Hyungsoo Kim and Ze’ev Ronai preview work from Colaluca et al. that reveals the function of Numb exon 3–containing isoforms in the formation of a fuzzy complex with Mdm2 and the regulation of p53 stability.
Abstract
Although numerous pathways are known to control the tumor suppressor protein p53, coordinated regulation of the p53–Notch axis by Numb may have an even more remarkable impact. In this issue, Colaluca at al. (2018. J. Cell Biol. https://doi.org/10.1083/jcb.201709092) reveal an unexpected role of a newly characterized Numb splice variant in the regulation of p53, which may have significant implications for therapeutic intervention in breast cancer.
The E3 ligase Mdm2 is a well-established master regulator of p53 stability through ubiquitin-dependent degradation, and is also an effective target for therapy. Inhibition of Mdm2 rescues p53 expression and its properties—from cell cycle inhibition to DNA repair and death programs. Mdm2 itself is subject to complex regulation and may offer not only important mechanistic but also potential therapeutic modalities for p53. Among the known regulators of Mdm2 is Numb, a protein implicated in asymmetric cell division (Morrison and Kimble, 2006; Knoblich, 2008). Numb directly regulates Notch availability and activity (Spana and Doe, 1996; Colaluca et al., 2008). Numb’s ability to balance p53 and Notch activity has major effects on stem cell homeostasis with concomitant loss of self-renewing and replication capacity—a proven recipe for tumorigenesis. Furthermore, Numb-mediated control of the p53–Notch axis is known to influence resistance of cancer to therapy (Cicalese et al., 2009; Takebe et al., 2011; Tosoni et al., 2015).
Given these properties, it is not surprising that changes in Numb expression and/or activity have multiple effects on tumor growth. For example, Numb expression is lost in as many as 50% of human mammary tumors, resulting in enhanced Notch signaling (Pece et al., 2004) and reduced p53 expression secondary to the loss of Mdm2 suppression (Colaluca et al., 2008). This effectively qualifies Numb as a tumor suppressor, consistent with the observation that it is inactivated or lost in several tumor types. Different Numb variants, generated by alternative splicing, are expected to elicit distinct activities that could rebalance Notch and Mdm2/p53 activity, with profound implications for cancer cell properties (Fig. 1).
Figure 1.
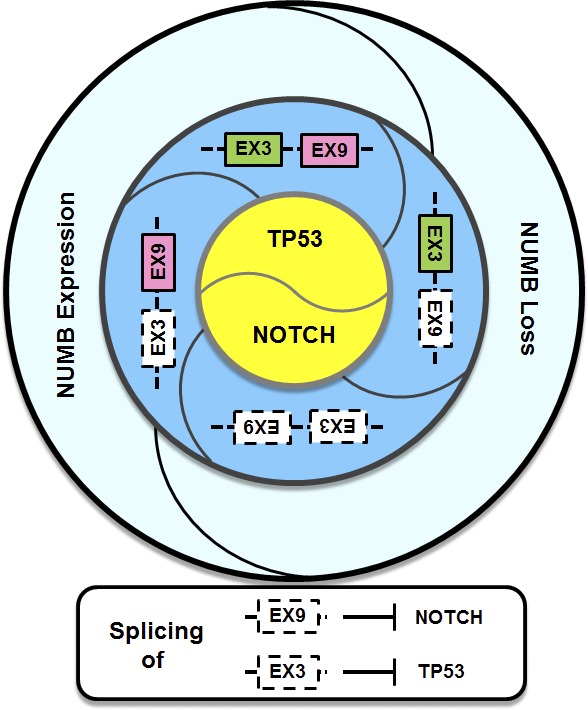
Numb’ing Notch and Mdm2. Numb activity can be regulated by loss of Numb expression seen in many cancers or by alternative splicing of Ex3 and/or Ex9. These changes will determine the expression/activity of TP53 and Notch, which, in turn, impact self-renewal capacity through the control of asymmetric replication and tumorigenesis.
Numb is controlled by alternative splicing of Exons 3 (Ex3) and 9 (Ex9) into four major isoforms (Bani-Yaghoub et al., 2007). Ex9 splicing is associated with Notch-dependent tumorigenesis (Bechara et al., 2013), as with impared development, differentiation, and replication (Kim et al., 2013; Tarn et al., 2016). In this issue, the study by Colaluca et al. has turned our attention to Ex3 splicing.
