Hansen et al. review the potential dual helpful and harmful roles of microglia in the development and progression of Alzheimer’s disease.
Abstract
Proliferation and activation of microglia in the brain, concentrated around amyloid plaques, is a prominent feature of Alzheimer’s disease (AD). Human genetics data point to a key role for microglia in the pathogenesis of AD. The majority of risk genes for AD are highly expressed (and many are selectively expressed) by microglia in the brain. There is mounting evidence that microglia protect against the incidence of AD, as impaired microglial activities and altered microglial responses to β-amyloid are associated with increased AD risk. On the other hand, there is also abundant evidence that activated microglia can be harmful to neurons. Microglia can mediate synapse loss by engulfment of synapses, likely via a complement-dependent mechanism; they can also exacerbate tau pathology and secrete inflammatory factors that can injure neurons directly or via activation of neurotoxic astrocytes. Gene expression profiles indicate multiple states of microglial activation in neurodegenerative disease settings, which might explain the disparate roles of microglia in the development and progression of AD pathology.
Introduction
With its aging populations, the world is facing a crisis of Alzheimer’s disease (AD). A progressive neurodegenerative disorder afflicting mainly the elderly, AD is the most common cause of dementia and a leading cause of death in the United States (James et al., 2014; Weuve et al., 2014). Alois Alzheimer first described the histopathology of AD (Alzheimer, 1907), which is characterized by brain atrophy, amyloid plaques (extracellular deposits of Aβ peptide aggregates), neurofibrillary tangles (composed largely of tau protein), loss of neurons and synapses, and dystrophic neurites. In addition, Alzheimer noted, “The glia have developed numerous fibers” (Alzheimer et al., 1995).
The reactive gliosis of AD histopathology reflects the abnormal morphology and proliferation of astrocytes and microglia. Microgliosis and astrogliosis are common features of many neurodegenerative diseases with distinct etiologies (Maragakis and Rothstein, 2006; Ransohoff and Perry, 2009; Glass et al., 2010), but it was uncertain whether these histopathological changes reflect a beneficial, detrimental, or inconsequential activity of glial cells in the neurodegenerative process. In recent years, however, biological advances stemming from human genetics data have removed any doubt that microglia play an important role in the pathogenesis of AD.
Microglia, the innate immune cells of the central nervous system (CNS), originate from erythromyeloid progenitor cells in the embryonic yolk sac and migrate into the brain around embryonic day 10.5 in mouse, after which they propagate, spread, and ramify throughout the brain parenchyma (Ginhoux et al., 2013). Their homeostasis and self-renewal is maintained by several factors, including TGF-β and CSF1R signaling (Ginhoux et al., 2010; Butovsky et al., 2014; Elmore et al., 2014). In the CNS, microglia serve as resident phagocytes that dynamically survey the environment, playing crucial roles in CNS tissue maintenance, injury response, and pathogen defense (Nayak et al., 2014; Colonna and Butovsky, 2017). Microglia also participate in the developmental sculpting of neural circuits by engulfment and removal of unwanted neurons and synapses (Schafer et al., 2012; Frost and Schafer, 2016). Parabiosis experiments in AD mouse models indicated that microglia account for the increased myeloid cell number observed in brains with plaque pathology, with minimal contribution of infiltrating macrophages (Wang et al., 2016).
In this review, we consider the genetic and cell biological data indicating that microglia have protective functions that restrain the toxic accumulation of β-amyloid and prevent the development of AD. We also review the evidence that, once activated, microglia can have harmful actions in AD, such as being the source of inflammatory factors and mediating the engulfment of neuronal synapses. Finally, we discuss recent advances in profiling the microglial transcriptome that help us better understand the role of microglia in mechanisms of AD pathology.
Human genetic evidence for microglia involvement in late-onset AD
The accumulation of Aβ peptides, and their aggregation and deposition in amyloid plaques, is believed to be a key pathogenic mechanism in AD. Occurring during the decade or two preceding dementia symptoms (Bateman et al., 2012; Fleisher et al., 2012; Villemagne et al., 2013), β-amyloidosis results from an imbalance in the production versus clearance of Aβ. The human genetics of familial AD highlighted excessive production of amyloidogenic Aβ as a cause of early onset AD; mutations in amyloid precursor protein (APP) or in its processing enzyme (γ-secretase, presenilin subunits PS1 or PS2) result in increased β-site cleavage of APP or favored production of longer, aggregation-prone variants of Aβ peptide (Selkoe and Hardy, 2016; Szaruga et al., 2017).
Familial AD is extremely rare, however; the vast majority of AD cases are “sporadic” and occur late in life. Late-onset AD appears to result mainly from a mixture of genetic and environmental factors, including aging, that impair the brain’s ability to clear Aβ (Mawuenyega et al., 2010; Wildsmith et al., 2013). In the last decade, human genetic studies, especially genome-wide association studies (GWASs) using single-nucleotide polymorphisms (SNPs), have identified over 20 genetic loci that robustly associate with AD risk (Lambert et al., 2013; Karch et al., 2014; Table 1).
Table 1. Microglial roles for AD risk genes identified in genome-wide associations.
| Gene/protein | Molecular function | Activities in microglial biology and AD |
|---|---|---|
| Lipid transport | ||
| APOE | Major apolipoprotein in brain HDL-like particles | Conveys Aβ to lipoprotein receptors for clearance. Microglial Apoe expression induced in neurodegenerative models. |
| CLU/apoJ | Another apolipoprotein in brain, usually in separate lipoparticles from apoE | Promotes Aβ solubility. Conveys Aβ to lipoprotein receptors for clearance (DeMattos et al., 2004). |
| Transmembrane proteins | ||
| SORL1 | Receptor for vesicular sorting of lipoproteins and various receptors | Binds Aβ and directs it to lysosome (Caglayan et al., 2014). Rare variant in this domain = familial AD. High microglial expression in human. |
| ABCA7 | ATP-binding cassette transporter; multipass transmembrane protein transports lipids | Localizes to phagocytic cup (Jehle et al., 2006); presumed role in membrane remodeling. Impaired Aβ phagocytosis in Abca7-null mice (Fu et al., 2016). |
| TREM2 | Binds anionic/lipophilic ligands; triggers DAP12 ITAM to recruit kinase Syk | Implicated in cell viability, chemotaxis, and phagocytosis. Disease mutations impair interactions with apoE and apoJ. |
| CD33 | Binds sialylated ligands; phosphorylated ITIM recruits phosphatase SHP-1 | Protective allele reduces surface CD33 levels and enhances Aβ uptake. AD association not replicated in meta-analysis. |
| MS4A6A | Four-pass transmembrane protein in MS4A family; function unknown | Likely involved in microglial receptor complex, like MS4A1 (CD20) in B cells and MS4A2 (FCER1B) in basophils/mast cells |
| CR1 | Complement receptor 1; binds C1q and C3b/C4b | Recognizes opsonized targets. Inactivates C3b/C4b. Variant with extra C3b/C4b-binding domain increases AD risk (Brouwers et al., 2012). |
| EPHA1 | Receptor tyrosine kinase for ephrin-A class ligands | Stimulates Pyk2 phosphorylation and migration in T lymphocytes (Aasheim et al., 2005). Not yet studied in microglia. |
| HLA-DRB1/5 | Major histocompatibility complex class II protein for extracellular antigen presentation | May serve as intracellular adaptors during the innate immune response (Liu et al., 2011) |
| IL1RAP | Coreceptor with IL1R1 for IL-1 signaling | Enables proinflammatory signal transduction that may mitigate plaque pathology but exacerbate Tau pathology (Ghosh et al., 2013) |
| Membrane and cytoskeletal dynamics | ||
| INPP5D/SHIP1 | SH3-containing inositol phosphatase, converts phosphatidylinositol (3,4,5)-trisphosphate to phosphatidylinositol (3,4)-bisphosphate | Interacts with DAP12, opposes PI3K recruitment, modulates receptor endocytosis, and curbs phagocytosis in macrophages |
| PLCG2 | Phospholipase activity cleaves phosphatidylinositol (4,5)-bisphosphate into IP3 and DAG second messengers | Acts downstream of SYK during ITAM signaling. IP3 and DAG effect calcium and PKC signaling. |
| CD2AP | Adaptor protein between membrane proteins and actin cytoskeleton | Interactions with SHIP1 and RIN3 (Bao et al., 2012; Rouka et al., 2015) suggest role in microglia endocytosis. Also implicated in neuronal APP trafficking and tau propagation. |
| BIN1 | Involved in membrane curvature and dynamin interaction | RIN3 interaction (Kajiho et al., 2003) suggests role in microglia endocytosis. Also implicated in neuronal APP trafficking and tau propagation. |
| RIN3-SLC24A4 | RIN3: a guanine nucleotide exchange factor for Rab5 and Rab31 | RIN3: interacts with BIN1 and CD2AP. Functions in the early endocytic pathway (Kajiho et al., 2011). |
| PICALM | Phosphatidylinositol-binding protein recruits clathrin and AP2 for vesicle assembly | Possible role in microglial endocytosis. Also implicated in neuronal APP trafficking and Aβ efflux via blood–brain barrier transcytosis. |
| PTK2B/Pyk2 | Non–receptor tyrosine kinase; homologue of focal adhesion kinase | Activated in microglia by fibrillar Aβ (Combs et al., 1999). Involved in chemotactic polarization and migration of macrophages (Okigaki et al., 2003). |
| CASS4 | Scaffold protein associated with focal adhesion kinases FAK, Pyk2 | Largely unstudied. Functions inferred by homology with p130Cas family. |
| ABI3 | Component of the Abi/WAVE complex involved in actin polymerization (Sekino et al., 2015) | Probably involved in microglial motility and/or phagocytosis |
| FERMT2 | Adaptor between membrane and actin cytoskeleton at extracellular matrix adhesion sites | Not expressed in microglia; no conjectured role in microglial function. Implicated in APP trafficking. |
| Transcription factors | ||
| SPI1/PU.1 | Important for myeloid and B cell lineages | Originally identified SNP was intronic rs10838725 in CELF1 gene. Protective SPI1 SNP = reduced PU.1 levels. |
| MEF2C | Widely studied in muscle cells and neurons | Microglial expression and calcium-dependent activation mechanisms (Lynch et al., 2005) suggest possible role downstream of TREM2. |
| Other | ||
| ZCWPW1 | Presumed epigenetic regulator through its chromatin-binding domains | SNPs across ∼10 genes show AD association; causal gene unknown. Neighboring genes encode paired immune receptors. |
| NME8 | Encodes protein with thioredoxin and nucleoside diphosphate kinase domains | Unknown cellular function |
AD-associated genes are described in terms of microglial functions known or speculated to govern AD pathogenesis. References are provided only for certain material not mentioned elsewhere in the text.
A significant fraction of the heritable risk for sporadic AD can be accounted for by the APOE gene, of which there are three common alleles encoding the apoE2, apoE3, and apoE4 variants of apolipoprotein E (Holtzman et al., 2012). Relative to the most common apoE3 variant, a single apoE4 or apoE2 allele confers an approximately threefold increased or approximately twofold reduced risk of developing AD, respectively (Corder et al., 1993, 1994; Strittmatter et al., 1993; Farrer et al., 1997). ApoE, the major protein component of high density lipoprotein (HDL)–like lipoprotein particles that transport lipids, cholesterol, and other hydrophobic molecules in the brain, is present in Aβ plaques (Namba et al., 1991; Wisniewski and Frangione, 1992; Zhan et al., 1995) and contributes to both the clearance and the amyloid deposition of Aβ peptide (Bales et al., 1997a; DeMattos et al., 2004; Bien-Ly et al., 2012; Holtzman et al., 2012). Relative to apoE3, apoE4 appears to reduce the clearance of Aβ and increase its deposition in plaques (Rebeck et al., 1993; Kok et al., 2009; Reiman et al., 2009; Castellano et al., 2011; Fleisher et al., 2013). The precise mechanisms by which apoE4 confers increased AD risk are not well understood.
