
Figure 4.
Glucose trimming from N-glycans and calnexin affected GPI-inositol deacylation. (A) Schematic of glucose trimming after protein N-glycosylation. ALG6, ALG8, and ALG10 are enzymes that add three glucoses (blue circles) one by one to the lipid-linked oligosaccharide Man9GlcNAc2 structure. MOGS and GANAB are α-glucosidase I and α-glucosidase II enzymes, removing the first and the other two glucose from N-glycans, respectively. CANX is calnexin, recognizing and binding to the monoglucosylated N-glycan structure. Green circles, mannose; blue squares, GlcNAc. (B) HEK293FF6 cells were treated with DNJ, an α-glucosidase inhibitor, or DMJ or KIF, α1,2-mannosidase inhibitors, for 11 d. Flow cytometry was used to analyze surface CD59 in cells with or without PIPLC treatment as described in Fig. 2 A. (C) MOGS-KO cells were transfected with plasmids expressing single guide (sg) ALG6 or sgALG8 RNAs to knock out those genes. Plasmid-positive cells were sorted and cultured for >10 d after transfection. Flow cytometry analysis of the surface CD59 in cells with or without PIPLC treatment was performed as described in Fig. 2 A. (D) Resistance of GPI-APs against PIPLC in CANX-KO cells was rescued by expression of WT but not lectin activity–deficient calnexin. WT cells and CANX-KO cells stably expressing WT CANX or mutant CANXs (Y164A or E216A) were treated with or without PIPLC. After treatment, cells were stained with anti-CD59 antibodies and analyzed by flow cytometry as described in Fig. 2 A. (E) Lysates of cells in D were analyzed by Western blotting. CANX and CALR were detected. Syntaxin 6 was used as the loading control. (F and G) WT, CANX-KO, CALR-KO, and CANX/CALR double-KO cells were treated with or without PIPLC. Flow cytometry analysis of surface CD59 was performed (F) as described in Fig. 2 A. CD59 levels remaining after PIPLC treatment are plotted. The remaining CD59 value in WT cells was set as 1. Relative values were calculated and are shown as means ± SD from three independent experiments. Lysates prepared from cells were analyzed by Western blotting (G).
We first assessed whether the α-glucosidase activities of MOGS and GANAB were specifically required for efficient GPI-inositol deacylation. Deoxynojirimycin (DNJ) is an inhibitor of α-glucosidases, whereas deoxymannojirimycin (DMJ) and kifunensine (KIF) are α-mannosidase-I inhibitors (Peyrieras et al., 1983; Vallée et al., 2000). Treatment with DMJ or KIF had no effects, whereas DNJ treatment caused PIPLC resistance of GPI-APs (Fig. 4 B). The results indicated that failure to trim glucose but not mannose from N-glycan impaired correct processing of GPI-APs. Even when exogenous Flag-rPGAP1 was stably expressed, DNJ treatment caused partial PIPLC resistance of GPI-APs (Fig. S3, A and B). This suggests that inhibition of ER α-glucosidases causes exposure of inositol-acylated GPI-APs, which were transported from the ER without processing by PGAP1.
Additions of three glucose residues to the N-glycan precursor, a lipid-linked oligosaccharide (Man9GlcNAc2) structure, were catalyzed stepwisely by ALG6, ALG8, and ALG10 (Fig. 4 A; Aebi, 2013). Thus generated, the Glc3Man9GlcNAc2 was transferred to proteins by the OST complex. Mutations in the MOGS gene inhibit removal of these glucose residues from N-glycans in the ER. We next addressed whether deletion of genes required for glucose addition would prevent the abnormal phenotype in MOGS-KO cells. In the human genome, two ALG10 genes (ALG10A and ALG10B) exist, whereas ALG6 and ALG8 are single genes. When the ALG6 or ALG8 gene was knocked out in a MOGS-KO background, sensitivity of GPI-APs to PIPLC was restored (Fig. 4 C). This suggests that the remaining glucose residues on the N-glycans by knocking out MOGS affected optimum GPI-inositol deacylation. In the ER, UGGT can transfer a glucose residue to the Man9GlcNAc2 structure, even with ALG6 knocked out. Newly synthesized glycoproteins cannot enter the calnexin cycle in MOGS-KO cells, whereas further mutations in ALG6 or ALG8 can bypass the effects of MOGS deletion, indicating that defects in entering the calnexin cycle caused inefficient GPI-inositol deacylation of GPI-APs.
Because calnexin possesses a lectin domain, we used two calnexin mutant constructs (Y164A or E216A in human calnexin), each defective in lectin activity, to analyze whether the glycan binding ability is required for the phenotype (Schrag et al., 2001). Compared with WT calnexin, PIPLC sensitivity of GPI-APs was not rescued or only slightly rescued when calnexin mutant constructs were expressed (Figs. 4, D and E), suggesting that lectin activity of calnexin was required for PIPLC sensitivity. Calreticulin is a homologue of calnexin in mammalian cells. Calnexin is a type I transmembrane protein, and calreticulin is a soluble protein (Helenius and Aebi, 2004). To examine selectivity of the PIPLC-sensitive phenotype, we generated cells with calreticulin (encoded by CALR) knocked out. CALR-KO did not affect the PIPLC sensitivity of GPI-APs, showing the selectivity of calnexin (Figs. 4, F and G). Because the PIPLC resistance of GPI-APs in CANX-KO cells was weaker than in MOGS- or GANAB-KO cells, we disrupted CALR in CANX-KO cells and analyzed the phenotype. The KO of CALR in CANX-KO cells increased PIPLC resistance of GPI-APs (Figs. 4, F and G). These results indicate that calnexin is the primary contributor to GPI-inositol deacylation, but they also indicate that calreticulin can partly compensate for this function when calnexin is absent. Because the protein parts of GPI-APs are located at the juxtamembrane region and GPI-inositol deacylation occurs on the membrane, a transmembrane form of calnexin would account for this regulation.
Chronic ER stress caused exposure of inositol-acylated GPI-APs
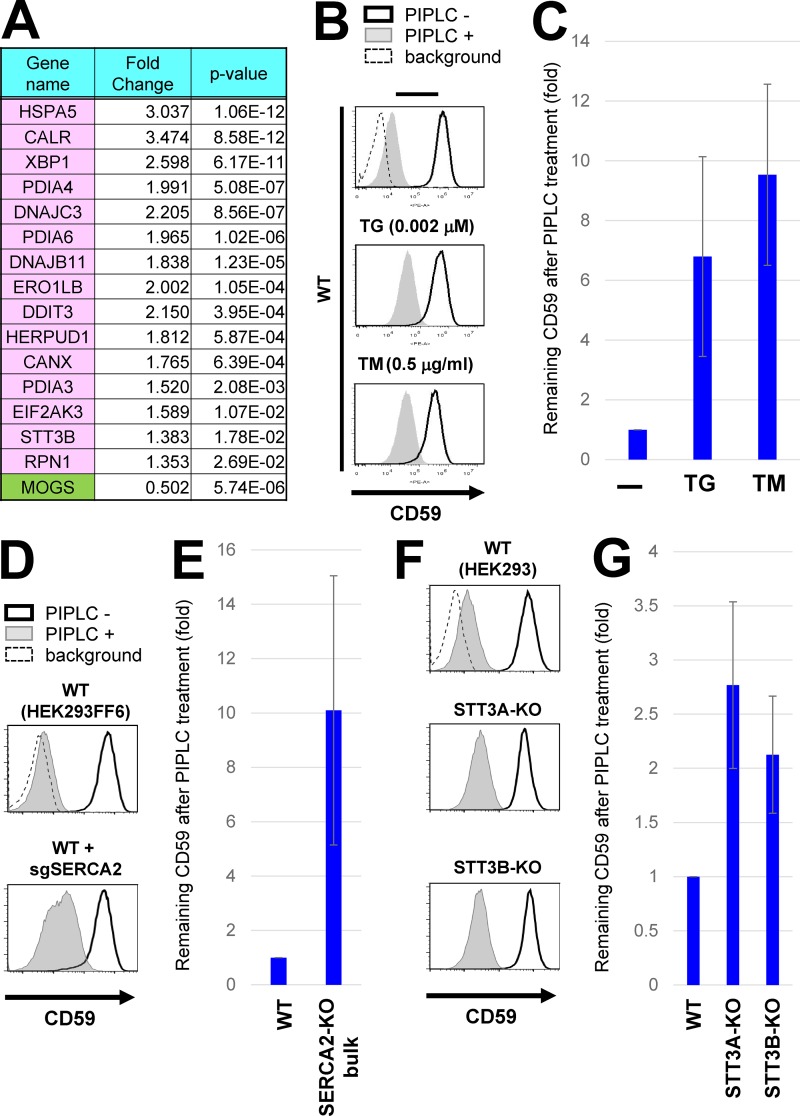
To understand the mechanisms involving N-glycan processing and calnexin in efficient GPI-inositol deacylation, we analyzed transcriptional changes in MOGS-KO cells compared with parental HEK293 cells using RNA sequencing analyses. In MOGS-KO cells, many genes involved in the unfolded protein response (UPR) were significantly up-regulated (Fig. S4 A and Table S4). Particularly, 15 genes such as HSPA5 (encoding BiP/GRP78), CALR (calreticulin) and XBP1, which are assigned in “protein processing in ER” in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, were up-regulated in MOGS-KO cells (Figs. 5 A and S4, B and C), suggesting that ER stress occurred under steady-state conditions in the MOGS-KO cells.
Figure 5.