Extensive biochemical and biophysical studies were performed by Colaluca et al. (2018) to characterize the importance of Ex3 in Numb-mediated control of Mdm2 and p53 stability. Numb requires Ex3 (Numb-1/2 isoforms) to inhibit Mdm2 and thus increase p53 availability. Accordingly, deletion of Numb-1/2, but not of Numb-3/4 (isoforms that lack Ex3), reduces p53 expression in an Mdm2-dependent manner. Likewise, the p53-stabilizing effects of genotoxic agents such as cisplatin are lost by deletion of Ex3-containing Numb, illustrating the importance of this exon for the DNA damage/repair response.
Colaluca et al. (2008) mapped the Numb–Mdm2 interaction sites to a stretch of 11 aromatic and positively charged amino acids in the phosphotyrosine-binding domain of Numb and to the unstructured acidic domain of Mdm2. The interaction mainly occurred through multiple hydrophobic and complementary polar interactions, constituting a fuzzy complex. Several intriguing questions are raised by this finding: Does the interaction require phosphorylation of Numb within the interaction domain? Is extracellular regulated kinase (ERK), which has been suggested to affect Numb splicing (Rajendran et al., 2016), involved in this event? Does ERK or an unidentified kinase provide another regulatory layer of Numb splicing or its interaction with Mdm2? Notably, binding of Mdm2 does not impair the ability of Numb to interact with and inhibit Notch activity. Thus, loss of Numb-mediated inhibition of Mdm2, while maintaining Notch inhibition, would result in loss of tumor suppression and gain of oncogenic signaling. This scenario suggests that loss of some isoforms might have different consequences and impact on tumorigenesis via alternate molecular mechanisms, as they would be also expected to affect asymmetric division and expansion of breast cancer (BCa) stem cells. Given the diverse pleiotropic functions of Numb in cell polarization, endocytosis, ubiquitination, and other processes (Pece et al., 2011), it seems clear that cataloging the role of Numb splice variants in each process will allow us to fully appreciate their importance and significance in other pathological conditions (Fig. 1).
Several important questions about Numb biology remain. For example, what is it that dictates splicing of Ex3 versus Ex9? Is it one of several splicing factors that have already been implicated, such as RBM4/5/6/10 or ASF/SF2 (Bechara et al., 2013; Rajendran et al., 2016; Tarn et al., 2016)? Could the expression and/or activity of this factor provide another prognostic marker for p53 WT BCa? Would the finding shown here for a subset of BCa be relevant to other p53 WT tumors? As noted in the current study, cancers where WT p53 is less frequent, such as lung cancer, would benefit not from loss of Ex3 but rather from loss of Ex9. Although the current findings are consistent with the anticipated role of the splicing machinery in oncogenesis (Bechara et al., 2013; Zong et al., 2014; Rajendran et al., 2016), the development of tools that enable mapping of specific splice variants in large transcriptomic-scale analyses is expected to enable their further characterization in select patient cohorts.
BCa provides a particularly striking demonstration of the physiological significance of Ex3-dependent Numb–Mdm2 interactions because expression of Ex3-deleted Numb correlates inversely with resistance to genotoxic agents. Furthermore, Numb-1/2low BCa cells can be resensitized to cisplatin by cotreatment with the Mdm2 inhibitor Nutlin-3. This observation not only substantiates the importance of Numb–Mdm2 interactions but also suggests a possible therapeutic mechanism to overcome cisplatin resistance. Earlier studies from this group linked the loss of Numb expression in BCa with poor prognosis (Pece et al., 2004). In the current study, Colaluca et al. (2008) investigate the possible implications of Ex3 loss in BCa etiology. Analysis of 890 BCa patients revealed that the expression of Ex3-deleted Numb in p53 WT tumors is associated with a high risk of metastasis. Accordingly, loss of Ex3 might be a useful predictive marker for metastasis in luminal BCa, which is predominantly p53 WT. Of interest, Nutlin-3 enabled correction of self-renewal properties of Numb-deficient cancer stem cells (CSC) and inhibited CSC expansion, with a marked effect on tumorigenicity and metastasis (Tosoni et al., 2017). Combining Nutlin-3 and chemotherapy prevented CSC-driven tumor relapse after removal of chemotherapy (Tosoni et al., 2017).
In conclusion, the current study defines a new layer of p53 control via Mdm2 and Numb, not only providing new insights into the fundamental regulation and function of p53 but also highlighting a mechanism by which tumor cell signaling can be rewired. Ex3 splicing of Numb provides another example of a central regulatory node that affects pro-tumorigenic properties via a loss of DNA repair and enhanced metastatic capacity (Colaluca et al., 2018). The search for the Ex3 splicing factor, its regulation, and its functions are expected to offer fundamental new insights for cancer rewiring control, within and beyond Numb.
Acknowledgments
National Cancer Institute grant R35 R35CA197465 (to Z.A. Ronai) is gratefully acknowledged.
The authors declare no competing financial interests.