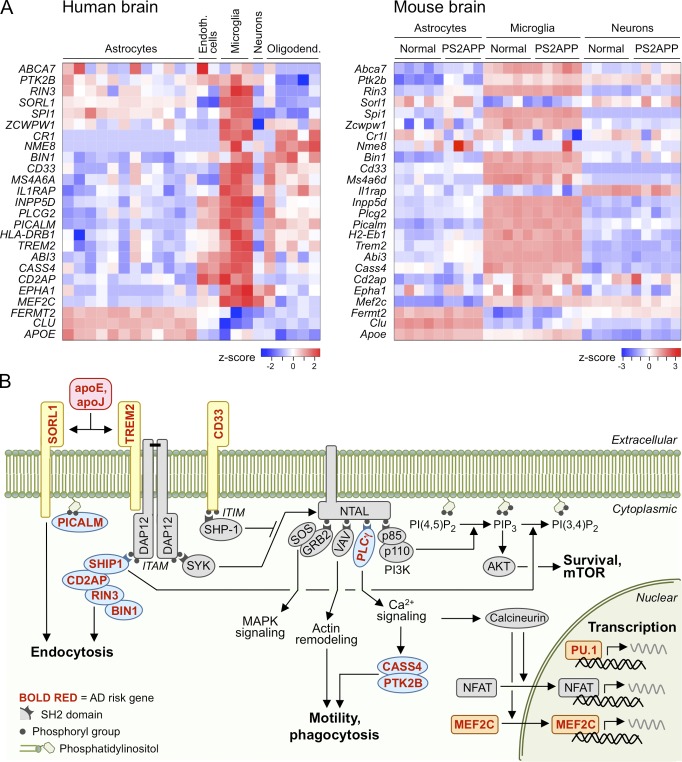
Other than APOE, the common genetic variants associated with AD confer only minor effects (∼10–20%) on AD risk (Lambert et al., 2013). Although the molecular-genetic mechanisms for most AD-associated loci remain to be elucidated, a striking feature of the identified risk genes is that the majority of them are expressed selectively or preferentially in microglia relative to other cell types in the brain (Fig. 1 A; Srinivasan et al., 2016; Zhang et al., 2016). For example, a common variant in SPI1 is associated with reduced AD risk and reduced SPI1 expression (Huang et al., 2017); SPI1 encodes the PU.1 transcription factor that is essential for microglial development (Schulz et al., 2012; Kierdorf et al., 2013). This broad trend implicates microglial dysfunction as a contributing factor, rather than an attendant feature, of AD pathogenesis.
Figure 1.
Expression and function of AD risk genes in microglia. (A) These heat maps depict relative expression levels of GWAS-identified AD risk genes among CNS cell types purified from human (Zhang et al., 2016) or mouse (Srinivasan et al., 2016) brain tissues and analyzed by RNA sequencing. Each column within a cell type represents one sample of those cells purified from a different brain. From human dataset GSE73721, samples derived from “normal” cortex are plotted, ranging in age from 8 to 63 yr. From mouse dataset GSE75431, samples from cortex of 13-mo PS2APP β-amyloid model or age-matched nontransgenic littermates are plotted. Z-score represents the number of standard deviations by which a sample’s expression level for a gene differs from the mean expression level for that gene across all samples. For human genes lacking clear mouse orthologues, suitable mouse homologues were selected. (B) This simplified schematic depicts how selected proteins encoded by AD risk genes (red, bold font) participate in pathways for microglial uptake and cellular activation. Lipoproteins containing apoE or apoJ may convey Aβ to microglia for uptake and degradation or may bind to TREM2 and stimulate ITAM-mediated cellular activation leading to chemotaxis, phagocytosis, survival, and transcription.
In addition to the common variants identified by GWASs, rare genetic variants associated with AD (found by gene sequencing or exome array) have also implicated microglia in determining the risk of AD (Guerreiro et al., 2013; Jonsson et al., 2013; Sims et al., 2017). Particularly momentous was the discovery of AD-associated variants in TREM2 (triggering receptor expressed in myeloid cells 2), a cell surface protein selectively and highly expressed by microglia in the brain, as well as by certain myeloid cells in the periphery. The TREM2 mutation most clearly associated with AD (R47H, carried by <0.5% of most populations) increases the risk of AD approximately threefold (Guerreiro et al., 2013; Jonsson et al., 2013). Because R47H appears to be a loss-of-function mutation that impairs TREM2-mediated microglial activation (see the following sections), the study of TREM2 has been instrumental in establishing the view that microglia normally operate in a protective capacity against AD (Jay et al., 2017b; Ulrich et al., 2017; Yeh et al., 2017).
Protective role of TREM2 and microglia in AD
TREM2 functions as a cell surface receptor on microglia; via its interaction with the activating adaptor protein DAP12 (encoded by the TYROBP gene), TREM2 stimulation initiates signal transduction pathways that promote microglial chemotaxis, phagocytosis, survival, and proliferation (Takahashi et al., 2005; Hsieh et al., 2009; Kleinberger et al., 2014; Poliani et al., 2015; Wang et al., 2015, 2016; Mazaheri et al., 2017; Zheng et al., 2017; Fig. 1 B). Extracellular ligands of TREM2 include a variety of phospholipids and glycolipids, lipoproteins (e.g., low density lipoprotein [LDL] and HDL), and apoptotic cells (Atagi et al., 2015; Bailey et al., 2015; Poliani et al., 2015; Wang et al., 2015; Yeh et al., 2016). Notably, TREM2 binds to apolipoproteins apoE and clusterin (CLU; also known as apoJ), which are themselves encoded by AD risk genes (Atagi et al., 2015; Bailey et al., 2015; Yeh et al., 2016). TREM2–ligand interactions are impaired by TREM2 variants that increase AD risk (Wang et al., 2015; Kober et al., 2016; Yeh et al., 2016; Song et al., 2017), implying that these AD variants are at least partial loss-of-function mutants. Homozygous null mutations in TREM2 cause Nasu–Hakola disease, characterized by early onset neurodegeneration (including white matter lesions) and bone abnormalities (Paloneva et al., 2002; Klünemann et al., 2005).
Phagocytosis and clearance of Aβ and cellular debris
An important aspect of tissue homeostasis by microglia is the engulfment and clearance of debris. TREM2 is required for microglial phagocytosis of a variety of substrates, including apoptotic neurons, bacteria, LDL and other lipoproteins, and Aβ (Takahashi et al., 2005; N’Diaye et al., 2009; Kleinberger et al., 2014; Atagi et al., 2015; Yeh et al., 2016). Aβ aggregates are much more efficiently taken up by microglia when Aβ is complexed with lipoproteins such as LDL, apoE, and CLU/apoJ (Terwel et al., 2011; Yeh et al., 2016). Importantly, TREM2-deficient microglia showed reduced uptake of Aβ-lipoprotein complexes in vitro (Yeh et al., 2016) and less evidence of Aβ internalization in vivo (Wang et al., 2016; Yuan et al., 2016). The interaction of TREM2 with its lipoprotein ligands (LDL, APOE, and CLU) was impaired by the AD-linked mutations of TREM2 (R47H, R62H, and D87N) and completely abolished by Nasu–Hakola disease–linked mutations (Y38C and T66M; Atagi et al., 2015; Bailey et al., 2015; Yeh et al., 2016). Blood monocyte–derived macrophages from human carriers of the TREM2 AD-linked R62H variant showed reduced uptake of Aβ–lipoprotein complexes (Yeh et al., 2016). Thus, impaired uptake and clearance of Aβ (perhaps as Aβ–lipoprotein complexes) could explain, at least in part, how TREM2 loss-of-function mutations increase the risk of developing AD (Fig. 2). Consistent with this notion, plaque accumulation in mouse models of β-amyloidosis (which typically overexpress mutant APP together with mutant PS1 or PS2) is exacerbated at later ages (8+ months) in Trem2 knockout mice (Wang et al., 2015; Jay et al., 2017a).
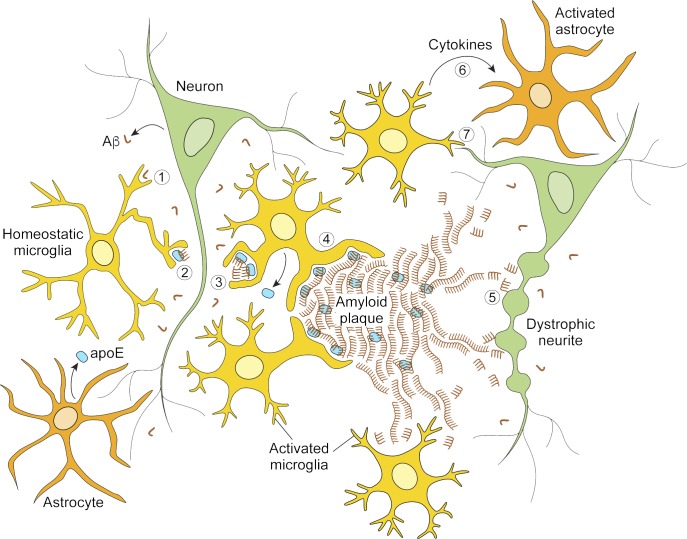
Figure 2.
Depiction of microglial cellular activities related to β-amyloid pathology. The left side illustrates protective microglial activities that limit disease progression. Microglia may clear Aβ peptides via macropinocytosis of soluble Aβ (1; Mandrekar et al., 2009), uptake of lipoprotein-associated Aβ (2), or phagocytosis of fibrillar Aβ aggregates (3). Microglia also help corral larger deposits of Aβ in plaques (4), minimizing damage to the adjacent neuropil. The right side illustrates disease states when microglial containment mechanisms are defective or outstripped. Aβ fibrils on the outskirts of plaque act as substrate for additional Aβ fibrillization and a reservoir of toxic Aβ species that induce neuritic dystrophy (5). Microglia can secrete factors that activate astrocytes (6) and participate in amyloid-dependent synapse loss (7). See also Fig. 3.
Beyond clearance of Aβ, another important TREM2-dependent function of microglia is to clean up debris from damaged or dying cells to promote a healthy brain environment, as observed in models of demyelination or ischemic stroke (Cantoni et al., 2015; Kawabori et al., 2015; Poliani et al., 2015). The interaction of TREM2 with apoE may facilitate microglial phagocytosis of apoptotic neurons (Atagi et al., 2015). We suggest that the TREM2–apoE axis plays a key role in the microglial clearance of a variety of extracellular and cellular detritus and thus may be generally important for minimizing bystander damage to neurons in neurodegenerative settings.
Congregation around amyloid plaques and barrier formation
In the normal brain, microglia dynamically extend and retract processes to probe the environment (Nimmerjahn et al., 2005). With β-amyloid deposition, however, some microglial processes become static, showing stable association with plaques over days or weeks, with TREM2, DAP12, and phosphotyrosine concentrated in those processes adjacent to plaques (Condello et al., 2015; Yuan et al., 2016). TREM2-deficient microglia fail to congregate or proliferate around plaques, lack the typical morphological changes of microglial activation, and show increased apoptosis (Jay et al., 2015; Wang et al., 2015, 2016; Mazaheri et al., 2017). The microglial gene expression profile induced by amyloid pathology (see later section on transcription) was broadly dampened in TREM2-deficient mice, indicating a key role of TREM2 in the microglial reaction to Aβ pathology (Wang et al., 2015; Keren-Shaul et al., 2017). A recent study suggested that the multifaceted microglial dysfunction observed in TREM2-deficient mice with β-amyloid pathology could stem from impaired mTOR signaling and a metabolic deficit (Ulland et al., 2017).
What is the role of reactive microglia around plaques? Recent evidence suggests that microglia form a protective barrier around amyloid deposits, compacting amyloid fibrils into a tightly packed and potentially less toxic form, preventing the accretion of new Aβ onto existing plaques, and reducing axonal dystrophy in the nearby neuropil (Condello et al., 2015). These protective “corralling” functions were more readily observed for small, early stage plaques and appeared compromised in mice lacking one or both copies of Trem2, as well as in human AD tissues from TREM2 R47H carriers (Condello et al., 2015; Wang et al., 2016; Yuan et al., 2016). A halo of soluble, oligomeric Aβ (the form currently thought to exert the most toxic effects on neurons) likely exists around amyloid plaques. Thus, the compaction of protofibrillary Aβ into a dense core plaque, which requires apoE and is promoted by TREM2 (Bales et al., 1997b, 1999; Bien-Ly et al., 2012; Wang et al., 2016; Yuan et al., 2016), could be a protective mechanism that limits neurotoxicity of amyloid deposits once they start to build up in the aging brain. Collectively, the TREM2 studies suggest several mechanisms by which microglia can protect from accumulation of toxic Aβ species and development of AD: uptake and clearance of soluble Aβ species, phagocytosis of insoluble fibrillar Aβ deposits, induction of the activated state and chemotaxis, and compaction and corralling of amyloid plaques (Fig. 2).
Potential involvement of other AD risk genes in microglial function
Many AD risk genes besides TREM2 are preferentially or selectively expressed in microglia (e.g., CD33, INPP5D, MS4A6A, and PLCG2) and therefore could impinge on the same microglial activities and pathways regulated by TREM2. Furthermore, several AD risk genes that have been studied in neurons in the context of APP trafficking, Aβ production, or tau pathology should also be considered for potential roles in microglia, given their pattern of microglial expression (i.e., SORL1, PICALM, CD2AP, BIN1, PTK2B, and ABCA7; Fig. 1 A and Table 1).
TREM2 signaling involves recruitment of tyrosine kinase SYK to the phosphorylated ITAM (immunoreceptor tyrosine-based activation motif) of DAP12 and thereby activates downstream effectors such as phosphoinositide 3-kinase (PI3K) and Ca2+ signaling (Fig. 1 B). INPP5D, which encodes the lipid phosphatase SHIP1, is another AD risk gene preferentially expressed in microglia. SHIP1 dephosphorylates phosphatidylinositol (3,4,5)-trisphosphate to phosphatidylinositol (3,4)-bisphosphate at the plasma membrane, altering the outcomes of PI3K activation. SHIP1 is involved in a clathrin-independent mode of receptor endocytosis (Boucrot et al., 2015) and inhibits phagocytosis in macrophages (Cox et al., 2001; Kamen et al., 2007). SHIP1 can interact with the phosphorylated ITAM of DAP12, so it may also moderate TREM2 signaling by competing with SYK and PI3K for ITAM occupancy (Peng et al., 2010). The AD-associated INPP5D SNP (rs35349669) increases INPP5D gene expression in whole blood (Jansen et al., 2017). If this relationship between genotype and expression level holds true in microglia, then it might explain the elevated AD risk, because higher SHIP1 level would dampen microglial activation and phagocytosis.