Chronic ER stress induced exposure of inositol-acylated GPI-APs on the cell surface. (A) Gene expression was analyzed in WT and MOGS-KO cells using RNA-seq analyses. Among genes altered in MOGS-KO cells compared with WT cells, genes categorized in the “Protein processing in endoplasmic reticulum” in the KEGG pathway were shown in pink (up-regulation) or green (down-regulation; see Fig. S4). The MOGS gene was significantly down-regulated in the MOGS-KO cells, probably as a result of nonsense-mediated mRNA decay. (B) WT HEK293FF6 cells were treated with or without the ER stress inducer TG (0.002 µM; middle) or TM (0.5 µg/ml; bottom) for 10 d. PIPLC-treated or -untreated cells were stained with anti-CD59 antibodies. Surface CD59 was detected by flow cytometry as described in Fig. 2 A. (C) CD59 levels remaining after PIPLC treatment are plotted. (D and E) WT HEK293FF6 cells were transfected with plasmids expressing sgSERCA2 sequences to knock out the gene. Plasmid-positive cells were sorted and cultured for 10 d after transfection. WT HEK293FF6 cells and cells with bulk KO of SERCA2 were treated with or without PIPLC. Surface CD59 was analyzed by flow cytometry (D). CD59 remaining after PIPLC treatment in SERCA2-KO cells is plotted in E. The value of remaining CD59 in WT cells was set as 1. (F and G) WT HEK293, STT3A-KO, and STT3B-KO cells were treated with or without PIPLC. Surface CD59 was analyzed by flow cytometry (F). CD59 remaining after PIPLC treatment is plotted in G. In all graphs, the remaining CD59 value in untreated cells was set as 1. Relative values were calculated and are shown as means ± SD from three independent experiments.
We hypothesized that chronic ER stress would lead to bypassing of GPI-inositol deacylation. To test this, we treated cells with reagents causing ER stress, thapsigargin (TG), and tunicamycin (TM). When treated with high concentrations (TG > 0.005 µM or TM > 1 µg/ml), HEK293 cells died during culture, probably through apoptosis induced by ER stress. Instead, HEK293 cells were then continuously cultured with low doses of TG and TM. After 10 d culture in the presence and absence of TG or TM, the PIPLC sensitivity of GPI-APs was analyzed (Fig. 5, B and C). Compared with untreated cells, those treated with TG or TM had GPI-APs with PIPLC resistance.
To confirm the phenomena caused by ER stress inducers, we knocked out the ATP2A2/SERCA2, STT3A, or STT3B genes. SERCA2, a sarco/endoplasmic reticulum Ca2+-ATPase, is a target of TG (Thastrup et al., 1990). We could not obtain clonal SERCA2-KO cell lines because the SERCA2 gene is believed to be essential for the survival of mammalian cells (Blomen et al., 2015). Instead, we analyzed the bulk population of cells after transfection with KO constructs for SERCA2. The SERCA2 gene was knocked out in a fraction of the cell population (Fig. S3 C). Under these conditions, PIPLC resistance was increased, consistent with the effects of TG treatment (Fig. 5, D and E). STT3A and STT3B are the catalytic subunits of the OST. STT3A and STT3B mainly function in cotranslational and posttranslocational protein N-glycosylation, respectively (Cherepanova et al., 2016). Glycosylation of the respective target proteins was impaired in KO cells (Fig. S3, D and E). The PIPLC resistance of CD59 was slightly increased in both STT3A-KO and STT3B-KO cells (Fig. 5, F and G). Because STT3s transfer N-glycan to CD59, we checked the glycosylation of CD59. Although the glycosylation of CD59 was not severely affected in those cells (Fig. S3 F), it would be still possible that the mild resistance to PIPLC might be caused by the glycosylation defects in CD59, not the general ER stress. These results indicate that chronic ER stress causes cell surface exposure of inositol-acylated GPI-APs.
Calnexin interacted with both GPI-APs and PGAP1 and facilitated GPI-inositol deacylation
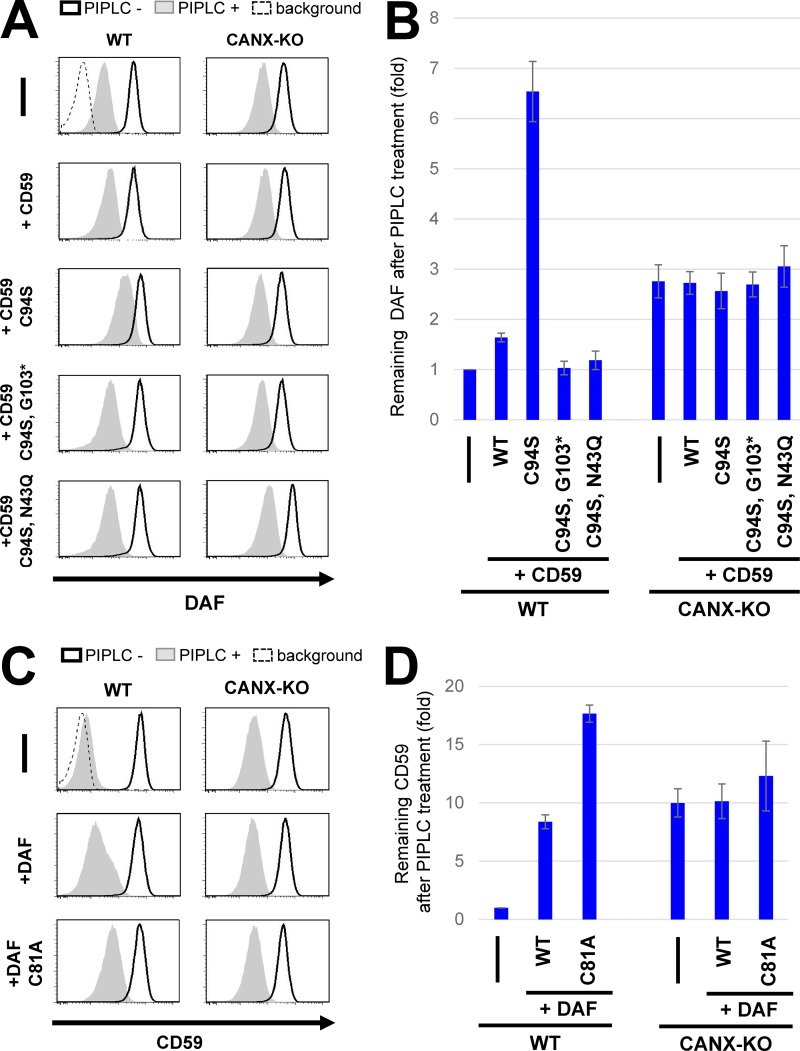
Under ER stress conditions, misfolded proteins including misfolded GPI-APs would accumulate in the ER. We expected that accumulation of misfolded proteins would inhibit the processing of normal GPI-APs. To investigate this possibility, we expressed misfolded GPI-APs in cells. As misfolded GPI-APs, EGFP-Flag-tagged (EGFP-F-) CD59 (C94S) and DAF (C81A) were used in which a cysteine residue was replaced with serine or alanine to prevent disulfide bond formation (Satpute-Krishnan et al., 2014). When the misfolded CD59 or DAF was expressed, genes induced under the UPR, such as GRP78 and GRP94, were up-regulated, suggesting that the misfolded proteins caused ER stress (Fig. S5 A). However, PGAP1 expression was not changed under these conditions. These mutant proteins were accumulated in the ER (Fig. S5, B and C). In cells expressing misfolded CD59, PIPLC resistance of endogenous DAF was increased (Fig. 6, A and B). Similarly, PIPLC resistance of endogenous CD59 was increased by expression of misfolded DAF (Fig. 6, C and D). To determine specificity, misfolded CD59 lacking GPI (C94S and G103stop*) or lacking an N-glycan (C94S and N43Q) were expressed, and the PIPLC sensitivity of endogenous DAF was analyzed. In contrast with cells expressing misfolded GPI-AP, increased PIPLC resistance was not observed in cells expressing other types of misfolded proteins (Fig. 6, A and B). To further confirm the specificity, we expressed a transmembrane type of misfolded CD59, CD59TM, whose GPI-attachment signal was replaced with a CD46 transmembrane domain (Hong et al., 2002) and a misfolded CD4, a type-I transmembrane protein (Fig. S5 D). We also expressed misfolded LY6D, a GPI-AP that possesses a Ly6 domain similar to CD59 but does not have N-glycosylation site. The expression of those WT and misfolded proteins did not change the PIPLC sensitivity of DAF (Fig. S5 E), suggesting that PIPLC sensitivity of normal GPI-APs was dependent on GPI and the N-glycans on misfolded GPI-APs. These results indicate that GPI-inositol deacylation of normal GPI-APs was affected by expression of misfolded GPI-APs. In CANX-KO cells, the GPI-APs were mildly resistant to PIPLC (Fig. 4). Interestingly, expression of misfolded GPI-APs in CANX-KO cells did not cause further PIPLC resistance of normal GPI-APs. However, it did in WT cells (Fig. 6, B and D), preventing the phenotype caused by CANX disruption. The results suggest that calnexin was responsible for the PIPLC-resistant phenotype caused by expression of misfolded GPI-APs.
Figure 6.