References
- Bani-Yaghoub M., Kubu C.J., Cowling R., Rochira J., Nikopoulos G.N., Bellum S., and Verdi J.M.. 2007. A switch in numb isoforms is a critical step in cortical development. Dev. Dyn. 236:696–705. 10.1002/dvdy.21072 [DOI] [PubMed] [Google Scholar]
- Bechara E.G., Sebestyén E., Bernardis I., Eyras E., and Valcárcel J.. 2013. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol. Cell. 52:720–733. 10.1016/j.molcel.2013.11.010 [DOI] [PubMed] [Google Scholar]
- Cicalese A., Bonizzi G., Pasi C.E., Faretta M., Ronzoni S., Giulini B., Brisken C., Minucci S., Di Fiore P.P., and Pelicci P.G.. 2009. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 138:1083–1095. 10.1016/j.cell.2009.06.048 [DOI] [PubMed] [Google Scholar]
- Colaluca I.N., Tosoni D., Nuciforo P., Senic-Matuglia F., Galimberti V., Viale G., Pece S., and Di Fiore P.P.. 2008. NUMB controls p53 tumour suppressor activity. Nature. 451:76–80. 10.1038/nature06412 [DOI] [PubMed] [Google Scholar]
- Colaluca I.N., Basile A., Freiburger L., D’Uva V., Disalvatore D., Vecchi M., Confalonieri S., Tosoni D., Cecatiello V., Malabarba M.G., et al. 2018. A Numb–Mdm2 fuzzy complex reveals an isoform-specific involvement of Numb in breast cancer. J. Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.K., Nam J., Mukouyama Y.S., and Kawamoto S.. 2013. Rbfox3-regulated alternative splicing of Numb promotes neuronal differentiation during development. J. Cell Biol. 200:443–458. 10.1083/jcb.201206146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J.A. 2008. Mechanisms of asymmetric stem cell division. Cell. 132:583–597. 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Morrison S.J., and Kimble J.. 2006. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 441:1068–1074. 10.1038/nature04956 [DOI] [PubMed] [Google Scholar]
- Pece S., Serresi M., Santolini E., Capra M., Hulleman E., Galimberti V., Zurrida S., Maisonneuve P., Viale G., and Di Fiore P.P.. 2004. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 167:215–221. 10.1083/jcb.200406140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S., and Confalonieri S., Romano R., and Di Fiore P.P.. 2011. NUMB-ing down cancer by more than just a NOTCH. Biochim. Biophys. Acta. 1815:26–43. [DOI] [PubMed] [Google Scholar]
- Rajendran D., Zhang Y., Berry D.M., and McGlade C.J.. 2016. Regulation of Numb isoform expression by activated ERK signaling. Oncogene. 35:5202–5213. 10.1038/onc.2016.69 [DOI] [PubMed] [Google Scholar]
- Spana E.P., and Doe C.Q.. 1996. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 17:21–26. 10.1016/S0896-6273(00)80277-9 [DOI] [PubMed] [Google Scholar]
- Takebe N., Harris P.J., Warren R.Q., and Ivy S.P.. 2011. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat. Rev. Clin. Oncol. 8:97–106. 10.1038/nrclinonc.2010.196 [DOI] [PubMed] [Google Scholar]
- Tarn W.Y., Kuo H.C., Yu H.I., Liu S.W., Tseng C.T., Dhananjaya D., Hung K.Y., Tu C.C., Chang S.H., Huang G.J., and Chiu I.M.. 2016. RBM4 promotes neuronal differentiation and neurite outgrowth by modulating Numb isoform expression. Mol. Biol. Cell. 27:1676–1683. 10.1091/mbc.E15-11-0798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosoni D., Zecchini S., Coazzoli M., Colaluca I., Mazzarol G., Rubio A., Caccia M., Villa E., Zilian O., Di Fiore P.P., and Pece S.. 2015. The Numb/p53 circuitry couples replicative self-renewal and tumor suppression in mammary epithelial cells. J. Cell Biol. 211:845–862. 10.1083/jcb.201505037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosoni D., Pambianco S., Ekalle Soppo B., Zecchini S., Bertalot G., Pruneri G., Viale G., Di Fiore P.P., and Pece S.. 2017. Pre-clinical validation of a selective anti-cancer stem cell therapy for Numb-deficient human breast cancers. EMBO Mol. Med. 9:655–671. 10.15252/emmm.201606940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong F.Y., Fu X., Wei W.J., Luo Y.G., Heiner M., Cao L.J., Fang Z., Fang R., Lu D., Ji H., and Hui J.. 2014. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 10:e1004289 10.1371/journal.pgen.1004289 [DOI] [PMC free article] [PubMed] [Google Scholar]