Opposing the activity of ITAM receptors and SYK are the inhibitory receptors, which contain ITIM (immunoreceptor tyrosine-based inhibitory motif) domains that recruit tyrosine phosphatases. One such ITIM-containing receptor is encoded by the AD risk gene CD33, a member of the SIGLEC (sialic acid–binding immunoglobulin-type lectins) family of receptors. The protective SNP alters CD33 mRNA splicing such that the extracellular sialic acid–binding domain is lost from the final protein (Malik et al., 2013, 2015; Raj et al., 2014). The CD33 genotype–phenotype relationship nicely aligns with that of TREM2: the TREM2 R47H mutation (which increases AD risk) reduces TREM2–ligand binding and ITAM signaling and impairs phagocytosis, whereas the CD33 variant (which reduces AD risk) prevents the CD33–ligand interaction and presumably ITIM signaling and promotes phagocytosis (Bradshaw et al., 2013; Griciuc et al., 2013).
Given the genetic evidence that microglia help reduce the incidence of AD, a critical question is whether microglia also restrain AD progression and at which stages of disease. A combined analysis of neuroimaging and genetic data found an association between a polymorphism in the IL1RAP gene and increased rate of Aβ accumulation in human brains of both AD and non-AD subjects (measured by Aβ-binding positron emission tomography tracer; Ramanan et al., 2015). Carriers of the IL1RAP polymorphism also showed reduced signal for microglial activation using a TSPO (translocator protein)-binding positron emission tomography tracer, as well as an increased rate of temporal lobe atrophy and higher likelihood of progression from mild cognitive impairment to AD. Another study that divided AD subjects into fast decliners and slow decliners measured a higher TSPO signal in the brains of slow decliners, consistent with a protective role for microglial activation even in stages of AD when dementia is evident (Hamelin et al., 2016). Human microglia in aged brains or in the vicinity of tau pathology may exhibit dystrophic, fragmented morphology, further suggesting that AD develops in the context of reduced neuroprotective microglial function (Streit et al., 2004, 2009).
Detrimental activities of microglia in AD
Although human genetic data argue that proper microglial function protects against AD, there is plentiful evidence that unbridled microglia activity can be harmful to neurons in neurodegenerative disease. Amyloid plaques appear a decade or two before clinical symptoms of AD, but it is tau pathology and synapse loss that correlate best with cognitive impairment during disease progression (Jack et al., 2010). In this section, we discuss how microglia can directly mediate synapse loss (Wu et al., 2015; Spangenberg and Green, 2017) and exacerbate tau pathology (Leyns and Holtzman, 2017). Moreover, activated microglia can secrete toxic factors to directly or indirectly injure neurons (Colonna and Butovsky, 2017; Liddelow et al., 2017).
Microglia, complement, and synapse engulfment
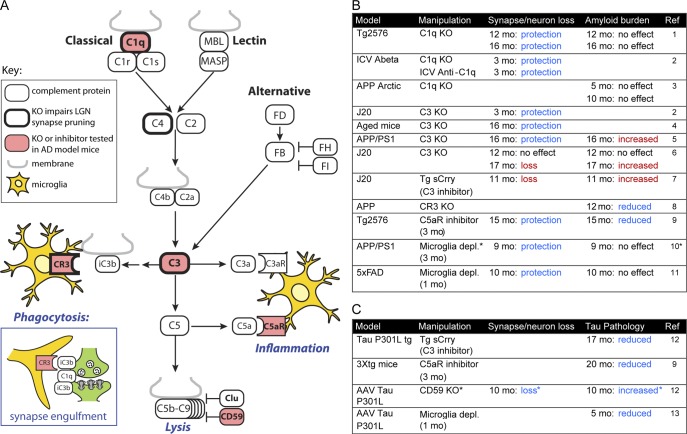
During normal brain development, mounting evidence indicates that microglia engulf synapses and sculpt synaptic connections via a novel role for complement, a component of the innate immune system that enhances the clearance of microbes or damaged cells by phagocytes (Boulanger, 2009; Stephan et al., 2012; Chung et al., 2015). Synapse pruning by microglia involves the classical pathway of complement, which normally functions to clear pathogens and apoptotic cells after binding of complement protein C1q (Fig. 3 A). Studies of C1q, C4, C3, and CR3 knockout mice suggest that the mechanism of developmental synapse pruning in the retinogeniculate system involves C1q tagging of synapses, opsonization of synapses by C3b, and subsequent phagocytosis of synapses by microglia (Stevens et al., 2007; Schafer et al., 2012; Sekar et al., 2016; Fig. 3 A). Synaptic material is observed within microglia by light microscopy, and this synapse engulfment is reduced in C3 or CR3 knockout mice (Schafer et al., 2012). Microglial engulfment of synapses likely varies with different developmental stages, brain regions, and disease states. For example, electron microscopy studies have detected engulfed synaptic material in the developing hippocampus (Paolicelli et al., 2011), but not in the hippocampus of adult mice undergoing synapse loss caused by prion disease (Sisková et al., 2009; Caleo et al., 2012).
Figure 3.
Summary of studies manipulating the complement system or depleting microglia in mouse models of AD. (A) Simplified schematic of the complement pathway illustrating selected proteins. The complement system can be initiated by the classical, lectin, or alternative pathways. Central to complement activation is the cleavage of C3. Effects downstream of C3 cleavage include (1) phagocytosis after recognition of C3b opsonized material by complement receptors, including CR3 (inset); (2) inflammatory signaling by C3a and C5a fragment activation of C3aR and C5aR; and (3) lysis via formation of the C5b-C9 membrane attack complex. In the brain, microglia (yellow) mediate phagocytosis and respond to inflammatory signaling, and are also the cell type that produces C1q. Complement proteins demonstrated to play a role in synapse removal during developmental refinement of retinal ganglion cell projections to the lateral geniculate nucleus using knockout mice are indicated with a heavy border. Proteins that have been studied using knockout mice or inhibitors in the context of AD model mice are highlighted in red. (B) Table indicating the amyloid mouse models that have been tested, manipulations that were tested (complement protein knockout or inhibition or microglia depletion), the resulting impact on synapse or neuronal loss (“protection” indicates rescue of amyloid model deficits, whereas “loss” indicates the manipulation causes deficits), and effects on amyloid load. Blue fonts indicate phenotypes that suggest a beneficial effect of reducing complement activation or microglial cell numbers, and red fonts indicate phenotypes that suggest undesirable effects of reducing complement activation. *Note that in this study, the authors claimed not to deplete microglia but to block microglial proliferation. (C) Similar table as in panel B, except showing models of tauopathy and impacts on tau pathology. *Note that CD59 is an inhibitor of complement pathway activity, so the synapse/neuron loss seen with CD59 knockout is consistent with a beneficial effect of reducing complement activation. Reference (Ref) 1, Fonseca et al., 2004; Ref 2, Hong et al., 2016; Ref 3, Fonseca et al., 2017; Ref 4, Shi et al., 2015; Ref 5, Shi et al., 2017a; Ref 6, Maier et al., 2008; Ref 7, Wyss-Coray et al., 2002; Ref 8, Czirr et al., 2017; Ref 9, Fonseca et al., 2009; Ref 10, Olmos-Alonso et al., 2016; Ref 11, Spangenberg et al., 2016; Ref 12, Britschgi et al., 2012; and Ref 13, Asai et al., 2015. FB (FD, FH, FI), complement factor B (D, H, I); KO, knockout; MASP, MBL-associated serine protease; MBL, mannose-binding lectin.
Given the emerging role of complement proteins in developmental synapse pruning, it is interesting that AD-associated genes include CR1 (complement receptor 1), which plays roles in phagocytosis, clearing of immune complexes, and inhibition of complement (Khera and Das, 2009; Fonseca et al., 2016), and CLU/apoJ, which reportedly can function as an inhibitor of the terminal complement complex (Murphy et al., 1989; Tschopp et al., 1993; McDonald and Nelsestuen, 1997). Interestingly, in knock-in mice expressing human apoE variants, the apoE4 risk allele increases C1q accumulation relative to apoE2 (Chung et al., 2016). The complement system is also implicated by human genetics as a cause of other nervous system disorders that feature neurodegeneration. A heightened risk of schizophrenia is associated with increased copy number and expression of complement factor C4A (Sekar et al., 2016), and genetic variants in complement proteins and regulators (most notably CFH) that lead to increased complement activity are highly associated with age-related macular degeneration (Klein et al., 2005; Boyer et al., 2017). Thus, human genetics point to excessive activation of the complement system as a cause of degeneration of neural tissue.
In human AD brain tissue, there is considerable immunohistochemical evidence for activation of complement, especially associated with plaques (C1q, C3, and C4) and to a lesser extent with neurofibrillary tangles and dystrophic neurites (C5b-C9; Zanjani et al., 2005). C1q is expressed in the adult brain, and protein levels rise steeply with aging, especially in the hippocampus (Stephan et al., 2013). Mouse models of β-amyloidosis exhibit elevated C1q levels, with increased synaptic localization of C1q even before plaques have formed (Hong et al., 2016). Although the binding partner for C1q at synapses that leads to activation of the C1 complex during synapse pruning remains unknown, it is noteworthy that C1q binding to Aβ can trigger activation of the classical complement cascade (Jiang et al., 1994; Rosen and Stevens, 2010). Genetic knockout of C1q or neutralizing antibodies against C1q protect against synapse loss observed in amyloid-bearing mice or induced by injected Aβ (Fonseca et al., 2004; Hong et al., 2016). Notably, C1q knockout, which affects the classical pathway of complement initiation, does not impact amyloid burden, so C1q appears to act downstream of Aβ (Fonseca et al., 2004, 2017). Knockout or inhibition of C3, which should block all pathways of complement activation, is also reported to provide neuroprotection (Shi et al., 2015, 2017a; Hong et al., 2016; for different results, see Wyss-Coray et al., 2002; Maier et al., 2008), though it appears to increase amyloid burden (Wyss-Coray et al., 2002; Maier et al., 2008; Shi et al., 2017a). Thus, in addition to mediating neuronal damage, C3 activation may be involved in clearance of plaque via classical pathway-independent mechanisms.
Microglia are likely key players in complement-mediated synapse loss in AD: they are the main source of C1q in the brain (Fonseca et al., 2017), they phagocytose synapses via CR3 during development (Schafer et al., 2012), and they express the C3a and C5a receptors that trigger inflammation in response to complement activation. Treatment with CSF1R inhibitors to deplete microglia or disable their proliferation protects against synapse loss and rescues behavioral deficits in amyloidosis mouse models (Olmos-Alonso et al., 2016; Spangenberg et al., 2016). Unlike with synapse loss, the formation and maintenance of β-amyloid plaques in amyloidosis models does not seem to be affected by CSF1R inhibition or other methods of microglia depletion (Grathwohl et al., 2009; Olmos-Alonso et al., 2016; Spangenberg et al., 2016). However, longer periods of depletion may be necessary to observe such an effect, and a study that used time-lapse imaging to track individual plaques in vivo reported that plaque size increased ∼13% over 1 wk in microglia-depleted brains (Zhao et al., 2017). Overall, the findings imply that synapse loss around plaques is dynamically regulated by microglia and can be potentially rescued by microglia depletion despite no change or possible increase in amyloid plaque size.
Thus, although there are conflicting results, most published studies in amyloidosis mouse models suggest that depleting microglia and blocking complement pathway activation may have beneficial effects in AD, at least in terms of synapse and neuron loss (Fig. 3 B). A harmful role for activated microglia may be a broad feature of neurodegenerative disease, as microglia and the complement pathway can also mediate synapse loss in other mouse models of neurodegeneration, including progranulin-deficient frontotemporal dementia (Lui et al., 2016), virus-induced cognitive impairment (Vasek et al., 2016), glaucoma (Stevens et al., 2007), macular degeneration (Ding et al., 2014), and CNS systemic lupus erythematosus (Bialas et al., 2017).
Microglia and tau pathology
Complement activation appears to exacerbate tau pathology in AD mouse models, though the mechanisms are unclear (Fig. 3 C). C3 inhibition by transgenic expression of a soluble form of the mouse complement inhibitory protein Crry (Britschgi et al., 2012) or C5aR antagonism (Fonseca et al., 2009) reduced the extent of tau pathology. Knockout of CD59, a protein that inhibits C5b-9 membrane attack complex formation, worsened pathology in a tau mouse model (Britschgi et al., 2012).