Overexpression of misfolded GPI-APs decreased deacylation efficiency of endogenous GPI-APs in WT but not CANX-KO cells. (A) WT HEK293FF6 or CANX-KO cells stably expressing WT EGFP-F–CD59, mutant CD59 (C94S and misfolded CD59), misfolded CD59 lacking GPI (C94S and G103stop*), or misfolded CD59 lacking an N-glycan (C94S and N43Q) were treated with or without PIPLC. After treatments, cells were stained for endogenous DAF and analyzed by flow cytometry. (B) DAF levels remaining after PIPLC treatment were plotted. The value for remaining DAF in WT cells without any exogenous expression was set to 1. (C) WT HEK293FF6 or CANX-KO cells stably expressing WT EGFP-F–DAF or mutant DAF (C81A, misfolded DAF) were treated with or without PIPLC. After treatment, cells were stained for endogenous CD59 and analyzed by flow cytometry. (D) CD59 levels remaining after PIPLC treatment are shown. The remaining CD59 value in WT cells without any exogenous expression was set to 1. Relative values were calculated and are represented as means ± SD from three independent experiments.
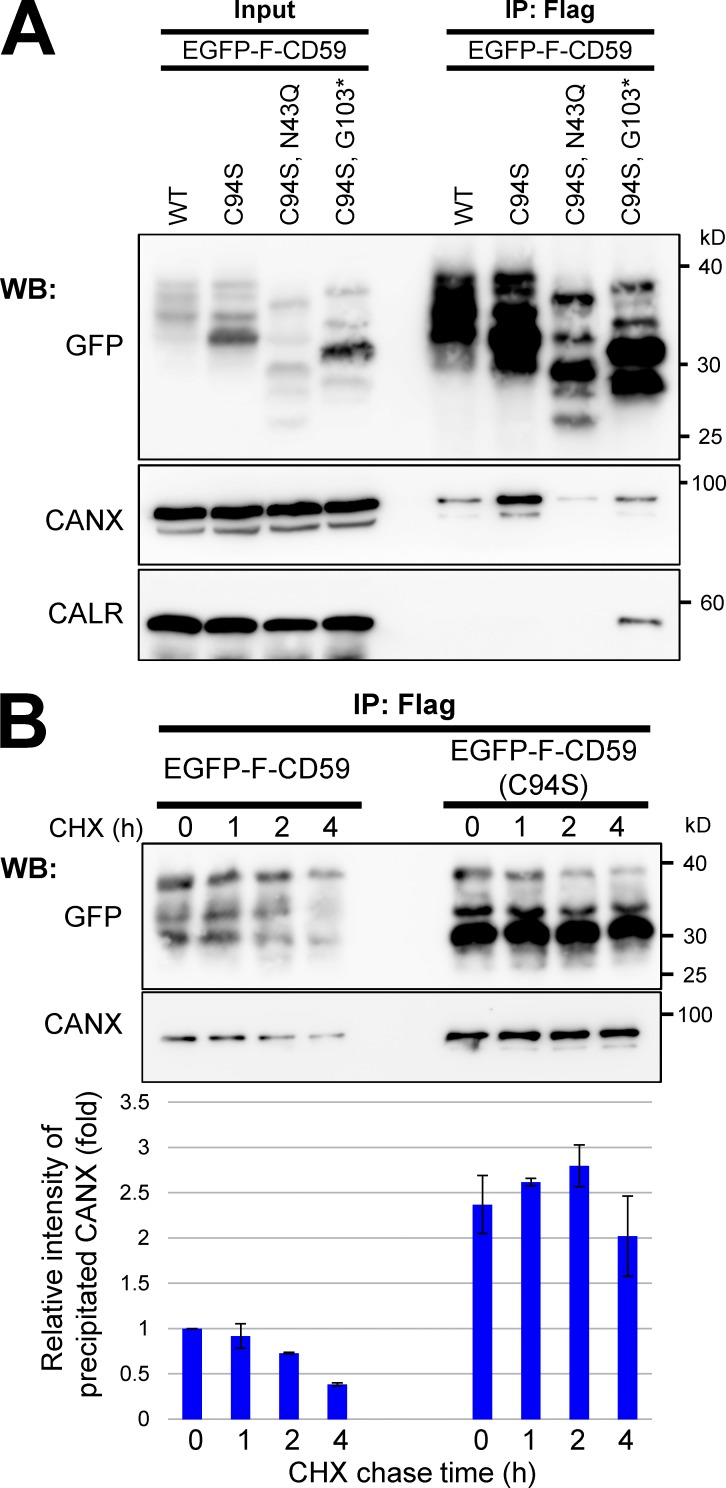
Misfolded GPI-APs inhibit efficient processing of normal GPI-APs. The inhibition was cancelled in CANX-KO cells. We next analyzed interactions between calnexin and GPI-APs. As GPI-APs, EGFP-F–CD59 and the mutant forms were used (Fig. S5, F and G). Calnexin, but not calreticulin, was associated with CD59. This interaction was greater with misfolded CD59 (Fig. 7 A). However, the binding disappeared or was weaker when CD59 lacked N-glycan or GPI. In this case, soluble misfolded CD59 (C94S; G103stop) was associated with calreticulin (Fig. 7 A). These results indicate that the interaction between calnexin and CD59 was dependent on N-glycan and GPI. This interaction was correlated with increased PIPLC resistance by expression of misfolded proteins, as shown in Fig. 6 B. When calnexin strongly interacted with misfolded GPI-APs, the efficiency of inositol-deacylation for normal GPI-APs was lower. These findings suggest that calnexin facilitates the inositol deacylation of GPI-APs by PGAP1, which is inhibited by misfolded GPI-APs. Calnexin preferentially interacted with misfolded versus normal GPI-APs. Such a stable interaction with misfolded GPI-APs would inhibit efficient processing of normal GPI-APs by PGAP1. To determine stability of the interaction between CD59 and calnexin, chase experiments were performed. After protein synthesis was inhibited by cycloheximide (CHX), WT CD59 started dissociation from calnexin in a time-dependent manner (Fig. 7 B). In contrast, misfolded CD59 was more strongly interacted with calnexin and maintained the interaction until at least 4 h after chase.
Figure 7.
Calnexin interacted with GPI-APs dependent on its N-glycan and GPI. (A) Cells were transiently transfected with WT EGFP-F–CD59, CD59 mutants (C94S and misfolded CD59), misfolded CD59 lacking GPI (C94S and G103stop*), or misfolded CD59 lacking an N-glycan (C94S and N43Q), and then they were lysed with lysis buffer containing 1% NP-40. The lysates were subjected to immunoprecipitation (IP) with anti-Flag beads. The precipitated proteins were released from the beads using Flag peptides. The input (10% of total protein) and immunoprecipitated fractions were analyzed by immunoblotting with the indicated antibodies. (B) Cells stably expressing EGFP-F–CD59 or –CD59 (C94S) were cultured. At the indicated time after protein synthesis was stopped by addition of CHX, EGFP-F–CD59 was precipitated, and coprecipitated calnexin was detected. The coprecipitated calnexin with WT CD59 at time 0 was set to 1. Relative values were calculated and are represented as means ± SEM from three independent experiments. WB, Western blot.
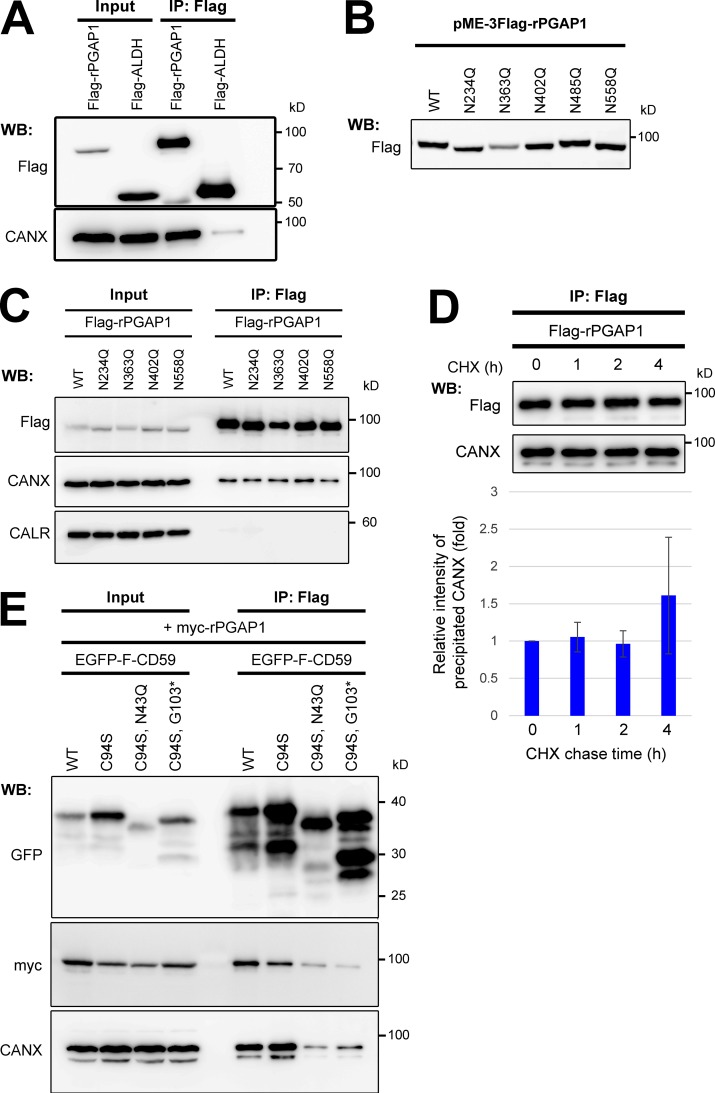
We next addressed the interaction between calnexin and PGAP1. PGAP1 but not another ER-resident protein, ALDH, coprecipitated with a fraction of calnexin (Fig. 8 A). Because PGAP1 is an N-glycosylated protein localized in the ER, the relationship between N-glycans on PGAP1 and calnexin was analyzed. Rat and human PGAP1s have five and four potential N-glycosylation sites, respectively. Each asparagine (N) residue on potential glycosylation sites of rat PGAP1 was replaced with glutamine (Q), and the size of the mutant PGAP1 protein was then determined. The protein was smaller in all N-glycosylation mutants except one (N485Q; Fig. 8 B). This result was not unexpected because the N-glycosylation site on N485 is not conserved in human PGAP1. Interaction between PGAP1 and calnexin was not changed in the N-glycan mutants compared with in the WT (Fig. 8 C), suggesting that at least calnexin did not interact with specific N-glycans on PGAP1. Because calnexin transiently interacts with newly synthesized glycoproteins for their folding, we determined whether the interaction with PGAP1 is temporal or not. A CHX chase experiment was performed under the same conditions as in Fig. 7 B. The interaction was stable until hour 4 of chase (Fig. 8 D), suggesting that a fraction of calnexin stably associated with PGAP1. Finally, we detected an interaction between GPI-APs and PGAP1 together with calnexin. Either WT CD59 or misfolded GPI-anchored CD59 interacted with both PGAP1 and calnexin, whereas misfolded CD59, lacking an N-glycan or GPI, did not (Fig. 8 E). These results indicated that GPI-APs, calnexin, and PGAP1 interacted with one another. It is still not certain whether they could make a ternary complex or interact individually. However, stable association of calnexin with PGAP1 would support efficient interaction of GPI-APs with PGAP1.
Figure 8.
PGAP1 associated with calnexin and GPI-APs. (A) Cells were transfected with plasmids expressing Flag-tagged rat PGAP1 (Flag-rPGAP1) or ALDH (Flag-ALDH) and then lysed with lysis buffer containing 1% digitonin and subjected to immunoprecipitation with anti-Flag beads. The input (10% of total protein) and immunoprecipitation (IP) fractions were analyzed by Western blotting (WB). (B) Asparagine (N) residues on potential N-glycosylation sites of rat PGAP1 were each replaced with glutamine (Q). Cells were transfected with plasmids expressing the mutant PGAP1, and Flag-rPGAP1 was then detected by immunoblotting. (C) Cells transfected with plasmids expressing Flag-rPGAP1 or N-glycan–deficient Flag-rPGAP1 (N234Q, N363Q, N402Q, or N558Q) were lysed with lysis buffer containing 1% digitonin. Flag-rPGAP1 was precipitated with anti-Flag beads. The input (10% of total protein) and immunoprecipitated fractions were analyzed by Western blotting. (D) Cells stably expressing Flag-rPGAP1 were cultured. At the indicated time after protein synthesis was stopped by addition of CHX, Flag-rPGAP1 was precipitated, and coprecipitated calnexin was detected. The coprecipitated calnexin with Flag-rPGAP1 at time 0 was set to 1. Relative values were calculated and are represented as means ± SEM from three independent experiments. (E) WT HEK293FF6 cells stably expressing WT EGFP-F–CD59 or CD59 mutants (C94S, C94S/N43Q, or C94S/G103stop*) were transfected with plasmids expressing myc-tagged rat PGAP1 (myc-rPGAP1). Cells were lysed in lysis buffer containing 1% digitonin and subjected to immunoprecipitation. The input (10% of total protein) and immunoprecipitation fractions were analyzed by Western blotting.
Discussion
N-Glycosylation and GPI anchoring of proteins occur in the ER. After being transferred to proteins, N-glycans and GPI anchors both undergo a series of processing steps before the proteins exit from the ER. N-Glycan processing is critical for protein folding and quality control (Tannous et al., 2015). GPI anchor remodeling is involved in efficient sorting of the modified proteins into transport vesicles (Fujita and Kinoshita, 2012; Muñiz and Riezman, 2016). In this study, we clarified the linkage between N-glycans and GPI to ensure folding and transport of GPI-APs. We identified seven genes involved in efficient GPI-inositol deacylation using mammalian haploid genetics. Among these were genes encoding α-glucosidases and calnexin. Glycoproteins cannot enter the calnexin cycle if these genes are defective. KO of these genes rendered GPI-APs partially resistant to PIPLC, and this phenotype was rescued by PGAP1 overexpression, suggesting inefficient GPI-inositol deacylation in these KO cells. These results indicate that N-glycans participate in quality control and processing of GPI-APs to ensure the correct folding in and exiting from the ER. Through this mechanism, properly folded and processed GPI-APs are sorted and transported from the ER in cells under physiological conditions. Once the system is disrupted by ER stress, unprocessed GPI-APs are exposed on the cell surface.
Calnexin would play two functional roles in the maturation of GPI-APs (Fig. 9). First, it binds with N-glycans on GPI-APs and promotes the protein folding. Second, calnexin retains GPI-APs in the ER and assists efficient inositol-deacylation of GPI-APs by PGAP1 for correct processing of the GPI moieties. The temporal trap of GPI-APs in the ER by calnexin provides time for PGAP1 to interact with substrate GPI-APs. Actually, there is a positive correlation between the ER-residence time and PIPLC sensitivity of GPI-APs. The ER-residence time of CD59 is much longer than that of DAF (Tanaka et al., 2004; Fujita et al., 2011). In this study, we showed the PIPLC sensitivity of CD59 (99.6%) was higher than that of DAF (96.5%) in WT HEK293 cells (Figs. 2 B and S2 A). In addition to the temporal ER retention of GPI-APs, interaction between calnexin and PGAP1 facilitates the efficient GPI-inositol deacylation through the substrate accessibility. So far, there is no evidence that PGAP1 itself recognizes the protein portion that is anchored by GPI. It is possible that the protein is recognized by calnexin, which associates with PGAP1. In the UniProt database, 138 human proteins are registered as GPI-APs. We added 10 proteins that are manually annotated as GPI-APs from the literature (Table S5). Of a total of 148 GPI-APs, ∼94.6% (140/148) have at least one potential N-glycosylation site, suggesting that most GPI-APs could use this monitoring system. Once GPI-inositol deacylation occurs, transport of GPI-APs from the ER is favored. Therefore, calnexin and PGAP1 act as gatekeepers, converting GPI-APs from a protein-folding phase in the ER to a transport phase. That is, there are multiple confirmatory steps during GPI-AP maturation in the ER by calnexin and PGAP1, to monitor protein folding and GPI processing.
Figure 9.
Summary models for folding and processing of GPI-APs regulated by calnexin and ER homeostasis. Top: under normal conditions, N-glycans and GPI are transferred to newly synthesized GPI-APs. The glucose residues on N-glycans are trimmed by α-glucosidases I and II, resulting in a Glc1Man9GlcNAc2 structure, which is recognized by calnexin. Calnexin assists protein folding and facilitates GPI-inositol deacylation by PGAP1 through the temporal ER retention of GPI-APs. Calnexin also interacts with PGAP1 to increase the efficiency of the GPI-inositol deacylation. Once the protein portion of each GPI-AP is folded, calnexin dissociates from the GPI-AP, which is further remodeled by PGAP5 and efficiently incorporated into transport vesicles by a signal-mediated pathway involving p24 proteins. Middle: in CANX-KO cells, GPI-APs cannot retain in the ER and decrease the efficient interaction with PGAP1, resulting in transport of some GPI-APs without processing. Calnexin-independent GPI-inositol deacylation also exists. Bottom: under chronic ER stress conditions, misfolded GPI-APs accumulate in the ER and occupy the calnexin and PGAP1. Availability of calnexin and PGAP1 is therefore decreased, and normal GPI-APs without processing leak into transport vesicles through a bulk flow pathway, resulting in expression of inositol-acylated GPI-APs on the cell surface.
In contrast, GPI-APs cannot stay in the ER until they are correctly processed by mutations in calnexin (Fig. 9, middle). As a result, some inositol-acylated GPI-APs would be transported from the ER through a bulk flow pathway. Because many GPI-APs are still processed, there are mechanisms involving a calnexin-independent pathway for GPI-inositol deacylation. Under ER stress conditions, misfolded GPI-APs accumulate in the ER and deplete the available calnexin and/or PGAP1 (Fig. 9, bottom). This situation impairs ER retention and access to PGAP1 of normal GPI-APs, resulting in the exit of normal GPI-APs from the ER without processing by PGAP1 through a bulk flow pathway.
In yeast, misfolded GPI-APs are degraded in both proteasomes and vacuoles (Hirayama et al., 2008; Sikorska et al., 2016). The PGAP1 homologue Bst1p and PGAP5 homologue Ted1p were required for efficient degradation of a misfolded GPI-AP (Fujita et al., 2006; Sikorska et al., 2016). It was recently reported that mammalian cells have a degradation pathway for misfolded GPI-APs under acute ER stress conditions, a process known as the rapid ER stress-induced export (RESET) pathway (Satpute-Krishnan et al., 2014). Under normal conditions, misfolded GPI-APs bind to calnexin and are retained in the ER. Under acute ER stress, misfolded GPI-APs are dissociated from calnexin, transported from the ER to the plasma membrane, and degraded in lysosomes. Our results also supported the model that misfolded GPI-APs are recognized by calnexin, which is dependent on N-glycan and GPI, for retention in the ER. Whereas the RESET pathway is induced by acute ER stress for transport of misfolded GPI-APs, we clarified the behavior of normally folded GPI-APs under physiological and chronic ER stress conditions. Under physiological conditions, GPI-APs are correctly processed and expressed on the cell surface, whereas levels of inositol-acylated GPI-APs are increased under chronic ER stress conditions.