Studies in a mouse model of tauopathy initiated via viral expression of mutant tau have implicated microglia in the cell-to-cell spread of tau pathology across the brain, possibly mediated by microglial uptake and exosomal release of tau (Asai et al., 2015). Deleting the microglial protein Cx3cr1 in transgenic tau models showed that genetically enhanced microglial activation accelerated the onset and progression of tau pathology (Bhaskar et al., 2010; Maphis et al., 2015). Interestingly, whereas a reduction in plaque load was observed with Cx3cr1 knockout in amyloid models (Lee et al., 2010; Liu et al., 2010), this was reportedly accompanied by increased proinflammatory cytokines, tau pathology, and functional deficits (Cho et al., 2011; Lee et al., 2014). Finally, a recent study in PS19 mice expressing the human tau P301S mutant transgene (Yoshiyama et al., 2007) and lacking murine Apoe showed that human apoE4 expression exacerbated tau pathology and heightened the microglial response compared with PS19 mice expressing apoE2, apoE3, or no apoE (Shi et al., 2017b). This suggested that besides the well-known effects of apoE4 on amyloid accumulation, apoE4 may contribute to AD pathogenesis by exacerbating neuroinflammation and tau pathology.
Recent studies of TREM2 function in mouse models of tau pathology reported seemingly conflicting roles, with Trem2 deletion appearing neuroprotective in the PS19 model (Leyns et al., 2017) but neurotoxic in the hTau model (Bemiller et al., 2017), which expresses the entire human MAPT gene from a P1-derived artificial chromosome in a Mapt-deficient background (Andorfer et al., 2003). However, the studies did not analyze similar stages of disease. The study in the hTau model looked at 6 mo (long before neuronal loss) and observed greater tau phosphorylation and aggregation in TREM2-deficient mice, arguing for an early protective role of TREM2 in restraining propagation of tauopathy (Bemiller et al., 2017). The PS19 study looked at 9 mo, when neurodegeneration is extensive, and observed preservation of piriform and entorhinal cortex volume in mice lacking TREM2, whereas tau phosphorylation and aggregation were unchanged (Leyns et al., 2017). The latter study suggested that TREM2 may enable the phagocytosis by microglia of stressed but viable neurons, a process termed phagoptosis (Brown and Neher, 2014).
Microglia and neurotoxic inflammatory signaling
In addition to damaging neurons through phagocytosis of synapses and worsening tau pathology, microglia can also react to protein aggregates and dying neurons in a proinflammatory fashion, thereby causing harm to neurons via release of inflammatory mediators. β-Amyloid aggregates/fibrils can act as disease-associated molecular patterns and stimulate Toll-like receptors (TLRs) and the NRLP3 inflammasome (Heneka et al., 2013, 2015), resulting in microglial production of TNFα, IL-1β, and other inflammatory cytokines. Consistent with a pathogenic role for cytokine release, the exacerbation of tau pathology in Cx3cr1 knockout mice was blocked with IL-1 antagonists (Bhaskar et al., 2010; Maphis et al., 2015), and harmful effects of apoE4 in the context of tau pathology were associated with increased TNFα production by microglia in vitro (Shi et al., 2017b). Genetic deletion of NLRP3, caspase-1 and TLRs have been reported to ameliorate Aβ deposition and cognitive deficits in amyloidosis mouse models, supporting the idea that “classical” inflammation exacerbates AD pathogenesis (Heneka et al., 2015). In this context, it is consistent that TREM2 signaling, which is protective against AD based on human genetics, curbs TLR signaling in macrophages (Hamerman et al., 2006; Turnbull et al., 2006). In contrast, the induction of Tnf and Il1b in the brains of amyloidosis mice is dampened in TREM2-deficient mice (Wang et al., 2015; Jay et al., 2017a), which is more in keeping with the idea that TREM2 is required for a full inflammatory response by microglia to β-amyloid.
Consistent with a deleterious role for microglia and inflammation, a recent study using transgenic mice showed that microglial proliferation and activation induced by a BRAF mutation restricted to erythromyeloid progenitors could drive late-onset neurodegeneration (Mass et al., 2017). Importantly, microglia proliferation preceded neuron loss, and microglia from the mutant mice as well as from brain tissue of patients with BRAF mutation–associated neurodegenerative disease showed increased inflammatory cytokine expression (Mass et al., 2017).
Microglia can also act in concert with astrocytes to cause neuronal injury. A triad of factors released by activated microglia (IL-1α, TNFα, and C1q) is necessary and sufficient to induce astrocytes into a neurotoxic state termed “A1” that causes neuronal death (Liddelow et al., 2017). A1 astrocytes are found in tau transgenic mice expressing human apoE4 (Shi et al., 2017b) and are reportedly found in CNS tissue from patients with various neurodegenerative diseases, including AD (Liddelow et al., 2017). Notably, A1 astrocytes show strongly induced expression of complement proteins C1r, C1s, C3, and C4 (Zamanian et al., 2012). Thus, astrocytes could cooperate with microglia to mediate complement-dependent neurotoxicity.
Microglial activation states defined by transcriptional profiling
Elucidating the different functional states of microglia, which may exist at different stages of AD or coexist in the same stage, is crucial to understanding the role of microglia in neurodegeneration. mRNA profiles of microglia sorted by flow cytometry from the brains of β-amyloid mouse models show marked changes in expression of hundreds of genes (Orre et al., 2014; Wang et al., 2015; Srinivasan et al., 2016; Keren-Shaul et al., 2017). These studies point to a disease-associated microglial (DAM) state in which expression of a “homeostatic” gene set is reduced (e.g., Cx3cr1, P2ry12, and Tmem119) and another set of “neurodegeneration” genes is highly induced (e.g., Apoe, Axl, Csf1, Clec7a, Cst7, Igf1, Itgax/CD11c, Lilrb4, Lpl, and some major histocompatibility complex class II genes). Analyzing activated microglia as a separate population or by single-cell RNA-sequencing also reveals subtle (two- to threefold) increases in Trem2 and Tyrobp mRNAs (Kamphuis et al., 2016; Keren-Shaul et al., 2017; Yin et al., 2017). Notably, DAM cells increase in number with progression of amyloidosis, lie spatially close to amyloid plaque, and show evidence of Aβ uptake. Informatics analysis of DAM genes highlights lysosomal, phagocytosis, lipid metabolism, and immune response pathways.
The DAM gene expression signature overlaps significantly with changes in microglial mRNA profiles observed in other neurodegeneration-related models including PS19 tau transgenic, SOD1-G93A transgenic, and aged mice (Chiu et al., 2013; Holtman et al., 2015; Friedman et al., 2017; Keren-Shaul et al., 2017). In β-amyloid models, the induction of DAM genes depends in large part on TREM2, arguing that the DAM profile reflects a protective state of microglia (Wang et al., 2015; Keren-Shaul et al., 2017); however, this is debatable, because TREM2 facilitates neurodegeneration in the PS19 model (Leyns et al., 2017) and is presumably required for the up-regulation of DAM genes in that model (Friedman et al., 2017). In fact, a recent transcriptomic study of the apoE–TREM2 microglial signaling axis posited that this neurodegeneration-related transcriptional response is detrimental (Krasemann et al., 2017). Further studies are needed to clarify whether the TREM2-dependent transcriptional response is helpful, damaging, or incidental in different disease contexts.
Single-cell RNA-sequencing analysis provides a powerful method to uncover the heterogeneity of functional states within a cell population. This method showed that that TREM2-deficient microglial cells in plaque-ridden brains displayed an intermediate state of activation between “homeostatic” microglia and fully activated DAM cells (Keren-Shaul et al., 2017). In a meta-analysis of microglial transcriptomic profiles from various models of CNS diseases (neurodegenerative, demyelinating, ischemic, infectious, inflammatory, and neoplastic), we identified several modules of coordinately regulated genes, including a neurodegeneration-associated module highly similar to the DAM genes mentioned previously (Friedman et al., 2017). Using these modules to probe the single-cell expression data of Keren-Shaul et al. (2017), we revealed a unique subset of microglia, distinct from the DAM cells, that expressed an interferon-related gene module and increased numerically in brains with β-amyloid pathology (Friedman et al., 2017). The functional role of this microglial subclass is unclear.
Because of practical challenges with postmortem human tissue, studies of microglial gene expression in purified microglia or at the single-cell level from AD brains have lagged behind. Nonetheless, the induction of neurodegeneration-associated genes found in AD mouse models has been observed in bulk human AD brain tissue, albeit more subtle in nature (Friedman et al., 2017). More interestingly, an LPS-specific gene cluster, which was not induced in mouse models of neurodegeneration, showed higher expression in AD brains than in control brains (Friedman et al., 2017), suggesting that microglia in human AD could have more severe inflammatory activation than in AD mouse models.
Expression profiles of microglia purified from human brains will be needed to understand the state of microglia in human AD, ideally at different stages of disease (including presymptomatic) and from different genotypes (e.g., APOE). RNA-sequencing studies of human microglia isolated from non-AD brain tissue (Galatro et al., 2017; Gosselin et al., 2017) showed broad similarity with mouse microglial gene expression profiles, although there were many differences such as higher expression of the C2 and C3 complement genes in human microglia. There was surprisingly limited overlap, however, in microglial genes regulated by aging in human versus mouse (Grabert et al., 2016; Galatro et al., 2017), which underscores the importance of studying human microglia to understand the pathomechanisms of AD.
Conclusion
Microglia have recently emerged as crucial players in the pathogenesis of late onset AD, but exactly how they are involved in the disease mechanism is not settled. The preponderance of human genetics evidence, exemplified by the large effect of loss-of-function TREM2 mutations on AD risk and on microglial function, argues that microglia have a protective function that lowers the incidence of AD. Conversely, there is also considerable evidence that microglia are responsible for neuronal damage in AD (albeit mostly in mouse models). In particular, microglia may engulf and remove synapses via a complement-dependent mechanism, and the induction of a microglial proinflammatory state may correlate with severity of neurodegeneration. Transcriptional profiling of microglial gene expression suggests that different states of microglial activation may occur during the course of AD, but more precise characterization—temporal, anatomical, and functional—is needed.
To synthesize the findings from disparate approaches, we propose the following hypothesis: Microglial function is normally protective in the brain, with microglia acting as housekeeping phagocytes to maintain tissue homeostasis and keep the extracellular space clean of Aβ, thereby preventing AD. When Aβ levels accumulate, microglia phagocytose and clear Aβ aggregates, and when outstripped in this activity, microglia compact Aβ aggregates in dense core plaques and shield them off from neurons. These latter protective activities involve activation of microglia to a DAM state, depend on TREM2, and are aided by apoE. Sometimes, because of aging or genetic susceptibility, microglial function becomes inadequate to prevent the onset and progression of AD. As toxic amyloid species accumulate, tau pathology accrues in stressed or damaged neurons, inducing microglia into a nonconstructive and inflammatory state in which they eat synapses, secrete neurotoxic cytokines that injure neurons and abet in the spread of tau pathology. In such a model of disease pathogenesis, microglia have two faces, one beneficial and one harmful, with the detrimental microglia population appearing later in the disease course and coinciding with synapse loss and symptomatic decline. If true, the double-edged sword of microglial function in AD will complicate therapeutic approaches that target microglia, because stimulation of microglial activity may be helpful at an early stage, to prevent AD before it is established, but become detrimental later, when the disease has reached a highly inflamed, neurodegenerative stage.
Acknowledgments
We thank Brad Friedman for computational support with gene expression data, Alison Bruce for artwork, and Menno van Lookeren Campagne, Kimberle Shen, Karpagam Srinivasan, Brad Friedman, Robby Weimer, Felix Yeh, and Michael Chang for comments on the manuscript. We apologize to authors whose work we could not discuss or cite due to space limitations.
The authors are full-time employees of Genentech, Inc., and are actively pursuing novel therapeutics for treatment or prevention of AD. The authors declare no further conflict of interest.