In the haploid genetic screens, we identified four genes (SELT, CLPTM1, SEC63, and C18orf32) other than MOGS, GANAB, and CANX. Their KO caused partial resistance of GPI-APs against PIPLC, suggesting that they were required for efficient GPI-inositol deacylation. SELT (or SELENOT) contains selenocysteine and has a thioredoxin fold (Dikiy et al., 2007). It is localized in the ER and involved in Ca2+ homeostasis (Grumolato et al., 2008). In mice, Selt plays important functions in insulin production or secretion in pancreatic β cells (Prevost et al., 2013) and is required to protect dopaminergic neurons against oxidative stress in brain (Boukhzar et al., 2016). It was reported recently that SELT acts as a component of the STT3A–OST complex (Hamieh et al., 2017), also supporting our proposal that N-glycans on GPI-APs contribute to efficient inositol deacylation. SEC63 is one of the components of the ER translocon complex, which works on translocation of selective substrates such as prion protein and ERj3 (Lang et al., 2012). CLPTM1 was identified as one of the candidate genes whose mutations can cause cleft lip and palate (Yoshiura et al., 1998). Among the clinical symptoms in some inherited GPI deficiencies are characteristic facial features, including a tented upper lip and cleft palate (Horn et al., 2011). Although the function of CLPTM1 is unknown, it associates with UBAC2 in an ER-associated degradation pathway (Christianson et al., 2011). C18orf32 is a small protein with 76 amino acids, and its molecular function has not been reported. We found that both CLPTM1 and C18orf32 proteins were localized in the ER (unpublished data), indicating that all seven factors identified in the genetic screen have functions in the ER. In their respective gene KO cells, PGAP1 expression and localization were not changed. It is unclear how these candidate genes are involved in efficient GPI-inositol deacylation. The issue will be addressed in future studies. Our screening results indicate that the structure and processing of GPI-APs are monitored and regulated by many factors.
Materials and methods
Cell lines, antibodies, and reagents
HAP1 cells were a gift from T.R. Brummelkamp (Netherlands Cancer Institute, Amsterdam, Netherlands; Carette et al., 2011). They were cultured in Iscove’s modified Dulbecco’s medium containing 10% FCS with antibiotics needed for selection. HEK293 and its derivative HEK293FF6 cells (Hirata et al., 2015) were cultured in DMEM containing 10% FCS.
Mouse monoclonal anti-CD59 (clone 5H8; Maeda et al., 2007), anti-DAF (clone IA10; Maeda et al., 2007), anti-Flag (F3165; M2; Sigma-Aldrich), anti-GFP (HT801-01; TransGen Biotech), anti-HA (H3663; HA-7; Sigma-Aldrich), anti-ERp57 (K0135-3; MBL), rabbit monoclonal antisyntaxin 6 (2869S; Cell Signaling Technology), anticalreticulin (12238S; Cell Signaling Technology), anti-myc (AM933; Beyotime), and polyclonal anticalnexin (C4731; Sigma-Aldrich) were used as primary antibodies. Phycoerythrin (PE)-conjugated goat anti–mouse IgG (12-4010-87; Thermo Fisher Scientific), HRP-conjugated anti–mouse IgG (HS211-01; TransGen Biotech), and anti–rabbit IgG (HS101-01; TransGen Biotech) were used as the secondary antibodies. DNJ (Cayman Chemical), DMJ (Cayman Chemical), KIF (Cayman Chemical), TG (Sigma-Aldrich), dithiothreitol (Sinopharm), and TM (Sigma-Aldrich) were used for drug treatments.
Plasmids
For the CRISPR-Cas9 system to knock out target genes, guide RNA sequences were designed using the E-CRISP website (Heigwer et al., 2014), and the resulting DNA fragments were ligated into the BbsI-digested pX330-EGFP (Hirata et al., 2015). All oligonucleotides used for gene KO in this study are listed in Table S1.
The fragments of rat PGAP1 and ALDH were digested with SalI and NotI from the mammalian expression plasmid pME-Py-GST-rPGAP1 (Tanaka et al., 2004) or pME-Flag-ALDH and were cloned into a pME-Hyg-3FLAG plasmid to generate pME-Hyg-3FLAG-rPGAP1 and pME-Hyg-3FLAG-ALDH. The N-glycan mutants of pME-Hyg-3FLAG-rPGAP1 were constructed by site-directed mutagenesis with primers (Table S2). The digested fragments of rat PGAP1 were also ligated to the plasmid pME-18Sf-N6myc to obtain pME-18Sf-N6myc-rPGAP1. Human PGAP1 was amplified, digested with SalI and NotI, and ligated to the plasmid pLIB2-Hyg to obtain pLIB2-Hyg-hPGAP1. To construct plasmids expressing candidate genes, DNA fragments corresponding to CANX, MOGS, GANAB, CLPTM1, and SEC63 were amplified from the human cDNAs with primers (Table S2) and cloned to the retroviral vector pLIB2-BSD, generating pLIB2-BSD-CANX, pLIB2-BSD-MOGS, pLIB2-BSD-GANAB, pLIB2-BSD-CLPTM1, and pLIB2-BSD-SEC63, respectively. Human SELT and C18orf32 were amplified from cDNA and ligated into pME-Hyg, generating pME-Hyg-SELT and pME-Hyg-C18orf32, respectively. The lectin mutant plasmids pLIB2-BSD-CANX (Y164A) and pLIB2-BSD-CANX (E216A) were constructed using site-directed mutagenesis with the primers (Table S2). The pPB–flippase recognition target (FRT)-PGKp-BSD plasmid, which contains a PGK promoter, a multiple cloning site, a bovine growth hormone polyadenylation signal, an SV40 promoter, and a blasticidin-resistant gene flanked by both piggyBac (PB) terminal repeat sequences and FRT at both ends was constructed by in-fusion cloning. To construct plasmids expressing misfolded CD59 and DAF, fragments containing corresponding mutations were amplified by PCR using primers (Table S2). Obtained fragments were introduced into pME-NeodH-mEGFP-Flag-CD59 plasmid (Hirata et al., 2013) cut by HindIII and NotI using in-fusion cloning, generating pME-NeodH-mEGFP-Flag-CD59 (C94S) and pME-NeodH-mEGFP-Flag-DAF (C81A). The DNA fragment of WT DAF was also amplified and cloned as described above, generating pME-NeodH-mEGFP-Flag-DAF. The DNA fragments coding mEGFP-Flag-CD59 and mEGFP-Flag-DAF were digested with EcoRI and NotI and cloned into pPB-FRT-PGKp-BSD to generate the pPB-FRT-PGKp-BSD-mEGFP-Flag-CD59 and pPB-FRT-PGKp-BSD-mEGFP-Flag-DAF plasmids, respectively. Constructs for misfolded CD59 and DAF were constructed from these plasmids using site-directed mutagenesis. For localization, the DNA fragment containing the GFP-KDEL sequence was amplified from plasmid pME-NeodH-mEGFP-Flag-CD59 and cloned to pME-Hyg to make the pME-Hyg-ssEGFP-KDEL. The pME-Zeo-mRFP-KDEL was previously described (Fujita et al., 2009). A DNA fragment of prosaposin was amplified and ligated into pME-3×Flag, generating pME-pSAP-3×Flag. The pcDNA3-CatCd234-HA was provided by R. Gilmore (University of Massachusetts Medical School, Worcester, MA).
Establishment of KO cell lines
To generate gene KO cell lines, HEK293 or HEK293FF6 cells were transiently transfected with pX330-EGFP plasmids bearing the target sequences. After 3 d, cells with EGFP were sorted with a cell sorter S3 (Bio-Rad Laboratories). The collected cells were cultured for ≥7 d and subjected to limiting dilution to obtain clonal KO cells. A clone with no WT allele was selected. DNA sequences were analyzed by the sequencing. The DNA sequence of each KO cell line is listed in Fig. S1.
Enrichment of the PIPLC-resistant HAP1 mutant population
A gene-trap virus was produced from the Plat-GP Retroviral Packaging Cell Line cultured in 10 15-cm dishes by transfecting with a mixture of pCMT-SApA-BSD and pLC-VSVG plasmids. The virus-containing supernatant was concentrated five times using a PEG-it solution (System Biosciences) and then mixed with 8 µg polybrene/ml. HAP1 cells (7 × 107 cells) were seeded into six-well plates containing 1.5 × 106 cells per well and infected. 2 d after infection, the cells were selected with 6 µg blasticidin/ml for 1 wk. HAP1 cells mutagenized with gene-trap vectors were harvested, treated with PIPLC (Thermo Fisher Scientific), and then stained with anti-CD59 antibody and goat anti–mouse IgG microbeads (130-048-401; Miltenyi Biotec). Cells with CD59 remaining on the cell surface were enriched three times by magnetic-activated cell sorting using a mass spectrometry column (Miltenyi Biotec).