References
- Aasheim H.C., Delabie J., and Finne E.F.. 2005. Ephrin-A1 binding to CD4+ T lymphocytes stimulates migration and induces tyrosine phosphorylation of PYK2. Blood. 105:2869–2876. 10.1182/blood-2004-08-2981 [DOI] [PubMed] [Google Scholar]
- Alzheimer A. 1907. Über eine eigenartige Erkrankung der Hirnrinde. Allgemeine Zeitschrift fur Psychiatrie und Psychisch-Gerichtliche Medizin. 64:146–148. [Google Scholar]
- Alzheimer A., Stelzmann R.A., Schnitzlein H.N., and Murtagh F.R.. 1995. An English translation of Alzheimer’s 1907 paper, “Uber eine eigenartige Erkankung der Hirnrinde”. Clin. Anat. 8:429–431. 10.1002/ca.980080612 [DOI] [PubMed] [Google Scholar]
- Andorfer C., Kress Y., Espinoza M., de Silva R., Tucker K.L., Barde Y.A., Duff K., and Davies P.. 2003. Hyperphosphorylation and aggregation of tau in mice expressing normal human tau isoforms. J. Neurochem. 86:582–590. 10.1046/j.1471-4159.2003.01879.x [DOI] [PubMed] [Google Scholar]
- Asai H., Ikezu S., Tsunoda S., Medalla M., Luebke J., Haydar T., Wolozin B., Butovsky O., Kügler S., and Ikezu T.. 2015. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat. Neurosci. 18:1584–1593. 10.1038/nn.4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atagi Y., Liu C.C., Painter M.M., Chen X.F., Verbeeck C., Zheng H., Li X., Rademakers R., Kang S.S., Xu H., et al. 2015. Apolipoprotein E Is a Ligand for Triggering Receptor Expressed on Myeloid Cells 2 (TREM2). J. Biol. Chem. 290:26043–26050. 10.1074/jbc.M115.679043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C.C., DeVaux L.B., and Farzan M.. 2015. The Triggering Receptor Expressed on Myeloid Cells 2 Binds Apolipoprotein E. J. Biol. Chem. 290:26033–26042. 10.1074/jbc.M115.677286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bales K.R., Verina T., Dodel R.C., Du Y., Altstiel L., Bender M., Hyslop P., Johnstone E.M., Little S.P., Cummins D.J., et al. 1997a Lack of apolipoprotein E dramatically reduces amyloid beta-peptide deposition. Nat. Genet. 17:263–264. 10.1038/ng1197-263 [DOI] [PubMed] [Google Scholar]
- Bales K.R., Verina T., Dodel R.C., Du Y., Altstiel L., Bender M., Hyslop P., Johnstone E.M., Little S.P., Cummins D.J., et al. 1997b Lack of apolipoprotein E dramatically reduces amyloid beta-peptide deposition. Nat. Genet. 17:263–264. 10.1038/ng1197-263 [DOI] [PubMed] [Google Scholar]
- Bales K.R., Verina T., Cummins D.J., Du Y., Dodel R.C., Saura J., Fishman C.E., DeLong C.A., Piccardo P., Petegnief V., et al. 1999. Apolipoprotein E is essential for amyloid deposition in the APP(V717F) transgenic mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 96:15233–15238. 10.1073/pnas.96.26.15233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao M., Hanabuchi S., Facchinetti V., Du Q., Bover L., Plumas J., Chaperot L., Cao W., Qin J., Sun S.C., and Liu Y.J.. 2012. CD2AP/SHIP1 complex positively regulates plasmacytoid dendritic cell receptor signaling by inhibiting the E3 ubiquitin ligase Cbl. J. Immunol. 189:786–792. 10.4049/jimmunol.1200887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman R.J., Xiong C., Benzinger T.L., Fagan A.M., Goate A., Fox N.C., Marcus D.S., Cairns N.J., Xie X., Blazey T.M., et al. Dominantly Inherited Alzheimer Network . 2012. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N. Engl. J. Med. 367:795–804. 10.1056/NEJMoa1202753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemiller S.M., McCray T.J., Allan K., Formica S.V., Xu G., Wilson G., Kokiko-Cochran O.N., Crish S.D., Lasagna-Reeves C.A., Ransohoff R.M., et al. 2017. TREM2 deficiency exacerbates tau pathology through dysregulated kinase signaling in a mouse model of tauopathy. Mol. Neurodegener. 12:74 10.1186/s13024-017-0216-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar K., Konerth M., Kokiko-Cochran O.N., Cardona A., Ransohoff R.M., and Lamb B.T.. 2010. Regulation of tau pathology by the microglial fractalkine receptor. Neuron. 68:19–31. 10.1016/j.neuron.2010.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialas A.R., Presumey J., Das A., van der Poel C.E., Lapchak P.H., Mesin L., Victora G., Tsokos G.C., Mawrin C., Herbst R., and Carroll M.C.. 2017. Microglia-dependent synapse loss in type I interferon-mediated lupus. Nature. 546:539–543. [DOI] [PubMed] [Google Scholar]
- Bien-Ly N., Gillespie A.K., Walker D., Yoon S.Y., and Huang Y.. 2012. Reducing human apolipoprotein E levels attenuates age-dependent Aβ accumulation in mutant human amyloid precursor protein transgenic mice. J. Neurosci. 32:4803–4811. 10.1523/JNEUROSCI.0033-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E., Ferreira A.P., Almeida-Souza L., Debard S., Vallis Y., Howard G., Bertot L., Sauvonnet N., and McMahon H.T.. 2015. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature. 517:460–465. 10.1038/nature14067 [DOI] [PubMed] [Google Scholar]
- Boulanger L.M. 2009. Immune proteins in brain development and synaptic plasticity. Neuron. 64:93–109. 10.1016/j.neuron.2009.09.001 [DOI] [PubMed] [Google Scholar]
- Boyer D.S., Schmidt-Erfurth U., van Lookeren Campagne M., Henry E.C., and Brittain C.. 2017. The Pathophysiology of Geographic Atrophy Secondary to Age-Related Macular Degeneration and the Complement Pathway as a Therapeutic Target. Retina. 37:819–835. 10.1097/IAE.0000000000001392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw E.M., Chibnik L.B., Keenan B.T., Ottoboni L., Raj T., Tang A., Rosenkrantz L.L., Imboywa S., Lee M., Von Korff A., et al. Alzheimer Disease Neuroimaging Initiative . 2013. CD33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nat. Neurosci. 16:848–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britschgi M., Takeda-Uchimura Y., Rockenstein E., Johns H., Masliah E., and Wyss-Coray T.. 2012. Deficiency of terminal complement pathway inhibitor promotes neuronal tau pathology and degeneration in mice. J. Neuroinflammation. 9:220 10.1186/1742-2094-9-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwers N., Van Cauwenberghe C., Engelborghs S., Lambert J.C., Bettens K., Le Bastard N., Pasquier F., Montoya A.G., Peeters K., Mattheijssens M., et al. 2012. Alzheimer risk associated with a copy number variation in the complement receptor 1 increasing C3b/C4b binding sites. Mol. Psychiatry. 17:223–233. 10.1038/mp.2011.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G.C., and Neher J.J.. 2014. Microglial phagocytosis of live neurons. Nat. Rev. Neurosci. 15:209–216. 10.1038/nrn3710 [DOI] [PubMed] [Google Scholar]
- Butovsky O., Jedrychowski M.P., Moore C.S., Cialic R., Lanser A.J., Gabriely G., Koeglsperger T., Dake B., Wu P.M., Doykan C.E., et al. 2014. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17:131–143. 10.1038/nn.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caglayan S., Takagi-Niidome S., Liao F., Carlo A.S., Schmidt V., Burgert T., Kitago Y., Füchtbauer E.M., Füchtbauer A., Holtzman D.M., et al. 2014. Lysosomal sorting of amyloid-β by the SORLA receptor is impaired by a familial Alzheimer’s disease mutation. Sci. Transl. Med. 6:223ra20 10.1126/scitranslmed.3007747 [DOI] [PubMed] [Google Scholar]
- Caleo M., Restani L., Vannini E., Siskova Z., Al-Malki H., Morgan R., O’Connor V., and Perry V.H.. 2012. The role of activity in synaptic degeneration in a protein misfolding disease, prion disease. PLoS One. 7:e41182 10.1371/journal.pone.0041182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantoni C., Bollman B., Licastro D., Xie M., Mikesell R., Schmidt R., Yuede C.M., Galimberti D., Olivecrona G., Klein R.S., et al. 2015. TREM2 regulates microglial cell activation in response to demyelination in vivo. Acta Neuropathol. 129:429–447. 10.1007/s00401-015-1388-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano J.M., Kim J., Stewart F.R., Jiang H., DeMattos R.B., Patterson B.W., Fagan A.M., Morris J.C., Mawuenyega K.G., Cruchaga C., et al. 2011. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci. Transl. Med. 3:89ra57 10.1126/scitranslmed.3002156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu I.M., Morimoto E.T., Goodarzi H., Liao J.T., O’Keeffe S., Phatnani H.P., Muratet M., Carroll M.C., Levy S., Tavazoie S., et al. 2013. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Reports. 4:385–401. 10.1016/j.celrep.2013.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.H., Sun B., Zhou Y., Kauppinen T.M., Halabisky B., Wes P., Ransohoff R.M., and Gan L.. 2011. CX3CR1 protein signaling modulates microglial activation and protects against plaque-independent cognitive deficits in a mouse model of Alzheimer disease. J. Biol. Chem. 286:32713–32722. 10.1074/jbc.M111.254268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W.S., Welsh C.A., Barres B.A., and Stevens B.. 2015. Do glia drive synaptic and cognitive impairment in disease? Nat. Neurosci. 18:1539–1545. 10.1038/nn.4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W.S., Verghese P.B., Chakraborty C., Joung J., Hyman B.T., Ulrich J.D., Holtzman D.M., and Barres B.A.. 2016. Novel allele-dependent role for APOE in controlling the rate of synapse pruning by astrocytes. Proc. Natl. Acad. Sci. USA. 113:10186–10191. 10.1073/pnas.1609896113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M., and Butovsky O.. 2017. Microglia Function in the Central Nervous System During Health and Neurodegeneration. Annu. Rev. Immunol. 35:441–468. 10.1146/annurev-immunol-051116-052358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs C.K., Johnson D.E., Cannady S.B., Lehman T.M., and Landreth G.E.. 1999. Identification of microglial signal transduction pathways mediating a neurotoxic response to amyloidogenic fragments of beta-amyloid and prion proteins. J. Neurosci. 19:928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condello C., Yuan P., Schain A., and Grutzendler J.. 2015. Microglia constitute a barrier that prevents neurotoxic protofibrillar Aβ42 hotspots around plaques. Nat. Commun. 6:6176 10.1038/ncomms7176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder E.H., Saunders A.M., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Small G.W., Roses A.D., Haines J.L., and Pericak-Vance M.A.. 1993. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 261:921–923. 10.1126/science.8346443 [DOI] [PubMed] [Google Scholar]
- Corder E.H., Saunders A.M., Risch N.J., Strittmatter W.J., Schmechel D.E., Gaskell P.C. Jr., Rimmler J.B., Locke P.A., Conneally P.M., Schmader K.E., et al. 1994. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat. Genet. 7:180–184. 10.1038/ng0694-180 [DOI] [PubMed] [Google Scholar]
- Cox D., Dale B.M., Kashiwada M., Helgason C.D., and Greenberg S.. 2001. A regulatory role for Src homology 2 domain-containing inositol 5′-phosphatase (SHIP) in phagocytosis mediated by Fc gamma receptors and complement receptor 3 (alpha(M)beta(2); CD11b/CD18). J. Exp. Med. 193:61–71. 10.1084/jem.193.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czirr E., Castello N.A., Mosher K.I., Castellano J.M., Hinkson I.V., Lucin K.M., Baeza-Raja B., Ryu J.K., Li L., Farina S.N., et al. 2017. Microglial complement receptor 3 regulates brain Aβ levels through secreted proteolytic activity. J. Exp. Med. 214:1081–1092. 10.1084/jem.20162011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMattos R.B., Cirrito J.R., Parsadanian M., May P.C., O’Dell M.A., Taylor J.W., Harmony J.A., Aronow B.J., Bales K.R., Paul S.M., and Holtzman D.M.. 2004. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron. 41:193–202. 10.1016/S0896-6273(03)00850-X [DOI] [PubMed] [Google Scholar]
- Ding J.D., Kelly U., Groelle M., Christenbury J.G., Zhang W., and Bowes Rickman C.. 2014. The role of complement dysregulation in AMD mouse models. Adv. Exp. Med. Biol. 801:213–219. 10.1007/978-1-4614-3209-8_28 [DOI] [PubMed] [Google Scholar]
- Elmore M.R., Najafi A.R., Koike M.A., Dagher N.N., Spangenberg E.E., Rice R.A., Kitazawa M., Matusow B., Nguyen H., West B.L., and Green K.N.. 2014. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron. 82:380–397. 10.1016/j.neuron.2014.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer L.A., Cupples L.A., Haines J.L., Hyman B., Kukull W.A., Mayeux R., Myers R.H., Pericak-Vance M.A., Risch N., and van Duijn C.M.. APOE and Alzheimer Disease Meta Analysis Consortium . 1997. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. JAMA. 278:1349–1356. 10.1001/jama.1997.03550160069041 [DOI] [PubMed] [Google Scholar]