Sequence analysis of gene-trap insertion sites
Gene-trap insertion sites were analyzed as described previously (Rong et al., 2015). Genomic DNA was isolated from 3 × 107 cells, either nonselected control or PIPLC-resistant cells, using the Wizard Genomic DNA purification kit (Promega) according to the manufacturer’s protocol. Genomic DNA (15 µg) was digested with HaeIII, followed by ligation with the splinkerette adapter. After ligation, the DNA fragments were digested with PvuII, which cleaves the vector sequence between the 3′LTR and the upstream HaeIII site, to prevent unwanted vector amplification. After column purification, the fragments were used as templates for splinkerette PCR using the SPl-P1 primer (5′-CGAATCGTAACCGTTCGTACGAGAA-3′) and the first LTR primer (5′-AGTGTATGTAAACTTCTGACCCACTGG-3′). The resulting DNA fragments were further amplified by nested PCRs using the SPl-P2 primer (5′-TCGTACGAGAATCGCTGTCCTCTCC-3′), the second LTR primer (5′-CTTGTGTCATGCACAAAGTAGATGTCC-3′), Rd1Tru-LTR (5′-acactctttccctacacgacgctcttccgatctGCTAGCTTGCCAAACCTACAGGTGGG-3′) and Rd2Tru-Splink (5′-gtgactggagttcagacgtgtgctcttccgatctGCTGTCCTCTCCAACGAGCCAAGG-3′). The lowercased sequences indicate the Illumina sequencing primers. Illumina P5 (5′-AATGATACGGCGACCACCG-3′) and P7 (5′-CAAGCAGAAGACGGCATACGA-3′) adapters and barcode sequences were attached to the products by six cycles of PCR with 10 ng of each initial PCR product as the template. Single-end sequencing (151-bp) was performed on the HiSeq (Illumina) system. The numbers of reads obtained from nonselected and PIPLC-resistant cells were ∼15 and 3.0 million, respectively.
Analysis of gene-trap insertions
FASTQ data files were analyzed using CLC Genomic Workbench software (version 7.0.4; QIAGEN) using a previously described method with some modifications (Rong et al., 2015). After quality trimming and removal of the common LTR sequence, the 50-bp reads were mapped onto the human genome (hg19). To exclude ambiguous alignments, mismatch reads were not allowed, and all nonspecific matched reads were ignored. To eliminate PCR amplification bias and determine the unique insertion sites, duplicate reads were removed and counted as one read (a unique insertion site). The total insertions were counted. In the analysis, ∼56,859 and 23,594 independent insertions were determined from nonselected control and PIPLC-resistant cells, respectively. The amount of enrichment of a particular gene in the screen was calculated by comparing the selected with the nonselected population. For each gene, a p-value and a p-value corrected for the false discovery rate (FDR) were calculated by the one-sided Fisher exact test using R software.
PIPLC treatment and flow cytometry
Cells (106) were harvested with trypsin/EDTA and treated with PIPLC. Cells were incubated with 5 U/ml PIPLC dissolved in PBS supplemented with 0.5% BSA, 5 mM EDTA, and 10 mM Hepes plus DMEM without FCS for 1.5 h at 37°C. After washing with PBS, cells were stained with anti-CD59 or anti-DAF (10 µg/ml) as the primary antibody for 25 min on ice. After incubation, cells were washed twice with FACS buffer (PBS containing 1% BSA and 0.1% NaN3). Phycoerythrin-conjugated goat anti–mouse IgG was incubated with secondary antibody for 25 min on ice. After staining, cells were washed two times with FACS buffer and analyzed using a Canto-II or Accuri C6 (BD). The resulting data were analyzed with an Accuri C6 and FlowJo software (BD). The amount of CD59 remaining was calculated from mean fluorescence intensity values for GPI-APs treated with or without PIPLC.
Immunofluorescence
To analyze PGAP1 localization, HEK293FF6 and gene-KO cells, which stably expressed Flag-tagged PGAP1, were transiently transfected with pME-ssEGFP-KDEL. After transfection, the cells were plated on glass coverslips, pretreated with 1% gelatin, and cultured for another 2 d. Cells were washed with PBS, fixed in 4% paraformaldehyde, washed with PBS, and incubated with 45 mM ammonium chloride. Then cells were permeabilized and blocked with blocking buffer A (1% BSA, 0.1% NaN3, and 0.1% Triton X-100 in PBS) for 1 h. Cells were immunostained with an anti-Flag antibody in blocking buffer B (2.5% goat serum, 0.05% NaN3, and 0.1% Triton X-100 in PBS) for 1 h at room temperature and washed three times with blocking buffer A followed by incubation with a secondary mouse Alexa Fluor 555 antibody (A31570; Thermo Fisher Scientific) for 1 h. Finally, the coverslips were mounted onto slides using mounting solution containing DAPI for 5 min. To detect EGFP-Flag–tagged WT CD59 (EGFP-F–CD59), mutant CD59 (EGFP-F–CD59 [C94S]), WT DAF (EGFP-F–DAF), and mutant DAF (EGFP-F–DAF [C81A]), HEK293FF6 cells were transfected with plasmids expressing these proteins together with pME-RFP-KDEL plasmid. Stained cells were visualized by a confocal microscope (C2si; Nikon) with a CFI Plan Apochromat VC oil objective lens (100× magnification and 1.4 NA) or FV1000 (Olympus) with a UPLSAPO oil lens (100× magnification and 1.4 NA).
RNA extraction and strand-specific RNA library preparation
Total RNA was extracted using the mirVana miRNA Isolation kit (Thermo Fisher Scientific) and was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies). The libraries were constructed using TruSeq Stranded mRNA LTSample Prep kit (Illumina). Then, these libraries were sequenced on the Illumina sequencing platform.
RNA-seq analysis
The original data were transferred into sequence data by base calling, which are defined as raw reads and saved as FASTQ files. The dirty raw reads were removed using the software FASTQC (Babraham Bioinformatics). Reads with adapters, reads in which unknown bases are >10%, and low-quality reads (the percentage of the low quality bases of quality value ≤5 is >50% in a read) were removed using the software NGS QC TOOLKIT (v2.3.3; Patel and Jain, 2012). Clean reads were mapped to reference sequences (human genome hg19) using TopHat (https://ccb.jhu.edu/software/tophat/index.shtml). Mismatches of no more than two bases were allowed in the alignment. Differential expression analysis was designed to identify differentially expressed genes in different samples. HTseq software (Anders et al., 2014) was used for obtaining the number of reads in genes, and the negative binomial distribution test in the DESeq software package (Anders and Huber, 2010) was used to calculate different expression of genes. The significance of the read number was assessed by a negative binomial distribution test, and the basemean expression was used to estimate the amount of gene expression (Table S2). The default filter of the different condition was set as P ≤ 0.05. Genes that significantly up-regulated in MOGS-KO cells were analyzed using Gene Ontology enrichment analysis and the KEGG pathway. The number of differential genes included in each GO or KEGG entry was calculated, and the significance of differential gene enrichment in each GO item was calculated by hypergeometric test.
Quantitative RT-PCR analysis
Approximately 24 h before experiments, cells were seeded in six-well plates at 0.5 × 106 cells per well. Cells were then harvested and washed with PBS. RNA was extracted according the manufacturer’s protocol (Promega), and total RNA was resuspended in 50 µl RNase-free water. To synthesize cDNA, 150 ng RNA was diluted in 45 µl RNase-free water, 5 µl 5× PrimeScript RT Master Mix for quantitative RT-PCR (Takara Bio Inc.) was added, and the reaction was incubated for 15 min at 37°C and then for 5 s at 85°C, and then it was maintained at 4°C. For real-time PCR, 2 µl cDNA was mixed with 10 µl SYBR Premix Ex Taq II (Tli RNaseH Plus; Takara Bio Inc.), 0.4 µl 6-carboxy-X-rhodamine Reference Dye II, and 6 µl RNase-free water containing 10 µM forward and reverse primers. Triplicate reactions for each cDNA and primer pair were run using standard parameters in a StepOnePlus Real-Time PCR system (Thermo Fisher Scientific).
Immunoprecipitation and Western blotting
HEK293FF6 cells cultured in 10-cm dishes were transfected with 16 µg of each plasmid using Lipofectamine 2000 (Invitrogen) by incubating for 48 h. Cells were harvested and washed in PBS at 4°C. A 600-µl aliquot of lysis buffer (25 mM Hepes, pH 7.4, 150 mM NaCl, 1% digitonin or 1% NP-40, protein inhibitor cocktail, and 1 mM PMSF) was added to each cell pellet, and samples were rotated at 4°C for 1 h. After incubation, tubes were centrifuged at 10,000 g for 10 min at 4°C. A portion of each supernatant was mixed with sample buffer, the other was transferred to a new tube, and then 20 µl of prewashed anti-Flag beads (A2220; Sigma-Aldrich) was added. After rotating tubes at 4°C for 2 h, the beads with tagged proteins were washed four times with lysis buffer. The proteins were then eluted from the beads with Flag peptide and mixed with sample buffer and then maintained at 4°C overnight. Samples were subjected to SDS-PAGE, and proteins were visualized by Western blotting.
CHX chase experiment
HEK293FF6 cells stably expressing 3×Flag-rPGAP1, EGFP-F–CD59, or EGFP-F–CD59 (C94S) were cultured in 10-cm dishes. The chase experiment was started by addition of 100 µg/ml CHX. Then, cells were harvested at the indicated times and immunoprecipitated with anti-Flag beads. Samples were subjected to SDS-PAGE, and proteins were visualized by Western blotting.
PNGase F and Endo H treatments and Triton X-114 partitioning
The samples of EGFP-F–CD59, EGFP-F–CD59 C94S, EGFP-F–CD59 C94S N43Q, and EGFP-F–CD59 C94S G103* immunoprecipitated with anti-Flag beads were mixed with denaturing sample buffer and kept at 4°C overnight.
For PNGase F and Endo H treatments, the reaction mixtures were prepared in 10 µl of total reaction mixture by adding 1 µl of 10× GlycoBuffer2, 1 µl of 10% NP-40, 1 µl of PNGase F (500 U; New England Biolabs, Inc.), and 7 µl of samples or by adding 1 µl of 10× GlycoBuffer3, 1 µl of Endo Hf (1,000 U; New England Biolabs, Inc.), and 8 µl of samples, respectively. The mixtures were incubated at 37°C for 3 h. Then, samples were subjected to SDS-PAGE, and proteins were visualized by Western blotting.