- Fleisher A.S., Chen K., Quiroz Y.T., Jakimovich L.J., Gomez M.G., Langois C.M., Langbaum J.B., Ayutyanont N., Roontiva A., Thiyyagura P., et al. 2012. Florbetapir PET analysis of amyloid-β deposition in the presenilin 1 E280A autosomal dominant Alzheimer’s disease kindred: a cross-sectional study. Lancet Neurol. 11:1057–1065. 10.1016/S1474-4422(12)70227-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleisher A.S., Chen K., Liu X., Ayutyanont N., Roontiva A., Thiyyagura P., Protas H., Joshi A.D., Sabbagh M., Sadowsky C.H., et al. 2013. Apolipoprotein E ε4 and age effects on florbetapir positron emission tomography in healthy aging and Alzheimer disease. Neurobiol. Aging. 34:1–12. 10.1016/j.neurobiolaging.2012.04.017 [DOI] [PubMed] [Google Scholar]
- Fonseca M.I., Zhou J., Botto M., and Tenner A.J.. 2004. Absence of C1q leads to less neuropathology in transgenic mouse models of Alzheimer’s disease. J. Neurosci. 24:6457–6465. 10.1523/JNEUROSCI.0901-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca M.I., Ager R.R., Chu S.H., Yazan O., Sanderson S.D., LaFerla F.M., Taylor S.M., Woodruff T.M., and Tenner A.J.. 2009. Treatment with a C5aR antagonist decreases pathology and enhances behavioral performance in murine models of Alzheimer’s disease. J. Immunol. 183:1375–1383. 10.4049/jimmunol.0901005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca M.I., Chu S., Pierce A.L., Brubaker W.D., Hauhart R.E., Mastroeni D., Clarke E.V., Rogers J., Atkinson J.P., and Tenner A.J.. 2016. Analysis of the Putative Role of CR1 in Alzheimer’s Disease: Genetic Association, Expression and Function. PLoS One. 11:e0149792 10.1371/journal.pone.0149792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca M.I., Chu S.H., Hernandez M.X., Fang M.J., Modarresi L., Selvan P., MacGregor G.R., and Tenner A.J.. 2017. Cell-specific deletion of C1qa identifies microglia as the dominant source of C1q in mouse brain. J. Neuroinflammation. 14:48 10.1186/s12974-017-0814-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman B.A., Srinivasan K., Ayalon G., Meilandt W.J., Lin H., Huntley M.A., Cao Y., Lee S.-H., Haddick P.C.G., Ngu H., et al. 2017. Diverse Brain Myeloid Expression Profiles Reveal Distinct Microglial Activation States and Aspects of Alzheimer’s Disease Not Modeled in Mice. Cell Reports. In Press. [DOI] [PubMed] [Google Scholar]
- Frost J.L., and Schafer D.P.. 2016. Microglia: Architects of the Developing Nervous System. Trends Cell Biol. 26:587–597. 10.1016/j.tcb.2016.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Hsiao J.H., Paxinos G., Halliday G.M., and Kim W.S.. 2016. ABCA7 Mediates Phagocytic Clearance of Amyloid-β in the Brain. J. Alzheimers Dis. 54:569–584. 10.3233/JAD-160456 [DOI] [PubMed] [Google Scholar]
- Galatro T.F., Holtman I.R., Lerario A.M., Vainchtein I.D., Brouwer N., Sola P.R., Veras M.M., Pereira T.F., Leite R.E.P., Möller T., et al. 2017. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat. Neurosci. 20:1162–1171. 10.1038/nn.4597 [DOI] [PubMed] [Google Scholar]
- Ghosh S., Wu M.D., Shaftel S.S., Kyrkanides S., LaFerla F.M., Olschowka J.A., and O’Banion M.K.. 2013. Sustained interleukin-1β overexpression exacerbates tau pathology despite reduced amyloid burden in an Alzheimer’s mouse model. J. Neurosci. 33:5053–5064. 10.1523/JNEUROSCI.4361-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M.F., Conway S.J., Ng L.G., Stanley E.R., et al. 2010. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 330:841–845. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Lim S., Hoeffel G., Low D., and Huber T.. 2013. Origin and differentiation of microglia. Front. Cell. Neurosci. 7:45 10.3389/fncel.2013.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C.K., Saijo K., Winner B., Marchetto M.C., and Gage F.H.. 2010. Mechanisms underlying inflammation in neurodegeneration. Cell. 140:918–934. 10.1016/j.cell.2010.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D., Skola D., Coufal N.G., Holtman I.R., Schlachetzki J.C.M., Sajti E., Jaeger B.N., O’Connor C., Fitzpatrick C., Pasillas M.P., et al. 2017. An environment-dependent transcriptional network specifies human microglia identity. Science. 356:356 10.1126/science.aal3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabert K., Michoel T., Karavolos M.H., Clohisey S., Baillie J.K., Stevens M.P., Freeman T.C., Summers K.M., and McColl B.W.. 2016. Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat. Neurosci. 19:504–516. 10.1038/nn.4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grathwohl S.A., Kälin R.E., Bolmont T., Prokop S., Winkelmann G., Kaeser S.A., Odenthal J., Radde R., Eldh T., Gandy S., et al. 2009. Formation and maintenance of Alzheimer’s disease beta-amyloid plaques in the absence of microglia. Nat. Neurosci. 12:1361–1363. 10.1038/nn.2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griciuc A., Serrano-Pozo A., Parrado A.R., Lesinski A.N., Asselin C.N., Mullin K., Hooli B., Choi S.H., Hyman B.T., and Tanzi R.E.. 2013. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 78:631–643. 10.1016/j.neuron.2013.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J.S., Younkin S., et al. Alzheimer Genetic Analysis Group . 2013. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 368:117–127. 10.1056/NEJMoa1211851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamelin L., Lagarde J., Dorothée G., Leroy C., Labit M., Comley R.A., de Souza L.C., Corne H., Dauphinot L., Bertoux M., et al. Clinical IMABio3 team . 2016. Early and protective microglial activation in Alzheimer’s disease: a prospective study using 18F-DPA-714 PET imaging. Brain. 139:1252–1264. 10.1093/brain/aww017 [DOI] [PubMed] [Google Scholar]
- Hamerman J.A., Jarjoura J.R., Humphrey M.B., Nakamura M.C., Seaman W.E., and Lanier L.L.. 2006. Cutting edge: inhibition of TLR and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. J. Immunol. 177:2051–2055. 10.4049/jimmunol.177.4.2051 [DOI] [PubMed] [Google Scholar]
- Heneka M.T., Kummer M.P., Stutz A., Delekate A., Schwartz S., Vieira-Saecker A., Griep A., Axt D., Remus A., Tzeng T.C., et al. 2013. NLRP3 is activated in Alzheimer’s disease and contributes to pathology in APP/PS1 mice. Nature. 493:674–678. 10.1038/nature11729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneka M.T., Golenbock D.T., and Latz E.. 2015. Innate immunity in Alzheimer’s disease. Nat. Immunol. 16:229–236. 10.1038/ni.3102 [DOI] [PubMed] [Google Scholar]
- Holtman I.R., Raj D.D., Miller J.A., Schaafsma W., Yin Z., Brouwer N., Wes P.D., Möller T., Orre M., Kamphuis W., et al. 2015. Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: a co-expression meta-analysis. Acta Neuropathol. Commun. 3:31 10.1186/s40478-015-0203-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman D.M., Herz J., and Bu G.. 2012. Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2:a006312 10.1101/cshperspect.a006312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Beja-Glasser V.F., Nfonoyim B.M., Frouin A., Li S., Ramakrishnan S., Merry K.M., Shi Q., Rosenthal A., Barres B.A., et al. 2016. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 352:712–716. 10.1126/science.aad8373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.L., Koike M., Spusta S.C., Niemi E.C., Yenari M., Nakamura M.C., and Seaman W.E.. 2009. A role for TREM2 ligands in the phagocytosis of apoptotic neuronal cells by microglia. J. Neurochem. 109:1144–1156. 10.1111/j.1471-4159.2009.06042.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.L., Marcora E., Pimenova A.A., Di Narzo A.F., Kapoor M., Jin S.C., Harari O., Bertelsen S., Fairfax B.P., Czajkowski J., et al. Alzheimer’s Disease Neuroimaging Initiative . 2017. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat. Neurosci. 20:1052–1061. 10.1038/nn.4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R. Jr., Knopman D.S., Jagust W.J., Shaw L.M., Aisen P.S., Weiner M.W., Petersen R.C., and Trojanowski J.Q.. 2010. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 9:119–128. 10.1016/S1474-4422(09)70299-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James B.D., Leurgans S.E., Hebert L.E., Scherr P.A., Yaffe K., and Bennett D.A.. 2014. Contribution of Alzheimer disease to mortality in the United States. Neurology. 82:1045–1050. 10.1212/WNL.0000000000000240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., Hottenga J.J., Nivard M.G., Abdellaoui A., Laport B., de Geus E.J., Wright F.A., Penninx B.W.J.H., and Boomsma D.I.. 2017. Conditional eQTL analysis reveals allelic heterogeneity of gene expression. Hum. Mol. Genet. 26:1444–1451. 10.1093/hmg/ddx043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay T.R., Miller C.M., Cheng P.J., Graham L.C., Bemiller S., Broihier M.L., Xu G., Margevicius D., Karlo J.C., Sousa G.L., et al. 2015. TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J. Exp. Med. 212:287–295. 10.1084/jem.20142322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay T.R., Hirsch A.M., Broihier M.L., Miller C.M., Neilson L.E., Ransohoff R.M., Lamb B.T., and Landreth G.E.. 2017a Disease Progression-Dependent Effects of TREM2 Deficiency in a Mouse Model of Alzheimer’s Disease. J. Neurosci. 37:637–647. 10.1523/JNEUROSCI.2110-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay T.R., von Saucken V.E., and Landreth G.E.. 2017b TREM2 in Neurodegenerative Diseases. Mol. Neurodegener. 12:56 10.1186/s13024-017-0197-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehle A.W., Gardai S.J., Li S., Linsel-Nitschke P., Morimoto K., Janssen W.J., Vandivier R.W., Wang N., Greenberg S., Dale B.M., et al. 2006. ATP-binding cassette transporter A7 enhances phagocytosis of apoptotic cells and associated ERK signaling in macrophages. J. Cell Biol. 174:547–556. 10.1083/jcb.200601030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Burdick D., Glabe C.G., Cotman C.W., and Tenner A.J.. 1994. beta-Amyloid activates complement by binding to a specific region of the collagen-like domain of the C1q A chain. J. Immunol. 152:5050–5059. [PubMed] [Google Scholar]
- Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P.V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A.I., Lah J.J., et al. 2013. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368:107–116. 10.1056/NEJMoa1211103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiho H., Saito K., Tsujita K., Kontani K., Araki Y., Kurosu H., and Katada T.. 2003. RIN3: a novel Rab5 GEF interacting with amphiphysin II involved in the early endocytic pathway. J. Cell Sci. 116:4159–4168. 10.1242/jcs.00718 [DOI] [PubMed] [Google Scholar]
- Kajiho H., Sakurai K., Minoda T., Yoshikawa M., Nakagawa S., Fukushima S., Kontani K., and Katada T.. 2011. Characterization of RIN3 as a guanine nucleotide exchange factor for the Rab5 subfamily GTPase Rab31. J. Biol. Chem. 286:24364–24373. 10.1074/jbc.M110.172445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamen L.A., Levinsohn J., and Swanson J.A.. 2007. Differential association of phosphatidylinositol 3-kinase, SHIP-1, and PTEN with forming phagosomes. Mol. Biol. Cell. 18:2463–2472. 10.1091/mbc.E07-01-0061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis W., Kooijman L., Schetters S., Orre M., and Hol E.M.. 2016. Transcriptional profiling of CD11c-positive microglia accumulating around amyloid plaques in a mouse model for Alzheimer’s disease. Biochim. Biophys. Acta. 1862:1847–1860. 10.1016/j.bbadis.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Karch C.M., Cruchaga C., and Goate A.M.. 2014. Alzheimer’s disease genetics: from the bench to the clinic. Neuron. 83:11–26. 10.1016/j.neuron.2014.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabori M., Kacimi R., Kauppinen T., Calosing C., Kim J.Y., Hsieh C.L., Nakamura M.C., and Yenari M.A.. 2015. Triggering receptor expressed on myeloid cells 2 (TREM2) deficiency attenuates phagocytic activities of microglia and exacerbates ischemic damage in experimental stroke. J. Neurosci. 35:3384–3396. 10.1523/JNEUROSCI.2620-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. 2017. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 169:1276–1290. [DOI] [PubMed] [Google Scholar]
- Khera R., and Das N.. 2009. Complement Receptor 1: disease associations and therapeutic implications. Mol. Immunol. 46:761–772. 10.1016/j.molimm.2008.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierdorf K., Erny D., Goldmann T., Sander V., Schulz C., Perdiguero E.G., Wieghofer P., Heinrich A., Riemke P., Hölscher C., et al. 2013. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 16:273–280. 10.1038/nn.3318 [DOI] [PubMed] [Google Scholar]