For Triton X-114 phase partitioning, 200 µl of immunoprecipitated samples were incubated with or without 0.5 µl PIPLC at 37°C for 1 h and cooled on ice for 3 min and then were resuspended in aqueous 2% (wt/vol) Triton X-114 and kept on ice for 10 min. Then, samples were warmed at 37°C for 3 min and centrifuged at 1,000 g for 10 min at 37°C. From the sample separated into two phase, the upper aqueous phase (A) and lower detergent phase (D) were carefully collected. The aqueous phase was further purified by adjusting the Triton X-114 concentration to 2% (wt/vol) and repeating the phase partitioning as above. Similarly, the detergent phase was further purified by adding an equal volume of lysis buffer and repeating the phase partitioning process. Finally, the sample was mixed with sample buffer and kept at 4°C overnight. Then, samples were subjected to SDS-PAGE, and proteins were visualized by Western blotting.
Quantification and statistical analysis
The type of statistical test is annotated in the figure legends and/or in the Materials and methods. In addition, statistical parameters such as the value of n, SD, and significance level are reported in the figures and/or in the figure legends. Higher p-values are sometimes displayed in the figures but are not referred to as significant. Statistical analyses were performed using Excel (Microsoft) or R as described in the Analysis of gene-trap insertions and PIPLC treatment and flow cytometry sections of Materials and methods.
Online supplemental material
Fig. S1 shows validation of gene KOs. Fig. S2 shows PIPLC sensitivity of DAF in KO cells and restoration of PIPLC sensitivity by expression of the responsible genes. Fig. S3 shows increased PIPLC resistance of GPI-APs with DNJ treatment and chronic ER stress. Fig. S4 shows RNA-seq analysis in WT and MOGS-KO cells. Fig. S5 shows effect of misfolded proteins on PIPLC sensitivity of GPI-APs. Table S1 shows oligonucleotides used for KO construction. Table S2 shows oligonucleotide primers used in this study. Table S3 shows enrichment of gene-trap insertions in the populations resistant to PIPLC. Table S4 shows differential expression of genes in MOGS-KO cells. Table S5 shows list of GPI-APs and their potential N-glycan sites.
Supplementary Material
Acknowledgments
We thank Drs. Thijn R. Brummelkamp and Reid Gilmore for reagents. We thank Drs. Yusuke Maeda, Hideki Nakanishi, Ning Wang, Zi-Jie Li, and Yun Hu for discussion.
This work was supported by grants-in-aid from the National Natural Science Foundation of China (31400693 and 31770853), the Natural Science Foundation of Jiangsu Province (BK20140141), the Young Thousand program (to M. Fujita), the Program of Introducing Talents of Discipline to Universities (111-2-06), and Japan Society for the Promotion of Science KAKENHI grant JP16H04756 (to T. Kinoshita).
The authors declare no competing financial interests.
Author contributions: Y.-S. Liu, X.-Y. Guo, and T. Hirata designed and performed all the experiments except those indicated below and contributed to writing the manuscript. Y. Rong established several KO cells used in this study. D. Motooka and S. Nakamura carried out deep sequencing analyses. T. Kitajima, Y. Murakami, and X.-D. Gao conducted biochemical experiments and edited the manuscript. T. Kinoshita cosupervised the project and edited the manuscript. M. Fujita supervised the project and wrote the manuscript.
References
- Aebi M. 2013. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta. 1833:2430–2437. 10.1016/j.bbamcr.2013.04.001 [DOI] [PubMed] [Google Scholar]
- Anders S., and Huber W.. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2014. HTSeq — A Python framework to work with high-throughput sequencing data. Bioinformatics. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen V.A., Májek P., Jae L.T., Bigenzahn J.W., Nieuwenhuis J., Staring J., Sacco R., van Diemen F.R., Olk N., Stukalov A., et al. . 2015. Gene essentiality and synthetic lethality in haploid human cells. Science. 350:1092–1096. 10.1126/science.aac7557 [DOI] [PubMed] [Google Scholar]
- Boukhzar L., Hamieh A., Cartier D., Tanguy Y., Alsharif I., Castex M., Arabo A., El Hajji S., Bonnet J.J., Errami M., et al. . 2016. Selenoprotein T Exerts an Essential Oxidoreductase Activity That Protects Dopaminergic Neurons in Mouse Models of Parkinson’s Disease. Antioxid. Redox Signal. 24:557–574. 10.1089/ars.2015.6478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J.E., Raaben M., Wong A.C., Herbert A.S., Obernosterer G., Mulherkar N., Kuehne A.I., Kranzusch P.J., Griffin A.M., Ruthel G., et al. . 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 477:340–343. 10.1038/nature10348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Walter E.I., Parker G., Lapurga J.P., Millan J.L., Ikehara Y., Udenfriend S., and Medof M.E.. 1998. Mammalian glycophosphatidylinositol anchor transfer to proteins and posttransfer deacylation. Proc. Natl. Acad. Sci. USA. 95:9512–9517. 10.1073/pnas.95.16.9512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanova N., Shrimal S., and Gilmore R.. 2016. N-linked glycosylation and homeostasis of the endoplasmic reticulum. Curr. Opin. Cell Biol. 41:57–65. 10.1016/j.ceb.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson J.C., Olzmann J.A., Shaler T.A., Sowa M.E., Bennett E.J., Richter C.M., Tyler R.E., Greenblatt E.J., Harper J.W., and Kopito R.R.. 2011. Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol. 14:93–105. 10.1038/ncb2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikiy A., Novoselov S.V., Fomenko D.E., Sengupta A., Carlson B.A., Cerny R.L., Ginalski K., Grishin N.V., Hatfield D.L., and Gladyshev V.N.. 2007. SelT, SelW, SelH, and Rdx12: genomics and molecular insights into the functions of selenoproteins of a novel thioredoxin-like family. Biochemistry. 46:6871–6882. 10.1021/bi602462q [DOI] [PubMed] [Google Scholar]
- Ellgaard L., and Helenius A.. 2003. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4:181–191. 10.1038/nrm1052 [DOI] [PubMed] [Google Scholar]
- Ferguson M.A.J., Kinoshita T., and Hart G.W.. 2009. Glycosylphosphatidylinositol Anchors. In Essentials of Glycobiology. Varki A., Cummings R.D., Esko J.D., Freeze H.H., Stanley P., Bertozzi C.R., Hart G.W., and Etzler M.E., editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY. 143–162. [PubMed] [Google Scholar]
- Fujita M., and Kinoshita T.. 2012. GPI-anchor remodeling: potential functions of GPI-anchors in intracellular trafficking and membrane dynamics. Biochim. Biophys. Acta. 1821:1050–1058. 10.1016/j.bbalip.2012.01.004 [DOI] [PubMed] [Google Scholar]
- Fujita M., Yoko-O T., and Jigami Y.. 2006. Inositol deacylation by Bst1p is required for the quality control of glycosylphosphatidylinositol-anchored proteins. Mol. Biol. Cell. 17:834–850. 10.1091/mbc.E05-05-0443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Maeda Y., Ra M., Yamaguchi Y., Taguchi R., and Kinoshita T.. 2009. GPI glycan remodeling by PGAP5 regulates transport of GPI-anchored proteins from the ER to the Golgi. Cell. 139:352–365. 10.1016/j.cell.2009.08.040 [DOI] [PubMed] [Google Scholar]
- Fujita M., Watanabe R., Jaensch N., Romanova-Michaelides M., Satoh T., Kato M., Riezman H., Yamaguchi Y., Maeda Y., and Kinoshita T.. 2011. Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI. J. Cell Biol. 194:61–75. 10.1083/jcb.201012074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzow M., Paramasivam N., Hinderhofer K., Fischer C., Chotewutmontri S., Kaufmann L., Evers C., Kotzaeridou U., Rohrschneider K., Schlesner M., et al. . 2015. Loss of function of PGAP1 as a cause of severe encephalopathy identified by Whole Exome Sequencing: Lessons of the bioinformatics pipeline. Mol. Cell. Probes. 29:323–329. 10.1016/j.mcp.2015.05.012 [DOI] [PubMed] [Google Scholar]
- Grumolato L., Ghzili H., Montero-Hadjadje M., Gasman S., Lesage J., Tanguy Y., Galas L., Ait-Ali D., Leprince J., Guérineau N.C., et al. . 2008. Selenoprotein T is a PACAP-regulated gene involved in intracellular Ca2+ mobilization and neuroendocrine secretion. FASEB J. 22:1756–1768. 10.1096/fj.06-075820 [DOI] [PubMed] [Google Scholar]
- Hamieh A., Cartier D., Abid H., Calas A., Burel C., Bucharles C., Jehan C., Grumolato L., Landry M., Lerouge P., et al. . 2017. Selenoprotein T is a novel OST subunit that regulates UPR signaling and hormone secretion. EMBO Rep. 18:1935–1946. 10.15252/embr.201643504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F., Kerr G., and Boutros M.. 2014. E-CRISP: fast CRISPR target site identification. Nat. Methods. 11:122–123. 10.1038/nmeth.2812 [DOI] [PubMed] [Google Scholar]
- Heinz D.W., Ryan M., Bullock T.L., and Griffith O.H.. 1995. Crystal structure of the phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with myo-inositol. EMBO J. 14:3855–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A., and Aebi M.. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73:1019–1049. 10.1146/annurev.biochem.73.011303.073752 [DOI] [PubMed] [Google Scholar]
- Hirata T., Fujita M., Kanzawa N., Murakami Y., Maeda Y., and Kinoshita T.. 2013. Glycosylphosphatidylinositol mannosyltransferase II is the rate-limiting enzyme in glycosylphosphatidylinositol biosynthesis under limited dolichol-phosphate mannose availability. J. Biochem. 154:257–264. 10.1093/jb/mvt045 [DOI] [PubMed] [Google Scholar]
- Hirata T., Fujita M., Nakamura S., Gotoh K., Motooka D., Murakami Y., Maeda Y., and Kinoshita T.. 2015. Post-Golgi anterograde transport requires GARP-dependent endosome-to-TGN retrograde transport. Mol. Biol. Cell. 26:3071–3084. 10.1091/mbc.E14-11-1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama H., Fujita M., Yoko-o T., and Jigami Y.. 2008. O-mannosylation is required for degradation of the endoplasmic reticulum-associated degradation substrate Gas1*p via the ubiquitin/proteasome pathway in Saccharomyces cerevisiae. J. Biochem. 143:555–567. 10.1093/jb/mvm249 [DOI] [PubMed] [Google Scholar]
- Hong Y., Ohishi K., Inoue N., Kang J.Y., Shime H., Horiguchi Y., van der Goot F.G., Sugimoto N., and Kinoshita T.. 2002. Requirement of N-glycan on GPI-anchored proteins for efficient binding of aerolysin but not Clostridium septicum alpha-toxin. EMBO J. 21:5047–5056. 10.1093/emboj/cdf508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D., Krawitz P., Mannhardt A., Korenke G.C., and Meinecke P.. 2011. Hyperphosphatasia-mental retardation syndrome due to PIGV mutations: expanded clinical spectrum. Am. J. Med. Genet. A. 155:1917–1922. 10.1002/ajmg.a.34102 [DOI] [PubMed] [Google Scholar]
- Kettwig M., Elpeleg O., Wegener E., Dreha-Kulaczewski S., Henneke M., Gärtner J., and Huppke P.. 2016. Compound heterozygous variants in PGAP1 causing severe psychomotor retardation, brain atrophy, recurrent apneas and delayed myelination: a case report and literature review. BMC Neurol. 16:74 10.1186/s12883-016-0602-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Fujita M., and Maeda Y.. 2008. Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: recent progress. J. Biochem. 144:287–294. 10.1093/jb/mvn090 [DOI] [PubMed] [Google Scholar]
- Lamriben L., Graham J.B., Adams B.M., and Hebert D.N.. 2016. N-Glycan-based ER Molecular Chaperone and Protein Quality Control System: The Calnexin Binding Cycle. Traffic. 17:308–326. 10.1111/tra.12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang S., Benedix J., Fedeles S.V., Schorr S., Schirra C., Schäuble N., Jalal C., Greiner M., Hassdenteufel S., Tatzelt J., et al. . 2012. Different effects of Sec61α, Sec62 and Sec63 depletion on transport of polypeptides into the endoplasmic reticulum of mammalian cells. J. Cell Sci. 125:1958–1969. 10.1242/jcs.096727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y., Tashima Y., Houjou T., Fujita M., Yoko-o T., Jigami Y., Taguchi R., and Kinoshita T.. 2007. Fatty acid remodeling of GPI-anchored proteins is required for their raft association. Mol. Biol. Cell. 18:1497–1506. 10.1091/mbc.E06-10-0885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean D.M., and Niswander L.. 2012. Defects in GPI biosynthesis perturb Cripto signaling during forebrain development in two new mouse models of holoprosencephaly. Biol. Open. 1:874–883. 10.1242/bio.20121982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon A.K. 1994. Structural analysis of glycosylphosphatidylinositol anchors. Methods Enzymol. 230:418–442. 10.1016/0076-6879(94)30027-5 [DOI] [PubMed] [Google Scholar]
- Muñiz M., and Riezman H.. 2016. Trafficking of glycosylphosphatidylinositol anchored proteins from the endoplasmic reticulum to the cell surface. J. Lipid Res. 57:352–360. 10.1194/jlr.R062760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y., Siripanyapinyo U., Hong Y., Kang J.Y., Ishihara S., Nakakuma H., Maeda Y., and Kinoshita T.. 2003. PIG-W is critical for inositol acylation but not for flipping of glycosylphosphatidylinositol-anchor. Mol. Biol. Cell. 14:4285–4295. 10.1091/mbc.E03-03-0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y., Tawamie H., Maeda Y., Büttner C., Buchert R., Radwan F., Schaffer S., Sticht H., Aigner M., Reis A., et al. . 2014. Null mutation in PGAP1 impairing Gpi-anchor maturation in patients with intellectual disability and encephalopathy. PLoS Genet. 10:e1004320 10.1371/journal.pgen.1004320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R.K., and Jain M.. 2012. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrieras N., Bause E., Legler G., Vasilov R., Claesson L., Peterson P., and Ploegh H.. 1983. Effects of the glucosidase inhibitors nojirimycin and deoxynojirimycin on the biosynthesis of membrane and secretory glycoproteins. EMBO J. 2:823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost G., Arabo A., Jian L., Quelennec E., Cartier D., Hassan S., Falluel-Morel A., Tanguy Y., Gargani S., Lihrmann I., et al. . 2013. The PACAP-regulated gene selenoprotein T is abundantly expressed in mouse and human β-cells and its targeted inactivation impairs glucose tolerance. Endocrinology. 154:3796–3806. 10.1210/en.2013-1167 [DOI] [PubMed] [Google Scholar]
- Rong Y., Nakamura S., Hirata T., Motooka D., Liu Y.S., He Z.A., Gao X.D., Maeda Y., Kinoshita T., and Fujita M.. 2015. Genome-Wide Screening of Genes Required for Glycosylphosphatidylinositol Biosynthesis. PLoS One. 10:e0138553 10.1371/journal.pone.0138553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpute-Krishnan P., Ajinkya M., Bhat S., Itakura E., Hegde R.S., and Lippincott-Schwartz J.. 2014. ER stress-induced clearance of misfolded GPI-anchored proteins via the secretory pathway. Cell. 158:522–533. 10.1016/j.cell.2014.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrag J.D., Bergeron J.J., Li Y., Borisova S., Hahn M., Thomas D.Y., and Cygler M.. 2001. The Structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol. Cell. 8:633–644. 10.1016/S1097-2765(01)00318-5 [DOI] [PubMed] [Google Scholar]
- Schröter S., Derr P., Conradt H.S., Nimtz M., Hale G., and Kirchhoff C.. 1999. Male-specific modification of human CD52. J. Biol. Chem. 274:29862–29873. 10.1074/jbc.274.42.29862 [DOI] [PubMed] [Google Scholar]
- Sikorska N., Lemus L., Aguilera-Romero A., Manzano-Lopez J., Riezman H., Muñiz M., and Goder V.. 2016. Limited ER quality control for GPI-anchored proteins. J. Cell Biol. 213:693–704. 10.1083/jcb.201602010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Maeda Y., Tashima Y., and Kinoshita T.. 2004. Inositol deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J. Biol. Chem. 279:14256–14263. 10.1074/jbc.M313755200 [DOI] [PubMed] [Google Scholar]
- Tannous A., Pisoni G.B., Hebert D.N., and Molinari M.. 2015. N-linked sugar-regulated protein folding and quality control in the ER. Semin. Cell Dev. Biol. 41:79–89. 10.1016/j.semcdb.2014.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thastrup O., Cullen P.J., Drøbak B.K., Hanley M.R., and Dawson A.P.. 1990. Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)-ATPase. Proc. Natl. Acad. Sci. USA. 87:2466–2470. 10.1073/pnas.87.7.2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treumann A., Lifely M.R., Schneider P., and Ferguson M.A.. 1995. Primary structure of CD52. J. Biol. Chem. 270:6088–6099. 10.1074/jbc.270.11.6088 [DOI] [PubMed] [Google Scholar]
- Ueda Y., Yamaguchi R., Ikawa M., Okabe M., Morii E., Maeda Y., and Kinoshita T.. 2007. PGAP1 knock-out mice show otocephaly and male infertility. J. Biol. Chem. 282:30373–30380. 10.1074/jbc.M705601200 [DOI] [PubMed] [Google Scholar]
- Vallée F., Karaveg K., Herscovics A., Moremen K.W., and Howell P.L.. 2000. Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases. J. Biol. Chem. 275:41287–41298. 10.1074/jbc.M006927200 [DOI] [PubMed] [Google Scholar]
- Walter E.I., Roberts W.L., Rosenberry T.L., Ratnoff W.D., and Medof M.E.. 1990. Structural basis for variations in the sensitivity of human decay accelerating factor to phosphatidylinositol-specific phospholipase C cleavage. J. Immunol. 144:1030–1036. [PubMed] [Google Scholar]
- Wong Y.W., and Low M.G.. 1992. Phospholipase resistance of the glycosyl-phosphatidylinositol membrane anchor on human alkaline phosphatase. Clin. Chem. 38:2517–2525. [PubMed] [Google Scholar]
- Yoshiura K., Machida J., Daack-Hirsch S., Patil S.R., Ashworth L.K., Hecht J.T., and Murray J.C.. 1998. Characterization of a novel gene disrupted by a balanced chromosomal translocation t(2;19)(q11.2;q13.3) in a family with cleft lip and palate. Genomics. 54:231–240. 10.1006/geno.1998.5577 [DOI] [PubMed] [Google Scholar]
- Zoltewicz J.S., Ashique A.M., Choe Y., Lee G., Taylor S., Phamluong K., Solloway M., and Peterson A.S.. 2009. Wnt signaling is regulated by endoplasmic reticulum retention. PLoS One. 4:e6191 10.1371/journal.pone.0006191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.