- Klein R.J., Zeiss C., Chew E.Y., Tsai J.Y., Sackler R.S., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T., et al. 2005. Complement factor H polymorphism in age-related macular degeneration. Science. 308:385–389. 10.1126/science.1109557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinberger G., Yamanishi Y., Suárez-Calvet M., Czirr E., Lohmann E., Cuyvers E., Struyfs H., Pettkus N., Wenninger-Weinzierl A., Mazaheri F., et al. 2014. TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci. Transl. Med. 6:243ra86 10.1126/scitranslmed.3009093 [DOI] [PubMed] [Google Scholar]
- Klünemann H.H., Ridha B.H., Magy L., Wherrett J.R., Hemelsoet D.M., Keen R.W., De Bleecker J.L., Rossor M.N., Marienhagen J., Klein H.E., et al. 2005. The genetic causes of basal ganglia calcification, dementia, and bone cysts: DAP12 and TREM2. Neurology. 64:1502–1507. 10.1212/01.WNL.0000160304.00003.CA [DOI] [PubMed] [Google Scholar]
- Kober D.L., Alexander-Brett J.M., Karch C.M., Cruchaga C., Colonna M., Holtzman M.J., and Brett T.J.. 2016. Neurodegenerative disease mutations in TREM2 reveal a functional surface and distinct loss-of-function mechanisms. eLife. 5:5 10.7554/eLife.20391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok E., Haikonen S., Luoto T., Huhtala H., Goebeler S., Haapasalo H., and Karhunen P.J.. 2009. Apolipoprotein E-dependent accumulation of Alzheimer disease-related lesions begins in middle age. Ann. Neurol. 65:650–657. 10.1002/ana.21696 [DOI] [PubMed] [Google Scholar]
- Krasemann S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O’Loughlin E., Xu Y., Fanek Z., et al. 2017. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity. 47:566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., et al. Cohorts for Heart and Aging Research in Genomic Epidemiology . 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45:1452–1458. 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Brott B.K., Kirkby L.A., Adelson J.D., Cheng S., Feller M.B., Datwani A., and Shatz C.J.. 2014. Synapse elimination and learning rules co-regulated by MHC class I H2-Db. Nature. 509:195–200. 10.1038/nature13154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Varvel N.H., Konerth M.E., Xu G., Cardona A.E., Ransohoff R.M., and Lamb B.T.. 2010. CX3CR1 deficiency alters microglial activation and reduces beta-amyloid deposition in two Alzheimer’s disease mouse models. Am. J. Pathol. 177:2549–2562. 10.2353/ajpath.2010.100265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyns C.E.G., and Holtzman D.M.. 2017. Glial contributions to neurodegeneration in tauopathies. Mol. Neurodegener. 12:50 10.1186/s13024-017-0192-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyns C.E.G., Ulrich J.D., Finn M.B., Stewart F.R., Koscal L.J., Remolina Serrano J., Robinson G.O., Anderson E., Colonna M., and Holtzman D.M.. 2017. TREM2 deficiency attenuates neuroinflammation and protects against neurodegeneration in a mouse model of tauopathy. Proc. Natl. Acad. Sci. USA. 114:11524–11529. 10.1073/pnas.1710311114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow S.A., Guttenplan K.A., Clarke L.E., Bennett F.C., Bohlen C.J., Schirmer L., Bennett M.L., Münch A.E., Chung W.S., Peterson T.C., et al. 2017. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 541:481–487. 10.1038/nature21029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhan Z., Li D., Xu L., Ma F., Zhang P., Yao H., and Cao X.. 2011. Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining activation of the kinase Btk. Nat. Immunol. 12:416–424. 10.1038/ni.2015 [DOI] [PubMed] [Google Scholar]
- Liu Z., Condello C., Schain A., Harb R., and Grutzendler J.. 2010. CX3CR1 in microglia regulates brain amyloid deposition through selective protofibrillar amyloid-β phagocytosis. J. Neurosci. 30:17091–17101. 10.1523/JNEUROSCI.4403-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui H., Zhang J., Makinson S.R., Cahill M.K., Kelley K.W., Huang H.Y., Shang Y., Oldham M.C., Martens L.H., Gao F., et al. 2016. Progranulin Deficiency Promotes Circuit-Specific Synaptic Pruning by Microglia via Complement Activation. Cell. 165:921–935. 10.1016/j.cell.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J., Guo L., Gelebart P., Chilibeck K., Xu J., Molkentin J.D., Agellon L.B., and Michalak M.. 2005. Calreticulin signals upstream of calcineurin and MEF2C in a critical Ca(2+)-dependent signaling cascade. J. Cell Biol. 170:37–47. 10.1083/jcb.200412156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier M., Peng Y., Jiang L., Seabrook T.J., Carroll M.C., and Lemere C.A.. 2008. Complement C3 deficiency leads to accelerated amyloid beta plaque deposition and neurodegeneration and modulation of the microglia/macrophage phenotype in amyloid precursor protein transgenic mice. J. Neurosci. 28:6333–6341. 10.1523/JNEUROSCI.0829-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik M., Simpson J.F., Parikh I., Wilfred B.R., Fardo D.W., Nelson P.T., and Estus S.. 2013. CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J. Neurosci. 33:13320–13325. 10.1523/JNEUROSCI.1224-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik M., Chiles J. III, Xi H.S., Medway C., Simpson J., Potluri S., Howard D., Liang Y., Paumi C.M., Mukherjee S., et al. 2015. Genetics of CD33 in Alzheimer’s disease and acute myeloid leukemia. Hum. Mol. Genet. 24:3557–3570. 10.1093/hmg/ddv092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrekar S., Jiang Q., Lee C.Y., Koenigsknecht-Talboo J., Holtzman D.M., and Landreth G.E.. 2009. Microglia mediate the clearance of soluble Abeta through fluid phase macropinocytosis. J. Neurosci. 29:4252–4262. 10.1523/JNEUROSCI.5572-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maphis N., Xu G., Kokiko-Cochran O.N., Jiang S., Cardona A., Ransohoff R.M., Lamb B.T., and Bhaskar K.. 2015. Reactive microglia drive tau pathology and contribute to the spreading of pathological tau in the brain. Brain. 138:1738–1755. 10.1093/brain/awv081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragakis N.J., and Rothstein J.D.. 2006. Mechanisms of Disease: astrocytes in neurodegenerative disease. Nat. Clin. Pract. Neurol. 2:679–689. 10.1038/ncpneuro0355 [DOI] [PubMed] [Google Scholar]
- Mass E., Jacome-Galarza C.E., Blank T., Lazarov T., Durham B.H., Ozkaya N., Pastore A., Schwabenland M., Chung Y.R., Rosenblum M.K., et al. 2017. A somatic mutation in erythro-myeloid progenitors causes neurodegenerative disease. Nature. 549:389–393. 10.1038/nature23672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawuenyega K.G., Sigurdson W., Ovod V., Munsell L., Kasten T., Morris J.C., Yarasheski K.E., and Bateman R.J.. 2010. Decreased clearance of CNS beta-amyloid in Alzheimer’s disease. Science. 330:1774 10.1126/science.1197623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaheri F., Snaidero N., Kleinberger G., Madore C., Daria A., Werner G., Krasemann S., Capell A., Trümbach D., Wurst W., et al. 2017. TREM2 deficiency impairs chemotaxis and microglial responses to neuronal injury. EMBO Rep. 18:1186–1198. 10.15252/embr.201743922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J.F., and Nelsestuen G.L.. 1997. Potent inhibition of terminal complement assembly by clusterin: characterization of its impact on C9 polymerization. Biochemistry. 36:7464–7473. 10.1021/bi962895r [DOI] [PubMed] [Google Scholar]
- Murphy B.F., Saunders J.R., O’Bryan M.K., Kirszbaum L., Walker I.D., and d’Apice A.J.. 1989. SP-40,40 is an inhibitor of C5b-6-initiated haemolysis. Int. Immunol. 1:551–554. 10.1093/intimm/1.5.551 [DOI] [PubMed] [Google Scholar]
- N’Diaye E.N., Branda C.S., Branda S.S., Nevarez L., Colonna M., Lowell C., Hamerman J.A., and Seaman W.E.. 2009. TREM-2 (triggering receptor expressed on myeloid cells 2) is a phagocytic receptor for bacteria. J. Cell Biol. 184:215–223. 10.1083/jcb.200808080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba Y., Tomonaga M., Kawasaki H., Otomo E., and Ikeda K.. 1991. Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer’s disease and kuru plaque amyloid in Creutzfeldt-Jakob disease. Brain Res. 541:163–166. 10.1016/0006-8993(91)91092-F [DOI] [PubMed] [Google Scholar]
- Nayak D., Roth T.L., and McGavern D.B.. 2014. Microglia development and function. Annu. Rev. Immunol. 32:367–402. 10.1146/annurev-immunol-032713-120240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A., Kirchhoff F., and Helmchen F.. 2005. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 308:1314–1318. 10.1126/science.1110647 [DOI] [PubMed] [Google Scholar]
- Okigaki M., Davis C., Falasca M., Harroch S., Felsenfeld D.P., Sheetz M.P., and Schlessinger J.. 2003. Pyk2 regulates multiple signaling events crucial for macrophage morphology and migration. Proc. Natl. Acad. Sci. USA. 100:10740–10745. 10.1073/pnas.1834348100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos-Alonso A., Schetters S.T., Sri S., Askew K., Mancuso R., Vargas-Caballero M., Holscher C., Perry V.H., and Gomez-Nicola D.. 2016. Pharmacological targeting of CSF1R inhibits microglial proliferation and prevents the progression of Alzheimer’s-like pathology. Brain. 139:891–907. 10.1093/brain/awv379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orre M., Kamphuis W., Osborn L.M., Jansen A.H.P., Kooijman L., Bossers K., and Hol E.M.. 2014. Isolation of glia from Alzheimer’s mice reveals inflammation and dysfunction. Neurobiol. Aging. 35:2746–2760. 10.1016/j.neurobiolaging.2014.06.004 [DOI] [PubMed] [Google Scholar]
- Paloneva J., Manninen T., Christman G., Hovanes K., Mandelin J., Adolfsson R., Bianchin M., Bird T., Miranda R., Salmaggi A., et al. 2002. Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am. J. Hum. Genet. 71:656–662. 10.1086/342259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli R.C., Bolasco G., Pagani F., Maggi L., Scianni M., Panzanelli P., Giustetto M., Ferreira T.A., Guiducci E., Dumas L., et al. 2011. Synaptic pruning by microglia is necessary for normal brain development. Science. 333:1456–1458. 10.1126/science.1202529 [DOI] [PubMed] [Google Scholar]
- Peng Q., Malhotra S., Torchia J.A., Kerr W.G., Coggeshall K.M., and Humphrey M.B.. 2010. TREM2- and DAP12-dependent activation of PI3K requires DAP10 and is inhibited by SHIP1. Sci. Signal. 3:ra38 10.1126/scisignal.2000500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliani P.L., Wang Y., Fontana E., Robinette M.L., Yamanishi Y., Gilfillan S., and Colonna M.. 2015. TREM2 sustains microglial expansion during aging and response to demyelination. J. Clin. Invest. 125:2161–2170. 10.1172/JCI77983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj T., Ryan K.J., Replogle J.M., Chibnik L.B., Rosenkrantz L., Tang A., Rothamel K., Stranger B.E., Bennett D.A., Evans D.A., et al. 2014. CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer’s disease susceptibility. Hum. Mol. Genet. 23:2729–2736. 10.1093/hmg/ddt666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan V.K., Risacher S.L., Nho K., Kim S., Shen L., McDonald B.C., Yoder K.K., Hutchins G.D., West J.D., Tallman E.F., et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI) . 2015. GWAS of longitudinal amyloid accumulation on 18F-florbetapir PET in Alzheimer’s disease implicates microglial activation gene IL1RAP. Brain. 138:3076–3088. 10.1093/brain/awv231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff R.M., and Perry V.H.. 2009. Microglial physiology: unique stimuli, specialized responses. Annu. Rev. Immunol. 27:119–145. 10.1146/annurev.immunol.021908.132528 [DOI] [PubMed] [Google Scholar]
- Rebeck G.W., Reiter J.S., Strickland D.K., and Hyman B.T.. 1993. Apolipoprotein E in sporadic Alzheimer’s disease: allelic variation and receptor interactions. Neuron. 11:575–580. 10.1016/0896-6273(93)90070-8 [DOI] [PubMed] [Google Scholar]
- Reiman E.M., Chen K., Liu X., Bandy D., Yu M., Lee W., Ayutyanont N., Keppler J., Reeder S.A., Langbaum J.B., et al. 2009. Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 106:6820–6825. 10.1073/pnas.0900345106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen A.M., and Stevens B.. 2010. The role of the classical complement cascade in synapse loss during development and glaucoma. Adv. Exp. Med. Biol. 703:75–93. 10.1007/978-1-4419-5635-4_6 [DOI] [PubMed] [Google Scholar]
- Rouka E., Simister P.C., Janning M., Kumbrink J., Konstantinou T., Muniz J.R., Joshi D., O’Reilly N., Volkmer R., Ritter B., et al. 2015. Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3). J. Biol. Chem. 290:25275–25292. 10.1074/jbc.M115.637207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D.P., Lehrman E.K., Kautzman A.G., Koyama R., Mardinly A.R., Yamasaki R., Ransohoff R.M., Greenberg M.E., Barres B.A., and Stevens B.. 2012. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 74:691–705. 10.1016/j.neuron.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C., Gomez Perdiguero E., Chorro L., Szabo-Rogers H., Cagnard N., Kierdorf K., Prinz M., Wu B., Jacobsen S.E., Pollard J.W., et al. 2012. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 336:86–90. 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- Sekar A., Bialas A.R., de Rivera H., Davis A., Hammond T.R., Kamitaki N., Tooley K., Presumey J., Baum M., Van Doren V., et al. Schizophrenia Working Group of the Psychiatric Genomics Consortium . 2016. Schizophrenia risk from complex variation of complement component 4. Nature. 530:177–183. 10.1038/nature16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekino S., Kashiwagi Y., Kanazawa H., Takada K., Baba T., Sato S., Inoue H., Kojima M., and Tani K.. 2015. The NESH/Abi-3-based WAVE2 complex is functionally distinct from the Abi-1-based WAVE2 complex. Cell Commun. Signal. 13:41 10.1186/s12964-015-0119-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe D.J., and Hardy J.. 2016. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 8:595–608. 10.15252/emmm.201606210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q., Colodner K.J., Matousek S.B., Merry K., Hong S., Kenison J.E., Frost J.L., Le K.X., Li S., Dodart J.C., et al. 2015. Complement C3-Deficient Mice Fail to Display Age-Related Hippocampal Decline. J. Neurosci. 35:13029–13042. 10.1523/JNEUROSCI.1698-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q., Chowdhury S., Ma R., Le K.X., Hong S., Caldarone B.J., Stevens B., and Lemere C.A.. 2017a Complement C3 deficiency protects against neurodegeneration in aged plaque-rich APP/PS1 mice. Sci. Transl. Med. 9:9 10.1126/scitranslmed.aaf6295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Yamada K., Liddelow S.A., Smith S.T., Zhao L., Luo W., Tsai R.M., Spina S., Grinberg L.T., Rojas J.C., et al. Alzheimer’s Disease Neuroimaging Initiative . 2017b ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature. 549:523–527. 10.1038/nature24016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims R., van der Lee S.J., Naj A.C., Bellenguez C., Badarinarayan N., Jakobsdottir J., Kunkle B.W., Boland A., Raybould R., Bis J.C., et al. GERAD/PERADES, CHARGE, ADGC, EADI . 2017. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 49:1373–1384. 10.1038/ng.3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisková Z., Page A., O’Connor V., and Perry V.H.. 2009. Degenerating synaptic boutons in prion disease: microglia activation without synaptic stripping. Am. J. Pathol. 175:1610–1621. 10.2353/ajpath.2009.090372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Hooli B., Mullin K., Jin S.C., Cella M., Ulland T.K., Wang Y., Tanzi R.E., and Colonna M.. 2017. Alzheimer’s disease-associated TREM2 variants exhibit either decreased or increased ligand-dependent activation. Alzheimers Dement. 13:381–387. 10.1016/j.jalz.2016.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenberg E.E., and Green K.N.. 2017. Inflammation in Alzheimer’s disease: Lessons learned from microglia-depletion models. Brain Behav. Immun. 61:1–11. 10.1016/j.bbi.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenberg E.E., Lee R.J., Najafi A.R., Rice R.A., Elmore M.R., Blurton-Jones M., West B.L., and Green K.N.. 2016. Eliminating microglia in Alzheimer’s mice prevents neuronal loss without modulating amyloid-β pathology. Brain. 139:1265–1281. 10.1093/brain/aww016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K., Friedman B.A., Larson J.L., Lauffer B.E., Goldstein L.D., Appling L.L., Borneo J., Poon C., Ho T., Cai F., et al. 2016. Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat. Commun. 7:11295 10.1038/ncomms11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan A.H., Barres B.A., and Stevens B.. 2012. The complement system: an unexpected role in synaptic pruning during development and disease. Annu. Rev. Neurosci. 35:369–389. 10.1146/annurev-neuro-061010-113810 [DOI] [PubMed] [Google Scholar]
- Stephan A.H., Madison D.V., Mateos J.M., Fraser D.A., Lovelett E.A., Coutellier L., Kim L., Tsai H.H., Huang E.J., Rowitch D.H., et al. 2013. A dramatic increase of C1q protein in the CNS during normal aging. J. Neurosci. 33:13460–13474. 10.1523/JNEUROSCI.1333-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B., Allen N.J., Vazquez L.E., Howell G.R., Christopherson K.S., Nouri N., Micheva K.D., Mehalow A.K., Huberman A.D., Stafford B., et al. 2007. The classical complement cascade mediates CNS synapse elimination. Cell. 131:1164–1178. 10.1016/j.cell.2007.10.036 [DOI] [PubMed] [Google Scholar]
- Streit W.J., Sammons N.W., Kuhns A.J., and Sparks D.L.. 2004. Dystrophic microglia in the aging human brain. Glia. 45:208–212. 10.1002/glia.10319 [DOI] [PubMed] [Google Scholar]
- Streit W.J., Braak H., Xue Q.S., and Bechmann I.. 2009. Dystrophic (senescent) rather than activated microglial cells are associated with tau pathology and likely precede neurodegeneration in Alzheimer’s disease. Acta Neuropathol. 118:475–485. 10.1007/s00401-009-0556-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter W.J., Saunders A.M., Schmechel D., Pericak-Vance M., Enghild J., Salvesen G.S., and Roses A.D.. 1993. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. USA. 90:1977–1981. 10.1073/pnas.90.5.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaruga M., Munteanu B., Lismont S., Veugelen S., Horre K., Mercken M., Saido T.C., Ryan N.S., De Vos T., Savvides S.N., et al. 2017. Alzheimer’s-Causing Mutations Shift Abeta Length by Destabilizing gamma-Secretase-Abetan Interactions. Cell. 170:443–456. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Rochford C.D., and Neumann H.. 2005. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med. 201:647–657. 10.1084/jem.20041611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwel D., Steffensen K.R., Verghese P.B., Kummer M.P., Gustafsson J.A., Holtzman D.M., and Heneka M.T.. 2011. Critical role of astroglial apolipoprotein E and liver X receptor-α expression for microglial Aβ phagocytosis. J. Neurosci. 31:7049–7059. 10.1523/JNEUROSCI.6546-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp J., Chonn A., Hertig S., and French L.E.. 1993. Clusterin, the human apolipoprotein and complement inhibitor, binds to complement C7, C8 beta, and the b domain of C9. J. Immunol. 151:2159–2165. [PubMed] [Google Scholar]
- Turnbull I.R., Gilfillan S., Cella M., Aoshi T., Miller M., Piccio L., Hernandez M., and Colonna M.. 2006. Cutting edge: TREM-2 attenuates macrophage activation. J. Immunol. 177:3520–3524. 10.4049/jimmunol.177.6.3520 [DOI] [PubMed] [Google Scholar]
- Ulland T.K., Song W.M., Huang S.C., Ulrich J.D., Sergushichev A., Beatty W.L., Loboda A.A., Zhou Y., Cairns N.J., Kambal A., et al. 2017. TREM2 Maintains Microglial Metabolic Fitness in Alzheimer’s Disease. Cell. 170:649–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich J.D., Ulland T.K., Colonna M., and Holtzman D.M.. 2017. Elucidating the Role of TREM2 in Alzheimer’s Disease. Neuron. 94:237–248. 10.1016/j.neuron.2017.02.042 [DOI] [PubMed] [Google Scholar]
- Vasek M.J., Garber C., Dorsey D., Durrant D.M., Bollman B., Soung A., Yu J., Perez-Torres C., Frouin A., Wilton D.K., et al. 2016. A complement-microglial axis drives synapse loss during virus-induced memory impairment. Nature. 534:538–543. 10.1038/nature18283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemagne V.L., Burnham S., Bourgeat P., Brown B., Ellis K.A., Salvado O., Szoeke C., Macaulay S.L., Martins R., Maruff P., et al. Australian Imaging Biomarkers and Lifestyle (AIBL) Research Group . 2013. Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer’s disease: a prospective cohort study. Lancet Neurol. 12:357–367. 10.1016/S1474-4422(13)70044-9 [DOI] [PubMed] [Google Scholar]
- Wang Y., Cella M., Mallinson K., Ulrich J.D., Young K.L., Robinette M.L., Gilfillan S., Krishnan G.M., Sudhakar S., Zinselmeyer B.H., et al. 2015. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 160:1061–1071. 10.1016/j.cell.2015.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ulland T.K., Ulrich J.D., Song W., Tzaferis J.A., Hole J.T., Yuan P., Mahan T.E., Shi Y., Gilfillan S., et al. 2016. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213:667–675. 10.1084/jem.20151948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weuve J., Hebert L.E., Scherr P.A., and Evans D.A.. 2014. Deaths in the United States among persons with Alzheimer’s disease (2010-2050). Alzheimers Dement. 10:e40–e46. 10.1016/j.jalz.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildsmith K.R., Holley M., Savage J.C., Skerrett R., and Landreth G.E.. 2013. Evidence for impaired amyloid β clearance in Alzheimer’s disease. Alzheimers Res. Ther. 5:33 10.1186/alzrt187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski T., and Frangione B.. 1992. Apolipoprotein E: a pathological chaperone protein in patients with cerebral and systemic amyloid. Neurosci. Lett. 135:235–238. 10.1016/0304-3940(92)90444-C [DOI] [PubMed] [Google Scholar]
- Wu Y., Dissing-Olesen L., MacVicar B.A., and Stevens B.. 2015. Microglia: Dynamic Mediators of Synapse Development and Plasticity. Trends Immunol. 36:605–613. 10.1016/j.it.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T., Yan F., Lin A.H., Lambris J.D., Alexander J.J., Quigg R.J., and Masliah E.. 2002. Prominent neurodegeneration and increased plaque formation in complement-inhibited Alzheimer’s mice. Proc. Natl. Acad. Sci. USA. 99:10837–10842. 10.1073/pnas.162350199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh F.L., Wang Y., Tom I., Gonzalez L.C., and Sheng M.. 2016. TREM2 Binds to Apolipoproteins, Including APOE and CLU/APOJ, and Thereby Facilitates Uptake of Amyloid-Beta by Microglia. Neuron. 91:328–340. 10.1016/j.neuron.2016.06.015 [DOI] [PubMed] [Google Scholar]
- Yeh F.L., Hansen D.V., and Sheng M.. 2017. TREM2, Microglia, and Neurodegenerative Diseases. Trends Mol. Med. 23:512–533. 10.1016/j.molmed.2017.03.008 [DOI] [PubMed] [Google Scholar]
- Yin Z., Raj D., Saiepour N., Van Dam D., Brouwer N., Holtman I.R., Eggen B.J.L., Möller T., Tamm J.A., Abdourahman A., et al. 2017. Immune hyperreactivity of Aβ plaque-associated microglia in Alzheimer’s disease. Neurobiol. Aging. 55:115–122. 10.1016/j.neurobiolaging.2017.03.021 [DOI] [PubMed] [Google Scholar]
- Yoshiyama Y., Higuchi M., Zhang B., Huang S.M., Iwata N., Saido T.C., Maeda J., Suhara T., Trojanowski J.Q., and Lee V.M.. 2007. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 53:337–351. 10.1016/j.neuron.2007.01.010 [DOI] [PubMed] [Google Scholar]
- Yuan P., Condello C., Keene C.D., Wang Y., Bird T.D., Paul S.M., Luo W., Colonna M., Baddeley D., and Grutzendler J.. 2016. TREM2 Haplodeficiency in Mice and Humans Impairs the Microglia Barrier Function Leading to Decreased Amyloid Compaction and Severe Axonal Dystrophy. Neuron. 90:724–739. 10.1016/j.neuron.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian J.L., Xu L., Foo L.C., Nouri N., Zhou L., Giffard R.G., and Barres B.A.. 2012. Genomic analysis of reactive astrogliosis. J. Neurosci. 32:6391–6410. 10.1523/JNEUROSCI.6221-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanjani H., Finch C.E., Kemper C., Atkinson J., McKeel D., Morris J.C., and Price J.L.. 2005. Complement activation in very early Alzheimer disease. Alzheimer Dis. Assoc. Disord. 19:55–66. 10.1097/01.wad.0000165506.60370.94 [DOI] [PubMed] [Google Scholar]
- Zhan S.S., Veerhuis R., Kamphorst W., and Eikelenboom P.. 1995. Distribution of beta amyloid associated proteins in plaques in Alzheimer’s disease and in the non-demented elderly. Neurodegeneration. 4:291–297. 10.1016/1055-8330(95)90018-7 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Sloan S.A., Clarke L.E., Caneda C., Plaza C.A., Blumenthal P.D., Vogel H., Steinberg G.K., Edwards M.S., Li G., et al. 2016. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 89:37–53. 10.1016/j.neuron.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Hu W., Tsai J., Li W., and Gan W.B.. 2017. Microglia limit the expansion of β-amyloid plaques in a mouse model of Alzheimer’s disease. Mol. Neurodegener. 12:47 10.1186/s13024-017-0188-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Jia L., Liu C.C., Rong Z., Zhong L., Yang L., Chen X.F., Fryer J.D., Wang X., Zhang Y.W., et al. 2017. TREM2 Promotes Microglial Survival by Activating Wnt/β-Catenin Pathway. J. Neurosci. 37:1772–1784. 10.1523/JNEUROSCI.2459-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]